Abstract
Rev1 is a member of the Y-family of DNA polymerases and is known for its deoxycytidyl transferase activity that incorporates dCMP into DNA and its ability to function as a scaffold factor for other Y-family polymerases in translesion bypass events. Rev1 also is involved in mutagenic processes during somatic hypermutation of immunoglobulin genes. In light of the mutation pattern consistent with dCMP insertion observed earlier in mouse fibroblast cells treated with a base excision repair-inducing agent, we questioned whether Rev1 could also be involved in base excision repair (BER). Here, we uncovered a weak 5′-deoxyribose phosphate (5′-dRP) lyase activity in mouse Rev1 and demonstrated the enzyme can mediate BER in vitro. The full-length Rev1 protein and its catalytic core domain are similar in their ability to support BER in vitro. The dRP lyase activity in both of these proteins was confirmed by NaBH4 reduction of the Schiff base intermediate and kinetics studies. Limited proteolysis, mass spectrometry and deletion analysis localized the dRP lyase active site to the C-terminal segment of Rev1's catalytic core domain. These results suggest that Rev1 could serve as a backup polymerase in BER and could potentially contribute to AID-initiated antibody diversification through this activity.
INTRODUCTION
Base excision repair (BER) plays an important role in mammalian cells by repairing DNA base lesions and single-strand breaks formed by endogenous and exogenous DNA-reactive agents (1–5). The BER process is multifaceted with many sub-pathways and variations (6,7). A useful working model for one of these sub-pathways, termed single-nucleotide or short patch BER, may be outlined as follows: The process for correcting a damaged base is initiated by spontaneous base loss or monofunctional DNA glycosylase activity (4,8). The resulting apurinic/apyrimidinic (AP) site is incised by AP endonuclease 1 (APE1) producing a single-nucleotide gap with 3′-OH and 5′-deoxyribose phosphate (5′-dRP) groups at the margins (9,10). DNA polymerase (pol) β, or pol λ, binds at the gap, removes the dRP group via 5′-dRP lyase activity (11–14) and then inserts a dNMP in accordance with Watson-Crick base pairing (15,16). In the final step, the BER intermediate containing a nick is ligated by DNA ligase I or III (17–20). Studies of mouse fibroblasts with a deletion in the pol β gene revealed that these cells displayed elevated MMS-induced mutagenesis (21), and this phenotype was later found to be strictly dependent on expression of Rev1 (22). The elevated mutagenesis phenotype was found in a genome-integrated phage lambda CII gene system, and the MMS-induced mutation spectrum was consistent with Rev1 dCMP insertion in a BER intermediate. An understanding of the mechanism of this Rev1 effect on MMS-induced mutagenesis in the pol β null background was not evaluated earlier, and this was a focus of the present study.
Rev1 was initially described as a specialized DNA polymerase with the ability to incorporate dCMP into DNA in an untemplated fashion (23–25). The enzyme also is known to be involved in mutagenesis in Saccharomyces cerevisiae; for example, disruption of the REV1 gene results in decreased UV-induced and MMS-induced mutagenesis in a strain lacking AP endonucleases (26), as well as in a wild-type strain (27,28). Yeast cells harboring a polymerase-dead mutant of REV1 continued to exhibit a mutagenesis phenotype, and it was found that this pro-mutagenic effect was due to Rev1 scaffolding properties carried in its carboxy-terminal domain, such that Y-family DNA polymerases could be recruited for lesion bypass (29–33). In addition, mice expressing catalytically dead Rev1 showed a reduction in C to G and G to C transversions, consistent with disruption of its deoxycytidyl transferase activity, but also showed reductions in C to T and G to A transitions and A:T transversions. These results suggested that Rev1 is involved in multiple mutagenic pathways through functional interaction with other DNA polymerases, and these pathways may include Ig gene hypermutations (34). In mouse fibroblast cells, down-regulation of the Rev1 level, by anti-sense RNA or siRNA, abrogates the MMS-induced mutagenesis phenotype (22,35,36). In chicken DT40 cells, Rev1 was implicated in immunoglobulin gene somatic hypermutation (SHM) (37), pointing to the notion that Rev1 plays a role in SHM through an error-prone DNA synthesis process. For instance, it was proposed that the U:G mismatch and AP site could be processed by mismatch repair and BER, respectively, resulting in mutations at sites distal to the U:G mismatch (38–42).
The involvement of low-fidelity DNA polymerase (s) mediated-BER during SHM is still under investigation. The major BER DNA polymerases pol β and pol λ appear to play minimal if any role for SHM (43,44). One of the important features for a DNA polymerase to participate in BER is possession of dRP lyase activity that is required for cleaning the 5′-end of the BER intermediate so that DNA ligation can occur. Several other DNA polymerases have been evaluated for the presence of the dRP lyase activity. Pol ι and pol θ have dRP lyase activity, but the participation of pol ι in SHM is less clear (45). Here, mouse Rev1 was examined for enzymatic properties consistent with a possible role in BER. The purified full-length enzyme was first tested for its ability to substitute for pol β in an in vitro assay of single-nucleotide BER. The results indicated Rev1 is capable of substituting for pol β. Rev1 was found to have 5′-dRP lyase activity, in addition to its well known insertion of dCMP into a single-nucleotide gapped substrate. Next, we cloned, expressed and purified the catalytic domain of Rev1 (residues 335 to 825), and further studies revealed this domain peptide is sufficient to support single-nucleotide BER. These results are discussed in the context of circumstances where Rev1 could be an important BER factor.
MATERIALS AND METHODS
Materials
Oligonucleotides were from Oligos Etc, Inc. (Wilsonville, OR, USA) and The Midland Certified Reagent Co. (Midland, TX, USA), Inc. [α-32P]dCTP and [α-32P]Cordycepin (3000 Ci/mmol), a substitute of ddATP, and [γ-32P]ATP (6000 Ci/mmol) were from PerkinElmer (Waltham, MS). Optikinase and terminal deoxynucleotidyl transferase were from USB Corp. (Cleveland, OH, USA) and Fermentas Inc. (Hanover, MD, USA), respectively. Protease inhibitor complete (EDTA-free) was from Roche Molecular Diagnostics (Pleasanton, CA, USA). Leupeptin, aprotinin, and phenylmethylsulfonyl fluoride were from Calbiochem (La Jolla, CA, USA). Recombinant human DNA pol β was overexpressed and purified as described previously (46). Human recombinant APE1, uracil-DNA glycosylase (UDG) with 84 amino acids deleted from the amino-terminus and DNA ligase I were purified as described previously (47–49).
