Abstract
Single molecule FISH (smFISH) allows studying transcription and RNA localization by imaging individual mRNAs in single cells. We present smiFISH (single molecule inexpensive FISH), an easy to use and flexible RNA visualization and quantification approach that uses unlabelled primary probes and a fluorescently labelled secondary detector oligonucleotide. The gene-specific probes are unlabelled and can therefore be synthesized at low cost, thus allowing to use more probes per mRNA resulting in a substantial increase in detection efficiency. smiFISH is also flexible since differently labelled secondary detector probes can be used with the same primary probes. We demonstrate that this flexibility allows multicolor labelling without the need to synthesize new probe sets. We further demonstrate that the use of a specific acrydite detector oligonucleotide allows smiFISH to be combined with expansion microscopy, enabling the resolution of transcripts in 3D below the diffraction limit on a standard microscope. Lastly, we provide improved, fully automated software tools from probe-design to quantitative analysis of smFISH images. In short, we provide a complete workflow to obtain automatically counts of individual RNA molecules in single cells.
INTRODUCTION
Transcription is an inherently stochastic process, and this leads to heterogeneity in mRNA production within cell populations and has a number of important consequences for living organisms (1). For many genes, the localization of mRNA within cells is also non-uniform, and this can lead to local protein synthesis, a phenomenon known to be involved in many biological processes (2). Characterizing these temporal and spatial heterogeneities is thus important for our understanding of gene function, and this is made possible by single-cell single-molecule approaches, in particular by single-molecule RNA fluorescence in situ hybridization (smFISH) (3,4). Here, individual mRNA molecules of a given gene are targeted with 10–50 fluorescently labelled probes. These mRNAs are subsequently visualized as bright, diffraction limited spots under a wide-field microscope, and they can be located and counted with dedicated image analysis methods (4,5). This analysis can be performed for individual cells, therefore providing the distribution of mRNA counts and localization across the cell population. The entire smFISH workflow encompasses probe design, the actual wet lab experiment, image acquisition and image analysis with cell segmentation and mRNA detection. While some of these steps are well established, we identified two bottlenecks, which we clear in this study.
The first bottleneck is the cost of smFISH, which mostly comes from the necessity to use a large number of fluorescent oligonucleotide probes (3,4). Labelling of the smFISH probes can be achieved during probe synthesis, or post-synthesis if a primary amine is incorporated in the oligonucleotides (6). In both cases, modified oligonucleotides are required, hence the high cost, which increases with the number of oligonucleotides used. It is important to note that this issue is not trivial and that it has direct consequences on signal quality. smFISH experiments always suffer from background due to non-specific binding of stray probes. This can yield false-positive and false-negative detections because the signal stemming from a true mRNA would not be always bright enough to be separated from this background signal. Minimizing these artefacts is usually achieved by using a larger number of probes, because this increases the signal of true mRNAs without much affect on the background signals. A higher number of probes thus results in higher signal-to-noise ratio and to a better separation of true positives from true negatives. The possibility to use more oligonucleotides probes can thus directly affect signal quality. Recently, alternative smFISH approaches have been developed that use unlabelled primary probes, which are detected by fluorescently labelled secondary probes (7,8). Although promising, these techniques involve sophisticated protocols using either branched DNAs, which leads to poor nuclear RNA detection, or complex oligonucleotide synthesis schemes tailored for high-content screening (7,8). We developed here an approach that also uses unlabelled primary probes, but in a simple design—termed single molecule inexpensive FISH (smiFISH)—that is well suited for standard smFISH experiments. Because of the low cost of the unlabelled primary probes, more probes per gene can be used, thus resulting in a substantial increase in signal quality.
The second bottleneck lies in the analysis of smFISH images. To obtain meaningful statistics on mRNA counts or localization in individual cells, hundreds to thousands of cells have to be included in the analysis. Such an analysis ideally uses fully automated RNA detection (5,9), but also requires an accurate segmentation of cells. Segmentation is routinely performed on 2D maximum intensity projections, even if images are acquired in 3D. We found that such projections can lead to blurry cell boundaries, reducing the segmentation quality and result in the loss of thin cellular extensions such as pseudopods. Here, we present a new focus-based projection approach that allows a better determination of cell boundaries. This approach can use the non-specific smFISH signal and can be combined with traditional 2D segmentation software (10,11) based on mathematical morphology and traditional filtering and thresholding techniques. We integrated this approach in the existing Matlab toolbox to analyse smFISH data (FISH-quant; (5)). FISH-quant can be fully controlled via graphical user interfaces and is hence easily accessible for non-specialist.
In summary, we present a complete and validated workflow for single molecule FISH. This workflow comes with several key advantages: (i) a reduced probe cost allowing the generation of more efficient probe sets, (ii) an optimized approach to determine good hybridizing sequences, (iii) a simple experimental protocol – including probe synthesis (Supplementary Protocol), (iv) a flexible probe design, allowing multi-colour smiFISH or super-resolution imaging with expansion microscopy (12), (v) software tools going from probe design to image analysis.
MATERIALS AND METHODS
Cells
Monoparental mouse embryonic stem cells (mES) lines were previously derived (13). mES cells were cultured on gelatin-coated dishes in feeder- and serum-free conditions in ESgro-complete-plus medium. A HeLa cell line with a stably integrated HIV reporter with MS2 repeats, and constitutive expression of MS2-GFP and the transactivator Tat was used (provided by Stéphane Emiliani, Institut Cochin, Paris). HeLa cells were grown in DMEM medium supplemented with 10% FCS. For neuronal cells preparation, hippocampi from E17-18 OF1 mice were dissected out in 1X phosphate buffered saline (PBS) supplemented with 3% glucose (PBS-Glu). Cells were enzymatically dissociated in PBS-Glu supplemented with 0.025% Trypsin during 5–10 min at 37°C and then physically dissociated using glass fire polished Pasteur pipette. After centrifugation (5 min at 200 x g), cells were resuspended in plating medium (Neurobasal medium supplemented with B27, Glutamax and 2% serum) and seeded at a density of 75 000 cells per well (P12). At DIV 4, 2/3 of the plating medium were removed and replaced by 2/3 maintain medium (Neurobasal medium supplemented with B27 and AraC 5 μM).
