Abstract
There is a growing perception that long non-coding RNAs (lncRNAs) modulate cellular function. In this study, we analyzed the role of the lncRNA HOTAIR in mesenchymal stem cells (MSCs) with particular focus on senescence-associated changes in gene expression and DNA-methylation (DNAm). HOTAIR binding sites were enriched at genomic regions that become hypermethylated with increasing cell culture passage. Overexpression and knockdown of HOTAIR inhibited or stimulated adipogenic differentiation of MSCs, respectively. Modification of HOTAIR expression evoked only very moderate effects on gene expression, particularly of polycomb group target genes. Furthermore, overexpression and knockdown of HOTAIR resulted in DNAm changes at HOTAIR binding sites. Five potential triple helix forming domains were predicted within the HOTAIR sequence based on reverse Hoogsteen hydrogen bonds. Notably, the predicted triple helix target sites for these HOTAIR domains were also enriched in differentially expressed genes and close to DNAm changes upon modulation of HOTAIR. Electrophoretic mobility shift assays provided further evidence that HOTAIR domains form RNA–DNA–DNA triplexes with predicted target sites. Our results demonstrate that HOTAIR impacts on differentiation of MSCs and that it is associated with senescence-associated DNAm. Targeting of epigenetic modifiers to relevant loci in the genome may involve triple helix formation with HOTAIR.
INTRODUCTION
Mesenchymal stem cells (MSCs) comprise multipotent cells capable of differentiation towards adipogenic, osteogenic and chondrogenic lineage (1). Isolation procedures of MSCs and expansion to relevant cell numbers necessitate in vitro culture. However, culture expansion is associated with continuous and dramatic changes of the isolated cells: they acquire large and flat morphology, lose differentiation potential, and ultimately enter proliferation arrest—a state commonly referred to as replicative senescence (2,3). Therefore the state of cellular aging needs to be considered for quality control of MSCs, especially if intended for clinical application.
Replicative senescence is also reflected by highly reproducible epigenetic modifications, particularly in the DNA methylation (DNAm) pattern (4). Some of these senescence-associated DNAm (SA-DNAm) changes are almost linearly acquired during culture expansion and can therefore be used as biomarker for the state of cellular aging (5–7). Notably, SA-DNAm changes are enriched in developmental genes, such as homeobox genes, and they can be reversed by reprogramming into induced pluripotent stem cells (iPSCs) (4,7,8). This indicates that SA-DNAm changes—and hence also the process of replicative senescence—are somehow regulated, but the underlying mechanism is still unclear. We have recently described that SA-DNAm is frequently observed close to specific transcription factor binding sites (e.g. EGR1, TFAP2A, and ETS1) (9). Therefore, it is conceivable that such proteins guide epigenetic modifiers to specific sites in the genome based on protein–DNA interaction to mediate senescence-associated molecular changes.
Epigenetic modifications can also be mediated by long non-coding RNAs (lncRNAs; > 200 nucleotides), which play major roles in regulation of gene transcription, chromatin structure, and mRNA stability during cell development and diseases (10–12). Thousands of lncRNAs have been reported, but their precise function remains largely unknown. Recently, the lncRNA PANDA, which interacts with polycomb repressive complexes (PRC1 and PRC2) and the nuclear transcription factor Y subunit A (NF-YA), was shown to either promote or suppress senescence (13). Another relevant example is HOTAIR, a lncRNA transcribed from the HOXC locus that acts as a scaffold for histone modification complexes to coordinately interact with PRC2 and lysine-specific demethylase 1 (LSD1) (14,15). Thereby HOTAIR may mediate site-specific epigenetic modifications, particularly modifications in the histone code. Direct binding of RNA to chromatin for regulation of multiple gene expression events has already been proposed almost half a century ago (16). One concept describing how lncRNAs might target specific sites in the genome is based on nucleic acid triple-stranded structures (17). These triple helices are complexes of three oligonucleotide strands that may be implicated in transcriptional regulation, chromatin organization, DNA repair, and RNA processing (18–20). Triple helix complexes are formed by interactions of DNA-binding sites within the RNA through Hoogsteen or reverse Hoogsteen hydrogen bonds (21,22). The third strand can bind to the DNA double helix in either parallel or antiparallel manner containing a pyrimidine or purine motif (23). For example, the lncRNA Fendrr, which is implicated in regulation of murine mesoderm differentiation, has recently been suggested to form triple helices on the promoter sequence of two target genes: Foxf1 and Pitx2 (24,25).
HOTAIR may be involved in regulation of cellular senescence: this lncRNA was shown to be upregulated upon induction of senescence by either radiation or downregulation of SV40 large-T antigen activity in a fibroblast cell line (26). On the other hand, siRNA mediated knockdown of HOTAIR reduced expression of senescence-associated beta galactosidase (SA-β-gal) and other senescence markers (26). HOTAIR expression is elevated in multiple cancer types (e.g. breast and ovarian cancer (27,28), colorectal cancer (29), hepatocellular carcinoma (30), gastrointestinal stromal cancer (31), pancreatic cancer (32), laryngeal squamous cell carcinoma (33), and nasopharyngeal carcinoma (34)), which is usually associated with poor prognosis (29,32,35). Therefore, it is conceivable that HOTAIR expression supports escape of malignant cells from replicative senescence.
In this study, we addressed the role of HOTAIR in MSCs. We demonstrate that overexpression and knockdown of HOTAIR impact on in vitro differentiation and modulate gene expression as well as DNAm profiles of MSCs. Furthermore, we provide evidence that HOTAIR potentially regulates genes by targeting specific sites in the genome via purine motif triple helix formation.
MATERIALS AND METHODS
Cell culture
Bone marrow derived MSCs were isolated from caput femoris upon hip replacement surgery after written consent according to the guidelines approved by the Ethic Committee of RWTH Aachen University (Permit Number EK300/13) as described in detail before (8,36,37). Cells were cultured in DMEM low glucose medium (PAA) supplemented with 2 mM l-glutamine, 100 U/ml penicillin/streptomycin, 5000 U/ml heparin and 10% human platelet lysate in a humidified atmosphere at 5% CO2. All cell preparations were characterized with regard to immunophenotype and in vitro differentiation potential towards osteogenic and adipogenic lineages as described before (38).
Retroviral overexpression of HOTAIR
The plasmids lzrs-HOTAIR and lzrs-GFP were purchased from Addgene (Cambridge, USA; #26110 and #21961, respectively), transduced into Escherichia coli, and isolated by NucleoBond Xtra Maxi Prep (Macherey-Nagel, Düren, Germany). Amphotrophic Phoenix cells were treated with a suspension of 50% HOTAIR-plasmid, 10% gagPol-plasmid (pVPack-GP; Stratagene, Agilent Technologies, Santa Clara, CA, USA; #217566) and 40% env-plasmid (pVPack-10A1; Stratagene; #217570) suspended in 2× HBS and Calcium chloride. After 72 h the virus-containing supernatant was harvested and transferred onto non-confluent MSCs at passage 4. After 24 h transfection efficiency of usually >50% was observed in the GFP transfected controls using an Evos FL fluorescence microscopy (Life Technologies, Darmstadt, Germany).
siRNA mediated knockdown
MSCs at 80% confluent growth were transfected with a final concentration of 0.1 μM siRNA using 3% HiPerfect transfection reagent (Qiagen, Hilden, Germany) in serum free DMEM. ANRIL siRNA (si04378563; Qiagen); HOTAIR siRNA (5′-GAACGGGAGUACAGAGAGA-3′; MWG, Huntsville, USA); and the AllStars negative siRNA (AF488, Qiagen, Hilden, Germany), which has no human target sequence and carries an AlexaFluor488 molecule, were used as indicated in the text. Knockdown efficiency was determined after 48 h by qRT-PCR. For in vitro differentiation experiments the transfections were repeated each week over the course of differentiation.
Proliferation analysis
Five thousand MSCs were seeded per cm2 and after 5 days the cells were harvested by trypsinisation and counted in a Neubauer cell chamber.