Preparation of substrates for dRP lyase and NaBH4 cross-linking assays
Preparation of the 3′-end labeled dRP lyase substrate was as described previously (50). The 32P-labeled duplex DNA was pretreated with UDG and APE1 to prepare the single-nucleotide gapped substrate that contained a 5′-dRP flap and a 3′-OH at the margins. For preparation 5′-end labeled substrate, dephosphorylated 17-mer oligodeoxyribonucleotide (5′-UGTS-SGGATCCCCGGGTACBiotin-3′) containing a uracil residue at the 5′-end, a disulfide bond (S-S) three nucleotide from the 5′-end, and biotin at the 3′-end was phosphorylated with Optikinase and [γ-32P]ATP. A 34-mer (5′-GTACCCGGGGATCCGTACGGCGCATCAGCTGCAG-3′) template was then annealed with a 15-mer (5′-CTGCAGCTGATGCGC-3′) and the 17-mer 32P-labeled oligonucleotides by heating the solution at 90°C for 3 min and allowing the solution to slowly cool to 25°C. The 32P-labeled duplex DNA was treated with UDG to generate the 32P-labeled deoxyribose sugar phosphate-containing single-nucleotide gapped substrate. The S-S bond was included in the substrate molecule to enable future studies on cross-linking within the dRP lyase active site.
dRP lyase assay
dRP lyase activity was measured essentially as described previously (50,51). Briefly, the reaction mixture (10 μl) contained 50 mM HEPES, pH 7.5, 20 mM KCl, 2 mM dithiothreitol, 1 mM EDTA, and 50 nM preincised 32P-labled AP site -containing DNA. The reaction was initiated by adding appropriate dilutions of either purified full-length Rev1, catalytically active DNA polymerase domain and referred to here as the core domain (CD), or pol β; the incubation was at 37°C as indicated in the figure legends. After the incubation, the reaction products were stabilized by addition of freshly prepared 1 M NaBH4 to a final concentration of 100 mM. Reaction mixtures then were transferred to 0–1°C (on ice), and incubation was continued for 30 min on ice. Next, after incubation at 75°C for 2 min, the reaction products were separated by electrophoresis in a 17% polyacrylamide gel containing 8 M urea in 89 mM Tris–HCl, pH 8.8, 89 mM boric acid and 2 mM EDTA. Imaging and data analysis were performed by PhosphorImager and ImageQuant software.
Covalent cross-linking assay
To prepare the covalent cross-linked protein–DNA complex, a NaBH4 trapping technique was utilized EDTA, 200 nM 5′ 32P-labeled UDG/APE1-treated duplex DNA, appropriate dilutions of Rev1/CD/pol β as indicated in figure legends, and 1 mM NaBH4. The reaction mixture was incubated for 60 min on ice and then 10 min at room temperature. After incubation, the reaction was terminated by addition of 10 μl of SDS-PAGE gel-loading buffer. NuPAGE Bis–Tris gel (10%) and MOPS running buffer system were used to separate protein–DNA cross-linked complexes. Typhoon PhosphorImager was used for scanning the gels.
Kinetic measurements of dRP lyase activity
Kinetic analysis of dRP lyase activity of the CD of Rev1 was performed essentially as described previously (51,52). For the kinetic measurements, a 34-bp duplex DNA was used that contained uracil at position 16 and a nick between positions 15 and 16. This DNA was prepared by annealing both a 15-mer oligonucleotide and a 19-mer oligonucleotide with uracil at the 5′-end and 6-FAM tag at the 3′-end to the 34-mer complementary DNA strand. The DNA substrate was pretreated with UDG as above. The reaction mixture contained 50 mM HEPES, pH 7.4, 20 mM KCl, 1 mM EDTA, 100 nM duplex DNA. For a time course determination, the reaction mixture (50 μl) was assembled at 0–1°C (on ice) in the above buffer. Reactions were initiated by adding 500 nM CD peptide or the dilution buffer (control) and reaction mixtures were incubated at 37°C. Aliquots (9 μl each) were withdrawn at different time intervals and transferred to 0–1°C to stop the reaction. DNA products were stabilized by addition of 100 mM NaBH4 and incubated 30 min on ice. Then, an equal volume of gel-loading buffer was added, and the reaction mixture was incubated at 75°C for 2 min. The reaction products were separated by electrophoresis in a 17% polyacrylamide gel containing 8 M urea in 89 mM Tris–HCl, pH 8.8, 89 mM boric acid, and 2 mM EDTA. To quantify the reaction products, gels were scanned on a PhosphorImager and the data were analyzed as above. The rate of dRP lyase activity of Rev1 was determined by plotting the amount of product formed as a function of time. The background obtained from reaction mixtures with dilution buffer for the respective time interval was subtracted. Data were fitted to the appropriate equation by nonlinear least squares methods.
In vitro BER
The repair reaction mixture (10–20 μl) was assembled on ice containing a 35-base pair oligonucleotide duplex DNA (250 nM) with uracil at position 15, 50 mM HEPES, pH 7.5, 0.5 mM EDTA, 2 mM dithiothreitol, 20 mM KCl, 4 mM ATP, 5 mM phosphocreatine, 100 μg/ml phosphocreatine kinase, 0.5 mM NAD, 5 mM MgCl2, 2.2 μM [α-32P]dCTP (specific activity, 1 × 106 dpm/pmol), 40 nM UDG and 20 nM APE1. The pretreated DNA substrate on ice was transferred to the reaction mixture, and it was supplemented either with purified full-length Rev1 (50–500 nM), CD (500 nM), or with pol β (10 nM) and 250 nM DNA ligase I, as indicated in the figure legends. The repair reaction was then initiated by transferring reaction mixtures at 37°C. Aliquots (4.5 μl) were withdrawn at the indicated time intervals. Reactions were terminated by the addition of an equal volume of DNA gel-loading buffer (95% formamide, 20 mM EDTA, 0.02% bromophenol blue, and 0.02% xylene cynaol). After incubation at 75°C for 2 min, the reaction products were separated by electrophoresis and the data were analyzed as above.
Limited proteolysis and biochemical analysis of tryptic fragments of the CD of Rev1
Limited proteolysis and amino-terminal sequencing of the CD (100 kDa) of Rev1 were performed as described previously (51). Briefly, the purified His-MBP-tagged CD (66 μg) and trypsin (0.66 μg) were mixed at a 1:50 weight ratio (trypsin:CD) in 100 mM Tris–HCl, pH 8.0, and the reaction mixture was incubated at 25°C. The final reaction mixture volume was 100 μl. Aliquots (20 μl each) were withdrawn at 0, 2, 5, 10 and 20 min time intervals. A portion of each sample (18 μl) was mixed immediately with 10 μl SDS-PAGE gel-loading buffer, boiled for 5 min, and separated by electrophoresis in a 12% NuPAGE Bis–Tris gel with a MOPS running buffer system. Proteins were electrophoretically transferred onto an Immun-Blot PVDF membrane (7 × 8.4 cm) (Bio-Rad) using a transfer buffer that contained 10 mM CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] and 10% methanol, pH 11.0 at 50 V (7 V/cm) for 1 h. The membrane was stained briefly with 0.2% (w/v) Coomassie blue R-250 in 45% methanol and 10% acetic acid and destained with 90% methanol and 10% acetic acid. The membrane was air-dried, and protein bands were cut with a scalpel. Amino-terminal sequencing was performed using a Model 492 Procise sequencing system (Applied Biosystems) at Wake Forest University, Winston-Salem, NC. The remaining portion of the trypsin digested sample (2 μl) was subjected to NaBH4 cross-linking to with a 5′-end labeled dRP lyase DNA substrate, as described above. The resulting gel was subjected to phosphorimaging, and then the same gel was stained with simlplyBlue SafeStain (Invitrogen) for detection of proteins. The area corresponding to the radioactive band in the phosphorimage and to ∼ 8-kDa peptide in the stained gel was excised and subjected to mass spectrometry analysis at the Mass Spectrometry Research and Support Group Facility, NIEHS-NIH.
