Abstract
Aberrant stem cell-like gene regulatory networks are a feature of leukaemogenesis. The ETS-related gene (ERG), an important regulator of normal haematopoiesis, is also highly expressed in T-ALL and acute myeloid leukaemia (AML). However, the transcriptional regulation of ERG in leukaemic cells remains poorly understood. In order to discover transcriptional regulators of ERG, we employed a quantitative mass spectrometry-based method to identify factors binding the 321 bp ERG +85 stem cell enhancer region in MOLT-4 T-ALL and KG-1 AML cells. Using this approach, we identified a number of known binders of the +85 enhancer in leukaemic cells along with previously unknown binders, including ETV6 and IKZF1. We confirmed that ETV6 and IKZF1 were also bound at the +85 enhancer in both leukaemic cells and in healthy human CD34+ haematopoietic stem and progenitor cells. Knockdown experiments confirmed that ETV6 and IKZF1 are transcriptional regulators not just of ERG, but also of a number of genes regulated by a densely interconnected network of seven transcription factors. At last, we show that ETV6 and IKZF1 expression levels are positively correlated with expression of a number of heptad genes in AML and high expression of all nine genes confers poorer overall prognosis.
INTRODUCTION
The differentiation of haematopoietic stem cells (HSCs) into lineage-committed haematopoietic cells is driven by gene expression patterns that are tightly controlled by transcription factors (1,2). The faithful, cell type-specific, expression of these transcription factors enables a proper execution of the normal haematopoietic differentiation program. However, aberrations in gene regulatory networks frequently occur in leukaemias, leading to impaired differentiation and an expansion of an immature cell population (3,4). Chromosome translocations involving transcription factors or alterations in the chromatin structure of gene regulatory regions upstream of genes encoding haematopoietic transcription factors are frequently observed in leukaemias (5). Furthermore, mutations within coding sequences of transcription factors with important roles in developmental haematopoiesis, such as RUNX1 and CEBPA, also occur in various forms of leukaemia (6). Intriguingly, the gene expression signatures of leukaemic cells overlap more with normal HSCs than with those of the more differentiated progenitor cells from which they arise (7). A number of oncogenes have been shown to promote self-renewal or confer a stem cell-like gene expression profile in leukaemic cells (8). However, it remains poorly understood whether there might be differences in the underlying mechanisms by which leukaemic cells initiate and/or maintain a stem cell-like phenotype. Uncovering these differences has significant clinical implications, as it would pave the way for therapies that could specifically target leukaemic cells whilst sparing normal cells by leveraging underlying differences (9).
One contributing factor for our lack of understanding of the regulation of transcriptional networks in leukaemic cells is the limitation of the most commonly used technique in studying transcription factor—DNA interactions, the chromatin immunoprecipitation (ChIP) assay. In ChIP, proteins are formaldehyde cross-linked to in vivo bound regions of chromatin and an antibody specific to the protein of interest is used to immunoprecipitate the protein and isolate regions of the genome that are bound by that protein. ChIP has been an invaluable tool in determining genome-wide binding of many transcription factors and has contributed much to efforts in re-constructing gene regulatory networks. However it has an inherent limitation as an a priori discovery method to identify new proteins bound to a regulatory region, as it requires the use of antibodies against pre-determined proteins. In addition, as there are only a limited number of highly-specific, ChIP-grade antibodies available against transcription factors, there is a constraint to the number of transcription factors that can be practically studied by ChIP. Furthermore, despite the development of modified ChIP techniques such as sequential ChIP, studying multi-protein complexes with these methodologies remains very challenging. For these reasons, utilizing a complementary discovery method to study factors involved in regulating gene expression in leukaemic cells would improve our knowledge of the drivers of the aberrant stem cell-like transcriptional program in leukaemias.
One alternative to ChIP is digital genomic footprinting (10), a technique that can reveal all the regions in the genome that are bound by proteins but which does not reveal the identities of these bound proteins. In silico methods may be combined with digital genomic footprinting to predict which transcription factors that might be bound at these regions based on the presence of conserved DNA sequence motifs. However, a lack of experimentally validated data and insufficient knowledge of the binding motifs for the majority of the ∼1200 transcription factors encoded by the human genome, limit the power of such methods. In addition, as multiple members within a family of closely related transcription factors can bind the same motif, it remains difficult to unequivocally determine which proteins might be bound at a given motif. Furthermore, these methods can only inform us of factors that directly bind DNA sequences but not of those that bind indirectly through protein-protein interactions. New technological advances in mass spectrometry (MS) provide a means to overcome many of these limitations (11,12). In recent years, a number of groups have coupled traditional DNA affinity chromatography with sensitive MS methods to study DNA–protein interactions (13–15). Methods incorporating stable isotope labelling by amino acids in culture (SILAC), along with DNA pull downs and MS have been developed to study proteins binding to DNA (13) and to chromatin (16). These approaches have been extended to identify allele-specific binding of transcription factors to single nucleotide polymorphisms (17) as well as to discover specific binders of methylated and hydroxymethylated DNA (18,19).
As a first step towards further elucidating differences in the gene regulatory networks driving a stem cell-like program in leukaemic cells, we adapted a SILAC-based proteomics method (13), hereafter referred to as reverseChIP, to characterize the transcriptional regulation of ERG in leukaemic cells. ERG is an important regulator of haematopoiesis (20,21) which is also highly expressed in leukaemias. Murine models over-expressing ERG develop T-ALL and acute myeloid leukaemia (AML) (22–24) while high ERG expression is an independent predictor of poor outcomes in subsets of T-ALL and AML patients (25–27). Thus, while necessary for normal haematopoiesis, the failure to properly regulate ERG transcription contributes to leukaemogenesis. Previous ChIP studies have revealed that the binding of a heptad of transcription factors to the ERG +85 stem cell enhancer upstream regulates high ERG expression in healthy haematopoietic stem and progenitor cells (HSPCs) (2,28) as well in leukaemic cells (22,29). However, it is unknown if the heptad represents the entire complement of factors contributing to dysregulated ERG expression in leukaemic cells or whether additional factors might also be involved. By performing reverseChIP on the ERG +85 enhancer in T-ALL and AML cells, we have identified two new factors, ETV6 and IKZF1, as previously unknown binders and regulators of this region in leukaemic cells. We confirmed that these factors bind the +85 enhancer in vivo by ChIP, and used siRNA-mediated knockdown experiments to show that they promote high ERG expression in leukaemic cells. Consistent with our findings, we also discovered a positive correlation between high ETV6 expression and ERG expression in AML patients. Extending our studies to healthy CD34+ HSPCs, we found by ChIP that both ETV6 and IKZF1 are enriched at a number of haematopoietic enhancers, including the +85 enhancer. In summary, by using a quantitative proteomics-based discovery approach that is complementary to ChIP, we have discovered new transcriptional regulators of ERG.
MATERIALS AND METHODS
Cell culture
MOLT-4 and KG-1 leukaemic cell lines were grown in RPMI 1640 medium containing 10% Fetal Bovine Serum (FBS), 1% Glutamax and 1% penicillin-streptomycin. For SILAC experiments, cells were cultured in RPMI 1640 media (Thermo Fisher Scientific) containing either unlabelled/natural lysine and arginine (‘light’) or with L-Arginine.HCl (13C6, 99%) and L-Lysine.2HCl (13C6, 99%), designated as ‘heavy’ media. Isotope-labelled amino acids were purchased from Cambridge Isotope Laboratories Inc. (MA, USA). Cells were cultured in ‘heavy’ media for at least six passages for SILAC experiments. For CD34+ HSPCs, mobilized apheresis samples from normal donors were obtained from the Prince of Wales Hospital. The CD34+ fraction was purified using an automated CliniMACS cells separation system (Miltenyi Biotec, Bergist Gladbach, Germany). Collection of HSPCs from normal donors was approved by the Ethics Committee at the Prince of Wales Hospital and endorsed by the Human Research Ethics committee at the University of New South Wales, Sydney, Australia.
ReverseChIP
After metabolic labelling of cells by SILAC, nuclear protein extracts were prepared from labelled (‘heavy’) or unlabelled (‘light’) cultures using the NE-PER Kit (Thermo Scientific). Heavy nuclear extracts were assessed by MS to ensure complete labelling of proteins. The wild-type and mutant DNA baits were generated by polymerase chain reaction (PCR) using a 5′-biotinylated forward primer and an unbiotinylated reverse primer from plasmids containing the wild-type ERG +85 enhancer or mutant constructs, which had been previously generated (22). PCR amplicons were purified using the Qiaquick PCR Purification kit (Qiagen). Plasmid and primer details are provided in Supplementary Table S1. The remaining steps were done essentially as previously described (13). The wild-type or mutant DNA baits were separately immobilized onto streptavidin-coated magnetic beads (Dynabeads M-280, Life Technologies, using 300 μg of beads per 1.5 μg of dsDNA) at room temperature for 3 h and washed with Buffer DW (Tris pH 8, 2M NaCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 0.03% NP-40) to remove unbound DNA. The DNA-conjugated streptavidin beads were then blocked with Blocking Buffer (20 mM HEPES-NaOH, 0.05 mg/ml bovine serum albumin, 0.3M KCl, 0.02% NP-40, 5 mg/ml polyvinylpyrrolidone, 0.05 mg/ml glycogen and 2.5 mM dithiothreitol, DTT) at room temperature for 1 h with rotation. Pre-clearing of the nuclear protein extracts, prior to incubation with DNA baits was done as follows: first, potassium glutamate (to a final concentration of 10 mM) and one volume of Buffer G (20 mM Tris pH 7.4, 10% Glycerol, 0.1 M KCl, 0.2 mM EDTA, 10 mM potassium glutamate, 0.04% NP-40, 2 mM DTT, supplemented with protease and phosphatase inhibitor cocktails, Roche) containing 0.2 mg/ml poly dA.dT or poly dI.dC were added to 0.5 mg of labelled- or unlabelled-nuclear extract. Then, the extracts were incubated separately with washed, unconjugated, streptavidin beads for 1 h at 4°C with rotation in order to remove any non-specific binders to the beads. The unconjugated beads were then removed by magnetic separation and the resulting pre-cleared nuclear extracts were incubated with the appropriate DNA-conjugated, blocked, streptavidin beads for 3 h at 4°C, with rotation. The wild-type or mutant DNA-conjugated beads, with bound proteins, were washed four times with Buffer G and the beads were then combined. DNA-bound proteins were eluted by boiling in NuPAGE LD buffer (Life Technologies) at 95°C before being separated on a sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Bands were visualized by coomassie staining and the entire lane was diced into 1 mm pieces and subjected to in-gel trypsin digestion. The digested peptides were eluted and analysed by MS.
