Abstract
A coordinated and faithful DNA damage response is of central importance for maintaining genomic integrity and survival. Here, we show that exposure of human cells to benzo(a)pyrene 9,10-diol-7,8-epoxide (BPDE), the active metabolite of benzo(a)pyrene (B(a)P), which represents a most important carcinogen formed during food preparation at high temperature, smoking and by incomplete combustion processes, causes a prompt and sustained upregulation of the DNA repair genes DDB2, XPC, XPF, XPG and POLH. Induction of these repair factors on RNA and protein level enhanced the removal of BPDE adducts from DNA and protected cells against subsequent BPDE exposure. However, through the induction of POLH the mutation frequency in the surviving cells was enhanced. Activation of these adaptive DNA repair genes was also observed upon B(a)P treatment of MCF7 cells and in buccal cells of human volunteers after cigarette smoking. Our data provide a rational basis for an adaptive response to polycyclic aromatic hydrocarbons, which occurs however at the expense of mutations that may drive cancer formation.
INTRODUCTION
Genotoxic stress causes DNA damage that can lead to mutations, carcinogenesis and/or cell death. To counteract these disastrous effects, DNA repair mechanisms have been evolved that remove or tolerate DNA lesions and thereby maintain genomic integrity. Most DNA repair pathways are complex, involving multiple proteins that work in coordinated and regulated steps. Erroneous repair might be a consequence of uncoordinated or imbalanced expression of DNA repair proteins. A major mechanism regulating DNA repair activity rests on the transcriptional activation of DNA repair genes (1).
To date, more than 20 mammalian DNA repair genes have been reported to be subject to transcriptional activation by genotoxic stress, mediated predominantly via the transcription factors p53 and AP-1 (1). However, only for some of them evidence was provided that transcriptional activation has a biological consequence. For example, p53-deficient cells are hypersensitive to ultraviolet (UV) light due to lack of induction of the NER (nucleotide excision repair) genes DDB2 and XPC (2,3). Similar to p53, also c-Fos deficient cells are UV hypersensitive (4,5). In this case c-Fos, being a component of AP-1, regulates the induction of the NER endonucleases XPF and XPG (6–8) and the exonuclease TREX1 (9,10), thus abrogating the block of transcription and replication following UV irradiation (6). An important biological consequence of transcriptional activation of DNA repair genes is the so-called adaptive response. In a narrow sense, adaptive response pertains to exposure to a low priming dose of a genotoxicant, which leads to enhanced protection against a subsequent higher challenging dose of the same genotoxicant. In a broader sense, the genotoxic adaptive response describes a protection of pre-exposed cells against a wide range of genotoxic agents. The adaptive response was first discovered in Escherichia coli upon exposure to the methylating agent N-methyl-N'-nitro-N-nitrosoguanidine (MNNG), where it was shown to be a consequence of the induction of DNA repair genes such as the alkyltransferase Ada (11–14). In mammalian cells, an adaptive response was shown following exposure to ionizing radiation (IR) (15), bleomycin, mitomycin C (16), methylnitrosourea (17,18) and ethylnitrosourea (19), however the question of whether this phenomena were caused by the induction of DNA repair has not yet been answered. Also upon UV-C exposure, an adaptive response was reported, which was associated with transcriptional upregulation of the NER genes XPF and XPG via the MAPK/AP-1 pathway (8). However, a direct proof for the importance of transcriptional activation in this process is still missing. Furthermore, it has not been shown that DNA repair can be upregulated on transcriptional level in response to environmental genotoxic stress and carcinogen exposures such as smoking and that the adaptive response plays a role in tumor prevention.
Tobacco smoking represents the major preventable cause of cancer, contributing to one in five deaths in developed countries. It was estimated that 50% of lifelong smokers die prematurely of smoking-related diseases (20). Smoking is causally linked to cancer of lung, bladder, renal pelvis, oral cavity, pharynx, larynx, esophagus, pancreas, liver, stomach, uterine, cervix, ovary, tongue, nasal cavity, bone marrow, colon and rectum (21). Among the various carcinogenic substances present in tobacco smoke, the most abundant are polycyclic aromatic hydrocarbons (PAHs) (22). The most relevant PAH is benzo(a)pyrene (B(a)P), representing the predominant product of incomplete combustion formed, among others, during tobacco smoking and food preparation. Due to its lipophilic nature, B(a)P has to be metabolized in order to be removed from the body. The metabolism is complex as it involves several enzymatic systems and generates reactive metabolites. In brief, in a first step B(a)P is metabolically activated by cytochrome P450 oxidase (subtypes CYP1A1 and CYP1B1) to 2,3-, 4,5-, 7,8- and 9,10-epoxides, which are further metabolized by the epoxide hydrolase to the corresponding 4,5-, 7,8- and 9,10-trans-dihydrodiols. Additional oxidation of the benzo(a)pyrene 7,8-dihydrodiol by cytochrome P450 results in the ultimate carcinogen 7,8-dihydrodiol-9,10-epoxide (BPDE). BPDE can bind to the exocyclic N2-position of guanine (23) thus forming a bulky adduct in the DNA. The binding occurs via the epoxide ring at the C-10 position, the so-called bay region of B(a)P (24). The N2-guanine adducts are formed dose-dependently (25) and, remaining unrepaired, block cell division and induce mutations (26). A genotoxic potential of BPDE has been reported in vivo and in vitro (27). The repair of BPDE-induced N2-guanine adducts is performed in a base pair conformation-dependent manner by NER (28).
The wide environmental distribution of B(a)P, its high carcinogenic potential and the manifold routes of exposures of humans lead to the question of whether protection mechanisms exist against this carcinogen and how efficient the protection is. Here, we addressed specifically the question of whether BPDE is able to upregulate DNA repair genes and whether this ameliorates the DNA repair capacity leading to an adaptive response. To answer this, we analyzed the regulation of genes involved in DNA repair, apoptosis and cell cycle. Further, we studied the NER activity, induction of cell death and mutations in human cells exposed to BPDE. We demonstrate that pre-exposure of cells to a low BPDE dose induces the transcription of the NER genes DDB2, XPC, XPF and XPG and ameliorates the repair of BPDE adducts induced by a subsequent challenge dose. This is the first evidence of an adaptive response induced by and directed against BPDE. We also show that killing protection occurred at the expense of a higher mutation frequency, which resulted from transcriptional activation of the translesion polymerase eta (POLH). We observed the induction of NER genes and PolH also in vivo, i.e. in human buccal cells obtained from human volunteers immediately after cigarette smoking, showing that the data can be translated to humans exposed to benzo(a)pyrene (e.g. following smoking). The observed induction of repair and TLS genes may result in an enhanced survival and at the same time an increased mutation frequency, which may drive a carcinogenic process in benzo(a)pyrene exposed cells.