Differentiation experiments
Cells were seeded at a density of 20 000 cells per cm2 and treated every 7 days with differentiation medium for 3 weeks (39). After 3 weeks cells were fixed with 4% paraformaldehyde and stained with Alizarin-red for analysis of osteogenic differentiation. Alizarin-red extraction was performed with acetic acid and ammoniumhydroxide and quantified at 405 nm on a Tecan plate reader (40). For adipogenic differentiation cells were fixed with 4% paraformaldehyde and stained with 10 μM diamidino-phenylindole (DAPI) and 1 μM boron-dipyrromethene (BODIPY) (Life Technologies; Thermo Fisher Scientific, Darmstadt, Germany). The percentage of cells that revealed fat droplets was determined using an Evos FL microscope as described before (41). To estimate statistical significance we utilized the paired Student's t-test.
RNA isolation and quantitative RT-PCR
Total RNA was extracted using NucleoSpin miRNA kit (Macherey-Nagel, Düren, Germany) and converted to cDNA using cDNA Reverse Transcription Kit (Applied Biosystems, Darmstadt, Germany). Quality of RNA was analyzed with the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, USA). Expression levels of HOTAIR and ANRIL were measured with a StepOnePlus Realtime PCR System (Applied Biosystems, Darmstadt, Germany) according to manufacturer's PCR program. Analyses were performed in duplicates using Taqman-probes for Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (hs02758991_g1, Life Technologies, Darmstadt, Germany), TATA Box Protein (TBP) (MWG Operon, Ebersberg, Germany), HOTAIR (5′ FAM-TTC TCT CGC CAA TGT GCA TAC TTA TAA G-3′BHQ1, MWG Operon) and ANRIL (hs03300534_m1, Life Technologies). Calculation of mRNA levels was performed using 2 deltaCT method versus the GAPDH or TBP.
Gene expression profiles
Five hundred ng of genomic RNA was analyzed with Human Gene 2.0 ST Array (Affymetrix, Santa Clara, USA). Data have been deposited at NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/; GSE69518). Preprocessing and quantile normalization (42) was performed with the oligo R package (43). Probes were mapped to Entrez gene IDs. In case of multiple mapped probes, we kept the probe ID with the highest variation according to its interquartile range. Initial differential expression analysis was performed with limma's moderated t-test (44) with an adjusted P-value of 0.05 (FDR), which indicated no differentially expressed genes. Alternatively, we selected genes with at least 1.2-fold differential expression (log2 fold change cutoff of 1.2). For gene set variation analysis, we used GSVA (45) with c2 collection (curated gene sets) from MsigDB (http://www.broadinstitute.org/gsea/msigdb)—a major advantage of this method is that it does not require a specific cutoff.
DNA methylation profiles
Genomic DNA was isolated from MSCs of early and late passage as described before (8). DNAm profiles were then analyzed with Infinium HumanMethylation450K BeadChips (Illumina, San Diego, USA) according to manufacturer′s instructions. Hybridization and initial data analysis was performed with BeadStudio Methylation Module at Life and Brain (Bonn, Germany). This platform represents >480 000 CpGs covering 99% of RefSeq genes and 96% of CGIs (46). Raw data have been deposited at GEO (GSE69518). Differentially methylated CpGs were identified using the minfi package (47). Preprocessing included quality control, removal of low quality samples (cutoff of 10.5), and quantile normalization. The moderated t-test (adjusted FDR < 0.05) revealed no significant differentially methylated CpGs and therefore we alternatively selected CpGs with at least 10% difference in DNAm level.
Genomic association analysis
We utilized 832 HOTAIR ChIRP-seq peaks that have been identified in MDA-MB-231 breast cancer cells (48), and SA-DNAm changes identified by 450k BeadChip technology in MSCs of early and late passage (8). To analyze whether SA-DNAm changes are enriched in the vicinity of HOTAIR ChIRP-Seq sites we calculated the number of differentially methylated CpGs within a maximal distance of 50, 250, 500, 1000, 1500, 2000, 2500, 3000, 6000 and 10 000 base pairs to at least one ChIRP-Seq peak and compared this to random distribution, which was obtained by counting the number of methylated sites in 10 000 random regions. Furthermore, we used the projection test (49) to evaluate the overlap between DNAm changes upon HOTAIR overexpression or knockdown with HOTAIR ChIRP-Seq regions. As background for the projection test we used all positions on the 450K BeadChips. P-values were corrected for multiple testing using a FDR < 0.05.
Association of DNAm changes with cancer datasets
DNAm profiles (all 450k Illumina BeadChip data) of 18 types of cancer with at least three samples of corresponding normal tissue (Supplementary Table S1) were obtained from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/). To determine association with DNAm changes upon modulation of HOTAIR, we tested if there was a significant difference in beta-value distribution between cancer and control samples at the CpGs that revealed 10% change in DNAm level upon either HOTAIR overexpression or knockdown. Statistical significance was evaluated with a Mann-Whitney test and a FDR of <0.05.
Detection of triple helix DNA binding domains
To find functionally enriched triple helix forming domains in HOTAIR, we used Triplex Domain Finder (TDF; http://www.regulatory-genomics.org/tdf/). For a given RNA and target DNA regions, TDF performs sequence based predictions of triple helices with Triplexator (50) (default parameters). As target DNA regions we analyzed: (i) HOTAIR ChIRP-Seq peaks, (ii) promoters of differentially expressed genes (1 kb upstream) or (iii) differentially methylated CpGs (±150 bp) after HOTAIR overexpression/knockdown. Next, TDF merges all RNA positions with triple helix binding sites to obtain candidate DNA binding domains. Significant enrichment (Fisher's Exact test) is then estimated in comparison to non-target regions (either all promoter regions or all positions on the 450K BeadChips, respectively). As there is no pre-defined background for ChIRP-Seq peaks, we randomly sampled 10 000 random genomic regions with same size as the ChIRP-Seq peaks. Then, we applied an empirical P-value to detect if the number of target regions of each predicted triple helix forming domain is higher than in random non-target regions (adjusted P-value < 0.05).
Electrophoretic mobility shift assay
Single nucleotide strands of DNA for predicted triple helix forming sites in PCDH7 and HOXB2, as well as relevant RNA strands with specific sequences of predicted binding domain II in HOTAIR, reverse sequences, or control RNA were purchased from Metabion (Steinkirchen Germany; Supplementary Table S2). For initial double strand hybridization complementary DNA single strands were incubated (1.1 equivalents were used of the pyrimidine-rich DNA strand in some experiments to further rule out the possibility of DNA:DNA-DNA triplex formation) in hybridization buffer (10 mM Tris–HCl with 50 mM NaCl and 10 mM MgCl2; pH 7.4) for 5 min at 95°C and then cooled down to room temperature. Triple helix formation was then performed by incubating 200 nM dsDNA with different concentrations of RNA single strands in hybridization buffer for 1 h at 60°C (if not indicated otherwise) and then cooling down to room temperature. For some experiments, we performed additional RNaseH treatment (RNaseH, New England BioLabs, Frankfurt am Main, Germany) after triplex formation at 30°C for 1 h. Inactivation of RNaseH was then performed by adding Proteinase K and incubation at 30°C for another 1 h. Four microliter of the reaction mixtures were analyzed on a 15% polyacrylamide gel and DNA was visualized by GelRed staining for 35 min.
