Abstract
Epigallocatechin-3-gallate (EGCG) is the major polyphenol in green tea. It has antimicrobial properties and disrupts the ordered structure of amyloid fibrils involved in human disease. The antimicrobial effect of EGCG against the opportunistic pathogen Pseudomonas aeruginosa has been shown to involve disruption of quorum sensing (QS). Functional amyloid fibrils in P. aeruginosa (Fap) are able to bind and retain quorum-sensing molecules, suggesting that EGCG interferes with QS through structural remodeling of amyloid fibrils. Here we show that EGCG inhibits the ability of Fap to form fibrils; instead, EGCG stabilizes protein oligomers. Existing fibrils are remodeled by EGCG into non-amyloid aggregates. This fibril remodeling increases the binding of pyocyanin, demonstrating a mechanism by which EGCG can affect the QS function of functional amyloid. EGCG reduced the amyloid-specific fluorescent thioflavin T signal in P. aeruginosa biofilms at concentrations known to exert an antimicrobial effect. Nanoindentation studies showed that EGCG reduced the stiffness of biofilm containing Fap fibrils but not in biofilm with little Fap. In a combination treatment with EGCG and tobramycin, EGCG had a moderate effect on the minimum bactericidal eradication concentration against wild-type P. aeruginosa biofilms, whereas EGCG had a more pronounced effect when Fap was overexpressed. Our results provide a direct molecular explanation for the ability of EGCG to disrupt P. aeruginosa QS and modify its biofilm and strengthens the case for EGCG as a candidate in multidrug treatment of persistent biofilm infections.
Keywords: amyloid, biofilm, fibril, natural product, Pseudomonas aeruginosa (P. aeruginosa), bacterial fibrils, epigallocatechin gallate (EGCG), fibrillation inhibition, nanoindentation, pyocyanin binding
Introduction
Biofilms are defined by the International Union of Pure and Applied Chemistry as aggregates of microorganisms in which cells, which are frequently embedded within a self-produced matrix of extracellular polymeric substance, adhere to each other and/or to a surface (1). The development of biofilms occurs in multiple stages, including an initial attachment to a substratum, formation of microcolonies, development of mature biofilms, and finally biofilm dispersal (2). This process is controlled through the excretion of so-called quorum-sensing (QS)2 signal molecules that induce transcriptional changes in the recipient cells (3, 4). QS allows the bacteria to collectively express specific genes in response to environmental cues, and disruption of the QS system shows promise as an antibiofilm and antipathogenic target (5). Biofilms show much higher resistance to antibiotics than do planktonic bacteria due to the physical protection afforded by the extracellular matrix (2). Accordingly, they can cause persistent bacterial infections, and one extensively studied biofilm-forming microorganism is the opportunistic pathogen Pseudomonas aeruginosa that forms persistent infections in the lungs of cystic fibrosis patients (6, 7). The extracellular matrix of biofilms consists of exopolysaccharides, extracellular DNA, and amyloid fibrils (8–10).
Amyloids are highly ordered insoluble fibrils formed by widely different proteins (11, 12). Some amyloids are associatedwith protein misfolding and diseases; however, natively amyloidogenic proteins have also evolved to serve functional roles in many organisms (13). In prokaryotes, functional amyloids were first discovered in Escherichia coli and termed curli (14), and several others were later discovered, e.g. TasA in Bacillus subtilis (15) and Fap in Pseudomonas (16, 17). These organisms all produce extracellular fibrils as part of the biofilm. Evolutionary studies of the curli and Fap amyloids have shown that these amyloid systems have spread to multiple bacterial phyla and classes (18, 19). Moreover, many other biofilm-forming prokaryotes stain positive for amyloids (10, 21, 22), highlighting the general importance of amyloids in biofilms.
In the Fap system, six proteins (FapA–FapF) in total are transcribed from the fap operon of which FapC is the major fibril component. Protein composition of the fibrils and sequence analysis suggests that these proteins have specific roles as a fibrillation nucleator (FapB), a β-barrel membrane pore used for secretion of FapC and FapB monomers (FapF), and a protease required for processing of the amyloid subunits (FapD) (16, 23). Fap was originally thought to provide only a structural component to the biofilm as rigid fibers; however, recent studies have shown a more complex role for amyloids (16, 17, 23, 24). Thus, overexpression of the fap operon in P. aeruginosa PAO1 leads to a highly aggregative and adherent phenotype that forms excessive amounts of biofilm. Analysis of this overexpressing strain showed that amyloid formation markedly changes the overall proteome (16, 23). In general, fap overexpression reduces the abundance of virulence factors and increases that of biofilm-associated proteins. Interestingly, these proteomic changes have several parallels to those observed in chronic infections of cystic fibrosis patients (23). Also, a transposon deletion mutant assay of a virulent and high QS molecule-producing P. aeruginosa strain identified the fapC deletion mutant as deficient in quorum sensing, more susceptible to leukocyte phagocytosis, and highly attenuated in killing the nematode Caenorhabditis elegans (25). Hence, FapC is considered an important factor in P. aeruginosa virulence. Recently, we also showed that FapC fibrils could bind QS molecules, suggesting amyloid involvement in information transfer in non-quiescent environments (24). In addition, Fap expression in the Fap model organism Pseudomonas sp. UK4 (26, 27) has been shown to increase biofilm hydrophobicity and mechanical strength.
Epigallocatechin-3-gallate (EGCG) is the major polyphenolic compound found in green tea at typical levels of 0.7 g/liter (1.5 mm) (28). EGCG has a broad antibacterial effect (29), possibly through interfering with QS as exposure of P. aeruginosa PAO1 to EGCG down-regulates both the las and pqs QS systems and reduces swarming (30). EGCG displays very promiscuous protein binding and inhibits the function of a broad range of proteins (31–36). Although such promiscuity complicates simple deductions about its effects in vivo, it arguably increases the impact of the compound on bacterial communities. Furthermore, EGCG can weakly bind to but not penetrate cellular membranes (37), so its targets will be limited to the proteins found in the extracellular matrix. Indeed, EGCG binds Staphylococcus aureus and E. coli cell walls (38), doubles S. aureus cell wall thickness (39), and increases P. aeruginosa membrane permeability (40). Suggested mechanisms for these effects include H2O2 production and blockage of outer membrane porins (41, 42). EGCG also inhibits fibrillation of a broad range of amyloidogenic proteins such as transthyretin (43), lysozyme (44), Sup35 prion (45), α-synuclein, and β-amyloid (33, 46). In these cases, EGCG induces formation of partially stable oligomers that do not form amyloids (33). The partially stable oligomers have been suggested to result from covalent reactions between EGCG and protein sulfhydryl and amino groups (31, 32). However, covalent modification is not a prerequisite for the fibrillation-inhibitory effect of EGCG (47, 48). Additionally, EGCG can rearrange preformed amyloid fibrils to amorphous aggregates, most likely via hydrophobic sites on the fibrils (47). The mechanism by which EGCG remodels fibrils into amorphous aggregates has not yet been elucidated.
Given the known effects of EGCG on amyloidogenic proteins associated with misfolding and disease, EGCG is likely to affect naturally occurring functional amyloids as well. Very recently, it was reported that EGCG interferes with biofilm formation in E. coli via both the curli and cellulose biosynthesis pathways (49). The authors elegantly showed that EGCG acts on the small RNA RybD to activate the RpoE response and thus reduce the expression of the regulator protein CsgD. This in turn down-regulates biosynthesis of both curli and cellulose. In addition, there was indirect evidence that EGCG prevents assembly of secreted CsgA as curli. However, no direct observations on biofilm properties, antibiotic resistance, or assembly properties of the amyloid-forming protein were reported. The objective of this study was to determine whether EGCG can inhibit Fap amyloid assembly; remodel existing Fap amyloids in P. aeruginosa PAO1 biofilms; and affect quorum sensing, biofilm strength, and antibiotic resistance. Here we show that EGCG inhibits fibrillation of FapC and structurally remodels fibrils formed in vitro. Furthermore, EGCG reduced the amyloid-specific signals from thioflavin T in biofilms in situ. The fibril remodeling affected binding of the QS molecule pyocyanin to FapC. Furthermore, in a P. aeruginosa PAO1 strain overexpressing functional amyloid, EGCG reduced the biofilm Young's modulus and increased biofilm susceptibility to the antibiotic tobramycin.
Results
EGCG and Doxycycline Remodel FapC Amyloid Fibrils
To investigate the possibility that functional amyloids in P. aeruginosa can be remodeled, we screened the effect of EGCG together with three other compound candidates that have shown antiamyloid effects on other proteins (proteins indicated in parentheses): silibinin (human islet amyloid polypeptide fibrils) (50), doxycycline (Aβ and β2-microglobulin fibrils) (51, 52), and curcumin (Aβ fibrils) (53).
Initially, we tested whether these compounds had an effect on the structure of functional bacterial amyloids in vitro. Our objective was to work with highly pure yet biologically representative fibril samples. However, native fibrils purified in bulk from P. aeruginosa PAO1 cultures often contain polysaccharides, membrane fragments, and residual SDS, which in turn may complicate spectral assays and transmission electron microscopy (TEM) (16, 23). In contrast, fibrils formed in vitro from highly purified and His-tagged FapC could differ from the original (authentic) fibrils formed in vivo as the monomer fibrillation involves self-propagation of a particular cross-β structure (54). This issue was resolved by seeding recombinantly expressed FapC with native fibrils purified as fibrils from P. aeruginosa PAO1 cultures, reasoning that the native fibrils would imprint their cross-β structure on monomeric FapC through an iterative fibril elongation (24, 55, 56). Briefly, after each fibril elongation step, the fibrils were diluted 20-fold and used as seeds in the next round of fibril elongation by monomeric FapC. The process was repeated three times, thereby reducing the original sample to 0.02% (w/w) and resulting in large amounts of highly pure but authentic fibrils.
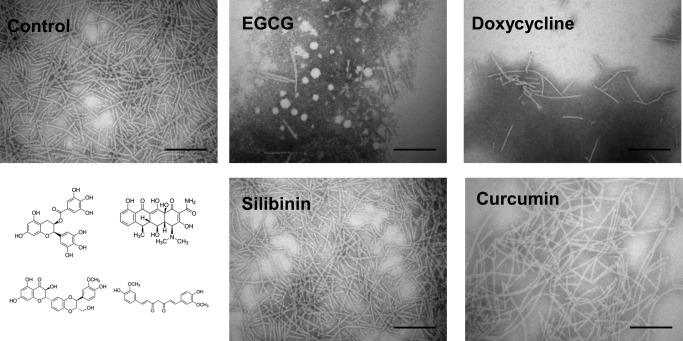
Using these authentic fibrils, the four potential amyloid modifiers were tested. Fibrils (0.5 mg/ml) were incubated in 100 μm solutions of the compounds for 24 h and examined by TEM (Fig. 1). In the absence of these compounds, the fibrils were long, unbranched, and slightly curved, similar to native amyloids purified from P. aeruginosa PAO1 cultures (16). However, incubation with EGCG or doxycycline reduced the amount of fibril material markedly and led to formation of diffuse aggregates located close to the fibrils. In contrast, neither silibinin nor curcumin induced any detectable changes in fibril morphology or the amount of fibrils.
FIGURE 1.
