Abstract
Using budding yeast, we investigated a negative interaction network among genes for tRNA modifications previously implicated in anticodon-codon interaction: 5-methoxy-carbonyl-methyl-2-thio-uridine (mcm5s2U34: ELP3, URM1), pseudouridine (Ψ38/39: DEG1) and cyclic N6-threonyl-carbamoyl-adenosine (ct6A37: TCD1). In line with functional cross talk between these modifications, we find that combined removal of either ct6A37 or Ψ38/39 and mcm5U34 or s2U34 results in morphologically altered cells with synthetic growth defects. Phenotypic suppression by tRNA overexpression suggests that these defects are caused by malfunction of tRNALysUUU or tRNAGlnUUG, respectively. Indeed, mRNA translation and synthesis of the Gln-rich prion Rnq1 are severely impaired in the absence of Ψ38/39 and mcm5U34 or s2U34, and this defect can be rescued by overexpression of tRNAGlnUUG. Surprisingly, we find that combined modification defects in the anticodon loops of different tRNAs induce similar cell polarity- and nuclear segregation defects that are accompanied by increased aggregation of cellular proteins. Since conditional expression of an artificial aggregation-prone protein triggered similar cytological aberrancies, protein aggregation is likely responsible for loss of morphogenesis and cytokinesis control in mutants with inappropriate tRNA anticodon loop modifications.
INTRODUCTION
tRNA molecules undergo post-transcriptional modifications many of which are conserved from yeast to man and thought to be important for structural integrity, stability, decoding efficiency and accuracy during mRNA translation. Eukaryotic tRNA modification enzymes have been extensively characterized in the model system Saccharomyces cerevisiae (1,2). Even though most modifications are dispensable for yeast growth, some are critical for proliferation or even essential for cell viability. One such modification is 5-methoxy-carbonyl-methyl-2-thio-uridine (mcm5s2U), which is found at uridines in the anticodon wobble position (U34) of tRNAGlnUUG, tRNALysUUU and tRNAGluUUC (3,4). These tRNAs decode the A-ending codons for the respective amino acids, whereas the G-ending codons for the same amino acids are read by isoacceptor tRNAs with a C in the anticodon wobble position. Functionally, the mcm5s2U modification appears to improve decoding of the A-ending codons for Glu (GAA), Gln (CAA) and Lys (AAA) (4,5).
mcm5s2U formation involves two genetically separable pathways: the Elongator pathway, which introduces the mcm5 or the related 5-carbamoylmethyl (ncm5) side chain at C5 of the uracil base (U34), and the Urm1 pathway, which mediates thiolation at C2 (6–8). Elongator is an evolutionary conserved complex that interacts with additional regulatory proteins and the Urm1 pathway utilizes an ancient ubiquitin-like protein modifier (Urm1) for sulfur transfer from cysteine to the C2 position of U34 (8–17). Simultaneous loss of both U34 modification pathways in yeast results in growth delay, reduced protein levels, increased protein aggregation due to translational slow down or even inviability depending on strain backgrounds (18–22). While phenotypically milder, the removal of either U34 modification pathway (Elongator or Urm1) alone already causes a wide range of detectable phenotypes, including drug and various stress sensitivities and cell cycle delay (19–21). Also, nutrient sensitive regulatory circuits such as target of rapamycin (TOR) or general amino acid control (GAAC) signaling pathways become deregulated in response to mcm5/s2U modification defects (22–24). It has been proposed that several of these pleiotropic phenotypes occur because of translational defects of mRNA transcripts enriched in A-ending codons (CAA, AAA, GAA, see above) that are efficiently read by mcm5s2U modified anticodons of the respective tRNAs (i.e. tRNAGlnUUG, tRNALysUUU and tRNAGluUUC) (5,21,25). Loss of the mcm5s2U modification not only impairs translation of mRNAs enriched in CAA and AAA codons but also induced a global protein aggregation phenomenon causing a reduced ability of cells to respond to proteotoxic stress (22).
Among other tRNA modifications important for normal growth in the yeast model are N6-threonyl-carbamoyl-adenosine at position 37 (t6A37) and pseudouridine at positions 38 and 39 (Ψ38/39). The latter is formed by pseudouridine synthase Deg1 (depressed growth), and genetic analyses suggest that the growth defects typical of deg1 mutants are mainly caused by a reduced function of tRNAGlnUUG (26,27). t6A is conserved in all three domains of life, and its absence generally causes slow growth or lethal phenotypes (28–32). It is present in tRNAs that decode ANN codons including the initiator tRNAMet and is required for reading frame maintenance and prevention of translational initiation downstream of start AUG codons (33,34). It was shown that t6A in yeast and bacteria is a hydrolysis product of a dehydrated t6A derivative, termed cyclic N6-threonyl-carbamoyl-adenosine (ct6A) which is converted to t6A during tRNA sample preparation, indicating that ct6A is the naturally occurring configuration of the tRNA modification in some organisms and likely to promote its decoding efficiency (35).
Here, we examined, based on synthetic genetic array (SGA) data from yeast, negative interactions among genes required for modification of U34 (ELP3, URM1/UBA4) (36) and other anticodon loop positions, Ψ38/39 (DEG1) and ct6A37 (TCD1/TCD2). Remarkably, synthetic phenotypes and translational defects that result from combined loss of these tRNA modifications can be suppressed by overexpression of specific tRNAs suggesting the growth defects are caused by malfunctional tRNA species that carry the modification defects in question. Surprisingly, we observe in these tRNA modification mutants a common major interference with cytoskeleton integrity and cytokinesis as well as nuclear segregation and cell polarity defects. Intriguingly, these morphological aberrancies are associated with increased aggregation of cellular proteins, and we demonstrate that conditional expression of a single aggregation-prone protein in wild type cells can induce a similar change in morphology. Thus, malfunction of distinct tRNAs due to inappropriate modifications in their anticodon stem loop evokes shared morphological phenotypes that are ascribable to defects in translation and improper protein synthesis, folding and solubility.
MATERIALS AND METHODS
Strains and general methods
Strains used in this study are listed in Supplementary Table S1. Standard methods were used for yeast growth and maintenance (37). Genomic deletions were generated by PCR (38) and oligonucleotides targeting ELP3, URM1 or DEG1 (Supplementary Table S2). Replacements were verified by PCR using fw/rv primers located outside of the target genes (Supplementary Table S2).
Plasmid construction and shuffling
To facilitate deletion of DEG1 in urm1Δ::KanMX4 or elp3Δ::KanMX4 strains, these were first transformed with pFF8 (ELP3; (19)) or pHA-URM1 (URM1; (39)). Subsequently, DEG1 was deleted by using a PCR generated deletion cassette (deg1Δ::SpHIS5) and URA3 plasmids pFF8 or pHA-URM1 eliminated by growth on minimal media containing uracil and 5-fluoro-orotate (1 ml/ml). Overexpression of tRNA species involved YEplac195(URA3)-, YEplac181(LEU2)-, or pRS423(HIS3)-based plasmids YEpQ (tRNAGlnUUG, pQ), pSZ16 (tRNAGluUUC, pE), pDJ82 (tRNALysUUU, pK) and pQKE (tRNAGlnUUG, tRNAGluUUC and tRNALysUUU) (19,40,41). The insert from YEplac195-based YEpQ containing the tRNAGlnUUG gene was subcloned into pRS425 using SalI and EcoRI, generating pRK55. Multi copy expression of tRNAGlnCUG (CDC65/SUP70) employed plasmid pSUP70-2μ (42). Rnq1 was expressed as a GFP fusion from plasmid p1332 (43). Expression of aggregating and non-aggregating variants of huntingtin (Htt103Q and Htt25Q) involved pYES2-HTT103Q-RFP and pYES2-HTT25Q-RFP constructs (44).