RESULTS
HOTAIR is associated with senescence-associated DNAm changes
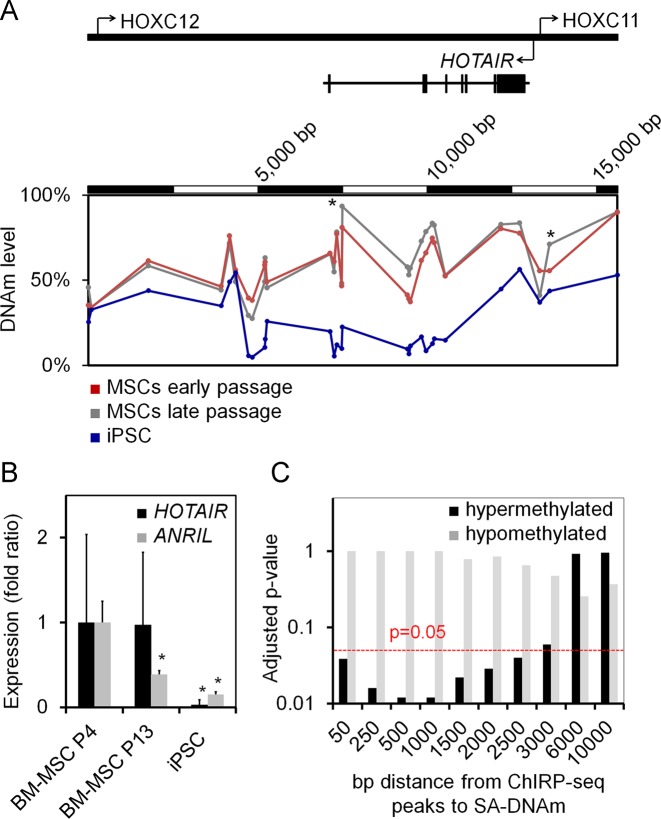
In our previous work, we have compared DNAm profiles of MSCs at early and late passage using 450k Illumina BeadChip technology (8). DNAm increased significantly at various CpG sites at the HOTAIR locus during culture expansion (Figure 1A). These findings are also in agreement with our MethylCap-seq analysis of fibroblasts at early versus late passage (9). However, these changes in DNAm were not very pronounced and not reflected on gene expression level: neither qRT-PCR analysis, nor RNA-sequencing analysis (9) demonstrated significant differential expression of HOTAIR in MSCs of early versus late passage, whereas HOTAIR expression was generally higher in MSCs than in fibroblasts (Supplementary Figure S1). For comparison we have analyzed ANRIL expression—a lncRNA previously shown to be downregulated upon replicative senescence (51)—and indeed, ANRIL expression decreased in MSCs of later passages (Figure 1B). Although HOTAIR did not reveal clear differential expression upon long-term culture we followed the hypothesis that it might be implicated in regulation of SA-DNAm changes. To select potential HOTAIR binding sites we utilized the previously published dataset based on chromatin isolation by RNA purification (ChIRP) in MDA-MB-231 breast cancer cells (48): 832 genomic regions were shown to be bound by HOTAIR and these were compared to our SA-DNAm changes in MSCs (8). In fact, HOTAIR binding sites in the ChIRP-seq dataset were significantly enriched around CpGs (±500 bp) with senescence-associated hypermethylation in comparison to randomly selected CpGs. In contrast, this enrichment was not observed at loci with senescence-associated hypomethylation (Figure 1C). However, it has to be taken into account that there may be cell-type specific differences in HOTAIR binding (52). These findings indicate that HOTAIR is associated with genomic regions that become hypermethylated during long-term culture of MSCs.
Figure 1.
HOTAIR binding is enriched close to senescence-associated hypermethylation. (A) DNA-methylation was measured in MSCs and iPSCs with Infinium Human Methylation450K BeadChips (n = 4) (8). These DNAm profiles revealed higher methylation levels in the HOTAIR locus in MSCs of later passage as compared to early passage, particularly at two CpGs (*P < 0.05; adjusted paired limma t-test; cg14691529 and cg06850442). (B) HOTAIR and ANRIL expression were further analyzed by qRT-PCR in MSCs of early (P4) and late (P13) passage (n = 3; normalized to MSC P4; *P < 0.05). (C) Senescence-associated DNAm changes in MSCs (hypo- and hypermethylated CpGs) were correlated with HOTAIR binding regions of a publically available ChIRP-sequencing dataset of MDA-MB-231 breast cancer cells (48). This analysis indicated that HOTAIR binding was significantly enriched in the vicinity of CpGs that are hypermethylated upon culture expansion in comparison to random selected CpGs (red line indicates P = 0.05 cut-off).
HOTAIR reduces in vitro differentiation of MSCs
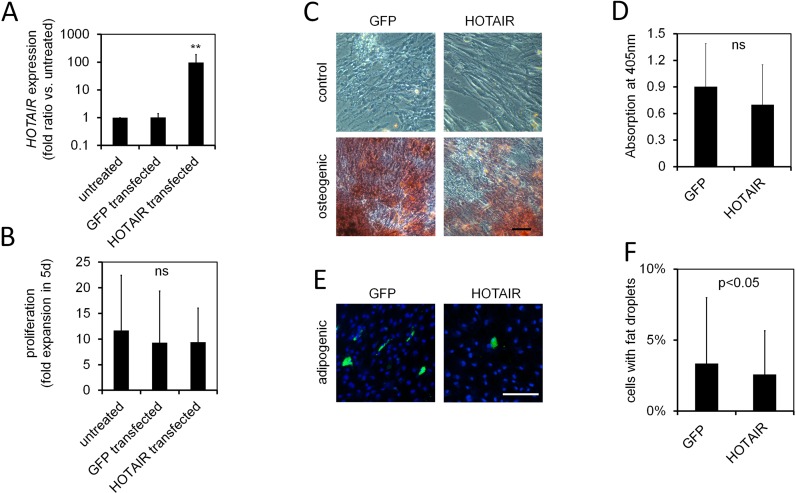
To further characterize the role of HOTAIR we overexpressed this lncRNA in MSCs at passage 4, using a retroviral system. Mean HOTAIR expression was 97-fold higher in quantitative RT-PCR as compared to either untreated MSCs or controls with corresponding GFP vector (n = 7; P < 0.005; Figure 2A). This overexpression did not have a significant impact on proliferation (Figure 2B) or cellular morphology (Supplementary Figure S2). Furthermore, it had no significant effect on osteogenic differentiation potential as determined by Alizarin-red staining after three weeks of in vitro differentiation (n = 11; Figure 2C and D). In contrast, adipogenic differentiation was reduced by HOTAIR overexpression: the proportion of cells with fat droplets (stained by the fluorescent dye BODIPY) was significantly lower than in the corresponding controls of the same MSC donor (n = 9; paired t-test: P < 0.05; Figure 2E and F).
Figure 2.
HOTAIR overexpression reduces adipogenic differentiation. (A) Retroviral overexpression of HOTAIR in MSCs of passage 4 revealed 97-fold upregulation by qRT-PCR analysis two weeks after transfection (GFP-insert was used for control; normalized to untreated control; n = 7). (B) Proliferation within 5 days (as determined by cell counting in a Neubauer cell chamber) was not significantly affected by overexpression (n = 5). (C and D) Osteogenic differentiation was not significantly affected by HOTAIR overexpression (Alizarin red extraction with acetic acid after 21 days; absorption measured on a plate reader; n = 11). (E and F) Adipogenic differentiation was reduced upon HOTAIR overexpression (percentage of BODIPY positive cells as compared to all nuclei counterstained with DAPI; n = 9). Paired t-test: *P < 0.05; **P < 0.01; Size bar = 100 μm.
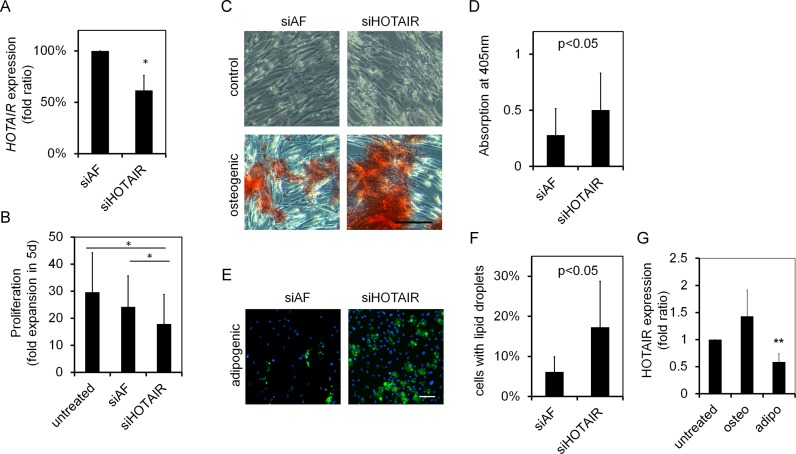
Alternatively, we used siRNAs to transiently knock down HOTAIR expression in MSCs. Quantitative RT-PCR analysis demonstrated that expression levels of HOTAIR decreased to 59% of non-specific controls (MSCs treated with siRNA containing an Alexa Fluor molecule; siAF; n = 9; P < 0.001; Figure 3A). HOTAIR knockdown significantly reduced proliferation (n = 5; P < 0.05; Figure 3B). Furthermore, osteogenic (n = 6; paired t-test: P = 0.041; Figure 3C and D) as well as adipogenic differentiation (n = 5; paired t-test: P < 0.005; Figure 3E and F) were increased by knockdown of HOTAIR. HOTAIR expression was subsequently analyzed upon in vitro differentiation of MSCs and it was reduced upon differentiation towards adipogenic lineage (n = 3; P < 0.05; Figure 3G). Taken together, HOTAIR exerts negative effects on in vitro differentiation of MSCs, particularly on differentiation toward adipogenic lineage.