RESULTS
In vitro BER activity of mouse Rev1
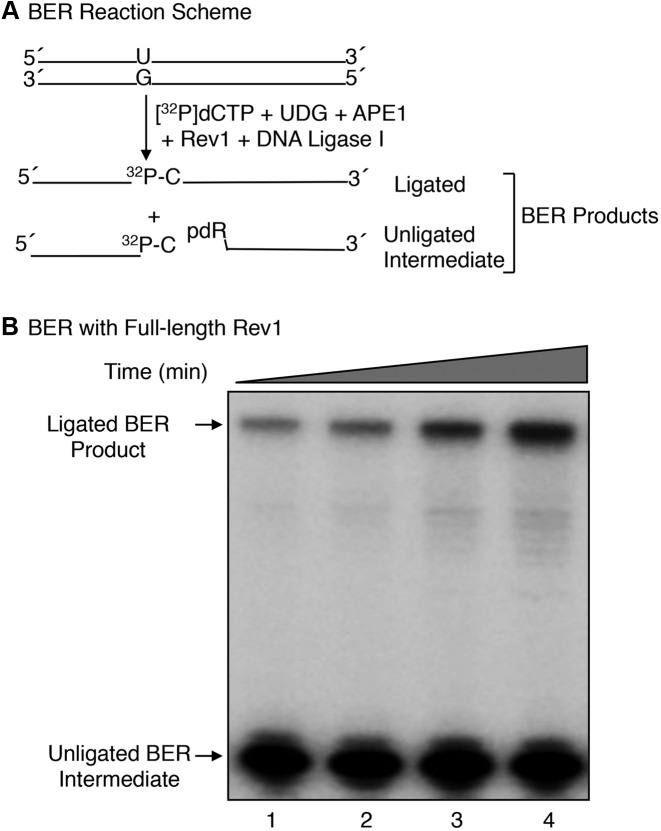
Mouse fibroblast cells lacking pol β have a DNA repair deficiency and a phenotype of elevated MMS-induced mutations (22,53). We had shown previously that Rev1 is strictly required for the elevated MMS-induced mutagenesis phenotype (22) observed in the pol β null background. To explore underlying mechanisms of the Rev1-mediated MMS-induced mutagenesis, we expressed and purified full-length mouse Rev1 (Supplementary Figure S1B) and tested its ability to support in vitro BER. Reaction mixtures were assembled with a 35-bp DNA substrate with uracil opposite guanine, purified BER enzymes including UDG, APE1, DNA ligase I and Rev1 (Figure 1A). Reactions were initiated by the adding Mg2+ and [α-32P]dCTP, and repair activity was monitored by insertion of [α-32P]dCMP in place of dUMP in the 35-bp substrate (Figure 1B). BER activity of Rev1 was observed, and the activity was dependent on Rev1 concentration and incubation time (Figure 1B, Supplementary Figure S1C). Since the complete ligated BER reaction product was observed, these results suggested Rev1 catalyzed both dRP removal and single-nucleotide gap-filling DNA synthesis.
Figure 1.
In vitro BER activity of full-length mouse Rev1. (A) A schematic representation of the in vitro BER substrate (34-base pair duplex DNA containing uracil opposite guanine) and the expected BER products after replacement of uracil with [32P]dCMP. (B) Phosphorimage of denaturing PAGE demonstrating the in vitro BER activity of full-length Rev1 is shown. BER activity of Rev1 was evaluated on a uracil-containing BER substrate by measuring incorporation of [α-32P]dCMP as a function of incubation time. Reaction conditions and product analysis were as described under ‘Materials and Methods.’ A 35-bp oligonucleotide duplex DNA (250 nM), pretreated with UDG and APE1, was used in a reaction mixture with 500 nM full-length Rev1 and 250 nM DNA ligase I. Aliquots (4.5 μl) were withdrawn at 7.5, 15, 30 and 60 min intervals. The reaction products were separated by 15% denaturing PAGE. The migration positions of ligated BER product and unligated BER intermediate are indicated.
However, ligation of the gap-filled BER intermediate in this experiment was much weaker than in reference reactions with pol β (Supplementary Figure S1C, lane 4); this was the case even though the reaction mixtures contained a high level of DNA ligase I. When the incubation was extended to 60 min, a larger fraction of the gap-filled BER intermediate was converted to the ligated BER product (Figure 1B, compare lanes 1 and 4). These initial results suggested that Rev1 has two BER enzymatic activities: Single-nucleotide gap-filling DNA synthesis and dRP lyase removal of the 5′-dRP group. Nevertheless, accumulation of the unligated DNA synthesis product during the incubations suggested that the dRP lyase activity of Rev1 was much weaker than the gap-filling DNA synthesis activity.
5′-dRP lyase activity of mouse Rev1
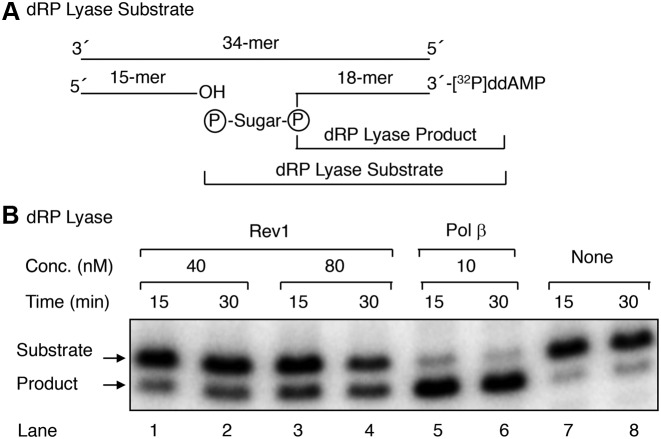
To characterize 5′-dRP lyase properties of purified Rev1, we used a BER oligonucleotide substrate that was 3′-end labeled with [32P]ddAMP. After annealing to the complementary strand, the duplex DNA then was pretreated with UDG and APE1 (Figure 2A). The resulting BER substrate contained a one-nucleotide gap with a 5′-dRP flap and the 32P-label at the 3′-end of the dRP-containing strand (Figure 2A). Removal of the 5′-dRP group from the 32P-labeled strand was monitored by electrophoresis in a denaturing gel. The substrate was incubated either with Rev1 or pol β, that was used here as a reference. The results demonstrated that Rev1 could remove the 5′-dRP group, and this was dependent on the concentration of Rev1 (Figure 2B). Yet, the Rev1 dRP lyase activity was much weaker than that of pol β (Figure 2B).
Figure 2.
dRP lyase activity of full-length Rev1. (A) A schematic representation of the dRP lyase substrate (18-mer + 32P-dRP) generated by pretreatment of 32P-labeled 34-bp DNA with UDG and APE1; the expected 32P-labeled 18-mer product formed as a result of Rev1 or pol β (positive control) excision of the 5′-dRP group. (B) Pre-incised DNA substrate (50 nM) was incubated either without enzyme (lanes 7 and 8) or with 40 nM Rev1 (lanes 1 and 2), 80 nM Rev1 (lanes 3 and 4), or 10 nM pol β (lanes 5 and 6) for 15 and 30 min. After the incubation, the DNA products were stabilized and analyzed as described under ‘Materials and Methods.’ A photograph of the phosphorimage, illustrating the reaction products, is shown. The positions of the substrate and product are indicated.