Protein isolation and western blotting
Protein extracts were obtained from cells grown to OD600 nm = 1 by disruption with glass beads (45) and quantified using Bradford assay (46). Equal amounts of total protein from different strains were used for loading of gels. Transfer and detection was done as described previously (20) and involved anti-GFP (Santa Cruz Biotechnology, USA) and anti-Cdc19 antibodies (kindly provided by Dr J. Thorner).
mRNA quantification using qPCR
Cultures were grown to OD600 nm = 1 and total RNA was isolated from cells using TRIzol reagent (Thermo Fisher, Germany). Four hundred nanogram of RNA was subjected to reverse transcription and labeling using the SensiFAST SYBR® No-ROX Kit (Bioline, Germany) and Mastercycler ep realplex (Eppendorf, Germany). Quantification of relative RNQ1 mRNA levels in different strains was done according to (47) and involved normalization to ACT1 mRNA levels and oligonucleotides listed in Supplementary Table S2.
Isolation of protein aggregates
Aggregated proteins were isolated from 50 ml YPD cultures grown to OD600 nm = 1 essentially as described (22,48). After breaking cells by sonification, protein content of cleared supernatants was determined using Bradford assay and for each strain, 4 mg total protein were subjected to centrifugation and washing (48). The entire aggregate pellet obtained was boiled in SDS sample buffer and separated on NuPAGE Bis–Tris 4–12% gradient gels. Equal amounts of total extracts (25 μg) were separated on identical gels for comparison.
Staining of dead cells and nuclei
For detection of dead cells, cultures were mixed with an equal volume of 0.01% methylene blue, 2% sodium–citrate dihydrate and analysed in bright field and phase contrast microscopy. For visualization of nuclei, cells were fixed in 70% ethanol and stained with 1 μg ml-1 4’,6-diamidino-2-phenylindole (DAPI) prior to observation in phase contrast and fluorescence optics using an Olympus BX53 microscope.
Actin patch and cable formation
Cells were fixed in 3.7% formaldehyde for 1 h, washed with phosphate-buffered saline (PBS), resuspended in PBS containing 1 μg ml-1 DAPI and incubated for 1 h at room temperature in the dark. Subsequently, cells were washed once with PBS and then resuspended in PBS containing 14 μM rhodamine-conjugated phalloidine. Cells were analysed in phase contrast as well as in the fluorescence mode using rhodamine and DAPI fluorescence filters.
Bud scar staining
Cells were washed once with PBS, resuspended in PBS containing 0.1 mg ml-1 calcoflour white and incubated for 1 h at room temperature. Subsequently, cells were washed twice with PBS and wet mounts analysed using phase contrast and the DAPI fluorescence filter.
Budding pattern of isolated cells
Unbudded phenotypically normal cells were micromanipulated to defined positions on an agar plate, allowed to form two to three daughter cells and positions of daughter and daughter's daughter cells determined by microscopic observation at hourly intervals using the Singer MSM400 dissection microscope in the replicative ageing mode. If second daughter cells appeared distal to or clearly separated from the first daughter or if the first daughter's daughter was formed opposite to the birth site, these were recorded as non-axial budding events.
Time lapse bud site selection
Liquid YPD agar was applied to a single well containing microscope slide and covered with a flat slide until the agar was cooled. The covering slide was removed, leaving the well filled with a thin YPD pad. 2.5 μl of log phase YPD culture were added, sealed with a coverslip and imaged at 30–60 min time intervals.
Cluster formation during replicative ageing
Single phenotypically normal and unbudded cells were micromanipulated to defined positions on a YPD agar plate using the Singer MSM400 micromanipulator in the replicative ageing mode. Cells were analysed in hourly intervals and after completion of the first budding event the mother cell was discarded from each position, generating the starting assembly of virgin mother cells (n = 32 to 45). After each cell cycle, daughter cells were removed by micromanipulation and counted until either cell division ceased or elongated daughter cells were formed that were non-separable from mother cells and started budding in a polar mode. The fraction of viable cells for each generation number was calculated by dividing the number of cells still forming daughters by the starting number of virgin mother cells that did form a first bud. The fraction of clustered cells for each generation number was calculated by dividing the number of cells that formed an elongated non-separable bud by starting number of virgin mother cells that did form a first bud.
RESULTS
Synthetic interactions between deletions of genes for mcm5s2U34 formation and other tRNA anticodon loop modifications
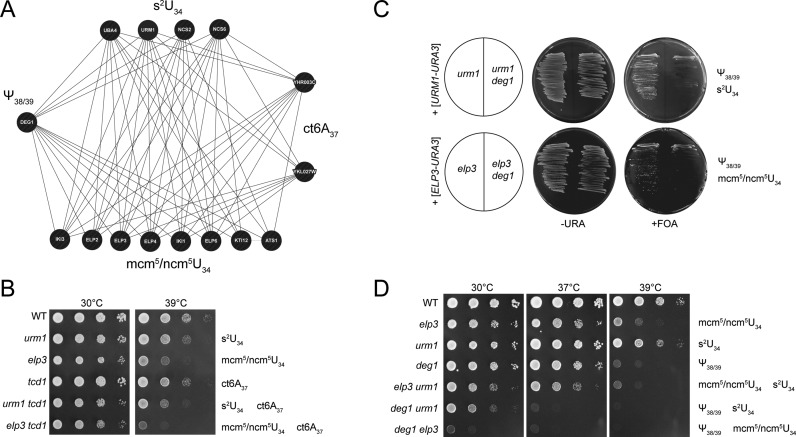
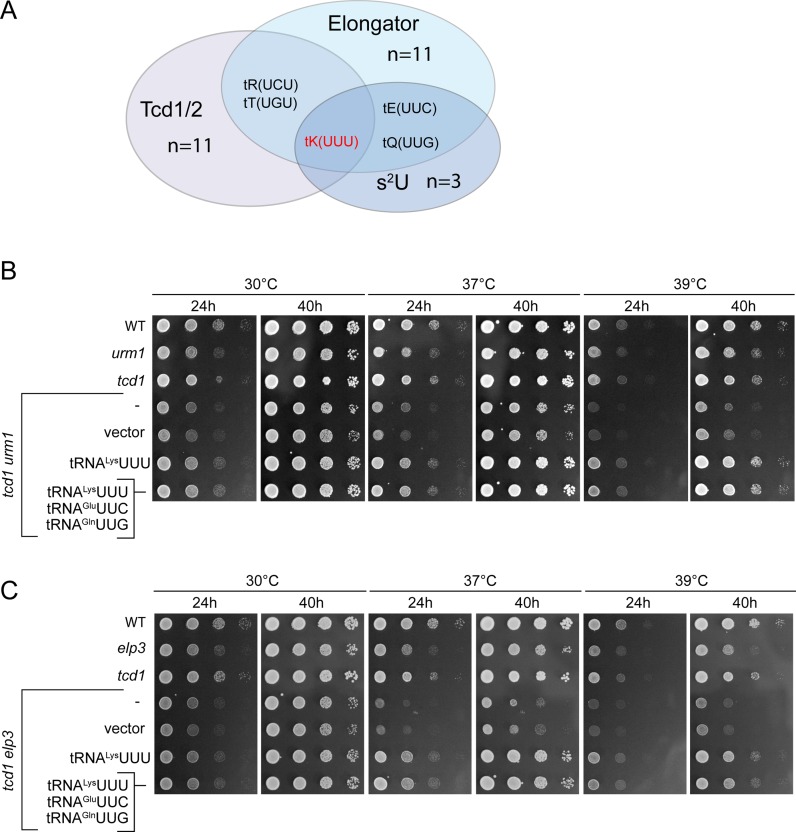
SGA data (36) revealed DEG1, YHR003 (TCD1) and YKL027W (TCD2) as genes maintaining negative genetic interactions with almost all of the genes required for either m/ncm5U34 or s2U34 formation (Figure 1A). TCD1 and TCD2 are both required for formation of ct6A (35) at tRNA position 37 and DEG1 is responsible for generation of pseudouridine (Ψ) at positions 38 and 39 (26,49). To verify and further characterize the impact of simultaneous absence of such modifications, we generated tcd1 urm1 and tcd1 elp3 double mutants and analysed their growth in response to elevated cultivation temperatures. In either case, the combination of both modification defects was found to enhance the fitness defect, confirming the negative genetic interactions revealed by SGA (Figure 1B).
Figure 1.
