Abstract
Extrasynaptic γ-aminobutyric acid type A receptors (GABAARs),which contribute generalized inhibitory tone to the mammalian brain, are major targets for general anesthetics. To identify anesthetic binding sites in an extrasynaptic GABAAR, we photolabeled human α4β3δ GABAARs purified in detergent with [3H]azietomidate and a barbiturate, [3H]R-mTFD-MPAB, photoreactive anesthetics that bind with high selectivity to distinct but homologous intersubunit binding sites in the transmembrane domain of synaptic α1β3γ2 GABAARs. Based upon 3H incorporation into receptor subunits resolved by SDS-PAGE, there was etomidate-inhibitable labeling by [3H]azietomidate in the α4 and β3 subunits and barbiturate-inhibitable labeling by [3H]R-mTFD-MPAB in the β3 subunit. These sites did not bind the anesthetic steroid alphaxalone, which enhanced photolabeling, or DS-2, a δ subunit-selective positive allosteric modulator, which neither enhanced nor inhibited photolabeling. The amino acids labeled by [3H]azietomidate or [3H]R-mTFD-MPAB were identified by N-terminal sequencing of fragments isolated by HPLC fractionation of enzymatically digested subunits. No evidence was found for a δ subunit contribution to an anesthetic binding site. [3H]azietomidate photolabeling of β3Met-286 in βM3 and α4Met-269 in αM1 that was inhibited by etomidate but not by R-mTFD-MPAB established that etomidate binds to a site at the β3+-α4− interface equivalent to its site in α1β3γ2 GABAARs. [3H]Azietomidate and [3H]R-mTFD-MPAB photolabeling of β3Met-227 in βM1 established that these anesthetics also bind to a homologous site, most likely at the β3+-β3− interface, which suggests a subunit arrangement of β3α4β3δβ3.
Keywords: allosteric regulation, anesthetic, Cys-loop receptor, GABA receptor, photoaffinity labeling
Introduction
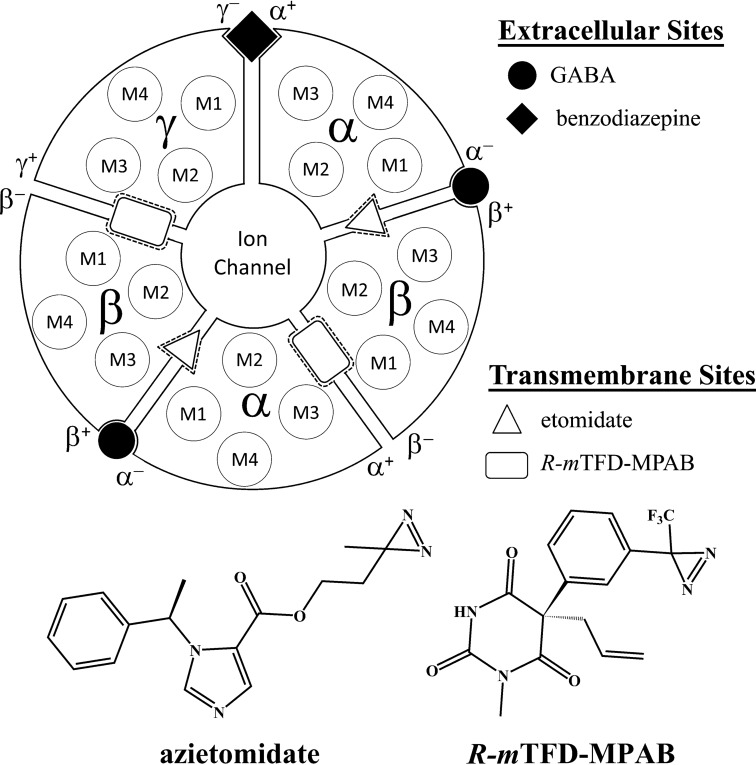
γ-Aminobutyric acid type A receptors (GABAARs)2 are the major inhibitory neurotransmitter receptors in the mammalian brain. They are members of the pentameric ligand-gated ion channel superfamily that consists of five homologous subunits, each of which has a large extracellular domain, a transmembrane domain of four transmembrane helices (M1–M4), and an intracellular domain connecting the third and fourth transmembrane helices. GABAARs, which are the target of many drugs, among them benzodiazepines and general anesthetics, are heteropentamers, and drug action often depends on the subunit composition. For example, at synaptic receptors, which commonly have a subunit composition of (α)2(β)2γ, arranged βαβαγ counterclockwise when viewed from the synaptic or extracellular side of the receptor, benzodiazepines act in the extracellular domain between α+-γ− subunits at a site homologous to the GABA binding sites at the two β+-α− subunit interfaces (Fig. 1) (1–3).
FIGURE 1.
Interface binding sites for GABA, benzodiazepines, etomidate, and the barbiturate R-mTFD-MPAB in an α1β3γ2 GABAAR and the chemical structures of photoreactive anesthetics used to identify anesthetic binding sites.
General anesthetics have long been known to bind to sites in the transmembrane domains of pentameric ligand-gated ion channels (reviewed in Refs. 4–7). Photolabeling of endogenous and heterologous GABAARs by [3H]azietomidate located the etomidate binding site in the two β+-α− subunit interfaces (8, 9), 50 Å from the GABA site and at a position later shown to overlap with the five ivermectin sites in the crystal structure of the homopentameric glutamate-gated chloride channel (GluCl) (10). More recently, a photoreactive, anesthetic barbiturate, R-mTFD-MPAB, has been shown to bind to sites in the γ+-β− and α+-β− subunit interfaces homologous to the etomidate binding sites, introducing the concept of subtype-dependent action of general anesthetics (11). Whereas etomidate and R-mTFD-MPAB bind with high selectivity to their sites, propofol, pentobarbital, and other barbiturates bind with much less selectivity to these two classes of sites.
The in vivo mechanism of action of etomidate has been firmly linked to the GABAAR. Heterologously expressed GABAARs that have an N256M mutation on the M2 helix of the β3 subunit (β+ surface of the interface) are relatively insensitive to etomidate (12), and sleep times in knock-in mice bearing the same mutation are much shorter than in wild-type mice (13). Azietomidate causes normal anesthesia in wild-type mice with the same potency as etomidate, and its action is similarly attenuated in the knock-in mouse (14). R-mTFD-MPAB also causes general anesthesia in mice and is equally potent in wild-type and N256M knock-in mice (15), consistent with the location of its binding sites at the β− subunit interfaces.
The contrasting subunit-selective actions of these two agents raise questions about the mechanism of general anesthesia itself, because there are 19 known GABA subunits, and which of the possible combinations occur in vivo is not yet fully defined. The state of anesthesia involves many behavioral components (16), so subunit-selective general anesthetics might be associated with specific subsets of the behavioral impairments experienced during anesthesia (17). Of particular interest are the relative contributions of phasic (synaptic) and tonic (extrasynaptic) inhibition actions (18, 19). The focus of this study is on the extrasynaptic α4β3δ GABAARs that are sensitive to endogenous neurosteroids and general anesthetics at concentrations lower than necessary to potentiate inhibitory postsynaptic currents (20–24). Expression studies in fibroblasts and oocytes establish that multiple combinations of α4, β, and δ subunits can combine to form functional receptors, which results in alternative subunit interfaces (25–31).
In this work, we photolabeled detergent-solubilized, purified heterologous α4β3δ GABAARs with [3H]azietomidate and [3H]R-mTFD-MPAB. Two distinct high affinity anesthetic sites were identified: 1) [3H]azietomidate photolabeling established that azietomidate and etomidate bind to a β3+-α4− interface site that does not bind R-mTFD-MPAB with high affinity; and 2) [3H]azietomidate and [3H]R-mTFD-MPAB share a common binding site with etomidate at a β3− subunit interface. DS2, a positive allosteric modulator selective for GABAARs containing a δ subunit (32), did not bind to these sites.
Results
Biochemical Characterization of the α4β3δ GABAAR
Comparison of [3H]muscimol binding to α4β3δ GABAAR in membranes and after purification in asolectin/CHAPS established that positive allosteric modulation was retained by etomidate and by DS2, a positive allosteric modulator selective for GABAARs containing the δ subunit (32) (Table 1). In contrast to α1β3γ2 GABAARs, which bound [3H]muscimol with similar affinity in membrane-bound (Keq = 50 nm) and purified (Keq = 80 nm) states (33), [3H]muscimol bound to α4β3δ GABAARs in membranes (Keq = 13 nm) with higher affinity than after purification in CHAPS/asolectin (Keq = 90 nm). After purification, etomidate (10 μm) and DS2 (30 μm) increased the specific binding of 2 nm [3H]muscimol by ∼30%.
TABLE 1.
Etomidate and DS2 enhancement of [3H]muscimol binding to membrane-bound and purified α4β3δ GABAAR
Anesthetic modulation was calculated as the ratio (%) of specific [3H]muscimol binding (2 nm) in the presence versus the absence of modulator. The results are the means ± S.D. from three independent experiments.
| Compound | Concentration | Membrane-bound GABAAR | Purified GABAAR |
|---|---|---|---|
| μm | % | % | |
| Etomidate | 10 | 147 ± 28 | 129 ± 8 |
| DS2 | 30 | 139 ± 8 | 129 ± 14 |
When samples of purified human α4β3δ GABAAR were fractionated by SDS-PAGE and visualized by Coomassie Blue stain, bands were readily visualized at 78 and 58 kDa, along with fainter bands at 72, 62, and 54 kDa (Fig. 2, lane 1). When extracted materials from in-gel tryptic digests of these bands were characterized by LC/MS/MS (Table 2), fragments of the GABAAR α4 subunit were most enriched in the 72 kDa band, consistent with the expected mobility of the mature subunit (58 kDa + 3 N-linked glycosylations). Fragments from the β3 subunit were concentrated in the 62 and 58 kDa bands, as found for β3 subunit from expressed α1β3γ2 GABAARs (11). Fragments from the δ subunit were broadly distributed in the 62, 58, and 54 kDa bands, with α4 subunit fragments also recovered from the 54 kDa band. However, in contrast to the recovery of α4 subunit fragments from the 72 kDa gel band, for the 54 kDa band, no fragments were recovered from the α4 cytoplasmic domain beginning about 30 amino acids after the end of the M3 helix (data not shown). This result suggests that the 54 kDa band contains an N-terminal fragment of the α4 subunit containing the M1–M3 helices that was probably produced by proteolytic cleavage during receptor purification. The major component in the 78 kDa band was identified as the chaperone heat shock 70-kDa protein 1A (HSP70-1).