MATERIALS AND METHODS
Cell lines and chemicals
The human diploid VH10tert foreskin fibroblast cell line, immortalized by stable transfection with the telomerase gene (TERT), was kindly provided by Prof. Mullenders (Department of Toxicogenetics at Leiden University Medical Centre, the Netherlands). Human primary bronchial epithelial cells (PBECs) were purchased from Provitro (Berlin) and cultivated in Airway epithelial cell growth medium containing 10% fetal bovine serum. VH10tert cells were cultivated in Dulbecco's minimal essential medium containing 10% fetal bovine serum. MCF7 breast cancer cells were obtained from CLS Cell Lines Service GmbH, Eppelheim, Germany. All cells were regularly checked for mycoplasma contamination and cultivated under nitrogen atmosphere (7% CO2, 5% O2) at 37°C. Experiments involving human individuals (healthy males 25–40 years), were approved by the ethic committee ‘Ethik-Kommision Rheinland Pfalz’ (837.198.15 (9966)) and a written consent from all individuals was obtained. B(a)P was purchased from SIGMA (B1760), activated r-7,t-8-Dihydroxy-t-9,10-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene (anti-BPDE; CAS no. 58917-67-2) was prepared from trans-7,8-dihydroxy-7,8-dihydrobenzo(a)pyrene (29) as described (30). Commercially available cigarettes used for the in vivo determination of DNA repair genes contained 0.6 mg nicotine, 7 mg tar and 9 mg carbon monoxide.
Overexpression and knockdown of DNA repair genes
For transient transfection, 1 × 106 cells were seeded per 10-cm dish. One day later, cells were transfected with 2 μg of pcDNA3.1 or the pcDNA3.1 vector containing the specific cDNA (coding for the DNA repair factor), using Effectene reagent (Qiagen). Knockdown of PolH was performed using the siRNA from Santa Cruz (sc-36289) and the Lipofectamine RNAiMAX Transfection Kit (Invitrogen).
Determination of clonogenic survival, apoptosis, metabolic competence
To monitor clonogenic survival, 500 cells per 6-cm dish were seeded and exposed to BPDE. Fourteen days later, colonies were fixed with methanol, stained with crystal violet (1%) and counted. To determine the apoptosis frequency, cells were incubated at different time points after BPDE exposure for 30 min with 0.1 mg/ml RNAse in phosphate buffered saline (PBS) and stained with propidium iodide (PI). The sub-G1 fraction was determined by flow cytometry as described previously (31). For monitoring drug-induced apoptosis and necrosis within the same cell population, annexin V-FITC/PI double staining combined with flow cytometry was applied. The protocol was conducted as described (32). The metabolic activity was determined by 3-(4,5-dimethyldiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as already described (33). In all cases, experiments were repeated at least three times; mean values ± standard deviation (SD) are shown.
Determination of cell cycle progression and replication
To determine cell cycle distribution, cells were incubated at different time points after BPDE exposure for 30 min with 0.1 mg/ml RNAse in PBS and stained with PI. The cell cycle distribution was determined by flow cytometry and quantified using ModFit LT3 software. To follow progression of cells through the cell cycle, cells were pulse-labeled with 10 μM BrdU for 30 min and thereafter the thymidine analog was washed out and cells were non-treated or treated with BPDE. After treatment, cells were fixed, incubated with mouse anti-BrdU antibody (BD Biosciences) and subsequently with Alexa Fluor 488-coupled anti-mouse antibody (Millipore), and stained with PI as described (32). Cells were analyzed by flow cytometry (FACS Calibur, BD Biosciences). DNA synthesis was measured via the colorimetric BrdU ELISA assay according to the manufacturer's protocol (Roche Diagnostics). Furthermore, cells were labeled with 10 μM BrdU 60 min before the end of BPDE exposure, collected, fixed, incubated with antibodies and stained with PI as described above. In all cases, experiments were repeated at least three times; mean values ± SD are shown.
Mutagenicity assay
Determination of the mutation frequency (hypoxanthin-guanin-phosphoribosyltransferase (HRPT) assay) was performed as described (34). In brief, cells were seeded and incubated with BPDE. Seven days later, the cells were collected. Five hundred cells per experimental point were reseeded in triplicates and the relative plating efficiency was determined using the clonogenic survival assay in non-selective medium. For the determination of mutation frequency, the remaining cells were reseeded at a concentration of 1 × 107 in selective medium (6-thioguanine, 10 μg/ml, Sigma). Two weeks later, colonies were fixed, stained and counted. Mutation frequency was determined by correcting mutant colony counts for plating efficiency. In all cases, experiments were repeated at least three times; mean values ± SD are shown.
Preparation of RNA, endpoint PCR, qRT-PCR and ChIP
Total RNA was isolated from cell cultures using the RNA II Isolation Kit (Machery and Nagel). For isolation of RNA from buccal cells, the cells were obtained using MasterAmp™ Buccal Brush (MB100SP, Epicentre) and directly lyzed in Trizol. Following Trizol/Chloroform isolation, the RNA was further purified by the RNA II Isolation Kit. RNA was stored at −80°C and cDNA synthesis was performed using the Verso cDNA Kit (Thermo Scientific). RNA concentration was determined using NanoDrop 1000 Spectrophotometer. Endpoint polymerase chain reaction (PCR) was performed using Red-Taq Ready Mix (Sigma-Aldrich) and qRT-PCR was performed using the SensiMix™ SYBR Green & Fluorescein Kit (Bioline, London, UK) and the CFX96 Real-Time PCR Detection System (Biorad). Chromatin immunoprecipitation (ChiP) was performed as described (9). qPCR was performed using specific primers flanking the p53 binding site of POLH. In all experiments, qRT-PCR was performed in technical triplicates, SD shows intra-experimental variation. Analysis was performed using CFX Manager™ Software. non template controls (NTCs) were included in each run, expression was normalized to gapdh and β-actin; the untreated control was set to one. The specific primers are depicted in Supplementary Table S1.
Preparation of protein extracts and western blot analysis
Whole-cell and nuclear extracts were prepared as previously described (35). For western blot analysis using phospho-specific antibodies, cells were directly lyzed in 1 × sodium dodecyl sulphate-polyacrylamide gel electrophoresis sample buffer and subsequently sonified. Mouse mAb were diluted 1:500-1:1000 in 5% bovine serum albumin (BSA), 0.1% Tween-TBS and incubated overnight at 4°C. Rabbit pAb were diluted 1:2000 and incubated 2 h at RT. The protein–antibody complexes were visualized by Pierce ECL Western Blotting Substrate (32106, Thermo Fisher). The specific antibodies are depicted in Supplementary Table S2.
Preparation of nuclear extracts and EMSA
Nuclear extracts were prepared as previously described (36) and subjected to electromobility shift assay (EMSA). The sequence of the oligonucleotides specific for the AP-1 binding site of the collagenase promoter was 5′-AGTGGTGACTCATCACT-3′.
Southwestern blot analysis
Genomic DNA was isolated by the use of the QIA(amp) blood mini kit (Qiagen, Hilden, Germany) and southwestern analysis was performed as already described (6). Monoclonal antibody against BPDE-N2-Guanine adducts was purchased from Santa Cruz (sc-52625) and diluted 1:200 in 5% BSA/0.1% Tween-TBS.
Immunofluorescence
Cells were seeded on cover slips. Following BPDE exposure, cells were washed with PBS and fixed with 4% paraformaldehyde. After washing with PBS 0.2% Tween, a second fixation step was performed using 100% methanol, followed by additional washing steps with PBS/0.2% Tween. The antibodies used were anti-γH2AX (Upstate, #05-164) and as secondary antibody Alexa Fluor 488 F(ab′) fragment of goat anti-rabbit IgG (Invitrogen, A11070). Nuclear staining was performed using TO-PRO-3 iodide (Invitrogen). Slides were mounted in Vectashield medium (Vector Laboratories Inc.) and analyzed using a confocal laser scanning microscope (LSM 710, Zeiss). 200 cells were scored for each time point. Data represent the mean of three independent experiments ± SD.