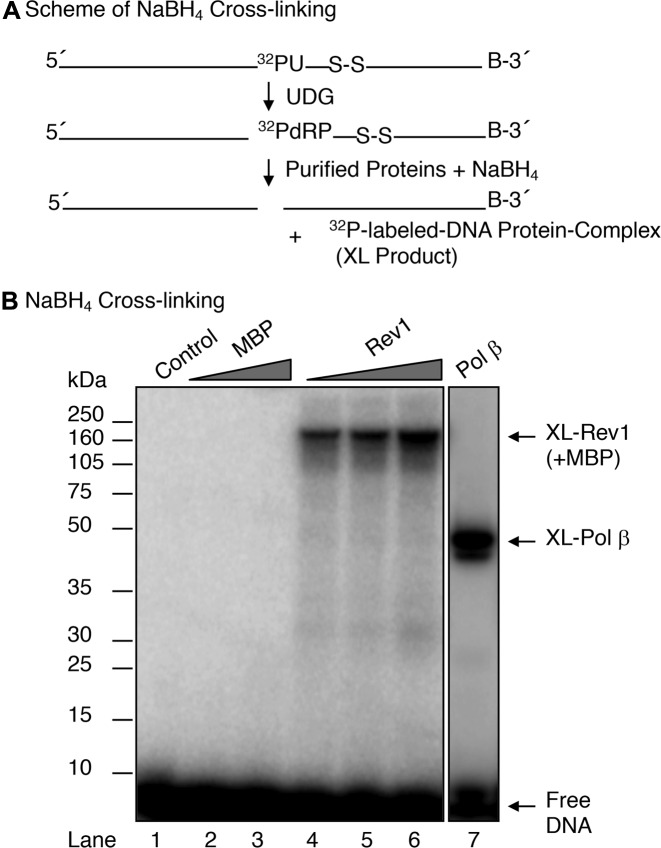
To verify that the dRP lyase activity observed in Rev1 was intrinsic to the enzyme, we performed NaBH4 cross-linking of Rev1 in complex with a 5′-dRP-containing DNA substrate (Figure 3A). In this experiment, the 5′-dRP group in the substrate reacts with a lysine ϵ-amine group in the enzyme and forms a Schiff base-containing intermediate; the Schiff base linkage is then be converted to a stable covalent bond by NaBH4 reduction (12,51). When MBP- Rev1 and reference pol β were subjected to the NaBH4 cross-linking protocol followed by SDS-PAGE analysis of the samples, protein–DNA complexes of ∼186-kDa and ∼46-kDa, respectively, were observed (Figure 3B, lanes 4 to 6, and lane 7). As a control, incubation of the substrate DNA with purified MBP alone failed to show any cross-linked product (Figure 3B, lanes 2–3). The electrophoretic mobilities of the ∼186-kDa and ∼46-kDa protein–DNA complexes were consistent with the molecular masses of MBP-Rev1 and pol β, respectively, in complex with the 32P-labeled 18-mer oligonucleotide (Figure 3B, lanes 4–6 and lane 7). These results are consistent with the interpretation that Rev1 has intrinsic dRP lyase activity.
Figure 3.
NaBH4 cross-linking of full-length Rev1. (A) A schematic representation of the cross-linking substrate (18-mer + 32P-dRP) generated by pretreatment of 5′- 32P-labeled 34-bp DNA with UDG and the expected 32P-labeled protein DNA complex product (XL product) are shown. (B) UDG pre-treated DNA substrate (200 nM) was used in a reaction mixture without enzyme (lane 1) or with 280 and 1000 nM MBP (lanes 2 and 3) or 140, 280 and 1000 nM REV1 (lanes 4, 5, 6), or 100 nM pol β (lane 7), and 1 mM NaBH4. The reaction mixtures were incubated on ice for 1 h followed by incubation at room temperature for 10 min. Covalently cross-linked DNA-protein products were separated by 10% NuPAGE Bis-Tris gel, and the gel was scanned on a PhosphorImager. The positions of cross-linked products and free DNA are indicated. Relative positions of protein markers are indicated on the left side of the phosphorimage.
Characterization of Rev1 domains in relation to BER activities
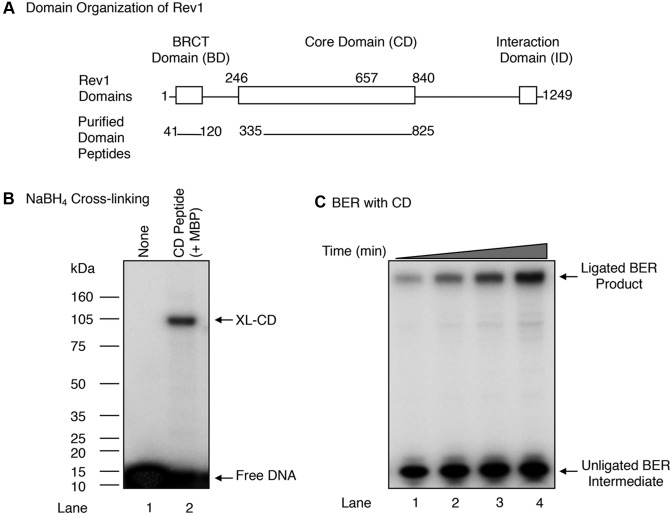
Full-length native mouse Rev1, a 137-kDa polypeptide, contains three main regions, namely: The N-terminal region that contains a sequence similar to the BRCT domain found in several DNA repair proteins, such as XRCC1, PARP-1 and DNA ligase III (54–56) and referred to here as the BD; the central region containing the DNA polymerase active site and referred to here as the CD; and the C-terminal region termed the interaction domain (ID) (Figure 4A and Supplementary Figure S2A) (30,33,57). Since the BD and CD domains are involved in DNA transactions (34,57), we decided to express, purify, and examine the corresponding peptides for their ability to conduct BER activities. First, DNA regions corresponding to the BD (residues 41–120) and CD (residues 335–825) of Rev1 were cloned into an expression vector for production of His-NusA and His-MBP tagged polypeptides, respectively. The proteins were overexpressed in Escherichia coli and purified (Supplementary Figure S2B). In the case of the BD polypeptide, the His-NusA tag was removed by TEV protease.
Figure 4.
In vitro BER and cross-linking (XL) activities of the CD of Rev1. (A) A schematic representation of mouse Rev1 sub-domains. Relative positions of the BRCT domain (BD), core domain (CD) and interaction domain (ID) are indicated. BD (residues 41 to 120) and CD (residues 335 to 825) purified peptides are shown. (B) NaBH4 cross-linking of the Rev1 CD was evaluated as in Figure 2B. UDG pre-treated DNA substrate (200 nM) was used in a reaction mixture without enzyme (lane 1) or with 400 nM purified CD (lane 2), and 1 mM NaBH4. The reaction products were processed as described under ‘Materials and Methods.’ The positions of XL-CD and free DNA are indicated. Relative positions of protein markers are indicated on the left side of the phosphorimage. (C) Phosphorimage of denaturing PAGE demonstrating the in vitro BER activity of the Rev1 CD. BER activity of the CD (500 nM) was evaluated using a uracil-containing BER substrate by measuring incorporation of [α-32P]dCMP as a function of incubation time, as in Figure 1B. A 35-bp oligonucleotide duplex DNA (250 nM) was added to a reaction mixture with 500 nM CD and 250 nM DNA ligase I. After incubation aliquots (4.5 μl) were withdrawn at 7.5, 15, 30 and 60 min. Reactions were terminated by addition of an equal volume of gel-loading buffer. The reaction products were analyzed as above. The migration positions of the ligated BER product and unligated BER intermediate are indicated.