Negative genetic interactions between tRNA modification genes. (A) Synthetic genetic array-derived interaction network of genes required for s2U, mcm5/ncm5U, Ψ38/39 and ct6A. The network diagram was constructed with GeneMANIA (genemania.com) and is based on the negative interaction dataset from Costanzo et al. (36). (B) Wild type (WT) and tRNA modification mutants were serially diluted and replica spotted on YPD plates and incubated at 30 or 39°C for 40 h. (C) Plasmid shuffle results. Indicated strains were streaked on –URA or FOA, resulting in loss of plasmids carrying ELP3 or URM1 wild type genes. Missing tRNA modifications are indicated to the right. (D) Wild type (WT) and tRNA modification mutants as indicated were serially diluted and replica spotted on YPD plates and incubated at 30, 37 or 39°C for 40h.
Double mutants elp3 deg1 or urm1 deg1 were generated by plasmid shuffle. Uncovering the double deletions elp3 deg1 and urm1 deg1 by removal of the plasmid with wild type ELP3 or URM1 genes revealed a major growth defect in both cases (Figure 1C). When urm1 deg1 and elp3 deg1 colonies were phenotypically analysed, a strong temperature sensitive (TS) phenotype exceeding the one of the elp3 urm1 double mutant in strength was observed. As with elp3 urm1 cells, urm1 deg1 and elp3 deg1 double mutants also display a growth defect at 30°C, which is most pronounced in the latter strain (Figure 1D). Comparison of elp3 urm1 (n/mcm5/s2U34 negative), urm1 deg1 (s2U34 and Ψ38/39 negative) and elp3 deg1 (n/mcm5U34 and Ψ38/39 negative) phenotypes indicates a gradual increase in growth defects and stress sensitivity in this order (Figure 1D). While this work was in progress, another study reported negative genetic interactions between KTI12 (required for n/mcm5 formation) or UBA4 and DEG1 (27), supporting our conclusion that simultaneous removal of either m/ncm5U or s2U together with Ψ38/39 affects cell proliferation.
Identification of affected tRNAs by TS suppressor analysis
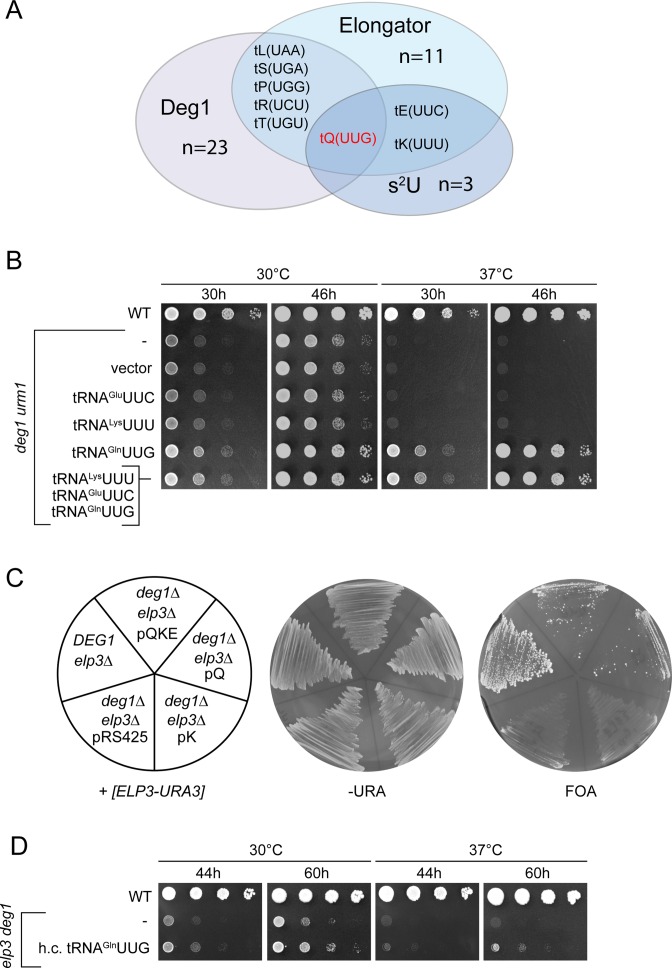
The synthetic phenotypes of the above tRNA modification mutants resemble traits of previously studied elp3 urm1 and elp3 uba4 strains (19); in the latter case, phenotypes were shown to be partly suppressible by overexpression of tRNALysUUU and more efficiently by combined overexpression of tRNALysUUU, tRNAGluUUC and tRNAGlnUUG, which are the three tRNA species normally carrying the mcm5s2U34 modification (19). Thus, absence of U34 modification was concluded to induce a functional defect in these three tRNAs, which can be compensated by overproducing them. We reasoned that in case the synthetic phenotypes of double mutants involving deg1 or tcd1 are similarly caused by reduced functionality of individual tRNA species, these should be identifiable on the basis of their capacity to suppress the mutant growth defects when overexpressed. To select tRNAs for this approach, we grouped all tRNAs carrying uridine at positions 38 or 39 (which is modified to Ψ in all sequenced yeast tRNAs) as a prerequisite for pseudouridylation at these sites and all tRNAs either known to carry t6A or carrying U36-A37 (which is modified to ct6A/t6A37 in all sequenced yeast tRNAs) as a prerequisite for ct6A formation. These two groups were overlapped with the known set of tRNAs carrying either m/ncm5U34 (11 species) or s2U34 (3 species) (4). While based on these criteria, there are 23 Deg1 targets and 11 Tcd1 targets, tRNALysUUU is the only tRNA in yeast that fulfills the sequence requirements to carry mcm5s2U34 and ct6A37 (Figure 2A) and tRNAGlnUUG is the only tRNA that can carry mcm5s2U34 and Ψ38 (Figure 3A). No tRNA meets the sequence requirements to carry all three modifications or the combination of mcm5s2U34 and Ψ39. Thus, tRNALysUUU and tRNAGlnUUG were analysed for their potential to suppress the above phenotypes of urm1 tcd1 and elp3 tcd1 (tRNALysUUU, Figure 2B, C) or urm1 deg1 and elp3 deg1 mutants (tRNAGlnUUG, Figure 3B, C). Indeed, higher-than-normal levels of tRNALysUUU suppressed the TS phenotype of urm1 tcd1 and elp3 tcd1 and the combined overexpression of tRNALysUUU together with tRNAGlnUUG and tRNAGluUUC was no more effective than overexpression of tRNALysUUU alone (Figure 2B, C). Likewise, overexpression of tRNAGlnUUG effectively rescued the growth defect and TS phenotype of the urm1 deg1 mutant and again, combined overexpression of tRNAGlnUUG together with tRNAGluUUC and tRNALysUUU was no more effective than tRNAGlnUUG overexpression alone (Figure 3B). For elp3 deg1, which shows the strongest growth defect, we tested the effect of overexpression of tRNAGlnUUG alone or in combination during generation of the double mutant via plasmid shuffle. Upon chase out of the ELP3-URA3 plasmid from the elp3 deg1 double mutant, colony formation was significantly improved by the presence of either tRNAGlnUUG overexpression (pQ) or the combined overexpression (pQKE) construct (Figure 3C). However, when elp3 deg1 pQ colonies are compared to the empty vector control in drop dilution assays, we detected only a modest improvement of growth (Figure 3D), suggesting that in such background, tRNAGlnUUG is less efficient in phenotypic suppression compared to the urm1 deg1 background (Figure 3B).
Figure 2.
Phenotypic rescue of tcd1 urm1 and tcd1 elp3 double mutants by tRNA overexpression. (A) Venn diagram indicating the number and identity of tRNAs potentially subject to Elongator, s2U and Tcd1/2 dependent tRNA modification. The set of Elongator and s2U dependent tRNAs is according to Johansson et al.(4) and the set of Tcd1/2 dependent tRNAs based on the sequence U36-A37, which is always modified to t6A37 (modomics.genesilico.pl). Note that ct6A is the natural configuration of t6A in yeast (35). (B and C) Wild type (WT) and tRNA modification mutants carrying no vector (–), empty vector (vector) or tRNA overexpression plasmids for tRNALysUUU alone or in combination with tRNAGluUUC and tRNAGlnUUG were serially diluted and replica spotted on YPD plates and incubated at 30, 37 or 39°C for 24 h, photographed, incubated for additional 16 h (40 h total) and photographed again.
Figure 3.
