Abstract
Self-renewal and pluripotency are two fundamental characteristics of embryonic stem cells (ESCs) and are controlled by diverse regulatory factors, including pluripotent factors, epigenetic regulators and microRNAs (miRNAs). Although histone methyltransferases are key epigenetic regulators, whether and how a histone methyltransferase forms a network with miRNAs and the core pluripotent factor system to regulate ESC stemness is little known. Here, we show that the protein arginine methyltransferase 7 (PRMT7) is a pluripotent factor essential for the stemness of mouse ESCs. PRMT7 repressed the miR-24-2 gene encoding miR-24-3p and miR-24-2-5p by upregulating the levels of symmetrically dimethylated H4R3. Notably, miR-24-3p targeted the 3′ untranslated regions (UTRs) of the major pluripotent factors Oct4, Nanog, Klf4 and c-Myc, whereas miR-24-2-5p silenced Klf4 and c-Myc expression. miR-24-3p and miR-24-2-5p also targeted the 3′UTR of their repressor gene Prmt7. miR-24-3p and miR-24-2-5p induced mouse ESC differentiation, and their anti-sense inhibitors substantially reversed spontaneous differentiation of PRMT7-depleted mouse ESCs. Oct4, Nanog, Klf4 and c-Myc positively regulated Prmt7 expression. These findings define miR-24-3p and miR-24-2-5p as new anti-pluripotent miRNAs and also reveal a novel epigenetic stemness-regulatory mechanism in which a double-negative feedback loop consisting of PRMT7 and miR-24-3p/miR24-2-5p interplays with Oct4, Nanog, Klf4 and c-Myc to control ESC stemness.
INTRODUCTION
Embryonic stem cells (ESCs) are characterized by their ability to self-renew and differentiate into many different cell types. These stemness characteristics are in principle subject to epigenetic regulation, which is governed largely by both chromatin modifications and molecular interaction of chromatin with chromatin-bound proteins and non-coding RNAs (1–4). Of chromatin modifications, covalent histone modifications, including histone methylation, acetylation, phosphorylation and ubiquitination, play a central role in dynamically regulating gene expression by directly or indirectly affecting chromatin structures (5–8).
Histone methylation is a critical player for epigenetic and transcriptional regulation of gene expression and commonly occurs at the positively charged lysine (K) and arginine (R) residues—two unusually abundant amino acids within histones. Histone lysine methylation is catalyzed at the epsilon nitrogen atom in the side chain of lysine residues by specific methyltransferases. This modification is coupled with activation or silencing of gene expression, depending on sites of methylation. Arginine methylation occurs at a unique guanidino nitrogen in the side chain of arginine residues and can be classified into three types (i.e. monomethylation, symmetric dimethylation and asymmetric dimethylation) (9). Arginine methylation is mediated by protein arginine methyltransferases (PRMTs). Type I PRMTs (e.g. PRMT1, 2, 3 and 6 and CARM1/PRMT4) catalyze asymmetric dimethylation whereas type II PRMTs (e.g. PRMT5 and 9) generate symmetrically dimethylated arginine (9–11). Type I and II PRMTs can also generate monomethylated arginine, but the type III PRMT (i.e. PRMT7) is considered to catalyze only monomethylation (9,10).
Certain histone methyltransferases are involved in the maintenance of self-renewal and pluripotency. The H3K9 methyltransferase ESET is required for the maintenance of ESCs (12). PRMT5 has been shown to maintain the self-renewal and pluripotency by repressing differentiation-specific genes via symmetric dimethylation of H2AR3 in ESCs (13). CARM1 contributes to blastomere fate determination process via arginine methylation (14). ESC pluripotency is also regulated by various microRNAs (miRNAs) (15). miRNA-mediated gene targeting represents a post-transcriptional regulation of gene expression, because miRNAs target mRNAs via base-pairing between miRNAs and their cognate mRNA sequences (16). In contrast to advances in the understanding of the individual roles of histone methyltransferases and miRNAs in ESC stemness, little is known about whether and how a histone methyltransferase modulate ESC stemness by networking miRNAs and the core pluripotent factors.
We previously reported that the H3K4 methyltransferase MLL4 is necessary for retinoic acid (RA)-induced neuronal differentiation of the embryonal carcinoma cell line NT2/D1 and that PRMT7, a transcriptional co-repressor, counteracts MLL4–mediated activation of differentiation-specific genes during NT2/D1 cell differentiation by increasing the levels of the repressive mark symmetrically dimethylated H4R3 (H4R3me2s) (17). This prompted us to investigate whether PRMT7 plays a role in maintaining the ESC stemness. In the current study, we identified PRMT7 as a new stemness factor indispensable for maintenance of mouse ESCs (mESCs). We found that PRMT7 epigenetically downregulated the miR-24-2 gene encoding miR-24-3p and miR-24-2-5p to maintain Oct4, Nanog, Klf4 and c-Myc levels in mESCs. Our data revealed miR-24-3p and miR-24-2-5p as new anti-pluripotent miRNAs that collectively silence the major pluripotency genes Oct4 (also known as Pou5f1), Nanog, Klf4 and c-Myc as well as their own repressor gene Prmt7. Prmt7 expression was directly activated by Oct4, Nanog, Klf4 and c-Myc in mESCs. These findings uncover a previously unknown stemness-regulatory mechanism in which a feedback loop between PRMT7 and miR-24-3p/miR24-2-5p is interactive with the major pluripotent factors Oct4, Nanog, Klf4 and c-Myc to maintain mESC stemness.
MATERIALS AND METHODS
Antibodies, plasmids and other reagents
Anti-PRMT7 antibodies were purchased from Epicypher (#13-1009), Santa Cruz Biotechnology (SC9882) and Millipore (#07-639). Anti-Nanog (#61419), anti-Sox2 (#39823) and anti-H4R3me2s (#61187) antibodies were from Active Motif. Anti-Oct4 (#2840), anti-c-Myc (#5605) and anti-Klf4 (#4038) antibodies were from Cell Signaling. Anti-β-actin antibody (A5441) was from Sigma-Aldrich. Anti-H4R3me1 antibody (PA5-27065) was from Thermo Scientific. Anti-H3 antibody (ab1791) was from Abcam. Anti-mouse Argonaute 2 (Ago2) antibody was from Wako Chemicals (292-67301) and Cell Signaling (2897P). The cDNA constructs of human (OpenBiosystem) and mouse PRMT7 (OpenBiosystem) were cloned into pCDH1-EF1-IRES-GFP vector (System Biosciences) using the cloning sites EcoRI and NotI. Human PRMT7's catalytic mutant (m. PRMT7) was generated from pCDH1-EF1-PRMT7-IRES-GFP by mutating several residues (E144A, E153A, E478A and H644A) using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). These residues are likely located in arginine binding pockets according to crystal structures of PRMT1 and Caenorhabditis elegans PRMT7 (18,19). Oligonucleotides used for cloning, ChIP-PCR, reverse-transcription (RT)-PCR, site-directed mutagenesis are listed in Supplementary Table S1.
Mouse ESC culture
mESCs were maintained in complete knockout Dulbecco's modified Eagle's medium (Life Technologies) containing 4.5 mg/ml glucose, 20% ES grade fetal bovine serum (Life Technologies), 2 mM L-glutamine, 50 μg/ml penicillin and 50 μg/ml streptomycin (Life Technologies), 0.1 mM β-mercaptoethanol, 0.1 mM Minimum Essential Medium (MEM) non-essential amino acids and 1000 U/ml leukemia inhibitory factor (LIF) with or without irradiated feeder cell layers (less than passage 5). Glutamine was freshly added when the medium was replaced daily. mESCs were incubated at 37°C in a 5% CO2 and 95% relative humidity chamber.
RA-induced differentiation of mouse ESCs
To generate embryoid body (EB), mESCs were trypsinized, transferred onto 10 cm petri dish and cultured in ESC media without LIF for 5 days. EBs were then transferred onto gelatin-coated tissue culture dishes and cultured in ESC media in the presence of RA (0.5 μM) without LIF for 5 days to induce cellular differentiation.
RNA interference
Mouse shPRMT7s (shPRMT7-4, TRCN0000097474; shPRMT7-5, TRCN0000097475; shPRMT7-6, TRCN0000097476; shPRMT7-7, TRCN0000097477; shPRMT7-8, TRCN0000097478), mouse Oct4 (shOct4, TRCN0000009613), mouse Nanag (shNanog, TRCN0000075335), mouse c-Myc (shc-Myc, TRCN0000234924) and mouse Klf4 (shKlf4, TRCN0000095370) in the puromycin-resistant PLKO.1 vector were purchased from Sigma-Aldrich. mESCs (5 × 105 cells in 0.8 ml) were transfected with 30 μg of plasmids using a Gene Pulser Xcell Electroporation System (500 μF, 250 volts; BioRad) according to the manufacturer's protocol and were plated on a 6 cm dish. The cells were treated with puromycin (1.0 μg/ml) 48 h later, cultured for 14 days and then harvested for further analysis.
Western blot analysis
Whole-cell lysates were prepared using lysis buffer (20 mM Tris–HCl, 137 mM NaCl, 1.5 mM MgCl2, 1 mM ethylenediaminetetraacetic acid, 10% glycerol, 1% Triton X-100, 0.2 mM Phenylmethylsulfonyl fluoride (PMSF), 1 mg/ml aprotinin, 2.5 mg/ml leupeptin and 1 mg/ml pepstatin, pH 8.0), subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with antibodies against PRMT7, Oct4, Nanog, Sox2, c-Myc or Klf4. β-actin was used as an internal control for western blot analysis.
Cell cycle analysis
Cells were washed with phosphate-buffered saline and fixed in cold 75% ethanol for 30 min at 4°C. Then, cells were again washed two times with phosphate-buffered saline and stained in a buffer containing 10 mM TRIS–HCl (pH 7.0), 5 mM MgCl2, 50 μg/ml propidium iodide and 25 μg/ml RNaseA for 30 min at 37°C. Propidium iodide-stained cells were analyzed on the basis of their DNA contents by fluorescence-activated cell sorting.