Probes
smiFISH primary probes and FLAPs (secondary probes, either fluorescent or conjugated to digoxigenin) were produced and purchased from Integrated DNA Technologies (IDT), with the following production details. The primary probes are produced using high-throughput oligonucleotides synthesis in 96-well plates. To make use of low-scale synthesis (25 nmol), the total length of primary probes (transcript-binding + FLAP-binding) should not exceed 60 nucleotides for cheaper synthesis. At this scale, oligonucleotides synthesis is possible at the price of ∼0.1 Euros per base at the time of the writing of the paper (∼150 Euros for 24 primary probes). The secondary probes are conjugated to two Cy3, Cy5 or digoxygenin moieties through 5′ and 3′ amino modifications. smFISH probes were synthesized by J.M. Escudier (SPCMIB, Toulouse, France) and labelled with Cy3 mono-reactive dye pack (GE Healthcare).
All primary probes sequences are available online at https://bitbucket.org/muellerflorian/fish_quant in the Oligostan folder. FLAP sequences are listed in Supplementary Note 1.
smiFISH and image analysis
smiFISH was performed according to the Supplementary Protocol. Three-dimensional image stacks were captured on a wide-field microscope (Zeiss Axioimager Z1/Apotome) equipped with a 100 × 1.4 NA objective and a CCD camera (Axiocam MRm 4) and controlled with Metamorph (Molecular Devices). For MS2 experiments, spots in the GFP and Cy3 channels were detected in 3D stacks with Imaris (Bitplane) and categorized as being labelled either with MS2-GFP, smiFISH probes or both. Because the MS2-GFP protein is nuclear, DAPI staining was used to define nuclear regions and restrain mRNA counts to nuclear spots only.
Expansion microscopy and image analysis
To perform expansion microscopy, we used secondary probes (FLAP-Y) from IDT with a 5′-acrydite modification and a 3′-Atto565 label. Primary probes (CRM1 or GAPDH), were pre-hybridized with secondary probes as described in the Supplementary Protocol. smFISH experiments were performed with the Stellaris RNA FISH buffers (Biosearch Technologies) according to the provided protocol (https://www.biosearchtech.com/support/resources/stellaris-protocols), except that the final mounting step with Vectashield was omitted. Expansion was conducted as described (12) (a more detailed version is available at http://expansionmicroscopy.org/), with some modifications as explained next. Cells were grown on 18 mm coverslips to facilitate the smFISH experiments. To cast the gel, coverslips were quickly air-dried after the final washing step of the smFISH protocol. As a mold, individual wells from non-adhesive silicon insulators with an inner diameter of 4.5 mm (Grace bio-labs, Product #664206) were cut, and gently pressed on the coverslip. Gels were poured with 30 μl of the monomer solution with a cross-linker concentration of 0.2%. After 1 h, coverslips were transferred to Nunc 2-well LabTek chambers (Thermo Scientific). Proteinase K treatment was performed for 4 h at 37°C and expansion was performed as described (12). Expanded samples were embedded in 2% low melting agarose, to avoid drift during acquisition.
Three-dimensional images were acquired on a Nikon Ti Eclipse, with a LED light-source (Lumencor Spectra X light engine), a 60 × 1.4 NA objective and an Orca flash 4.0 LT sCMOS camera. Before expansion 41 z-slices with a spacing of 300 nm were acquired, after expansion 40 slices with a spacing of 600 nm. DAPI images were acquired by excitation with 390 nm at 4% for 100 ms, smFISH images at 560 nm at 40% and 500 ms.
Nuclear area was measured in 2D maximum intensity projections of DAPI images with CellProfiler (11) after automated segmentation. mRNA detection was performed with FISH-quant (5) with local-maximum detection after Laplacian of Gaussian filtering. Signal-to-noise ratio (SNR) was calculated for individual cells as the ratio of the mean amplitude of the fitted 3D Gaussian to the standard deviation of the background in a region without cells. In Supplementary Note 3, we provide carefully validated guidelines to select the best mRNA detection method in FISH-quant.
Specimen free gels were cast in 6 mm silicon tubes with varying concentrations of the cross-linker. Gels were expanded as described above for cells, but without the digestion. Diameter of expanded gels were measured, and ratio to unexpanded gels reported as expansion factor.
Code availability
The source code for Oligostan and FISH-quant is available together with test data and detailed tutorials at https://bitbucket.org/muellerflorian/fish_quant
RESULTS
smiFISH – principle and validation
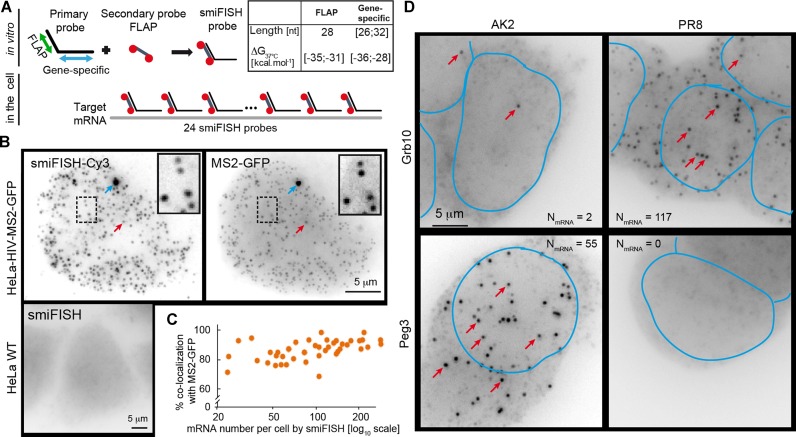
smiFISH combines two types of probes: (i) 12 to 48 unlabelled primary probes containing both a shared sequence (FLAP) and a gene-specific targeting sequence; (ii) a secondary probe labelled with two fluorophores, which is pre-hybridized in vitro to the primary probes via the FLAP sequence (Figure 1A, Supplementary Note 1). This approach is cost effective because unlabelled primary probes are cheap, while the secondary FLAP probe is used for all primary probes and can thus be synthesized at large scale, thereby greatly reducing its cost.