Phenotypic rescue of deg1 urm1 and deg1 elp3 double mutants by tRNA overexpression. (A) Venn diagram indicating the number and identity of tRNAs potentially subject to Elongator, s2U and Deg1 dependent tRNA modification. The set of Elongator and s2U dependent tRNAs (4) and the set of potentially Deg1 dependent tRNAs is derived from tRNA species carrying a uridine in position 38 or 39, which are modified in all sequenced tRNAs to ψ (modomics.genesilico.pl). (B) Wild type (WT) and tRNA modification mutants carrying no vector (–), empty vector pRS425 (vector) or tRNA overexpression plasmids for tRNAGlnUUG alone (pQ) or in combination with tRNAGluUUC and tRNALysUUU (pQKE) were serially diluted and replica spotted on YPD plates, which were incubated at 30, 37 or 39°C for 30 h, photographed, incubated for additional 16 h (46 h total) and photographed again. (C) Plasmid shuffle result. Indicated strains carrying a plasmid borne wild type copy of ELP3 were streaked on –URA or FOA. (D) Wild type and elp3 deg1 mutants carrying no plasmid (–) or the overexpression plasmid for tRNAGlnUUG were serially diluted and replica spotted on YPD plates, which were incubated at 30°C or 37°C for 44 h, photographed, incubated for an additional 16 h (60 h total) and photographed again.
Translational defect caused by tRNAGlnUUG in combined absence of mcm5/s2U and Ψ38/39
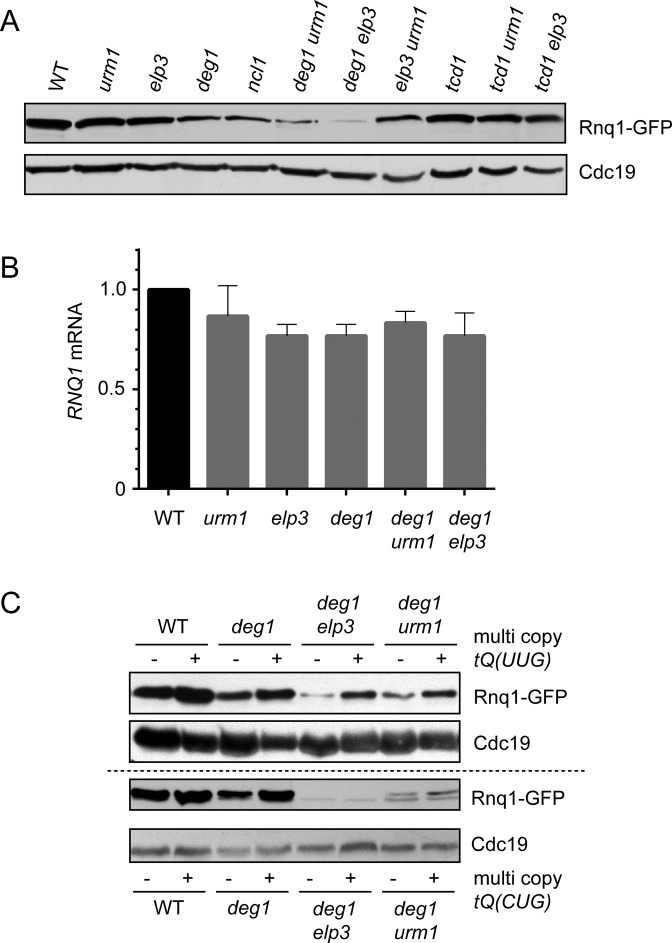
To analyse a potential translational defect of tRNA in the combined absence of modifications, we investigated expression of the N- and Q-rich prion Rnq1 (50) in various tRNA modification mutants. Rnq1 is among the most Q-rich proteins in yeast (19% Gln, 77 of 405 aa, the eighth rank in terms of relative Gln abundance) and the RNQ1 gene displays a mixed CAA/CAG (Gln) codon usage of 53/24 (yeastmine and SGD data). In contrast, Rnq1 contains only seven Lys codons (1.7%, the 5855th rank in terms of relative Lys abundance) and displays at the gene level an AAA/AAG (Lys) codon usage ratio of 3/4. Hence, translation of RNQ1 mRNA should be sensitive to functional disturbance of tRNAGlnUUG rather not tRNALysUUU. To test this, we expressed a GFP tagged variant of Rnq1 in wild type, single and double tRNA modification mutants and analysed protein levels of Rnq1-GFP as well as Cdc19 as a control by Western blotting. Other than Rnq1, Cdc19 is not enriched in Gln codons (2% Gln, 10 of 501 aa, 5421th rank in terms of Gln abundance). We observed a clear downregulation of Rnq1-GFP protein levels in deg1 and a much stronger effect in urm1 deg1 and elp3 deg1 double mutants (Figure 4A). At the same time, no fluctuation of Cdc19 levels was observable. Combinations of urm1 tcd1 or elp3 tcd1, which target tRNALysUUU function instead of tRNAGlnUUG (Figure 2), did not change expression levels of Rnq1-GFP compared to wild type (Figure 4A). To check whether downregulation of Rnq1 protein levels in tRNA modification mutants is due to inefficient translation or due to transcriptional repression, we quantified RNQ1 mRNA levels by qPCR. We observed only a moderate (∼25%) downregulation of transcription (Figure 4B) that is insufficient to explain the severe, near complete elimination of Rnq1 production in urm1 deg1 and elp3 deg1 (Figure 4A). Thus, a significant translational inefficiency of the RNQ1 mRNA exists in cells deficient in ψ38/39 and m/ncm5U or s2U. To explore whether tRNA overexpression, which was shown to suppress growth defects in these mutants (Figure 3) also rescues this translational defect, we analysed Rnq1 protein levels in urm1 deg1 and elp3 deg1 cells carrying overexpression constructs for tRNAGlnUUG, tRNAGlnCUG or empty vector controls. As shown in Figure 4C, the severe reduction of Rnq1 signals in urm1 deg1 and elp3 deg1 is efficiently rescued by tRNAGlnUUG but not by tRNAGluCUG overexpression. Thus, urm1 deg1 and elp3 deg1 mutations induce a translational defect of the RNQ1 mRNA which can be attributed to reduced function of tRNAGlnUUG and consequently to inappropriate decoding of CAA codons.
Figure 4.
Translational defect of the Q/N-rich prion Rnq1 in combined absence of ψ38/39 and either mcm5U or s2U. (A) Western blot of total extracts from wild type (WT) or indicated tRNA modification mutants carrying p1332 (Rnq1-GFP). GFP and Cdc19 signals from identical extracts are shown. (B) Quantification of RNQ1 mRNA (relative to ACT1) in wild type (WT) or the indicated tRNA modification mutants by qPCR. (C) Rescue of translational defects by overexpression of tRNAGlnUUG. Wild type or tRNA modification mutants carrying p1332 and additionally either empty vector (–), multi copy tQ(UUG) (+, upper panel) or multi copy tQ(CUG) (+, lower panel) were used to prepare total protein extracts and detect Rnq1-GFP or Cdc19 signals.