Figure 3.
HOTAIR knockdown impairs proliferation and enhances in vitro differentiation. (A) Quantitative RT-PCR validation of siRNA mediated HOTAIR knockdown in MSCs after 48 h (normalized to controls with non-specific siRNA with Alexa Fluor (siAF); n = 9). (B) Proliferation of MSCs, as determined by cell counting after 5 days, was significantly reduced by HOTAIR knockdown (n = 5). In vitro differentiation towards osteogenic (C and D) and adipogenic (E and F) lineages was analyzed after 24 days (with repeated siRNA transfections at days 1, 8, 15 and 22). Alizarin red analysis indicated increased osteogenic differentiation upon HOTAIR knockdown (n = 6). Furthermore, the percentage of BODIPY-positive cells with fat droplets increased upon HOTAIR knockdown (n = 5). (G) HOTAIR expression in MSCs was significantly reduced upon differentiation of MSCs towards adipogenic lineage (n = 3; normalized to untreated MSCs). Paired t-test: *P < 0.05; **P < 0.01; size bar = 100 μm.
HOTAIR has a modulating effect on gene expression profiles
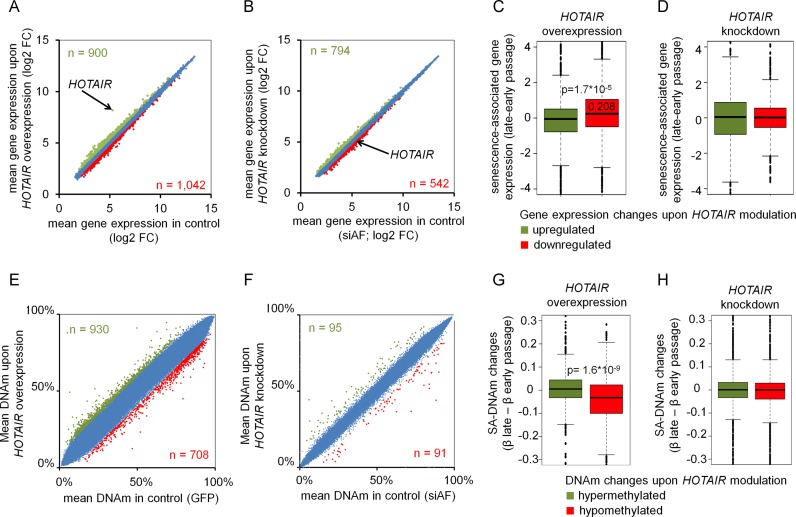
Effects of HOTAIR overexpression or knockdown in MSCs were subsequently analyzed using Affymetrix microarrays (2.0 ST Arrays; three biological replicas for each comparison). Overall, gene expression changes were very moderate and none of the genes reached statistical significance in adjusted paired limma t-test. Alternatively, we therefore applied a low cutoff (log2 fold change of 1.2) to estimate differential gene expression on a wider basis: HOTAIR overexpression resulted in slight upregulation of 900 genes and downregulation of 1042 genes (n = 3; compared to GFP control vector; Figure 4A). HOTAIR knockdown entailed slight upregulation of 794 genes and downregulation of 542 genes (n = 3; compared to siAF; Figure 4B). We observed consistent changes also in the individual replicas (Supplementary Figure S3A), but the differences were too small to be validated on single gene level (data not shown). HOTAIR revealed highest upregulation upon overexpression indicating that the data are reliable. Furthermore, gene expression changes upon overexpression or knockdown revealed a complementary tendency (Chi square test = 4.6 × 10−10; Supplementary Figure S3B), indicating that they are mildly associated with HOTAIR. We then performed gene set variation analysis (GSVA) to study the underlying gene sets (i.e. pathways and gene ontology terms) with expression changes (45). This method does not require lists of differentially expressed genes and can capture gene sets with moderate but coordinated expression changes. In fact, HOTAIR overexpression and knockdown revealed complementary association with PRC2 pathways (Supplementary Figure S4A). For example, HOTAIR overexpression was negatively associated with all H3K27me3, SUZ12 or PRC2 target gene sets, whereas the opposite effect was observed upon HOTAIR knockdown (P < 0.05). Furthermore, female cancer gene sets with lower grading, better prognosis and lower relapse were negatively associated with gene expression upon HOTAIR overexpression and positively associated with HOTAIR knockdown (Supplementary Figure S4B). Since HOTAIR is known to interact with PRC2 (28,53) and to be associated with poor prognosis in cancer (27–29,32,35), these findings provide further evidence that modulation of HOTAIR governs regulation of corresponding gene sets.
Figure 4.
HOTAIR modulates gene expression patterns and DNA methylation. Scatter plots demonstrate gene expression changes of MSCs upon either HOTAIR overexpression (A) or knockdown (B) in comparison to corresponding controls (means of three replicas). Genes with moderate upregulation and downregulation (log fold-change > 1.2) are indicated in green and red, respectively. HOTAIR revealed highest upregulation in overexpression data but was only moderately downregulated upon siRNA knockdown (indicated by arrows). (C) Genes that were either upregulated (900 genes; green) or downregulated (1042 genes, red) upon HOTAIR overexpression were analyzed in RNA-seq data of replicative senescent MSCs. Downregulated genes were significantly higher expressed at later passage (P = 1.7 × 10−5). (D) In contrast, genes that were either upregulated (794; red) or downregulated (542; green) upon HOTAIR knockdown revealed no significant changes in expression during replicative senescence. DNA methylation profiles of MSCs (P4) were analyzed upon HOTAIR overexpression or knockdown with Infinium Human Methylation450K BeadChips (Illumina, San Diego, USA). Scatter plots reveal DNAm changes upon (E) overexpression (compared to GFP control) and (F) knockdown (compared to fluorescence labelled negative antisense control; siAF). CpGs with >10% hypermethylation (green) and hypomethylation (red) are highlighted. (G) CpGs that were hypomethylated upon HOTAIR overexpression (708 CpGs) were overall significantly higher methylated in early versus later passages of MSCs (P = 1.6 × 10−9) (8). (H) CpGs with differential DNAm upon HOTAIR knockdown did not reveal a clear trend with senescence-associated DNAm.
To estimate whether differentially expressed genes upon either overexpression (Figure 4C) or knockdown of HOTAIR (Figure 4D) were associated with senescence-associated gene expression changes we analyzed up- and downregulated genes in mRNA-sequencing data of MSCs at early versus late passage (9). In fact, genes that were downregulated by HOTAIR overexpression were significantly higher expressed in MSCs of later passage as compared to MSCs of early passage (P-value 1.7 × 10−5; Mann–Whitney's test). In contrast, knockdown of HOTAIR or randomly selected groups of genes are not associated to gene expression changes during senescence (Supplementary Figure S5). Overall, these results reinforce previous findings that targets of HOTAIR are differentially expressed upon replicative senescence.