Rescue experiments by PRMT7 expression
DNA (30 μg) was electroporated into mESCs (5 × 105 cells) using a Gene Pulser Xcell Electroporation System (500 μF, 250 volts; BioRad) according to the manufacturer's protocol. Plasmids encoding shPRMT7s were co-electroporated with expression plasmids (pCDH1-EF1-IRES-GFP) encoding both green fluorescent protein (GFP) and either PRMT7 or its catalytic mutant. Cells were plated on a 6 cm dish and treated with puromycin (1.0 μg/ml) 48 h later. Then, cells were cultured for 14 days and harvested for further analysis.
Quantitative PCR for mRNA and miRNA expression
Total RNAs were isolated using TRIzol reagent (Life Technologies) according to the manufacturer's instructions. For mRNA quantitation, total RNAs were reverse-transcribed with the iScript cDNA Synthesis Kit (BioRad) and the iQ SYBR Green Supermix (BioRad) was used for quantitative polymerase chain reaction (PCR). GAPDH mRNAs or 18s rRNA levels were used as internal normalization controls for mRNAs. miRNA levels were quantified using the qScript miRNA Quantification System (Quanta Biosciences) according to the manufacturer's instructions and normalized to sno47 and sno66. Quantitative PCR was carried out using the CFX384 real-time PCR detection system (BioRad). The relative mRNA or miRNA levels indicate the fold change compared to the control.
Microarray analysis for miRNA expression
miRNA microarray assay was performed in duplicate using about 0.2 μg of total RNA samples by a service provider (LC Sciences). Data were analyzed by subtracting background and then normalizing signals using a locally weighted regression (20). The ratio of detected signals (log2 transformed, balanced) between control and PRMT7-depleted groups was calculated to determine fold changes.
Luciferase assays
The 3′UTRs of mouse Oct4, Nanog, Klf4 and c-Myc genes were cloned into pMIR-REPORT (Ambion) vector containing a firefly luciferase gene using the cloning sites HindIII and SpeI. The potential targeting sites of miR-24-3p in their 3′UTRs were mutated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). The premature miR-24-2 sequence was synthesized and cloned into pMDH1-PGK-GFP vector. HEK293T cells (1 × 105 cells per well in a 12-well plate) were transfected with 200 ng of the pMIR-REPORT (Ambion) vector containing wild-type (WT) or mutant 3′UTRs, 20 ng of the transfection control Renilla luciferase vector (pRLTK, Promega) and 1 μg of the expression vector (pMDH1-PGK-GFP) encoding premature miR-24-2 (or 5 pmol of miRNA mimics) using Lipofectamine 2000 (Life Technologies). For luciferase assay using mESCs, mESCs (5 × 105 cells) were first electro-transfected with 30 μg of the shPRMT7-7 plasmid. Twelve days later, cells (1 × 105 cells per well in a 12-well plate) were re-transfected with 200 ng of the pMIR-REPORT containing WT or mutant 3′UTRs, 20 ng of pRLTK and 5 pmol of LNA-miRNA-24-3p (Exiqon) using Lipofectamine 3000 (Life Technologies). Cells were harvested 2 days after transfection.
A miR-24-2 promoter region (2.0 kb) and its deletion mutants (0.5, 1.0 and 1.6 kb) were cloned into pGL2-Basic (Promega) vector containing a firefly luciferase gene using the cloning sites XhoI and HindIII. mESCs (0.5 × 105 cells per well in a 12-well plate) were transfected with 200 ng of the pGL2-Basic vector containing the miR-24-2 promoter (or its deletion mutants) and 20 ng of pRLTK using Lipofectamine 3000 (Life Technologies). mESCs were harvested 2 days after transfection.
Luciferase activities were measured using Dual-Luciferase Reporter assay system (Promega) according to the manufacturer's instructions. The firefly luciferase activities were normalized to Renilla luciferase activities. All the experiments were performed in triplicate.
Ago2 immunoprecipitation
To examine whether the interaction of Ago2 with Oct4, Nanog, Klf4 and c-Myc mRNAs is increased by miR-24-3p and miR-24-2-5p mimics, mESCs were transfected with miR-24-3p or miR-24-2-5p mimics using Lipofectamine 3000 and harvested 48 h later. To also examine whether the interaction of Ago2 with Oct4, Nanog, Klf4 and c-Myc mRNA is augmented by ‘endogenous’ miR-24-3p and miR-24-2-5-p levels that were upregulated by PRMT7 knockdown, PRMT7-depleted ESCs were harvested 16 days after transfection of mESCs (5 × 105 cells) with 30 μg of the shPRMT7-7 plasmid. Ago2 IP was performed according to Wako Chemicals’ instructions. In brief, cells were lysed and cell extracts were incubated with 50 μl of anti-Ago2 beads (Wako Chemicals) for 2 h. IgG/protein A beads were used as a control. The beads were extensively washed and eluted, and total RNA was extracted using TRIzol reagent.
Transfections of LNA oligonucleotides and miRNA mimics
Mouse ESCs (5 × 105 cells) were transfected with 150 pmole of miR-24-3p mimic (Ambion), miR-24-2-5p mimic (Ambion), LNA-miR-24-3p (Exiqon) or LNA-miR-24-2-5p oligonucleotides (Exiqon) using Gene Pulser Xcell Electroporation Systems (BioRad) according to the manufacturer's instructions and were transferred in 3 ml of media in a 6 cm dish. Cells transfected with miRNA mimics were incubated for 2 or 4 days whereas LNA-miRNA-transfected cells were incubated for 4 days. Cells were then harvested for further analysis.
ChIP assay
ChIP assay was carried out as previously described (17). In brief, DNA was purified from chromatin immunoprecipitates for PRMT7 and the indicated histone marks using the Qiagen PCR purification kit. Then, DNA was amplified by quantitative PCR and normalized to input.
Statistical analysis
Data are presented as the mean ± standard deviation (SD) of at least three independent experiments (except miRNA microarray data). The statistical significance between the two groups was analyzed by a two-tailed unpaired student's t-test using Prism software (GraphPad Software). P (probability) < 0.05 (*), P < 0.01 (**) and P < 0.001(***) indicate statistically significant changes.
RESULTS
PRMT7 is essential for the self-renewal and pluripotency of mouse ESCs
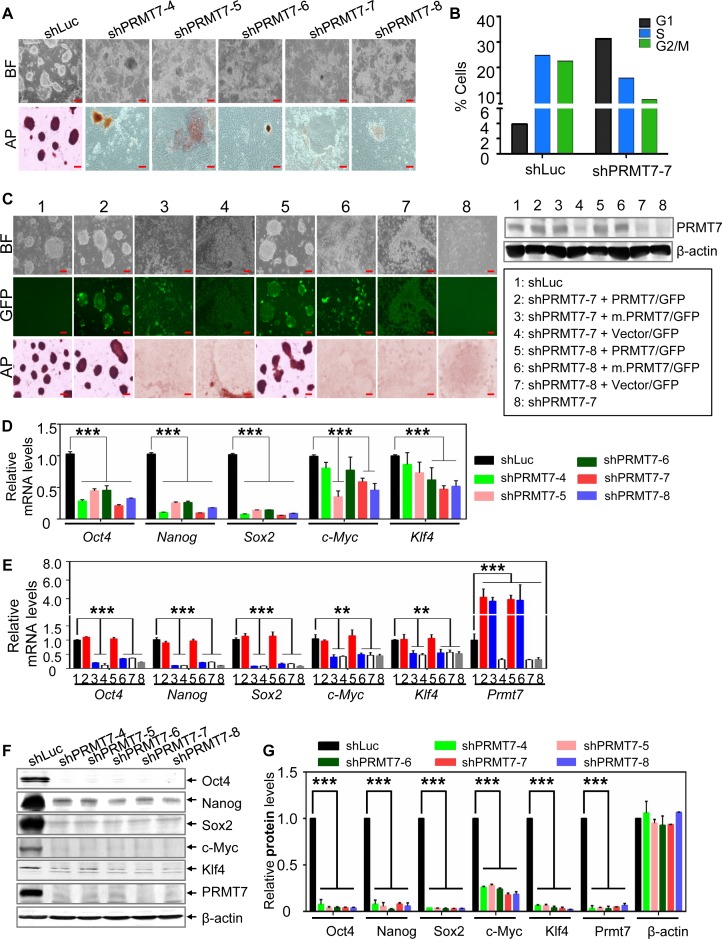
To assess the role of PRMT7 in the self-renewal and pluripotency of ESCs, we depleted PRMT7 in V6.5 mESCs using short hairpin (sh) RNA against PRMT7 (Supplementary Figure S1A and B). PRMT7 depletion resulted in spontaneous differentiation of V6.5 mESCs, as recognized by both morphological changes (i.e. the loss of their ball-like shape) and weak alkaline phosphatase (AP) staining (Figure 1A). Notably, all of five different shPRMT7s induced similar patterns of spontaneous differentiation (Figure 1A). Consistent with spontaneous differentiation of PRMT7-depleted mESCs, cell cycle analysis of these cells revealed an increase in cells in G1 phase and a decrease in cells in S and G2/M phase (Figure 1B and Supplementary Figure S1C). Interestingly, PRMT7 levels were reduced during mESC differentiation that was induced by RA treatment of EBs, suggesting the importance of PRMT7 levels in stemness maintenance (Supplementary Figure S1D). As in V6.5 mESCs, PRMT7 knockdown in another mESC line R1 caused spontaneous differentiation (Supplementary Figure S2A and B).
Figure 1.