The dRP-NaBH4 cross-linking of these polypeptides was assayed using a 32P-labeled dRP lyase substrate. The BD failed to show cross-linking to the dRP lyase substrate (data not shown), whereas the CD had retained cross-linking activity (Figure 4B, lane 2). Since the CD of Rev1 has polymerase and lyase activities (as shown here), we next investigated whether the purified CD could support BER (Figure 4C). BER reaction mixtures were assembled under similar conditions as above, and the in vitro BER activity of the CD was monitored (Figure 4C). The results indicated that the CD was able to mediate BER, involving dRP removal and one-nucleotide gap-filling activities (Figure 4C); therefore, CD was selected for more detailed studies of the dRP lyase activity.
Kinetics measurements of dRP lyase activity of the Rev1 CD
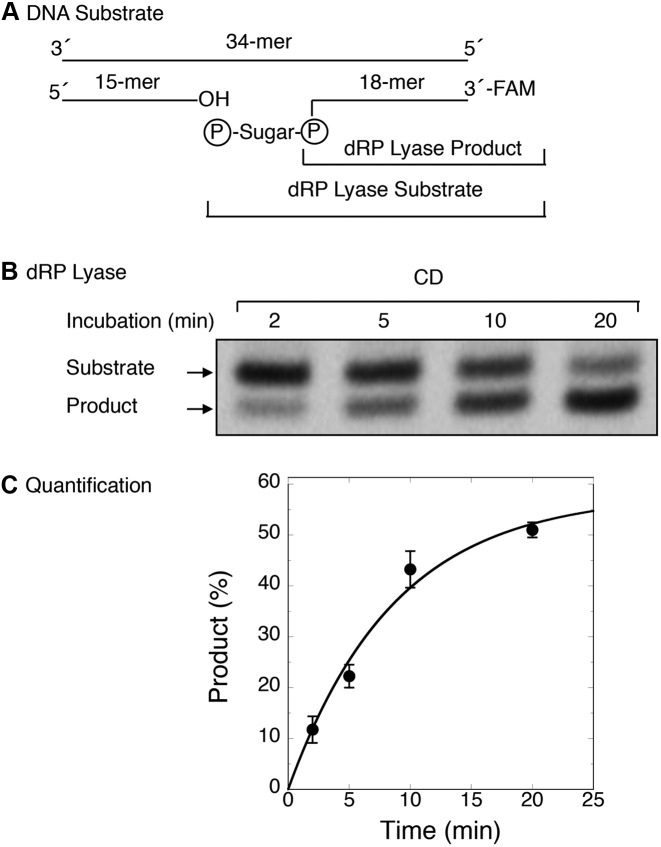
To determine kinetic parameters of 5′-dRP lyase activity of the Rev1 CD, we prepared a 34-bp duplex DNA substrate by annealing three DNA strands (Figure 5A). The substrate contained an 18-mer DNA strand with 6-FAM at the 3′-end and 5′-RP flap at the margin of a single-nucleotide gap (Figure 5A). The rate of dRP removal by the CD was determined under single turnover kinetics conditions (i.e., ratio of enzyme/DNA = 5). The results revealed that the CD released the 5′-dRP group, as monitored by product (labeled 18-mer DNA) formation (Figure 5B), with an observed rate of ∼ 0.10 per min (Figure 5C).
Figure 5.
Kinetic measurements of dRP lyase activity of the CD of Rev1. The release of dRP from the incised AP-site containing DNA substrate was examined as a function of incubation time, and the rate of dRP removal by CD was determined under single turnover conditions (i.e. enzyme/DNA = 5). The reaction was performed with purified CD as described under ‘Materials and Methods’. (A) Schematic representation of the DNA substrate prepared by annealing a 15-mer oligonucleotide, a 18-mer oligonucleotide containing uracil at the 5′-end in the nick and FAM at the 3′-end, and a 34-mer oligonucleotide template. (B) The duplex DNA (100 nM) was added to a reaction mixture with 500 nM CD. Aliquots were withdrawn at the indicated times, and the products were stabilized by the addition of 20 mM NaBH4 and incubation on ice for 30 min. Then, an equal volume of gel-loading dye buffer was added, and the reaction products were analyzed as in Figure 2B. A representative phosphorimage of the polyacrylamide gel illustrates product formation at various times. (C) Time course data were analyzed using ImageQuant software and fitted to an exponential equation by using nonlinear least squares methods to determine the reaction rate (kobs = 0.10 per minute). Average values of relative product formed (%) from four experiments were plotted against incubation time (min). Standard error bars are shown.
Domain mapping of the CD by limited proteolysis
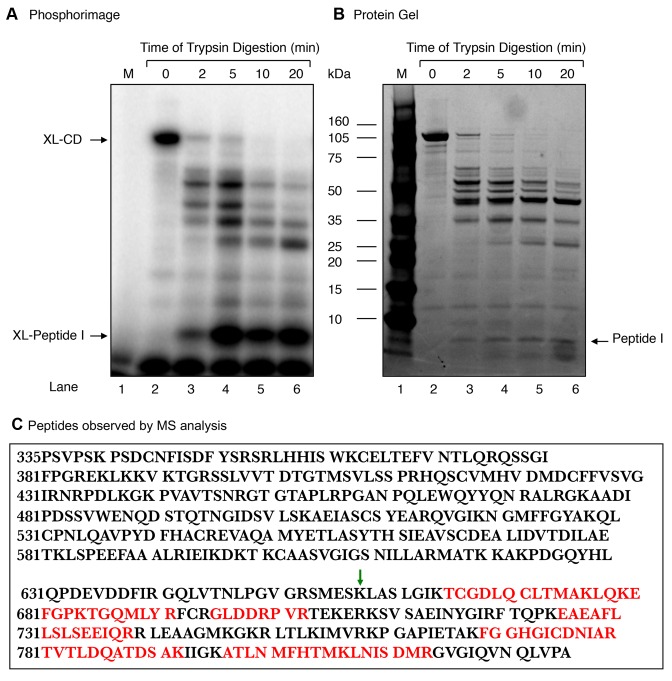
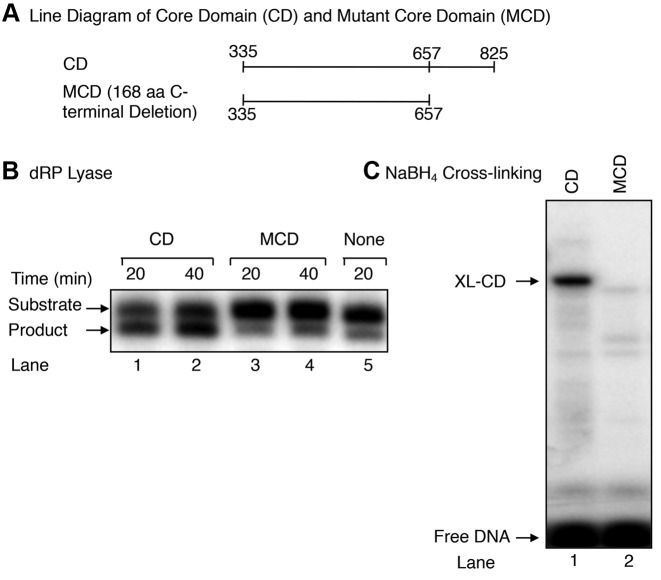
Experiments were conducted to further localize the active site of the Rev1 dRP lyase activity. The CD was subjected to limited tryptic digestion, and then the incubation mixtures were subjected to NaBH4 cross-linking (Figure 6A). With 5 min digestion, one major labeled peptide (termed Peptide I) of ∼ 8-kDa and minor higher molecular mass peptides were observed (Figure 6A). The labeled ∼ 8-kDa peptide persisted during longer incubations, i.e., 10 and 20 min (Figure 6A, lanes 5 and 6). When the same gel was stained for peptides, a band of ∼ 8-kDa was observed (Figure 6B). A gel slice corresponding to the ∼ 8-kDa area of the gel was recovered; the peptide was subjected to mass spectrometric analysis (Figure 6B). Several peptides (shown in red) were observed corresponding to the ∼ 170 amino acid segment of the carboxy-terminus of the CD (Figure 6C). The identity of the ∼ 8-kDa peptide (I) (Figure 6C) also was verified by N-terminal sequencing. Therefore, we cloned and expressed a deletion mutant (residues 335–657) lacking the 168-amino acid segment the carboxy-terminus of the CD, and we refer to this deletion mutant as the ‘mutant core domain’ (MCD). Next, we tested for dRP lyase and NaBH4 cross-linking activities of the MCD. The results revealed the MCD was devoid of both dRP lyase and cross-linking activities (Figure 7B and C). Thus, these results revealed that the dRP lyase active site resides in the 168-amino acid segment of the carboxy-terminus of the Rev1 CD.