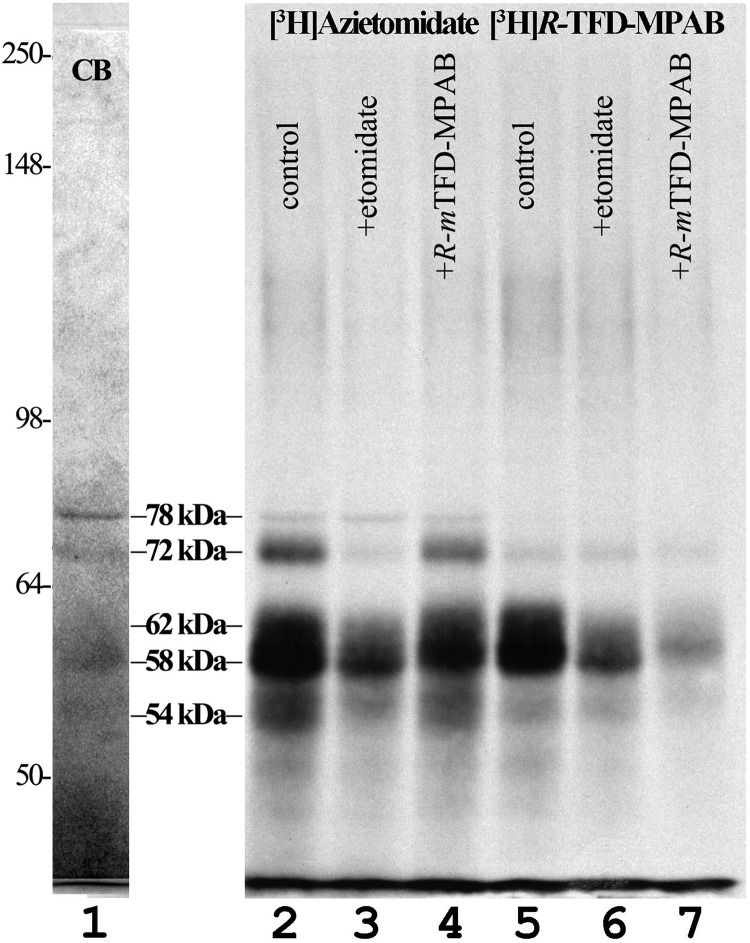
FIGURE 2.
Photolabeling of α4β3δ GABAARs by [3H]azietomidate and [3H]R-mTFD-MPAB. Aliquots of α4β3δ GABAAR (8 pmol of [3H]muscimol sites/aliquot) were photolabeled with 3 μm [3H]azietomidate (lanes 2–4) or 1 μm [3H]R-mTFD-MPAB (lanes 5–7) in the absence (lanes 2 and 5) or presence of 1 mm etomidate (lanes 3 and 6) or 60 μm R-mTFD-MPAB (lanes 4 and 7), and subunits were resolved by SDS-PAGE. After Coomassie Blue staining (CB, representative image in lane 1), the gel was prepared for fluorography (lanes 2–7). Migration positions of molecular mass standards (in kDa) are denoted to the left of lane 1.
TABLE 2.
LC/MS/MS identification of major peptides in α4β3δ GABAAR SDS-polyacrylamide gel bands
Shown are search results from a Homo sapiens protein database: HSPA1A, heat shock 70-kDa protein 1A/1B; PRMT5, arginine N-methyltransferase 5; GABRA4, GABAAR α4 subunit; LMNB1, lamin-B1; GABRD, GABAAR δ subunit; IGF2BP1, insulin-like growth factor 2 mRNA-binding protein 1; ABCD3, ATP-binding cassette subfamily D member 3; GABRB3, GABAAR β3 subunit; TUBA1A, tubulin α-1A chain; TUBB2A, tubulin β-2A chain; DDX47, probable ATP-dependent RNA helicase DDX47.
| Gel band | Protein (gene name) | Peptides detected | MS/MS scans | Average intensity | Coverage |
|---|---|---|---|---|---|
| % | |||||
| 78 kDa | HSPA1A | 68 | 911 | 1,450,000 | 70 |
| PRMT5 | 40 | 166 | 597,000 | 48 | |
| GABRA4 | 27 | 127 | 393,000 | 46 | |
| LMNB1 | 29 | 65 | 253,000 | 48 | |
| 72 kDa | GABRA4 | 57 | 1,005 | 756,000 | 55 |
| GABRD | 15 | 60 | 209,000 | 29 | |
| IGF2BP1 | 17 | 42 | 173,000 | 31 | |
| ABCD3 | 17 | 36 | 118,000 | 23 | |
| 62 kDa | GABRD | 29 | 487 | 924,000 | 38 |
| GABRB3 | 33 | 282 | 723,000 | 41 | |
| GABRA4 | 32 | 127 | 500,000 | 45 | |
| TUBA1A | 24 | 118 | 336,000 | 46 | |
| 58 kDa | GABRB3 | 35 | 304 | 1,970,000 | 40 |
| GABRD | 27 | 421 | 1,400,000 | 39 | |
| TUBA1A | 34 | 272 | 956,000 | 61 | |
| GABRA4 | 28 | 115 | 712,000 | 42 | |
| TUBB2A | 47 | 476 | 655,000 | 59 | |
| 54 kDa | GABRA4 | 26 | 140 | 1,180,000 | 41 |
| GABRD | 25 | 393 | 892,000 | 40 | |
| GABRB3 | 15 | 52 | 639,000 | 30 | |
| DDX47 | 13 | 31 | 176,000 | 35 |
When material eluted from the 72 kDa band was characterized by Edman degradation, the primary sequence identified (XXLNXXPGQNQXXXXL …) matched a region near the predicted N terminus of the human α4 GABAAR subunit (VCLNESPGQNQKEEKL …). Multiple amino acids were detected at similar levels at each cycle of Edman degradation of the 62- and 58-kDa samples, which precluded de novo identification of the subunits present. Sequence analysis of material from the 54 kDa band identified a primary sequence (XNDIXXYKXD …) matching the N terminus region of the FLAG-tagged human δ GABAAR subunit sequence (MNDIGDYKDDDDK …, with the underline denoting the FLAG peptide sequence). The N termini of the α4 and δ subunits identified by Edman degradation are those predicted to be the N termini of the mature subunits by the signal sequence cleavage site prediction program P-signal (34). No N-terminal sequence was detected from the 78-kDa material, consistent with the fact that the N-terminal alanine of 70-kDa heat shock protein is acetylated (35), preventing Edman degradation.
Labeling Human α4β3δ GABAAR with Photoreactive Anesthetics
α4β3δ GABAARs were photolabeled at anesthetic concentrations with [3H]azietomidate or [3H]R-mTFD-MPAB in the absence or presence of etomidate at 1 mm or non-radioactive R-mTFD-MPAB at 60 μm, concentrations at which they each bind selectively to the β+ or β− intersubunit sites in α1β3γ2 GABAARs (11). When 3H incorporation was determined by fluorography after SDS-PAGE (Fig. 2, lanes 2–7), 3H incorporation was highest in the 58/62 kDa gel region for both photoreactive anesthetics. The 72 kDa band (α4) was labeled prominently only by [3H]azietomidate, and that photolabeling was inhibitable by etomidate but not by R-mTFD-MPAB. [3H]Azietomidate photolabeling in the 54/58/62 kDa gel bands was inhibited to a greater extent by etomidate than by R-mTFD-MPAB, and, conversely, [3H]R-mTFD-MPAB photolabeling of 58/62 kDa gel bands was inhibited to a greater extent by R-mTFD-MPAB than by etomidate. These findings suggested that 1) there is an etomidate/azietomidate binding site associated with the α4 subunit that does not bind R-mTFD-MPAB with high affinity; 2) azietomidate, etomidate, and R-mTFD-MPAB share a common binding site associated with the 58/62 kDa gel band; and 3) there may be an R-mTFD-MPAB binding site associated with the 58/62 kDa gel band that does not bind etomidate.
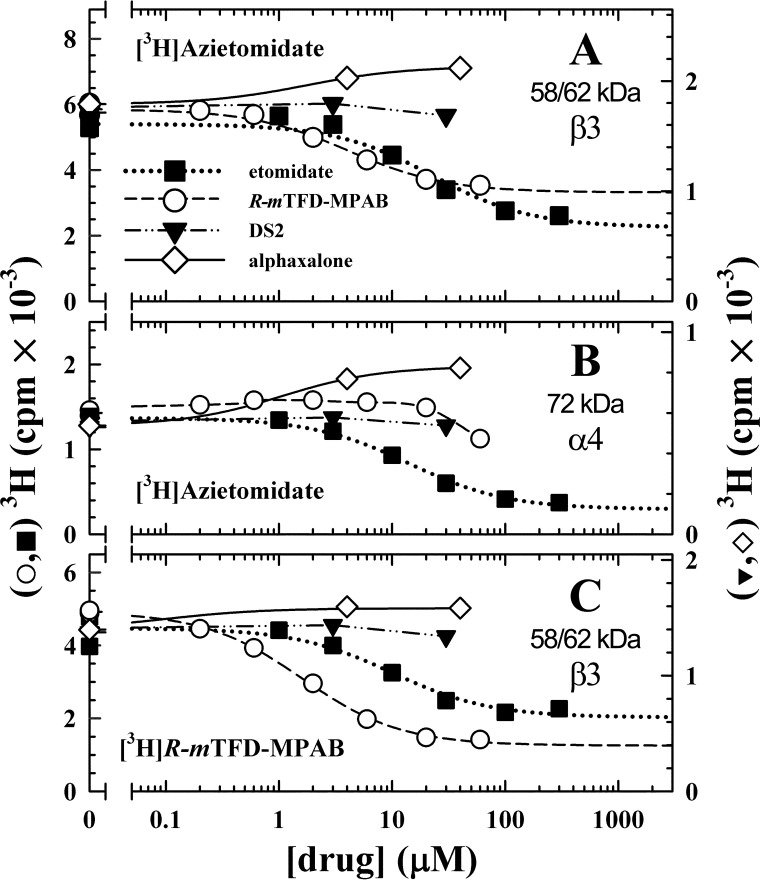
To further characterize the pharmacological specificity of [3H]azietomidate and [3H]R-mTFD-MPAB incorporation at the subunit level, photolabelings were performed on an analytical scale in the presence of various concentrations of etomidate, R-mTFD-MPAB, the neuroactive steroid alphaxalone, or DS2, with 3H incorporation into the gel bands quantified by liquid scintillation counting (Fig. 3). Etomidate inhibited [3H]azietomidate photoincorporation into the 72 kDa (α4) and 58/62 kDa bands with IC50 values of ∼15 μm, with high concentrations producing maximal inhibition of subunit photolabeling by 80 and 60%, respectively. Etomidate also inhibited [3H]R-mTFD-MPAB photolabeling in the 58/62 kDa bands with similar potency. R-mTFD-MPAB inhibited [3H]R-mTFD-MPAB photolabeling in the 58/62 kDa bands with an IC50 of 2 μm and a maximal inhibition of 75%. R-mTFD-MPAB also inhibited [3H]azietomidate photolabeling in the 58/62 kDa bands with similar potency, but it inhibited [3H]azietomidate labeling in the 72 kDa (α4) band only at the highest concentration tested (60 μm, ∼30% inhibition).