Mass spectrometry
All samples were analysed using an Orbitrap Velos (Thermo Electron, Bremen, Germany) mass spectrometer. The digested peptides were separated by nano-LC using an Ultimate 3000 RSLC and autosampler system (Dionex, Amsterdam, The Netherlands). Samples were concentrated and desalted onto a micro C18 precolumn (500 μm × 2 mm, Michrom Bioresources, Auburn, CA, USA) with H2O:CH3CN (98:2, 0.05% Trifluoroacetic acid (TFA) at 15 μl/min. After a 4 min wash the pre-column was switched (Valco 10 port valve, Dionex) into line with a fritless nano column (75 μm × ∼12 cm) containing C18 media (3 μ, 200 Å Magic, Michrom or 1.9 μ, 120 Å Dr Maisch, Ammerbuch-Entringen Germany). Peptides were eluted using a linear gradient of H2O:CH3CN (98:2, 0.1% formic acid) to H2O:CH3CN (64:36, 0.1% formic acid) at 200 nl/min over 36 min. High voltage (2000 V) was applied to low volume union (Upchurch Scientific) and the column tip positioned ∼0.5 cm from the heated capillary (T = 275°C). Positive ions were generated by electrospray and the Orbitrap operated in data dependent acquisition mode. A survey scan m/z 350–1750 was acquired in the Orbitrap (Resolution = 30 000 at m/z 400, with an accumulation target value of 1 000 000 ions) with lockmass enabled. Up to the ten most abundant ions (>5000 counts) with charge states > +2 were sequentially isolated and fragmented within the linear ion trap using collision induced dissociation with an activation q = 0.25 and activation time of 10 ms at a target value of 30 000 ions. M/z ratios selected for MS/ MS were dynamically excluded for 30 s.
Mass spectrometry data analysis
All LC-MS/MS data were anaylsed using MaxQuant (version 1.3.0.5 Max Planck Institute of Biochemistry, Martinsried, Germany). The analysis of protein identification and quantifications was carried out as described by the software user guide. Briefly, RAW files generated by Thermo mass spectrometer were uploaded into MaxQuant software and analysed using the following search parameter: (i) variable modifications: Oxidation (M), Acetyl (Protein N-term); (ii) fixed modification: Carbamidomethyl (C); (iii) labels: Arginine (R) Label: 13C6, Lysine (K) label: 13C6; (iv) enzyme specified was trypsin and allowing maximum two missed cleavages. All spectra were search using Uniprot database (UniProtKB/Swiss-Prot, release 2013) with common contaminants included. A one sample t—statistic for each protein was generated by first evaluating the heavy/light peptide log-ratios matched to the heavy/light protein ratio count, where ratio count is the number of peptides used for quantitation (30). The peptide log-ratio is then divided by the standard error, where ratio variability % is the standard deviation of peptides. All the ratio values were generated in MaxQuant. The following formula was used to yield the t—statistic:
 |
Under the null hypothesis that there is no change in abundance of the protein between the two populations and under the assumption that the distribution of the peptide log-ratios are normal. The P-value of each protein was then calculated using the Student-t distribution. For protein abundance—gene expression correlations, absolute protein amounts were calculated as the sum of all peptide peak intensities divided by the number of theoretically observable tryptic peptides (31). The log2 value of the normalized protein intensity was used to plot against microarray gene expression. Microarray gene expression data from leukaemic cell lines samples were obtained from CellLineNavigator (32). The raw expression files were pre-processed including background subtractions, quantile normalization, log2 transformation and batch effect removal using Partek Genomics Suite software (Partek Inc). Expression of genes was selected based on the corresponding probe with the maximum average expression across all samples. Correlations between gene expression values and quantitative MS abundance measurements were performed in ‘R’ and the line of best fit obtained by performing linear modelling using the ‘lm’ package in ‘R’.
Chromatin immunoprecipitation
ChIP assays were performed as previously described (29) using the following antibodies: ETV6 (Santa Cruz, sc-8546x), IKZF1 (BD AF4984), PU.1 (Santa Cruz, sc-352X) or, as control, non-specific goat IgG (Sigma, 15256). Quantitative PCR was performed to measure the level of enrichment at the ERG +85 enhancer region. A full list of primers used in ChIP-qPCR is provided in Supplementary Table S1. Data analysis was done using MxPro qPCR software (Agilent Techchologies) and relative expression levels were calculated using a standard curve (range: 0.4–50 ng of the input material). Enrichment values were normalized to IgG.
Stable transfection assays
Transfections of MOLT-4 and KG-1 cells (5–10 × 106 cells/pulse) were performed using standard electroporation conditions (900 μF / 220 V on a BioRad Gene Pulser Xcell™). Cells were co-transfected with 10 μg of linearized plasmid DNA (wild-type pGL2p ERG+85 or mutants) and 1 μg pGKNeo encoding the Neomycin resistant gene. Cells were cultured under normal conditions. Geneticin (G418) (Life Technologies) was added to stable transfected cells 48 h after electroporation at a concentration of 750 μg/m of media. After 15 days, 1 × 106 cells were harvested and luciferase activity was measured using a microplate luminometer (GloMax96, Promega) as previously described (29).
Gene knockdown experiments and gene expression analyses
siRNA knockdowns of ETV6 and IKZF1 were performed in a KG-1 cell line stably transfected with a pGL2p ERG +85 reporter. siRNA transfections were performed using the Neon® transfection system (Life Technologies). All siRNA (purchased from Qiagen) were labelled with Cy3 using the Silencer siRNA labelling kit (Life Technologies) and 0.3 μM of Cy3-labelled siRNA was used per transfection. One hour after electroporation, Cy3-positive cells were isolated using a BD Influx™ cell sorter and allowed to grow for 72 h. RNA was extracted using the RNeasy Mini kit (Qiagen) and complementary DNA synthesis was performed using standard methods as previously described (29). Knockdown of target genes was measured by quantitative real time-PCR. Relative expression levels were calculated using a standard curve (range: 0–50 ng) for each gene and normalized against housekeeping genes as per established guidelines (33).
Lentiviral-mediated shRNA knockdown vectors were purchased from Sigma and the hairpins were subcloned into a modified pLKO.1 vector in which the puromycin resistance cassette was replaced with green fluorescent protein (GFP). Lentiviral particles were produced by co-transfecting 293T cells with the shRNA clone and packaging plasmids (34). Viral supernatants were generated by culturing the 293T producer cells in medium appropriate for the target cells for 16 h and 0.45 μm—filtered viral supernatants were then used immediately for target cell transduction. For MOLT-4 transductions, two rounds of transduction were performed on sequential days. Cells were resuspended in viral supernatant supplemented with 8 μg/ml protamine and incubated at 37°C until the following day. After two rounds of transduction, cells were washed and resuspended in RPMI supplemented with 10% FBS, 2 mM glutamine and 100 U/ml penicillin/streptomycin and incubated at 37°C for a further 72 h prior to RNA extraction. The transduction efficiency, as measured by the percentage of GFP+ cells, was generally 95% or more.
Primary CD34+ isolation, transduction and OP-9 stromal co-culture were done essentially as previously described (35–37). Cord blood CD34+ cells were thawed and pooled, then cultured overnight in IMDM supplemented with 20% FBS, 2 mM glutamine, 50 μg/ml gentamycin, 100 ng/ml human stem cell factor (SCF, Peprotech), 50 ng/ml hrombopoietin (TPO, Peprotech) and 100 ng/ml FLT3 ligand (FLT3L, Peprotech). Two rounds of transduction were performed on sequential days. Cells were resuspended in viral supernatants supplemented with 8 μg/ml protamine, 100 ng/ml SCF, 100 ng/ml TPO and 100 ng/ml FLT3L and spun on RetroNectin (Takara) coated plates at 250 g for 90 min at 30°C. Plates were then incubated at 37°C until the following day. After two rounds of transduction, cells were washed and resuspended in fresh medium containing cytokines and incubated at 37°C for a further 72 h. GFP positive cells were sorted using a FACS Aria IIIu (Becton Dickinson) and RNA was extracted immediately after sorting. For assessment of proliferation potential of ETV6 knockdown cells in culture, transduced cells were grown in a cytokine-driven medium essentially as previously described (37). Aliquots of cells were collected at different time points during a 6-day period and GFP expression status was assessed by flow cytometry.
For CD34+ transduction experiments followed by OP-9 stromal co-culture, a lentiviral shETV6 vector (Sigma-Aldrich) was used to knock down ETV6. As a non-targeting shRNA control, MISSION pLKO.1-puro Non-Mammalian shRNA Control Plasmid was used. Virus production was performed in HEK293TN cells using JetPEI polyplus with pMD2.G (envelope plasmid), psPAX2 (packaging plasmid) and MISSION control shRNA or MISSION shETV6 vector (target plasmid) in 0.1/0.9/1 ratios. CD34+ cord blood cells were brought into culture and transduced the next day with the lentiviral viruses. Forty-eight hours after transduction, GFP expressing (i.e. transduced) cells were co-cultured with OP9 stromal cells. Another 72 h later, transduced cord blood cells were sorted based on GFP and hCD45 positivity on a BD FACSAria II sorter.
Western blots
Proteins were separated on SDS-PAGE gels and transferred onto nitrocellulose membranes using the iBlot® Gel Transfer Device (Life Technologies) at 20V for 8 min. Western blot analysis was performed using the following primary antibodies: ERG (Santa Cruz, sc-354x), ETV6 (Abcam, ab64909), IKZF1 (BD, AF4984), PU.1 (Santa Cruz, sc-352x), GATA2 (Santa Cruz, sc-9008x), RUNX1 (Abcam, ab23980) and TOPO1 (BD Pharmingen, 556597). The following secondary antibodies were used: polyclonal goat anti-rabbit HRP (Darko, P0448), polyclonal rabbit anti-mouse HRP (Darko, P0260), polyclonal rabbit anti-goat HRP (Darko, P0449). Band detection and quantification were performed using an ImageQuant LAS4000 machine (GE Healthcare Life Sciences, Uppsala, Sweden).
AML patient data analysis
The raw expression data for cytogenetically normal AML patients (38) was extracted and pre-processed using Partek Genomics Suite (v6.6) including background subtractions, quantile normalization and log2 transformation. The normalized expression levels were processed in Matlab (R2015b) and Pearson's linear correlation coefficient were computed between different probesets/genes. The normalized expression levels were further processed in Matlab (R2015b) and Prism (v6.07) to calculate overall survival differences using the Kaplan–Meier estimator. In case of single gene analysis (i.e. ETV6 or IKZF1 alone), patient subgroups were first stratified by the mean expression of the analysed gene. In case of multi-gene analysis, patient subgroups were first partitioned using the k-means clustering algorithm with k = 2. Patient subgroups were then compared using the Log-rank (Mantel-Cox) test and Kaplan–Meier curves were generated using Prism.