PRMT7 and its catalytic activity are indispensable for the stemness of mESCs, and Oct4, Nanog, Klf4 and c-Myc protein levels are more reduced than their mRNA levels in PRMT7-depleted mESCs. (A) Microscopic and AP-staining images of shLuc-treated and PRMT7-depleted V6.5 mESCs. Five shPRMT7s (shPRMT7-4, -5, -6, -7 and -8) were used to deplete PRMT7. BF, bright field; AP, alkaline phosphatase; red scale bar, 100 μm. (B) The percentages of shLuc- or shPRMT7-treated V6.5 mESCs in G1, S and G2/M phase of the cell cycle. shLuc-treated cells were used as a control. (C) Rescue experiments of PRMT7-depleted mESCs with PRMT7 or its catalytic mutant (m.PRMT7). shPRMT7-7 or shPRMT7-8 plasmids were co-electroporated with expression plasmids encoding both GFP and either PRMT7 or its catalytic mutant. The GFP-expressing vector pCDH1-EF1-IRES-GFP (vector/GFP) was used to clone PRMT7 cDNA. Microscopic, green-fluorescent and AP staining images of indicated group of cells are shown (left panel). Western blotting was used to analyze the expression levels of PRMT7 or its mutant in eight different experimental conditions (right panel). (D) Analysis of Oct4, Nanog, Sox2, c-Myc and Klf4 mRNA levels in shLuc-treated and PRMT7-depleted V6.5 mESCs using quantitative RT-PCR. (E) The effect of ectopic expression of PRMT7 or its catalytic mutant (m.PRMT7) on Oct4, Nanog, Sox2, c-Myc, Klf4 and Prmt7 mRNA levels in PRMT7-depleted mESCs. Relative Oct4, Nanog, Sox2, c-Myc, Klf4 and Prmt7 mRNA levels were examined in eight different experimental conditions (see panel C for lane description). Red scale bar, 100 μm. (F and G) Western blot analysis of Oct4, Nanog, Sox2, c-Myc, Klf4 and PRMT7 levels in shLuc-treated and PRMT7-depleted V6.5 mESCs. Oct4, Nanog, Sox2, c-Myc, Klf4 and PRMT7 protein levels in shLuc-treated and PRMT7-depleted V6.5 mESCs were quantified (G). Data are presented as the mean ± SD of three independent experiments. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) indicate statistically significant changes.
To determine the specificity of shPRMT7s and a requirement of PRMT7's enzymatic activity for stem cell maintenance, we carried out a rescue experiment by depleting PRMT7 using shPRMT7-7 or shPRMT7-8 plasmids and simultaneously expressing PRMT7 or its catalytic mutant (m.PRMT7) in mESCs (GFP was also expressed via an internal ribosome entry site). Ectopic expression of PRMT7 but not m.PRMT7 in PRMT7-depleted cells restored ESC morphology (Figure 1C). These results indicate that PRMT7 is essential for the self-renewal of mESCs and that the effect of PRMT7 on ESC stemness depends largely on its enzymatic activity.
ESCs can be induced to differentiate into three germ layers: endoderm, mesoderm and ectoderm. To determine which germ cell layers may be derived from PRMT7-depleted cells, we compared mRNA expression profiles between PRMT7-depleted (shPRMT7-7 and shPRMT7-8) and shLuciferase (control)-treated V6.5 mESCs. Our results showed that most genes regulated by shPRMT7-7 overlapped those modulated by shPRMT7-8 (Supplementary Figure S3A). PRMT7 depletion highly upregulated many endoderm and mesoderm marker genes but had a weak effect on ectoderm marker genes (Supplementary Figure S3B). Consistent with these results, our quantitative RT-PCR data also demonstrated that several genes associated with endoderm and mesoderm fate determination, including bone morphogenetic protein 2 (Bmp2), Gata4, Gata6 and Sox17, were remarkably increased by PRMT7 depletion in V6.5 mES cells (Supplementary Figure S3C). It has been shown that individual ectopic expression of Bmp2, Gata4, Gata6 and Sox17 determines differentiation of ES cells into endoderm or mesoderm (21–26). Expression of BMP2 drives ESCs into cardiac progenitor cells (21). Constitutive expression of SOX17 predisposes human ESCs to definitive endoderm progenitors (26). Expression of Gata4 or Gata6 induces differentiation program for extraembryonic endoderm from mESCs (24). Therefore, our results suggest that PRMT7 suppresses the differentiation of mESCs into endoderm and mesoderm germ layers.
Oct4, Nanog, Klf4 and c-Myc protein levels are reduced to a greater degree than their mRNA levels in PRMT7-depleted cells
Consistent with differentiation of PRMT7-depleted cells, our expression profile analysis showed that Oct4, Nanog, Sox2, Klf4 and other pluripotent genes were downregulated by PRMT7 knockdown (Supplementary Figure S3B). Oct4, Nanog, Sox2, c-Myc and Klf4 are the well-characterized major pluripotent factors that are important for maintenance of ESC stemness (27–29). Thus, we sought to confirm whether expression of these factors are decreased in PRMT7-depleted cells using quantitative RT-PCR. Our results showed that Oct4, Nanog and Sox2 mRNA levels were commonly downregulated by all five shPRMT7s (Figure 1D). c-Myc and Klf4 mRNA levels were reduced by shPRMT7-7 and -8 (Figure 1D). Similar to this, Oct4, Nanog, Sox2, c-Myc and Klf4 mRNA levels in R1 mESCs were decreased by PRMT7 knockdown (Supplementary Figure S3D). Exogenous expression of PRMT7 but not the catalytic mutant m.PRMT7 in PRMT7-depleted cells rescued the expression of Oct4, Nanog, Sox2, c-Myc and Klf4 genes (Figure 1E).
We next compared Oct4, Nanog, Klf4, Sox2 and c-Myc mRNA levels to their protein levels in PRMT7-depleted cells. Interestingly, protein levels of Oct4, Nanog, Klf4 and c-Myc were decreased to a greater degree than their mRNA levels (Figure 1D versus Figure 1F and G). Sox2 protein levels appeared to be slightly more decreased than its mRNA levels, although both protein and mRNA levels of Sox2 were highly reduced (Figure 1D versus Figure 1F and G). These results indicate that these factors are post-transcriptionally regulated.
The precursor miRNA miR-24-2 processed as miR-24-3p and miR-24-2-5p targets the 3′UTRs of Oct4, Nanog, Klf4 and c-Myc
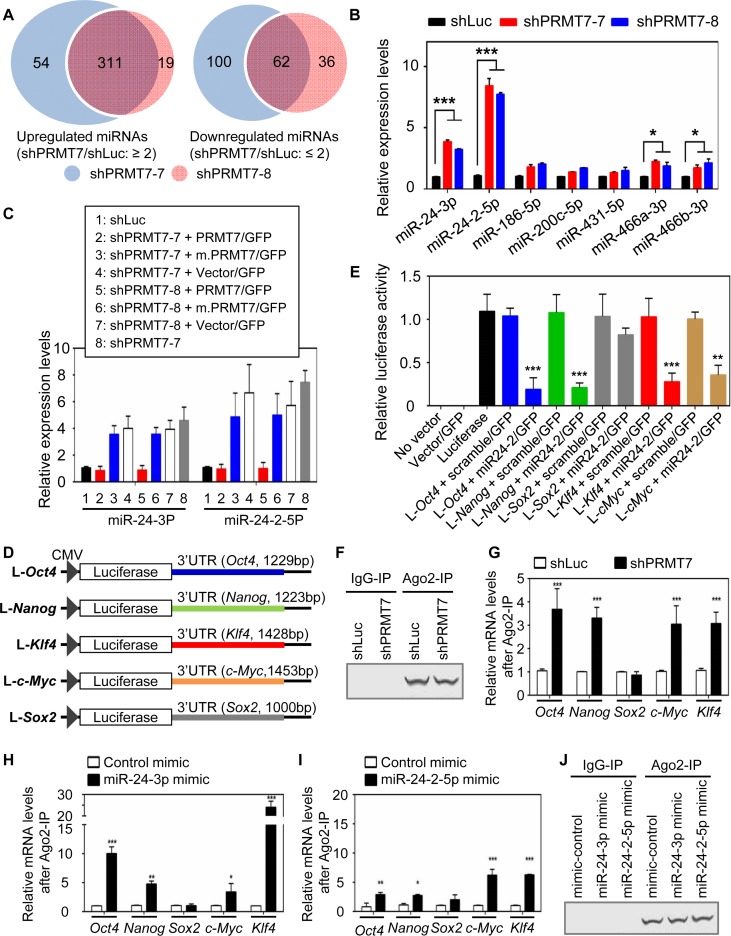
We next sought to assess how the protein levels of Oct4, Nanog, Klf4, c-Myc and/or Sox2 are post-transcriptionally regulated. Because miRNAs are a key modulator for the post-transcriptional regulation (30), we reasoned that PRMT7 may maintain ESC stemness by downregulating miRNAs that target Oct4, Nanog, Klf4, c-Myc and/or Sox2. To identify PRMT7-downregulated miRNAs, we compared the miRNA expression profiles between PRMT7-depleted cells (shPRMT7-7- or shPRMT7-8-treated cells) and shLuciferase-treated cells using miRNA microarray experiments (Supplementary Figure S4A and Supplementary Table S2). This analysis showed that a substantial number of miRNAs were upregulated or downregulated at least 2-fold upon PRMT7 depletion (Figure 2A), whereas expression levels of many other miRNAs, including miR-24-1-5p, miR-23a-3p, miR-23a-5p, miR-27a-3p and miR-27a-5p, were unchanged or weakly changed by PRMT7 knockdown (Supplementary Figure S4B).
Figure 2.