FIGURE 3.
Pharmacological specificity [3H]azietomidate and [3H]R-mTFD-MPAB photoincorporation into α4β3δ GABAAR subunits. Aliquots of α4β3δ GABAAR were equilibrated with 1 μm [3H]azietomidate or 0.4 μm [3H]R-mTFD-MPAB in the presence of 300 μm GABA and various concentrations of etomidate (■), R-mTFD-MPAB (○), DS2 (▾), or alphaxalone (♢, -GABA). After UV irradiation, the samples were separated by SDS-PAGE, the stained GABAAR subunit gel bands were excised, and 3H incorporation was determined by liquid scintillation counting for the 58/62 kDa (β3) (A) and 72 kDa (α4) (B) bands labeled with [3H]azietomidate and for the 58/62 kDa (β3) band (C) labeled with [3H]R-mTFD-MPAB. Different receptor preparations were used for each drug tested, with each drug assayed in parallel with [3H]azietomidate and [3H]R-mTFD-MPAB, with receptors also photolabeled in the presence of 300 μm etomidate or 60 μm R-mTFD-MPAB to define nonspecific photolabeling. Due to limited quantities of receptor, competition assays were done only once, and the S.E. value given are from the least-squares fits. A and B, etomidate inhibited [3H]azietomidate labeling of β3 and α4 with IC50 values of 23 ± 7 and 14 ± 1 μm, respectively. R-mTFD-MPAB inhibited [3H]azietomidate labeling of β3 with an IC50 of 4.1 ± 0.8 μm. R-mTFD-MPAB had no effect on α4 labeling up to 20 μm, whereas it inhibited ∼30% of the specific labeling at 60 μm. DS2 had no effect on [3H]azietomidate labeling, whereas alphaxalone potentiated labeling 40–74%. C, R-mTFD-MPAB inhibited [3H]R-mTFD-MPAB labeling of β3 with an IC50 of 1.6 ± 0.1 μm. Etomidate inhibited [3H]R-mTFD-MPAB labeling of β3 with an IC50 of 10 ± 5 μm. DS2 had no effect on [3H]R-mTFD-MPAB labeling, whereas alphaxalone increased labeling by 50%.
As seen for [3H]azietomidate and [3H]R-mTFD-MPAB photolabeling of α1β3γ2 GABAARs (11), in the absence of GABA, the neuroactive steroid alphaxalone at concentrations up to 30 μm potentiated photoincorporation into the GABAAR subunit bands, maximally by ∼50%. This result establishes that alphaxalone does not bind to the sites in the purified α4β3δ GABAAR photolabeled by [3H]azietomidate or [3H]R-mTFD-MPAB but that there is positive allosteric linkage between alphaxalone and azietomidate/R-mTFD-MPAB binding. At concentrations up to 30 μm, DS2 had little or no effect on photolabeling by [3H]azietomidate or [3H]R-mTFD-MPAB in the presence of GABA.
Localization of α4β3δ GABAAR Residues Photolabeled by [3H]Azietomidate
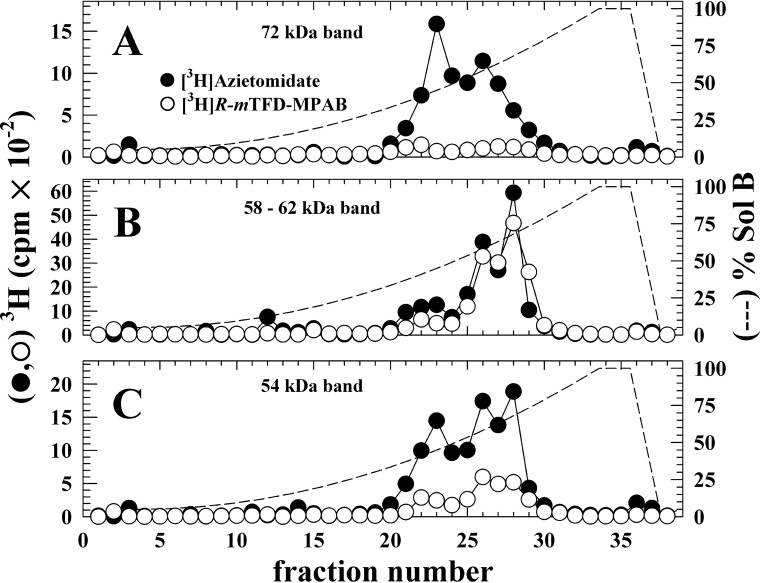
To provide an initial characterization of the locations of photolabeled amino acids, we fractionated by reversed phase HPLC (rpHPLC) EndoLys-C digests of subunit bands isolated from α4β3δ GABAARs photolabeled with [3H]azietomidate or [3H]R-mTFD-MPAB (Fig. 4). For both photoreactive anesthetics, the digests of the 58/62 kDa band (β3 and δ subunits) contained peaks of 3H in hydrophobic fractions (∼55 and 70% organic solvent) where fragments beginning at the N termini of the βM3 and βM1 helices are known to elute (36). For the 72 kDa band (α4) labeled by [3H]azietomidate, the 3H eluted in two peaks, a broad hydrophobic peak (55% organic solvent), which for digests of α1 subunits contains fragments beginning at the N termini of the M1 and M3 helices, and a peak at 40% organic solvent, where fragments from the α1 subunit extracellular domain elute (36). For the 54 kDa band, for each drug, there were peaks of 3H at 40, 55, and 70% organic solvent, corresponding to the peaks seen in either of the higher molecular weight gel bands.
FIGURE 4.
rpHPLC fractionation of EndoLys-C digests of [3H]azietomidate (●) or [3H]R-mTFD-MPAB (○) photolabeled α4β3δ GABAAR subunit bands. 3H elution profiles, determined by liquid scintillation counting of 10% aliquots, are shown for EndoLys-C-digested subunits isolated from 72 kDa (A, α4), 58/62 kDa (B, β3/δ), and 54 kDa (C) gel bands. The elution gradient (% Organic Solvent) is indicated by the dashed lines.
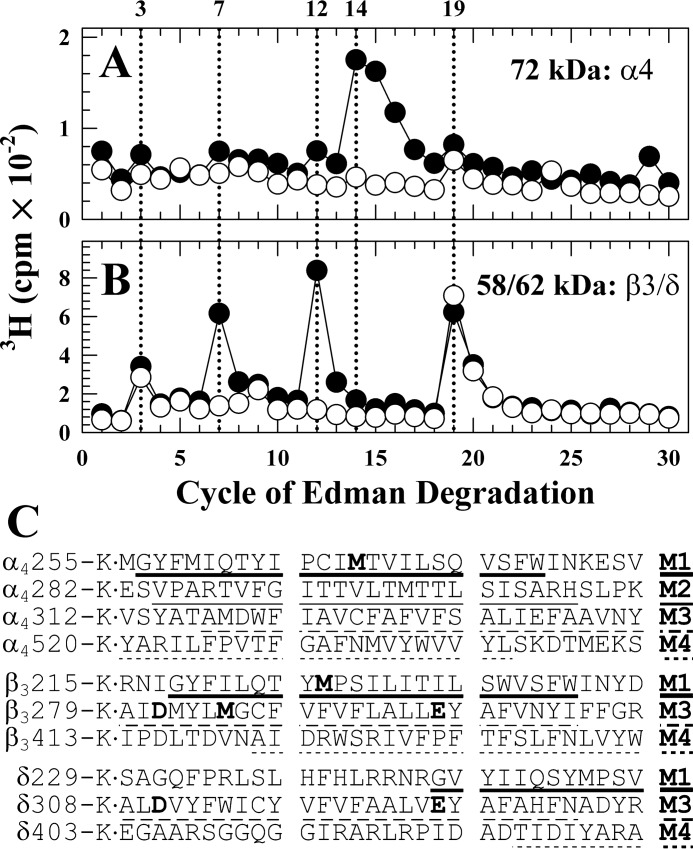
Etomidate Inhibits [3H]Azietomidate Photolabeling of α4Met-269 (αM1), β3Met-227 (βM1), and β3Met-286 (βM3)
Aliquots were sequenced of unfractionated EndoLys-C digests from the 72 and 58/62 kDa gel bands from GABAARs photolabeled with [3H]azietomidate in the absence and presence of non-radioactive etomidate (Fig. 5). For the 72 kDa band, there was a major peak of etomidate-inhibitable 3H release in cycle 14 (Fig. 5A). For the digest from the 58/62 kDa band (Fig. 5B), there were peaks of etomidate-inhibitable 3H release in cycles 7 and 12 (pharmacologically specific photolabeling) and peaks of 3H release in cycles 3 and 19 that were not inhibited by etomidate (nonspecific labeling). The 72 kDa gel band digest will contain all possible α4 subunit proteolytic fragments, including fragments beginning near the N termini of the M1–M4 helices. For the 58/62 kDa band, digests will include fragments beginning near the N termini of the M1, M3, and M4 helices of the β3 and δ subunits (Fig. 5C). However, the etomidate-inhibitable peak of 3H release in cycle 14 for the 72 kDa band digest occurs in the cycle predicted to contain α4Met-269, the residue homologous to α1Met-236 in the α1 subunit M1 helix that was photolabeled by [3H]azietomidate (8, 9, 11). Similarly, the peaks of etomidate-inhibitable 3H release in cycles 7 and 12 for the 58/62 kDa band digest occur in the sequencing cycles that will contain β3Met-286 in βM3 and β3Met-227 in βM1, respectively, residues also photolabeled by [3H]azietomidate in α1β3 or α1β3γ2 GABAAR (9, 11), as well as the residues from δM3 (δTrp-315 and δPhe-320). The peaks of release in cycles 3 and 19 that were not inhibited by etomidate occur in cycles that contain Asp or Glu near the N and C termini of the β3 and δ subunit M3 helices.
FIGURE 5.