RESULTS
Identifying binders of the ERG +85 enhancer in MOLT-4 cells using reverseChIP
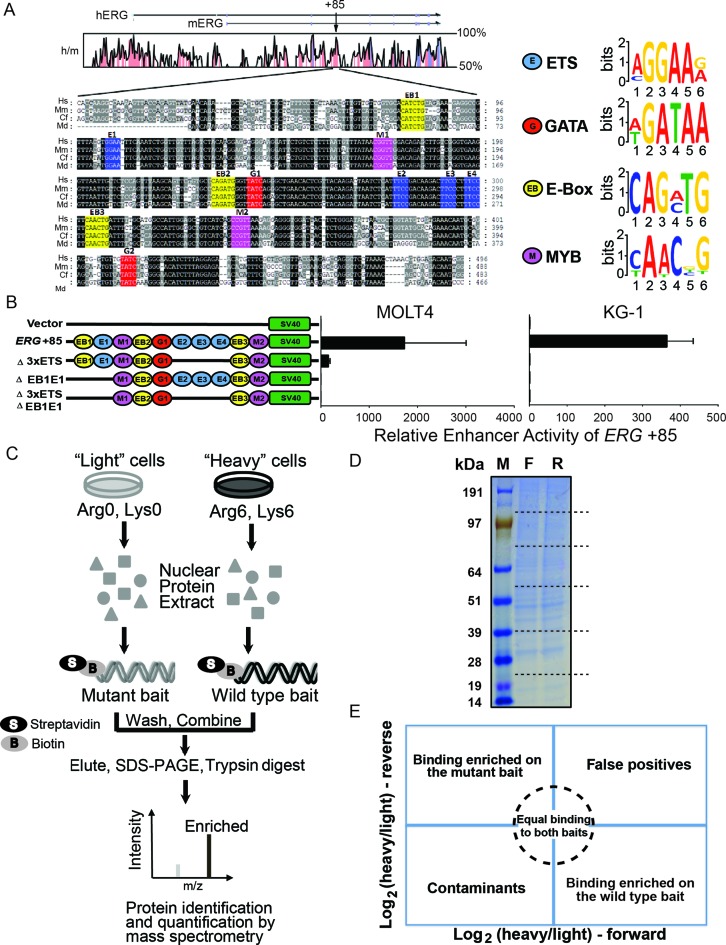
The highly conserved ERG +85 stem cell enhancer, henceforth referred to as the +85 enhancer, contains consensus motifs for a number of different transcription factors (Figure 1A) and strongly drives the expression of ERG in healthy HSPCs (28) as well as in T-ALL (22) and AML (29). Mutational studies presented here (Figure 1B) and previously (22,29), have identified specific regions within the +85 enhancer that are necessary for full transcriptional activity in T-ALL and AML cells. Prior ChIP studies have revealed that a heptad of transcription factors (FLI1, ERG, GATA2/3, RUNX1, SCL, LYL1 and LMO2) binds the +85 enhancer in healthy HSPCs as well as in T-ALL and AML cells, including in the cell lines MOLT-4 and KG-1 (2,22,28,29). However, the identities of the specific factors that bind within the sub-regions identified as important for transcriptional activity of the +85 enhancer is unknown. Furthermore, it remains unknown whether any additional factors beyond the heptad also bind the +85 enhancer and might be involved in regulating high ERG expression in leukaemic cells.
Figure 1.
The ERG +85 stem cell enhancer and reverseChIP methodology. (A) Multiple sequence alignment of the highly conserved ERG +85 enhancer, with human (Hs), mouse (Mm), dog (Cf) and opossum (Md) sequences, adapted from (22,29), is shown. The position of the +85 enhancer is shown relative to the start of ERG for both human (hERG) and mouse (mERG), along with human and mouse conservation, at the top. The sequences highlighted in black have 100% sequence conservation while the grey regions are less conserved. Consensus motifs for different transcription factors are also indicated, including ETS (blue, E1, E2, E3 and E4), GATA (red, G1, G2), E-Box (yellow, EB1, EB2 and EB3) and MYB (magenta, M1 and M2). Predicted sequence motifs are depicted on the right. (B) Mutational analysis of the +85 enhancer. The schematic on the left depicts the transcription factors motifs which were present on the different +85 enhancer constructs that were tested in stable transfection assays in MOLT-4 (left) and KG-1 (right) cells. (C) Schematic of the reverseChIP method. Metabolically labelled (‘heavy’, Arginine-6 and Lysine-8) or unlabelled (‘light’) cells were grown and nuclear protein extracts prepared. The extracts were separately incubated with biotin-labelled wild-type or mutated DNA baits and conjugated onto streptavidin beads. The beads were then washed, combined and bound proteins were eluted. Eluates were run on a sodium dodecyl sulphate-polyacrylamide gel electrophoresis gel, the lane cut into multiple pieces and proteins digested in-gel with trypsin. Peptides were recovered and analysed by mass spectrometry (MS). (D) A representative gel indicating equal protein yields in the eluates from forward and reverse experiments. The lane was cut into slices (putative cut sites indicated as dotted lines). (E) A schematic illustrating enrichment biplots generated from parallel, order-swapped, reverseChIP experiments with the location of binders based on specificity of binding.
In order to shed more light on these unknowns, we adapted a recently developed, SILAC-based, quantitative MS method (13) to interrogate the +85 enhancer. We modified the original protocol at various steps in order to improve the sensitivity and workflow of the technique. First, by using a nuclear protein extraction kit that efficiently extracts nuclear proteins in a native, non-denatured form, thereby preserving their functionality, we were able to circumvent the laborious nuclei preparation steps or a need to dialyse away detergents and renature proteins that were employed in the original protocol. Next, by using biotin-labelled PCR primers to generate amplicons, we could quickly generate biotinylated baits from a variety of genomic templates without requiring the in vitro re-annealing of single-stranded DNA fragments that has been previously employed. As presented later in this manuscript, this innovation also enabled us to easily generate large baits from the entire +85 enhancer in order to identify binders to the entire enhancer region. At last, by directly eluting DNA-bound proteins off streptavidin beads, we averted the need to perform restriction digests to release DNA (and DNA-bound proteins) from the beads, a critical last step that is very sensitive to buffer conditions and could adversely decrease elution yields. As outlined in Figure 1C, the method we have refined (henceforth ‘reverseChIP’) involves generating biotinylated wild-type or mutated DNA fragments of the +85 enhancer by PCR using biotin-tagged primers, followed by immobilization of the amplicons onto streptavidin-coated magnetic beads. The wild-type or mutant DNA-conjugated beads are then incubated separately with metabolically-labelled (‘heavy’) or unlabelled (‘light’) nuclear protein extracts, followed by stringent washes to remove non-specific background proteins. The beads are then combined, DNA-bound proteins eluted and fractionated by SDS-PAGE, entire gel lanes cut into multiple pieces and an in-gel trypsin digestion is performed. The digested peptides are then analysed by quantitative MS to identify specific binders to each DNA fragment (Figure 1C).
As a proof-of-principle, we initially interrogated a region of the +85 enhancer containing three tandem ETS-binding motifs (E2, E3, E4, collectively 3×ETS), which when mutated abrogates transcriptional activity of the enhancer in MOLT-4 T-ALL cells (Figure 1B and (22)). Using PCR primers flanking the 3×ETS motifs, we generated a 91 bp wild-type fragment and a 75 bp mutant bait lacking the 3×ETS motifs (designated as ‘Δ3×ETS’, Figure 1B) from appropriate templates (listed in Supplementary Table S1) to perform reverseChIP. To provide an additional level of stringency in distinguishing truly specific binding from spurious enrichment, we ran two parallel experiments: a forward experiment where ‘heavy’ proteins were incubated with wild-type bait and ‘light’ proteins were incubated with the mutant bait; and a reverse experiment where the order was swapped (i.e. ‘light’ proteins with wild-type bait and ‘heavy’ proteins with mutant bait). Approximately equal quantities of proteins were eluted in forward and reverse experiments, as can be seen in a representative gel image (Figure 1D). To easily visualize binders of each bait, a two-dimensional logarithmic plot of enrichment values for the different binders in a forward and reverse experiment can be plotted (Figure 1E). Factors enriched on the wild-type fragment would have a log2 (‘heavy’/‘light’) ratio > 0 in the forward experiment and a ratio < 0 in the reverse experiment, thus falling within the bottom right quadrangle (Figure 1E); non-specific binders would have approximately equal ‘heavy’/‘light’ ratios in both experiments and will therefore be present around the origin, within the circled region; while factors that are enriched on the mutant bait would have a ratio < 0 in the forward experiment and >0 in the reverse experiment, thereby falling within the top left quadrangle in Figure 1E. Externally contaminating proteins, such as keratin, would always be unlabelled (i.e. ‘light’ in both forward and reverse experiments) and consequently lie in the bottom left quadrangle while false positives would lie in the top right quadrangle.
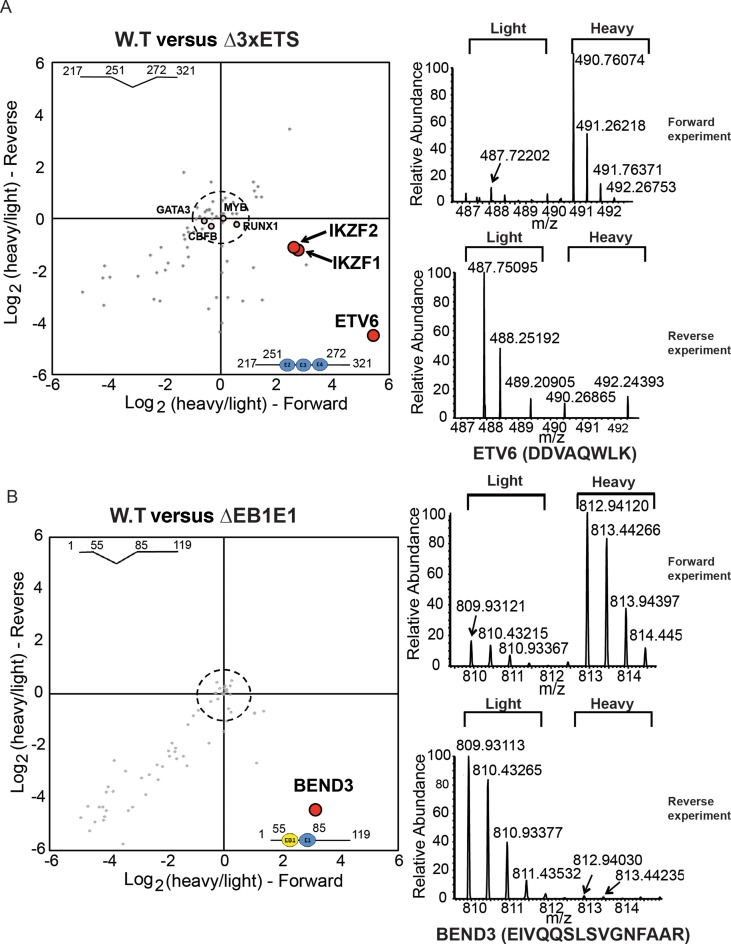
Focusing on factors with known roles in transcriptional regulation, we have identified ETV6, an ETS-family transcription factor that is an essential regulator of normal haematopoiesis (39,40) but which is also mutated in leukaemias including in an aggressive form of T-ALL (41,42), as a specific binder of the 3×ETS region in MOLT-4 cells (with log2 enrichment values 5.44 and −4.51 in the forward and reverse experiments respectively and a P-value for enrichment: 1.23×10−7, Figure 2A). This is the first report to identify ETV6 binding at the +85 enhancer. We have also identified the Ikaros zinc finger transcription factor, IKZF1, another critical regulator of haematopoiesis (43) which is also frequently mutated in ALL patients with a high risk of relapse (44), as a factor enriched on the wild-type bait (Figure 2A). Additionally, we detected the binding of a closely related family member, Helios/IKZF2, on the wild-type bait. Closer sequence analysis of the 3×ETS fragment showed the previously missed presence of the IKZF1 core binding motif (GGGAA) within the region disrupted on the mutant bait. IKZF1 and IKZF2 are known co-interactors which bind DNA together in a complex (45,46), and the nearly identical enrichment values for IKZF1 and IKZF2 on the wild-type bait (log2 enrichment values for IKZF1: 2.75 and −1.22 and IKZF2: 2.60 and −1.10, and a P-value for enrichment: 1.06 × 10−5, Figure 2A) are consistent with this. We also detected the binding of other previously known haematopoietic regulators, such as RUNX1, CBFB, GATA3 and MYB. However, unlike ETV6 and IKZF1 or IKZF2, these other factors were equally enriched on both the wild-type and mutant baits and are therefore situated near the origin (Figure 2A), indicating that the deletion of the 3×ETS motifs has not abrogated their binding sites. RUNX1 and CBFB bind DNA together as a heterodimeric transcriptional complex termed the core binding factor, with RUNX1 involved in direct DNA binding (47,48). Consistent with this, we observed both these factors binding with very similar enrichment values in our reverseChIP experiment. The full list of binding proteins to the 3×ETS bait is provided in Supplementary Table S2.