PRMT7 downregulates the levels of miR-24-3p and miR-24-2-5p, which collectively target the 3′UTRs of Oct4, Nanog, Klf4 and c-Myc mRNAs. (A) Venn diagrams of miRNAs that were significantly upregulated or downregulated by two shPRMT7s (shPRMT7-7 and shPRMT7-8). (B) Comparison of cellular levels of multiple miRNAs between shLuc-treated and PRMT7-depleted V6.5 mESCs using quantitative, miRNA-specific PCR. (C) The effect of ectopic expression of PRMT7 or its catalytic mutant (m.PRMT7) on miR-24-3p and miR24-2-5p levels in PRMT7-depleted mESCs. (D) Schematic representation of luciferase reporter constructs containing Oct4-3′UTR, Nanog-3′UTR, Sox2-3′UTR, Klf4-3′UTR or c-Myc-3′UTR. (E) Relative luciferase activities of reporter constructs containing Oct4-3′UTR, Nanog-3′UTR, Sox2-3′UTR, Klf4-3′UTR or c-Myc-3′UTR in the absence or presence of miR24-2 expression. The reporter constructs, alone or together with a miR-24-2 expression plasmid encoding miR-24-3p and miR24-2-5p, were transfected into HEK293T cells. Firefly luciferase activities were normalized to the internal transfection control Renila luciferase. (F and G) Comparison of endogenous association of Ago2 with Oct4, Nanog, Sox2, Klf4 and c-Myc mRNAs between shLuc-treated cells and PRMT7-depleted cells. Ago2 IP was performed (F) and IP eluates were analyzed using quantitative RT-PCR (G). (H−J) Analysis of the association of Ago2 with Oct4, Nanog, Sox2, Klf4 and c-Myc mRNAs after transient transfection of control mimic, miR-24-3p mimic (H) or miR-24-2-5p mimic (I) in mESCs. Following transfection of mimic RNAs, cells were incubated for 48 h, lysed and used for Ago2 IP (J). IP eluates were analyzed using quantitative RT-PCR (H and I). Data are presented as the mean ± SD of three independent experiments. P < 0.05 (*), P < 0.01 (**) and P <0.001 (***) indicate statistically significant changes.
Base pairing between a miRNA and its target mRNAs is required for miRNA-based mRNA targeting and primarily depends on the complementarity between the nucleotide positions 2−8 (called seed sequences) of the miRNA and its corresponding mRNA sequences (31). Although they are also localized in the 5′ UTRs and open reading frames (31), the miRNA target sites are frequently located in the 3′ untranslated regions (UTRs). Using these criteria, we searched miRNAs that may target the 3′UTRs of Oct4, Nanog, Klf4, Sox2 and c-Myc genes among shPRMT7-upregulated miRNAs. Our screening using 10 different miRNA target prediction programs resulted in the identification of multiple candidate miRNAs that may target at least one of these pluripotent genes (Supplementary Figure S4C and Supplementary Table S3). In particular, our analysis, including manual examination, uncovered putative targeting sites of miR-24-3p and miR-24-2-5p in Oct4, Nanog, Klf4, c-Myc and Sox2 3′UTRs (Supplementary Table S4). In addition, quantitative mature miRNA-specific PCR of these miRNAs confirmed that miR-24-3p and miR-24-2-5p were highly upregulated by PRMT7 knockdown (Figure 2B). For these reasons, we focused on analyzing the roles of miR-24-3p and miR-24-2-5p in targeting Oct4, Nanog, Klf4, c-Myc and Sox2 mRNAs.
To determine whether miR-24-3p and miR-24-2-5p levels are increased specifically by shPRMT7s and are regulated in a manner dependent on PRMT7's enzymatic activity, we examined the effect of ectopic expression of PRMT7 and its catalytic mutant on miR-24-3p and miR-24-2-5p levels in PRMT7-depleted mESCs. Exogenous expression of PRMT7 but not m.PRMT7 in these cells repressed miR-24-3p and miR-24-2-5p levels (Figure 2C). These results indicate that PRMT7 specifically downregulates miR-24-3p and miR-24-2-5p levels and that PRMT7-mediated repression of these miRNAs requires PRMT7's enzymatic activity.
miR-24-3p is associated with increased susceptibility of Dicer-deficient mice to vesicular stomatitis virus (32) and is known to inhibit the proliferation of hematopoietic cells (33). It may target the histone H2A variant H2AFX and the anti-apoptotic factor BCL2 (34). miR-24-2-5p may target protein kinase C-alpha and suppress cell survival in breast cancer cell line MCF7 (35). Nevertheless, the roles of miR-24-3p and miR-24-2-5p in ESC self-renewal and pluripotency are not clear. To assess whether miR-24-2 targets Oct4, Nanog, Klf4, Sox2 and c-Myc 3′UTRs, we individually cloned their 3′UTR parts containing putative targeting sites of miR-24-3p and miR-24-2-5p into a luciferase reporter plasmid (Figure 2D). We then determined the effect of ectopic expression of a miR-24-2 expression plasmid encoding miR-24-3p and miR-24-2-5p on luciferase reporter activities of these 3′UTRs. Ectopic expression of miR-24-2 but not scramble miRNA strongly reduced the reporter activities of Oct4-3′UTR, Nanog-3′UTR, Klf4-3′UTR and c-Myc-3′UTR (Figure 2E). In contrast, the reporter activity of Sox2-3′UTR was not affected by miR-24-2 expression (Figure 2E). These results suggest that miR-24-3p and/or miR-24-2-5p processed from their precursor miRNA miR-24-2 silences the expression of Oct4, Nanog, Klf4 and c-Myc, but not Sox2.
miR-24-3p targets Oct4, Nanog, Klf4 and c-Myc 3′UTRs, whereas miR-24-2-5p targets Klf4 and c-Myc 3′UTRs
During their targeting processes, miRNAs bind their target mRNAs in Ago2, a component of the RNA-inducing silencing complex. If miR-24-2-mediated targeting of Oct4, Nanog, Klf4 and c-Myc mRNAs is augmented in PRMT7-depleted cells compared to control cells, the association of Ago2 with Oct4, Nanog, Klf4 and c-Myc mRNAs is likely higher in PRMT7-depleted cells than in control cells. To test this, we performed Ago2 immunoprecipitation (IP) experiments using control cells and PRMT7-depeleted cells. Ago2 recovery between these two IP experiments was similar (Figure 2F). Our quantitative PCR results of Ago2 IP eluates showed that PRMT7 knockdown increased the endogenous interaction of Ago2 with Oct4, Nanog, Klf4 and c-Myc mRNAs but not with Sox2 mRNA (Figure 2G). In support of this result, transfection of miR-24-3p mimic (small double-stranded RNAs corresponding to endogenous miR-24-3p) increased the association of Ago2 with Oct4, Nanog, Klf4 and c-Myc mRNAs but not with Sox2 mRNA in mESCs (Figure 2H). Transfection of miR-24-2-5p mimic also enhanced the interaction of Ago2 with Klf4 and c-Myc mRNAs and to a less extent Oct4 and Nanog mRNAs (Figure 2I). It should be noted that Ago2 recovery was similar among IP experiments using different miRNA mimics (control, miR-24-3p mimic or miR-24-2-5p mimic) (Figure 2J). These results suggest that miR-24-3p silences Oct4, Nanog, Klf4 and c-Myc expression whereas miR-24-2-5p targets preferentially Klf4 and c-Myc mRNAs.
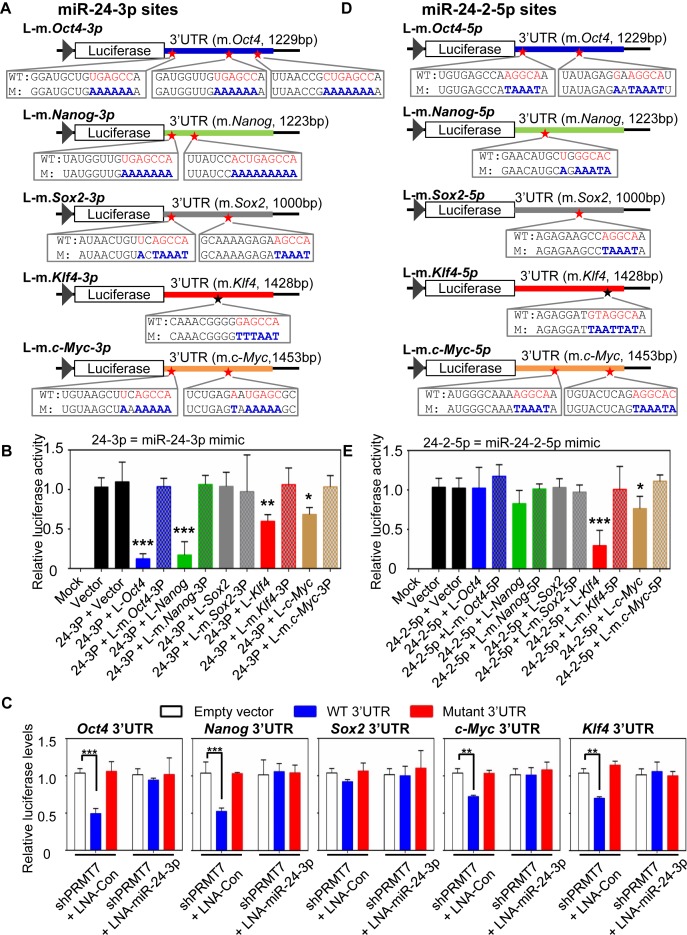
To determine whether miR-24-3p specifically targets 3′UTRs of Oct4, Nanog, Klf4 and c-Myc, we first introduced mutations into putative targeting sites of miR-24-3p in their 3′UTR reporters (Figure 3A). We then compared the effect of miR-24-3p mimic on the reporter activities between WT and mutant 3′UTRs. Transient transfection of miR-24-3p mimic greatly decreased the luciferase activities of the Oct4-3′UTR and Nanog-3′UTR reporters while substantially reducing the activities of the Klf4-3′UTR and c-Myc-3′UTR reporters (Figure 3B). In contrast, mutations in all putative miR-24-3p target sites in Oct4-3′UTR, Nanog-3′UTR, Klf4-3′UTR and c-Myc-3′UTR reporters nullified the effects of miR-24-3p mimic on their reporter activities (Figure 3B). In line with the above results using the miR-24-2 expression plasmid, miR-24-3p mimic had no significant effect on the reporter activities of Sox2-3′UTR and its mutant. Interestingly, it has been shown that miR-24-3p targets the 3′UTR of human c-Myc mRNAs in hematopoietic cells via uncanonical complementarity that does not involve a seed sequence match (33). However, such targeting mechanism of human c-Myc mRNAs by miR-24-3p may not be relevant to mouse c-Myc mRNAs, because the target sequences of miR-24-3p in human c-Myc mRNA are not conserved in mousec-Myc mRNA. Nevertheless, these results suggest that miR-24-3p is a miRNA that silences the expression of Oct4, Nanog, Klf4 and c-Myc by targeting their mRNA 3′UTRs.