3H release during N-terminal sequencing of EndoLys-C digests of 72 kDa (α4) and 62 kDa (β3) subunit bands. Subunit digests from purified human α4β3δ GABAAR photolabeled with 3 μm [3H]azietomidate in the absence (●) or presence (○) of 1 mm etomidate were loaded directly onto sequencing filter without prior purification by rpHPLC. In this experiment, 7,620 (●) and 3,620 (○) cpm were loaded for the 72 kDa (A) band, 22,020 cpm (●) and 17,210 (○) cpm for the 58/62 kDa band (B), and five-sixths of the material from each cycle of Edman degradation was collected for determination of released 3H. Included in C are the subunit fragment sequences containing transmembrane helices that can be sequenced after EndoLys-C digestion.
R-mTFD-MPAB Inhibits [3H]Azietomidate Photolabeling of β3Met-227 but Not α4Met-269
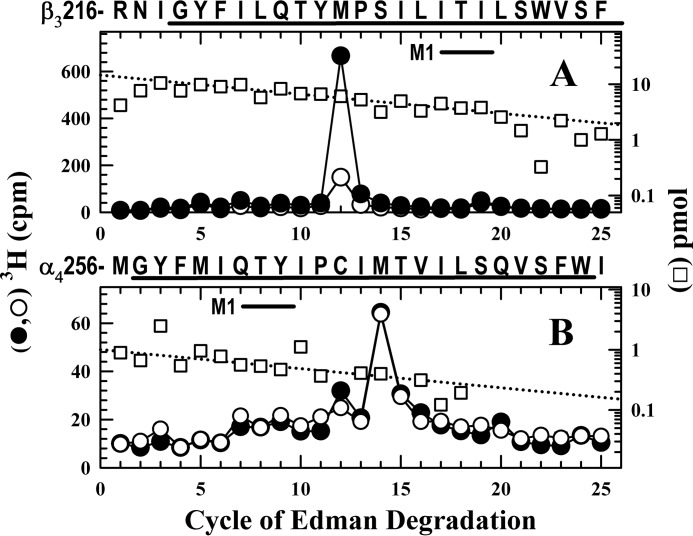
To confirm that [3H]azietomidate photolabeled α4Met-269 and β3Met-227, samples were sequenced after rpHPLC fractionation of EndoLys-C digests of material from the 72, 58/62, and 54 kDa gel bands. To determine whether R-mTFD-MPAB also inhibited photolabeling of these residues, receptors were photolabeled in the absence or the presence of 20 μm R-mTFD-MPAB, a concentration sufficient to occupy ∼90% of its high affinity binding sites based upon the inhibition of photolabeling at the subunit level (Fig. 3). When fractions from the 58/62 kDa band were sequenced that contained the fragment beginning at β3Arg-216 at ∼15 pmol (Fig. 6A), the peak of 3H release in cycle 12 confirmed labeling of β3Met-227 (220 cpm/pmol), and R-mTFD-MPAB inhibited that labeling by ∼80%. When fractions from the 72 kDa (α4) gel band were sequenced (Fig. 6B) that contained the fragment beginning at α4Met-256 (1 pmol), there was a single peak of 3H release in cycle 14, consistent with photolabeling of α4Met-269 in α4M1 at 230 cpm/pmol in the absence or presence of R-mTFD-MPAB.
FIGURE 6.
R-mTFD-MPAB inhibits [3H]azietomidate photolabeling of β3Met-227 (βM1) but not α4Met-269 (αM1). A and B, 3H (●, ○) and pmol of PTH-derivatives (□) released during sequencing of subunit fragments beginning at β3Arg-216 and at α4Met-256 isolated by rpHPLC from EndoLys-C digests of subunits in the 58/62 kDa (A, β3/δ) or 72 kDa (B, α4) gel bands isolated by SDS-PAGE from α4β3δ GABAARs (110 pmol of muscimol sites per condition) photolabeled with 3.5 μm [3H]azietomidate in the absence (●, □) or presence (○) of 20 μm R-mTFD-MPAB. rpHPLC fractions 28 and 29 (A) and 25–28 (B) were sequenced. A, the primary sequence began at β3Arg-216 (I0 = 14 pmol, both conditions), and the peak of 3H release in cycle 12 indicated labeling of β3Met-227 at 220 cpm/pmol in the absence and at 50 cpm/pmol in the presence of R-mTFD-MPAB. A secondary sequence was present beginning at β3Ala-280 (β3M3, ∼1 pmol). B, the fragment beginning at α4Met-256 (I0 = 1 pmol) was present, along with fragments beginning at α4Val-313 (α4M3, ∼2.6 pmol) and α4Ser-238 (∼1 pmol). The peak of 3H release in cycle 14 was consistent with labeling of α4Met-269 (230 cpm/pmol) unaffected by R-mTFD-MPAB.
R-mTFD-MPAB Does Not Inhibit [3H]Azietomidate Photolabeling of β3Met-286
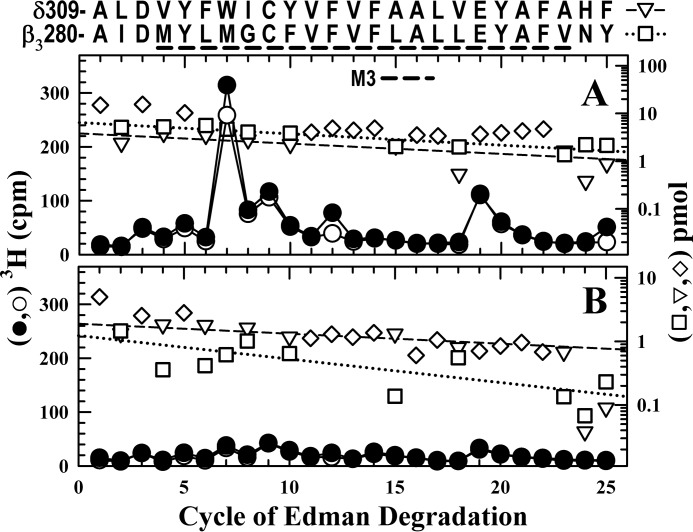
To determine whether [3H]azietomidate was photolabeling amino acids in βM3 and/or δM3, we sequenced rpHPLC fractions from EndoLys-C digests of the 58/62 kDa gel band enriched in βM3 and from the 54 kDa gel band enriched in δM3. When fractions from the 58/62 kDa gel band were sequenced containing fragments beginning at β3Ala-280 (∼6 pmol) and at δAla-309 (4 pmol) (Fig. 7A), there was a peak of 3H release in cycle 7 (280 cpm), which R-mTFD-MPAB inhibited by <15%. In contrast, when fractions from the 54 kDa gel band were sequenced containing fragments beginning at β3Ala-280 (1 pmol) and at δAla-309 (2 pmol), the peak of 3H release in cycle 7 was 25 cpm (Fig. 7B). Because the 3H releases in cycle 7 correlated well with the amount of the β3Ala-280 fragment but not with the amount of the δM3 fragment, the peak of 3H release in cycle 7 indicated photolabeling of β3Met-286 (130 cpm/pmol) rather than δTrp-315. That [3H]azietomidate labeling of β3Met-286 (βM3) and α4Met-269 (αM1) was inhibited by etomidate (Fig. 5), but not by R-mTFD-MPAB, indicates that etomidate and azietomidate bind to a site at the β3+-α4− interface that does not bind R-mTFD-MPAB with high affinity and is homologous to their binding site at the β3-α1 interface.
FIGURE 7.
R-mTFD-MPAB does not inhibit [3H]azietomidate photolabeling of β3Met-286 (βM3). 3H (●, ○) and pmol of PTH-derivatives (□, ▿, ♢) released during sequencing of subunit fragments beginning near the N termini of βM3 and δM3 isolated by rpHPLC from EndoLys-C digests of material eluted from SDS-polyacrylamide gel bands migrating at 58/62 kDa (A) or 54 kDa (B) from the α4β3δ GABAAR photolabeling of Fig. 6 in the absence (●, □, ▿, ♢) or presence (○) of R-mTFD-MPAB. rpHPLC fractions 25–27 (A) and 26 and 27 (B) were sequenced. A, the primary sequence began at β3Ala-280 (I0 = 6 pmol; □, residues unique to β3M3) with the secondary sequence beginning at δAla-309 (I0 = 4 pmol; ▿, residues unique to δM3). The detected pmol of residues common to both β3M3 and δM3 (♢) were not used for the repetitive yield fits. The peak of 3H release in cycle 7, if originating from β3M3, indicated labeling of β3Met-286 at 130 cpm/pmol, and R-mTFD-MPAB inhibited that labeling by <15%. B, the primary sequence began at δAla-309 (I0 = 2 pmol; ▿, residues unique to δM3) with the secondary sequence beginning at β3Ala-280 (I0 = 1 pmol; □, residues unique to δM3). The peaks of 3H release in cycle 7 (A, 300 cpm; B, 25 cpm) correlate well with the amounts of the β3Ala-280 fragment (A, 6 pmol; B, 1 pmol) but not with those of the δAla-309 fragment (A, 4 pmol; B, 2 pmol).
[3H]R-mTFD-MPAB Photolabels β3Met-227 (βM1) and β3Met-286/Phe-289 (βM3)
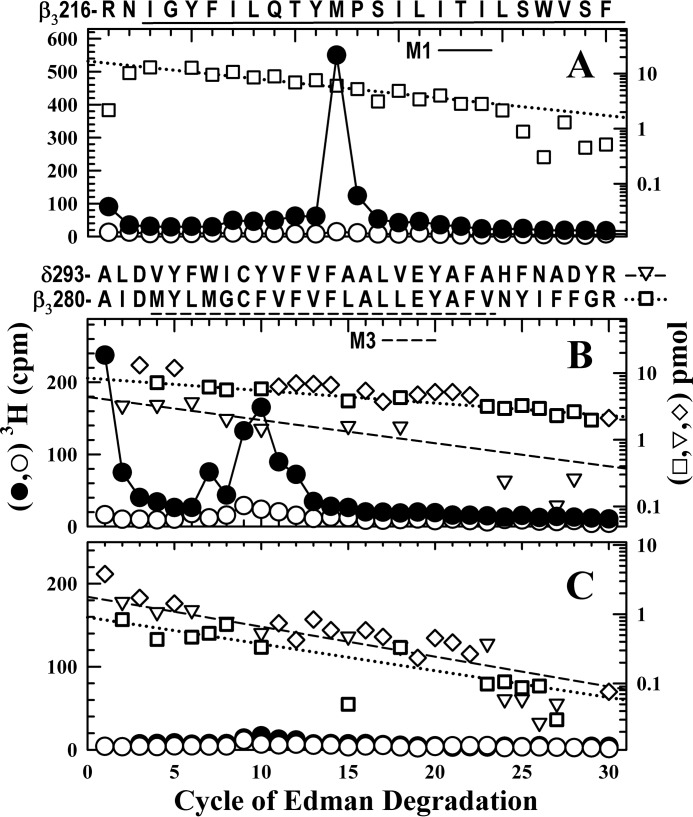
Amino acids photolabeled by [3H]R-mTFD-MPAB were identified by sequencing appropriate fractions from rpHPLC fractionations of EndoLys-C digests of material from the 58/62 and 54 kDa gel bands from α4β3δ GABAARs photolabeled with 0.5 μm [3H]R-mTFD-MPAB in the absence or presence of 60 μm R-mTFD-MPAB (Fig. 8). When fractions were sequenced containing the fragment beginning at β3Arg-216 (15 pmol), the single peak of 3H release in cycle 12 indicated photolabeling of β3Met-227 (160 cpm/pmol) that R-mTFD-MPAB inhibited by >95% (Fig. 8A). When rpHPLC fractions from the 58/62 kDa digest were sequenced that contained the M3 fragments, there were prominent peaks of 3H release in cycles 7 (50 cpm), 9 (90 cpm), and 10 (75 cpm) (Fig. 8B), whereas for the 54 kDa band, peaks of 3H release were <7 cpm (Fig. 8C). As for labeling by [3H]azietomidate, the peaks of 3H release correlated well with the amounts of the β3Ala-280 fragment (Fig. 8, B (8 pmol) and C (1 pmol)) but not the δAla-309 fragment (Fig. 8, B (4 pmol) and C (2 pmol)). The peaks of 3H release in cycles 7 and 10 indicate photolabeling in β3M3 of β3Met-286 (16 cpm/pmol), β3Cys-288 (30 cpm/pmol), and β3Phe-289 (34 cpm/pmol). Based upon the calculated efficiencies of photolabeling (cpm/pmol) in the absence and presence of 60 μm R-mTFD-MPAB (see the legend to Fig. 8), the labeling of β3Met-286 and β3Phe-289 was inhibited by >90%, whereas photolabeling of βCys-288 was inhibited by ≤50%.