Figure 2.
ReverseChIP on two regions within the ERG +85 enhancer in MOLT-4 cells. (A) The enrichment biplot on the left shows the results from parallel reverseChIP experiments on the Δ3×ETS region in MOLT-4 cells. Representative mass spectra for ETV6 (for the exemplary peptide: DDVAQWLK) is shown on the right to illustrate specific enrichment on the wild-type bait in both forward and reverse experiments. (B) ReverseChIP results of the ΔEB1E1 region in MOLT-4 cells. The representative mass spectra on the right (for an exemplary peptide: EIVQQSLSVGNFAAR) illustrates specific enrichment for BEND3 on the wild-type bait.
Another region within the +85 enhancer that is essential for full transcriptional activity contains an E-box and an ETS-motif (‘EB1’ and ‘E1’ respectively, EB1E1 collectively). By PCR, we generated a 116 bp wild-type bait and a 92 bp mutant bait lacking the EB1E1 motifs (‘ΔEB1E1’, Figure 1B), which we used in reverseChIP. We have identified, for the first time, the BEN-domain containing protein BEND3 as a highly enriched binder of the wild-type bait (P-value for enrichment: 2.83 × 10−14, Figure 2B). BEND3 has been recently discovered to recruit the PRC2 complex to pericentromeric regions (49) and is implicated in establishing higher-order chromatin structure (50). While a DNA binding consensus motif (TCYAATHRGAA) has been identified for a Drosophila melanogaster BEN-domain containing nuclear co-repressor, Insensitive (51), no analogous consensus motifs are known for BEND3 nor could we find any matches to the Drosophila consensus motif within the EB1E1 region. BEND3 has never previously been implicated in regulating transcription of haematopoietic genes. The full list of binding proteins to the EB1E1 bait is provided in Supplementary Table S2.
Interrogating the entire ERG +85 enhancer region with reverseChIP
Having established that the reverseChIP method is capable of discovering new transcription factors binding to small regions of interest within the ERG +85 enhancer, we explored whether the method could be extended to study the full length enhancer, a genetically complex region larger than 300 bp containing binding sites for a number of transcription factors (Figure 1A). In order to limit non-specific binding of proteins to DNA probes of interest, previous MS-based studies of DNA binding factors had restricted the analyses to 40–60 bp DNA probes (13,17,19,52,53). Extending the capability of the method to study larger genomic fragments would not only facilitate our characterization of the entire repertoire of factors binding the ERG +85 enhancer, but would also lay the groundwork for future studies of other less-well characterized genomic regions.
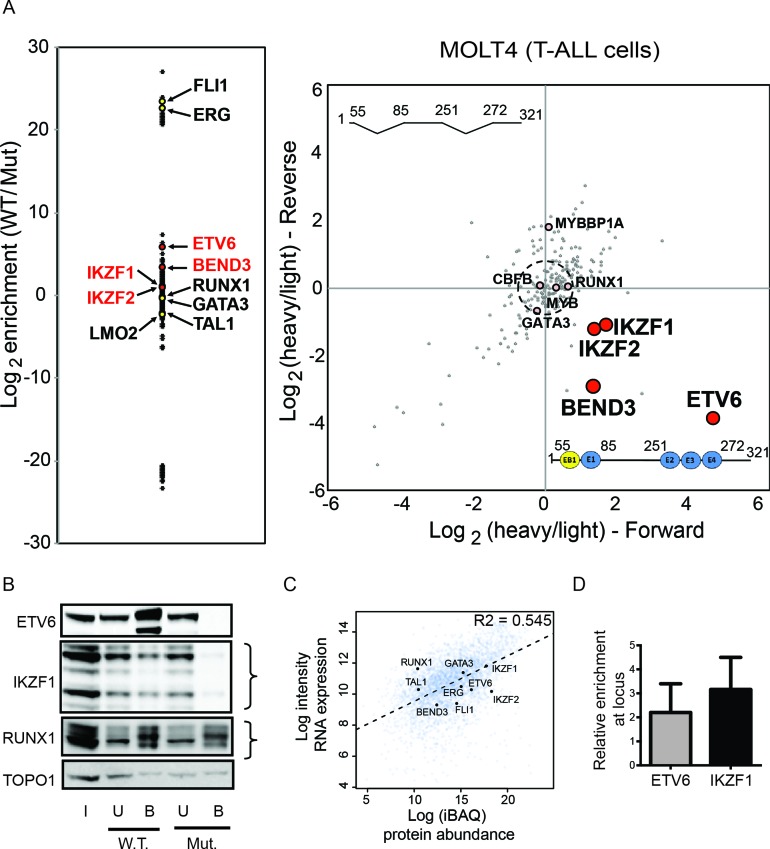
As a first step towards this, we generated biotinylated wild-type and mutant baits of the entire ERG +85 enhancer region by PCR. ReverseChIP was performed using a 327 bp wild-type bait, spanning from EB1 to G2 (Figure 1A) and a 286 bp mutant fragment lacking the 3×ETS and EB1E1 motifs (Δ3×ETS ΔEB1E1, Figure 1B). We sought to determine whether the identity of the specific binders previously identified on the individual small fragments would remain consistent in the context of many more potential binders. As seen in Figure 3A, we have identified most of the previously known binders (22) of the ERG +85 enhancer in MOLT-4 cells originally discovered by conventional ChIP (including SCL/TAL1, LMO2, ERG, FLI1, RUNX1 and GATA3). In order to better distinguish specific binders from spurious enrichment, we again performed parallel forward and reverse reverseChIP experiments. In addition to the previously known binders above, we have also identified all of the novel specific binders of the wild-type bait that we identified by doing reverseChIP on smaller baits (ETV6, IKZF1, IKZF2 and BEND3, all P < 0.01, Figure 3A). This result shows that the binding of specific factors to the wild-type bait were unaffected by the binding of other factors on the large bait. In addition, the signal of specific binders enriched on the wild-type bait comes through clearly, despite the increased possibility for noise emanating from non-specific binders.
Figure 3.
Identification of binders of the composite 321 bp ERG +85 enhancer fragment in MOLT-4 cells. (A) The enrichment plot on the left plots log2 ratios for the abundance of the different factors bound to the wild-type (W.T.) bait compared to the mutant Δ3×ETSΔEB1E1 (Mut.) bait. The enrichment biplot on the right shows log2 (heavy/light) ratios of binders from parallel, order-swapped, reverseChIP experiments. (B) Western blot analyses of factors bound to the W.T. and Mut. enhancer fragments. Input (I), Unbound (U) and Bound (B) fractions are shown. Curly brackets indicate differently migrating isoforms for the relevant proteins. (C) Scatterplot of protein abundance from quantitative MS versus microarray gene expression levels from CellLineNavigator (32) for all identified proteins in MOLT-4 nuclear extracts. The dotted line shows a line of best fit and an R2 value from Spearman correlation is also indicated. The identities of selected transcription factors identified as binders of the +85 enhancer in MOLT-4 cells is also shown. (D) ChIP-qPCR showing enrichment, relative to IgG, of ETV6 and IKZF1 at the +85 enhancer locus in vivo. Error bars represent standard deviation from replicates.
To verify the MS findings by an orthogonal method, we performed DNA affinity capture of nuclear proteins using the wild-type or mutant +85 enhancer fragments as baits and probed the eluted proteins using primary antibodies against some of the specific proteins identified by MS. Consistent with the MS results, we detect specific enrichment for ETV6 and IKZF1 on the wild-type bait compared to the mutant bait, whereas RUNX1 binds equally to both baits (Figure 3B). To rule out that reverseChIP is biased towards detecting the binding of only the most abundantly expressed transcription factors, we performed quantitative MS to determine the abundance of all proteins in the nucleoplasm, the input material for our reverseChIP experiments. We assessed the correlation between protein abundance and gene transcription, using publicly available microarray data from MOLT-4 cells in CellLineNavigator (32). As shown in Figure 3C, there is generally a good correlation between protein levels and gene expression (R2 = 0.545). However, there is no correlation between the identity of factors detected as binders of the ERG +85 enhancer by reverseChIP and their abundance in nucleoplasm (Figure 3C). For example, BEND3 is a factor that is clearly enriched on the wild-type 3×ETS bait and 22 unique peptides were detected in MS (Supplementary Table S2), however its abundance is relatively low compared to the other detected binders (Figure 3C). ETV6, another factor enriched on the wild-type bait for which 17 unique peptides were detected in MS, is only moderately abundant compared to other binders (Figure 3C). Conversely, none of the most abundant nuclear proteins were identified by reverseChIP as specific binders of the +85 enhancer. Our data therefore indicates that the reverseChIP assay is pulling down factors that specifically bind the region of interest rather than merely identifying abundant transcription factors.
While the reverseChIP assay identifies potential binders of a naked DNA sequence in vitro, the chromatin environment around the cognate region can influence the binding of a transcription factor inside cells. We therefore performed a conventional ChIP assay to ascertain whether ETV6 and IKZF1 bind the +85 enhancer within MOLT-4 cells. Consistent with our reverseChIP data, we find enrichment for both ETV6 (2.2×-fold) and IKZF1 (3.16×-fold) at the +85 enhancer (Figure 3D), showing for the first time that these factors bind the ERG enhancer within cells.
ETV6 and IKZF1 are also novel binders of the ERG +85 enhancer in AML cells
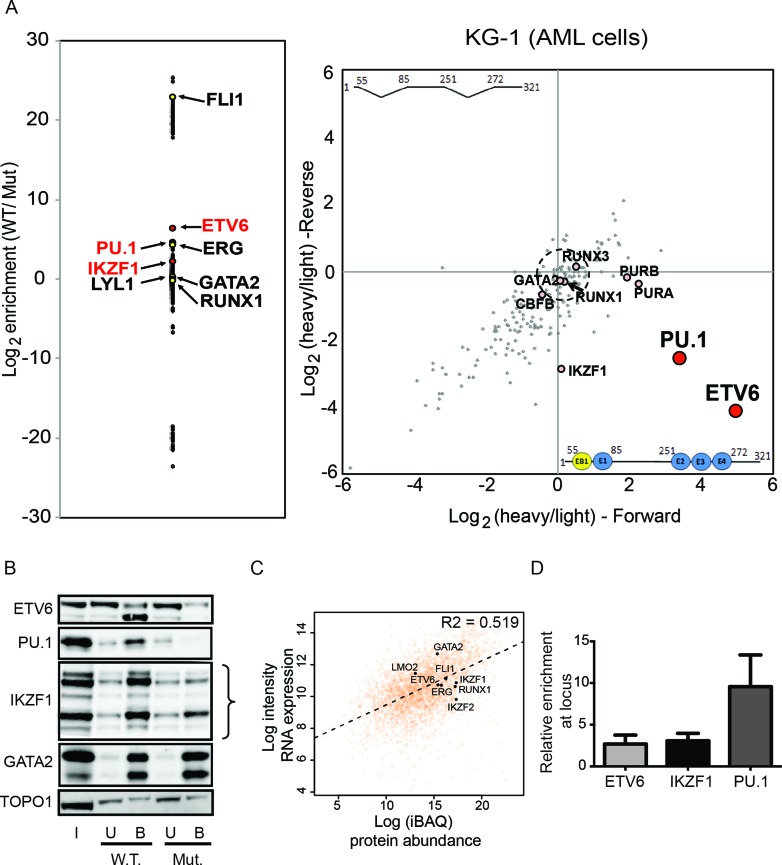
ERG is also a potent oncogene in AML and we have previously shown that its high expression in AML cells is dependent on the binding of a heptad of transcription factors to the +85 enhancer (29). However, it is unknown if additional factors might also bind this region in AML cells. Having established that we could interrogate the full-length +85 enhancer fragment with reverseChIP, we proceeded to identify the complement of transcription factors that can bind this region in AML cells. As the 3×ETS and EB1E1 regions are also necessary for full transcriptional activity of the +85 enhancer in the KG-1 AML cell line (Figure 1B and (29)), we performed reverseChIP using the same 327 bp wild-type +85 enhancer and the 286 bp mutant Δ3×ETS ΔEB1E1 fragments as before, using SILAC-labelled (‘heavy’) and unlabelled (‘light’) nuclear protein extracts prepared from KG-1 cells.