Synthetic cellular phenotypes of mutants with combined tRNA modification defects
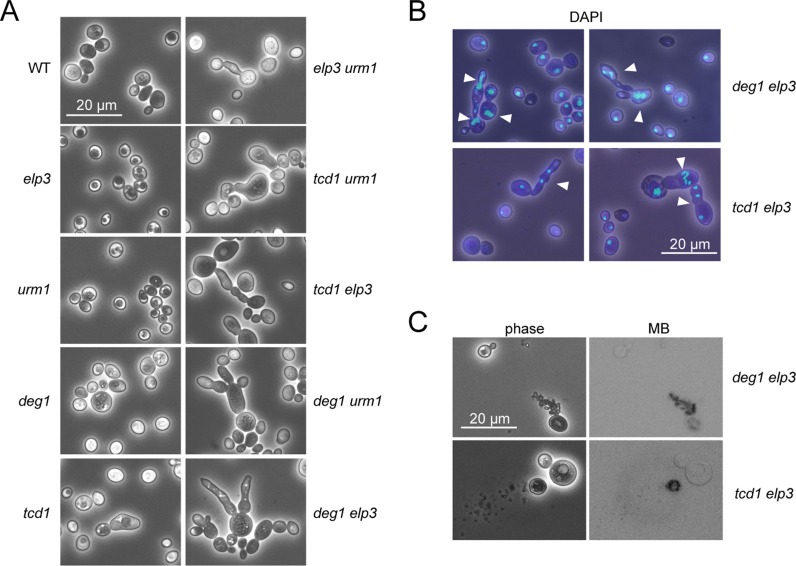
We have previously shown that complete loss of mcm5s2U34 results in the formation of abnormally elongated, multi-budded and lysed cells (19). Inspection of the double mutants generated in this study revealed that combinations of tcd1 or deg1 with either elp3 or urm1 mutations result in similar phenotypic abnormalities and spontaneous cell lysis (Figure 5A–C). In all cases, phenotypically normal cells exist together with the abnormal (elongated, multibudded and lysed) cell types, which can represent a fraction of 11–25% of the culture (Supplementary Table S3). None of the single mutants or the wild type displays these phenotypic abnormalities. The tcd1 mutant was found to form slightly bulky, but not largely elongated or lysed cells (Figure 5A). The degree of change from normal to abnormal cell types and morphology was found to be most severe in the tcd1 elp3 and deg1 elp3 double mutants, which both tend to form clustered aggregates containing several cells that apparently failed to separate, indicative of a cytokinesis defect (Figure 5A). DAPI staining of nuclear DNA revealed multiple nuclei in many of the phenotypically abnormal cell types, consistent with a severe post-mitotic nuclear segregation defect (Figure 5B). To check whether morphological alterations and cytokinesis defects are suppressible by tRNA overexpression, we quantified numbers of cells displaying elongated morphology or cell lysis in tRNA modification mutants carrying tRNA overexpression constructs or empty vector controls. We analysed elp3 deg1 and urm1 deg1 mutants with overexpression of tRNAGlnUUG and elp3 tcd1 mutants with overexpression of tRNALysUUU grown at standard (30°C) or elevated (37–39°C) temperature. We found that both formation of abnormal cell types and lysis is largely suppressible by tRNALysUUU in elp3 tcd1 and by tRNAGlnUUG in urm1 deg1 and elp3 deg1 (Supplementary Table S3). In addition, elevated temperature increased the frequency of morphological defects in urm1 deg1 and cell lysis in the latter and elp3 tcd1 mutants. Both effects are clearly suppressible by tRNA overexpression (Supplementary Table S3), however, they are less effectively suppressed in elp3 deg1 compared to urm1 deg1 (Supplementary Table S3).
Figure 5.
Cytological phenotype of tRNA modification mutants. (A) Wild type (WT) and indicated tRNA modification mutants were analysed by phase contrast microscopy. (B) Nuclear staining (DAPI) of deg1 elp3 and tcd1 elp3 double mutants. (C) Methylene-blue staining of deg1 elp3 and tcd1 elp3 double mutants.
Since double mutant cultures analysed in this study always contain a mixture of phenotypically normal and highly aberrant cell types, (11–25% at 30°C, Supplementary Table S3), we investigated for one strain (elp3 deg1) in detail at which point during the life span of individual cells morphological alteration occurs. We analysed a set of 32 or 45 virgin mother cells of the wild type and elp3 deg1, respectively, and followed their replicative lifespan by microdissection and counting all daughter cells from each of the virgin mothers until either senescence or cluster formation occurred. The wild type displayed an average lifespan of 21.3 generations and only one of the mother cells (3.2%) formed a non-separable cluster after 24 normal cell divisions (Supplementary Figure S1). For elp3 deg1, we found that 39 (86.6%) mother cells formed a cluster after varying numbers (1–7) of normal cell divisions (Supplementary Figure S1). The remaining six cells that did not form a cluster ceased growth after only 1–8 generations. Due to the non-separable nature of clusters and continuous growth of daughter cells, it is not possible to follow replicative lifespan of the initially observed mother cells once the transition into a cluster occurred. However, the majority of elp3 deg1 cells (>50%) formed a cluster after 8 or less generations. Thus, loss of different tRNA modifications results in a common phenotype including slow growth, thermosensitivity and cytokinesis defects, the latter becoming acute after varying numbers of normal cytokinesis events.
Actin patch and cable formation in mutants with combined tRNA modification defects
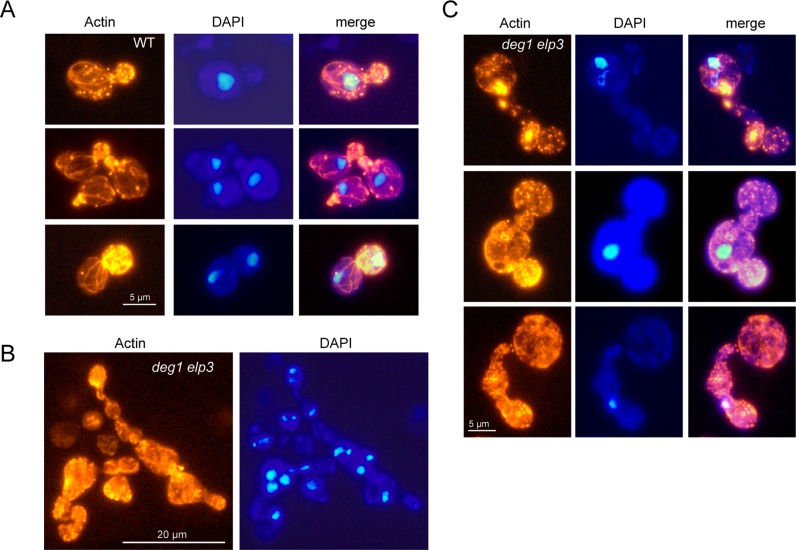
Since the polarization of actin cytoskeleton is crucial in the processes of budding and cytokinesis (51,52), the observed abnormal cell morphologies suggested the possibility of actin cytoskeleton defects due to inappropriate tRNA modifications. To test this option, we analysed actin polarization and cable formation in the various tRNA modification mutants (Figure 6). In wild type cells, we observed normally polarized cell types, where actin patches accumulate in small buds of single nucleus containing cells and detected actin cable structures that run mostly parallel to the mother bud axis (Figure 6A, Supplementary Figure S2). In elp3 deg1 double mutants, abnormal cell types are characterized by a massive breakdown of actin cables and by multiple sites of actin aggregation or extensive patch formation in the absence of polarization (Figure 6B and C, Supplementary Figure S2). Very similar effects on actin cable formation and aggregation were observed in the tcd1 elp3 mutant as well (Supplementary Figure S2), identifying common aberrances in the actin cytoskeleton of mutants with defects in different tRNA anticodon stem loop modifications.
Figure 6.
Actin cytoskeleton defects. Exponentially growing wild type (WT) (A) or deg1 elp3 double mutant cells (B and C) were fixed and actin was visualized using rhodamine-conjugated phalloidine. Simultaneously, positions and numbers of nuclei were detected using DAPI.
Bud site selection is abnormal in mutants with combined tRNA modification defects
The abnormal mutant cell types are characterized by elongated or deformed buds that appear on both cell poles or at random positions (Figure 5, Supplementary Figure S2), suggesting that the regular axial bud site selection pattern of haploid yeast cells (51,52) becomes disturbed in the combined absence of m/ncm5U34 and ct6A37 or Ψ38/39 modifications. To verify this and to investigate whether the defect also applies to morphologically normal cell types in these genetic backgrounds, we analysed bud scar positioning. In haploid control cells, we observe the expected arrangement of bud scars in chains on the cell surface (axial budding pattern) (52). In isogenic diploid control cells, bud scars are located on both cell poles, but not randomly distributed over the surface (52) (Supplementary Figure S3A). In both elp3 tcd1 and elp3 deg1 mutants, however, phenotypically normal cells often display a mixed-up bud site selection pattern, where some scars are arranged in chains and others appear randomly on the cell surface or on both the cell poles (Supplementary Figure S3B). To quantify these effects, unbudded cells (>200) were isolated by micromanipulation and bud positioning during the first 3 generations was observed microscopically for each cell. While all wild type cells displayed axial budding, 9.4 and 24.5% of tcd1 elp3 and deg1 elp3 cells displayed non-axial bud positioning (Supplementary Table S4). Abnormal cell types are also characterized by a random bud scar appearance and in addition display strong, partly delocalized chitin deposition at sites where septum formation would be expected (at the junction between mother and bud that failed to complete cytokinesis) (Supplementary Figure S3C). Using time lapse microscopy, we find that both elp3 tcd1 and elp3 deg1 cells display a mixture of polar and normal, axial bud site selection (Supplementary Figures S3D and S4). In particular, elongated cell types that had undergone polar budding typically switch back to axial budding for the next division cycles (Supplementary Figure S4), discriminating the observed mutant phenotype from the filamentous growth mode, which is characterized by unipolar budding (53). It should be noted, however, that tRNA modification defects induced by Elongator mutations can modulate (suppress) filamentous growth and pseudouridylation of small nuclear RNA U6 by Pus1 is induced during induction of filamentous growth (54,55). In this study, we analysed mutant phenotypes in the non-filamentous strain BY4741 and observe a phenotype of tRNA modification mutants that resembles filamentous growth but is distinct from this growth pattern in several aspects (see above). Our results indicate that a normal and proper haploid specific bud site selection process depends on proper formation of tRNA anticodon stem loop modifications including n/mcm5/s2U, Ψ38/39 and ct6A.