FIGURE 8.
[3H]R-mTFD-MPAB specifically photolabels β3Met-227, β3Met-286, and β3Phe-289 in human α4β3δ GABAAR. A–C, 3H (●, ○) and pmol of PTH-derivatives (□, ▿, ♢) released during sequencing of subunit fragments beginning near the N termini of βM1 (A), βM3 (B), or δM3 (C) isolated by rpHPLC from EndoLys-C digests of subunits in the 58/62 kDa (A and B) or 54 kDa (C) gel bands isolated by SDS-PAGE from α4β3δ GABAARs (50 pmol of muscimol sites per condition) photolabeled with 0.5 μm [3H]R-mTFD-MPAB in the absence (●, □, ▿, ♢) or presence (○) of 60 μm R-mTFD-MPAB. rpHPLC fractions 28 and 29 (A) and 25–27 (B and C) were sequenced. A, the primary sequence began at β3Arg-216 (□, I0 = 17/8 pmol, without/with R-mTFD-MPAB), and the peak of 3H release in cycle 12 indicated photolabeling of β3Met-227 (160 cpm/pmol) that R-mTFD-MPAB inhibited by >95%. B, the primary sequence began at β3Ala-280 (β3M3) (□, residues unique to β3M3; I0 = 8/4 pmol, without/with R-mTFD-MPAB) with the secondary sequence beginning at δAla-309 (▿, residues unique to δM3; I0 = 4/2 pmol, without/with R-mTFD-MPAB). The detected pmol of residues common to β3M3 and δM3 (♢) were not used for the repetitive yield fits. The 3H release in cycles 7 and 10, if originating from β3M3, indicated labeling of β3Met-286 and β3Phe-289 at 16 and 34 cpm/pmol, respectively, with >90% inhibition by R-mTFD-MPAB. The 3H release in cycle 9 indicates photolabeling of β3Cys-288 (31/17 cpm/pmol without/with R-mTFD-MPAB) and/or δCys-317 (84/40 cpm/pmol without/with R-mTFD-MPAB). C, the primary sequence began at δAla-309 (▿, residues unique to δM3; I0 = 2/3 pmol, without/with R-mTFD-MPAB) with the secondary sequence beginning at β3Ala-280 (□, residues unique to β3M3,I0 = 1 pmol, both conditions). In B and C, the peaks of 3H release in cycles 7, 9, and 10 correlate well with the amounts of the β3Ala-280 fragment (B, 8 pmol; C, 1 pmol) but not with those of the δAla-309 fragment (B, 4 pmol; C, 2 pmol).
Discussion
In this report, we provide a first characterization of the locations of anesthetic binding sites in a GABAAR subtype expressed extrasynaptically in the CNS. We photolabeled purified human α4β3δ GABAARs with [3H]azietomidate and [3H]R-mTFD-MPAB, photoreactive anesthetics that have been used previously to identify two homologous but pharmacologically distinct classes of anesthetic binding sites in α1β3γ2 GABAARs (11). Based upon the identification of photolabeled amino acids and the results of competition photolabeling assays carried out at the level of intact subunits, we demonstrate that etomidate, but not R-mTFD-MPAB, binds with high affinity to a site at the β+-α− subunit interface in α4β3δ GABAARs that is equivalent to its binding site in α1β3γ2 GABAARs. In contrast to α1β3γ2 GABAARs, which bind R-mTFD-MPAB, but not etomidate, with high affinity to sites at the α+/γ+-β− interfaces in proximity to β3Met-227 in βM1, we find that etomidate as well as R-mTFD-MPAB bind with high affinity to a site in α4β3δ GABAARs containing β3Met-227. As discussed below, this site is most likely to be at a β+-β− subunit interface. The sites identified by photolabeling with [3H]azietomidate and [3H]R-mTFD-MPAB are distinct from the binding sites for alphaxalone, an anesthetic steroid, or DS-2, a δ subunit-selective positive allosteric modulator (32), because neither drug inhibited photolabeling.
α4β3δ GABAAR Composition
Based upon mass spectrometry and Edman degradation, the affinity-purified α4β3δ GABAARs used in this work contain α4 and β3 subunits as well as the δ subunit, whose presence is assured because the FLAG epitope used for purification is attached near the δ subunit N terminus. However, we do not know whether the preparation is characterized by a single dominant subunit composition. Whereas receptors having a β3α4β3α4δ subunit arrangement (counterclockwise when viewed from the extracellular side) with two β3+-α4− interfaces containing the agonist sites and a δ subunit replacing the γ subunit have been reported to be strongly favored in transiently transfected HEK cells (27, 28, 37), other studies indicate that subunit stoichiometry can be variable and dependent upon the subunit cDNA transfection ratios (26). Also, studies using concatenated subunits provide evidence that the δ subunit can assume multiple positions in a receptor pentamer and can contribute to a β+-δ− agonist binding site (25, 27, 30).
In the absence of independent definition of the subunit composition and arrangement in our purified α4β3δ GABAARs, consideration of our photolabeling results suggests a β3α4β3δβ3 or β3δβ3α4β3 organization for the stably transfected cell line used in our studies. We favor these stoichiometries because 1) they have a β3-β3 interface required for the shared azietomidate/etomidate/R-mTFD-MPAB binding site, and 2) they have three β3 subunits to every one α4 subunit, consistent with the similar levels of [3H]azietomidate incorporation (cpm/pmol) at the amino acid level in the β3 and α4 subunits (Fig. 6) in the presence of a higher level of 3H incorporation in the β3 gel band than in the α4 band (Fig. 2). However, a β2α4δα4β2 pentameric concatemer, containing a β-β interface, also forms a functional receptor (30).
An Etomidate Binding Site at the β3+-α4− Interface
[3H]Azietomidate photolabeled β3Met-286 in β3M3 (β3+ side of an interface) and α4Met-269 in α4M1 (α4− side), with etomidate inhibiting labeling by >90% and R-mTFD-MPAB by <15%. Because [3H]azietomidate also photolabeled β3Met-286 in α1β3γ2 GABAARs and α4Met-269 is homologous to α1Met-236 that was also photolabeled (11), the simplest interpretation of these results is that there is an etomidate/azietomidate binding site at a β3+-α4− interface homologous to the etomidate site at the β3+-α1− interfaces in α1β3γ2 GABAARs. This conservation of etomidate binding sites between α4β3δ and α1β3γ2 GABAARs is not unexpected, in view of the strong conservation of amino acids in the regions of the α4 and α1 subunit M1 and M2 helices that contribute to the α− surface of the etomidate binding sites (Fig. 9) and the fact that etomidate produces similar allosteric modulation in α1β3δ and α1β3γ2 GABAARs (38).
FIGURE 9.
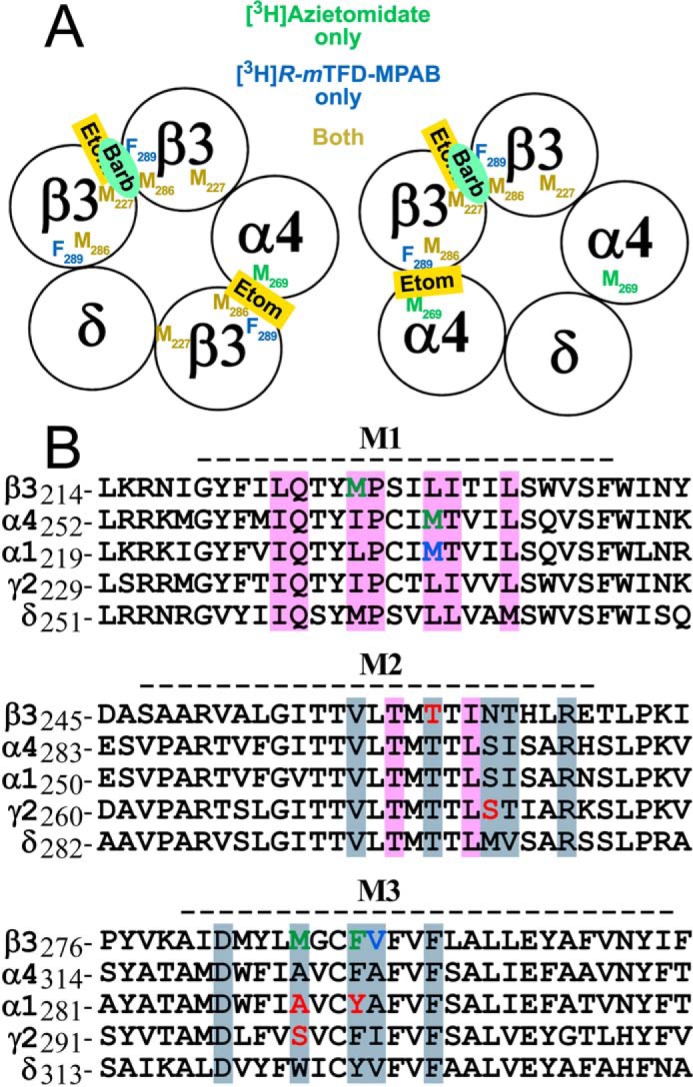
Sequence alignment of the transmembrane helices of extrasynaptic α4β3δ and synaptic α1β3γ2 GABAAR subunits: conserved anesthetic binding sites. A, schematic representation of two possible α4β3δ GABAAR subunit arrangements with the interface locations identified of the residues photolabeled in an anesthetic-inhibitable manner by [3H]azietomidate (green), [3H]R-mTFD-MPAB (blue), or both (gold). The photolabeling results are consistent with etomidate (Etom) binding at the β3+-α4− interface and both etomidate and R-mTFD-MPAB (Barb) binding to a common site at the β3+-β3− interface. B, alignments of the transmembrane M1, M2, and M3 helices with residues column-color-coded by subunit surface (+face, blue/gray; −face, pink) to indicate interface residues within 6 Å of etomidate docked in the β+-α− intersubunit pocket in an α1β3γ2 GABAAR homology model (36) based upon the β3 monomeric GABAAR crystal structure (Protein Data Bank code 4COF). Residues specifically labeled by [3H]azietomidate and/or [3H]R-mTFD-MPAB in the α4β3δ GABAAR are shown in green. Residues labeled by [3H]azietomidate or [3H]TDBzl-etomidate in the α1β3γ2 GABAAR β+-α− intersubunit pocket are shown in blue, whereas residues at the homologous α+/γ+-β− pocket photolabeled by the barbiturate probes [3H]R-mTFD-MPAB and/or [3H]S-mTFD-MPPB are in red (8, 9, 11, 36). The dashed line above each alignment denotes the extent of α-helices in the GABAAR structure.