Figure 1.
mRNA detection using smiFISH. (A) Principle of smiFISH. 24 primary probes are pre-hybridized in vitro with the secondary probe via the FLAP sequence. Resulting duplexes are subsequently hybridized in cells. Length (nt: nucleotides) and ΔG37°C are indicated. Red circles: Cy3 moieties. (B and C) Dual-colour labelling of HIV transcripts with smiFISH-Cy3 and MS2-GFP in HeLa-HIV-MS2-GFP cells and parental HeLa cells (negative control). (B) Red arrows indicate examples of individual mRNA molecules. Blue arrows indicate active transcription site. (C) Percentage of smiFISH spots that co-localized with a MS2 spot. Each dot corresponds to one cell, plotted as a function of the number of smiFISH spots per cell (N = 50 cells). (D) Androgenetic (AK2) and parthenogenetic (PR8) mouse embryonic stem cells mES cells were hybridized with smiFISH probes targeting either Grb10 or Peg3. Red arrows indicate examples of individual mRNA molecules. Number of detected mRNAs are reported for each image. Nuclei manually drawn from DAPI images (not shown) are outlined in blue.
We first tested smiFISH with a reporter gene allowing co-localization analysis in different colours, such that the specificity and efficiency of smiFISH could be investigated (Figure 1B, C and Supplementary Methods; (14)). This reporter gene was derived from HIV-1 and was stably integrated in HeLa cells. It contains MS2 stem-loops in an intron that allows to detect the RNA using GFP (15). The MS2 stem-loops are specifically recognized by the coat protein of the bacteriophage MS2 (MCP), and by expressing MCP fused to GFP, the mRNA can be visualized though the MCP–GFP fusion. Images show numerous dimmer nucleoplasmic spots corresponding to single molecules of the tagged pre-mRNA and frequently one brighter nuclear spot corresponding to the transcription site of the reporter (14). We then designed a set of 20 primary smiFISH probes against this reporter and counted 20 to over 200 pre-mRNA per cell (N = 50 cells), while no mRNA was detected in a control cell line lacking the MS2 reporter (Figure 1B and C). These experiments also showed that the smiFISH protocol left most of the GFP signal intact, most likely because mild hybridization and washing conditions were used (37°C; 15% formamide and 1xSSC; see also Supplementary Figure S1). The comparison of the Cy3 and GFP signals yielded 80% of co-localization, independently of the expression level of the reporter RNA (Figure 1B and C). A co-localization percentage of 80% is in line with previous studies for dual-colour labelling of other genes in various organisms (5,16,17), thus indicating specific and sensitive single mRNA detection obtained with smiFISH.
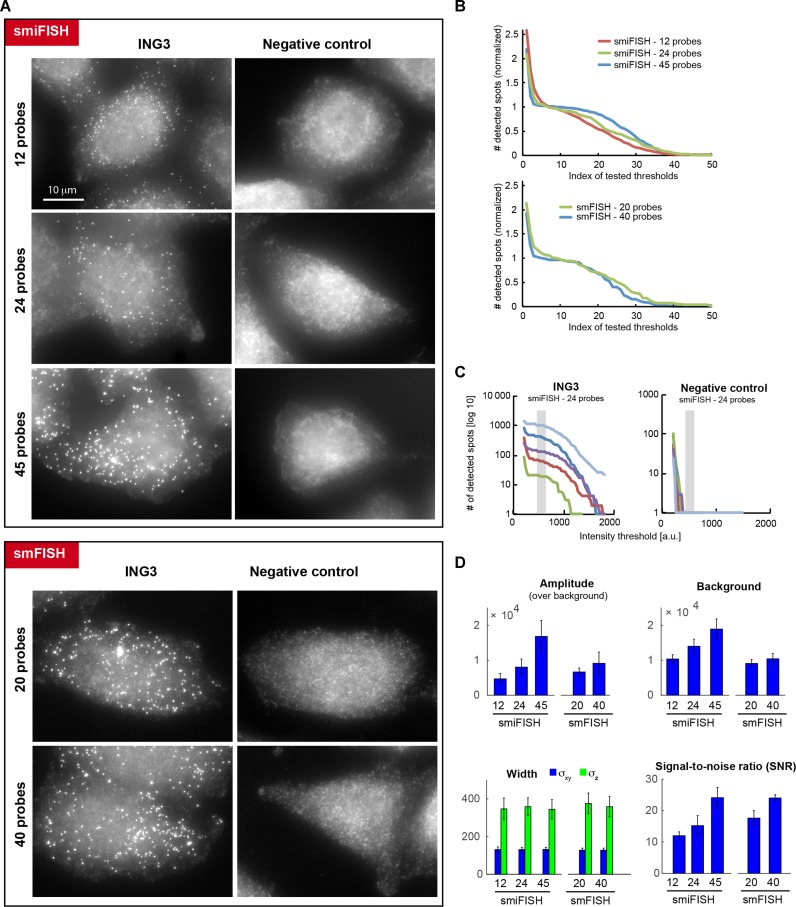
We then compared smiFISH to regular smFISH and evaluated the impact of using different numbers of probes. For these experiments, we used a cell line expressing a GFP-tagged version of the ING3 protein, which was expressed from a bacterial artificial chromosome (BAC) carrying the entire ING3 gene and stably integrated into the genome of a HeLa cell line (Hela-ING3-GFP cells; (18)). We designed a pool of 40 fluorescent smFISH oligonucleotide probes that hybridized against the GFP tag (carrying up to 4 fluorophores each; (3)), as well as a pool of 45 smiFISH probes (carrying 2 fluorophores each; Figure 2). We then performed in situ hybridization on HeLa-ING3-GFP cells and on cells lacking the ING3 BAC as control. We analysed the impact of changing the number of used probes on signal quality (12, 24, 45 for smiFISH; 20, 40 for smFISH). Bright cytoplasmic spots corresponding to single mRNAs were detected in HeLa-ING3-GFP cells and were absent from control cells (Figure 2A). We first counted the number of detected spots as a function of a threshold with increasing intensity (Figure 2B). The number of detected spots decreased with increasing threshold intensities, but a plateau appeared in the middle of the curve when using 24 or 45 smiFISH probes (or 20 to 40 smFISH probes). This plateau separates spots of low and high intensities. Control cell line lacked the spots of high intensities (Figure 2C), indicating the low and high intensity spots corresponded to false positive (background) and true positive (RNA molecules) spots, respectively. As reported before (4,5), this plateau corresponds hence to a range of intensity values that yield an optimal detection. Smaller intensity values lead to an over-detection, larger intensity values to an under-detection. Thus, we concluded that the use of 24 to 45 smiFISH probes allowed a proper separation between true positive and false positive detections, and that a higher number of probes yielded a better separation between them. Next, we fit each detected RNA spot with a 3D Gaussian function (5). We found that spot intensities (Amplitude of the Gaussian) increased with the number of probes (Figure 2C). Finally, we calculated the signal-to-noise ratio (SNR) as the ratio of amplitude and standard deviation of the background, and found that the SNR also increased with probe number, and that smiFISH and smFISH yielded similar SNRs when a similar number of probes were used (Figure 2D). Altogether, these data indicated that smiFISH performed similarly to smFISH, and that using more probes yielded a better separation between the signal of true RNA molecules from false detections. However, more probes also mean higher cost, and 24 probes provide a good compromise between cost and signal quality since it readily allowed to separate true RNA signals from background (Figure 2C).