EGCG and doxycycline can remodel FapC amyloid fibrils. Shown are TEM images of 0.5 mg/ml fibrils incubated with 100 μm concentrations of the indicated compounds for 24 h. Scale bars, 200 nm. The chemical structures of compounds are shown to the bottom left with the same spatial arrangement as the four images of the compound-treated fibrils.
Doxycycline Is Cytotoxic, and EGCG Reduces Growth Rate
Doxycycline and EGCG appeared promising as potential modifiers of Fap fibrils. However, before testing this possibility in situ on biofilms containing FapC fibrils, the potential antibacterial properties of the two compounds had to be examined given the use of doxycycline as a broad spectrum antibiotic. The effect of the compounds on the growth rate of the non-biofilm-forming E. coli DSM429 strain (57) was investigated (Fig. 2). Consistent with reported minimum inhibitory concentrations of 0.5–32 mg/liter for E. coli isolates (58), doxycycline was cytotoxic and arrested culture growth at a concentration of 7 μm (3 mg/liter). In contrast, EGCG did not arrest cell growth across the concentration range used (up to 500 μm), but it did reduce the growth rate by 36% with a half-maximum effective concentration (EC50) of 4.7 μm. The cytotoxicity of doxycycline makes it unsuitable for experiments in situ as effects other than that on amyloids may dominate at the concentrations required for testing fibril remodeling. On the contrary, EGCG only had a moderate effect on cell growth, and for this reason we limited our subsequent investigations to EGCG.
FIGURE 2.
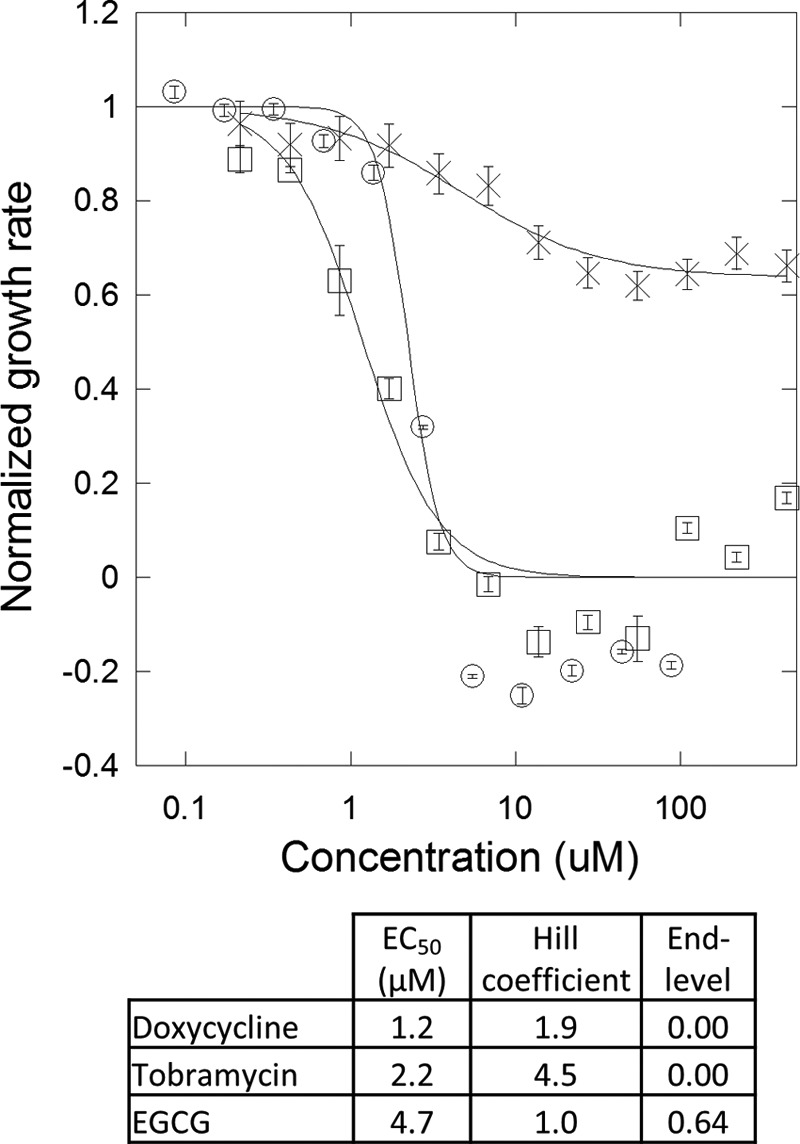
Doxycycline stops growth of non-biofilm forming E. coli DSM429, whereas EGCG reduces growth rates by 35%. Growth rates of E. coli DSM429 with different concentrations of doxycycline (□), tobramycin (○), and EGCG (×) are shown. Doxycycline and EGCG are with 1% DMSO, and tobramycin is with 1% ddH2O. Error bars denotes S.D. (n = 3). The Hill equation was fitted to the data. Fitting values are shown in the table.
EGCG Affects Conversion of Monomer into Amyloid Fibril
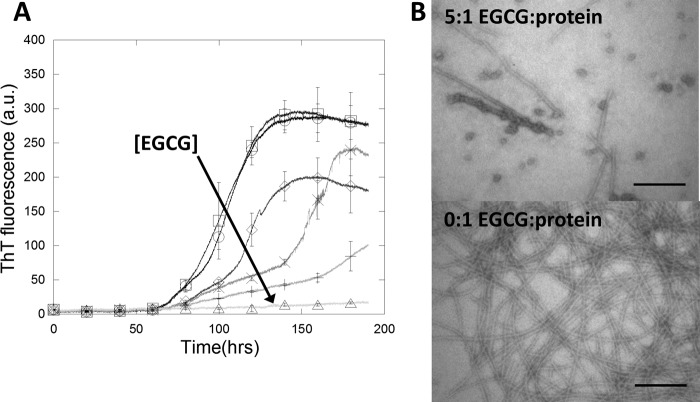
To understand how EGCG can inhibit the formation of new fibrils, its effect on the conversion of FapC monomers into amyloid fibrils was investigated. FapC monomer (0.5 mg/ml or 15 μm) was incubated with varying amounts of EGCG and monitored with the amyloid-binding dye thioflavin T (ThT) with agitation (Fig. 3A). Without EGCG, the lag time before fibrillation was ∼60 h, and the elongation phase contained two parts: an initial slower rise followed by a more rapid increase in ThT fluorescence, i.e. a steeper slope of the curve. For EGCG:FapC ratios of 0.5:1, 1:1, and 2:1, the lag time was unchanged, whereas the duration of the slower phase increased from 25 h without EGCG to 50 and 110 h at 1:1 and 2:1, respectively. The transition from the slow phase to the fast phase occurred at ∼60 absorbance units of ThT fluorescence up to a 2:1 EGCG:FapC ratio. The duration of the fast phase was ∼30 h before a plateau was reached (for molar ratios up to 1:1). Taken together, these results indicated that for EGCG:FapC ratios ≤2:1 EGCG affected the slower phase of protein fibrillation but not the duration of the lag time preceding the slow phase or the fast phase. At a 5:1 EGCG:FapC ratio, there was only a very slow increase in ThT fluorescence beginning at 100 h. The ThT fluorescence end point correlated within error with the increased EGCG concentration. In theory, fibril-binding compounds such as EGCG could affect the fluorescent signal of ThT by quenching or competitive binding, thereby biasing the results (59). However, quenching would affect the magnitude of the signal only and not the binding kinetics, so we attribute the change in lag time observed to a genuine effect on the fibrillation process.
FIGURE 3.
EGCG inhibits FapC fibrillation. A, protein fibrillation of 0.5 mg/ml (≈15 μm) recombinant FapC in the presence of 40 μm ThT in PBS with 1% DMSO. ThT fluorescence increases markedly when binding to amyloids. Increasing EGCG:FapC molar ratio is indicated by lighter gray scale and ○ (0:1), □ (0.2:1), ♢ (0.5:1), × (1:1), + (2:1), and ▵ (5:1). The direction of the arrow indicates higher EGCG concentration. Error bars denote S.D. (n = 3) and are shown for every 20 hour data point. B, TEM images of samples with or without a 5:1 molar ratio of EGCG:FapC monomer. Scale bars, 200 nm. a.u., arbitrary units.
EGCG Binds to Monomeric FapC and Forms Oligomers
Incubation of FapC with a 5× molar eq. of EGCG nearly abolished the fibrillation of FapC (Fig. 3A). However, the question remained whether incubation with EGCG yielded the same amorphous aggregates as observed when incubating preformed fibrils with EGCG. TEM images of samples without EGCG showed fibrils highly similar to the native-like fibrils formed by repeated seeding (compare Fig. 1, Control, with Fig. 3B, lower panel). Addition of EGCG apparently reduced the amount of fibrils present, which was consistent with the near absence of ThT fluorescence (Fig. 3, A and B). No amorphous aggregate was observed; rather, besides a few short linear fibrils, spherical oligomers of 29 ± 7 nm (S.D., n = 12) in diameter were observed. Similar oligomers have previously been observed when incubating the amyloidogenic α-synuclein with EGCG (33). These observations combined with those from incubating preformed fibrils with EGCG supported two different effects of EGCG: 1) EGCG remodels preformed FapC fibrils into large amorphous aggregates, but 2) EGCG incubation with monomeric FapC leads to more well defined oligomers instead of amorphous aggregates.
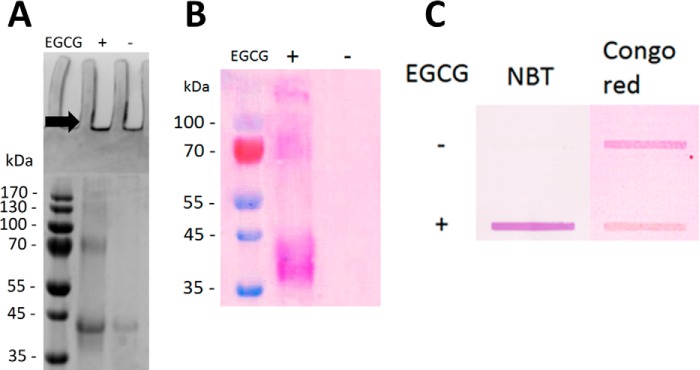
Different methods were used to probe how these oligomers were formed. According to denaturing gel electrophoresis (SDS-PAGE), fibrillated FapC incubated without EGCG was largely unable to enter the gel and produced only a faint protein band with an apparent molecular mass of 40 kDa (Fig. 4A). With EGCG, a more intense protein band at the same 40-kDa position was detected along with a series of protein bands at 80 kDa, 120 kDa, and upward. This series of protein bands indicated higher order species of stable oligomers as also observed for incubation of α-synuclein and amyloid-β with EGCG (33). Quinones such as EGCG can convert nitro blue tetrazolium (NBT) into formazan, which can be visually detected, and NBT staining of protein bands demonstrated that EGCG was indeed part of the FapC oligomers (60) (Fig. 4B). Because a large part of the total FapC amount did not enter the gel, the FapC incubated with or without EGCG was blotted onto a nitrocellulose membrane and NBT-stained (Fig. 4C). Formazan was clearly observed for samples incubated with EGCG but not those without. Staining the samples with Congo red, another amyloid-binding dye, revealed that non-EGCG-treated FapC contained amyloids, whereas the EGCG-treated FapC stained markedly less, which strongly supports that EGCG inhibited amyloid formation.