Implication of protein aggregates in phenotypes of tRNA modification mutants
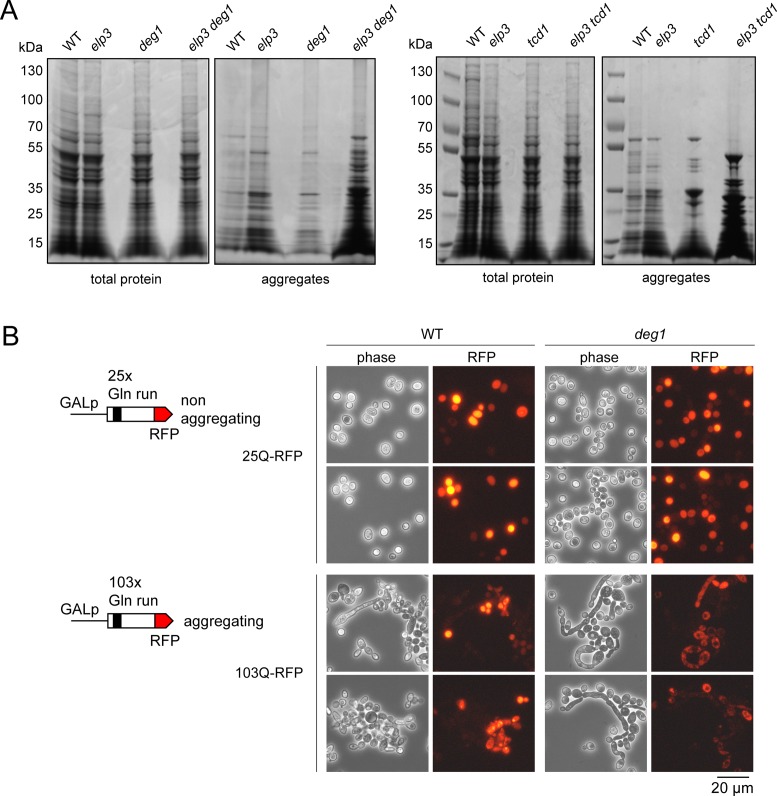
The complete absence of mcm5s2U and loss of ct6A were recently shown to induce a widespread protein aggregation phenomenon in yeast that associates with ribosomal slow down and stalling due to inefficient codon recognition (22,32). Since elongated and lysed cells were found to be induced in elp3 uba4, a mutant also lacking mcm5s2U (19), we analysed whether shared phenotypes of tRNA modification mutants could possibly be attributed to the formation of protein aggregates. We found the single mutations in ELP3, TCD1 and DEG1 induced a moderate increase in protein aggregates compared to the wild type. Combinations of modification defects in elp3 deg1 and elp3 tcd1, however, strongly increased the amount of detectable protein aggregates compared to either single mutant (Figure 7A) alone. Thus, the negative synthetic interactions between tRNA modification gene deletions analysed in this study go hand-in-hand with strongly increased formation of protein aggregates.
Figure 7.
Protein aggregation in tRNA modification mutants and severe cytokinesis defects in cells with conditional expression of an aggregating protein. (A) Total protein or aggregates were isolated from wild type (WT) and indicated tRNA modification mutants as described in the methods section and analysed by PAGE and Coomassie staining. Identical amounts of total extract were subjected to isolation and the entire aggregate pellet was loaded. (B) Two constructs were used to express either the non-aggregating Htt25Q-RFP or the aggregating Htt103Q-RFP in wild type (WT) or deg1 single mutants. Morphology changes and expression of RFP fusion proteins were detected by phase contrast or fluorescence microscopy, respectively.
To test whether induction of cellular protein aggregation by other means is capable of inducing cytokinesis defects similar to the ones observed in tRNA modification mutants, we utilized the well characterized yeast huntingtin model system (44) to artificially express an aggregation-prone protein. Two different constructs allowing the conditional expression of a huntingtin-RFP fusion with either 25× (Htt25Q, non-aggregating) or 103× (Htt103Q, aggregating) Gln repeats (44) were introduced into wild type and deg1 mutants and subjected to prolonged (4 days) induction on solid galactose medium. Cells were recovered from the plates and analysed for cytokinesis defects and RFP signal distribution. Expression (RFP signal) was detectable in all strains and displayed uniform distribution in the cytoplasm for the Htt25Q variant in both wild type and deg1 mutants. All cells showed normal unbudded or single budded ellipsoid morphology (Figure 7B, Supplementary Figure S5A). With the construct inducing Htt103Q, RFP expression was observed in both strains and signal distribution was grainy, consistent with the established tendency of Htt103Q-RFP to form aggregates. Strikingly, however, strongly elongated and clustered cell types were observable for both wild type and deg1 mutant cells (Figure 7B, Supplementary Figure S5). In the latter, elongated morphology is further enhanced compared to the wild type (Supplementary Figure S5A) with some buds being ∼10 times as long as the mother cell. DAPI staining revealed the occurrence of multiple nuclei (Supplementary Figure S5B), indicating a nuclear segregation defect induced by Htt103Q-RFP in wild type and deg1 cells that goes along with clustered and strongly elongated morphology. This situation is highly reminiscent of the pleiotropic defects observed with our tRNA anticodon modification mutants (Figures 1, 5 and 6) strongly suggesting that protein aggregation (due to defects in protein folding and/or solubilty) may be the common denominator for aberrant cell morphology, polarity and cytokinesis defects.
DISCUSSION
Since mcm5s2U is a composite modification generated by two distinct pathways that independently introduce mcm5 and s2 at the same U34 base, and loss of both induces a synthetic growth defect, they both apparently cooperate in tRNA functioning (18,19). A similar negative interaction between genes for synthesis of mcm5s2U (elp3; kti12; uba4 urm1) and Ψ (deg1) was observed (8,27,36), indicating that mcm5/s2 parts of the wobble base modification may also cooperate with Ψ in position 38 or 39 in proper tRNA functioning. Consistently, we identified synthetic sick interactions in elp3/deg1 and urm1/deg1 combinations, suggesting that any mutation affecting either mcm5U or s2U synthesis is deleterious in tandem with loss of Ψ38/39. Since in yeast, tRNAGlnUUG is the only species that carries mcm5s2U34 together with Ψ38, its overexpression is able to partly rescue the growth defects triggered in the double mutants (Figure 3). In comparison, however, overexpression of tRNAGlnUUG is much more efficient in the rescue of urm1 deg1 compared to elp3 deg1 (Figure 3). Whereas in the former case tRNAGlnUUG is the only tRNA predicted to be affected by both modification defects, additional tRNA targets can be anticipated for the elp3 deg1 mutant background, since Elongator is instrumental in the U34 modification of a total of 11 tRNA species, six of which (tRNAGlnUUG, tRNALeuUAA, tRNASerUGA, tRNAProUGG, tRNAArgUCU and tRNAThrUGU) are verified or potential substrates for Ψ synthase Deg1 (4), (modomics.genesilico.pl; Figure 3). Indeed, overexpression of one of them (tRNAProUGG) was shown to increase the thermo-resistance of a deg1 kti12 mutant already overexpressing tRNAGlnUUG (27). However, rescue was still far from wild type growth levels, suggesting that tRNA species other than tRNAGlnUUG and tRNAProUGG do suffer from the joint absence of mcm5/ncm5U34 and Ψ38/39.