Figure 2.
Comparison of smiFISH and standard smFISH against a GFP-tagged version of ING3. Negative control is a cell line not expressing the tagged ING3. (A) Representative images for smFISH and smiFISH performed with varying numbers of probes. Images were rescaled such that cellular background is comparable. The granular background in the negative control likely stems from non-specifically bound stray probes. Same scale bar for all images. (B) Number of (normalized) detected spots shown as a function of different (normalized) intensity thresholds. The number of detected spots were renormalized such that the actually detected number is 1, in order to compare cells with different numbers of detected mRNAs. The tested intensity thresholds are not listed with their intensity values, but with an increasing index, in order to compare images with different intensity values. (C) Number of detected spots as a function of different tested intensity thresholds for smiFISH with 24 probes. Panel on the left shows results for 5 cells with different expression levels for ING3, panel on the right for 5 cells of the negative control. Gray vertical bar indicates manually determined plateau for mRNA detection. Note that in the negative control no spots are detected with this threshold. (D) Estimated parameters after fitting detected spots with a 3D Gaussian function (mean ±/- standard deviation). For each experiment, 5 cells with a total of 500± spots were considered. Increasing the probe number leads to brighter amplitude but also more background. Estimated width of the Gaussian stays, however, unchanged. Signal-to-noise ratio (SNR) relates the intensity of the mRNA molecules to fluctuations in background intensity. SNR was calculated as μ/σ, where μ is the average estimated amplitude after fitting individual mRNA molecules with a Gaussian function with FISH-quant (5), σ is the standard deviation of the background intensity (measured in parts of the cells with no mRNA molecules). Reported values are the mean ±/- standard deviation for the same cells used in (C). Using increasing probe numbers leads to a higher SNR for smiFISH and smFISH.
Next, we optimized the method to determine the best hybridization sequences. Traditionally, sequences are chosen such that they have a similar melting temperature (Tm). However, hybridization and washing are performed at 37°C, far lower than the typical Tm. The standard free energy of binding (ΔG) varies in a sequence-dependent manner with temperature (19), and oligonucleotides with different sequences may thus have an identical ΔG at the Tm, but different ΔG at 37°C. We thus developed a script—Oligostan—to identify hybridization sequences with a common ΔG37°C, and thus with an identical target affinity at this temperature (Supplementary Note 2). To further improve probe design, we also incorporated empirical criteria for optimal determination of hybridization sequences, which were determined previously in large-scale hybridization experiments (20). We tested Oligostan on three genes with different expression levels: GAPDH, CTNNA1 and CTNNB1. In each case, we obtained images with clearly identifiable mRNA molecules (Supplementary Figure S2). A direct comparison of 14 probe sets (24 oligonucleotides each), designed with Oligostan or using only the Tm further showed that Oligostan indeed yielded frequently higher SNR and thus better better mRNA detection (Supplementary Table S1).
We then applied smiFISH to detect two endogenous genes—Grb10 and Peg3—in mES. These genes are subject to genomic imprinting (21), an epigenetic phenomenon in which parentally inherited DNA methylation marks mediate parent-of-origin specific monoallelic expression. The mouse gene Grb10 (Growth factor receptor-bound protein 10) is mostly expressed from the maternal allele (22,23), whereas Peg3 (Paternally expressed gene 3) is mainly expressed from the paternal allele (24,25). As expected, we observed many mRNA molecules for Grb10 in parthenogenetic (PR8) cells and for Peg3 in androgenetic (AK2) cells (Figure 1D). In contrast, we detected only few molecules for each gene in its respective imprinted cell line (Figure 1D, Supplementary Figure S3A), consistent with the very low expression detected by RNA-Seq (Supplementary Figure S3B), and in-line with the regulation of these genes by genomic imprinting. Finally, we used smiFISH to successfully detect β-actin mRNAs in neurons. The results indicated that smiFISH properly detects mRNA while preserving cell's morphology, even long and fragile cellular extensions (Supplementary Figure S4).
Taken together, these experiments underline the sensitivity and specificity of the smiFISH approach in different cell lines, as well as its ability to preserve cell's morphology and other fluorescent labels such as GFP.