FIGURE 4.
EGCG forms SDS-stable oligomers with FapC. Shown are samples from Fig. 2 without EGCG or with a 5:1 EGCG:FapC monomer molar ratio. A, SDS-PAGE. B, SDS-polyacrylamide gel electroblotted onto PVDF membrane and stained with NBT. C, slot-blotting of 10 μl of sample in each slot and stained with either NBT or Congo red. Contrast was adjusted using GiMP version 2.8.
EGCG Remodels Fap Fibrils in a Dose-dependent Manner
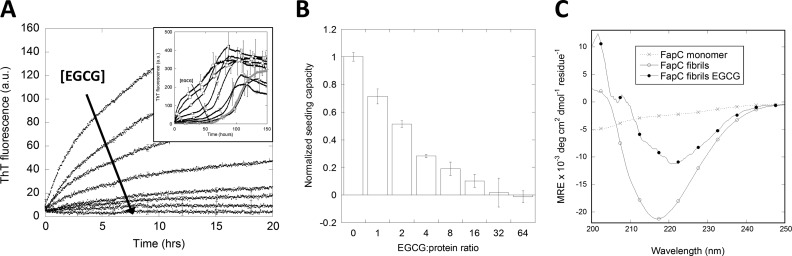
Addition of EGCG to Fap fibrils leads to a rapid, dose-dependent reduction in ThT fluorescence (data not shown). However, it was not clear whether this was due to effects on fibril structure or competitive displacement of ThT. ThT has previously been shown to inhibit EGCG remodeling of amyloids through competitive binding; likewise, EGCG can displace ThT, thereby reducing ThT fluorescence (61). We attempted to remove EGCG from solution by centrifugation at 100,000 × g for 30 min, but this led to loss of significant (15–30%) amounts of protein. To minimize EGCG-ThT interactions, we instead developed an assay to estimate fibril seeding capacity following incubation with EGCG. The seeding capacity was measured as the initial rate of fibrillation, i.e. the initial ThT fluorescence slope of incubating fibril seeds with monomeric FapC (Fig. 5A). This seeding capacity of sonicated fibrils was expected to decrease as incubation with EGCG transforms seeds from amyloids into amorphous aggregates; the concentration of EGCG was kept sufficiently low to avoid significant interference with ThT fluorescence. Hence, the effect of EGCG on the fibril seeds could be extrapolated from the effects on the seeding capacity. In practice, EGCG was incubated with FapC seeds and afterward diluted 20-fold into a FapC monomeric solution for another round of fibrillation monitored by ThT fluorescence. The seeding capacity was halved when using an EGCG:FapC molar ratio of 2:1 during incubation, whereas a 64:1 incubation ratio was needed to abolish the seeding potential (Fig. 5B). The final EGCG concentrations were very low at most of these molar ratios after 20-fold dilution of the seeds, allowing us to attribute essentially all the observed effect to seed remodeling rather than direct interference with ThT fluorescence. At the highest EGCG:FapC ratio used for seed incubation (64:1), the subsequent 20-fold dilution leads to a 3.2:1 EGCG:FapC molar ratio, which may still interfere with ThT fluorescence as well as the fibrillation process itself. However, compared with the absence of EGCG and seeds, the final ThT fluorescence signal was only 33% lower and followed similar fibrillation kinetics (Fig. 5 inset), supporting a negligible interference with ThT fluorescence. Based on Fig. 3, an inhibition of monomeric fibrillation with 3.2:1 EGCG:FapC would be expected; however, this was not observed. We attribute this to preincubation of EGCG, which allows it to bind to the fibril seeds as well as to undergo oxidation and polymerization. Hence, the effect of EGCG observed on the seeding capacity could be ascribed primarily to EGCG remodeling fibrils seeds as opposed to EGCG affecting the fibrillation process itself. Taken together, a few molar excess of EGCG was sufficient to remodel the majority of amyloid fibrils into non-seeding aggregates, whereas a large molar excess was needed for complete fibril seed remodeling in vitro.
FIGURE 5.
EGCG significantly reduces the seeding capacity of FapC fibrils. Seeds were preincubated with EGCG at an EGCG:protein molar ratio of 0, 1, 2, 4, 8, 16, 32, and 64. Subsequently, 5% seeds were added to 95% FapC monomer to a total concentration of 0.5 mg/ml and incubated with ThT. A, ThT fluorescence assay. The direction of the arrow indicates higher EGCG concentration. The panel shows the full time scale of 150 h including a non-seeded control (gray). B, initial slope of ThT fluorescence normalized to the signal from the sample without EGCG. Error bars denote S.D. (n = 3). C, far-UV CD spectra of FapC monomer and FapC fibrils incubated with and without EGCG showing a marked change in the FapC fibril structure upon incubation with EGCG. a.u., arbitrary units; MRE, mean residue ellipticity; deg, degrees.
Complementary evidence for this structural rearrangement was provided by far-UV CD spectra of FapC fibrils before and after incubation with EGCG (Fig. 5C). A marked reduction in the CD signal as well as a shift from a minimum around 217 to 221–222 nm confirmed that structural changes indeed accompanied this proposed remodeling.
EGCG Binds to Fibrils and Affects Their Binding of Pyocyanin
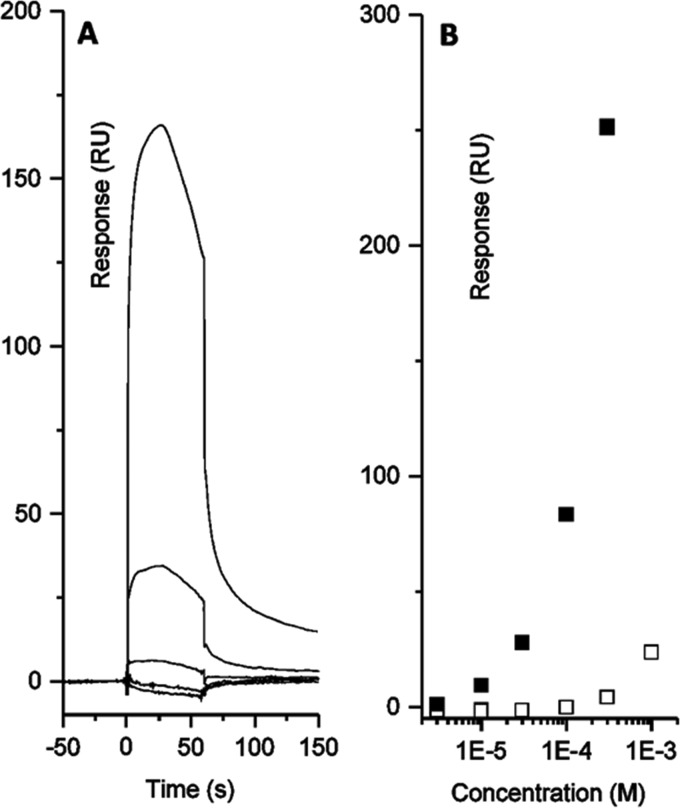
We previously showed that Fap fibrils are able to bind and retain QS molecules (24), and we reasoned that this binding may be affected by EGCG remodeling the fibrils. To test this hypothesis, the binding of pyocyanin, a central QS molecule and virulence factor, to immobilized Fap fibrils was measured using surface plasmon resonance (SPR). Pyocyanin has the added benefit of eliciting a strong response in the SPR signal (24). Immobilization of fibrils to the SPR chip surface was confirmed using ThT, which binds with low affinity, leading to an apparent dissociation constant Kd of ∼48 μm, consistent with the ∼100 μm estimated in our previous study (data not shown) (24). For EGCG, a rapid association and slow dissociation response was observed, indicating binding of EGCG to the amyloid fibrils (Fig. 6A). The more gradual dissociation compared with the QS molecules was likely due to the fact that EGCG is a larger, bulkier molecule than for example ThT or pyocyanin. Because of the chemical instability of EGCG, deviations from normal binding kinetics could be attributed to EGCG oxidation. EGCG binding to Aβ fibrils comprises both hydrophobic and hydrogen-binding contributions without specific binding sites (1, 2). A similar binding is hence expected for EGCG and FapC fibrils. Upon EGCG remodeling of FapC fibrils, pyocyanin elicits a much higher SPR response (Fig. 6B). We cannot conclude exactly how this signal increase occurred, but it indicates an altered mode of binding. It was not possible to measure the affinity of EGCG or pyocyanin for fibrils by isothermal titration calorimetry due to poor data quality (data not shown), but we confirmed the increased binding of pyocyanin to EGCG-remodeled fibrils as follows. Fibrils with or without pretreatment with 500 μm EGCG were incubated with 500 μm pyocyanin, and the amount of free (unbound) pyocyanin was determined by filtering the solution and measuring the concentration of pyocyanin remaining in the filtrate. Consistent with our SPR results, EGCG-free fibrils did not bind detectable amounts of pyocyanin, but the EGCG-remodeled fibrils bound 168 ± 12 μm pyocyanin. Hydrophobic interactions also seem to drive Fap fibril-QS molecule interactions (3), and we attribute the signal increase to changes in fibril structure that increased pyocyanin binding capacity, possibly through exposure of hydrophobic protein structure, although this must remain speculative.
FIGURE 6.
EGCG binds fibrils and affects pyocyanin binding. A, surface plasmon resonance sensorgram with different concentrations of EGCG. B, dose-response curves for pyocyanin and FapC fibrils before (white) and after (black) EGCG incubation of FapC fibrils. All experiments were performed in triplicate. Errors are smaller than the size of the symbols. RU, resonance units.
EGCG Can Remodel Fibrils in P. aeruginosa PAO1 Biofilm
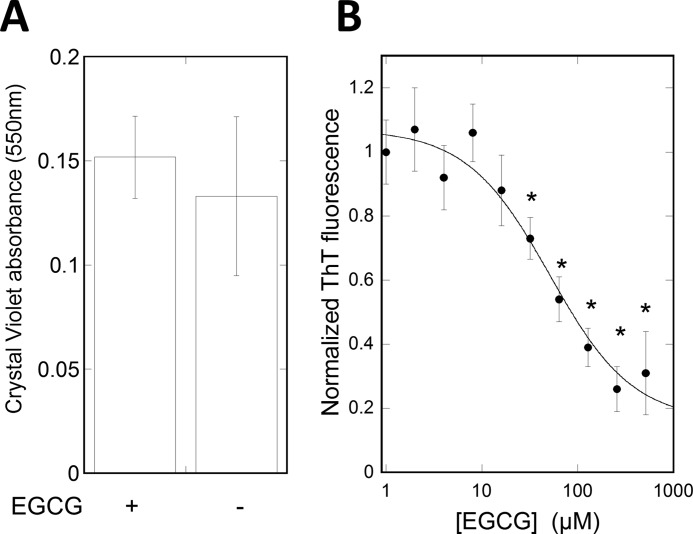
In P. aeruginosa PAO1 biofilms, the Fap amyloids are embedded in the biofilm extracellular matrix along with exopolysaccharides and extracellular DNA (8, 62). Motivated by the effect of EGCG on fibrils in vitro, we investigated whether EGCG can access and remodel the Fap fibrils when they are embedded in the biofilm. P. aeruginosa PAO1 biofilms were grown in a microtiter assay, exposed to EGCG, washed, and sonicated to release the biofilm. The amount of amyloid present was then detected by ThT fluorescence. Crystal violet staining of the biofilm followed by visual inspection confirmed that the biofilm was released from the wells after sonication. Absorbance measurements of released crystal violet showed no significant difference in biofilm amount with and without EGCG (Fig. 7A). A reduction in ThT fluorescence was detected with increasing concentrations of EGCG. A maximum reduction in fluorescence was reached at 250 μm EGCG, and EGCG showed an EC50 of 49 μm (Fig. 7B). Hence, EGCG can access and remodel amyloid fibrils in biofilms at a concentration range that overlaps with the concentration used in studies on EGCG inhibition of bacterial growth (0.1–1.1 mm) (29, 41). However, it cannot be ruled out that Fap amyloid fibrils were removed by the bacteria due to EGCG-induced transcriptional changes. Approximately 20% of the ThT fluorescence signal remained even after EGCG treatment. We attribute this to amyloid inaccessibility, reduced efficacy toward remaining amyloid (as seen in vitro), ThT binding to non-amyloid structures, or a combination thereof.