HOTAIR impacts on DNAm profiles
HOTAIR is implicated in regulation of the histone code, whereas it is largely unclear if HOTAIR is also an epigenetic modifier on DNAm level. Therefore, we analyzed DNAm profiles using 450k Illumina Bead Chips upon either HOTAIR overexpression or knockdown in MSCs. We applied a cutoff of 10% difference in mean DNAm level of two biological replicas: HOTAIR overexpression led to hypermethylation in 930 CpGs and hypomethylation in 708 CpGs (Figure 4E; Supplementary Figure S6). HOTAIR knockdown resulted only in 95 hypermethylated CpGs and 91 hypomethylated CpGs (Figure 4F). Enrichment of these differentially methylated CpGs in gene regions or with regard to CpG islands was analyzed with a hypergeometric test. Overall, HOTAIR overexpression and knockdown presented complementary associations: HOTAIR overexpression entailed hypomethylation in 3′ untranslated regions (3′UTR; 1.85-fold; P < 0.00005), whereas HOTAIR knockdown was rather associated with hypermethylation in 3′UTR (2.01-fold; P < 0.05). An inverted tendency was also observed for promoter regions, particularly within 200 bp upstream of the transcription start. CpGs not associated with CpG islands (open sea) were hypermethylated upon HOTAIR overexpression (1.4-fold; P = 1.2 × 10−23) and hypomethylated upon HOTAIR knockdown (1.52-fold; P < 0.0005; Supplementary Figure S7).
Next, we analyzed if HOTAIR-induced DNAm changes were enriched in HOTAIR binding sites (ChIRP-seq data (48)) using a projection test (49). Notably, there is a small but significant overlap of hypermethylated CpGs upon HOTAIR overexpression and HOTAIR binding regions in ChIRP-seq data (adjusted P-value = 0.021; Supplementary Figure S8; Table S3). These results support the notion that HOTAIR binding is associated with site specific methylation of DNA. Subsequently, we analyzed if DNAm changes upon modulation of HOTAIR impact on senescence-associated differentially methylated regions (SA-DMRs). DNAm levels of hypomethylated sites upon HOTAIR overexpression significantly decrease during cellular senescence (P = 1.6×10−9; Mann–Whitney's test; Figure 4G). DNAm changes upon knockdown of HOTAIR were not related to senescence-associated DNAm changes (Figure 4H). Overall, these results indicate that reduced HOTAIR binding to specific chromosomal regions upon replicative senescence may contribute to site-specific hypomethylation.
HOTAIR has been implicated in tumor progression. Accordingly we analyzed whether DNAm changes upon HOTAIR modulation are also specifically affected in cancer development. DNAm profiles of 18 different types of cancer as well as corresponding control tissue were obtained from The Cancer Genome Atlas (TCGA; Supplementary Table S1). Overall, CpGs that were hypomethylated upon HOTAIR overexpression revealed also lower DNAm levels in bladder urothelial carcinoma, lung squamous cell carcinoma, pheochromozytome, and sarcoma (adjusted P-value < 0.05; two way Mann-Whitney's test). CpGs that were hypermethylated upon HOTAIR overexpression were also higher methylated in cervical carcinoma, kidney renal papillary cell carcinoma, and prostate adenocarcinoma, (adjusted P-value < 0.05; two way Mann–Whitney's test). The association of DNAm changes upon HOTAIR overexpression with several types of cancer indicates that HOTAIR contributes to modification of DNAm patterns during cancer development.
Triple helix potential of HOTAIR
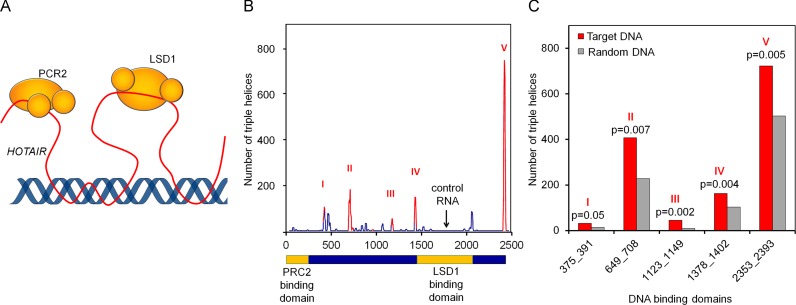
We hypothesized that site-specific modification of DNAm patterns via HOTAIR might be mediated by triple helix formation (Figure 5A). To address this hypothesis, we applied Triplex Domain Finder (TDF) to identify DNA-binding domains in HOTAIR. This new approach is based on triple helix predictions from Triplexator (50) in DNA sequences, i.e. genomic regions of HOTAIR ChIRP-Seq peaks (48). TDF calculates an empirical P-value to identify regions within the RNA that are more likely to form triple helices in the target DNA regions than in random DNA regions through Hoogsteen (parallel orientation) or reverse Hoogsteen (antiparallel orientation) hydrogen bonding. Triple helix prediction of the HOTAIR sequence within DNA regions of HOTAIR ChIRP-Seq peaks revealed five regions within the HOTAIR sequence that are significantly enriched for potential triple helix formation (adjusted P-value < 0.05; Figure 5B and C). Notably, none of these potential triplex forming domains overlapped with the protein binding domains for PRC2 (54) or LSD1 (53) as these may be spared for protein-DNA interaction. Particularly the DNA sequences that were predicted to interact with HOTAIR domains revealed enrichment for an A/G rich motif that is related to the de novo motifs previously described in HOTAIR bound DNA regions (Supplementary Figure S9) (48,53).
Figure 5.
Triple Helix forming potential of the HOTAIR sequence. (A) RNA might bind to a DNA double helix through reverse Hoogsteen hydrogen bonds and form a triple helix. (B) ChIRP-Seq-data of HOTAIR and the HOTAIR sequence were used for analysis with Triplex Domain Finder; Red peaks show significant DNA binding domains within the HOTAIR-sequence (P < 0.05); yellow boxes mark regions of RNA interaction with PRC2 and LSD1. (C) Number of triple helix binding sites for each significant DNA binding domain on ChIRP-Seq and random regions.
Subsequently, we analyzed whether similar triple helix forming domains are enriched either in promoter regions of differentially expressed genes or close to DMRs upon HOTAIR modulation. Therefore, we considered a region 1kb upstream of the transcription start site of differentially expressed genes or a 150 bp window around CpGs with DNAm changes, respectively. Based on these DNA regions the same triple helix forming domains were identified as calculated for ChiRP-Seq-peaks (Table 1). Notably, this analysis highlighted the same five regions that were predicted based on HOTAIR ChIRP-seq data. The most significant predictions for triple helix formations of HOTAIR were calculated for hypermethylated regions upon HOTAIR overexpression. In addition triple helix formation was calculated to occur more frequently in promoter regions of downregulated genes upon HOTAIR overexpression than in those being upregulated (Table 1). Thus, potential triple helix formation of HOTAIR seems to be associated with hypermethylation and downregulation of HOTAIR target genes, indicating that site specific DNAm changes might be targeted via triple helix formation.
Table 1. P-values for predicted triple helices within HOTAIR sequence.
| Predicted triplex binding domains (region in HOTAIR sequence) | ChIRP-Seq peaks | Up-regulated genes upon HOTAIR overexpression | Down-regulated genes upon HOTAIR overexpression | Up-regulated genes upon HOTAIR knockdown | Down-regulated genes upon HOTAIR knockdown | Hypermethylated regions upon HOTAIR overexpression | Hypomethylated regions upon HOTAIR overexpression | Hypermethylated regions upon HOTAIR knockdown | Hypomethylated regions upon HOTAIR knockdown |
|---|---|---|---|---|---|---|---|---|---|
| Domain I (375–391) | 0.04738 | 2.76E-06 | 1.12E-05 | ||||||
| Domain II (649–708) | 0.0069 | 1.27E-07 | 1.48E-15 | 1.68E-07 | 9.83E-19 | ||||
| Domain III (1123–1149) | 0.00115 | 0.00012 | |||||||
| Domain IV (1378–1402) | 0.0034 | 2.35E-25 | 1.11E-12 | 0.12952 | 3.80E-20 | 0.039 | |||
| Domain V (2353–2393) | 0.0046 | 8.35E-61 | 5.94E-49 | 2.54E-32 | 7.99E-40 |
This table demonstrates predicted triple helix forming domains within the HOTAIR sequence for various genomic regions (promoter regions of differentially expressed genes, or differentially methylated CpGs upon modulation of HOTAIR expression). This is in analogy to the previously described analysis with HOTAIR ChIRP-seq data. The domains I to V represent regions predicted to form triple helices on ChIRP-seq data (Figure 5B and C). Values in the table represent the P-value of the enrichment estimated with Triplex Domain Finder. Only significant P-values (<0.05) are depicted.