An Etomidate/R-mTFD-MPAB Binding Site at a β3− Interface
The most prominently labeled residue in the β3 subunit for both [3H]azietomidate and [3H]R-mTFD-MPAB was β3Met-227. Whereas in α1β3γ2 GABAARs, etomidate enhanced [3H]R-mTFD-MPAB photolabeling of this residue in α+-β− and/or γ+-β− intersubunit sites (11), etomidate inhibited this photolabeling by >90% in the α4β3δ GABAAR, where β3Met-227 can potentially contribute to anesthetic binding sites at the α4+-β3−, δ+-β3−, or β3+-β3− subunit interfaces.
Several lines of evidence indicate that the β3+-β3− interface is the most likely interface for the site binding etomidate, azietomidate, and R-mTFD-MPAB with high affinity. 1) Photolabeling studies with expressed α1β3 GABAARs establish that etomidate, [3H]azietomidate, and [3H]R-mTFD-MPAB all bind with high affinity to the β3+-β3− interface pocket that is present in α1β3 but not in α1β3γ2 GABAARs (9, 39). 2) Examination of the amino acid residues that would contribute to the three alternative binding pockets (Fig. 9) identifies non-conservative substitutions contributing to the (+)-surface of the binding pocket that are expected to prevent the high affinity binding of etomidate in an α4+-β3− or δ+-β3− intersubunit pocket. In α1β2/3γ2 GABAARs, β2/3Asn-265 (βM2–15′) is known to be a major determinant of etomidate binding affinity, and in vitro and in vivo mutational analyses establish that replacement by Ser (α4M2–15′) or Met (δM2–15′) reduces etomidate potency by >10-fold (12, 13, 40–42). Similarly, substitution of β3Met-286 by Trp (the δ residue in the β3Met-286 position) also inhibits the effects of etomidate (40, 43). Therefore, it is unlikely that etomidate can bind with high affinity at either the α4+-β3− or δ+-β3− interface.
Contributions of δ Subunit Residues to Etomidate/Barbiturate Binding Sites
In our study, we did not identify any δ subunit amino acids photolabeled in an anesthetic-inhibitable manner by [3H]azietomidate or [3H]R-mTFD-MPAB. Based upon sequence analyses of samples containing variable amounts of βM3 and δM3, any pharmacologically specific photolabeling in δM3 is at <15% the level of βM3. It is possible that [3H]azietomidate does bind in a pocket containing δM3 residues that is homologous to the β+-α− site without photolabeling any residue in δM3, because the pocket would lack the methionine side chains favored by azietomidate's photoreactive intermediate. However, it is unlikely that R-mTFD-MPAB binds without photolabeling any residues in its vicinity because both it and S-mTFD-MPPB, another closely related trifluoromethylphenyldiazirine, have been found to react broadly in GABAARs with aliphatic as well as aromatic and nucleophilic side chains (11, 36). δ subunit fragmentation with EndoLys-C produced a cleavage 19 amino acids before the NH2 terminus of δM1, which was not close enough to allow high sensitivity sequence analysis. In fact, δM1 may contribute to a barbiturate binding site, because studies with receptors containing α1, β3, and chimeric γ/δ subunits indicated that pentobarbital sensitivity determinants were contained within a fragment containing the amino terminus and the first 3 amino acids of δM1 (44). Further studies will be necessary to clarify whether general anesthetics also bind with high affinity in the pocket at the β+-δ− interface in the cell line used in this study or at the α+-δ− interface in βαβαδ GABAARs.
Functional Significance of the Identified Binding Sites
Photolabeling studies provided a first definition of two classes of pharmacologically distinct binding sites for intravenous general anesthetics at subunit interfaces in the α1β3γ2 GABAAR transmembrane domain (8, 11, 36) that overlap with the binding sites for ivermectin (45). Mutational analyses of the residues identified by photoaffinity labeling as well as neighboring residues in the shared subunit interface pockets have demonstrated their contributions to GABAAR gating and as determinants of anesthetic efficacy (12, 40, 43, 46, 47). In addition, the capacity of anesthetics to protect against modification of substituted cysteines has expanded the definition of residues contributing to anesthetic binding sites (48, 49). Mutational analyses will be necessary to determine, for example, whether the β+-α− site and β+-β− sites identified by photoaffinity labeling are equally important for etomidate enhancement of GABA responses in an α4β3δ GABAAR. However, in view of the difficulty of expressing α4β3δ GABAARs with defined subunit stoichiometry and subunit arrangement, these studies should be carried out using pentameric concatenated receptors.
Experimental Procedures
Materials
[3H]Muscimol (36 Ci/mmol) was from PerkinElmer Life Sciences. The detergents n-dodecyl β-d-maltopyranoside and CHAPS were from Anatrace-Affymetrix (anagrade quality). R-mTFD-MPAB and [3H]R-mTFD-MPAB (38 Ci/mmol) were prepared previously (50), as was [3H]azietomidate (19.3 Ci/mmol) (39). Soy bean asolectin, R-etomidate, and GABA were from Sigma. DS2 and alphaxalone were from Tocris. EndoLys-C was from Roche Applied Sciences.
Purification of α4β3δ GABAAR
A detailed description of the expression and affinity purification of α4β3δ GABAARs will be presented elsewhere. As described previously for α1β3γ2 GABAARs (33), a stably transfected, tetracycline-inducible HEK293-TetR cell line expressing human GABAAR subunits α4, β3 (splice variant 2), and δ containing a FLAG tag near its N terminus (between δGly-29 and δAsp-30) was induced and grown for 2–3 days, and then membranes were harvested, flash-frozen in liquid N2, and stored at −80 °C until use. GABAARs were solubilized with 30 mm n-dodecyl β-d-maltopyranoside and affinity-purified as described (11), using a FLAG M2 antibody column. Columns were washed with purification buffer supplemented with 200 μm asolectin and 5 mm CHAPS and then eluted with 1.5 mm FLAG peptide in the wash buffer. Aliquots of the eluate fractions were assayed for [3H]muscimol binding, and eluate fractions were flash-frozen in liquid N2 and stored at −80 °C until use. Membranes harvested from 60 15-cm plates contained ∼5–10 nmol of [3H]muscimol binding sites (15–20 pmol of sites/mg of membrane protein), and the eluate fractions from the purifications used for photolabeling contained 50–70 nm [3H]muscimol sites. Based upon [3H]muscimol binding, the receptor was purified at 10–25% yield from the starting membranes. Because the receptor was eluted in the presence of 1.5 mm FLAG peptide, it was not possible to estimate purity in terms of pmol of muscimol binding/mg of protein. Based upon analyses by SDS-PAGE and LC/MS/MS (see “Results”), GABAAR subunits were the dominant polypeptides in the preparation.
Radioligand Binding Assays
[3H]Muscimol binding to purified GABAAR was measured by filtration after precipitation with polyethylene glycol (8). The total concentration of sites in eluate fractions was determined at 250 nm [3H]muscimol with 1 mm GABA to determine nonspecific binding. Allosteric modulation of 2 nm [3H]muscimol binding was determined as described (9, 11).
Sequence Numbering
For α4, residue 1 is the predicted signal sequence Met; for β3, residue 1 is the predicted N terminus of the mature protein (splice variant 1, QSNVD …), with β3Met-286 at the 15th position in the M2 helix (M2–15′); and for δ, the numbering begins with the signal sequence Met and excludes the inserted FLAG sequence (DYKDDDDK). The primary structure locations of transmembrane helices M1–M4 in the figures correspond to the extent of the individual α-helices in the β3 monomeric GABAAR crystal structure (Protein Data Bank code 4COF).
Analysis of the α4β3δ GABAAR Preparation by LC/MS and N-terminal Sequencing
Three aliquots (24 pmol of [3H]muscimol sites each) of α4β3δ GABAAR were separated by SDS-PAGE. Based upon Coomassie Blue staining, bands migrating at 78, 72, 62, 58, and 54 kDa were excised. The bands from one lane were submitted to the Harvard Medical School Taplin Mass Spectrometry Facility for reduction and alkylation, in-gel trypsin digestion, and peptide extraction for microcapillary LC/MS/MS analysis. The material from the equivalent gel bands from the other two lanes was eluted and subjected to N-terminal sequence analysis.
GABAAR Photolabeling
Aliquots of purified α4β3δ GABAARin elution buffer were photolabeled at analytical or preparative scale (150–200 μl or 1–2 ml of GABAAR per condition, respectively) to characterize photoincorporation at the subunit level or to identify individual photolabeled amino acids by protein microsequencing. Aliquots of [3H]azietomidate or [3H]R-mTFD-MPAB were dried under a gentle argon stream and resuspended with GABAAR solutions for 30 min on ice with gentle vortexing. For preparative photolabeling, non-radioactive drugs were added directly to this resuspension, whereas for analytical photolabeling, drug aliquots were added by the use of a 1-μl syringe (Hamilton 86200) to 10 μl of purified GABAAR, which was then combined with 90–150 μl of GABAAR equilibrated with radioligand. With the exception of studies with alphaxalone, all photolabeling was carried out in the presence of 300 μm GABA. Samples were transferred to 96-well plastic plates or 3.5-cm diameter Petri dishes (Corning catalogue numbers 2797 and 3001) for analytical or preparative scale labeling and irradiated on ice with a 365-nm UV lamp (Spectroline EN-280L) for 30 min at a distance of <1 cm. Samples were then denatured by mixing 2 parts sample with 1 part SDS-PAGE sample buffer, incubated for ∼30 min, and fractionated by modified Laemmli SDS-PAGE (11).