Figure 3.
miR-24-3p's targeting sites are located at Oct4, Nanog, Klf4 and c-Myc 3′UTRs, whereas miR24-2-5p's sites are localized at Klf4 and c-Myc 3′UTRs. (A) Schematic representation of luciferase reporter constructs containing mutations in miR-24-3p target sites in Oct4-3′UTR, Nanog-3′UTR, Sox2-3′UTR, Klf4-3′UTR and c-Myc-3′UTR. WT, wild-type; M, mutant. (B) The effect of miR-24-3p mimic on reporter activities of WT Oct4-3′UTR, Nanog-3′UTR, Sox2-3′UTR, Klf4-3′UTR, c-Myc-3′UTR and their mutants. The reporter constructs, together with miR-24-3p mimic, were transfected into HEK293T cells. Mock indicates non-transfected cells, and vector denotes a reporter plasmid without any 3′UTR. Firefly luciferase activities were normalized to an internal transfection control (Renila luciferase). (C) The effect of endogenous miR-24-3p on the reporter activities of Oct4-3′UTR, Nanog-3′UTR, Sox2-3′UTR, Klf4-3′UTR and c-Myc-3′UTR. mESCs (5 × 105 cells) were first electroporated with 30 μg of the shPRMT7-7 plasmid. Twelve days later, PRMT7-depleted mESCs were co-transfected with both WT luciferase-3′UTR (or luciferase-mutant 3′UTR) reporters and control LNA (or LNA-miR-24-3p [a strong anti-sense inhibitor of miR-24-3p]). Cells were harvested 2 days after transfection. (D) Schematic representation of luciferase reporter constructs bearing mutations in miR-24-2-5p target sites in Oct4-3′UTR, Nanog-3′UTR, Sox2-3′UTR, Klf4-3′UTR and c-Myc-3′UTR. WT, wild-type; M, mutant. (E) The effect of miR-24-2-5p mimic on reporter activities of WT Oct4-3′UTR, Nanog-3′UTR, Sox2-3′UTR, Klf4-3′UTR, c-Myc-3′UTR and their mutants. Data are presented as the mean ± SD of three independent experiments. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) indicate statistically significant changes.
To vigorously determine whether miR-24-3p targets Oct4, Nanog, Klf4 and c-Myc 3′UTRs, we co-transfected both WT luciferase-3′UTR (or mutant luciferase-3′UTR) reporter and locked-nucleic-acid (LNA) control (or LNA-modified miR-24-3p, i.e. a strong anti-sense inhibitor of miR-24-3p) into PRMT7-depleted mESCs that contain the increased endogenous levels of miR-24-3p. As shown in Figure 3C, PRMT7 knockdown decreased the luciferase activities of WT Oct4-3′UTR, Nanog-3′UTR, Klf4-3′UTR and c-Myc-3′UTR but not those of their mutant 3′UTRs. Such effects of PRMT7 knockdown on the reporter activities of these WT 3′UTRs were nullified by LNA-miR-24-3p. These results indicate that endogenous miR-24-3p targets Oct4, Nanog, Klf4 and c-Myc 3′UTRs.
To assess whether miR-24-2-5p inhibits the activities of Oct4-3′UTR, Nanog-3′UTR, Klf4-3′UTR and c-Myc-3′UTR reporters, we produced their mutant reporters bearing mutations in putative target sites of miR-24-2-5p (Figure 3D) and determine the effect of miR-24-2-5p mimic on activities of WT and mutant 3′UTR reporters. Transient transfection of miR-24-2-5p mimic significantly decreased the luciferase activities of Klf4-3′UTR and c-Myc-3′UTR reporters. In contrast, their reporter constructs bearing mutations in putative miR-24-2-5p target sites were resistant to the inhibitory action of miR-24-2-5p mimic (Figure 3E). miR-24-2-5p mimic had no significant effect on the reporter activities of Oct4-3′UTR, Nanog-3′UTR and Sox2-3′UTR as well as their mutants (Figure 3E). These results point out miR-24-2-5p as a miRNA that represses expression of Klf4 and c-Myc.
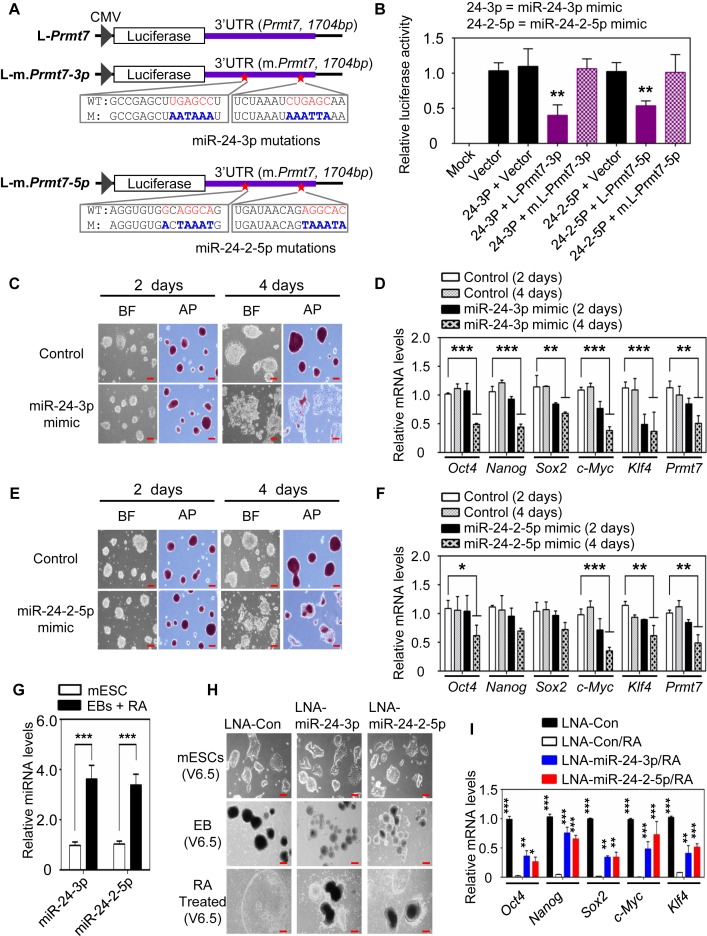
miR-24-3p and miR-24-2-5p target the 3′UTR of their transcriptional repressor gene Prmt7
Interestingly, PRMT7 protein levels were also decreased to a greater degree than PRMT7 mRNA levels in PRMT7-depleted mESC cells, suggesting a post-transcriptional regulation of PRMT7 levels (Figure 1D versus Figure 1F and G). In fact, our analysis suggested that miR-24-3p and miR-24-2-5p might also target the 3′UTR of their transcriptional repressor gene Prmt7 (Figure 4A and Supplementary Table S5). To test this possibility, we cloned a Prmt7 3′UTR containing putative miR-24-3p and miR-24-2-5p target sites into a luciferase reporter plasmid and also generated its mutant reporters containing mutations in these target sites (Figure 4A). miR-24-3p and miR-24-2-5p mimics decreased the activities of WT Prmt7-3′UTR reporter but did not affect the reporter activities of Prmt7-3′UTR mutants, pointing out that these miRNAs can target Prmt7-3′UTR (Figure 4B). These results, together with the above results, indicate that miR-24-3p/ miR-24-2-5p and PRMT7 constitute a double negative feedback loop.
Figure 4.
miR-24-3p and miR-24-2-5p target Prmt7 3′UTR and impede the stemness of mESCs. (A) Schematic representation of luciferase reporter constructs containing WT Prmt7-3′UTR or its mutants. (B) Relative luciferase activities of reporter constructs containing Prmt7-3′UTR or its mutants in HEK293T cells after transfection of miR-24-3p and miR-24-2-5p mimics. (C and E) Microscopic and AP staining images of V6.5 mESCs after treatment with miR-24-3p (C) or miR-24-2-5p (E) mimic. V6.5 mESCs were treated with miR-24-3p and miR-24-2-5p mimics and incubated for 2d or 4d. (D and F) Quantitative analysis of Oct4, Nanog, Sox2, Klf4, c-Myc and Prmt7 mRNA levels in V6.5 mESCs after treatment with miR-24-3p (D) or miR-24-2-5p (F) mimic. (G) Quantitative RT-PCR analysis of miR-24-3p and miR-24-2-5p levels in WT and differentiated V6.5 mESCs (EBs + RA). (H and I) The effect of LNA-miRNAs (a type of anti-sense microRNAs) against miR-24-3p or miR-24-2-5p on RA-induced mESC differentiation. mESCs were transfected with LNA-miRNAs (H). Oct4, Nanog, Sox2, c-Myc and Klf4 mRNA levels were measured using quantitative RT-PCR (I). In G−I, mESCs were induced to form EBs for 5 days and then treated with RA for another 5 days. Data are presented as the mean ± SD of three independent experiments. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) indicate statistically significant changes.
miR-24-3p and miR-24-2-5p impede mESC stemness and are required for mESC differentation
To determine whether miR-24-3p inhibits the stemness of mESCs, we transfected V6.5 mESCs with miR-24-3p mimic. Our results showed that miR-24-3p mimic caused spontaneous differentiation of mESCs while decreasing both mRNA and protein levels of PRMT7 and several other pluripotent transcription factors (Figure 4C and D; Supplementary Figure S4D). Similar to miR-24-3p mimic, miR-24-2-5p mimic also induced spontaneous differentiation of mESCs while reducing the expression of several pluripotent factors (Figure 4E and F; Supplementary Figure S4D).