Figure 6.
Limited proteolysis of the Rev1 CD with trypsin. Purified 100-kDa CD (66 μg) was digested at 25°C with trypsin (0.66 μg) at a weight ratio of 1:100 (trypsin:CD) in 100 mM Tris–HCl, pH 8.0. Aliquots were withdrawn at 2, 5, 10 and 20 min, as indicated at the top of the photograph. (A) A portion (1/10th) of each digested sample was incubated with 5′-end labeled dRP lyase DNA substrate and then subjected to NaBH4 cross-linking as in Figure 2. The gel was scanned on a PhosphorImager. The positions of XL-CD and the ∼XL-peptide (I) (∼8-kDa) are indicated. (B) A photograph of the SimplyBlue stained gel in (A) is shown. (C) The radiolabeled fragment corresponding to the ∼ 8-kDa peptide (I) from the stained gel in (B) was incised from the gel and subjected to MS analysis. The tryptic peptide amino acid sequences found in MS/MS analysis, corresponding to the ∼170 amino acid segment of the carboxy-terminus of the CD of Rev1, are indicated in red. The green arrow indicates the deletion site in the mutant CD (MCD) of Rev1 (see Figure 7).
Figure 7.
dRP lyase and NaBH4 cross-linking activities of the CD and MCD of Rev1. (A) A line diagram illustrating the amino acid segments of CD and MCD of Rev1 (B) dRP lyase activity of the CD and MCD of Rev1. UDG/APE1 pre-treated DNA substrate (50 nM) was incubated either with 200 nM CD (lanes 1 and 2), 200 nM MCD (lanes 3 and 4), or without enzyme (lane 5). After incubation of 20 and 40 min, the DNA products were stabilized and analyzed as in Figure 2B. The positions of substrate and the product are indicated. (C) NaBH4 cross-linking (XL) activity of the CD and MCD. UDG/APE1 pre-treated DNA substrate (200 nM) as in Figure 4B was mixed either with dilution buffer (lane 1), 400 nM CD (lane 2), or 400 nM MCD (lane 3) and 1 mM NaBH4. The reaction conditions and product analysis were as in Figure 4B. The positions of XL-CD and free DNA are indicated.
DISCUSSION
We reported earlier that Rev1 is required for the MMS-induced mutagenesis phenotype observed in a pol β null background in mouse fibroblast cells (22). Our interpretations of this phenotype were that Rev1 substituted for pol β during BER and processed intermediates of BER in an error-prone manner or that Rev1 recruited another DNA polymerase to the BER intermediates, possibly an error-prone Y-family DNA polymerase that had already been implicated in BER. In the current study to explore the possibility that Rev1 itself may contribute enzymatic roles in BER, we examined purified Rev1 for activities involved in BER, including gap-filling DNA synthesis and 5′-dRP lyase. When Rev1 was substituted for pol β, the complete ligated repair product was observed (Figure 1). Rev1 insertion of dCMP into a gap opposite deoxyguanine in the template was not surprising since Rev1 is well known for its specific deoxycytidyl transferase activity (58). This result suggested Rev1 has a new activity capable of removing the 5′-dRP group at the margin of the DNA gap formed by APE1 incision of the AP site in DNA. We confirmed this activity of Rev1 by direct assays for dRP lyase activity and by Schiff base reduction experiments to trap the β-elimination intermediate Further, we found that both the dRP lyase and gap-filling activities reside in the ∼57 kDa region of the catalytic domain of Rev1, spanning residues 335–825, and an expressed protein corresponding to this region was sufficient to perform BER in a fashion similar to that of full-length Rev1 (Figures 1 and 4). Using limited proteolysis, mass spectrometry and sodium borohydride cross-linking methods, we demonstrated that the dRP lyase activity resides in the 168 residue C-terminal segment of the ∼57-kDa region of Rev1.
Characterization of enzymatic properties of the Rev1 dRP lyase activity revealed that the rate constant from single-turnover analysis (0.1/min) was orders of magnitude lower than that of pol β. Yet, the steady-state rate (0.1/min) was only 45-fold lower than that of pol β (4.5/min) (50). As expected for a weak single-turnover rate constant, the steady-state rate was similar.
From biochemical and genetic studies, it was proposed that the N-terminal region of Rev1 is responsible for recruiting Rev1 to DNA damaged through the BD that mediates protein–protein interactions. Interestingly, deletion of the BD of Rev1 in ES cells showed hypersensitivity to DNA damaging agents. These results were consistent with a regulatory role of the BD of Rev1 in TLS (59). The central region of Rev1, the catalytic domain, is responsible for the DNA polymerase activity that primarily incorporates dCMP opposite deoxyguanines and AP sites. The extreme C-terminal region of Rev1 was shown to play a role in recruiting Y-family DNA polymerases and repair proteins to sites of DNA damage (33,60). Mice harboring catalytically inactive Rev1 showed reductions of not only C to G and G to C transversions, but also C to T and G to A transitions and A:T transversions, suggesting regulation of these mutations through functional interactions with other DNA polymerases (34). In this study, the authors suggested that Rev1 participates in multiple mutagenetic pathways (34), and our previous observation of Rev1-dependance in MMS-induced mutagenesis was consistent with the notion of Rev1 involvement in multiple pro-mutagenesis pathways. In that study, a subset of the mutations observed in pol β null MEFs could have originated from Rev1-mediated BER, apart from the expected contribution of lesion bypass DNA polymerases to the mutagenesis (22).
During SHM in immunoglobulin genes and class switch recombination, AP sites are generated after activation-induced cytidine deaminase (AID) action (61). AID deaminates cytosines to uracils and the AP sites are formed by UDG removal of uracils. AP sites also are formed spontaneously as a result of hydrolytic cleavage of the N-glycosylic bond or through DNA glycosylase catalyzed damaged base removal, and these AP sites are common intermediates in several DNA transactions in mammalian cells, including SHM, TLS and BER (1,5,61–63). While some of the AP sites are processed through other repair pathways, a portion of these AP sites might be processed through BER after APE1 incision (39,61,63).