Multi-colour smiFISH
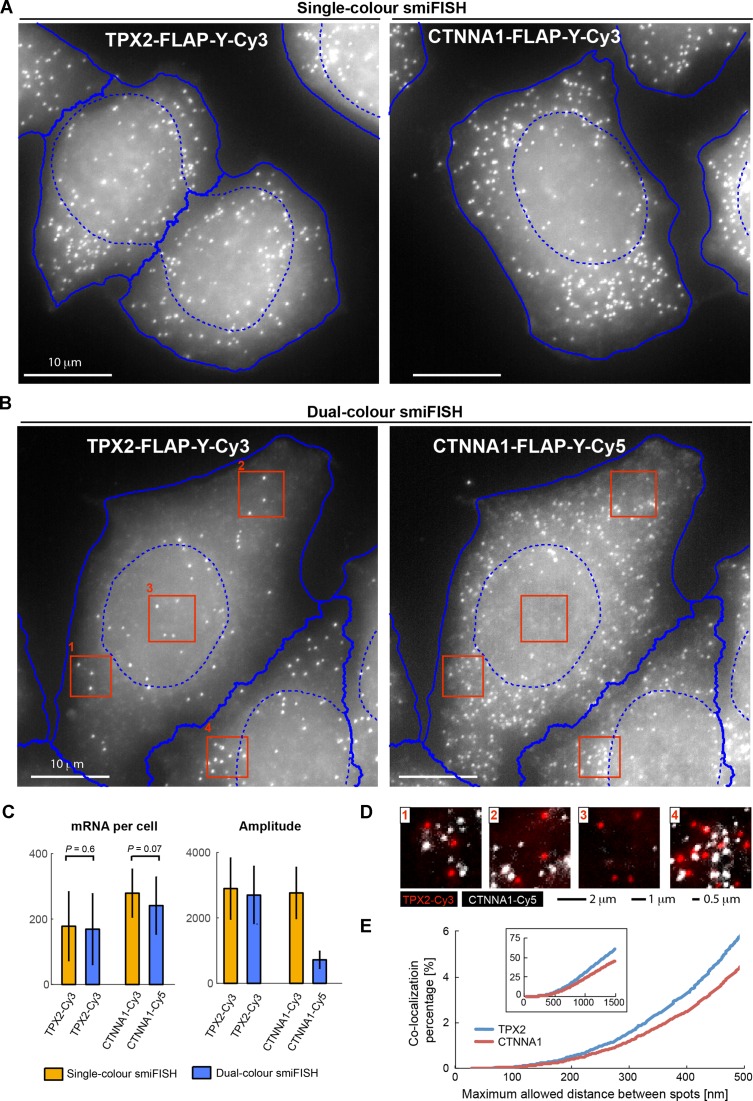
We then adapted smiFISH to the simultaneous detection of two different mRNAs, by labelling the primary probes for the different mRNAs in spectrally distinct colours. Importantly, this does not require having different FLAP sequences on the primary probes. In the smiFISH protocol, primary probes are pre-hybridized to the secondary probe before being used in cells, and each primary probe set can thus be labelled in a separate reaction with the desired fluorescent secondary probe (Figure 1A). We found that this allows the simultaneous use of different primary probe sets carrying the same FLAP sequence. We illustrate this with single- and dual-colour smiFISH against TPX2 and CTTNA1 (Figure 3A and B). The estimated mRNA number in single-colour and dual-colour experiments were similar (Figure 3C), and a co-localization analysis revealed no co-localization between the different channels, ruling out cross-hybridization (Figure 3D and E). The pre-hybridization step of the smiFISH protocol therefore avoids synthesizing primary probes with different FLAP sequences, since different fluorophores can be combined with existing libraries of primary probes.
Figure 3.
Detection of CTNNA1 and TPX2 mRNA by two-colour smiFISH in HeLa cells. Single-colour smiFISH performed with Cy3 against either gene. Dual-colour smiFISH performed with Cy3 against TPX2, and Cy5 against CTNNA1. Both probe-sets contain FLAP-Y, and were prehybridized separately with the secondary probes before the actual experiment. (A and B) Maximum intensity projections of representative smiFISH images for (A) single-colour and (B) dual-colour experiments. For automated segmentation, nuclei were marked with DAPI, and cells with HCS CellMaskTM Green (Molecular Probes). Segmentation was performed on focus-projected images (see last section of results) with CellCognition (10): nuclei shown with dashed blue lines, cells with solid blue lines. (C) Left: estimated mRNA number per cell. Reported P-value from Wilcoxon rank sum test for equal medians (Matlab function ranksum). Right: amplitude of 3D Gaussian fit to each detected mRNA molecule. TPX2 mRNA detection was similar in single-colour and dual-colour experiments with respect to estimated mRNA counts and mRNA spot intensity. Labelling of CTNNA1 mRNA with Cy5 leads to substantially dimmer spots, which makes mRNA detection more challenging and leads to a lower P-value for the comparison of mRNA counts. Number of cells per condition: NCTTNA1 = 143, NTPX2 = 115; NCTTNA1,TPX2 = 150. (D) Dual-colour smiFISH (TPX2 in red, and CTNNA1 in white). Images are zoom-ins indicated by red rectangles in (B). For better visualization, images were background corrected with the Subtract Background function in Fiji (Rolling ball radius of 50 pixels). (E) Co-localization analysis for dual-colour smiFISH experiment in (C). mRNAs were detected in 3D images with FISH-quant (5) and co-localization determined with the Matlab function munkres using the Hungarian Algorithm for linear assignment problems, which is available on Matlab File Exchange. Plots show the co-localization percentage for TPX2 (blue, 27 178 mRNAs) and CTNNA1 (red, 35 623 mRNAs) as a function of the maximum allowed distance between spots to be still considered to be co-localized. For distances smaller than 500 nm, co-localization is smaller than 5%. Larger allowed distances lead to a significant increase of co-localization percentage, since neighbouring spots are erroneously considered to co-localize (especially in denser area such as zoom-in 4).