FIGURE 7.
EGCG can remodel Fap fibrils in biofilms. A, determination of biofilm amount using crystal violet assay after treatment with or without 500 μm EGCG for 6 h. B, biofilm ThT fluorescence after 6-h treatment with different EGCG concentrations. All experiments were performed in triplicate. Measurements were fitted with a dose-response curve. Error bars denote 95% confidence intervals, and * denotes a p value <0.0001 compared with 0 μm EGCG.
EGCG Affects the Structural Stability of Biofilms with Fap Overexpression
The data in Fig. 7 indicated that EGCG reduced the amyloid content of biofilm, although the total amount of biofilm remained unchanged. To investigate whether the amyloid reduction affects the mechanical properties of the biofilm, nanoindentation was performed using a large microbead attached to an atomic force microscopy (AFM) cantilever. The indentation curve at a constant loading force (2 nanonewtons) was used to calculate the Young's modulus of the biofilm, as a measure of biofilm stiffness, following the same procedure that we have previously demonstrated on Pseudomonas sp. UK4 biofilm (Fig. 8) (26). Two P. aeruginosa strains were used: the PAO1 wild-type strain (PAO1 WT) and the Fap-overproducing strain (PAO1 pFap) (16). Overexpression of pFap led to a ∼2.5-fold decrease in biofilm stiffness, which is opposite that observed for Pseudomonas sp. UK4 biofilm. However, this is consistent with the observation that Fap overexpression in P. aeruginosa PAO1 leads to a mucoid phenotype due to the increased alginate production (16, 23). This also suggested that Fap might have different interactions and lead to different responses in various Pseudomonas species. Addition of EGCG to PAO1 WT did not result in a significantly different stiffness in agreement with the low levels of amyloid in this strain and the consequent inability to carry out fibrillar remodeling (Fig. 8). In contrast, a ∼2-fold reduction in biofilm stiffness was observed in PAO1 pFap biofilms.
FIGURE 8.
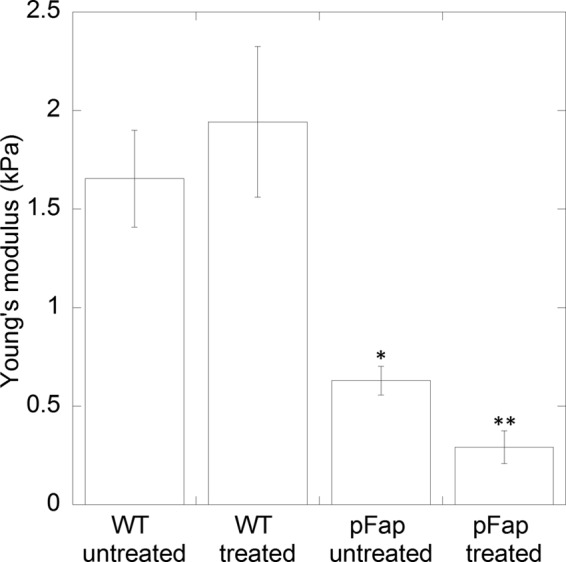
EGCG reduces biofilm stiffness in fap overexpression strain. Young's modulus was calculated by indentation curves of a microbead attached to an AFM cantilever as in Zeng et al. (26). The two P. aeruginosa strains used were PAO1 WT and PAO1 pFap (Fap overexpression) untreated or treated with EGCG for 5 h. Error bars denote 95% confidence intervals, and * and ** denote p values <0.001 compared with PAO1 WT untreated and PAO1 pFap untreated, respectively. kPa, kilopascals.
EGCG Increases the Antibiotic Potency of Tobramycin
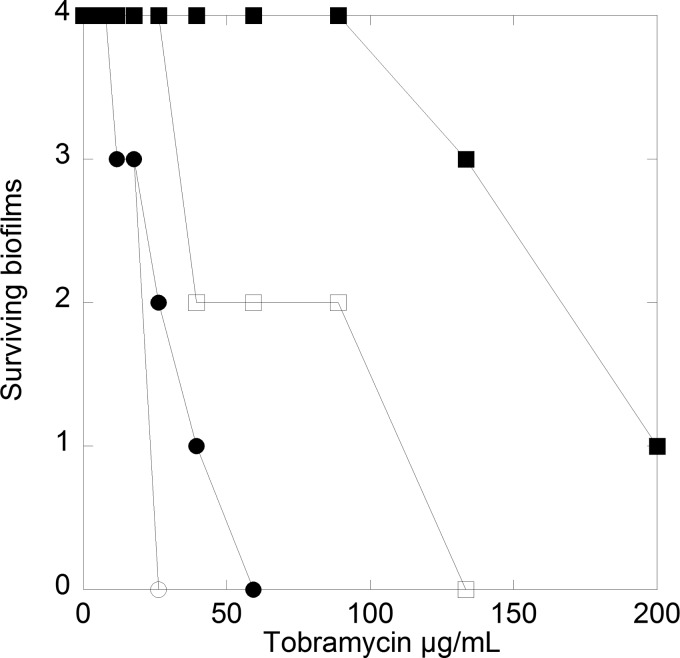
It is a salient feature of biofilms that they increase the resistance of P. aeruginosa toward antibiotics and that the aminoglycoside tobramycin is frequently used to treat P. aeruginosa infections in cystic fibrosis patients (6, 63). Based on the effects of EGCG on amyloids in the P. aeruginosa PAO1 biofilm, the effect on biofilm strength, and amyloid binding to pyocyanin, we hypothesized that EGCG could disrupt biofilm and/or QS, thereby increasing bacterial susceptibility to antibiotics. Accordingly, P. aeruginosa PAO1 biofilms were subjected to a dilution series of tobramycin to determine the minimal bactericidal eradication concentration (MBEC) with and without 500 μm EGCG (Fig. 9). Both the PAO1 WT and PAO1 pFap strain were used, and MBECs were measured under culturing conditions with biofilm formation. Overexpression of the fap operon led to biofilms with much higher tobramycin resistance than that of PAO1 WT biofilms. PAO1 WT biofilms were partially eradicated at 10 mg/liter tobramycin and completely eradicated at 59 mg/liter, whereas as much as 200 mg/liter tobramycin failed to completely eradicate PAO1 pFap biofilms. The markedly lower antibiotic susceptibility of the PAO1 pFap were likely due to the ability of this strain to produce excessive amounts of biofilm (16, 23). Addition of EGCG during tobramycin challenge of biofilms increased the antibiotic susceptibility of both PAO1 WT and PAO1 pFap. However, the effect was most pronounced for PAO1 pFap where EGCG reduced the amount of tobramycin needed to halve biofilm survival by a factor of ∼3 (from (<150 to <50 μg/ml); for PAO1 WT, this fell from ∼30 to ∼25 μg/ml (Fig. 9). These results indicate that the presence of EGCG during the tobramycin challenge strongly reduces the resistance of P. aeruginosa PAO1 biofilms to antibiotics when amyloid production is part of the resistance mechanism.
FIGURE 9.
EGCG induces tobramycin susceptibility in PAO1 biofilms. Shown are MBECs of PAO1 WT (○) and PAO1 pFap (□) (fap-overproducing strain). Four biofilms were tested for each tobramycin concentration with (open symbols) and without 500 μm EGCG (filled symbols). Survival of the biofilm was measured by allowing the biofilm to regrow in medium for 24 h and detected by growth on agar plates.
Discussion
EGCG Remodels FapC Fibrils in Vitro and in Situ
This study provides strong evidence that EGCG can both inhibit fibrillation and remodel already formed Fap fibrils in vitro. The mechanism of fibrillation inhibition is similar to the effect of EGCG shown for other amyloid-forming proteins (33, 43–46). The FapC monomers are directed into oligomers that are at least partially stable toward heat and SDS (consistent with indirect observations on EGCG solubilization of CsgA in E. coli (49)), and preformed fibrils are turned into amorphous aggregates with a change in far-UV CD structure that is significantly larger than the effect reported on e.g. islet amyloid polypeptide fibrils (47). Other studies support that EGCG is covalently cross-linked to proteins in these SDS-stable oligomers, and EGCG binding to transthyretin has been confirmed by mass spectroscopy (43). Formation of Fap amyloids in vivo does not follow the simple nucleation-dependent fibrillation that occurs in vitro; e.g. the other five fap proteins and potentially other proteins are involved in regulating functional amyloid formation (17). FapB is a homologue of FapC and likely nucleates FapC in vivo analogously to the CsgA/CsgB pair in E. coli curli (17, 64). Whether EGCG can inhibit fibrillation of FapC in vivo is unknown but may be irrelevant in view of the fact that EGCG can remodel mature fibrils in vitro and in situ, i.e. rearrange them to non-amyloid aggregates. Using a novel method of measuring fibril remodeling by their seeding potential, we were able to measure the effect of EGCG at very high concentrations while minimizing effects on ThT fluorescence quenching and displacement from amyloids. We propose that this method can be useful for screening for fibril disrupters while reducing false positives from fluorescence quenching and/or competitive binding. Two molar equivalents of EGCG were able to halve the fibril seeding potential, although complete abolishment required an ∼64-fold excess. The FapC concentration in situ is unknown, and such high molar EGCG concentrations cannot be probed as they are toxic to planktonic bacteria (41). Nevertheless, even a halving of fibrillation potential can have significant effects. A dose-response relationship in situ was observed with a concentration of 49 μm for half-maximum effect. In this relationship, 20% of the ThT signal is unaccounted for and could be due to the same diminished effect as observed in vitro.