SGA network analysis further revealed that deletions of TCD1 and TCD2 genes, which are required for conversion of t6A to ct6A (35), negatively interact with most of the mcm5/s2U genes. We confirmed this SGA result and showed that tcd1 urm1 and tcd1 elp3 synthetic phenotypes could be suppressed by overexpression of tRNALysUUU, which represents the only tRNA species in yeast that carries mcm5s2U34 and ct6A37modifications (4) (modomics.genesilico.pl). Thus, this result and previous suppression of elp3 urm1 and elp3 uba4 phenotypes by joint overexpression of tRNAGluUUC, tRNAGlnUUG and tRNALysUUU (19) suggest that combined loss of distinct anticodon loop modifications causes functional defects in the very tRNAs that normally carry both the modifications and that higher-than-normal levels of the affected tRNA can routinely suppress the malfunction. Interestingly, a synthetic growth defect was also observed in E. coli mutants lacking ct6A and s2U34 (35) suggesting functional cooperation between ct6A and the modified wobble uridine is conserved between bacteria and yeast, a notion that further adds up to the significance of cross-talk between distinct modifications in the tRNA anticodon loop.
We assume that combined tRNA modification defects analysed in here impose a defect on individual tRNA species that result in a disturbance of decoding and translational elongation. In support of this option, loss of ncm5/mcm5U and/or s2U modifications has already been implicated in reduced ribosomal A-site binding due to weakened anticodon codon interactions and shown to induce ribosome pausing at CAA and AAA codons (5,22) and subsequent ribosomal frameshifting due to an A-site effect (56). Here, we have demonstrated a strong translational defect of mRNA coding for the Q/N rich prion Rnq1 specifically when mcm5U or s2U is missing in tandem with Ψ38/39. The latter is due to a malfunction of tRNAGlnUUG, since overexpression of this tRNA (but not the tRNAGlnCUG isoacceptor) not only rescues growth and morphological phenotypes but also re-establishes translational efficiency (Figure 4C). This result indicates a cross talk between the mcm5s2U and ψ38 modifications that is critical for tRNAGlnUUG decoding and translational efficiency. Presumably, independently of each other, ψ38 and mcm5s2U may promote related functions of tRNAGlnUUG, such as mediating efficient ribosomal A-site entry or A-site binding. Hence, a scenario, where loss of either modification results in a rather mild disturbance of A-site binding but combined loss leading to a severe defect in the same essential process seems plausible and is consistent with the observed genetic interactions between elp3/urm1 and deg1.
Since we have previously shown that loss of mcm5s2U triggers formation of cells with elongated or multiple buds (19), we examined how synthetic combinations of tRNA modification gene deletions relate to bud site selection, cell morphology and cytokinesis. We find that all of them share highly aberrant cell types, carrying elongated multiple buds appearing on various cell poles. Apparently, haploid specific axial bud site selection is disturbed in the different tRNA modification mutants, but in all cases analysed, cells display a mixture of polar and random or axial budding. Since aberrant cells exhibit clear signs of actin cytoskeleton defects (Figure 6) and because the dynamic remodeling of the actin cytoskeleton is fundamental for the establishment of cell polarity and cytokinesis (52), morphological defects of tRNA modification mutants might be caused in part by actin cytoskeleton breakdown. The latter could also be involved in the observed nuclear segregation defects, since actin cables are important for capture of cytoplasmic microtubules and Myo2 dependent transport of the nucleus to the bud neck before cytokinesis (57). As the modification defects apparently affect distinct tRNA species reading different codons (see above), the observed common phenotypes are unlikely caused by inefficient translation of mRNAs enriched for particular codons. Interestingly, a recent study identifying widespread protein aggregation in a strain lacking mcm5s2U (22) shows that among the detected 610 proteins forming aggregates, 41 are associated with the gene ontology (GO) term cytoskeleton organization. These include several CCT genes, which encode subunits of the cytosolic chaperonin complex required for folding of actin subunits (58). Importantly, the aggregating proteins in the mcm5s2U deficient strain are not enriched on their mRNA levels for mcm5s2U dependent codons (22), stressing that aggregation might be induced by translational slow down and ribosomal stalling and subsequently involves folding defects in proteins not directly affected by the translational defect. Global protein aggregation could be triggered by destabilizing effects of initial aggregates on protein complexes (22) or by affecting the efficiency or availability of chaperones for folding of proteins other than those being synthesized slowly. In both situations, the global protein aggregation would be initiated by slow translation but would not necessarily be linked to the identity of codon(s) inducing the slow-down. Thus, ribosomal-slow down at any codon could in theory induce such effect and possibly explain why defects of distinct tRNAs induce a common phenotype, as observed in this study. Because Deg1 can also modify uridines in mRNA (59), loss of this function could in principle also contribute to double mutant phenotypes. However, with efficient phenotypic suppression by multi copy tRNA and without a known mRNA modification role of the Urm1/Elongator pathways synthetic phenotypes are likely induced mainly due to loss of tRNA function.
Since the mcm5s2U defective elp3 urm1 mutant used in this study also forms elongated multi-budded cells (19) (Figure 4A), aggregation of proteins important for cytoskeleton organization could represent a common basis of the cellular defects that we observe in here. Possibly, tRNA modification mutant cultures experience cumulative protein damage, resulting in the onset of morphological alteration when a certain threshold is reached, explaining why genetically identical mutant cells exist as a mixture of phenotypically normal and highly aberrant cell types. In support of this assumption, we showed for the elp3 deg1 mutant in a replicative ageing experiment that the majority of mother cells do form aberrant and clustered cell types after a comparatively low number of normal cell divisions. Further, the combination of elp3/urm1 mutations with defects in tcd1 or deg1 strongly aggravated the formation of protein aggregates and ectopic expression of aggregating Htt103-RFP, but not the non-aggregating Htt25Q-RFP induced cytokinesis- and nuclear segregation defects highly similar to those observed with our various tRNA modification mutants. Thus, the induction of shared pleiotropic and severe phenotypes under conditions where distinct tRNAs become compromised due to the loss of modifications in their anticodon stem loops is highly likely correlated with a common induction of protein aggregation.
Supplementary Material
Acknowledgments
We thank Dr R. Wickner (Bethesda, USA), Dr F. Giorgini (Leicester, UK) and Dr I. Stansfield (Aberdeen, UK) for providing plasmids.