One particular advantage of smiFISH is its flexibility with respect to the used labels, since only the secondary probes are labelled. This labelling is not limited to fluorophores. In a second dual-colour smFISH experiment, we tagged secondary probes with digoxigenin, which was then detected with fluorescein-labelled anti-digoxigenin antibodies (Supplementary Figure S5). Each antibody was labelled with 3–4 fluoresceins, and hence providing brighter signal than can be obtained by directly coupling secondary probes with a green fluorescent dye.
Super-resolution imaging with smiFISH
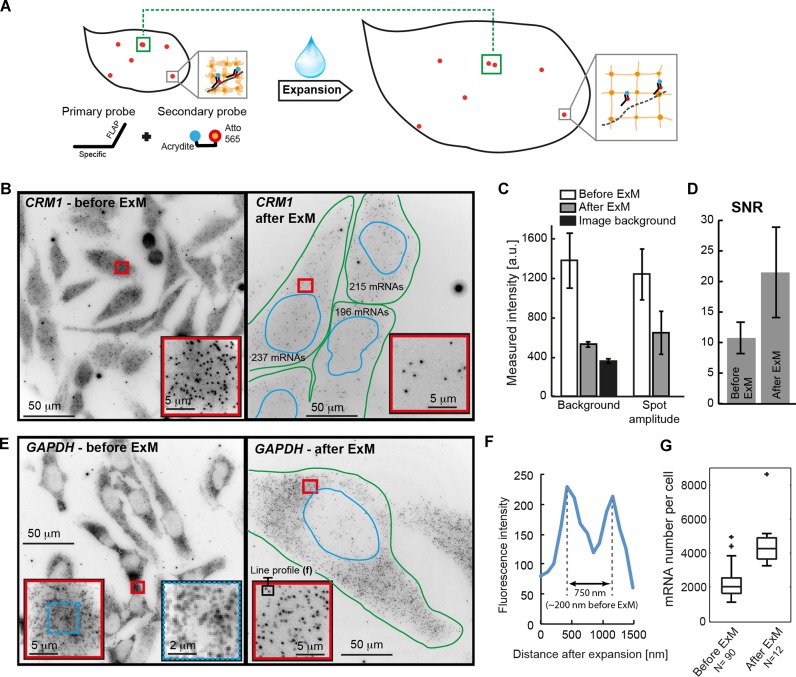
Recently, expansion microscopy (ExM) has been introduced to obtain higher resolution by physically enlarging samples (12). In the original ExM protocol, a protein of interest is targeted with antibodies labelled with DNA probes carrying both a fluorophore and an acrydite modification. The acrydite then integrates into a swellable polymer network, which is established within the cell. After proteolysis and addition of water, the polymer—and thereby the embedded fluorescently labelled probes—expand isotropically. The physically enlarged sample allows features previously located under the diffraction limit to become resolvable through standard microscopy. We reasoned that the flexible design of smiFISH should make it readily amenable for ExM by replacing one of the fluorophores on the secondary probes with an acrydite (Figure 4A). To test if ExM is compatible with smiFISH, we first examined the moderately expressed gene CRM1 using such secondary probes. Before expansion, the typical smFISH signal was visible, with bright mRNA molecules over a non-specific background (Figure 4B, left). We then performed ExM (See Material and Methods). It has been reported that the polymerization leads to photobleaching of certain fluorophores (12), but after expansion, the DAPI signal was still present (Supplementary Figure S6A and B), although at reduced intensity. The increase of the nuclear area (roughly 14-fold, Supplementary Figure S6C) indicates an expansion factor of ∼3.7, in agreement with measurements of expansion in specimen free gels (Supplementary Figure S6D). Remarkably, we could also observe individual mRNA molecules after expansion (Figure 4B and Supplementary Figure S6E). While these spots were ∼2 times dimmer after expansion (in agreement with the previously observed reduction of fluorescence after expansion; (12)), the cellular background was almost completely removed (Figure 4C). The latter effect likely results from dilution of non-specifically bound probes in the expanded volume and a reduction of cellular autofluorescence due to proteolysis. Importantly, this strong background reduction more than compensated for the decreased mRNA signal intensity, resulting in a substantial (∼2-fold) increase in spot SNR after expansion (Figure 4D), improving mRNAs detection and localization precision. Because the expansion increases the available volume for spot detection, we hypothesized that ExM could help for highly expressed genes, where individual transcripts are often too dense to be resolved by conventional microscopy. We therefore applied smiFISH with ExM to GAPDH – a housekeeping gene routinely used as an expression control (Supplementary Figure S2). Before expansion, individual GAPDH molecules were often too close to be readily distinguished (Figure 4E, left). After expansion, individual mRNA could be distinguished also in dense regions (Figure 4E, right and Figure 4F). As a consequence, mRNA counts after expansion were substantially higher (Figure 4G) reflecting the improved ability to spatially resolve transcripts. In summary, combining smiFISH with ExM provides better SNR and allows resolving spatially dense transcripts.
Figure 4.
Expansion microscopy with smiFISH. (A) Principle of ExM with smiFISH. Secondary probes carry an acrydite modification that anchors them into the polymer. After addition of water, the polymer swells and the cell will physically increase in size. Close spots (green rectangle) will be separated more, and can be distinguished under a regular microscope. Grey rectangles show zoom-in to illustrate how the secondary probes get anchored in the polymer network. (B) smiFISH against CRM1 before (left) and after (right) ExM. Numbers in plot after ExM indicate estimated number of mature mRNA molecules. In the image before ExM 186±/-70 mRNAs were detected (Supplementary Figure S8). Images show entire field of view (220 × 220 μm). (C) Bar plots show estimated background and amplitude after fitting detected mRNAs with FISH-quant. N = 5 cells. Black bar shows imaging background without cells. (D) Bar plot shows SNR with standard deviation before and after ExM. (E) As in (B), but for GAPDH. (F) Intensity profile through two mRNA spots after ExM highlighted in inset of (E). Scaled distance obtained by dividing by estimated expansion factor. (G) Estimated mRNA number with FISH-quant before and after ExM with identical detection settings. Boxplot generated with Matlab. Central mark indicates the median, the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers (indicated by crosses).