FapC Fibril Remodeling by EGCG Affects Quorum-sensing Molecule Binding
As QS controls the expression of virulence factors, disruption of the QS system signaling is a valuable target for antimicrobial agents in chronic infections (65). EGCG attenuates the swarming mobility of P. aeruginosa (30, 66). Using QS reporter strains, the loss of swarming mobility has been suggested to originate from QS inhibition by EGCG in P. aeruginosa as well as other bacterial strains (30, 67). Mutations in the enoyl-(acyl-carrier-protein) reductase in P. aeruginosa lead to a significant reduction in the expression of the las QS system (68). In E. coli, EGCG inhibits reductase enzymes involved in type II fatty acid synthesis in vitro and reduces fatty acid biosynthesis in vivo (69). Based on homology modeling, this has also been suggested for P. aeruginosa (30). However, enzyme reductase overexpression did not rescue cells, suggesting that the reductases are not an important biological target for EGCG. Rather, we have shown that the amyloid content in P. aeruginosa biofilm was reduced upon incubation with EGCG, and we suggest that this may have consequences for the quorum system. We have previously shown that Fap amyloid fibrils can bind and retain QS molecules (24), and now we have found a 20-fold increase in relative SPR light reflection upon pyocyanin binding after incubation with EGCG. This is supported by data from our simple binding-and-filtration assay. In our view, the simplest explanation for this observation is that the remodeling into amorphous aggregates increases the binding capacity for pyocyanin by the FapC protein by exposing hydrophobic protein structure, but we emphasize that this must remain speculative. Indirect support for this interpretation is that EGCG binding has been shown to involve hydrophobic interactions for other proteins (70–73), and hydrophobic interactions also seem to drive QS molecule interaction with Fap fibrils (24). QS molecules and EGCG may compete for the same fibril binding sites, but once the fibrils have been remodeled new binding sites for pyocyanin may be exposed. The FapC remodeling could possibly affect the QS system in two different ways. 1) By changing the binding kinetics of QS molecules, the dynamics of information flow between bacteria can be altered. 2) By increasing the binding capacity for QS molecules, the remodeled FapC molecules can “soak” up QS molecules and thereby block the information transfer required to maintain the biofilm. Other explanations for the in vivo effects of EGCG may also be considered, e.g. effects on transcriptional activity.
EGCG Affects Biofilm Structural Integrity
Fap has been implicated as an important virulence factor (25), but the amount of Fap produced by P. aeruginosa in vivo remains unknown. The PAO1 WT strain produces little Fap under laboratory conditions, although amyloid could still be detected using ThT fluorescence (16). However, we have recently shown that Fap is expressed by PAO1 WT in vivo in murine infection models using a novel amyloid-targeting fluorescent probe (74). This observation is further supported by a recent transcriptomics study, which showed that transcription of the fap operon was highly up-regulated in murine models of acute burn and chronic surgical wound infections when compared with traditional laboratory conditions (75). Thus, when the effect of EGCG was investigated on P. aeruginosa biofilm stiffness and tobramycin susceptibility, we turned to a fap overexpression strain (PAO1 pFap). Because of the mechanical strength of the amyloid fibril, functional amyloid is important for biofilm stiffness (26, 76). Overexpression of fap leads to a 2.5-fold reduction in biofilm stiffness. This contrasts with previous results from Pseudomonas sp. UK4. However, it is known that overexpression of Fap induces large proteomic changes in PAO1, leading to a mucoid phenotype due to increased alginate production (16, 23). This in turn may explain the difference in biofilm stiffness between the PAO1 WT and PAO1 pFap. Importantly, addition of EGCG did not change the biofilm stiffness of PAO1 WT but induced a 2-fold reduction in stiffness of PAO1 pFap biofilms. Hence, EGCG remodeling of functional amyloid in P. aeruginosa PAO1 affects the mechanistic properties of the biofilm and might compromise the structural integrity of the biofilm. Moreover, these results support a structural role for Fap amyloids in P. aeruginosa PAO1 biofilms in addition to their effect on QS.
EGCG Increases P. aeruginosa Biofilm Susceptibility to Antibiotics
EGCG increased PAO1 WT and PAO1 pFap susceptibility toward tobramycin. Although the effect on the PAO1 WT strain was modest, it was increased by Fap overexpression in PAO1 pFap. Thus, for clinical strains with higher expression of Fap, EGCG could potentially help alleviate the persistent biofilm infection. Fap overexpression leads to a global proteomic effect with several parallels to chronic infections of cystic fibrosis patients, indicating that Fap might have a more significant role in infections than under laboratory settings, e.g. murine wound infections (23, 75).
Expectorated sputum from cystic fibrosis patients after tobramycin solution inhalation has been found to range from a peak concentration of 314.5 μg/ml (25th and 75th percentiles are 124.4 and 409.5 μg/ml, respectively) to a trough concentration of 11.5 μg/ml (25th and 75th percentiles are 6.6 and 68.1 μg/ml, respectively) (77). Although the peak concentration is many times higher than the MBEC for PAO1 WT, the trough concentration is lower. Addition of EGCG reduces the MBEC from 59 to 26 μg/ml, thereby increasing the likelihood of total sputum PAO1 WT eradication. The MBEC of PAO1 pFap, however, was >200 μg/ml, and this strain could therefore survive even in some patients' peak concentrations. Addition of EGCG significantly reduces the MBEC with some biofilms being eradicated at 59 μg/ml and full eradication at 133 μg/ml. Based on the sputum concentrations of tobramycin, the MBEC change with EGCG could result in a more efficient tobramycin treatment.
Based on the observed changes to QS molecule binding and biofilm mechanistic properties, we propose a 2-fold mechanism for the increased tobramycin susceptibility from exposure to EGCG in amyloid-producing strains. 1) Functional amyloid remodeling by EGCG changes QS molecule binding to the amyloid fibril, thus disrupting the QS system and thereby increasing antibiotic susceptibility. 2) Functional amyloid remodeling by EGCG disrupts the biofilm structural integrity, leading to decreased protection against antibiotics. This may occur through increased diffusion through a biofilm network with increased porosity and/or physically less compact internal structure.
EGCG as Part of a Multidrug Treatment in Cystic Fibrosis
Oral ingestion of EGCG is generally well tolerated at least up to 1600 mg in a single dose (78). Unfortunately, this only leads to a peak free plasma concentration of 6.4 μm, which is well below the antiamyloidogenic concentration needed in the in situ experiments. Instead, EGCG might be directly administered to cystic fibrosis patients through nebulization. To our knowledge, no studies have been conducted to measure a patient's tolerance of nebulized EGCG. Gargling of a polyphenol solution, containing mostly EGCG, showed no adverse effects on the mouth or upper respiratory tract, indicating that nebulized administration of EGCG is possible (79), but systematic studies are needed to probe potential effects.
EGCG has many roles as a bactericidal molecule. Indeed, in E. coli alone, it can both directly affect CsgA assembly into curli fibrils and interfere with the biosynthesis of this protein and other biopolymers such as cellulose (49). How much each role accounts for in the combined bactericidal effect is difficult to elucidate. In this study, we have shown that the antiamyloid effect of EGCG remodels the functional amyloids in P. aeruginosa biofilms in the concentration range usually used to study the antibactericidal effects of EGCG. How this correlates with the other roles remains to be seen, but we submit that the amyloid effect should be taken into account. With both structurally destabilizing and QS-modifying capabilities, EGCG remains a strong interesting candidate for multidrug treatment of persistent biofilm infections.
Experimental Procedures
Chemicals
All chemicals were from Sigma-Aldrich except nitroblue tetrazolium (Biomol, Hamburg, Germany), Bacto casamino acids (BD Biosciences), and boric acid (J. T. Baker).
Purification of Native Fibrils from P. aeruginosa PAO1
Authentic Fap fibrils were purified from the PAO1 pFap strain as described (16).
Purification of Recombinant His-tagged FapC
Transfected E. coli BL21(DE3) cells carrying the pET32b(+) vector (Novagen) encoding His-tagged FapC were spread on LB agar plates and grown overnight (24). A colony was transferred to 1 liter of LB medium in a shake flask, and the culture was again grown overnight with 120 rpm shaking. To each of six shake flasks containing 2 liters of LB medium with 0.1% glucose was added a 50-ml overnight culture, and the new cultures were grown to an absorbance at 600 nm (A600) of 1 before induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cultures were grown for another 3 h before cells were harvested by centrifugation at 4000 × g for 20 min. Pellets were resuspended in 100 ml of extraction buffer (50 mm Tris-HCl, 8 m guanidinium chloride, pH 8.0) and sonicated on ice using a Q500 sonicator (Qsonica, Newtown, CT) with a 6-mm tip at 25% power for 30 cycles (5-s sonication/5-s pause). Lysed cell solution was centrifuged for 20 min at 10,000 × g, and the supernatant was incubated with nickel-nitrilotriacetic acid beads for >1 h at 4 °C. The beads were washed with 50 ml of extraction buffer, 50 ml of extraction buffer with 30 mm imidazole, and finally 50 ml of extraction buffer. Proteins were eluted from beads with 50 ml of extraction buffer supplemented with 150 mm EDTA. Immediately prior to use, the protein was desalted into the desired buffer using PD-10 desalting columns (GE Healthcare) according to the manufacturer's protocol. Protein purity was assayed using SDS-PAGE, and concentration was measured by UV absorption at 280 nm (extinction coefficient, 0.701 liter/g·cm). All culturing and inoculation were at 37 °C and with medium containing 100 mg/liter ampicillin.
Protein Fibrillation Monitored Using ThT
20 mm ThT (in ethanol) was added to desalted FapC monomers in PBS to a final composition of 40 μm ThT, 0.5 mg/ml FapC, 1% DMSO, and varying concentrations of EGCG. Each sample was mixed to a volume of 500 μl, and 150 μl were added in triplicate to a 96-well clear bottom black polystyrene plate (catalog number 265301, Thermo Scientific Nunc, Waltham, MA) and covered with clear sealing tape (catalog number 232701, Nunc). The plate fluorescence from ThT was measured in a Genius Pro plate reader (Tecan, Männedorf, Switzerland) with the following settings: temperature, 37 °C; excitation, 448 nm; emission, 485 nm; gain, 40; and integration time, 40 μs. For each measurement, linear shaking for 30 s at 180 rpm was applied before a 5-s settle time, and then 10 single reads were averaged for each well. Measurements were done every 91 s. This fibrillation procedure was used to produce native-like fibrils (see below) as well as to investigate the effect of EGCG on fibrillation.
Authentic Fap Fibril Preparation
To prepare fibrils imprinted with the authentic in vivo amyloid structure, fibrils purified from P. aeruginosa PAO1 were used to seed fibrillation of His-tagged FapC (24). The seeds were produced by sonicating fibrils on ice using a Q500 sonicator with a 1-mm tip at 25% power for six cycles (5-s sonication/5-s pause). Seeds and His-tagged FapC monomers were mixed in a 1:19 mass ratio for a total concentration of 0.5 mg/ml in 50 mm Tris-HCl, pH 7.4, and incubated as described above. Three wells with and without seeds contained 40 μm ThT to follow the seeded fibrillation. Other wells without ThT were pooled and used for reseeding a new generation of fibrils. Sonication, dilution with fresh monomer, and fibrillation was repeated three times over. Furthermore, buffer exchange from 50 mm Tris-HCl to PBS was done by centrifuging second generation fibrils for 30 min at 100,000 × g, washing with PBS, recentrifuging, and resuspending in PBS. Seeds made from these fibrils in PBS were used to make the third generation authentic fibrils.
TEM Imaging of Protein Fibrils
10 mm silibinin, EGCG, doxycycline, and curcumin were prepared in DMSO. 0.5 mg/ml authentic fibrils were added to 1% (v/v) compound (final concentrations, 100 μm compound and 1% (v/v) DMSO). Samples were incubated as described for the ThT assay (without ThT) for 24 h and visualized by TEM as described previously (24). Samples from in vitro protein fibrillation were frozen in liquid nitrogen, stored at −20 °C, and then imaged by TEM.