Similar to T-ALL cells, we found that ETV6 binding was strongly enriched on the wild-type enhancer fragment in AML cells (log2 enrichment values of 3.39 and −2.57 in the forward and reverse experiments respectively, P-value for enrichment: 1.4 × 10−9, Figure 4A). We also observed enrichment for IKZF1 and PU.1 on the wild-type fragment, and identified most of the previously known binders of the +85 enhancer identified by ChIP (29), including ERG, FLI1, LYL1, RUNX1 and GATA2 (all P < 0.01). Consistent with the MS findings, affinity capture of nuclear proteins followed by western blots against specific proteins show enrichment in binding for ETV6 and IKZF1 on the wild-type probe, whereas a non-specific binder such as GATA2 bound equally to both baits (Figure 4B). Additionally, we tested for any correlation between binders identified by reverseChIP and protein abundance. We once again performed quantitative MS of nuclear protein extracts from KG-1 cells and determined that there was an overall good correlation with gene expression levels in KG-1 cells from CellLineNavigator (R2 = 0.519, Figure 4C). However, as in MOLT-4 cells, we did not find any correlation between the identity of binders by reverseChIP and their abundance (Figure 4C), indicating once again that we are not simply identifying the most abundant proteins but rather specific potential binders of the +85 enhancer in KG-1 nuclear extracts. Lastly, by conventional ChIP, we validated the binding of ETV6 and IKZF1 at the ERG +85 locus within cells, thereby identifying for the first time that these factors are true binders of this region within AML cells (Figure 4D).
Figure 4.
Identification of binders of the ERG +85 enhancer in KG-1 cells. (A) The enrichment plot on the left plots log2 ratios for the abundance of the different factors bound to the wild-type (W.T.) bait compared to the mutant Δ3×ETSΔEB1E1 (Mut.) bait. The enrichment biplot on the right shows log2 (heavy/light) ratios of binders from parallel, order-swapped, reverseChIP experiments. (B) Western blot analyses of factors bound to the wild-type (W.T.) and mutant (Mut.) enhancer fragments. Input (I), Unbound (U) and Bound (B) fractions are shown. Curly brackets indicate differently migrating isoforms for the relevant proteins. (C) Scatterplot of protein abundance from quantitative MS versus microarray gene expression levels from CellLineNavigator (32) for all identified proteins in KG-1 nuclear extracts. The dotted line shows a line of best fit and an R2 value from Spearman correlation is also indicated. The identities of selected transcription factors identified as binders of the +85 enhancer in KG-1 cells is also shown. (D) ChIP-qPCR enrichment of factors, relative to IgG, at the +85 enhancer locus in vivo. Error bars represent standard deviation from replicates.
ETV6 and IKZF1 are bona fide transcriptional regulators of heptad target genes, including ERG, in leukaemic cells and in healthy HSPCs
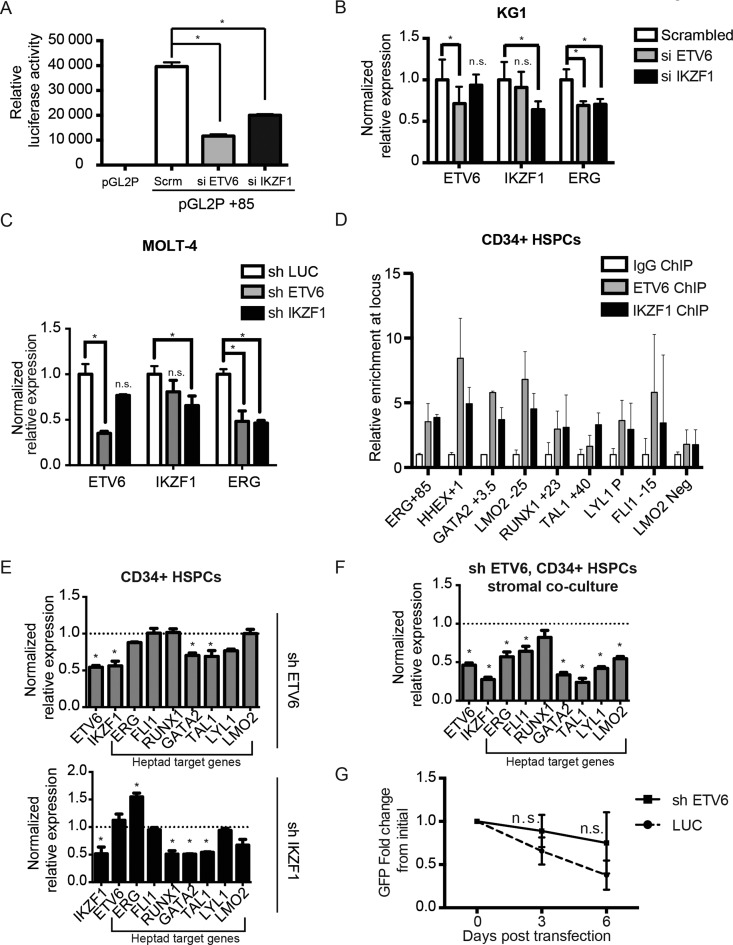
To determine whether ETV6 and IKZF1 regulate high ERG expression in leukaemic cells, we performed siRNA-mediated knockdown experiments of both factors in KG-1 cells. We generated a KG-1 reporter cell line wherein the transcription of the luciferase gene was controlled by the ERG +85 enhancer. Cells were transfected with Cy3-labelled siRNA against ETV6, IKZF1 or a scrambled control, FACS sorted for Cy3-positive cells and used to determine luciferase activity and gene expression by quantitative PCR. siRNA knockdown of either ETV6 or IKZF1 led to >50% decrease in luciferase activity compared to control cells treated with scrambled siRNA (P < 0.05, Figure 5A). Additionally, the expression of the endogenous ERG gene was decreased by ∼30% in cells where ETV6 or IKZF1 were knocked down, compared to scrambled control cells (P < 0.05, Figure 5B). Additionally, shRNA mediated stable knockdowns in MOLT-4 cells of ETV6 and IKZF1 also reduced ERG transcription (Figure 5C), indicating for the first time that ETV6 and IKZF1 are transcriptional regulators of ERG in leukaemic cells. Leukaemic cells are especially sensitive to the modulation of ERG levels, with knockdowns leading to increased cell death in MOLT-4 (22), Jurkat (54) and REH cells (54). We therefore suspect the results here might be an underestimation of the actual decrease in ERG mRNA levels upon knockdown of ETV6 and IKZF1.
Figure 5.
ETV6 and IKZF1 regulate ERG expression. (A and B) KG1 cells were transfected with Cy3-labelled scrambled siRNA (control), ETV6 siRNA (si ETV6) or IKZF1 siRNA (si IKZF1). Cy3-positive cells were flow sorted after transfection, grown for 72 h and used in assays. (A) Stable transfection assay, measuring luciferase activity in vector only (pGL2P) or in conjunction with the +85 enhancer (pGL2P +85). *: P < 0.05, two-tailed t-test, in comparison with scrambled control (Scr.). (B) Examination of endogenous ERG expression levels in KG1 cells treated with siRNA by quantitative real-time PCR. *: P < 0.05, n.s.: P > 0.05, two-tailed t-test, compared to control and error bars represent standard deviation from technical replicates. (C) Examination of endogenous ERG expression levels in MOLT-4 cells following shRNA-mediated knockdowns of ETV6 (top panel) and IKZF1 (bottom panel) by quantitative real-time PCR. *: P < 0.05, n.s.: P > 0.05, two-tailed t-test, compared to control and error bars represent standard deviation from technical replicates. (D) ChIP-qPCR enrichment of ETV6 and IKZF1 in healthy CD34+ HSPCs at well characterized hematopoietic enhancers that are also heptad targets. LMO2 neg is a negative control region that it not bound by the heptad. Error bars represent standard deviation from replicates. (E) Normalized expression values from quantitative real-time PCR measurement of heptad target genes in primary cord blood CD34+ HSPCs following shRNA-mediated knockdowns of ETV6 (top panel) or IKZF1 (bottom panel). * denotes genes with P < 0.05, two-tailed t-test, compared to control luciferase knockdown. All other genes were non-significant (i.e. P > 0.05) and error bars represent standard deviation from technical replicates. (F) Normalized expression values from quantitative real-time PCR measurement of heptad target genes in GFP+CD45+ sorted primary cord blood CD34+ HSPCs following 5 days of stromal co-culture after shRNA-mediated knockdown of ETV6. * denotes genes with P < 0.05, two-tailed t-test, compared to control luciferase knockdown. All other genes were non-significant (i.e. P > 0.05) and error bars represent standard deviation from technical replicates. (G) Flow cytometry-based assessment of the maintenance of GFP+ primary cord blood CD34+ HSPCs in cytokine-containing media following shRNA-mediated knockdown of ETV6. n.s.: P > 0.05, two-tailed t-test, compared to control luciferase knockdown and error bars represent standard deviation from technical replicates.
Previous studies have shown that the binding of the heptad of transcription factors to the +85 enhancer is also important for high ERG expression in healthy HSPCs (2,28). We sought to determine if ETV6 and IKZF1 might represent additional, as yet unknown, regulators of ERG transcription alongside the heptad in healthy HSPCs. We performed ChIP experiments of ETV6 and IKZF1 in primary human CD34+ HSPCs. We observed enrichment for both factors at the ERG +85 enhancer compared to a negative control region (LMO2 Neg, Figure 5D). The +85 stem cell enhancer is one of a number of haematopoietic enhancers that are bound by the heptad in HSPCs (2,55). Other haematopoietic enhancers bound by the heptad include HHEX +1 (56), GATA2 +3.5 (57), LMO2 -25 (58), RUNX1 +23 (59), TAL1 +40 (60), FLI1 -15 (56) and the LYL1 promoter (61). As with the ERG +85 enhancer, we observed an enrichment for ETV6 and IKZF1 at these regions (Figure 5D). To validate that ETV6 and IKZF1 regulate the transcription of other heptad genes, we performed shRNA-mediated stable knockdowns of ETV6 and IKZF1 in primary HSPCs. A two-fold reduction in ETV6 resulted in reduced transcription of a subset of heptad target genes we tested (GATA2 and TAL1, Figure 5E) as well as a 50% reduction in IKZF1 (Figure 5E). Two-fold reduction of IKZF1 also led to the transcriptional down-regulation of a subset of heptad target genes, including GATA2, RUNX1 and TAL1 (Figure 5E). However, unlike in leukemic cells, we only observed a slight reduction in ERG transcription (∼12.3% compared to a control luciferase knockdown, Figure 5E) in healthy HSPCs upon ETV6 knockdown and conversely, an increase in ERG transcripts upon IKZF1 knockdown (Figure 5E).