Statistical analysis
Data were statistically analyzed using Student's unpaired two-sample t-test on the basis of a difference between sample means. Calculations were performed using the GraphPad Prism software (*P < 0.05, **P < 0.005).
RESULTS
Toxicity of BPDE, activation of p53 and AP-1 and upregulation of NER genes
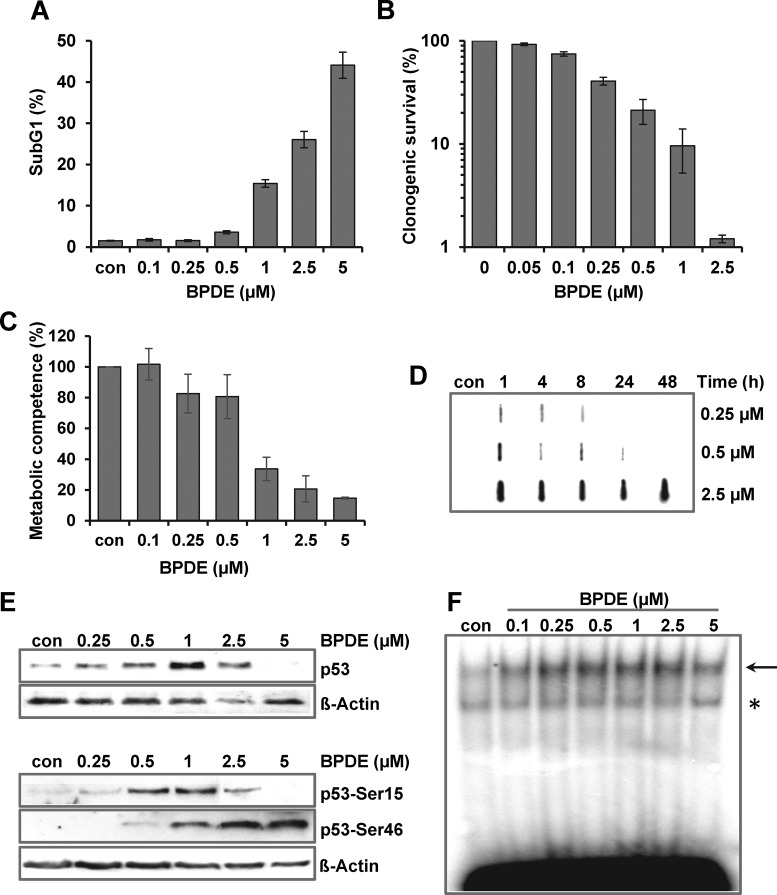
To analyze the complex network of transcriptional alterations upon BPDE, we first determined the cytotoxicity of BPDE in human diploid fibroblasts (VH10tert). VH10tert cells showed a cytotoxic response with BPDE concentrations >1 μM, as indicated by the level of apoptosis (Figure 1A), clonogenic survival (Figure 1B) and metabolic competence (Figure 1C). Concentrations <0.1 μM were non-toxic while 0.25 and 0.5 μM BPDE showed a modest level of toxicity. The low toxicity of 0.25 and 0.5 μM BPDE was verified at early (24 h) and late (96–144 h) time points after BPDE pulse-treatment (Supplementary Figure S1A).
Figure 1.
Consequences of BPDE exposure in human VH10tert fibroblasts. (A–C) Dose- dependent toxicity of BPDE was detected by sub-G1 measurement (A, 72 h), clonogenic survival (B, 14 days) and metabolic competence (C, 72 h). (D) Formation and repair of BPDE adducts was measured 24 h upon treatment with 0.25, 0.5 or 2.5 μM BPDE. At different time points, genomic DNA was isolated and subjected to southwestern analysis using a BPDE-adducts specific antibody. (E) Activation of p53 was analyzed by immunodetection using p53 antibodies in nuclear extracts (upper blot) or phospho-specific antibodies targeting Ser15 and Ser46 in whole cell extracts (lower blot) 24 h upon exposure to different BPDE concentrations. ß-Actin was used as loading control. (F) Activation of AP-1 was analyzed by EMSA 24 h upon exposure to different BPDE concentrations. Arrow indicates the specific band and asterisk indicates an unspecific band.
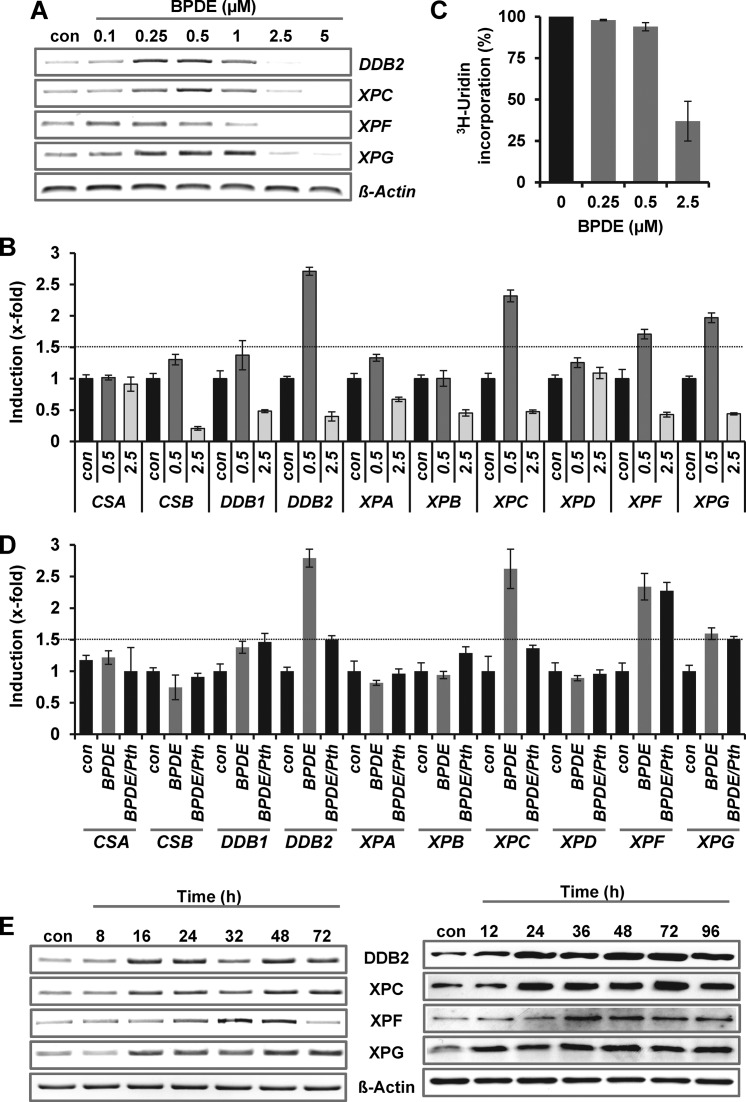
Next, we assessed the repair of BPDE adducts. In cells treated with 0.25 and 0.5 μM BPDE, adducts were removed within 8–24 h (Figure 1D). In contrast, cells were unable to remove BPDE adducts following treatment with higher concentrations (2.5 μM). In the same concentration range, activation of p53 (Figure 1E) and AP-1 (Figure 1F) was observed, peaking with doses of 0.5 and 1 μM BPDE. Interestingly, phosphorylation of p53 at Ser15 occurred already in the low dose range and was strongly reduced following high-dose BPDE (2.5 and 5 μM), whereas phosphorylation at Ser46 was enhanced. The level of AP-1 was enhanced over the whole concentration range, as determined in bandshift experiments (EMSA) (Figure 1F). The specificity of the AP-1 EMSA was verified by competition and supershift assays (Supplementary Figure S1B). Furthermore transcriptional and post-translational activation of the AP-1 component c-FOS was observed upon BPDE (Supplementary Figure S1C and D). A concentration-dependent response was also found for the transcriptional upregulation of the NER genes DDB2, XPC, XPF and XPG. Induction was observed only in a very narrow window, ranging from 0.25 to 1 μM BPDE (Figure 2A). At higher BPDE concentrations the expression of the repair genes was not further enhanced, it was rather strongly reduced. The reduced expression following 2.5 μM BPDE was also observed for other NER genes (CSB, DDB1, XPA and XPB) but not for CSA and XPD (Figure 2B) and was associated with a global transcriptional arrest (Figure 2C).