SDS-PAGE and Electroblotting of FapC Incubated with EGCG
Samples from the fibrillation assay with monomer FapC incubated with EGCG were separated using SDS-PAGE in a 12% (w/v) gel, and samples were loaded alongside a prestained protein ladder (catalog number 26616, Thermo Scientific). Gels were stained with Coomassie, and the gel bands were visualized in a Gel Doc EZ system (Bio-Rad). An unstained gel was electroblotted onto an Immobilon P transfer PVDF membrane (Merck Millipore, Darmstadt, Germany) at 200 mA for 30 min using a V10-SDB semidry blotter (Scie-Plas Ltd., Cambridge, UK).
Slot-blotting of FapC Incubated with EGCG
Slot-blotting was done using a Bio-Dot slot format microfiltration unit (Bio-Rad) according to the manufacturer's protocol onto a nitrocellulose membrane. 20-μl protein samples from ThT-monitored fibrillations were diluted into 380 μl of TBS buffer (20 mm Tris-HCl, 500 mm NaCl, pH 7.5). 200 μl were added to 2 wells for each sample, and wells were then washed with 2 ml of TBS buffer.
NBT and Congo Red Staining of Electro- and Slot-blotted FapC
PVDF and nitrocellulose membranes from electro- and slot-blotting, respectively, were stained by submersion in NBT solution (0.6 mg/ml NBT, 2 m potassium glycinate, pH 10) in the dark for 45 min. NBT solution was removed, and the membranes were washed twice in 0.64 m boric acid, pH 9.0, and soaked overnight. Membranes were washed with ddH2O water and imaged using an image scanner. Image contrast was adjusted using GiMP version 2.8.
Separate nitrocellulose membranes were stained with 2.5 mg/ml Congo red for 1 h. The membrane was washed with ddH2O and destained overnight. The membrane was imaged by an image scanner, and image contrast was adjusted using GiMP version 2.8.
Effect of EGCG and Antibiotics on E. coli Growth Rate
E. coli was grown overnight in LB medium (37 °C at 120 rpm), diluted, grown to an A600 of 1, and then further diluted to an A600 of 0.4. Doxycycline, tobramycin, and EGCG stocks were 2-fold serially diluted in either ddH2O or 2% DMSO (EGCG) and mixed 1:1 with the culture, and culture growth was followed using A600. Cultures were mixed in quadruplicates in a 96-well clear bottom black polystyrene plate with 100 μl in each well, and A600 was measured every 15 min in a Varioskan Flash plate reader (Thermo Scientific) using a bandwidth of 5 nm and 100-ms integration time. Growth rate was determined as the slope of the first logarithm of A600 as a function of time between 15 and 75 min. Addition of 1% DMSO did not affect the growth rate. Measured growth rates were normalized to the growth rate without compound and plotted as a function of compound concentration, and the measurements were fitted to a dose-response curve as follows.
| (Eq. 1) |
where End is the end level (0 for tobramycin and doxycycline), EC50 is the half-maximum effective concentration, and n is the Hill coefficient.
Removal of Unbound EGCG Using Spin-down Assay
100-μl samples from FapC fibrils incubated in the presence of EGCG were diluted 1:1 in PBS and centrifuged for 30 min at 100,000 × g, and supernatants were saved. Pellets were resuspended in 200 μl of PBS and recentrifuged, again saving the supernatants. Pellets were resuspended in 100 μl of PBS with 40 μm ThT and added to a 96-well clear bottom plate, and ThT fluorescence was measured on a Varioskan Flash plate reader with excitation at 448 nm, emission at 480 nm, 12-nm bandwidth, and 100-ms integration time. Measured ThT fluorescence from each well was normalized to the samples without EGCG.
Protein concentrations were measured using the CB-X assay (catalog number 786-12X, G-Biosciences) according to the manufacturer's protocol. Absorption at 595 nm of the supernatant, wash supernatant, and resuspended pellets (see section above) were measured in triplicate in a 96-well clear bottom plate on a Varioskan Flash plate reader at a bandwidth of 5 nm with a measurement time of 100 ms. Each well was measured three times and averaged. Buffer and non-pelleted authentic fibrils were used for normalization as 0 and 0.5 mg/ml, respectively, and determined concentrations were adjusted for dilution during washing.
Remodeling of Authentic Fibril Seeds by EGCG and Measurement of Pyocyanin Binding
Eight samples of 100 μl of 0.5 mg/ml authentic FapC fibril seeds, produced as in authentic fibril production, were incubated with 1% DMSO at up to a 64:1 molar ratio EGCG for 60 h at the same conditions as used for protein fibrillation. 25 μl of each sample were diluted to 100 μl with PBS buffer and sonicated as described earlier. 400 μl of FapC monomer solution with ThT were then added to final concentrations of 0.5 mg/ml and 40 μm, respectively, and incubated in triplicates as described for protein fibrillation. The initial fibrillation rates were calculated as the slope between 1 and 2 h of ThT fluorescence as a function of time. The 1st h was not used due to temperature effects on ThT fluorescence. To measure binding of pyocyanin to EGCG-treated fibrils, 1 mg/ml FapC fibrils, produced as in authentic fibril production, were incubated for 24 h at 37 °C while shaking with 500 μm EGCG (corresponding to a molar ratio of 1 FapC:19 EGCG). Subsequently, 500 μm pyocyanin was added (from a 10 mm stock in 10% ethanol), and the solution was incubated for 1 h at 37 °C. Afterward, the solutions were spun through a 10-kDa Centricon Plus-70 centrifugal filter (Millipore Merck Life Science), and HCl was added to 0.2 m. The pyocyanin concentration was determined using an extinction coefficient of the acidic form at 520 nm of 17.072 cm−1 (mg/ml)−1 (80). CD spectra of FapC fibrils and FapC fibrils remodeled with EGCG (prepared as above) were recorded on a J-810 spectrometer (Jasco) in a 1-mm quartz cuvette from 250 to 190 nm with 0.2-nm data pitch, 1-s integration time, 1-s bandwidth, and six accumulations. Only data points with detector voltages below 600 V were used.
Quorum-sensing Molecule Binding to Authentic FapC Fibrils Measured by SPR
Binding of the QS molecule pyocyanine to the native-like FapC fibrils was measured using SPR as described previously (24). Fibril immobilization on the SPR chip was verified via ThT binding before measuring binding of pyocyanin and lastly EGCG to immobilized fibrils. To allow for EGCG remodeling of immobilized FapC fibrils, the SPR chip was removed from the instrument and submerged in 500 μm EGCG in HEPES buffer overnight. The chip was thoroughly washed with buffer before pyocyanin binding was measured again.
EGCG Remodeling of Amyloid Fibrils in Biofilm in Situ Followed Using ThT Fluorescence
P. aeruginosa PAO1 was obtained from the P. aeruginosa PAO1 transposon mutant library at Washington University (Manoil laboratory) (81). PAO1 was grown overnight on LB agar, and a single colony was transferred to 10 ml of LB medium in a 50-ml tube and grown overnight at 120 rpm. 1% overnight culture was added to M63 medium supplemented with 1 mm MgSO4, 0.2% glucose, and 0.5% casamino acids as described previously (20). A 96-well clear, flat bottom plate (Sterilin microtiter plate, catalog number 611F96, Thermo Scientific) was sterilized by >0.5-h submersion in 70% ethanol. Subsequently, 100 μl of M63 culture were added to 80 wells, closed with a lid, sealed with Parafilm, and incubated for 24 h. Supernatant was carefully removed, and 120 μl of M63 medium supplemented with a 2-fold dilution series of EGCG in DMSO were added to wells. The final DMSO concentration was 1%, and the highest final EGCG concentration was 500 μm for each sample in eight replicas. The plate was closed with a lid, sealed with Parafilm, and incubated for 6 h at 120 rpm. Cell suspensions were carefully removed, and wells were washed with M63 medium. 150 μl of M63 medium supplemented with 100 μm ThT were added to each well, and the plate was sonicated in a Sonorex Digitec ultrasonic bath (Bandelin, Berlin, Germany) for 30 min, which completely dislodged the formed biofilm. After sonication, 100 μl of the now dislodged biofilm suspension from each well were moved to a black 96-well flat bottom plate (catalog number 655900, Greiner Bio One, Frickenhausen, Germany), and ThT fluorescence was measured as described for the spin-down assay, and five measurements per well were averaged. Measurements were fitted to a dose-response curve as described above. All incubations were at 37 °C. 95% confidence intervals were calculated using a T-distribution, and p values were calculated using a two-tailed, unequal variance t test.
MBEC of P. aeruginosa in the Presence of EGCG
All solutions used were sterilized by autoclaving or filtering except solutions in pure DMSO. MBEC was measured using the MBEC Biofilm 96-well inoculator with non-coated peg lids (Innovotech, Edmonton, Canada) according to the manufacturer's protocol. PAO1 WT and PAO1 pFap (16) were grown in 1 ml of LB medium overnight, and 1% of the overnight cultures were added to a total of 15 ml of mM63 medium supplemented with magnesium and casamino acids as in O'Toole (20) and 1 mm isopropyl 1-thio-β-d-galactopyranoside. 150 μl of PAO1 WT or PAO1 pFap culture were added to all wells of a sterile clear 96-well plate (catalog number 269787, Thermo Scientific) except for one row, which was used to inspect solution sterility in each protocol step. Biofilm was grown on pegs for 24 h at 110 rpm. In two challenge plates, 20 μl of tobramycin in ddH2O and 180 μl of supplemented M63 medium and 1% DMSO with or without 500 μm EGCG were added to each well in quadruplicates. The peg lid was moved from the incubation plate to the challenge plate and incubated for 24 h at 37 °C at 110 rpm. To four plates, 200 μl of supplemented M63 buffer were added in each well. Peg lids were washed in one plate for 10 s, moved to a second plate, and incubated for 24 h at 37 °C. Peg lids were removed and carefully pressed against colonization factor antigen (CFA) agar plates for 10 s, and agar plates were then incubated for 24 h. Biofilm survival was confirmed if bacterial colonies were detected by visual inspection. All incubations were at 37 °C, and medium used for PAO1 pFap was supplemented with 50 mg/liter tetracycline for plasmid stabilization until the challenge plate.
AFM Nanoindentation on Biofilm
PAO1 WT and pFap were prepared as above (overnight growth in LB medium and 100× dilution into supplemented M63 medium). PAO1 WT and pFap were incubated on two slides each of Superfrost Ultra Plus tissue attachment enhanced glass slides (Thermo Fisher Scientific Gerhard Menzel) for 24 h. Biofilms were dried in air and rehydrated in PBS for 30 min. Force curves were obtained, and Young's modulus was calculated using a glass microbead glued to an AFM cantilever as described (26). Young's modulus was calculated for five different locations on each coupon by averaging 10 force curves per location. Coupons were incubated with 500 μm EGCG in PBS for 5 h, and the force measurements was repeated. 95% confidence intervals were calculated using a T-distribution (df = 9), and p values were calculated using a two-tailed, unequal variance Student's t test (df = 18).