To assess whether miR-24-3p and miR-24-2-5p affect mESC differentiation, we first compared their expression levels between WT mESC and differentiated mESCs generated by RA treatment of EBs. Consistent with the notion that miR-24-3p and miR-24-2-5p inhibit mESC stemness, miR-24-3p and miR-24-2-5p levels were increased during RA-induced differentiation (Figure 4G). We next inhibited the action of miR-24-3p and miR-24-2-5p during RA-induced mESC differentiation using their LNA-miRNAs. The treatment of LNA-miR-24-3p or LNA-miR-24-2-5p impeded RA-induced differentiation of mESCs and restored the expression of several pluripotent genes (Figure 4H and I). These results indicate that miR-24-3p and miR-24-2-5p are necessary for proper mESC differentiation.
PRMT7-mediated repression of miR-24-3p and miR-24-2-5p levels is indispensable for maintaining mESC stemness
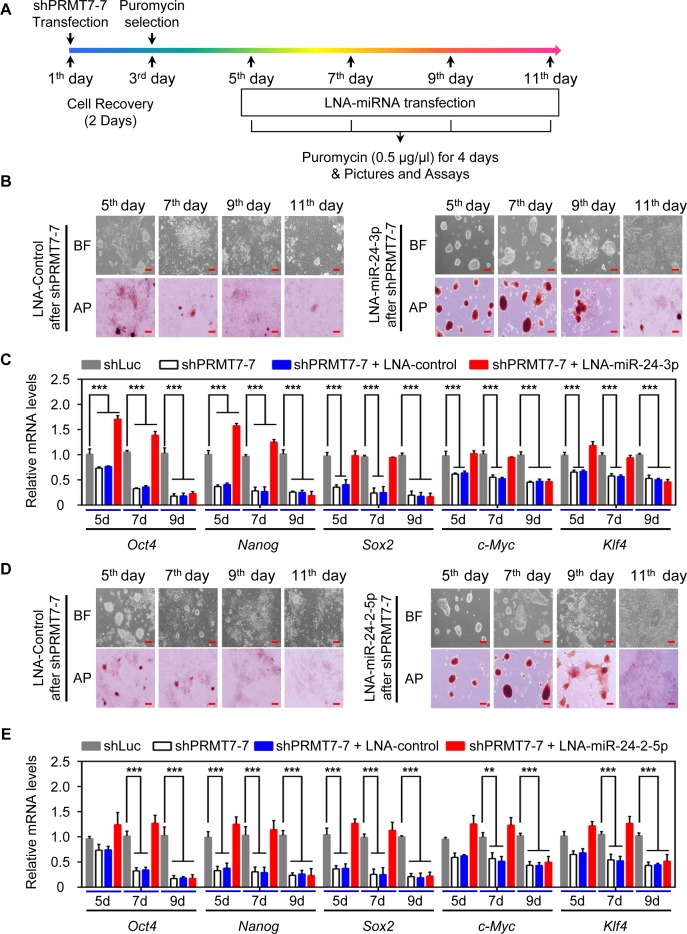
To determine whether PRMT7-mediated repression of miR-24-3p levels is necessary for the stemness of mESCs, we blocked the action of miR-24-3p in PRMT7-depleted V6.5 mESCs using LNA-miR-24-3p. LNA-miR-24-3p was transfected into PRMT7-depleted cells at the indicated time points (i.e. 5th, 7th, 9th or 11th day) after shPRMT7 transfection (Figure 5A). Transient transfection of LNA-miR-24-3p but not LNA control at the 5th or 7th day restored the morphology and AP staining of PRMT7-depleted cells close to those of WT mESCs (Figure 5B). In addition, quantitative RT-PCR results showed that although Prmt7 mRNA levels were still low at the 5th or 7th day, LNA-miR-24-3p restored the expression of several pluripotent transcription factors at the mRNA and protein levels (Figure 5C; Supplementary Figure S5A and B). In contrast, LNA-miR-24-3p transfection at the 9th or 11th day insignificantly reversed differentiation of PRMT7-depleted cells and expression of Oct4, Nanog, Klf4 and c-Myc (Figure 5B and C), suggesting that these time points may be too late to reverse shPRMT7-induced spontaneous differentiation. Similar to LNA-miR-24-3p, transient transfection of LNA-miR-24-2-5p at the 5th or 7th day after PRMT7 transfection largely rescued the morphology and AP staining of PRMT7-depleted cells as well as the expression of several pluripotent genes (Figure 5D and E; Supplementary Figure S5B and C). These results indicate that PRMT7-mediated repression of miR-24-3p and miR-24-2-5p is required for the stemness maintenance of mESCs.
Figure 5.
PRMT7-mediated repression of miR-24-3p/miR-24-2-5p levels is required for maintaining self-renewal and pluripotency. (A) The outline for treatment of PRMT7-depleted cells with LNA-control and LNA-miRNAs. (B and D) Microscopic and AP staining images of PRMT7-depleted V6.5 mESCs at the 5th, 7th, 9th, or 11th day after treatment with LNA-control or LNA-miR-24-3p (B) (LNA-miR-24-2-5p in [D]). Red scale bar, 100 μm. (C and E) Quantitative analysis of Oct4, Nanog, Sox2, c-Myc and Klf4 mRNA levels in PRMT7-depleted V6.5 mESCs at the 5th, 7th, 9th, or 11th day after treatment with LNA-control or LNA-miR-24-3p (C) (LNA-miR-24-2-5p in [E]). Data are presented as the mean ± SD of three independent experiments. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) indicate statistically significant changes.
PRMT7 downregulates the miR-24-2 gene
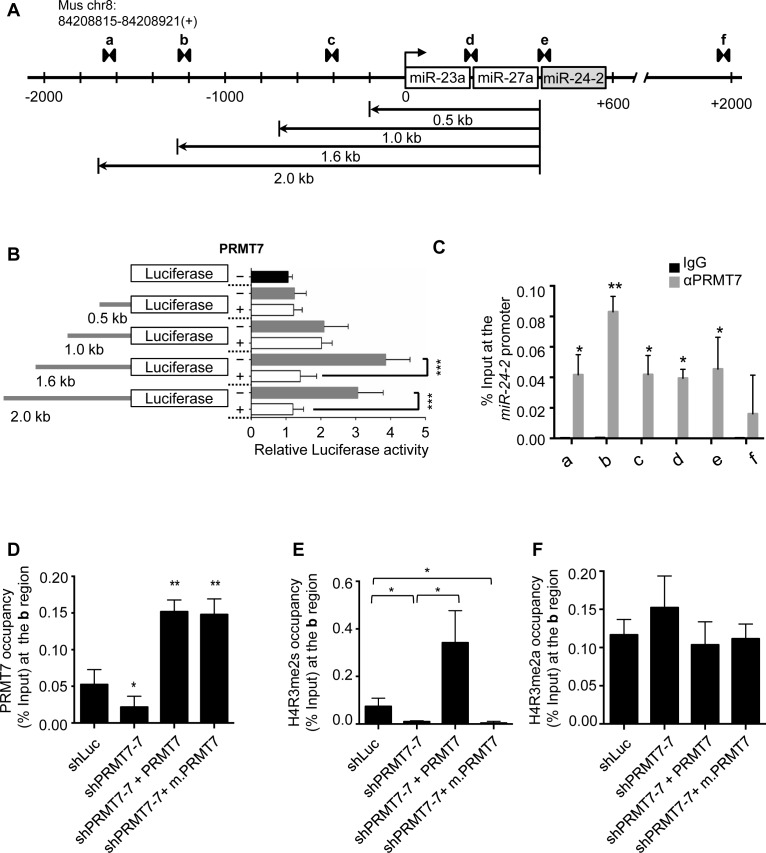
To understand how PRMT7 downregulates miR-24-3p and miR-24-2-5p levels, we sought to determine what promoter region in the miR-24-2 gene is responsible for PRMT7-mediated repression. We cloned a miR-24-2 promoter region into a luciferase-expressing plasmid and generated several deletion constructs of the promoter region (Figure 6A and B). Our reporter gene assays showed that a region between −1 and −1.6 kb in the miR-24-2 promoter was responsive to PRMT7-mediated repression (Figure 6B). In support of this finding, quantitative chromatin immunoprecipitation (ChIP) experiments showed that PRMT7 was highly enriched in the ‘b’ region between −1 and −1.6 kb than in other regions (Figure 6C).
Figure 6.
PRMT7 occupies the miR-24-2 gene and negatively regulates its expression via H4R3 methylation. (A) Schematic representation of the miR-24-2 gene. A set of arrow heads indicate a ChIP PCR amplicon. (B) Luciferase activities of the miR-24-2 promoter and its deletion mutants with or without ectopic expression of PRMT7 in V6.5 mESCs. V6.5 mESCs were transfected using Lipofectamine 3000 and incubated for 48 h. (C) Analysis of PRMT7 occupancy at the miR-24-2 promoter using quantitative ChIP. (D−F) Analysis of PRMT7 (D), H4R3me2s (E) and H4R3me2a (F) levels at the region ‘b’ in the miR-24-2 promoter after rescue experiments of PRMT7-depleted mESCs. Quantitative ChIP assays were performed using four groups of V6.5 mESCs: (i) shLuc-treated cells, (ii) PRMT7-depleted cells, (iii) PRMT7-depleted cells with ectopic expression of PRMT7 and (iv) PRMT7-depleted cells with ectopic expression of a catalytic mutant of PRMT7 (m.PRMT7).
In line with PRMT7's methyltransferase activity, PRMT7 knockdown decreased H4R3me2s levels at the b region in the miR-24-2 gene promoter (Figure 6D and E). Moreover, H4R3me2s level in the b region was rescued by PRMT7 but not its catalytic mutant (Figure 6D and E). In contrast, the activating mark H4R3me2a was not affected by PRMT7 (Figure 6F). These results indicate that PRMT7 directly represses miR-24-2 expression, at least in part, by upregulating H4R3me2s levels.