Stock solutions of non-radioactive R-mTFD-MPAB (60 mm), etomidate (60 mm), and alphaxalone (8 mm) were prepared in methanol. For these drugs, all samples during photolabeling contained methanol at a final concentration of 0.5% (v/v). DS2 was prepared at 6 mm in 90% methanol, 10% DMSO. For assays with DS2, samples during photolabeling contained methanol/DMSO at final concentrations of 0.45%/0.05% (v/v). To minimize losses of hydrophobic drugs due to adsorption on plastic surfaces, glass syringes, capillary pipettes, and vials were used for all material transfers up to the equilibration with the purified GABAAR in detergent/lipid.
After electrophoresis, gels were stained with Coomassie Brilliant Blue. In analytical scale experiments, 3H incorporation into subunits was determined either by fluorography or by liquid scintillation counting of excised gel bands as described (11). In preparative scale experiments, material was eluted from the excised stained bands as described (11) and resuspended in gel digestion buffer (15 mm Tris, 0.5 mm EDTA, and 0.1% SDS, pH 8.4) for further analysis. Results from three preparative photoaffinity labelings of purified human α4β3δ GABAARs are presented in this work: 1) GABAAR (145 pmol of muscimol sites per condition) photolabeled with 3 μm [3H]azietomidate in the presence of 300 μm GABA with or without 1 mm etomidate; 2) GABAAR (110 pmol of muscimol sites per condition) photolabeled with 3.5 μm [3H]azietomidate in the presence of 300 μm GABA with or without 20 μm R-mTFD-MPAB; and 3) GABAAR (50 pmol of muscimol sites per condition) photolabeled with 0.5 μm [3H]R-mTFD-MPAB in the presence of 300 μm GABA with or without 60 μm R-mTFD-MPAB.
To determine the relative binding affinity for anesthetics at the [3H]azietomidate or [3H]R-mTFD-MPAB binding sites, aliquots of α4β3δ GABAAR were photolabeled in the presence of various concentrations of a drug, and subunit gel slice counts from these aliquots were fit to the equation,
| (Eq. 1) |
where B(x) represents the gel slice 3H cpm at total inhibitor concentration x, B0 is the gel slice 3H cpm in the absence of competitor, Bns is nonspecific 3H cpm incorporation in the presence of maximal concentration of a competitor, and IC50 is the total drug concentration producing 50% inhibition. Data were fit using Sigma Plot version 11.0 (Systat Software, Inc.) with IC50 and Bns as adjustable parameters; B0 was fixed at the experimentally observed value. Due to limited quantities of receptor, competition assays were done only once, and the S.E. values given are from the least-squares fits.
Proteolysis, Reversed-phase HPLC, and N-terminal Sequence Analysis
Aliquots of labeled subunits isolated from gel bands were digested (2 weeks, 20 °C, 0.3–1 units/sample) with EndoLys-C (Roche Applied Science). Digests were fractionated by rpHPLC as described (51), except that the gradient began at 95% aqueous solvent (0.08% TFA) and 5% organic solvent (60% acetonitrile, 40% isopropyl alcohol, 0.05% TFA) and progressed to 100% organic in 75 min by approximating (in 5-min intervals) the quadratic growth curve, f(x) = 5 + 0.017 × x2, where x is time in minutes and f(x) is percentage of organic solvent. The flow rate was 200 μl/min, and fractions were collected every 2.5 min, with 10% assayed for 3H. Fractions of interest were pooled and drop-loaded onto glass fiber filters for N-terminal sequence analysis on an Applied Biosystems Procise 492 protein sequencer modified so that two-thirds of each cycle were injected for PTH-derivative detection and quantification, whereas one-third was collected for scintillation counting. Some digested samples were sequenced without rpHPLC separation by loading them onto Applied Biosystems ProSorbTM PVDF filters by diluting the samples 10-fold into 0.1% TFA. The pmol of PTH-derivatives detected were calculated by using rpHPLC peak heights at 269 nm compared with a standard injection.
Photolabeling in α4M1 or α4M3 was determined by sequencing appropriate rpHPLC fractions from digests of the 72 kDa gel band. Labeling in β3M1 and β3M3 was identified by sequencing fractions from the 58/62 kDa gel bands. In preliminary studies, we established that that fragments containing δM1 and δM3 were present at the highest level in the fractions containing βM3 from the 58/62 kDa gel band, where they were present at ∼50% the level of the βM3 fragment. The δM3 fragment was present in the equivalent fractions from the 54 kDa gel band at ∼200% the level of the βM3 fragment. The N termini of βM3 (β3Ala-280) and δM3 (δAla-309) were each at the first cycle of Edman degradation. However, comparison of 3H release profiles and relative amounts of βM3 and δM3 during sequence analyses of fractions from the 54 and 58/62 kDa gel bands established that the anesthetic-inhibitable peaks of 3H release originated from residues in βM3 rather than δM3. Sequencing through δM1 began only after 19 cycles of Edman degradation, at which point PTH-derivative and 3H releases were too low to allow characterization of photolabeling in δM1.
The detected sequences were quantitated by fitting the background-subtracted pmol of the detected peptide to the equation,
| (Eq. 2) |
where M(x) represents the pmol in cycle x, I0 is the initial amount of the peptide, and R is the repetitive yield. Cys, Trp, Ser, and His were omitted from the fits due to known problems with their quantitations. The 3H incorporation E(x), the efficiency of photolabeling (in cpm/pmol) of the amino acid in cycle x, was calculated by the following equation.
| (Eq. 3) |
Author Contributions
J. B. C. and K. W. M. conceived and coordinated the study. D. C. C. and J. B. C. designed and analyzed the experiments illustrated in Figs. 2–8 that were performed by D. C. C. Y. J. created the cell line expressing α4β3δ GABAAR, and X. Z. expressed, purified, and assayed the GABAAR under the guidance of K. W. M. P. Y. S. and K. S. B. synthesized the photoreactive anesthetics used in the study. J. B. C., D. C. C., and K. W. M. wrote the paper with input from all authors. All authors approved the final version of the manuscript.
Acknowledgment
We thank Selwyn S. Jayakar for assisting in the composition of Fig. 1 and for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-58448. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GABAAR
- γ-aminobutyric acid type A receptor
- etomidate
- ethyl 3-[(1R)-1-phenylethyl]imidazole-5-carboxylate
- azietomidate
- 2-(3-methyl-3H-diazirin-3-yl)ethyl (R)-1-(1-phenylethyl)-1H-imidazole-5-carboxylate
- R-mTFD-MPAB
- (R)-5-allyl-1-methyl-5-(m-trifluoromethyl-diazirinphenyl)barbituric acid
- S-mTFD-MPPB
- (S)-1-methyl-5-propyl-5-(m-trifluoromethyl-diazirinylphenyl)barbituric acid
- EndoLys-C
- Lysobacter enzymogenes endoproteinase Lys-C
- rpHPLC
- reversed-phase high performance liquid chromatography
- DS2
- δ-selective compound 2 (4-chloro-N-[2-(2-thienyl)imidazo[1,2-a]pyridine-3-yl]benzamide).