Author Contributions
M. S. and D. E. O. conceived and designed the experiments. M. S., M. S. D., G. Z., B. S. V., M. T. S., G. C., and T. S. performed the experiments. M. S., G. Z., G. C., and D. E. O. analyzed the data. M. S. D., S. G.-S., R. L. M., S. K., and P. H. N. contributed reagents, materials, and analysis tools. M. S. and D. E. O. wrote the manuscript.
Acknowledgments
We are grateful to Jørgen Kjems (iNANO, Aarhus University) for the use of the Gel Doc EZ system, Brian S. Vad (iNANO, Aarhus University) for discussion of results and experimental setup, and Chuah York Wieo for assistance with surface plasmon resonance assays. We also appreciate constructive suggestions by Per Hammarström and Niels Heegaard. The Singapore Centre on Environmental Life Sciences Engineering is supported by Singapore's National Research Foundation, the Ministry of Education, Nanyang Technological University (NTU), and National University of Singapore (NUS) and hosted by NTU in partnership with NUS.
This work was supported by the Sino-Danish Centre for Education and Research (to M. S.), Aalborg University and the Innovation Fund Denmark project NomiGas (to M. S. D.), Danish Council for Independent Research (DFF) Sapere Aude Starting Grant 0602-02130B (to G. Z. and R. L. M.), and DFF Technology and Production Grant 6111-00241B (to B. S. V. and D. E. O.). The authors declare that they have no conflicts of interest with the contents of this article.
- QS
- quorum-sensing
- EGCG
- epigallocatechin-3-gallate
- Fap
- functional amyloid fibrils in P. aeruginosa
- TEM
- transmission electron microscopy
- EC50
- half-maximum effective concentration
- ThT
- thioflavin T
- NBT
- nitro blue tetrazolium
- SPR
- surface plasmon resonance
- AFM
- atomic force microscopy
- MBEC
- minimum bactericidal eradication concentration
- ddH2O
- double distilled H2O.
References
- 1.Vert M., Doi Y., Hellwich K. H., Hess M., Hodge P., Kubisa P., Rinaudo M., and Schue F. (2012) Terminology for biorelated polymers and applications (IUPAC recommendations 2012). Pure Appl. Chem. 84, 377–408 [Google Scholar]
- 2.Hall-Stoodley L., Costerton J. W., and Stoodley P. (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 [DOI] [PubMed] [Google Scholar]
- 3.McDougald D., Rice S. A., Barraud N., Steinberg P. D., and Kjelleberg S. (2011) Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50 [DOI] [PubMed] [Google Scholar]
- 4.Williams P., Winzer K., Chan W. C., and Cámara M. (2007) Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 1119–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen T. B., and Givskov M. (2006) Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 296, 149–161 [DOI] [PubMed] [Google Scholar]
- 6.Costerton J. W., Stewart P. S., and Greenberg E. P. (1999) Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 [DOI] [PubMed] [Google Scholar]
- 7.Hassett D. J., Korfhagen T. R., Irvin R. T., Schurr M. J., Sauer K., Lau G. W., Sutton M. D., Yu H., and Hoiby N. (2010) Pseudomonas aeruginosa biofilm infections in cystic fibrosis: insights into pathogenic processes and treatment strategies. Expert Opin. Ther. Targets 14, 117–130 [DOI] [PubMed] [Google Scholar]
- 8.Flemming H. C., and Wingender J. (2010) The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 [DOI] [PubMed] [Google Scholar]
- 9.Dueholm M. S., Nielsen P. H., Chapman M., and Otzen D. (2013) Functional amyloids in bacteria, in Amyloid Fibrils and Prefibrillar Aggregates (Otzen D. E., ed) pp. 411–438, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 10.Larsen P., Nielsen J. L., Dueholm M. S., Wetzel R., Otzen D., and Nielsen P. H. (2007) Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9, 3077–3090 [DOI] [PubMed] [Google Scholar]
- 11.Nelson R., and Eisenberg D. (2006) Structural models of amyloid-like fibrils. Adv. Protein Chem. 73, 235–282 [DOI] [PubMed] [Google Scholar]
- 12.Fändrich M. (2007) On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell. Mol. Life Sci. 64, 2066–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles T. P., Vendruscolo M., and Dobson C. M. (2014) The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 [DOI] [PubMed] [Google Scholar]
- 14.Chapman M. R., Robinson L. S., Pinkner J. S., Roth R., Heuser J., Hammar M., Normark S., and Hultgren S. J. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero D., Aguilar C., Losick R., and Kolter R. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dueholm M. S., Søndergaard M. T., Nilsson M., Christiansen G., Stensballe A., Overgaard M. T., Givskov M., Tolker-Nielsen T., Otzen D. E., and Nielsen P. H. (2013) Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen 2, 365–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dueholm M. S., Petersen S. V., Sønderkær M., Larsen P., Christiansen G., Hein K. L., Enghild J. J., Nielsen J. L., Nielsen K. L., Nielsen P. H., and Otzen D. E. (2010) Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020 [DOI] [PubMed] [Google Scholar]
- 18.Dueholm M. S., Albertsen M., Otzen D., and Nielsen P. H. (2012) Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One 7, e51274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dueholm M. S., Otzen D., and Nielsen P. H. (2013) Evolutionary insight into the functional amyloids of the pseudomonads. PLoS One 8, e76630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Toole G. A. (2011) Microtiter dish biofilm formation assay. J. Vis. Exp. e2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen P., Nielsen J. L., Otzen D., and Nielsen P. H. (2008) Amyloid-like adhesins produced by floc-forming and filamentous bacteria in activated sludge. Appl. Environ. Microbiol. 74, 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordal P. B., Dueholm M. S., Larsen P., Petersen S. V., Enghild J. J., Christiansen G., Højrup P., Nielsen P. H., and Otzen D. E. (2009) Widespread abundance of functional bacterial amyloid in mycolata and other gram-positive bacteria. Appl. Environ. Microbiol. 75, 4101–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbst F. A., Søndergaard M. T., Kjeldal H., Stensballe A., Nielsen P. H., and Dueholm M. S. (2015) Major proteomic changes associated with amyloid-induced biofilm formation in Pseudomonas aeruginosa PAO1. J. Proteome Res. 14, 72–81 [DOI] [PubMed] [Google Scholar]
- 24.Seviour T., Hansen S. H., Yang L., Yau Y. H., Wang V. B., Stenvang M. R., Christiansen G., Marsili E., Givskov M., Chen Y., Otzen D. E., Nielsen P. H., Geifman-Shochat S., Kjelleberg S., and Dueholm M. S. (2015) Functional amyloids keep quorum-sensing molecules in check. J. Biol. Chem. 290, 6457–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiehlmann L., Munder A., Adams T., Juhas M., Kolmar H., Salunkhe P., and Tümmler B. (2007) Functional genomics of Pseudomonas aeruginosa to identify habitat-specific determinants of pathogenicity. Int. J. Med. Microbiol. 297, 615–623 [DOI] [PubMed] [Google Scholar]
- 26.Zeng G., Vad B. S., Dueholm M. S., Christiansen G., Nilsson M., Tolker-Nielsen T., Nielsen P. H., Meyer R. L., and Otzen D. E. (2015) Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front. Microbiol. 6, 1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dueholm M. S., Danielsen H. N., and Nielsen P. H. (2014) Complete genome sequence of Pseudomonas sp. UK4, a model organism for studies of functional amyloids in Pseudomonas. Genome Announc. 2, e00898-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang C. S., and Wang Z. Y. (1993) Tea and cancer. J. Natl. Cancer Inst. 85, 1038–1049 [DOI] [PubMed] [Google Scholar]
- 29.Steinmann J., Buer J., Pietschmann T., and Steinmann E. (2013) Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br. J. Pharmacol. 168, 1059–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L., Liu Y., Sternberg C., and Molin S. (2010) Evaluation of enoyl-acyl carrier protein reductase inhibitors as Pseudomonas aeruginosa quorum-quenching reagents. Molecules 15, 780–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii T., Mori T., Tanaka T., Mizuno D., Yamaji R., Kumazawa S., Nakayama T., and Akagawa M. (2008) Covalent modification of proteins by green tea polyphenol (−)-epigallocatechin-3-gallate through autoxidation. Free Radic. Biol. Med. 45, 1384–1394 [DOI] [PubMed] [Google Scholar]
- 32.Ishii T., Ichikawa T., Minoda K., Kusaka K., Ito S., Suzuki Y., Akagawa M., Mochizuki K., Goda T., and Nakayama T. (2011) Human serum albumin as an antioxidant in the oxidation of (−)-epigallocatechin gallate: participation of reversible covalent binding for interaction and stabilization. Biosci. Biotechnol. Biochem. 75, 100–106 [DOI] [PubMed] [Google Scholar]
- 33.Ehrnhoefer D. E., Bieschke J., Boeddrich A., Herbst M., Masino L., Lurz R., Engemann S., Pastore A., and Wanker E. E. (2008) EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 15, 558–566 [DOI] [PubMed] [Google Scholar]
- 34.Lorenzen N., Nielsen S. B., Yoshimura Y., Vad B. S., Andersen C. B., Betzer C., Kaspersen J. D., Christiansen G., Pedersen J. S., Jensen P. H., Mulder F. A., and Otzen D. E. (2014) How epigallocatechin gallate can inhibit alpha-synuclein oligomer toxicity in vitro. J. Biol. Chem. 289, 21299–21310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jankun J., Selman S. H., Swiercz R., and Skrzypczak-Jankun E. (1997) Why drinking green tea could prevent cancer. Nature 387, 561. [DOI] [PubMed] [Google Scholar]
- 36.Liang Y. C., Lin-shiau S. Y., Chen C. F., and Lin J. K. (1997) Suppression of extracellular signals and cell proliferation through EGF receptor binding by (−)-epigallocatechin gallate in human A431 epidermoid carcinoma cells. J. Cell. Biochem. 67, 55–65 [DOI] [PubMed] [Google Scholar]
- 37.Sun Y., Hung W. C., Chen F. Y., Lee C. C., and Huang H. W. (2009) Interaction of tea catechin (−)-epigallocatechin gallate with lipid bilayers. Biophys. J. 96, 1026–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama M., Shigemune N., Tsugukuni T., Tokuda H., and Miyamoto T. (2011) Difference of EGCG adhesion on cell surface between Staphylococcus aureus and Escherichia coli visualized by electron microscopy after novel indirect staining with cerium chloride. J. Microbiol. Methods 86, 97–103 [DOI] [PubMed] [Google Scholar]
- 39.Bikels-Goshen T., Landau E., Saguy S., and Shapira R. (2010) Staphylococcal strains adapted to epigallocathechin gallate (EGCG) show reduced susceptibility to vancomycin, oxacillin and ampicillin, increased heat tolerance, and altered cell morphology. Int. J. Food Microbiol. 138, 26–31 [DOI] [PubMed] [Google Scholar]
- 40.Yi S. M., Zhu J. L., Fu L. L., and Li J. R. (2010) Tea polyphenols inhibit Pseudomonas aeruginosa through damage to the cell membrane. Int. J. Food Microbiol. 144, 111–117 [DOI] [PubMed] [Google Scholar]