Figure 2.
BPDE-induced induction of NER genes. (A) Dose-dependent expression of NER genes was analyzed 24 h upon exposure to BPDE (0.1–5 μM) by endpoint PCR. (B) Expression of NER genes was analyzed 24 h upon exposure to BPDE (0.5 or 2.5 μM) by qRT-PCR. For quantification, the expression was normalized to gapdh and ß-actin and the untreated control was set to 1. (C) Transcriptional activity was analyzed 24 h upon BPDE (0.25, 0.5 or 2.5 μM) exposure by 3H-uridine incorporation. (D) Cells were pre-treated with 30 μM Pifithrin α (Pth) for 1 h and thereafter exposed to 0.25 μM BPDE for 24 h. Expression was analyzed by qRT-PCR. For quantification, the expression was normalized to β-Actin and SCLY and the untreated control was set to 1. (E) Time-dependent expression of NER genes (left panel) and protein (right panel) was analyzed upon exposure to 0.5 μM BPDE by endpoint PCR and immunodetection, respectively.
To analyze the impact of p53 on the regulation of the NER genes, cells were pre-treated with 30 μM pifithrin α for 1 h and thereafter exposed to BPDE (Figure 2D). The data revealed that upon BPDE exposure only DDB2 and XPC are regulated by p53. With a dose of 0.5 μM BPDE, a long-lasting increase in DDB2, XPC, XPF and XPG expression was observed on RNA (Figure 2E, left panel) and protein level (Figure 2E, right panel). Interestingly, low-dose BPDE exposure (0.25 or 0.5 μM) did not only transiently enhance the expression of DDB2, XPC, XPF and XPG, but also maintained their expression at high level following a subsequent treatment with 2.5 μM BPDE (Supplementary Figure S2A).
BPDE causes activation of an adaptive response pathway
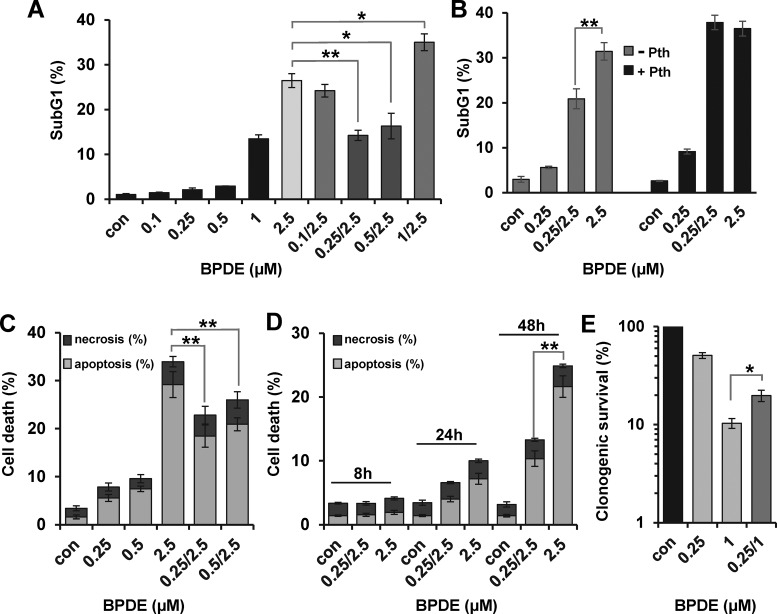
To investigate a putative adaptive response, VH10tert cells were pre-exposed to different concentrations of BPDE and 24 h later challenged with 2.5 μM BPDE. Pre-exposure to 0.25 and 0.5 μM clearly protected cells from toxicity induced by the subsequent higher challenging dose, as determined by apoptosis induction (Figure 3A). In contrast, lower doses (0.1 μM), which were not able to activate p53 and AP-1, and higher doses (1 μM) that were toxic by themselves did not elicit an adaptive response. This indicates that, similar to the induction of the NER genes, the adaptive response was only observed in a very narrow dose window. Furthermore, the adaptive response was not influenced by an effect of low-dose BPDE exposure on overall transcriptional activity, as measured by 3H-uridine incorporation (Supplementary Figure S2B).
Figure 3.
BPDE-induced adaptive response. (A and B) To monitor the impact of pre-exposure on sensitivity to BPDE, VH10tert cells were either not pre-exposed or pre-exposed to 0.1–1 μM BPDE and 24 h later exposed to 1 or 2.5 μM BPDE. Seventy-two hours later, cells were stained with PI and the sub-G1 fraction was determined by flow cytometry. (B) To analyze the impact of p53 on adaptive response, cells were additionally pre-treated with 30 μM Pifithrin α (Pth) for 1 h. (C and D) To monitor the impact of pre-exposure on apoptosis and necrosis, VH10tert cells were either not pre-exposed or pre-exposed to 0.25 and 0.5 μM BPDE and 24 h later exposed to 2.5 μM BPDE. At different time points, cells were stained with annexin V/PI and analyzed by flow cytometry. (E) Clonogenic survival was determined 14 days after BPDE exposure with or without low-dose pre-exposure (0.25/1). (A–E) Experiments were repeated three times; mean values ± SD are shown. Data were statistically analyzed using Student's unpaired two-sample t-test (*P < 0.05, **P < 0.005).
To analyze the involvement of p53 in the adaptive response, cells were pre-treated with 30 μM pifithrin α (PTHα) for 1 h (Figure 3B). Upon inhibition of p53, BPDE exposure resulted in increased toxicity, pointing to a protective effect of p53 and there was no adaptive response.