Triple helix formation in vitro
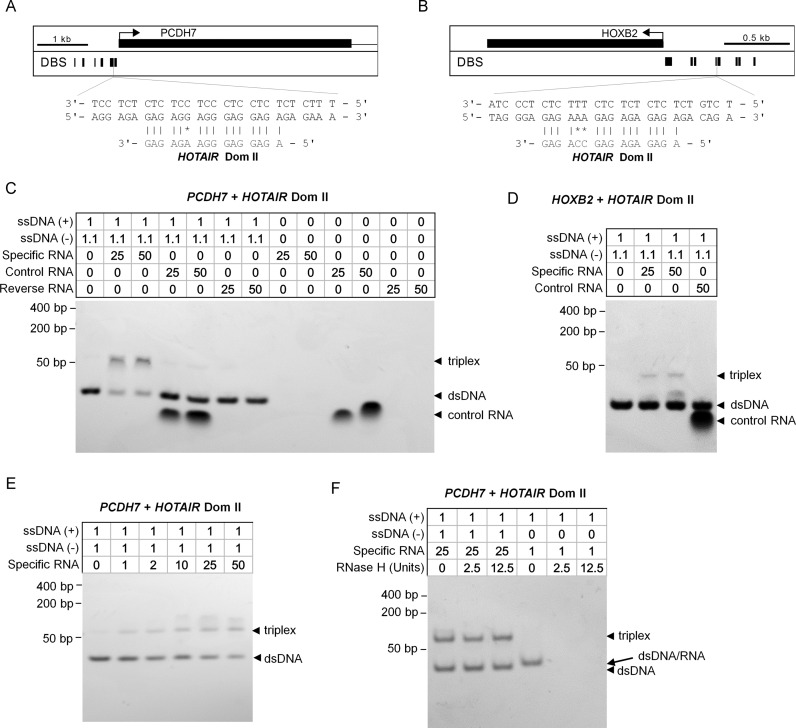
To further analyze triple helix forming potential of HOTAIR under in vitro conditions we performed an electrophoretic mobility shift assay (EMSA) (18,19,55,56). We chose two genes which are downregulated upon HOTAIR overexpression: protocadherin 7 (PCDH7) and homeobox B2 (HOXB2). PCDH7 was also demonstrated to be upregulated upon treatment with siHOTAIR in fibroblasts (57), and associated with reduced LSD1 binding (53). The latter study also indicates a loss of SUZ12 binding in the vicinity of HOXB2 promoter. For EMSA verifications we have chosen the largest triple helix binding sites in the promoter of these two genes and both of these were predicted to be associated to HOTAIR domain II (Figure 6A and B). For both double stranded target DNAs reduced mobility was particularly observed upon incubation with RNA of HOTAIR domain II, but not with RNA of the reverse sequence or additional control RNAs within the HOTAIR sequence (Figure 6C and D). Hybridization of dsDNA with RNA was performed at 60°C, but triplex formation was also observed if hybridization was performed at 50°C or 40°C, albeit less efficient (Supplementary Figure S10A). This is probably due to more efficient melting of the secondary structure of RNA or very slow kinetics of triplex formation. Additionally, we validated the melting temperature of the dsDNA by thermal denaturation experiments. In both cases the melting temperature was higher than the incubation temperature used for triple helix formation (PCDH7 = 79°C; HOXB2 = 74°C; Supplementary Figure S10B). Triplex formation increased with RNA concentrations (Figure 6E). Furthermore, we applied RNaseH to the complexes to demonstrate that the RNA strand is integrated into the triple helix through Hoogsteen hydrogen bonding and not Watson–Crick base pairing. A corresponding control shows digestion of the DNA-RNA duplex, which runs slightly slower than the dsDNA (Figure 6F). These EMSA assays support the notion that the predicted target regions in PCDH7 and HOXB2 form triplex structures with HOTAIR domain II in vitro.
Figure 6.
Electrophoretic mobility shift assays support triple helix formation. Computationally predicted triple helix forming sites in the promoter of (A) PCDH7 and (B) HOXB2 that were considered for in vitro validation. (C and D) Electrophoretic mobility shift assay of predicted binding domains (PCDH7 and HOXB2). Complementary oligodeoxynucleotides were preincubated to form double stranded DNA and then incubated with either specific RNA of predicted triplex binding domain II in HOTAIR, reverse RNA, or non-specific control RNA (the corresponding region of the control RNA is indicated in Figure 5B; notably, this RNA was also detected by Gel Red staining possibly due to hairpin formation; RNA was applied in 2-fold, 10-fold, 25-fold or 50-fold excess; 1.1 equivalents were used of the pyrimidine-rich DNA strand to reduce the possibility of DNA:DNA-DNA triplex formation. A mobility shift that indicates triplex formation was only observed with the specific sequences of HOTAIR domain II in both predicted target regions. (E) Triplex formation was then analyzed using different concentrations of RNA. (F) To rule out that conventional Watson-Crick hybridization between RNA and DNA results in mobility shift we treated the complexes with RNaseH. As expected, RNA in triplexes was protected from digestion, whereas it was digested in RNA:DNA double helices.
DISCUSSION
Functional analysis of specific lncRNAs is not trivial because their function is not directly related to a specific protein and they may simultaneously impact on a multitude of potential targets in a cell type dependent manner. This might explain why overall the molecular sequel of HOTAIR modulation with regard to specific genes or genomic regions was rather moderate. It has to be taken into account that 97-fold overexpression of HOTAIR is not physiologic and GFP could affect the gene expression profiles, too. Furthermore, siRNA mediated knockdown may evoke off-target effects and the impact on HOTAIR expression level is notoriously heterogeneous within MSC preparations. Other authors have indicated that modulation of HOTAIR evokes pronounced differences in gene expression (27,52), which might be attributed to the fact that these groups compared individual malignant cell lines rather than biological replicas. Differences between studies might also be due to tissue and cell type-dependent effects of HOTAIR (52). Furthermore, HOTAIR expression was even shown to differ in fibroblasts from different anatomical locations (58). Although the gene expression changes in this study were only very small and can hardly be validated on single gene level, they are overall significantly associated with senescence-associated gene expression, relevant GSVA categories, and predicted HOTAIR target regions.
So far, the functional role of HOTAIR was particularly addressed in cancer cell lines where HOTAIR knockdown has been correlated with: (i) cell cycle arrest through decrease of cells in S phase and gain of those in G2 phase (59), (ii) highly increased levels of apoptotic cells (26), and (iii) loss of proliferation rates (60). In human foreskin fibroblasts HOTAIR knockdown demonstrated loss of PRC2 occupancy and increased levels of HOXD (15,53). Homozygous Hotair knockout mice were viable and fertile, but revealed frequent abnormal skeletal phenotypes in spine and wrist, indicating that Hotair is also important for embryonic patterning of the skeletal system in vivo (61). Furthermore, Hotair seemed to be required for repression of HoxD genes and several imprinted loci such as Dlk1-Meg3 and Igf2-H19, without affecting imprinting choice (61). In analogy to our study, it has been shown that knockdown of HOTAIR impairs proliferation (62). The results of our study support the notion that HOTAIR is also important to modulate proliferation and differentiation of MSCs.
Several lncRNAs have recently been suggested to be involved in regulation of senescence, such as PANDA (13) and the mitochondrial lncRNA ASncmtRNA-2 (63). A recent study demonstrated upregulation of HOTAIR upon radiation-induced senescence in human fibroblasts (26). In our study, HOTAIR was not differentially expressed upon replicative senescence of MSCs – even though the HOTAIR locus revealed significant hypermethylation during long-term culture. This discrepancy may be attributed to the fact that DNAm is not generally reflected on gene expression level (64). Even if HOTAIR is not differentially expressed upon senescence, it may still be functionally relevant for senescence-associated DNAm changes: the activity of HOTAIR might be changed by alternative splicing, subcellular localization, or availability of other relevant interaction partners. On the other hand, several findings of this study substantiate that HOTAIR is involved in regulation of senescence-associated changes: (i) HOTAIR binding sites in ChIRP-seq data were significantly enriched in the vicinity of senescence-associated hypermethylation – even though these ChIRP-seq data were generated in a different cell line; (ii) genes that were downregulated by HOTAIR overexpression were associated with senescence-associated gene expression changes and (iii) overexpression of HOTAIR led to hypomethylation in CpGs that become hypomethylated during culture expansion. The loss of differentiation potential might also be associated with senescence-associated changes, but we did not observe typical morphological changes upon HOTAIR overexpression and no decay of proliferation. Furthermore, we tested our Epigenetic-Senescence-Signature that is based on DNAm levels at six CpGs (7) and the predictions of passage numbers were not affected by modulation of HOTAIR (results not shown). Taken together, these results suggest that HOTAIR is involved in regulation of senescence-associated DNAm changes—although it does not seem to be a unique trigger governing all processes of cellular senescence.