- 41.Cui Y., Oh Y. J., Lim J., Youn M., Lee I., Pak H. K., Park W., Jo W., and Park S. (2012) AFM study of the differential inhibitory effects of the green tea polyphenol (−)-epigallocatechin-3-gallate (EGCG) against Gram-positive and Gram-negative bacteria. Food Microbiol. 29, 80–87 [DOI] [PubMed] [Google Scholar]
- 42.Nakayama M., Shimatani K., Ozawa T., Shigemune N., Tsugukuni T., Tomiyama D., Kurahachi M., Nonaka A., and Miyamoto T. (2013) A study of the antibacterial mechanism of catechins: isolation and identification of Escherichia coli cell surface proteins that interact with epigallocatechin gallate. Food Control 33, 433–439 [Google Scholar]
- 43.Ferreira N., Cardoso I., Domingues M. R., Vitorino R., Bastos M., Bai G., Saraiva M. J., and Almeida M. R. (2009) Binding of epigallocatechin-3-gallate to transthyretin modulates its amyloidogenicity. FEBS Lett. 583, 3569–3576 [DOI] [PubMed] [Google Scholar]
- 44.He J., Xing Y. F., Huang B., Zhang Y. Z., and Zeng C. M. (2009) Tea catechins induce the conversion of preformed lysozyme amyloid fibrils to amorphous aggregates. J. Agric. Food Chem. 57, 11391–11396 [DOI] [PubMed] [Google Scholar]
- 45.Roberts B. E., Duennwald M. L., Wang H., Chung C., Lopreiato N. P., Sweeny E. A., Knight M. N., and Shorter J. (2009) A synergistic small-molecule combination directly eradicates diverse prion strain structures. Nat. Chem. Biol. 5, 936–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bieschke J., Russ J., Friedrich R. P., Ehrnhoefer D. E., Wobst H., Neugebauer K., and Wanker E. E. (2010) EGCG remodels mature α-synuclein and amyloid-β fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 107, 7710–7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palhano F. L., Lee J., Grimster N. P., and Kelly J. W. (2013) Toward the molecular mechanism(s) by which EGCG treatment remodels mature amyloid fibrils. J. Am. Chem. Soc. 135, 7503–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao P., and Raleigh D. P. (2012) Analysis of the inhibition and remodeling of islet amyloid polypeptide amyloid fibers by flavanols. Biochemistry 51, 2670–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serra D. O., Mika F., Richter A. M., and Hengge R. (2016) The green tea polyphenol EGCG inhibits E. coli biofilm formation by impairing amyloid curli fibre assembly and down-regulating the biofilm regulator CsgD via the σ-dependent sRNA RybB. Mol. Microbiol. 101, 136–151 [DOI] [PubMed] [Google Scholar]
- 50.Young L. M., Cao P., Raleigh D. P., Ashcroft A. E., and Radford S. E. (2014) Ion mobility spectrometry-mass spectrometry defines the oligomeric intermediates in amylin amyloid formation and the mode of action of inhibitors. J. Am. Chem. Soc. 136, 660–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giorgetti S., Raimondi S., Pagano K., Relini A., Bucciantini M., Corazza A., Fogolari F., Codutti L., Salmona M., Mangione P., Colombo L., De Luigi A., Porcari R., Gliozzi A., Stefani M., Esposito G., Bellotti V., and Stoppini M. (2011) Effect of tetracyclines on the dynamics of formation and destructuration of β2-microglobulin amyloid fibrils. J. Biol. Chem. 286, 2121–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forloni G., Colombo L., Girola L., Tagliavini F., and Salmona M. (2001) Anti-amyloidogenic activity of tetracyclines: studies in vitro. FEBS Lett. 487, 404–407 [DOI] [PubMed] [Google Scholar]
- 53.Yang F., Lim G. P., Begum A. N., Ubeda O. J., Simmons M. R., Ambegaokar S. S., Chen P. P., Kayed R., Glabe C. G., Frautschy S. A., and Cole G. M. (2005) Curcumin inhibits formation of amyloid β oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 280, 5892–5901 [DOI] [PubMed] [Google Scholar]
- 54.Smirnovas V., Kim J. I., Lu X., Atarashi R., Caughey B., and Surewicz W. K. (2009) Distinct structures of scrapie prion protein (PrPSc)-seeded versus spontaneous recombinant prion protein fibrils revealed by hydrogen/deuterium exchange. J. Biol. Chem. 284, 24233–24241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petkova A. T., Leapman R. D., Guo Z., Yau W. M., Mattson M. P., and Tycko R. (2005) Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science 307, 262–265 [DOI] [PubMed] [Google Scholar]
- 56.Andreasen M., Nielsen S. B., Runager K., Christiansen G., Nielsen N. C., Enghild J. J., and Otzen D. E. (2012) Polymorphic fibrillation of the destabilized fourth fasciclin-1 domain mutant A546T of the transforming growth factor-β-induced protein (TGFBIp) occurs through multiple pathways with different oligomeric intermediates. J. Biol. Chem. 287, 34730–34742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng G., Müller T., and Meyer R. L. (2014) Single-cell force spectroscopy of bacteria enabled by naturally derived proteins. Langmuir 30, 4019–4025 [DOI] [PubMed] [Google Scholar]
- 58.Kronvall G. (2010) Normalized resistance interpretation as a tool for establishing epidemiological MIC susceptibility breakpoints. J. Clin. Microbiol. 48, 4445–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hudson S. A., Ecroyd H., Kee T. W., and Carver J. A. (2009) The thioflavin T fluorescence assay for amyloid fibril detection can be biased by the presence of exogenous compounds. FEBS J. 276, 5960–5972 [DOI] [PubMed] [Google Scholar]
- 60.Paz M. A., Flückiger R., Boak A., Kagan H. M., and Gallop P. M. (1991) Specific detection of quinoproteins by redox-cycling staining. J. Biol. Chem. 266, 689–692 [PubMed] [Google Scholar]
- 61.Suzuki Y., Brender J. R., Hartman K., Ramamoorthy A., and Marsh E. N. (2012) Alternative pathways of human islet amyloid polypeptide aggregation distinguished by 19F nuclear magnetic resonance-detected kinetics of monomer consumption. Biochemistry 51, 8154–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiang W. C., Nilsson M., Jensen P. Ø., Høiby N., Nielsen T. E., Givskov M., and Tolker-Nielsen T. (2013) Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 57, 2352–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee D., and Stableforth D. (2000) The treatment of respiratory pseudomonas infection in cystic fibrosis: what drug and which way? Drugs 60, 1053–1064 [DOI] [PubMed] [Google Scholar]
- 64.DePas W. H., and Chapman M. R. (2012) Microbial manipulation of the amyloid fold. Res. Microbiol. 163, 592–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bjarnsholt T., Tolker-Nielsen T., Hoiby N., and Givskov M. (2010) Interference of Pseudomonas aeruginosa signalling and biofilm formation for infection control. Expert Rev. Mol. Med. 12, e11. [DOI] [PubMed] [Google Scholar]
- 66.O'May C., Ciobanu A., Lam H., and Tufenkji N. (2012) Tannin derived materials can block swarming motility and enhance biofilm formation in Pseudomonas aeruginosa. Biofouling 28, 1063–1076 [DOI] [PubMed] [Google Scholar]
- 67.Huber B., Eberl L., Feucht W., and Polster J. (2003) Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Z. Naturforsch. C 58, 879–884 [DOI] [PubMed] [Google Scholar]
- 68.Hoang T. T., and Schweizer H. P. (1999) Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 181, 5489–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y. M., and Rock C. O. (2004) Evaluation of epigallocatechin gallate and related plant polyphenols as inhibitors of the FabG and FabI reductases of bacterial type II fatty-acid synthase. J. Biol. Chem. 279, 30994–31001 [DOI] [PubMed] [Google Scholar]
- 70.Lopez del Amo J. M., Fink U., Dasari M., Grelle G., Wanker E. E., Bieschke J., and Reif B. (2012) Structural properties of EGCG-induced, nontoxic Alzheimer's disease Aβ oligomers. J. Mol. Biol. 421, 517–524 [DOI] [PubMed] [Google Scholar]
- 71.Grelle G., Otto A., Lorenz M., Frank R. F., Wanker E. E., and Bieschke J. (2011) Black tea theaflavins inhibit formation of toxic amyloid-β and α-synuclein fibrils. Biochemistry 50, 10624–10636 [DOI] [PubMed] [Google Scholar]
- 72.Maiti T. K., Ghosh K. S., and Dasgupta S. (2006) Interaction of (−)-epigallocatechin-3-gallate with human serum albumin: fluorescence, Fourier transform infrared, circular dichroism, and docking studies. Proteins 64, 355–362 [DOI] [PubMed] [Google Scholar]
- 73.Miyata M., Sato T., Kugimiya M., Sho M., Nakamura T., Ikemizu S., Chirifu M., Mizuguchi M., Nabeshima Y., Suwa Y., Morioka H., Arimori T., Suico M. A., Shuto T., Sako Y., Momohara M., Koga T., Morino-Koga S., Yamagata Y., and Kai H. (2010) The crystal structure of the green tea polyphenol (−)-epigallocatechin gallate-transthyretin complex reveals a novel binding site distinct from the thyroxine binding site. Biochemistry 49, 6104–6114 [DOI] [PubMed] [Google Scholar]
- 74.Kim J. Y., Sahu S., Yau Y. H., Wang X., Shochat S. G., Nielsen P. H., Dueholm M. S., Otzen D. E., Lee J., Delos Santos M. M., Yam J. K., Kang N. Y., Park S. J., Kwon H., Seviour T., Yang L., Givskov M., and Chang Y. T. (2016) Detection of pathogenic biofilms with bacterial amyloid targeting fluorescent probe, CDy11. J. Am. Chem. Soc. 138, 402–407 [DOI] [PubMed] [Google Scholar]
- 75.Turner K. H., Everett J., Trivedi U., Rumbaugh K. P., and Whiteley M. (2014) Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 10, e1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Knowles T. P., and Buehler M. J. (2011) Nanomechanics of functional and pathological amyloid materials. Nat. Nanotechnol. 6, 469–479 [DOI] [PubMed] [Google Scholar]
- 77.Ruddy J., Emerson J., Moss R., Genatossio A., McNamara S., Burns J. L., Anderson G., and Rosenfeld M. (2013) Sputum tobramycin concentrations in cystic fibrosis patients with repeated administration of inhaled tobramycin. J. Aerosol Med. Pulm. Drug Deliv. 26, 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ullmann U., Haller J., Decourt J. P., Girault N., Girault J., Richard-Caudron A. S., Pineau B., and Weber P. (2003) A single ascending dose study of epigallocatechin gallate in healthy volunteers. J. Int. Med. Res. 31, 88–101 [DOI] [PubMed] [Google Scholar]
- 79.Yamada H., Takuma N., Daimon T., and Hara Y. (2006) Gargling with tea catechin extracts for the prevention of influenza infection in elderly nursing home residents: a prospective clinical study. J. Altern. Complement. Med. 12, 669–672 [DOI] [PubMed] [Google Scholar]
- 80.Essar D. W., Eberly L., Hadero A., and Crawford I. P. (1990) Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172, 884–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., Will O., Kaul R., Raymond C., Levy R., Chun-Rong L., Guenthner D., Bovee D., Olson M. V., and Manoil C. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100, 14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]