The adaptive response seems to reduce specifically the BPDE-induced apoptosis. Thus no difference was observed in the induction of necrosis, which occurred at low levels upon BPDE (Figure 3C). To exclude that in the pre-exposed population highly damaged cells were eliminated before determination of apoptosis at 72 h, additional experiments were performed, showing that in the post-treatment period of 8–48 h no elimination of cells occurred. At all time points analyzed, the pre-treated cells showed a lower apoptotic frequency (Figure 3D). An adaptive response was also observed when cell death was measured by clonogenic survival. In this assay, lower BPDE doses (pre-exposure to 0.25 μM and challenge with 1 μM) had to be applied. However, also in this sensitive assay, BPDE pre-exposure protected against cell death provoked by a subsequent challenging dose (Figure 3E). Interestingly, concentrations of BPDE showing only a weak apoptotic effect (0.25 and 0.5 μM) strongly reduced colony formation of VH10tert cells. Colony formation is the most sensitive indicator, accumulating different changes that have an impact on cell growth. The discrepancy indicates that, besides cell death by apoptosis, BPDE activates other pathways like senescence and/or mitotic catastrophe, leading to reduced colony formation.
To characterize the mechanism underlying BPDE-induced apoptosis, we determined the expression of various apoptosis-regulating factors, known to be targets of p53. Interestingly, induction of most pro-apoptotic genes was only observed at concentrations insufficient to trigger apoptosis (Supplementary Figure S2C and D). At apoptosis-inducing concentrations, most of these genes were not expressed at all. Only the pro-apoptotic gene NOXA was strongly expressed upon high-dose exposure to 2.5 and 5 μM BPDE. In line with the data showing that NOXA is a specific target of p53Ser46 (37), only a weak induction of p53Ser15, but a strong and sustained induction of p53Ser46 occurred at apoptosis-inducing concentrations (Figure 1E and Supplementary Figure S2E). At low concentrations, predominant induction of p53Ser15 and only transient expression of p53Ser46 were observed. The data indicates that upon BPDE exposure p53Ser46-dependent induction of NOXA may be involved in triggering apoptosis and shows that BPDE-induced DNA damage signaling may vary between low and high-dose exposure.
NER upregulation elicits an adaptive response
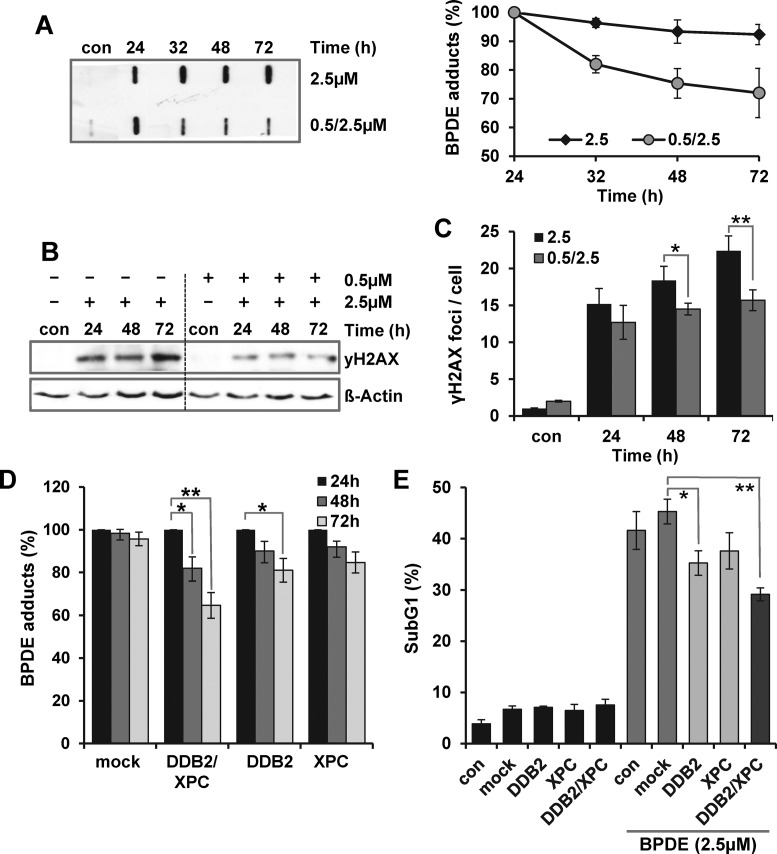
Next, we analyzed whether enhanced expression of the NER genes DDB2, XPC, XPF and XPG was sufficient to enhance the NER capacity. Therefore, VH10tert cells were pre-exposed to 0.5 μM BPDE and 24 h later challenged with 2.5 μM BPDE. Twenty-four to seventy-two hours later the amount of BPDE adducts was determined. As shown in Figure 4A, 24 h after pre-treatment (con 0.5/2.5) BPDE adducts were still detectable, indicating that they were not completely repaired. However, despite the higher level of BPDE adducts in the pre-exposed cells, their amount was similar 24 h after exposure to 2.5 μM BPDE. At later time points (32–72 h) an increased removal of the BPDE adducts was observed in the pre-exposed cells, whereas non-pre-exposed cells were unable to remove the BPDE adducts. Of note, the enhanced removal of the BPDE adducts is not caused by proliferation-induced dilution since high-dose exposure nearly completely arrested replication during the next 72 h (Supplementary Figure S3A). The replication stop was observed in pre-treated and non-pre-treated cells. In addition to adduct removal, the impact of pre-exposure on the formation of double-strand breaks was analyzed, detecting the phosphorylation of H2AX as well as the persistence of γH2AX foci. Also in this case a significant protective effect of low-dose pre-exposure was observed 48 and 72 h after treatment with 2.5 μM BPDE (Figure 4B and C). As mentioned above, p53 signaling is important for the adaptive response and DDB2 and XPC represent the only NER genes upregulated in dependence on p53. To further prove that induction of these NER genes is sufficient to enhance the NER capacity and to induce cellular protection, XPC and DDB2 were transiently overexpressed in VH10tert cells. Verification of separate (XPC or DDB2) and combined (XPC and DDB2) overexpression was performed by qRT-PCR (Supplementary Figure S3B). Twenty-four hours upon transfection, the cells were exposed to 2.5 μM BPDE and the removal of BPDE adducts as well as cytotoxicity were analyzed. The data indicate that combined overexpression of DDB2 and XPC is required for enhancing the NER capacity (Figure 4D for quantification and Supplementary Figure S3C for a representative experiment) and for ameliorating survival (Figure 4E).
Figure 4.
Impact of NER activation on the adaptive response. (A–C) VH10tert cells were exposed to 2.5 μM BPDE. Twenty-four hours earlier, the cells were either not pre-exposed (2.5) or pre-exposed to 0.5 μM BPDE (0.5/2.5). Twenty-four to seventy-two hours after exposure to 2.5 μM BPDE, (A) genomic DNA was isolated and subjected to southwestern analysis using a BPDE-adducts specific antibody (left panel: representative blot, right panel: quantification of three independent experiments), (B) whole-cell extracts were isolated and subjected to immunodetection using γH2AX specific antibody (ß-Actin was used as loading control), (C) formation of γH2AX foci was analyzed using immunofluorescence. (D and E) VH10tert cells were transiently transfected with plasmids coding for DDB2 and/or XPC and 24 h later treated with 2.5 μM BPDE. (D) Twenty-four to seventy-two hours thereafter, genomic DNA was isolated and subjected to southwestern analysis using a BPDE-adducts specific antibody. (E) Cytotoxicity was detected by sub-G1 measurement. (A, C–E) Mean values ± SD of three independent experiments are shown. Data were statistically analyzed using Student's unpaired two-sample t-test (*P < 0.05, **P < 0.005).