Triple helix formation of lncRNAs is one potential mechanism how epigenetic modifiers target specific sites in the genome (19,24,65,66). Our computational analysis based on a tool recently developed by our group (TDF; http://www.regulatory-genomics.org/tdf/) revealed 5 potential triple helix forming domains within the HOTAIR sequence, which do not overlap with the protein binding domains for PRC2 and LSD1. These findings are in line with the theory that lncRNA functionality is driven by their ability to combine distinct interaction domains in a single molecule (67,68). The predicted targets were significantly enriched in promoter regions of differentially expressed genes or close to differentially methylated CpGs. Furthermore, particularly the predicted triple helix binding domain II of HOTAIR has a binding motif starting with GAGA. As already indicated by other authors before, the GAGA motif is often found in Polycomb response elements – and hence, HOTAIR may be involved in recruitment of Polycomb to this motif (48). Last but not least, a band shift in EMSA indicates triple helix formation between two predicted target sites and HOTAIR domain II. These results provide evidence that triple helix formation is implicated in this process. It has to be noted that already more than 20 years ago it is been proposed that RNA sequences are not tolerated in purine motif triple helices (22,69). On the other hand, other authors more recently demonstrated that the lncRNA MEG3 (70) and microRNAs (71) form triple helices with purine motifs. In electromobility shift assays the possibility of strand rearrangement has to be taken into account. To further rule out misinterpretation of the band shifts we performed RNaseH treatment and thermal denaturation assays of the DNA double helices. The ultimate validation of triple helix formation, e.g. transfecting biotinylated lncRNA, cross-linking the cells and analyzing the DNA-lncRNA complexes under in vivo conditions (19), remains to be proven.
The molecular mechanism by which HOTAIR mediates epigenetic and transcriptomic changes still remains unclear. In principle, effects of HOTAIR on the DNAm pattern might be mediated by direct interaction with DNA methyltransferases—as also observed for other lncRNAs and rRNAs (19,72). On the other hand, we have recently observed that senescence-associated DNAm patterns are stochastically acquired in subpopulations of MSCs—they are not coherently modified at neighboring CpGs within the same MSC clone (Franzen et al., in revision). Therefore, it appears that at least senescence-associated DNAm patterns are rather changed indirectly, possibly mediated by chromatin changes evoked by the histone code. As mentioned above, HOTAIR is well known to interact with PRC2 and LSD1 (14,15). In fact, LSD1 may also modify the activity of DNMT1 (73). In ribosomal genes de novo CpG methylation by DNMT3B was shown to be guided by triple helix formation (19). PRC2 and LSD1 are rather involved in gene silencing and this is in line with the fact that triple helix predictions of HOTAIR domains were particularly observed in HOTAIR downregulated genes. Yet, they were also observed in some moderately upregulated genes and it has been shown that lncRNAs may also indirectly activate gene expression (70,74). Given the complexity of epigenetic regulation, it may be expected that HOTAIR can mediate different effects on higher order chromatin that are ultimately reflected in up- and downregulation of gene sets.
CONCLUSIONS
HOTAIR is involved in regulation of MSC functions, including proliferation and differentiation. Yet, overexpression or knockdown of HOTAIR evoked relatively few specific changes—it rather modulated gene expression patterns and DNAm profiles in a complementary way that reflected the previously described functional relevance. Furthermore, our results indicate that HOTAIR expression might contribute to regulation of cellular aging as it is associated with senescence-associated changes in gene expression and DNAm. Targeting of HOTAIR to specific sites in the genome seems to be mediated by triple helix formation. It is well known that HOTAIR is relevant for cancer development—this process may therefore involve triple helix mediated epigenetic modifications, particularly of genomic regions that are relevant for malignant transformation and escape of replicative senescence.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Else Kröner-Fresenius Stiftung [2010_A96; 2014_A193]; German Research Foundation (DFG) [WA 1706/2-1 and WE 1453/9-1]; German Ministry of Education and Research [BMBF; OBELICS]; Interdisciplinary Center for Clinical Research (IZKF); Start Program within the faculty of Medicine at the RWTH Aachen University as well as by the University College London Hospitals NHS Foundation Trust (UCLH), which received a proportion of its funding from the Department of Health NIHR Biomedical Research Centres (BRC) funding scheme. Funding for open access charge: University Hospital Aachen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 2.Wagner W., Horn P., Castoldi M., Diehlmann A., Bork S., Saffrich R., Benes V., Blake J., Pfister S., Eckstein V., et al. Replicative senescence of mesenchymal stem cells—a continuous and organized process. PLoS ONE. 2008;5:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roobrouck V.D., Ulloa-Montoya F., Verfaillie C.M. Self-renewal and differentiation capacity of young and aged stem cells. Exp. Cell Res. 2008;314:1937–1944. doi: 10.1016/j.yexcr.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Bork S., Pfister S., Witt H., Horn P., Korn B., Ho A.D., Wagner W. DNA Methylation Pattern Changes upon Long-Term Culture and Aging of Human Mesenchymal Stromal Cells. Aging Cell. 2010;9:54–63. doi: 10.1111/j.1474-9726.2009.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koch C.M., Wagner W. Epigenetic biomarker to determine replicative senescence of cultured cells. Methods Mol. Biol. 2013;1048:309–321. doi: 10.1007/978-1-62703-556-9_20. [DOI] [PubMed] [Google Scholar]
- 6.Schellenberg A., Mauen S., Koch C.M., Jans R., de W.P., Wagner W. Proof of principle: quality control of therapeutic cell preparations using senescence-associated DNA-methylation changes. BMC. Res. Notes. 2014;7:254. doi: 10.1186/1756-0500-7-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch C.M., Joussen S., Schellenberg A., Lin Q., Zenke M., Wagner W. Monitoring of cellular senescence by DNA-methylation at specific CpG sites. Aging Cell. 2012;11:366–369. doi: 10.1111/j.1474-9726.2011.00784.x. [DOI] [PubMed] [Google Scholar]
- 8.Koch C.M., Reck K., Shao K., Lin Q., Joussen S., Ziegler P., Walenda G., Drescher W., Opalka B., May T., et al. Pluripotent stem cells escape from senescence-associated DNA methylation changes. Genome Res. 2013;23:248–259. doi: 10.1101/gr.141945.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hänzelmann S., Beier F., Gusmao E.G., Koch C.M., Hummel S., Charapitsa I., Joussen S., Benes B., Brümmendorf T.H., Reid G., et al. Replicative senescence is associated with nuclear reorganization and with DNA methylation at specific transcription factor binding sites. Clin. Epigenet. 2015;4:19. doi: 10.1186/s13148-015-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA. Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi X., Sun M., Liu H., Yao Y., Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Postepska-Igielska A., Giwojna A., Gasri-Plotnitsky L., Schmitt N., Dold A., Ginsberg D., Grummt I. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 2015;60:626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Puvvula P.K., Desetty R.D., Pineau P., Marchio A., Moon A., Dejean A., Bischof O. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat. Commun. 2014;5:5323. doi: 10.1038/ncomms6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai M.C., Spitale R.C., Chang H.Y. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Britten R.J., Davidson E.H. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 17.Felsenfeld G., Davies D.R., Rich A. Formation of a three-stranded polynucleotide molecule. J Am. Chem. Soc. 1957;79:2023–2024. [Google Scholar]
- 18.Martianov I., Ramadass A., Serra B.A., Chow N., Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 19.Schmitz K.M., Mayer C., Postepska A., Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanak M., Alseiari M., Balasubramanian P., Addanki K., Aggarwal M., Noorali S., Kalsum A., Mahalingam K., Pace G., Panasik N., et al. Triplex-forming MicroRNAs form stable complexes with HIV-1 provirus and inhibit its replication. Appl. Immunohistochem. Mol. Morphol. 2010;18:532–545. doi: 10.1097/PAI.0b013e3181e1ef6a. [DOI] [PubMed] [Google Scholar]
- 21.Hoogsteen K. The structure of crysals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1959;12:822–823. [Google Scholar]