As the heptad is known to form a robust and highly stable network that is resistant to noisy perturbations in hematopoietic stem cells (62), we reasoned that the effects of knockdowns might require a longer time to manifest more pronouncedly. Since ETV6 knockdowns for 72 h hinted at a slight reduction in ERG transcription (Figure 5E), we investigated whether prolonged ETV6 knockdown would result in a more marked effect. We performed lentiviral-mediated shRNA knockdown of ETV6 in primary healthy HSPCs and maintained the transduced cells for five days using an OP9-based stromal co-culture setup (35,36), before sorting GFP+ CD45+ cells for analyses. We observed that sustained knockdown of ETV6 led to a robust decrease in ERG and in five of the remaining six heptad members (FLI1, GATA2, TAL1, LYL1 and LMO2, Figure 5F), as well as of IKZF1. Additionally, we also assessed whether prolonged ETV6 knockdown might have any proliferative consequences on healthy HSPCs. Using a competitive culture assay in which ETV6 knockdown (GFP+) cells were cultured together with control (GFP−) cells containing wild-type levels of ETV6, we were unable to observe any statistically significant changes in the proportions of GFP+ cells compared to control knockdown (Figure 5G). Our data shows that ETV6 knockdown per se does not impair the proliferation of cells. However, despite multiple attempts, we were unable to achieve a >50% reduction of ETV6. Auto-regulatory networks are bistable and can reversibly shift from OFF to ON and ON to OFF by varying expression levels of individual components below or above a threshold of expression (62). It is possible that the inability to propagate cells with lower levels of ETV6, which is essential for the survival of hematopoietic stem or progenitor cells (40) was due to switching OFF of this auto-regulatory stem cell network and impaired cell proliferation.
In summary, we have discovered that ETV6 and IKZF1 are bona fide transcriptional regulators of a number of heptad-regulated haematopoietic genes, including ERG, in leukemic cells as well as in healthy HSPCs.
Increased ETV6 and IKZF1 expression is associated with poor prognosis in AML
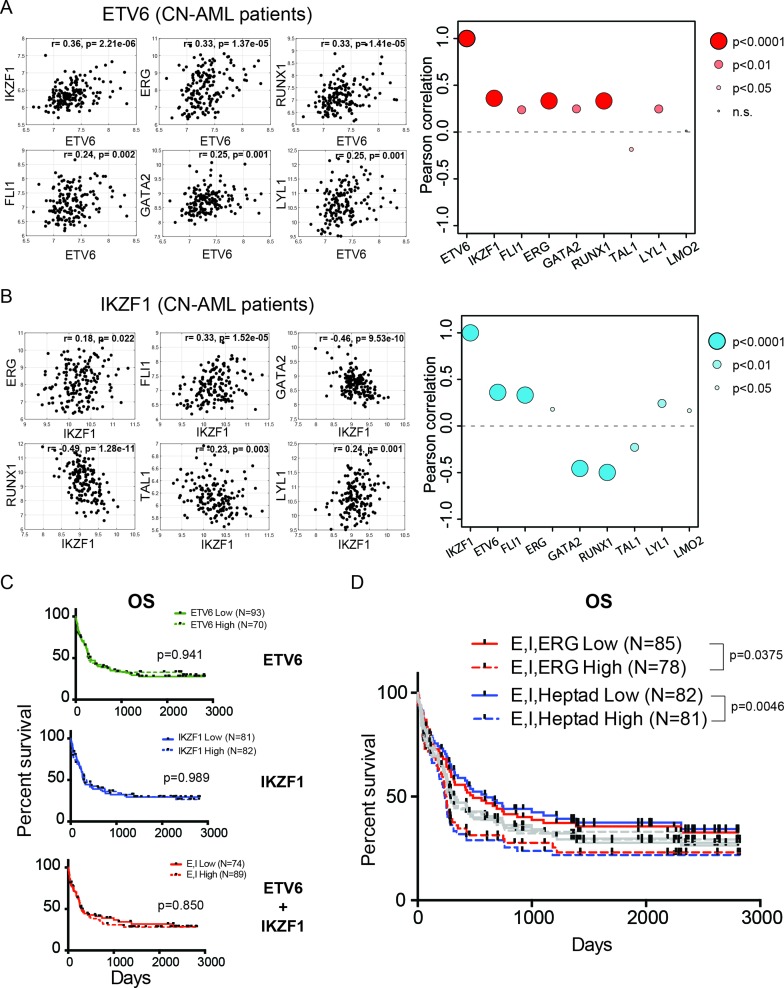
We have previously determined that an open chromatin state of the ERG +85 enhancer and high expression of the heptad factors that bind within this region are predictive of poor clinical outcomes in cytogenetically normal AML (CN-AML) patients (29). We investigated the correlation in expression between ETV6, IKZF1 and members of the heptad in this cohort of CN-AML patients, for whom gene expression and matched clinical data were both available (38). We observed that there was positive correlation (r: 0.24–0.36), with high statistical significance (P < 0.01), between the expression of ETV6 and five members of the heptad (ERG, FLI1, GATA2, RUNX1 and LYL1), as well as IKZF1 in these patients (Figure 6A). There was a slight anti-correlation with the expression of TAL1 and no correlation with LMO2 expression (Figure 6A). IKZF1 expression, on the other hand, is positively correlated with ETV6 and four heptad factors (ERG, FLI1, LYL1 and LMO2, r: 0.18–3), while being negatively correlated with GATA2, RUNX1 and TAL1 (Figure 6B).
Figure 6.
The roles of ETV6 and IKZF1 in leukaemogenesis. (A) Representative gene expression correlation plots between ETV6 and a number of heptad genes in cytogenetically normal AML (CN-AML) patients. Pearson correlation values (r) and associated P-values are also indicated. The bubble plots on the right summarize pairwise correlations between ETV6 and all members of the heptad as well as IKZF1, with the corresponding Pearson correlation indicated on the y-axis and the size of bubble indicating the P-value. Points above the dotted line indicate positive correlation, while points below suggest negative correlation. (B) Representative gene expression correlation plots and bubble plot between IKZF1 and heptad genes. (C) Kaplan–Meier survival curves for overall survival (OS) in cytogenetically normal AML patients segregated based on high or low expression of ETV6 alone (top panel), IKZF1 alone (middle panel) or ETV6 and IKZF1 combined (bottom panel). The indicated P-values were generated from a log-rank test. (D) Kapla–Meier survival curves for overall survival (OS) in cytogenetically normal AML patients segregated based on high or low expression of ETV6, IKZF1 and ERG only (E, I, ERG) or for ETV6, IKZF1 and the heptad (E, I, Heptad). The grey bars indicate all the survival curves illustrated in (C), overlaid for comparison. The indicated P-values were generated from a log-rank (Mantel–Cox) test.
Given the correlation in expression between ETV6/IKZF1 and the heptad, we tested the clinical implications of high expression of ETV6 and IKZF1 on overall survival of AML patients. We stratified patients based on their relative expression of ETV6 alone, IKZF1 alone or ETV6 and IKZF1 in combination and investigated differences in overall survival. As shown in Figure 6C, the relative expression of neither of these two factors, singly or together, were sufficient to segregate patients. However, the combination of high ETV6, IKZF1 and ERG expression stratified patients efficiently, with high expression of all three factors associated with poorer overall survival (E, I, ERG, P-value = 0.0375, Figure 6D). Further factoring in the high expression of all seven heptad factors together with ETV6 and IKZF1 additionally improves the stratification (E, I, Heptad: P-value = 0.0046, Figure 6D). Our data therefore indicates that ETV6 and IKZF1 are previously unknown components of a densely interconnected network of heptad transcription factors that regulate the expression of a number of important hematopoietic genes and whose high expression in AML is associated with poor prognosis.
DISCUSSION
In order to determine the full complement of transcriptional regulators mediating high expression of ERG in leukaemic cells, we adapted and refined a recently developed quantitative proteomics-based method (13). By applying reverseChIP to identify potential binders of the ERG +85 stem cell enhancer in T-ALL and AML cells, we discovered two new factors, ETV6 and IKZF1. We have used conventional ChIP to validate that these potential binders truly interact with the +85 enhancer within cells. Our data also indicates that these factors promote high ERG expression in leukaemic cells, as the knockdown of each factor reduced endogenous ERG transcription. In addition, we also demonstrated that these factors regulate the expression of a number of genes of the highly interconnected heptad of transcription factors in healthy CD34+ HSPCs. Finally, we have shown that the high expression of ETV6 and IKZF1, in combination with the heptad, confers poor overall prognosis in AML patients. In doing so, we have thus established the utility of the reverseChIP method as an effective means to discover novel transcriptional regulators of genes of disease relevance. Furthermore, we have extended the capability of the method beyond small (<60 bp) DNA fragments (13,17,19,52,53), using it to successfully interrogate the full-length ERG +85 enhancer, a complex 321 bp region containing binding sites for multiple transcription factors.
The reverseChIP method we present here is general enough to be translatable to other genomic regions of interest, including enhancers, promoters or mutated genomic regions identified in genome-wide studies. The recent development of high-throughput variations of the technique will also enable it to be applied to simultaneously screen a large number of regions of interest, such as multiple single nucleotide polymorphisms (SNPs) identified in GWAS studies (17,52). The SILAC-based quantitative proteomics we employed here requires culturing of cells in media containing labelled amino acids, which limits its direct utilization for studying primary cells or other cell types that are difficult to maintain in culture. However, alternative chemical labelling methods such as isobaric tags for relative and absolute quantitation/iTRAQ (63) or stable isotope dimethyl labelling (64), which don't require culturing of cells in specialized media could be utilized in such cases (52). Recent advances in label-free quantitative proteomics also make it feasible to quantify the abundance of specific binding proteins from extracts of primary cells without necessitating any labelling (65). Despite its advantages, we envisage the reverseChIP method as a complement to, rather than as a competitor of, ChIP. Both methods have their relative strengths, affording each its rightful place in the molecular toolkit used in studying protein–DNA interactions. While we have been able to optimize nuclear protein extractions and MS detection, sample consumption remains high and we require ∼20 million cells to reliably perform reverseChIP. In contrast, highly sensitive ChIP methods such as iChIP (66) and nanoChIP (67) enable binding of factors to be detected in as few as 500 cells, permitting ChIP to be applied to studying very rare populations of cells. Furthermore, as reverseChIP relies on the binding of factors to naked DNA in vitro, we cannot exclude the possibility that factors whose DNA binding is influenced by specific chromatin modifications and/or DNA modifications might be missed being detected (i.e. false negatives). As it is performed in vivo within the context of chromatin, ChIP does not have this limitation. Techniques that incorporate chromatin-assembled DNA baits (16) or methods capable of isolating specific regions of in vivo chromatin along with bound proteins, such as PICh (68) and CHAP-MS (69), could also circumvent these limitations but are less straightforward to perform than reverseChIP.