Impact of cell cycle alterations on BPDE adaptation
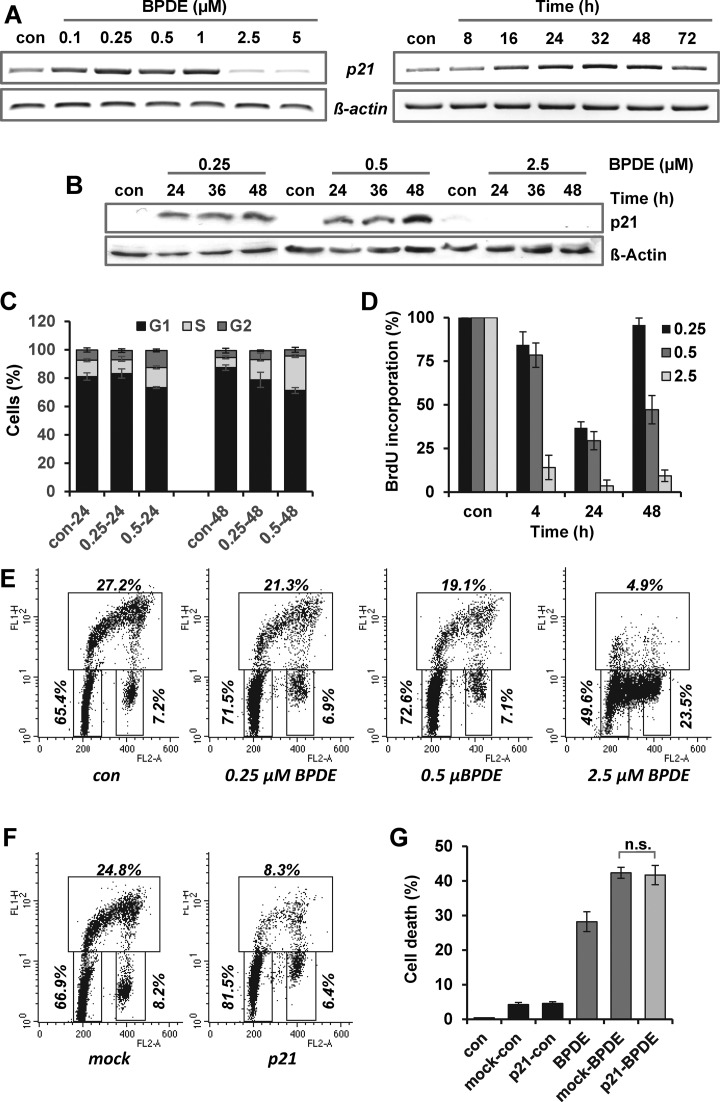
One of the most prominent target genes of p53 is the cyclin dependent kinase inhibitor 1A (CDKN1a, p21), which is the main factor in inducing G1/S-arrest. Since cell cycle blockage may impact survival, we addressed whether p21 is also induced upon BPDE exposure and whether a potential G1/S-arrest may contribute to the adaptive response. Similar to the NER genes, p21 was only upregulated after treatment with BPDE concentrations between 0.1 and 1 μM (Figure 5A) and the enhanced expression was observed 16–72 h after exposure. The most prominent difference was observed on protein level (Figure 5B). Whereas p21 was not detectable in non-exposed cells (con), treatment with 0.25 and 0.5 μM BPDE led to pronounced p21 protein expression; however using 2.5 μM BPDE no p21 expression was observed. In addition, low-dose BPDE pre-exposure (0.5 μM) was able to maintain the p21 expression upon challenge with 2.5 μM BPDE on mRNA and protein level (Supplementary Figure S4A). However, although strong induction of p21 occurred, only marginal changes in the cell cycle distribution (e.g. a slightly enhanced fraction of cells within the S and G2 phase) were observed 48 h following 0.25 and 0.5 μM BPDE (Figure 5C and Supplementary Figure S4B). The accumulation of cells in the S-phase was accompanied by a reduced DNA synthesis, as determined by BrdU incorporation (Figure 5D), indicating a slow-down of replication. Reduced replication and, in addition to this, a slightly increased amount of cells in the G1-phase were also observed 24 h upon BPDE treatment using PI/BrdU double staining (Figure 5E). Furthermore, within the 24 h post-exposure period, the cells were able to replicate the damaged DNA and progressed into G2 and further to G1 (Supplementary Figure S5A). Whereas low doses of BPDE had a rather weak effect on replication, the high dose of 2.5 μM BPDE immediately and completely blocked replication (Figure 5 D and E; Supplementary Figure S4A). Overall the data indicate that pre-exposure to low BPDE-concentrations induces only a transient slow-down of replication, but no pronounced cell cycle arrest.
Figure 5.
Impact of p21 activation on the adaptive response. (A) Expression of p21 mRNA was analyzed 24 h upon exposure to BPDE (0.1–5 μM, left panel) and 8–72 h upon treatment with 0.5 μM BPDE (right panel) by endpoint PCR. (B) Expression of p21 protein was analyzed 24–48 h upon exposure to BPDE (0.25, 0.5 or 2.5 μM) by immunodetection. ß-Actin was used as loading control. (C) VH10tert cells were exposed to 0.25 or 0.5 μM BPDE. After 24 and 48 h, cell cycle distribution was measured by FACS and analyzed using the ModFIT LT3 software. (D) VH10tert cells were exposed to 0.25, 0.5 or 2.5 μM BPDE. After 4–48 h, replication was measured by BrdU assay. (E) VH10tert cells were exposed to 0.25, 0.5 or 2.5 μM BPDE for 24 h. (F) VH10tert cells were transiently transfected with plasmids coding for p21 and incubated for 24 h. (E and F) Sixty minutes before the end of the exposure, cells were pulse-labeled with 10 μM BrdU to monitor cell cycle distribution. Representative graphs and the mean frequency from three independent experiments are shown. (G) VH10tert cells were transiently transfected with plasmids coding for p21 and 24 h later exposed to 2.5 μM BPDE. Seventy-two hours later, cytotoxicity was detected by sub-G1 measurement. (D and E) Mean values ± SD of three independent experiments are shown. Data were statistically analyzed using Student's unpaired two-sample t-test (n.s. = non-significant).
To further test the assumption that p21-mediated cell cycle perturbations might impact the cytotoxic response, we transiently overexpressed p21 in VH10tert cells. Overexpression was verified by qRT-PCR and immunodetection (Supplementary Figure S5B and C). The impact of p21 overexpression on cell cycle distribution and progression was analyzed using PI/BrdU double staining (Figure 5F), showing induction of G1-arrest, which was stronger than the arrest provoked by low dose BPDE pre-exposure. Twenty-four hours upon transfection, the cells were exposed to 2.5 μM BPDE and toxicity was determined by sub-G1 analysis. The data shows that overexpression of p21 had no impact on BPDE-induced apoptosis, indicating that induction of p21 is not sufficient to enhance the level of cellular resistance to BPDE (Figure 5G).