The pol β-mediated, relatively error-free, BER may not be advantageous during Ig genes hypermutation, and we and others have shown that the expression of pol β in B cells abolishes mutagenesis; these results were in agreement with those obtained by Rajeswky and associates in a mouse model in which they showed a lack of requirement of pol β in SHM (44,64,65). Consistent with these observations, Stavnezer and associates showed that deficiency of either pol β or pol λ or both polymerases had no direct role in SHM in a mouse model (43). Notably, we observed a significant level of pol β-independent BER in B cells, and this was consistent with involvement of error-prone BER during SHM mutagenesis. Therefore some of the mutations may have resulted from BER mediated by lesion bypass DNA polymerases (44). These results also suggested that Rev1 might participate during Ig gene SHM through its BER activities. Interestingly, DNA polymerase θ, a low-fidelity DNA polymerase that has been implicated in SHM, possesses weak dRP lyase and single-nucleotide gap-filling activities similar to Rev1 (52,66). Taken together, these results point to the possibility that Rev1 may process some of the AP sites through error-prone BER. It is also interesting that a mutational signature of APOBEC family of cytidine deaminases was found in breast and several other types of cancers genomic DNA (67–70). This suggests that APOBEC-mediated cytosine deaminase activity and Rev1 dCMP insertion could be involved in cancer genome mutagenesis, in addition to SHM.
Finally, it will be interesting in future experiments to identify the nucleophile (s) responsible for the dRP lyase β-elimination reaction and to examine the biological effects of alterations in this residue (s) on BER and Ig genes SHM. These experiments, however, were beyond the scope of the present study.
Supplementary Material
Acknowledgments
We thank W.A. Beard and Yesenia Rodriguez for critical reading of the manuscript; Dr. Robert Petrovich and Mrs. Lori Edwards, NIEHS Protein Expression Core Laboratory, for protein expression; Dr. Jason Williams and Mrs. Katina Johnson, NIEHS Mass Spectrometry Research and Support Group, for mass spectrometry; and Dr. Mark Lively, Wake Forest University, for protein sequencing.
Footnotes
Present address: Vladimir Poltoratsky, St. Johns University, 8000 Utopia Parkway Queens, New York, NY 11439, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences [Z01ES050158 and Z01ES050159]. Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences [Z01ES050158 and Z01ES050159].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. DNA repair enzymes. Annu. Rev. Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat. Res. 2000;462:129–135. doi: 10.1016/s1383-5742(00)00024-7. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T., Wood R.D. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 4.Seeberg E., Eide L., Bjoras M. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 5.Wilson S.H. Mammalian base excision repair and DNA polymerase beta. Mutat. Res. 1998;407:203–215. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 6.Fortini P., Pascucci B., Parlanti E., Sobol R.W., Wilson S.H., Dogliotti E. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry. 1998;37:3575–3580. doi: 10.1021/bi972999h. [DOI] [PubMed] [Google Scholar]
- 7.Fortini P., Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair. 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Krokan H.E., Bjoras M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doetsch P.W., Cunningham R.P. The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res. 1990;236:173–201. doi: 10.1016/0921-8777(90)90004-o. [DOI] [PubMed] [Google Scholar]
- 10.Doetsch P.W., Helland D.E., Haseltine W.A. Mechanism of action of a mammalian DNA repair endonuclease. Biochemistry. 1986;25:2212–2220. doi: 10.1021/bi00356a054. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto Y., Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 12.Piersen C.E., Prasad R., Wilson S.H., Lloyd R.S. Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction. J. Biol. Chem. 1996;271:17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 13.Prasad R., Beard W.A., Wilson S.H. Studies of gapped DNA substrate binding by mammalian DNA polymerase beta. Dependence on 5′-phosphate group. J. Biol. Chem. 1994;269:18096–18101. [PubMed] [Google Scholar]
- 14.Beard W.A., Prasad R., Wilson S.H. Activities and mechanism of DNA polymerase beta. Methods Enzymol. 2006;408:91–107. doi: 10.1016/S0076-6879(06)08007-4. [DOI] [PubMed] [Google Scholar]
- 15.Singhal R.K., Prasad R., Wilson S.H. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J. Biol. Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- 16.Sobol R.W., Horton J.K., Kuhn R., Gu H., Singhal R.K., Prasad R., Rajewsky K., Wilson S.H. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 17.Tomkinson A.E., Chen L., Dong Z., Leppard J.B., Levin D.S., Mackey Z.B., Motycka T.A. Completion of base excision repair by mammalian DNA ligases. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:151–164. doi: 10.1016/s0079-6603(01)68097-8. [DOI] [PubMed] [Google Scholar]
- 18.Prasad R., Singhal R.K., Srivastava D.K., Molina J.T., Tomkinson A.E., Wilson S.H. Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 19.Caldecott K.W., Tucker J.D., Stanker L.H., Thompson L.H. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prigent C., Satoh M.S., Daly G., Barnes D.E., Lindahl T. Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol. Cell. Biol. 1994;14:310–317. doi: 10.1128/mcb.14.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobol R.W., Wilson S.H. Mammalian DNA beta-polymerase in base excision repair of alkylation damage. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:57–74. doi: 10.1016/s0079-6603(01)68090-5. [DOI] [PubMed] [Google Scholar]
- 22.Poltoratsky V., Horton J.K., Prasad R., Wilson S.H. REV1 mediated mutagenesis in base excision repair deficient mouse fibroblast. DNA Repair. 2005;4:1182–1188. doi: 10.1016/j.dnarep.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Lin W., Xin H., Zhang Y., Wu X., Yuan F., Wang Z. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda Y., Kamiya K. Biochemical properties of the human REV1 protein. FEBS Lett. 2002;520:88–92. doi: 10.1016/s0014-5793(02)02773-4. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence C.W., Maher V.M. Eukaryotic mutagenesis and translesion replication dependent on DNA polymerase zeta and Rev1 protein. Biochem. Soc. Trans. 2001;29:187–191. doi: 10.1042/0300-5127:0290187. [DOI] [PubMed] [Google Scholar]
- 26.Johnson R.E., Torres-Ramos C.A., Izumi T., Mitra S., Prakash S., Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glassner B.J., Rasmussen L.J., Najarian M.T., Posnick L.M., Samson L.D. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monti P., Ciribilli Y., Russo D., Bisio A., Perfumo C., Andreotti V., Menichini P., Inga A., Huang X., Gold B., et al. Rev1 and Polzeta influence toxicity and mutagenicity of Me-lex, a sequence selective N3-adenine methylating agent. DNA Repair. 2008;7:431–438. doi: 10.1016/j.dnarep.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haracska L., Kondratick C.M., Unk I., Prakash S., Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 30.Ross A.L., Simpson L.J., Sale J.E. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sale J.E., Ross A.L., Simpson L.J. Analysis of DNA replication damage bypass and its role in immunoglobulin repertoire development. Subcell. Biochem. 2006;40:271–294. doi: 10.1007/978-1-4020-4896-8_16. [DOI] [PubMed] [Google Scholar]