The ability of transcription factors to modulate gene expression is regulated at a number of levels, including accessibility to their cognate binding site on DNA, interactions with specific co-factors and post-translational modifications on the proteins. Here, we have discovered that ETV6 and IKZF1 both contribute to high ERG expression in leukaemic cells. IKZF1 is a dual-functional transcriptional regulator that can activate or repress genes, as well as create favourable or repressive chromatin environments, depending on the genomic locus (70). In AML and T-ALL cells, our knockdown experiments indicate that IKZF1 acts as a transcriptional activator of ERG, while in healthy HSPCs it acts as a repressor. As IKZF1's opposing transcriptional roles have been linked to its interactions with specific co-factors, including the chromatin remodelling complex NuRD (71,72), it is possible that its association with different co-factors in leukaemic cells modulates its function aberrantly. In this context, it is interesting to note that in B-cell precursor ALL, concurrent intragenic deletion of ERG in the background of IKZF1 mutations confers better patient outcomes (73), while in BCR-ABL B-cell precursor ALL, IKZF1 mutations confer worse prognosis (74). Our data would predict that IKZF1 mutations in B-ALL lead to increased ERG expression, in line with our observations in healthy HSPCs, thereby conferring poor prognosis. However, a concurrent intragenic deletion of ERG would compensate for the resultant transcriptional upregulation, leading to the better outcomes reported.
ETV6, on the other hand, has a well-studied role in normal haematopoiesis as a transcriptional repressor of genes (75), though it functions as a transcriptional activator of ERG and other heptad target genes. However, like IKZF1, ETV6 too interacts with a wide array of co-factors including the MYST-family histone acetyltransferase TIP60/KAT5 (76), a known co-activator of genes (77–79). It is therefore entirely feasible that, just like IKZF1, ETV6 could be a context-dependent transcriptional regulator with opposing activating and repressing roles at different genes. Such a scenario is not inconceivable: the well-studied transcriptional activator MYB, for example, directly repress almost as many genes as it activates in myeloid cells (80). Intriguingly, transcriptional repression by MYB is additionally reliant on its interaction with p300, a known co-activator of MYB (80). We have previously shown that the stem cell-like transcriptional program driven by the heptad is active in NPM1 mutation negative CD34+ AMLs (29) and is mutually exclusive with the HOX/MYC signatures of NPM1 mutant and MLL-mutated leukaemias (81,82). Whereas our data support a role for functional ETV6 in cooperating with the heptad, it is noteworthy that loss of function ETV6 mutations (∼ 1.1% of all AML) is frequently associated with NPM1 mutations (∼ 23%, (83)) suggesting they arise in a very distinct subset of AML. It also does not preclude the emergence of leukaemia in the context of germ-line loss of function ETV6 mutations (in pre-B ALL, (84)), which may cooperate with other mutations to generate their own network structure that drives self-renewal of leukemic stem cells. Furthermore, our findings that ETV6 binds strongly to the 3×ETS motif of the ERG +85 enhancer raises an interesting question as to why this particular factor is enriched, when potentially a number of different transcription factors all belonging to the ETS family could all bind there. It is likely that the local chromatin environment, presence of other bound factors, specific DNA sequence around the bound site and post-translational protein modifications could all be further potential determinants for which particular factor binds at a given genomic locus. Future genome-wide studies of ETV6 and IKZF1 will be required to determine the exact role these factors play, together with the heptad, in initiating and/or maintaining stem cell-like transcriptional regulatory networks in healthy stem cells and in leukaemic cells.
Supplementary Material
Acknowledgments
The authors wish to thank Amy Daoud (Qiagen, Australia), Marc Hogan (Thermo Fisher Scientific, Australia) and members of the Flow Cytometry Facility, UNSW Mark Wainwright Analytical Centre (Emma Johansson Beves and Christopher Brownlee) for outstanding technical assistance rendered.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council of Australia [APP1102589, APP1100495 to J.E.P., APP1007911 to K.L.M.]; Cure Cancer Australia Foundation [APP1057921 to J.W.H.W.]; Australian Research Council [FT13010096 to J.W.H.W.]; Leukaemia Foundation (to A.U.); Anthony Rothe Memorial Trust (to J.A.I.T., D.B., A.U.); UNSW Australia Prince of Wales Clinical School Post-Graduate Research Scholarship (to Y.F.G.); South Eastern Area Laboratory Services (SEALS) (to J.E.P.); Fund for Scientific Research Flanders through the Odysseus program (to P.V.V., T.T.); Project Grants [GA00113N, 3G065614, G.0C47.13N, 31500615W to P.V.V., G0B2913N, G037514N, 3G002711 to T.T.); PhD Grant (to S.P.); Postdoctoral Grant (to I.V.de W.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Pimanda J.E., Gottgens B. Gene regulatory networks governing haematopoietic stem cell development and identity. Int. J. Dev. Biol. 2010;54:1201–1211. doi: 10.1387/ijdb.093038jp. [DOI] [PubMed] [Google Scholar]
- 2.Beck D., Thoms J.A., Perera D., Schutte J., Unnikrishnan A., Knezevic K., Kinston S.J., Wilson N.K., O'Brien T.A., Gottgens B., et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:e12–e22. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- 3.Bereshchenko O., Mancini E., Moore S., Bilbao D., Mansson R., Luc S., Grover A., Jacobsen S.E.W., Bryder D., Nerlov C. Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell. 2009;16:390–400. doi: 10.1016/j.ccr.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 4.Viatour P., Somervaille T.C., Venkatasubrahmanyam S., Kogan S., McLaughlin M.E., Weissman I.L., Butte A.J., Passegue E., Sage J. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 2008;3:416–428. doi: 10.1016/j.stem.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huntly B.J.P., Gilliland D.G. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat. Rev. Cancer. 2005;5:311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Eng. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J., et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 8.Kvinlaug B.T., Chan W.I., Bullinger L., Ramaswami M., Sears C., Foster D., Lazic S.E., Okabe R., Benner A., Lee B.H., et al. Common and overlapping oncogenic pathways contribute to the evolution of acute myeloid leukemias. Cancer Res. 2011;71:4117–4129. doi: 10.1158/0008-5472.CAN-11-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misaghian N., Ligresti G., Steelman L.S., Bertrand F.E., Basecke J., Libra M., Nicoletti F., Stivala F., Milella M., Tafuri A., et al. Targeting the leukemic stem cell: the Holy Grail of leukemia therapy. Leukemia. 2009;23:25–42. doi: 10.1038/leu.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hesselberth J.R., Chen X., Zhang Z., Sabo P.J., Sandstrom R., Reynolds A.P., Thurman R.E., Neph S., Kuehn M.S., Noble W.S., et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox J., Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 12.Cravatt B.F., Simon G.M., Yates J.R., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 13.Mittler G., Butter F., Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2009;19:284–377. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himeda C.L., Ranish J.A., Angello J.C., Maire P., Aebersold R., Hauschka S.D. Quantitative proteomic identification of six4 as the trex-binding factor in the muscle creatine kinase enhancer. Mol. Cell Biol. 2004;24:2132–2143. doi: 10.1128/MCB.24.5.2132-2143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaneva M., Tempst P. Affinity capture of specific DNA-binding proteins for mass spectrometric identification. Anal. Chem. 2003;75:6437–6448. doi: 10.1021/ac034698l. [DOI] [PubMed] [Google Scholar]
- 16.Bartke T., Vermeulen M., Xhemalce B., Robson S.C., Mann M., Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butter F., Davison L., Viturawong T., Scheibe M., Vermeulen M., Todd J.A., Mann M. Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. PLoS Genet. 2012;8:e1002982. doi: 10.1371/journal.pgen.1002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartels S.J., Spruijt C.G., Brinkman A.B., Jansen P.W., Vermeulen M., Stunnenberg H.G. A SILAC-based screen for Methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence-specific binding protein. PLoS One. 2011;6:e25884. doi: 10.1371/journal.pone.0025884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W., Bauer C., Munzel M., Wagner M., Muller M., Khan F., et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Taoudi S., Bee T., Hilton A., Knezevic K., Scott J., Willson T.A., Collin C., Thomas T., Voss A.K., Kile B.T., et al. ERG dependence distinguishes developmental control of hematopoietic stem cell maintenance from hematopoietic specification. Genes Dev. 2011;25:251–262. doi: 10.1101/gad.2009211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loughran S.J., Kruse E.A., Hacking D.F., de Graaf C.A., Hyland C.D., Willson T.A., Henley K.J., Ellis S., Voss A.K., Metcalf D., et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat. Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- 22.Thoms J.A., Birger Y., Foster S., Knezevic K., Kirschenbaum Y., Chandrakanthan V., Jonquieres G., Spensberger D., Wong J.W., Oram S.H., et al. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood. 2011;117:7079–7089. doi: 10.1182/blood-2010-12-317990. [DOI] [PubMed] [Google Scholar]
- 23.Salek-Ardakani S., Smooha G., de Boer J., Sebire N.J., Morrow M., Rainis L., Lee S., Williams O., Izraeli S., Brady H.J. ERG is a megakaryocytic oncogene. Cancer Res. 2009;69:4665–4673. doi: 10.1158/0008-5472.CAN-09-0075. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg L., Tijssen M.R., Birger Y., Hannah R.L., Kinston S.J., Schutte J., Beck D., Knezevic K., Schiby G., Jacob-Hirsch J., et al. Genome-scale expression and transcription factor binding profiles reveal therapeutic targets in transgenic ERG myeloid leukemia. Blood. 2013;122:2694–2703. doi: 10.1182/blood-2013-01-477133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baldus C.D., Burmeister T., Martus P., Schwartz S., Gokbuget N., Bloomfield C.D., Hoelzer D., Thiel E., Hofmann W.K. High expression of the ETS transcription factor ERG predicts adverse outcome in acute T-lymphoblastic leukemia in adults. J. Clin. Oncol. 2006;24:4714–4720. doi: 10.1200/JCO.2006.06.1580. [DOI] [PubMed] [Google Scholar]
- 26.Metzeler K.H., Dufour A., Benthaus T., Hummel M., Sauerland M.C., Heinecke A., Berdel W.E., Buchner T., Wormann B., Mansmann U., et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J. Clin. Oncol. 2009;27:5031–5038. doi: 10.1200/JCO.2008.20.5328. [DOI] [PubMed] [Google Scholar]
- 27.Marcucci G., Maharry K., Whitman S.P., Vukosavljevic T., Paschka P., Langer C., Mrozek K., Baldus C.D., Carroll A.J., Powell B.L., et al. High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B Study. J. Clin. Oncol. 2007;25:3337–3343. doi: 10.1200/JCO.2007.10.8720. [DOI] [PubMed] [Google Scholar]
- 28.Wilson N.K., Foster S.D., Wang X., Knezevic K., Schutte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E., et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 29.Diffner E., Beck D., Gudgin E., Thoms J.A., Knezevic K., Pridans C., Foster S., Goode D., Lim W.K., Boelen L., et al. Activity of a heptad of transcription factors is associated with stem cell programs and clinical outcome in acute myeloid leukemia. Blood. 2013;121:2289–2300. doi: 10.1182/blood-2012-07-446120. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen H.D., Wood I., Hill M.M. A robust permutation test for quantitative SILAC proteomics experiments. J. Integr. OMICS. 2012;2:80–93. [Google Scholar]