Induction of POLH by BPDE leads to enhanced mutation frequency
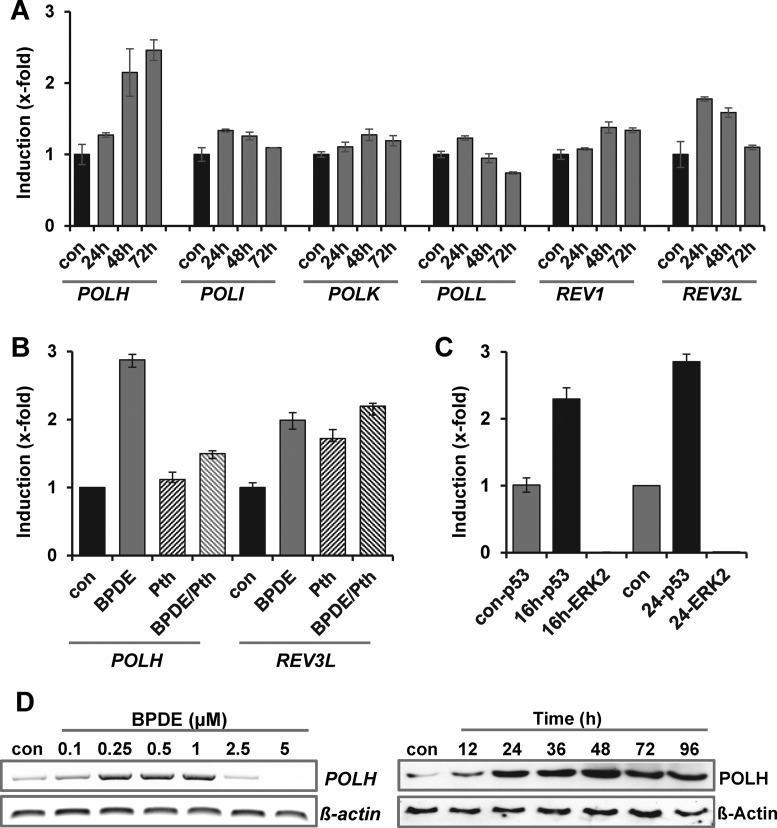
Multiple reports indicated that translesion synthesis (TLS) is involved in the bypass of BPDE adducts. To analyze whether activation of TLS polymerases is involved in the adaptive response to BPDE, the expression level of TLS-polymerases POLH, POLI, POLK, POLL, REV1 and REV3L was analyzed following BPDE using qRT-PCR (Figure 6A) and endpoint-PCR (Supplementary Figure S6A). The results indicate that only POLH (polymerase eta) and, to a lower extend, REV3L (catalytic subunit of POL zeta) were upregulated by BPDE (0.5 μM). To analyze the impact of p53 on the regulation of POLH and REV3L, cells were pre-treated with 30 μM pifithrin α for 1 h and thereafter exposed to BPDE (Figure 6B). Induction of POLH, but not REV3L was abrogated by p53 inhibition. Furthermore, binding of p53 to the POLH promoter was observed using ChIP (Figure 6C), clearly indicating that POLH is part of the p53 response to BPDE. Since POLH has previously been shown to be involved in bypass of BPDE adducts (38), we analyzed the role of POLH in the BPDE-induced adaptive response in more detail. In analogy to DDB2, XPC and p21, POLH was transcriptionally upregulated in a dose-range between 0.1 and 1 μM BPDE, and on protein level in a time- frame of 12–96 h (Figure 6D). POLH expression maintained on mRNA and protein level when cells were subsequently challenged with a dose of 2.5 μM BPDE (Supplementary Figure S6B).
Figure 6.
BPDE-induced induction of TLS polymerases. (A) VH10tert cells were exposed to 0.5 μM BPDE. Twenty-four hours later, expression of TLS polymerases was analyzed by qRT-PCR. For quantification, the expression was normalized to gapdh and ß-actin and the untreated control was set to 1. (B) Cells were pre-treated with 30 μM Pifithrin α (Pth) for 1 h and thereafter exposed to 0.5 μM BPDE for 24 h. Expression was analyzed by qRT-PCR. For quantification, the expression was normalized to β-Actin and SCLY and the untreated control was set to 1. (C) ChIP analysis was performed in untreated VH10tert cells and cells exposed to 0.5 μM BPDE for 16 or 24 h. Protein–DNA complexes were immuno-precipitated with anti-p53 or anti-ERK1/2 (neg. control) antibody and qRT-PCR was performed using primers flanking the p53-responsive element within the polH promoter. (D) Expression of POLH mRNA was analyzed 24 h upon exposure to BPDE (0.1–5 μM, left panel) by endpoint PCR, expression of POLH protein was analyzed 12–96 h upon exposure to 0.5 μM BPDE by immunodetection (right panel). ß-Actin was used as loading control.
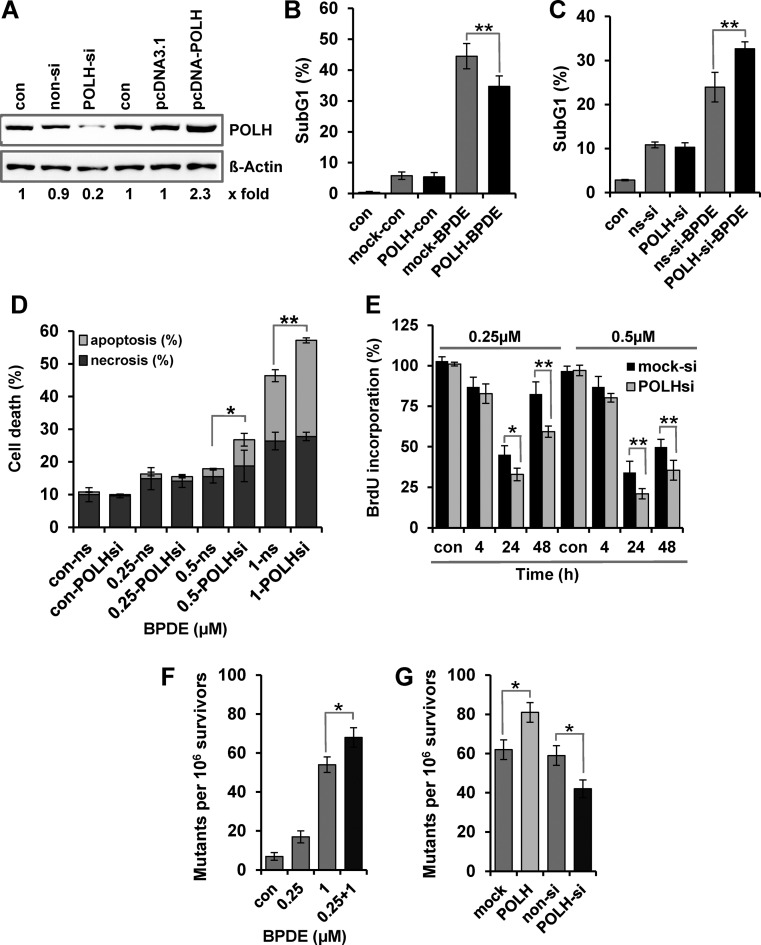
To analyze the impact of POLH on BPDE-induced cytotoxicity, POLH was transiently overexpressed (Supplementary Figure S6C, for qRT-PCR-based verification) or knocked down through siRNA treatment (Figure 7A, for immunodetection). Apoptosis was determined by sub-G1 analysis (Figure 7B and C) and annexin V/PI double staining (Figure 7D). The data clearly indicate that POLH overexpression could partially protect the cells against high-dose BPDE exposure (Figure 7B), whereas POLH knockdown sensitized the cells against BPDE (Figure 7C and D). In addition, proliferation and viability was determined in real time by impedance measurement (Supplementary Figure S6D), further indicating that PolH is involved in protection against BPDE. This protection seems to be caused by reduction of the BPDE-induced replication arrest. Thus, as measured by the BrdU assay, knockdown of POLH significantly reduced DNA synthesis, further showing the importance of POLH-associated TLS upon BPDE exposure (Figure 7E).