Automated image analysis for cell segmentation and mRNA detection
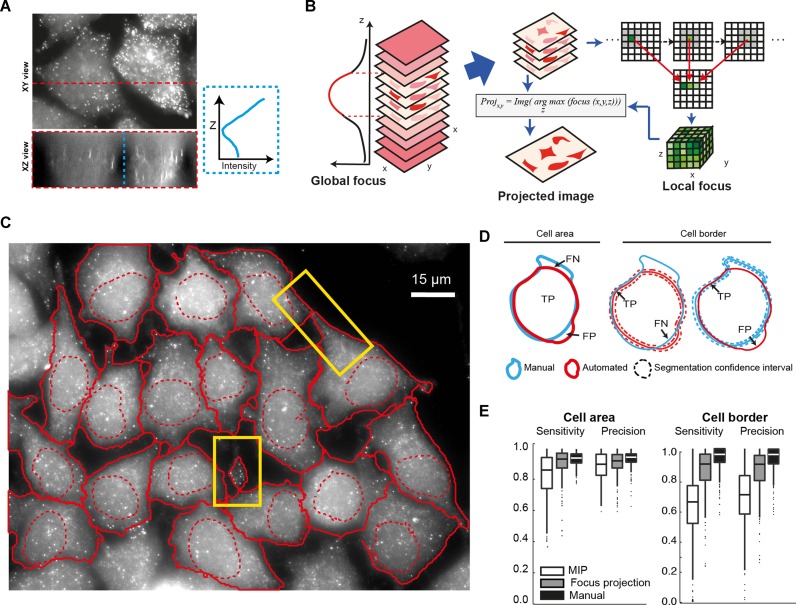
As smFISH experiments become increasingly powerful and used in high-throughput imaging efforts, the need grows for fully automated quantification of mRNA in single cells. Therefore, in addition to identification and counting of single mRNA molecules (5), accurate segmentation of cells is required (10,11). Segmentation can be performed with dedicated cell markers, but in the following we use the non-specific smFISH background signal to determine cell boundaries. While there are imaging conditions (confocal microscopy, use of an appropriate membrane marker), which allow in principle 3D segmentation of cells, this is rarely compatible with large scale screening approaches. As it is common in this field, we therefore performed segmentation on 2D intensity projections. However, we found that the projection used can have a strong impact on the segmentation quality. smFISH images—as other wide-field images—typically have large contributions of out-of-focus signal (Figure 5A). As a consequence, performing a standard maximum intensity projection along z often yields a good signal for bright cellular structures, but also results in blurred cellular boundaries, making cell segmentation less reliable with the frequent loss of thin cell extensions (Figure 5C and Supplementary Note 3). We therefore developed a new projection method that uses global and local focus measurements to circumvent this problem (Figure 5B). In short, we first used a global focus measurement to automatically remove out-of-focus slices. Second, for the remaining slices, we calculated a local focus measurement for each lateral position (x,y). The z-projection is then performed for each (x,y) position using voxels with the highest local focus values, rather than highest intensity value. These pixels are then included in an averaging operation that yields the final 2D image. This 2D image has sharper boundaries than a standard maximum-intensity projection (Figure 5C and Supplementary Note 3). In order to test if our projection approach improves cell segmentation, we used a fairly traditional workflow for cell segmentation based on prefiltering, global thresholding and watershed transformation, and we integrated it to the open-source software CellCognition (10). By comparison with a manually defined ground truth, we found that focus-based projections significantly improved segmentation quality relative to standard maximum-intensity projections (Figure 5D and E). Importantly, this allows to capture more complex cellular outlines as for instance small protrusions (Figure 5C; Supplementary Note 3). Note that this projection method is also successful for different organisms such as yeast and ES cells, and yields substantially sharper projections even when dedicated cell markers are used (Supplementary Note 3). We integrated this projection approach in our software FISH-quant (5), which allows the quantification of mature and nascent mRNA molecules, to provide an integrated image analysis package for smFISH.
Figure 5.
Improved image segmentation with focus projection. (A) Images show maximum intensity projections of smFISH against KIF5B in HeLa cells along the indicated axis. Plot on the right shows averaged pixel intensity along the dashed blue line. (B) Schematic of focus based projection. Out-of-focus slices are removed based on a global focus calculation. Local focus measurements are done on remaining images and a projection is performed by choosing for each XY position the pixel-intensity in Z with the maximum focus value. Focus is calculated with HELM operator (27), which computes the intensity ratio between pixels and their neighbourhood. (C) 2D cell segmentation after focus-projection (see also Supplementary Note 3). Eighteen cells were segmented in this image, please note that cells whose nucleus touches the image border were automatically excluded. Yellow rectangle indicate protrusions that were only properly segmented after focus-projection but not maximum intensity projection (See Supplementary Note 3, Figure 5). (D and E) Evaluation of segmentation quality by comparing automatic segmentation to manually segmented ground truth for cell area and cell border. Used abbreviations: TP = true positives, FP = false positives, FN = false negatives, FP = false positives. Sensitivity = TP/(TP±FN), or the proportion of ground truth area (or border) that is correctly segmented. Precision = TP/(TP±FP), or the proportion of the automatically segmented area (or border) belonging to the ground truth. A confidence zone of 36 pixels width is used when comparing cell borders.
DISCUSSION
We present smiFISH – an easy-to-use, cost-effective and reliable smFISH workflow going from probe design to image analysis. A key advantage of smiFISH is an optimized probe design that combines unlabelled specific primary probes with labelled secondary probes. This design has several advantages. First, probes are substantially cheaper, and this allows to use more probes per mRNA and thus to obtain a higher SNR and a better efficiency of RNA detection. Second, it provides large flexibility in labelling primary probes, to perform for instance dye-swap, multi-colour experiments and ExM without the need of generating new probes sets. We further provide fully automated software solution from probe design to image analysis. In particular, we implemented a focus-based projection method providing sharper 2D projections from 3D stacks. This tool yields reliable cellular segmentation and is widely applicable. Together with the automated mRNA detection in FISH-quant, large data sets can now be analysed with minimal user intervention.