Prmt7 expression is positively regulated by Oct4, Nanog, Klf4 and c-Myc
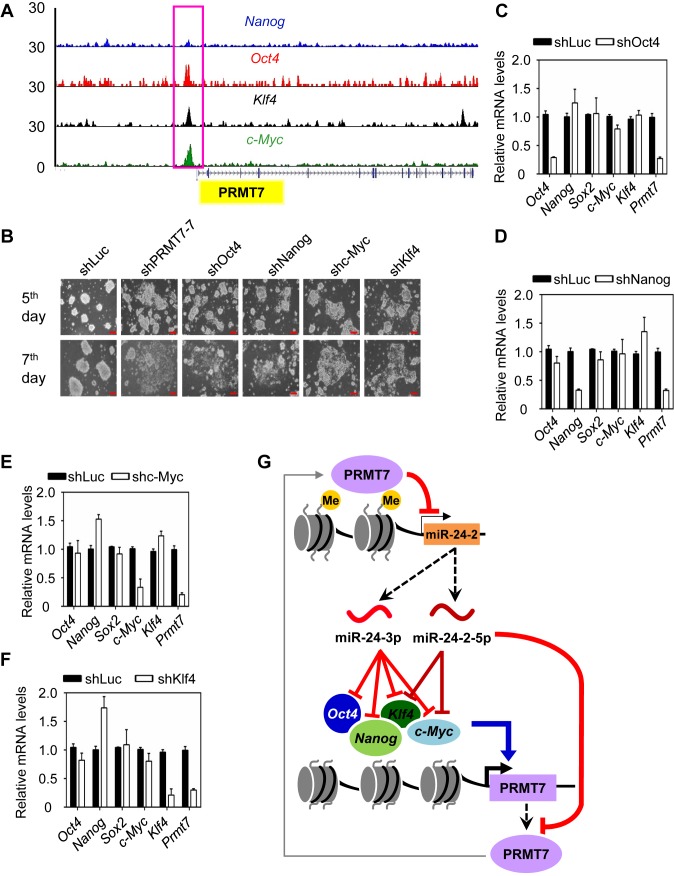
Several core pluripotent transcription factors, including Oct4, Nanog, Klf4 and c-Myc, regulate one another in the pluripotency-regulatory networks (27,28). Therefore, we tested the possibility that PRMT7 may be regulated by the other pluripotent factors Oct4, Nanog, Klf4 and c-Myc. In fact, analysis of a ChIP-Seq database (36) showed that Oct4, Nanog, Klf4 and c-Myc occupied the Prmt7 promoter (Figure 7A). We next examined the effect of knockdown of Oct4, Nanog, Klf4 or c-Myc on Prmt7 expression in V6.5 mESCs. Individual depletion of these pluripotent factors strongly or weakly induced differentiation phenotypes of mESCs (Figure 7B) and also decreased Prmt7 mRNA and protein levels (Figure 7C−F; Supplementary Figure S5D). These results indicate that Oct4, Nanog, Klf4 and c-Myc directly activate Prmt7 expression.
Figure 7.
The Prmt7 gene is activated by Oct4, Nanog, c-Myc and Klf4. (A) Analysis of Oct4, Nanog, c-Myc and Klf4 levels at the Prmt7 promoter using the publicly available ChIP-Seq database. (B) Microscopic images of V6.5 mESCs treated with shPRMT7-7, shOct4, shc-Myc, shKlf4 or shNanog. Red scale bar, 100 μm. (C−F) Effects of individual knockdown of Oct4 (C), Nanog (D), c-Myc (E), and Klf4 (F) on Prmt7 mRNA levels. V6.5 mESCs were electroporated with shOct4, shc-Myc, shKlf4, or shNanog. Four days later, cells were harvested. Expression levels were analyzed using quantitative RT-PCR. (G) A proposed model depicting a role for PRMT7 in maintaining the self-renewal and pluripotency of mESCs. miR-24-3p can silence the expression of Oct4, Nanog, Klf4 and c-Myc, whereas miR-24-2-5p can target Klf4 and c-Myc 3′UTRs. In addition, miR-24-3p and miR-24-2-5p target Prmt7 3′UTR. In mESCs, PRMT7 represses the miR-24-2 gene encoding miR-24-3p and miR-24-2-5p via H4R3 methylation. Therefore, PRMT7 antagonizes the anti-pluripotent effects of miR-24-3p and miR-24-2-5p against Oct4, Nanog, Klf4 and c-Myc in mESCs and positively regulates Oct4, Nanog, Klf4, and c-Myc levels to maintain mESC stemness. During differentiation, increased miR-24-3p and miR-24-2-5p levels may reduce PRMT7, Oct4, Nanog, Klf4, and c-Myc levels, facilitating mESC differentiation. The regulatory loop involving PRMT7 and miR-24-3p/miR-24-2-5p is interactive with the major pluripotent system containing Oct4, Nanog, Klf4, and c-Myc to fine-tune mESC stemness.
DISCUSSION
In this study, we identified PRMT7 as a new stemness factor that plays a critical role in maintaining Oct4, Nanog, Klf4 and c-Myc levels in mESCs. To our knowledge, we showed for the first time that a histone methyltransferase positively modulates cellular levels of these four major pluripotent transcription factors via epigenetic repression of anti-pluripotent miRNAs. PRMT7 represses the miR-24-2 gene encoding the new anti-pluripotent miRNAs miR-24-3p and miR-24-2-5p via upregulation of symmetric dimethylation of H4R3, thereby counteracting miR-24-3p/miR-24-2-5p-mediated silencing of Oct4, Nanog, Klf4 and c-Myc expression. In addition, Prmt7 expression is positively modulated by Oct4, Nanog, Klf4 and c-Myc in mESCs. Therefore, our results suggest a unique stemness-regulatory mechanism in which PRMT7 maintains the self-renewal and pluripotency of mESCs by epigenetically antagonizing miRNA-directed silencing of Oct4, Nanog, Klf4 and c-Myc expression while being activated by the same pluripotent factors (Figure 7G and Supplementary Figure S5E).
PRMT7 has been linked to several biological functions. It was reported that PRMT7 interacted with the BRG1-containing hSWI/SNF chromatin remodeling complex (37). PRMT7, together with the adenosine triphosphate-dependent helicase BRG1, downregulates DNA-repair-associated genes and PRMT7 decreases cellular resistance to genotoxic stress caused by DNA-damaging drugs (37). Seemingly contradictory to this finding, PRMT7 may be associated with increased cellular resistance to camptothecin, an inhibitor of DNA topoisomerase (an enzyme that removes torsion constraints during replication and transcription) (38). PRMT7 cooperates with the CCCTC-binding zinc finger protein CTCF-like to promote DNA methylation in the H19 imprinting control region (39). Distinct from these functions of PRMT7, results reported here showed that PRMT7 is required for mESC stemness. Several histone methylation modifiers often regulate biological functions independent of their catalytic activities. For example, the H3K27 demethylase UTX does not need its demethylase activity to control mesoderm differentiation (40). In contrast, our results showed that PRMT7-mediated H4R3 methylation at the miR-24-2 promoter was accompanied with the repression of miR-24-2 by PRMT7. In addition, PRMT7's catalytic mutant did not rescue spontaneous differentiation induced by PRMT7 knockdown (Figure 1C). Therefore, these results indicate that the enzymatic activity of PRMT7 is required for the maintenance of mESCs.
Several pluripotent factors are associated with oncogenic events (41). Oct4 is overexpressed in most, if not all, human germ cell tumors and its overexpression transforms non-tumorigenic Swiss 3T3 fibroblast cells to tumorigenic cells (42). c-Myc is a well-known oncoprotein that causes malignant transformation and tumor progression. The c-Myc regulatory network is common between ESCs and cancer cells (43). Similar to these pluripotent factors, PRMT7 promotes epithelial-to-mesenchymal transition by repressing the expression of the E-cadherin gene via cooperation with HDAC3 (44). Moreover, PRMT7 levels are higher in basal-like breast cancer (an aggressive subtype) than in luminal breast cancer and are increased in the histologic grade III compared to the grades I and II (44). Thus, PRMT7 also possesses oncogenic properties while acting as a pluripotent factor.
miRNAs are associated with ESC maintenance and differentiation (15). Pro-pluripotent miRNAs may target anti-pluripotent genes or cell cycle inhibitors and include miR-290/295 and miR-302/367 clusters (45,46). Anti-pluripotent miRNAs include miR-145, let-7, miR-21 and miR-34a, which regulate pluripotent gene expression (47–50). Specifically, our results revealed miR-24-3p as an anti-pluripotent miRNA that targets all the 3′UTRs of Oct4, Nanog, Klf4 and c-Myc mRNAs while establishing miR-24-2-5p as a silencer of Klf4 and c-Myc expression. In agreement with their roles in anti-pluripotency, miR-24-3p and miR-24-2-5p mimics induced spontaneous differentiation of mESCs (Figure 4C–F). In addition, anti-sense inhibitors of miR-24-3p and miR-24-2-5p largely prevented spontaneous differentiation of PRMT7-depleted mESCs and RA-induced differentiation of mESCs (Figures 4H and 5). Previous studies showed that Oct4, Nanog, Klf4 and c-Myc form the interconnected transcriptional regulatory networks involving numerous transcriptional factors (27,28). It is plausible that co-silencing of these core pluripotent factors by miR-24-3p and miR-24-2-5p may effectively disrupt the pluripotent factor networks during mESC differentiation, whereas PRMT7-mediated epigenetic repression of miR-24-3p and miR-24-2-5p is critical in maintaining such networks for the stemness of mESCs.