- 22.Escude C., Francois J.C., Sun J.S., Ott G., Sprinzl M., Garestier T., Helene C. Stability of triple helices containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res. 1993;21:5547–5553. doi: 10.1093/nar/21.24.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thuong N.T., Helene C. Sequence-specific recognition and modification of double-helical DNA by oligonucleotides. Angew. Chem. Int. Ed. Engl. 1993;32:666–690. [Google Scholar]
- 24.Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., Macura K., Blass G., Kellis M., Werber M., et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grote P., Herrmann B.G. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013;10:1579–1585. doi: 10.4161/rna.26165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon J.H., Abdelmohsen K., Kim J., Yang X., Martindale J.L., Tominaga-Yamanaka K., White E.J., Orjalo A.V., Rinn J.L., Kreft S.G., et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teschendorff A.E., Lee S.H., Jones A., Fiegl H., Kalwa M., Wagner W., Chindera K., Evans I., Dubeau L., Orjalo A., et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 2015;7:108. doi: 10.1186/s13073-015-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S., et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 30.Geng Y.J., Xie S.L., Li Q., Ma J., Wang G.Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int. Med. Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 31.Niinuma T., Suzuki H., Nojima M., Nosho K., Yamamoto H., Takamaru H., Yamamoto E., Maruyama R., Nobuoka T., Miyazaki Y., et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 32.Kim K., Jutooru I., Chadalapaka G., Johnson G., Frank J., Burghardt R., Kim S., Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D., Feng J., Wu T., Wang Y., Sun Y., Ren J., Liu M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am. J. Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 34.Nie Y., Liu X., Qu S., Song E., Zou H., Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z., Zhou L., Wu L.M., Lai M.C., Xie H.Y., Zhang F., Zheng S.S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 36.Koch C., Suschek C.V., Lin Q, Bork S, Goergens M, Joussen S, Pallua N., Ho A.D., Zenke M, Wagner W. Specific Age-associated DNA methylation changes in human dermal fibroblasts. PLoS ONE. 2011;6:e16679. doi: 10.1371/journal.pone.0016679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohmann M., Walenda G., Hemeda H., Joussen S., Drescher W., Jockenhoevel S., Hutschenreuter G., Zenke M., Wagner W. Donor age of human platelet lysate affects proliferation and differentiation of mesenchymal stem cells. PLoS ONE. 2012;7:e37839. doi: 10.1371/journal.pone.0037839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horn P., Bokermann G., Cholewa D., Bork S., Walenda T., Koch C., Drescher W., Hutschenreuther G., Zenke M., Ho A., et al. Comparison of individual platelet lysates for isolation of human mesenchymal stromal cells. Cytotherapy. 2010;12:888–898. doi: 10.3109/14653249.2010.501788. [DOI] [PubMed] [Google Scholar]
- 39.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 40.Gregory C.A., Gunn W.G., Peister A., Prockop D.J. An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal. Biochem. 2004;329:77–84. doi: 10.1016/j.ab.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Schellenberg A., Stiehl T., Horn P., Joussen S., Pallua N., Ho A., Wagner W. Population dynamics of mesenchymal stromal cells during culture expansion. Cytotherapy. 2012;14:401–411. doi: 10.3109/14653249.2011.640669. [DOI] [PubMed] [Google Scholar]
- 42.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 43.Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L., et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Favorov A., Mularoni L., Cope L.M., Medvedeva Y., Mironov A.A., Makeev V.J., Wheelan S.J. Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput. Biol. 2012;8:e1002529. doi: 10.1371/journal.pcbi.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buske F.A., Bauer D.C., Mattick J.S., Bailey T.L. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372–1381. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yap K.L., Li S., Munoz-Cabello A.M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M.J., Zhou M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heubach J., Monsior J., Deenen R., Niegisch G., Szarvas T., Niedworok C., Schulz W.A., Hoffmann M.J. The long noncoding RNA HOTAIR has tissue and cell type-dependent effects on HOX gene expression and phenotype of urothelial cancer cells. Mol. Cancer. 2015;14:108. doi: 10.1186/s12943-015-0371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu L., Murat P., Matak-Vinkovic D., Murrell A., Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry. 2013;52:9519–9527. doi: 10.1021/bi401085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fadloun A., Le G.S., Jost B., Ziegler-Birling C., Takahashi H., Gorab E., Carninci P., Torres-Padilla M.E. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- 56.O'Leary V.B., Ovsepian S.V., Carrascosa L.G., Buske F.A., Radulovic V., Niyazi M., Moertl S., Trau M., Atkinson M.J., Anastasov N. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015;11:474–485. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 57.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea M.D., Thomas K., Presser A., Bernstein B.E., van O.A., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rinn J.L., Bondre C., Gladstone H.B., Brown P.O., Chang H.Y. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS. Genet. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu J.J., Wang Y., Ding J.X., Jin H.Y., Yang G., Hua K.Q. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp. Cell Res. 2015;333:238–248. doi: 10.1016/j.yexcr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 60.Huang J., Ke P., Guo L., Wang W., Tan H., Liang Y., Yao S. Lentivirus-mediated RNA interference targeting the long noncoding RNA HOTAIR inhibits proliferation and invasion of endometrial carcinoma cells in vitro and in vivo. Int. J. Gynecol. Cancer. 2014;24:635–642. doi: 10.1097/IGC.0000000000000121. [DOI] [PubMed] [Google Scholar]
- 61.Li L., Liu B., Wapinski O.L., Tsai M.C., Qu K., Zhang J., Carlson J.C., Lin M., Fang F., Gupta R.A., et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang B., Su Y., Yang Q., Lv D., Zhang W., Tang K., Wang H., Zhang R., Liu Y. Overexpression of Long Non-Coding RNA HOTAIR Promotes Tumor Growth and Metastasis in Human Osteosarcoma. Mol. Cells. 2015;38:432–440. doi: 10.14348/molcells.2015.2327. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Bianchessi V., Badi I., Bertolotti M., Nigro P., D'Alessandra Y., Capogrossi M.C., Zanobini M., Pompilio G., Raucci A., Lauri A. The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in Endothelial Cells. J. Mol. Cell Cardiol. 2015;81:62–70. doi: 10.1016/j.yjmcc.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 64.Wagner J.R., Busche S., Ge B., Kwan T., Pastinen T., Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15:R37. doi: 10.1186/gb-2014-15-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bacolla A., Wang G., Vasquez K.M. New perspectives on DNA and RNA triplexes as effectors of biological activity. PLoS Genet. 2015;11:e1005696. doi: 10.1371/journal.pgen.1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li T., Mo X., Fu L., Xiao B., Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7:8601–8612. doi: 10.18632/oncotarget.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson R., Guigo R. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA. 2014;20:959–976. doi: 10.1261/rna.044560.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Semerad C.L., Maher L.J. III. Exclusion of RNA strands from a purine motif triple helix. Nucleic Acids Res. 1994;22:5321–5325. doi: 10.1093/nar/22.24.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E., et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paugh S.W., Coss D.R., Bao J., Laudermilk L.T., Grace C.R., Ferreira A.M., Waddell M.B., Ridout G., Naeve D., Leuze M., et al. MicroRNAs form triplexes with double stranded DNA at sequence-specific binding sites; a eukaryotic mechanism via which microRNAs could directly alter gene expression. PLoS Comput. Biol. 2016;12:e1004744. doi: 10.1371/journal.pcbi.1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di R.A., Ebralidze A.K., Benoukraf T., Amabile G., Goff L.A., Terragni J., Figueroa M.E., De Figueiredo Pontes L.L., Alberich-Jorda M., Zhang P., et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J., Hevi S., Kurash J.K., Lei H., Gay F., Bajko J., Su H., Sun W., Chang H., Xu G., et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 74.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.