- 31.Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 32.Krupp M., Itzel T., Maass T., Hildebrandt A., Galle P.R., Teufel A. CellLineNavigator: a workbench for cancer cell line analysis. Nucleic Acids Res. 2013;41:D942–D948. doi: 10.1093/nar/gks1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanawa H., Kelly P.F., Nathwani A.C., Persons D.A., Vandergriff J.A., Hargrove P., Vanin E.F., Nienhuis A.W. Comparison of various envelope proteins for their ability to pseudotype lentiviral vectors and transduce primitive hematopoietic cells from human blood. Mol. Ther. 2002;5:242–251. doi: 10.1006/mthe.2002.0549. [DOI] [PubMed] [Google Scholar]
- 35.Van de Walle I, Davids K., Taghon T. Characterization and isolation of human T cell progenitors. Methods Mol. Biol. 2016;1323:221–237. doi: 10.1007/978-1-4939-2809-5_19. [DOI] [PubMed] [Google Scholar]
- 36.Dolens A.C., Van de Walle I., Taghon T. Approaches to study human T cell development. Methods Mol. Biol. 2016;1323:239–251. doi: 10.1007/978-1-4939-2809-5_20. [DOI] [PubMed] [Google Scholar]
- 37.Tursky M.L., Beck D., Thoms J.A., Huang Y., Kumari A., Unnikrishnan A., Knezevic K., Evans K., Richards L.A., Lee E., et al. Overexpression of ERG in cord blood progenitors promotes expansion and recapitulates molecular signatures of high ERG leukemias. Leukemia. 2014;4:819–827. doi: 10.1038/leu.2014.299. [DOI] [PubMed] [Google Scholar]
- 38.Metzeler K.H., Hummel M., Bloomfield C.D., Spiekermann K., Braess J., Sauerland M.C., Heinecke A., Radmacher M., Marcucci G., Whitman S.P., et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L.C., Swat W., Fujiwara Y., Davidson L., Visvader J., Kuo F., Alt F.W., Gilliland D.G., Golub T.R., Orkin S.H. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 1998;12:2392–2402. doi: 10.1101/gad.12.15.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hock H., Meade E., Medeiros S., Schindler J.W., Valk P.J., Fujiwara Y., Orkin S.H. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18:2336–2341. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S.L., Payne-Turner D., Easton J., Chen X., Wang J., Rusch M., et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Vlierberghe P., Ambesi-Impiombato A., Perez-Garcia A., Haydu J.E., Rigo I., Hadler M., Tosello V., Della Gatta G., Paietta E., Racevskis J., et al. ETV6 mutations in early immature human T cell leukemias. J. Exp. Med. 2011;208:2571–2579. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida T., Ng S.Y., Zuniga-Pflucker J.C., Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullighan C.G., Su X., Zhang J., Radtke I., Phillips L.A., Miller C.B., Ma J., Liu W., Cheng C., Schulman B.A., et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Eng. J. Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley C.M., Ikeda T., Koipally J., Avitahl N., Wu L., Georgopoulos K., Morgan B.A. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr. Biol. 1998;8:508–515. doi: 10.1016/s0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- 46.Hahm K., Cobb B.S., McCarty A.S., Brown K.E., Klug C.A., Lee R., Akashi K., Weissman I.L., Fisher A.G., Smale S.T. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Speck N.A., Terryl S. A new transcription factor family associated with human leukemias. Crit. Rev. Eukaryot. Gene Expr. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 48.Melnikova I.N., Crute B.E., Wang S., Speck N.A. Sequence specificity of the core-binding factor. J. Virol. 1993;67:2408–2411. doi: 10.1128/jvi.67.4.2408-2411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saksouk N., Barth T.K., Ziegler-Birling C., Olova N., Nowak A., Rey E., Mateos-Langerak J., Urbach S., Reik W., Torres-Padilla M.E., et al. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol. Cell. 2014;56:580–594. doi: 10.1016/j.molcel.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Sathyan K.M., Shen Z., Tripathi V., Prasanth K.V., Prasanth S.G. A BEN-domain-containing protein associates with heterochromatin and represses transcription. J. Cell Sci. 2011;124:3149–3163. doi: 10.1242/jcs.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai Q., Ren A., Westholm J.O., Serganov A.A., Patel D.J., Lai E.C. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev. 2013;27:602–614. doi: 10.1101/gad.213314.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hubner N.C., Nguyen L.N., Hornig N.C., Stunnenberg H.G. A quantitative proteomics tool to identify DNA-protein interactions in primary cells or blood. J. Proteome Res. 2015;14:1315–1329. doi: 10.1021/pr5009515. [DOI] [PubMed] [Google Scholar]
- 53.Spruijt C.G., Baymaz H.I., Vermeulen M. Identifying specific protein-DNA interactions using SILAC-based quantitative proteomics. Methods Mol. Biol. 2013;977:137–157. doi: 10.1007/978-1-62703-284-1_11. [DOI] [PubMed] [Google Scholar]
- 54.Tsuzuki S., Taguchi O., Seto M. Promotion and maintenance of leukemia by ERG. Blood. 2011;117:3858–3868. doi: 10.1182/blood-2010-11-320515. [DOI] [PubMed] [Google Scholar]
- 55.Chacon D., Beck D., Perera D., Wong J.W., Pimanda J.E. BloodChIP: a database of comparative genome-wide transcription factor binding profiles in human blood cells. Nucleic Acids Res. 2014;42:D172–D177. doi: 10.1093/nar/gkt1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donaldson I.J., Chapman M., Kinston S., Landry J.R., Knezevic K., Piltz S., Buckley N., Green A.R., Gottgens B. Genome-wide identification of cis-regulatory sequences controlling blood and endothelial development. Hum. Mol. Genet. 2005;14:595–601. doi: 10.1093/hmg/ddi056. [DOI] [PubMed] [Google Scholar]
- 57.Khandekar M., Brandt W., Zhou Y., Dagenais S., Glover T.W., Suzuki N., Shimizu R., Yamamoto M., Lim K.C., Engel J.D. A Gata2 intronic enhancer confers its pan-endothelia-specific regulation. Development. 2007;134:1703–1712. doi: 10.1242/dev.001297. [DOI] [PubMed] [Google Scholar]
- 58.Landry J.R., Bonadies N., Kinston S., Knezevic K., Wilson N.K., Oram S.H., Janes M., Piltz S., Hammett M., Carter J., et al. Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood. 2009;113:5783–5792. doi: 10.1182/blood-2008-11-187757. [DOI] [PubMed] [Google Scholar]
- 59.Nottingham W.T., Jarratt A., Burgess M., Speck C.L., Cheng J.F., Prabhakar S., Rubin E.M., Li P.S., Sloane-Stanley J., Kong A.S.J., et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogilvy S., Ferreira R., Piltz S.G., Bowen J.M., Gottgens B., Green A.R. The SCL +40 enhancer targets the midbrain together with primitive and definitive hematopoiesis and is regulated by SCL and GATA proteins. Mol. Cell Biol. 2007;27:7206–7219. doi: 10.1128/MCB.00931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan W.Y., Follows G.A., Lacaud G., Pimanda J.E., Landry J.R., Kinston S., Knezevic K., Piltz S., Donaldson I.J., Gambardella L., et al. The paralogous hematopoietic regulators Lyl1 and Scl are coregulated by Ets and GATA factors, but Lyl1 cannot rescue the early Scl-/- phenotype. Blood. 2007;109:1908–1916. doi: 10.1182/blood-2006-05-023226. [DOI] [PubMed] [Google Scholar]
- 62.Narula J., Smith A.M., Gottgens B., Igoshin O.A. Modeling reveals bistability and low-pass filtering in the network module determining blood stem cell fate. PLoS Comput. Biol. 2010;6:e1000771. doi: 10.1371/journal.pcbi.1000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ross P.L., Huang Y.N., Marchese J.N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Boersema P.J., Raijmakers R., Lemeer S., Mohammed S., Heck A.J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 65.Wong J.W., Cagney G. An overview of label-free quantitation methods in proteomics by mass spectrometry. Methods Mol. Biol. 2010;604:273–283. doi: 10.1007/978-1-60761-444-9_18. [DOI] [PubMed] [Google Scholar]
- 66.Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S., et al. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adli M., Bernstein B.E. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat. Protoc. 2011;6:1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dejardin J., Kingston R.E. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Byrum S.D., Raman A., Taverna S.D., Tackett A.J. ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep. 2012;2:198–205. doi: 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwickert T.A., Tagoh H., Gultekin S., Dakic A., Axelsson E., Minnich M., Ebert A., Werner B., Roth M., Cimmino L., et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 2014;15:283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hahm K.B., Cho K., Lee C., Im Y.H., Chang J., Choi S.G., Sorensen P.H., Thiele C.J., Kim S.J. Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat. Genet. 1999;23:222–227. doi: 10.1038/13854. [DOI] [PubMed] [Google Scholar]
- 72.Sridharan R., Smale S.T. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J. Biol. Chem. 2007;282:30227–30238. doi: 10.1074/jbc.M702541200. [DOI] [PubMed] [Google Scholar]
- 73.Clappier E., Auclerc M.F., Rapion J., Bakkus M., Caye A., Khemiri A., Giroux C., Hernandez L., Kabongo E., Savola S., et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia. 2014;28:70–77. doi: 10.1038/leu.2013.277. [DOI] [PubMed] [Google Scholar]
- 74.van der Veer A., Zaliova M., Mottadelli F., De Lorenzo P., Te Kronnie G., Harrison C.J., Cave H., Trka J., Saha V., Schrappe M., et al. IKZF1 status as a prognostic feature in BCR-ABL1-positive childhood ALL. Blood. 2014;123:1691–1698. doi: 10.1182/blood-2013-06-509794. [DOI] [PubMed] [Google Scholar]
- 75.Bohlander S.K. ETV6: a versatile player in leukemogenesis. Semin. Cancer Biol. 2005;15:162–174. doi: 10.1016/j.semcancer.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 76.Putnik J., Zhang C.D., Archangelo L.F., Tizazu B., Bartels S., Kickstein M., Greif P.A., Bohlander S.K. The interaction of ETV6 (TEL) and TIP60 requires a functional histone acetyltransferase domain in TIP60. Biochim. Biophys. Acta. 2007;1772:1211–1224. doi: 10.1016/j.bbadis.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 77.Sapountzi V., Logan I.R., Robson C.N. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 78.Brady M.E., Ozanne D.M., Gaughan L., Waite I., Cook S., Neal D.E., Robson C.N. Tip60 is a nuclear hormone receptor coactivator. J. Biol. Chem. 1999;274:17599–17604. doi: 10.1074/jbc.274.25.17599. [DOI] [PubMed] [Google Scholar]
- 79.Jeong K.W., Kim K., Situ A.J., Ulmer T.S., An W., Stallcup M.R. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat. Struct. Mol. Biol. 2011;18:1358–1365. doi: 10.1038/nsmb.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao L., Glazov E.A., Pattabiraman D.R., Al-Owaidi F., Zhang P., Brown M.A., Leo P.J., Gonda T.J. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vassiliou G.S., Cooper J.L., Rad R., Li J., Rice S., Uren A., Rad L., Ellis P., Andrews R., Banerjee R., et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 2011;43:470–475. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Somerville T.D., Wiseman D.H., Spencer G.J., Huang X., Lynch J.T., Leong H.S., Williams E.L., Cheesman E., Somervaille T.C. Frequent derepression of the mesenchymal transcription factor gene FOXC1 in acute myeloid leukemia. Cancer Cell. 2015;28:329–342. doi: 10.1016/j.ccell.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 83.Haferlach C., Bacher U., Schnittger S., Alpermann T., Zenger M., Kern W., Haferlach T. ETV6 rearrangements are recurrent in myeloid malignancies and are frequently associated with other genetic events. Genes Chromosomes Cancer. 2012;51:328–337. doi: 10.1002/gcc.21918. [DOI] [PubMed] [Google Scholar]
- 84.Moriyama T., Metzger M.L., Wu G., Nishii R., Qian M., Devidas M., Yang W., Cheng C., Cao X., Quinn E., et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: a systematic genetic study. Lancet Oncol. 2015;16:1659–1666. doi: 10.1016/S1470-2045(15)00369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.