There are multiple modes of miRNA regulation. A mode of miRNA regulation involves the targeting efficiency of miRNAs that can be controlled by RNA-binding proteins and Ago-interacting proteins (51). For example, LIN41, a TRIM-NHL family of protein with E3 ubiquitin ligase activity, directly associates with Ago, targets Ago for ubiquitination-mediated protein degradation and subsequently decreases the targeting efficiency of miRNAs (52). In addition, miRNA stability can contribute to miRNA regulation. An example is that increased expression of Ago proteins upregulates mature miRNA levels by hindering exonucleolytic degradation of miRNAs (53). Furthermore, miRNA expression can be modulated by transcriptional factors and epigenetic factors. For instance, the transcriptional repressor REST regulates miRNA expression (e.g. miR-21) (50) and the histone deacetylase SIRT1 represses miR-134 expression (54). In the current study, we show a mode of epigenetic regulation of miRNAs in which miR-24-2 expression is repressed by the epigenetic modifier PRMT7. PRMT7-mediated repression of miRNA levels can be considered a distinct type of epigenetic regulation of miRNAs, because miRNA expression is controlled by PRMT7-regulated H4R3 methylation.
Interestingly, our results demonstrated that miR-24-3p and miR-24-2-5p target their own repressor gene Prmt7 (Figure 4A and B). This finding points out an epigenetic double-negative feedback loop in which PRMT7 represses miR-24-3p and miR-24-2-5p expression while being downregulated by the same miRNAs. Although PRMT7-mediated repression of miR-24-3p/miR-24-2-5p occurs mainly in mESCs, the silencing of PRMT7 by these miRNAs may occur largely during mESC differentiation. In fact, our data showed that miR-24-3p and miR-24-2-5p levels were increased during RA-induced mESC differentiation and that mESCs were spontaneously differentiated upon treatment of miR-24-3p or miR-24-2-5p mimics (Figure 4C–F). Therefore, the feedback regulatory loop comprising PRMT7 and miR-24-3p/miR-24-2-5p may fine-tune Oct4, Nanog, Klf4 and c-Myc levels to tip the balance between self-renewal and differentiation of mESCs. Because we also showed that Prmt7 expression is regulated by Oct4, Nanog, Klf4 and c-Myc, our results support the notion that this feedback regulatory loop is integrated with the pluripotent transcription factor system in a complicated epigenetic, stemness-regulatory circuit (Figure 7G and Supplementary Figure S5E).
Supplementary Material
Acknowledgments
We are thankful to Dr Mark Bedford for his comments and Dr Li Ma, Dr Zhenbo Han, Dr Xin Lin and Mr Su Zhang for their technical assistance and reagents. We also thank Markeda Wade for the manuscript editing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (NIH) [R01 GM095659, R01 CA157919 to M.G.L.]; Cancer Prevention and Research Institute of Texas [RP110183, RP140271 to M.G.L.]; Center for Cancer Epigenetics at MD Anderson Cancer Center [a seed grant to M.G.L.]; Center for Cancer Epigenetics at MD Anderson Cancer Center Scholar Fellowship (to S.S.D.); NIH [R01 HG007538 to W.L.]; Cancer Prevention and Research Institute of Texas [RP110471 to W.L.]. Funding for open access charge: NIH [R01 GM095659, R01 CA157919].
Conflict of interest statement. None declared.
REFERENCES
- 1.Orkin S.H., Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 3.Spivakov M., Fisher A.G. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 4.Gu B., Lee M.G. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013;3:39–53. doi: 10.1186/2045-3701-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Berger S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 8.Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Bedford M.T., Clarke S.G. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gayatri S., Bedford M.T. Readers of histone methylarginine marks. Biochim. Biophys. Acta. 2014;1839:702–710. doi: 10.1016/j.bbagrm.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y., Hadjikyriacou A., Xia Z., Gayatri S., Kim D., Zurita-Lopez C., Kelly R., Guo A., Li W., Clarke S.G., et al. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 2015;6:6428–6440. doi: 10.1038/ncomms7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan P., Han J., Guo G., Orlov Y.L., Huss M., Loh Y.H., Yaw L.P., Robson P., Lim B., Ng H.H. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tee W.W., Pardo M., Theunissen T.W., Yu L., Choudhary J.S., Hajkova P., Surani M.A. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24:2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torres-Padilla M.E., Parfitt D.E., Kouzarides T., Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huo J.S., Zambidis E.T. Pivots of pluripotency: the roles of non-coding RNA in regulating embryonic and induced pluripotent stem cells. Biochim. Biophys. Acta. 2013;1830:2385–2394. doi: 10.1016/j.bbagen.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babiarz J.E., Blelloch R. StemBook, editor. Stem Cell Res. Community. Harv. Stem Cell Inst. Cambridge: 2008. [Google Scholar]
- 17.Dhar S.S., Lee S.H., Kan P.Y., Voigt P., Ma L., Shi X., Reinberg D., Lee M.G. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012;26:2749–2762. doi: 10.1101/gad.203356.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa M., Toma-Fukai S., Kim J.D., Fukamizu A., Shimizu T. Protein arginine methyltransferase 7 has a novel homodimer-like structure formed by tandem repeats. FEBS Lett. 2014;588:1942–1948. doi: 10.1016/j.febslet.2014.03.053. [DOI] [PubMed] [Google Scholar]
- 19.Rust H.L., Zurita-Lopez C.I., Clarke S., Thompson P.R. Mechanistic studies on transcriptional coactivator protein arginine methyltransferase 1. Biochemistry. 2011;50:3332–3345. doi: 10.1021/bi102022e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 21.Leschik J., Stefanovic S., Brinon B., Puceat M. Cardiac commitment of primate embryonic stem cells. Nat. Protoc. 2008;3:1381–1387. doi: 10.1038/nprot.2008.116. [DOI] [PubMed] [Google Scholar]
- 22.Holtzinger A., Rosenfeld G.E., Evans T. Gata4 directs development of cardiac-inducing endoderm from ES cells. Dev. Biol. 2010;337:63–73. doi: 10.1016/j.ydbio.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turbendian H.K., Gordillo M., Tsai S.Y., Lu J., Kang G., Liu T.C., Tang A., Liu S., Fishman G.I., Evans T. GATA factors efficiently direct cardiac fate from embryonic stem cells. Development. 2013;140:1639–1644. doi: 10.1242/dev.093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujikura J., Yamato E., Yonemura S., Hosoda K., Masui S., Nakao K., Miyazaki Ji J., Niwa H. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 2002;16:784–789. doi: 10.1101/gad.968802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niakan K.K., Ji H., Maehr R., Vokes S.A., Rodolfa K.T., Sherwood R.I., Yamaki M., Dimos J.T., Chen A.E., Melton D.A., et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seguin C.A., Draper J.S., Nagy A., Rossant J. Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell. 2008;3:182–195. doi: 10.1016/j.stem.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J., Chu J., Shen X., Wang J., Orkin S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 31.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otsuka M., Jing Q., Georgel P., New L., Chen J., Mols J., Kang Y.J., Jiang Z., Du X., Cook R., et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Lal A., Navarro F., Maher C.A., Maliszewski L.E., Yan N., O'Day E., Chowdhury D., Dykxhoorn D.M., Tsai P., Hofmann O., et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to ‘seedless’ 3′UTR microRNA recognition elements. Mol. Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srivastava N., Manvati S., Srivastava A., Pal R., Kalaiarasan P., Chattopadhyay S., Gochhait S., Dua R., Bamezai R.N. miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: a potential for therapeutic intervention. Breast Cancer Res. 2011;13:R39–R50. doi: 10.1186/bcr2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin E.C., Elliott S., Rhodes L.V., Antoon J.W., Fewell C., Zhu Y., Driver J.L., Jodari-Karimi M., Taylor C.W., Flemington E.K., et al. Preferential star strand biogenesis of pre-miR-24-2 targets PKC-alpha and suppresses cell survival in MCF-7 breast cancer cells. Mol. Carcinog. 2014;53:38–48. doi: 10.1002/mc.21946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karkhanis V., Wang L., Tae S., Hu Y.J., Imbalzano A.N., Sif S. Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase delta catalytic subunit gene, POLD1. J. Biol. Chem. 2012;287:29801–29814. doi: 10.1074/jbc.M112.378281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verbiest V., Montaudon D., Tautu M.T., Moukarzel J., Portail J.P., Markovits J., Robert J., Ichas F., Pourquier P. Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins. FEBS Lett. 2008;582:1483–1489. doi: 10.1016/j.febslet.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 39.Jelinic P., Stehle J.C., Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C., Lee J.E., Cho Y.W., Xiao Y., Jin Q., Liu C., Ge K. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl. Acad. Sci. U.S.A. 2012;109:15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suva M.L., Riggi N., Bernstein B.E. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gidekel S., Pizov G., Bergman Y., Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 43.Kim J., Woo A.J., Chu J., Snow J.W., Fujiwara Y., Kim C.G., Cantor A.B., Orkin S.H. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao R., Jiang H., Ma Y., Wang L., Wang L., Du J., Hou P., Gao Y., Zhao L., Wang G., et al. PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer Res. 2014;74:5656–5667. doi: 10.1158/0008-5472.CAN-14-0800. [DOI] [PubMed] [Google Scholar]
- 45.Judson R.L., Babiarz J.E., Venere M., Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Baskerville S., Shenoy A., Babiarz J.E., Baehner L., Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu N., Papagiannakopoulos T., Pan G., Thomson J.A., Kosik K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 48.Choi Y.J., Lin C.P., Ho J.J., He X., Okada N., Bu P., Zhong Y., Kim S.Y., Bennett M.J., Chen C., et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E.E., Nitsch R., Wulczyn F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 50.Singh S.K., Kagalwala M.N., Parker-Thornburg J., Adams H., Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasquinelli A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 52.Rybak A., Fuchs H., Hadian K., Smirnova L., Wulczyn E.A., Michel G., Nitsch R., Krappmann D., Wulczyn F.G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 53.Diederichs S., Haber D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 54.Gao J., Wang W.Y., Mao Y.W., Graff J., Guan J.S., Pan L., Mak G., Kim D., Su S.C., Tsai L.H. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.