- 32.Jansen J.G., Langerak P., Tsaalbi-Shtylik A., van den Berk P., Jacobs H., de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo C., Fischhaber P.L., Luk-Paszyc M.J., Masuda Y., Zhou J., Kamiya K., Kisker C., Friedberg E.C. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda K., Ouchida R., Li Y., Gao X., Mori H., Wang J.Y. A critical role for REV1 in regulating the induction of C:G transitions and A:T mutations during Ig gene hypermutation. J. Immunol. 2009;183:1846–1850. doi: 10.4049/jimmunol.0901240. [DOI] [PubMed] [Google Scholar]
- 35.Clark D.R., Zacharias W., Panaitescu L., McGregor W.G. Ribozyme-mediated REV1 inhibition reduces the frequency of UV-induced mutations in the human HPRT gene. Nucleic Acids Res. 2003;31:4981–4988. doi: 10.1093/nar/gkg725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibbs P.E., Wang X.D., Li Z., McManus T.P., McGregor W.G., Lawrence C.W., Maher V.M. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson L.J., Sale J.E. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martomo S.A., Yang W.W., Gearhart P.J. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J. Exp. Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rada C., Di Noia J.M., Neuberger M.S. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Rada C., Ehrenstein M.R., Neuberger M.S., Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 41.Peled J.U., Kuang F.L., Iglesias-Ussel M.D., Roa S., Kalis S.L., Goodman M.F., Scharff M.D. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 42.Schaetzlein S., Chahwan R., Avdievich E., Roa S., Wei K., Eoff R.L., Sellers R.S., Clark A.B., Kunkel T.A., Scharff M.D., et al. Mammalian Exo1 encodes both structural and catalytic functions that play distinct roles in essential biological processes. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E2470–E2479. doi: 10.1073/pnas.1308512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrader C.E., Linehan E.K., Ucher A.J., Bertocci B., Stavnezer J. DNA polymerases beta and lambda do not directly affect Ig variable region somatic hypermutation although their absence reduces the frequency of mutations. DNA Repair. 2013;12:1087–1093. doi: 10.1016/j.dnarep.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poltoratsky V., Prasad R., Horton J.K., Wilson S.H. Down-regulation of DNA polymerase beta accompanies somatic hypermutation in human BL2 cell lines. DNA Repair. 2007;6:244–253. doi: 10.1016/j.dnarep.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bebenek K., Tissier A., Frank E.G., McDonald J.P., Prasad R., Wilson S.H., Woodgate R., Kunkel T.A. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science. 2001;291:2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 46.Beard W.A., Wilson S.H. Purification and domain-mapping of mammalian DNA polymerase beta. Methods Enzymol. 1995;262:98–107. doi: 10.1016/0076-6879(95)62013-3. [DOI] [PubMed] [Google Scholar]
- 47.Strauss P.R., Beard W.A., Patterson T.A., Wilson S.H. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Biol. Chem. 1997;272:1302–1307. doi: 10.1074/jbc.272.2.1302. [DOI] [PubMed] [Google Scholar]
- 48.Slupphaug G., Eftedal I., Kavli B., Bharati S., Helle N.M., Haug T., Levine D.W., Krokan H.E. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y.C., Burkhart W.A., Mackey Z.B., Moyer M.B., Ramos W., Husain I., Chen J., Besterman J.M., Tomkinson A.E. Mammalian DNA ligase II is highly homologous with vaccinia DNA ligase. Identification of the DNA ligase II active site for enzyme-adenylate formation. J. Biol. Chem. 1994;269:31923–31928. [PubMed] [Google Scholar]
- 50.Prasad R., Beard W.A., Strauss P.R., Wilson S.H. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J. Biol. Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 51.Prasad R., Bebenek K., Hou E., Shock D.D., Beard W.A., Woodgate R., Kunkel T.A., Wilson S.H. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase iota by controlled proteolysis. J. Biol. Chem. 2003;278:29649–29654. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- 52.Prasad R., Longley M.J., Sharief F.S., Hou E.W., Copeland W.C., Wilson S.H. Human DNA polymerase theta possesses 5′-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sobol R.W., Watson D.E., Nakamura J., Yakes F.M., Hou E., Horton J.K., Ladapo J., Van Houten B., Swenberg J.A., Tindall K.R., et al. Mutations associated with base excision repair deficiency and methylation-induced genotoxic stress. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6860–6865. doi: 10.1073/pnas.092662499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor R.M., Wickstead B., Cronin S., Caldecott K.W. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr. Biol. 1998;8:877–880. doi: 10.1016/s0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- 55.Eustermann S., Wu W.F., Langelier M.F., Yang J.C., Easton L.E., Riccio A.A., Pascal J.M., Neuhaus D. Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol. Cell. 2015;60:742–754. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dulic A., Bates P.A., Zhang X., Martin S.R., Freemont P.S., Lindahl T., Barnes D.E. BRCT domain interactions in the heterodimeric DNA repair protein XRCC1-DNA ligase III. Biochemistry. 2001;40:5906–5913. doi: 10.1021/bi002701e. [DOI] [PubMed] [Google Scholar]
- 57.Guo C., Sonoda E., Tang T.S., Parker J.L., Bielen A.B., Takeda S., Ulrich H.D., Friedberg E.C. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006;23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 58.Nelson J.R., Lawrence C.W., Hinkle D.C. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 59.Jansen J.G., Tsaalbi-Shtylik A., Langerak P., Calleja F., Meijers C.M., Jacobs H., de Wind N. The BRCT domain of mammalian Rev1 is involved in regulating DNA translesion synthesis. Nucleic Acids Res. 2005;33:356–365. doi: 10.1093/nar/gki189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohashi E., Murakumo Y., Kanjo N., Akagi J., Masutani C., Hanaoka F., Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 61.Neuberger M.S., Di Noia J.M., Beale R.C., Williams G.T., Yang Z., Rada C. Somatic hypermutation at A.T pairs: polymerase error versus dUTP incorporation. Nat. Rev. Immunol. 2005;5:171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- 62.McCullough A.K., Dodson M.L., Lloyd R.S. Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 63.Saribasak H., Gearhart P.J. Does DNA repair occur during somatic hypermutation. Semin. Immunol. 2012;24:287–292. doi: 10.1016/j.smim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Esposito G., Texido G., Betz U.A., Gu H., Muller W., Klein U., Rajewsky K. Mice reconstituted with DNA polymerase beta-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1166–1171. doi: 10.1073/pnas.97.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu X., Stavnezer J. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1677–1689. doi: 10.1084/jem.20070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshimura M., Kohzaki M., Nakamura J., Asagoshi K., Sonoda E., Hou E., Prasad R., Wilson S.H., Tano K., Yasui A., et al. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol. Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akre M.K., Starrett G.J., Quist J.S., Temiz N.A., Carpenter M.A., Tutt A.N., Grigoriadis A., Harris R.S. Mutation Processes in 293-Based Clones Overexpressing the DNA Cytosine Deaminase APOBEC3B. PLoS One. 2016;11:e0155391. doi: 10.1371/journal.pone.0155391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Borresen-Dale A.L., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krokan H.E., Saetrom P., Aas P.A., Pettersen H.S., Kavli B., Slupphaug G. Error-free versus mutagenic processing of genomic uracil–relevance to cancer. DNA Repair. 2014;19:38–47. doi: 10.1016/j.dnarep.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 70.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C., et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.