References
- 1.Sigel E. (2005) The benzodiazepine recognition site on GABAA receptors. Med. Chem. Rev. 2, 251–256 [DOI] [PubMed] [Google Scholar]
- 2.Miller P. S., and Smart T. G. (2010) Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol. Sci. 31, 161–174 [DOI] [PubMed] [Google Scholar]
- 3.Sieghart W. (2015) Allosteric modulation of GABAA receptors via multiple drug-binding sites. Adv. Pharmacol. 72, 53–96 [DOI] [PubMed] [Google Scholar]
- 4.Hemmings H. C. Jr., Akabas M. H., Goldstein P. A., Trudell J. R., Orser B. A., and Harrison N. L. (2005) Emerging molecular mechanisms of general anesthetic action. Trends Pharmacol. Sci. 26, 503–510 [DOI] [PubMed] [Google Scholar]
- 5.Forman S. A., Chiara D. C., and Miller K. W. (2015) Anesthetics target interfacial transmembrane sites in nicotinic acetylcholine receptors. Neuropharmacology 96, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauguet L., Shahsavar A., and Delarue M. (2015) Crystallographic studies of pharmacological sites in pentameric ligand-gated ion channels. Biochim. Biophys. Acta 1850, 511–523 [DOI] [PubMed] [Google Scholar]
- 7.Puthenkalam R., Hieckel M., Simeone X., Suwattanasophon C., Feldbauer R. V., Ecker G. F., and Ernst M. (2016) Structural studies of GABA-A receptor binding sites: which experimental structure tells us what? Front. Mol. Neurosci. 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G.-D., Chiara D. C., Sawyer G. W., Husain S. S., Olsen R. W., and Cohen J. B. (2006) Identification of a GABAA receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. J. Neurosci. 26, 11599–11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiara D. C., Dostalova Z., Jayakar S. S., Zhou X., Miller K. W., and Cohen J. B. (2012) Mapping general anesthetic binding site(s) in human α1β3 γ-aminobutyric acid type A receptors with [3H]TDBzl-etomidate, a photoreactive etomidate analogue. Biochemistry 51, 836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hibbs R. E., and Gouaux E. (2011) Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiara D. C., Jayakar S. S., Zhou X., Zhang X., Savechenkov P. Y., Bruzik K. S., Miller K. W., and Cohen J. B. (2013) Specificity of intersubunit general anesthetic-binding sites in the transmembrane domain of the human α1β3γ2 γ-aminobutyric acid type A (GABAA) receptor. J. Biol. Chem. 288, 19343–19357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belelli D., Lambert J. J., Peters J. A., Wafford K., and Whiting P. J. (1997) The interaction of the general anesthetic etomidate with the γ-aminobutyric acid type A receptor is influenced by a single amino acid. Proc. Natl. Acad. Sci. U.S.A. 94, 11031–11036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurd R., Arras M., Lambert S., Drexler B., Siegwart R., Crestani F., Zaugg M., Vogt K. E., Ledermann B., Antkowiak B., and Rudolph U. (2003) General anesthetic actions in vivo strongly attenuated by a point mutation in the GABAA receptor β3 subunit. FASEB J. 17, 250–252 [DOI] [PubMed] [Google Scholar]
- 14.Liao M., Sonner J. M., Husain S. S., Miller K. W., Jurd R., Rudolph U., and Eger E. I. 2nd (2005) R(+) etomidate and the photoactivable R(+) azietomidate have comparable anesthetic activity in wild-type mice and comparably decreased activity in mice with a N265M point mutation in the γ-aminobutyric acid receptor β3 subunit. Anesth. Analg. 101, 131–135, table of contents [DOI] [PubMed] [Google Scholar]
- 15.Amlong C. A., Perkins M. G., Houle T. T., Miller K. W., and Pearce R. A. (2016) Contrasting effects of the γ-aminobutyric acid type A receptor β3 subunit N265M mutation on loss of righting reflexes induced by etomidate and the novel anesthetic barbiturate R-mTFD-MPAB. Anesth. Analg. 123, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel P. M., Patel H. H., and Roth D. (2011) General anesthetics and therapeutic gases. In Goodman and Gilman's The Pharmacological Basis of Experimental Therapeutics (Brunton L., Chabner B., and Knollman B., eds) pp. 527–564, McGraw-Hill, New York [Google Scholar]
- 17.Drexler B., Antkowiak B., Engin E., and Rudolph U. (2011) Identification and characterization of anesthetic targets by mouse molecular genetics approaches. Can. J. Anaesth. 58, 178–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrant M., and Nusser Z. (2005) Variations on an inhibitory theme: phasic and tonic activation of GABAA receptors. Nat. Rev. Neurosci. 6, 215–229 [DOI] [PubMed] [Google Scholar]
- 19.Brickley S. G., and Mody I. (2012) Extrasynaptic GABAA receptors: their function in the CNS and implications for disease. Neuron 73, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stell B. M., Brickley S. G., Tang C. Y., Farrant M., and Mody I. (2003) Neuroactive steroids reduce neuronal excitability by selectively enhancing tonic inhibition mediated by δ subunit-containing GABA-A receptors. Proc. Natl. Acad. Sci. U.S.A. 100, 14439–14444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert J. J., Cooper M. A., Simmons R. D. J., Weir C. J., and Belelli D. (2009) Neurosteroids: endogenous allosteric modulators of GABA-A receptors. Psychoneuroendocrinology 34, S48–S58 [DOI] [PubMed] [Google Scholar]
- 22.Jia F., Yue M., Chandra D., Homanics G. E., Goldstein P. A., and Harrison N. L. (2008) Isoflurane is a potent modulator of extrasynaptic GABAA receptors in the thalamus. J. Pharmacol. Exp. Ther. 324, 1127–1135 [DOI] [PubMed] [Google Scholar]
- 23.Bieda M. C., Su H., and Maciver M. B. (2009) Anesthetics discriminate between tonic and phasic γ-aminobutyric acid receptors on hippocampal CA1 neurons. Anesth. Analg. 108, 484–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kretschmannova K., Hines R. M., Revilla-Sanchez R., Terunuma M., Tretter V., Jurd R., Kelz M. B., Moss S. J., and Davies P. A. (2013) Enhanced tonic inhibition influences the hypnotic and amnestic actions of the intravenous anesthetics etomidate and propofol. J. Neurosci. 33, 7264–7273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaur K. H., Baur R., and Sigel E. (2009) Unanticipated structural and functional properties of δ-subunit-containing GABAA receptors. J. Biol. Chem. 284, 7889–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagoner K. R., and Czajkowski C. (2010) Stoichiometry of expressed α4β2δ γ-aminobutyric acid type A receptors depends on the ratio of subunit cDNA transfected. J. Biol. Chem. 285, 14187–14194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eaton M. M., Bracamontes J., Shu H. J., Li P., Mennerick S., Steinbach J. H., and Akk G. (2014) γ-Aminobutyric acid type A α4, β2, and δ subunits assemble to produce more than one functionally distinct receptor type. Mol. Pharmacol. 86, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel B., Mortensen M., and Smart T. G. (2014) Stoichiometry of δ subunit containing GABAA receptors. Br. J. Pharmacol. 171, 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartiadi L. Y., Ahring P. K., Chebib M., and Absalom N. L. (2016) High and low GABA sensitivity α4β2δ GABAA receptors are expressed in Xenopus laevis oocytes with divergent stoichiometries. Biochem. Pharmacol. 103, 98–108 [DOI] [PubMed] [Google Scholar]
- 30.Wongsamitkul N., Baur R., and Sigel E. (2016) Toward understanding functional properties and subunit arrangement of α4β2δ γ-aminobutyric acid, type A (GABAA) receptors. J. Biol. Chem. 291, 18474–18483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botzolakis E. J., Gurba K. N., Lagrange A. H., Feng H. J., Stanic A. K., Hu N., and Macdonald R. L. (2016) Comparison of γ-aminobutyric acid, type A (GABAA), receptor αβγ and αβδ expression using flow cytometry and electrophysiology: evidence for alternative subunit stoichiometries and arrangements. J. Biol. Chem. 291, 20440–20461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen M. L., Wafford K. A., Brown A. R., Belelli D., Lambert J. J., and Mirza N. R. (2013) A study of subunit selectivity, mechanism and site of action of the δ selective compound 2 (DS2) at human recombinant and rodent native GABA-A receptors. Br. J. Pharmacol. 168, 1118–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dostalova Z., Zhou X., Liu A., Zhang X., Zhang Y., Desai R., Forman S. A., and Miller K. W. (2014) Human α1β3γ2L γ-aminobutyric acid type A receptors: high-level production and purification in a functional state. Protein Sci. 23, 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 35.Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J. R., and Mohammed S. (2009) Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal. Chem. 81, 4493–4501 [DOI] [PubMed] [Google Scholar]
- 36.Jayakar S. S., Zhou X., Savechenkov P. Y., Chiara D. C., Desai R., Bruzik K. S., Miller K. W., and Cohen J. B. (2015) Positive and negative allosteric modulation of an α1β3γ2 γ-aminobutyric acid type A (GABA-A) receptor by binding to a site in the transmembrane domain at the γ+-β− interface. J. Biol. Chem. 290, 23432–23446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrera N. P., Betts J., You H., Henderson R. M., Martin I. L., Dunn S. M. J., and Edwardson J. M. (2008) Atomic force microscopy reveals the stoichiometry and subunit arrangement of the α4β3δ GABAA receptor. Mol. Pharmacol. 73, 960–967 [DOI] [PubMed] [Google Scholar]
- 38.Feng H. J., Jounaidi Y., Haburcak M., Yang X., and Forman S. A. (2014) Etomidate produces similar allosteric modulation in α1β3δ and α1β3γ2L GABA(A) receptors. Br. J. Pharmacol. 171, 789–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jayakar S. S., Zhou X., Chiara D. C., Dostalova Z., Savechenkov P. Y., Bruzik K. S., Dailey W. P., Miller K. W., Eckenhoff R. G., and Cohen J. B. (2014) Multiple propofol-binding sites in a γ-aminobutyric acid type A receptor (GABAAR) identified using a photoreactive propofol analog. J. Biol. Chem. 289, 27456–27468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegwart R., Jurd R., and Rudolph U. (2002) Molecular determinants for the action of general anesthetics at recombinant α2β3γ2 γ-aminobutyric acid(A) receptors. J. Neurochem. 80, 140–148 [DOI] [PubMed] [Google Scholar]
- 41.Reynolds D. S., Rosahl T. W., Cirone J., O'Meara G. F., Haythornthwaite A., Newman R. J., Myers J., Sur C., Howell O., Rutter A. R., Atack J., Macaulay A. J., Hadingham K. L., Hutson P. H., Belelli D., et al. (2003) Sedation and anesthesia mediated by distinct GABA-A receptor isoforms. J. Neurosci. 23, 8608–8617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart D. S., Pierce D. W., Hotta M., Stern A. T., and Forman S. A. (2014) Mutations at β N265 in γ-aminobutyric acid type A receptors alter both binding affinity and efficacy of potent anesthetics. PLoS One 9, e111470, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart D., Desai R., Cheng Q., Liu A., and Forman S. A. (2008) Tryptophan mutations at azi-etomidate photoincorporation sites on α1 or β2 subunits enhance GABA-A receptor gating and reduce etomidate modulation. Mol. Pharmacol. 74, 1687–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng H. J., and Macdonald R. L. (2010) Barbiturates require the N terminus and first transmembrane domain of the δ subunit for enhancement of α1β3δ GABA-A receptor currents. J. Biol. Chem. 285, 23614–23621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Estrada-Mondragon A., and Lynch J. (2015) Functional characterization of ivermectin binding sites in α1β2γ2L GABA(A) receptors. Front. Mol. Neurosci. 8, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krasowski M. D., Nishikawa K., Nikolaeva N., Lin A., and Harrison N. L. (2001) Methionine 286 in transmembrane domain 3 of the GABAA receptor β subunit controls a binding cavity for propofol and other alkylphenol general anesthetics. Neuropharmacology 41, 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maldifassi M. C., Baur R., and Sigel E. (2016) Functional sites involved in modulation of the GABAA receptor channel by the intravenous anesthetics propofol, etomidate and pentobarbital. Neuropharmacology 105, 207–214 [DOI] [PubMed] [Google Scholar]
- 48.Stewart D. S., Hotta M., Li G. D., Desai R., Chiara D. C., Olsen R. W., and Forman S. A. (2013) Cysteine substitutions define etomidate binding and gating linkages in the α-M1 domain of γ-aminobutyric acid type A (GABAA) receptors. J. Biol. Chem. 288, 30373–30386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nourmahnad A., Stern A. T., Hotta M., Stewart D. S., Ziemba A. M., Szabo A., and Forman S. A. (2016) Tryptophan and cysteine mutations in M1 helices of α1β3γ2L γ-aminobutyric acid type A receptors indicate distinct intersubunit sites for four intravenous anesthetics and one orphan site. Anesthesiology 125, 1144–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savechenkov P. Y., Zhang X., Chiara D. C., Stewart D. S., Ge R., Zhou X., Raines D. E., Cohen J. B., Forman S. A., Miller K. W., and Bruzik K. S. (2012) Allyl m-trifluoromethyldiazirine mephobarbital: an unusually potent enantioselective and photoreactive barbiturate general anesthetic. J. Med. Chem. 55, 6554–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziebell M. R., Nirthanan S., Husain S. S., Miller K. W., and Cohen J. B. (2004) Identification of binding sites in the nicotinic acetylcholine receptor for [3H]azietomidate, a photoactivatable general anesthetic. J. Biol. Chem. 279, 17640–17649 [DOI] [PubMed] [Google Scholar]