Footnotes
Present address: Johanna Funk, Max-Planck-Institut für molekulare Physiologie, Dortmund, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG (Deutsche Forschungsgemeinschaft) [SCHA750/15, SCHA750/18 to R.S.]; DFG Priority Program SPP1784 ‘Chemical Biology of Native Nucleic Acid Modifications’ [SCHA750/20-1 to R.S., KL2937/1-1 to R.K.]. ZFF (Zentrale Forschungsförderung Universität Kassel) to R.K. [ZFF-PROJEKT 1906]. Funding for open access charge: Third party funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Phizicky E.M., Hopper A.K. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El Yacoubi B., Bailly M., de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 3.Huang B., Johansson M.J., Byström A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansson M.J., Esberg A., Huang B., Björk G.R., Byström A.S. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rezgui V.A., Tyagi K., Ranjan N., Konevega A.L., Mittelstaet J., Rodnina M.V., Peter M., Pedrioli P.G. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12289–12294. doi: 10.1073/pnas.1300781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang B., Lu J., Byström A.S. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noma A., Sakaguchi Y., Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leidel S., Pedrioli P.G., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 9.Jablonowski D., Butler A.R., Fichtner L., Gardiner D., Schaffrath R., Stark M.J. Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics. 2001;159:1479–1489. doi: 10.1093/genetics/159.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fichtner L., Frohloff F., Bürkner K., Larsen M., Breunig K.D., Schaffrath R. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol. Microbiol. 2002;43:783–791. doi: 10.1046/j.1365-2958.2002.02794.x. [DOI] [PubMed] [Google Scholar]
- 11.Fichtner L., Jablonowski D., Schierhorn A., Kitamoto H.K., Stark M.J.R., Schaffrathk R. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol. Microbiol. 2003;49:1297–1307. doi: 10.1046/j.1365-2958.2003.03632.x. [DOI] [PubMed] [Google Scholar]
- 12.Bär C., Zabel R., Liu S., Stark M.J., Schaffrath R. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol. Microbiol. 2008;69:1221–1233. doi: 10.1111/j.1365-2958.2008.06350.x. [DOI] [PubMed] [Google Scholar]
- 13.Zabel R., Bär C., Mehlgarten C., Schaffrath R. Yeast alpha-tubulin suppressor Ats1/Kti13 relates to the Elongator complex and interacts with Elongator partner protein Kti11. Mol. Microbiol. 2008;69:175–187. doi: 10.1111/j.1365-2958.2008.06273.x. [DOI] [PubMed] [Google Scholar]
- 14.Mehlgarten C., Jablonowski D., Breunig K.D., Stark M.J., Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol. Microbiol. 2009;73:869–881. doi: 10.1111/j.1365-2958.2009.06811.x. [DOI] [PubMed] [Google Scholar]
- 15.Mehlgarten C., Jablonowski D., Wrackmeyer U., Tschitschmann S., Sondermann D., Jäger G., Gong Z., Byström A.S., Schaffrath R., Breunig K.D. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 2010;75:1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdel-Fattah W., Jablonowski D., Di Santo R., Scheidt V., Hammermeister A., ten Have S.M., Thüring K.L., Helm M., Schaffrath R., Stark M.J.R. Phosphorylation of Elp1 by Hrr25 is required for Elongator-dependent tRNA modification in yeast. PLoS Genet. 2015;11:e1004931. doi: 10.1371/journal.pgen.1004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jüdes A., Ebert F., Bär C., Thüring K.L., Harrer A., Klassen R., Helm M., Stark M.J., Schaffrath R. Urmylation and tRNA thiolation functions of ubiquitin-like Uba4·Urm1 systems are conserved from yeast to man. FEBS Lett. 2015;589:904–909. doi: 10.1016/j.febslet.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 18.Björk G.R., Huang B., Persson O.P., Byström A.S. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klassen R., Grunewald P., Thüring K.L., Eichler C., Helm M., Schaffrath R. Loss of anticodon wobble uridine modifications affects tRNALys function and protein levels in Saccharomyces cerevisiae. PLoS One. 2015;10:e0119261. doi: 10.1371/journal.pone.0119261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frohloff F., Fichtner L., Jablonowski D., Breunig K.D., Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esberg A., Huang B., Johansson M.J.O., Byström A.S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 22.Nedialkova D.D., Leidel S.A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheidt V., Jüdes A., Bär C., Klassen R., Schaffrath R. Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling. Microb. Cell. 2014;12:416–424. doi: 10.15698/mic2014.12.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinshteyn B., Gilbert W.V. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013;9:e1003675. doi: 10.1371/journal.pgen.1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer F., Matsuyama A., Candiracci J., Dieu M., Scheliga J., Wolf D.A, Yoshida M., Hermand D. Translational control of cell division by Elongator. Cell Rep. 2012;1:424–433. doi: 10.1016/j.celrep.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecointe F., Simos G., Sauer A., Hurt E.C., Motorin Y., Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J. Biol. Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- 27.Han L., Kon Y., Phizicky E.M. Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA. 2015;21:188–201. doi: 10.1261/rna.048173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Yacoubi B., Lyons B., Cruz Y., Reddy R., Nordin B., Agnelli F., Williamson J.R., Schimmel P., Swairjo M.A., de Crécy-Lagard V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009;37:2894–2909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Yacoubi B., Hatin I., Deutsch C., Kahveci T., Rousset J.P., Iwata-Reuyl D., Murzin A.G., de Crécy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011;30:882–893. doi: 10.1038/emboj.2010.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiaville P.C., El Yacoubi B., Köhrer C., Thiaville J.J., Deutsch C., Iwata-Reuyl D., Bacusmo J.M., Armengaud J., Bessho Y., Wetzel C., et al. Essentiality of threonylcarbamoyladenosine (t6A), a universal tRNA modification, in bacteria. Mol. Microbiol. 2015;98:1199–1221. doi: 10.1111/mmi.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rojas-Benitez D., Thiaville P.C., de Crécy-Lagard V., Glavic A. The Levels of a Universally Conserved tRNA Modification Regulate Cell Growth. J Biol Chem. 2015;290:18699–18707. doi: 10.1074/jbc.M115.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiaville P.C., Legendre R., Rojas-Benítez D., Baudin-Baillieu A., Hatin I., Chalancon G., Glavic A., Namy O., de Crécy-Lagard V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell. 2015;3:29–45. doi: 10.15698/mic2016.01.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daugeron M.C., Lenstra T.L., Frizzarin M., El Yacoubi B., Liu X., Baudin-Baillieu A., Lijnzaad P., Decourty L., Saveanu C., Jacquier A., et al. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res. 2011;39:6148–6160. doi: 10.1093/nar/gkr178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiaville P.C., Iwata-Reuyl D., de Crécy-Lagard V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)A), a universal modification of tRNA. RNA Biol. 2014;11:1529–1539. doi: 10.4161/15476286.2014.992277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyauchi K., Kimura S., Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013;9:105–111. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- 36.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S., et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 38.Gueldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furukawa K., Mizushima N., Noda T., Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 2000;275:7462–7465. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 40.Klassen R., Paluszynski J.P., Wemhoff S., Pfeiffer A., Fricke J., Meinhardt F. The primary target of the killer toxin from Pichia acaciae is tRNA(Gln) Mol. Microbiol. 2008;69:681–697. doi: 10.1111/j.1365-2958.2008.06319.x. [DOI] [PubMed] [Google Scholar]
- 41.Jablonowski D., Zink S., Mehlgarten C., Daum G., Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006;59:677–688. doi: 10.1111/j.1365-2958.2005.04972.x. [DOI] [PubMed] [Google Scholar]
- 42.Kemp A.J., Betney R., Ciandrini L., Schwenger A.C., Romano M.C., Stansfield I. A yeast tRNA mutant that causes pseudohyphal growth exhibits reduced rates of CAG codon translation. Mol. Microbiol. 2013;87:284–300. doi: 10.1111/mmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayashiki T., Kurtzman C.P., Edskes H.K., Wickner R.B. Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mason R., Giorgini F. Modeling Huntington disease in yeast. Prion. 2001;5:269–276. doi: 10.4161/pri.5.4.18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zachariae W., Shin T.H., Galova M., Obermaier B., Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274:1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]
- 46.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Chem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 47.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koplin A., Preissler S., Ilina Y., Koch M., Scior A., Erhardt M., Deuerling E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 2010;189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lecointe F., Namy O., Hatin I., Simos G., Rousset J.P., Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 2002;277:30445–30453. doi: 10.1074/jbc.M203456200. [DOI] [PubMed] [Google Scholar]
- 50.Sondheimer N., Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 51.Madden K., Snyder M. Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 52.Bi E., Park H.O. Cell polarization and cytokinesis in budding yeast. Genetics. 2012;191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cullen P.J., Sprague G.F., Jr The regulation of filamentous growth in yeast. Genetics. 2012;190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdullah U., Cullen P.J. The tRNA modification complex elongator regulates the Cdc42-dependent mitogen-activated protein kinase pathway that controls filamentous growth in yeast. Eukaryot Cell. 2009;8:1362–1372. doi: 10.1128/EC.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basak A., Query C.C. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep. 2014;8:966–973. doi: 10.1016/j.celrep.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tükenmez H., Xu H., Esberg A., Byström A.S. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2016;43:9489–9499. doi: 10.1093/nar/gkv832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hwang E., Kusch J., Barral Y., Huffaker T.C. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stoldt V., Rademacher F., Kehren V., Ernst J.F., Pearce D.A., Sherman F. Review: the Cct eukaryotic chaperonin subunits of Saccharomyces cerevisiae and other yeasts. Yeast. 1996;12:523–529. doi: 10.1002/(SICI)1097-0061(199605)12:6%3C523::AID-YEA962%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 59.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.