Importantly, we demonstrate that smiFISH can be used in expansion microscopy to obtain super-resolved images on a standard microscope. Since these images are acquired in full 3D, this capability increases the potential of smFISH for analysing the spatial dimension of the transcriptome (26), for instance in multiplex smFISH (8) where many different mRNA species are imaged simultaneously. By combining this approach with protein labelling, it will be possible to study protein–mRNA complexes at high resolution in situ, and possibly down to the single molecule level.
Taken together, smiFISH and FISH-quant provide a complete, validated and flexible workflow for single molecule FISH from initial probe-design to final image analysis. We therefore believe that smiFISH has the potential to become an important routine technique to study mRNA biology at the single cell level.
Supplementary Material
Acknowledgments
The authors thank Benjamin King, Robert Feil, David Llères, Michael Girardot and Thierry Forné (IGMM, Montpellier, France) for helpful discussions, Stéphane Emiliani (Institut Cochin, Paris, France) for the HeLa-HIV-MS2-GFP cell line, Anne Debant and Susanne Schmidt (CRBM, Montpellier, France) for neuron samples.
Footnotes
Present address: Nikolay Tsanov, Genome Stability Lab, National University of Ireland, Galway, Ireland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Scholarship from Lebanese National Council for Scientific Research (LNCSR) [to R.C.]; Agence Nationale de la Recherche [ANR-11-BSV8-018-02, ANR-14-CE10-0018-01]; Institut Pasteur, Fondation pour la Recherche Médicale (FRM); Agence Nationale de Recherche sur le Sida (ANRS). Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
REFERENCES
- 1.Li G.-W., Xie X.S. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buxbaum A.R., Haimovich G., Singer R.H. In the right place at the right time: Visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Femino A.M., Fay F.S., Fogarty K., Singer R.H. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 4.Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller F., Senecal A., Tantale K., Marie-Nelly H., Ly N., Collin O., Basyuk E., Bertrand E., Darzacq X., Zimmer C. FISH-quant: Automatic counting of transcripts in 3D FISH images. Nat. Methods. 2013;10:277–278. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
- 6.Raj A., Tyagi S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 2010;472:365–386. doi: 10.1016/S0076-6879(10)72004-8. [DOI] [PubMed] [Google Scholar]
- 7.Battich N., Stoeger T., Pelkmans L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods. 2013;10:1127–1133. doi: 10.1038/nmeth.2657. [DOI] [PubMed] [Google Scholar]
- 8.Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coassin S.R., Orjalo A.V., Semaan S.J., Johansson H.E. Simultaneous detection of nuclear and cytoplasmic RNA variants utilizing Stellaris® RNA fluorescence in situ hybridization in adherent cells. Methods Mol. Biol. Clifton NJ. 2014;1211:189–199. doi: 10.1007/978-1-4939-1459-3_15. [DOI] [PubMed] [Google Scholar]
- 10.Held M., Schmitz M.H.A., Fischer B., Walter T., Neumann B., Olma M.H., Peter M., Ellenberg J., Gerlich D.W. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods. 2010;7:747–754. doi: 10.1038/nmeth.1486. [DOI] [PubMed] [Google Scholar]
- 11.Kamentsky L., Jones T.R., Fraser A., Bray M.-A., Logan D.J., Madden K.L., Ljosa V., Rueden C., Eliceiri K.W., Carpenter A.E. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen F., Tillberg P.W., Boyden E.S. Optical imaging. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen N.D., Barton S.C., Hilton K., Norris M.L., Surani M.A. A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development. 1994;120:1473–1482. doi: 10.1242/dev.120.6.1473. [DOI] [PubMed] [Google Scholar]
- 14.Tantale K., Mueller F., Kozulic-Pirher A., Lesne A., Victor J.-M., Robert M.-C., Capozi S., Chouaib R., Bäcker V., Mateos-Langerak J., et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 2016;7:12248. doi: 10.1038/ncomms12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 16.Batish M., van den Bogaard P., Kramer F.R., Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4645–4650. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou Y., Vafabakhsh R., Doğanay S., Gao Q., Ha T., Palese P. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9101–9106. doi: 10.1073/pnas.1206069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poser I., Sarov M., Hutchins J.R.A., Hériché J.-K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A.W., et al. BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat. Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu P., Nakano S., Sugimoto N. Temperature dependence of thermodynamic properties for DNA/DNA and RNA/DNA duplex formation. Eur. J. Biochem. FEBS. 2002;269:2821–2830. doi: 10.1046/j.1432-1033.2002.02970.x. [DOI] [PubMed] [Google Scholar]
- 20.Xu Q., Schlabach M.R., Hannon G.J., Elledge S.J. Design of 240, 000 orthogonal 25mer DNA barcode probes. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2289–2294. doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girardot M., Feil R., Llères D. Epigenetic deregulation of genomic imprinting in humans: Causal mechanisms and clinical implications. Epigenomics. 2013;5:715–728. doi: 10.2217/epi.13.66. [DOI] [PubMed] [Google Scholar]
- 22.Arnaud P., Monk D., Hitchins M., Gordon E., Dean W., Beechey C.V., Peters J., Craigen W., Preece M., Stanier P., et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 2003;12:1005–1019. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
- 23.Hikichi T., Kohda T., Kaneko-Ishino T., Ishino F. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 2003;31:1398–1406. doi: 10.1093/nar/gkg232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroiwa Y., Kaneko-Ishino T., Kagitani F., Kohda T., Li L.L., Tada M., Suzuki R., Yokoyama M., Shiroishi T., Wakana S., et al. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nat. Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- 25.Perera B.P.U., Teruyama R., Kim J. Yy1 gene dosage effect and bi-allelic expression of Peg3. PloS One. 2015;10:e0119493. doi: 10.1371/journal.pone.0119493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crosetto N., Bienko M., van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet. 2015;16:57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 27.Pertuz S., Puig D., Garcia M.A. Analysis of focus measure operators for shape-from-focus. Pattern Recognit. 2013;46:1415–1432. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.