Figure 7.
Impact of POLH activation on the adaptive response. (A) Expression of POLH protein was analyzed by immunodetection in VH10tert cells 24 h after transfection with plasmids coding for POLH or POLH specific siRNA. (B and C) VH10tert cells were transiently transfected with plasmids coding for POLH (B) or POLH specific siRNA (C). Twenty-four hours later, cells were treated with 2.5 μM BPDE and additional 72 h later, cytotoxicity was detected by sub-G1 measurement. (D) VH10tert cells were transiently transfected with POLH specific and non-silencing siRNA. Twenty-four hours later, cells were treated with 0.25, 0.5 or 1 μM BPDE and 72 h later, apoptosis and necrosis, were measured by annexin V/PI staining. (E) VH10tert cells were transiently transfected with POLH specific siRNA. Twenty-four hours later, cells were treated with 0.25 or 0.5 μM BPDE. After 4–48 h, replication rate was detected by BrdU incorporation. (F) VH10tert cells were exposed to 0.25 or 1 μM BPDE. In addition cells were exposed to 0.25 μM BPDE and 24 h later exposed to 1 μM BPDE. The mutation frequency was determined by HRPT assay. (G) VH10tert cells were transiently transfected with plasmids coding for POLH or POLH-specific siRNA. Twenty-four hours later cells were treated with 1 μM BPDE and mutation frequency was determined by HRPT assay. (B–G) Mean values ± SD of three independent experiments are shown. Data were statistically analyzed using Student's unpaired two-sample t-test (*P < 0.05, **P < 0.005).
An intriguing question is whether, apart from cell death, also mutagenesis is affected by POLH upregulation. To answer this question, VH10tert cells were treated with 0.25 and 1 μM BPDE. They were also exposed to 0.25 μM BPDE followed by exposure to 1 μM BPDE, which occurred 24 h later (0.25/1). The cells were left in culture for additional seven days, reseeded and the mutation frequency was analyzed by the HRPT assay 14 days later (Figure 7F). Our data clearly show that the low-dose pre-exposure not only protected the cells from cell death induced by the challenging dose, but also enhanced the mutation frequency. In a control experiment, POLH was silenced or transiently overexpressed and 24 h later cells were treated with 1 μM BPDE. The HRPT assay revealed that the mutation frequency was reduced when POLH was knocked down and enhanced when POLH was transiently overexpressed (Figure 7G). This clearly supports a role of POLH in BPDE-induced mutagenesis.
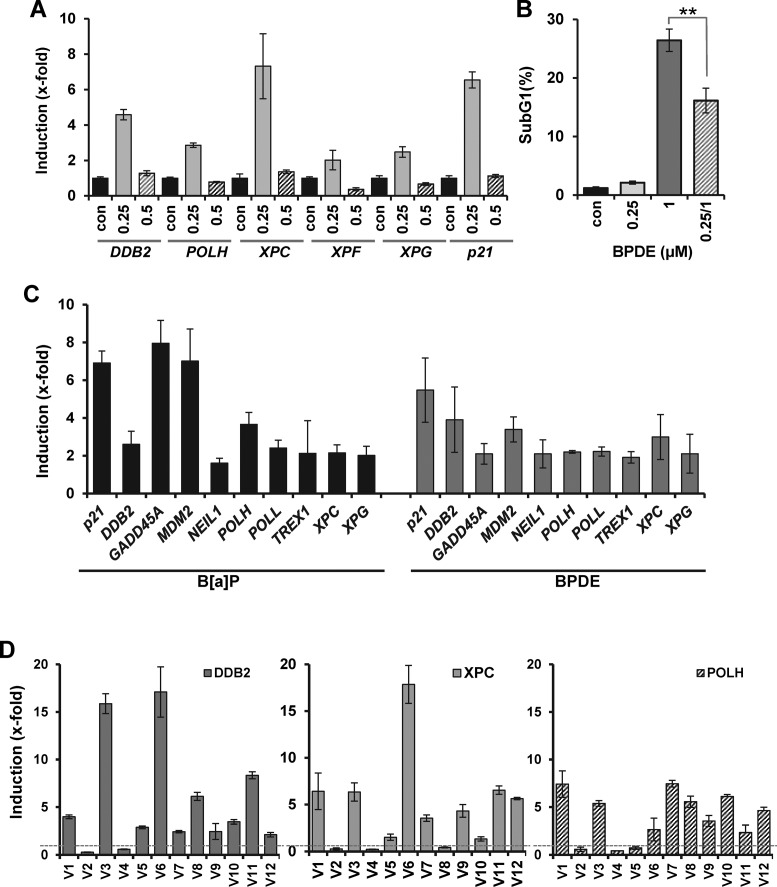
Induction of DNA repair genes in primary human lung epithelial cells treated with BPDE and metabolically competent MCF7 cells treated with B(a)P
B(a)P is present in tobacco smoke, targeting the mouth, trachea and lung epithelium. Therefore, we aimed to study whether induction of NER genes following BPDE treatment also occurs in primary bronchial epithelial cells (PBECs). Treating these cells with BPDE, we observed a dose-dependent killing effect, indicating a slightly higher sensitivity than in VH10tert cells (Supplementary Figure S7A). Induction of the repair genes DDB2, XPC, XPF and XPG and the TLS polymerase POLH was clearly observed following BPDE treatment (Figure 8A). The time-course of induction of DDB2, XPC, XPF, XPG and POLH revealed a peak at 24 h (Supplementary Figure S7B). Pre-treating PBECs with BPDE (0.25 μM) and challenging them with a higher dose (1 μM), we observed an increased survival in the challenged population (Figure 8B), indicating BPDE evoked an adaptive response in these cells.
Figure 8.
BPDE induces expression of DNA repair genes in primary human cells, MCF7 cells and in buccal cells of smokers. (A) Induction of DDB2, XPC, XPF, XPG, POLH and p21 was analyzed in primary human bronchial epithelial cells (PBECs) 24 h upon exposure to BPDE (0.25 and 0.5 μM) by qRT-PCR. (B) To monitor the impact of pre-exposure on sensitivity to BPDE, PBECs were either not pre-exposed or pre-exposed to 0.25 μM BPDE and 24 h later exposed to 1 μM BPDE. Seventy-two hours later, cytotoxicity was detected by sub-G1 measurement. (C) MCF7 cells were exposed to 0.5 μM BPDE or 1 μM B(a)P. Twenty-four hours later, expression of 108 DNA repair genes was analyzed by qRT-PCR. Data for all genes showing an induction level above 2-fold is shown. For quantification, the expression was normalized to gapdh and ß-actin and the untreated control was set to 1. (D) qRT-PCR expression of DDB2, XPC and POLH in buccal cells of 12 male non-smokers (V1-V12) 24 h upon exposure to three cigarettes. For quantification, the expression was normalized to gapdh and ß-actin and the untreated control was set to 1.
To analyze whether the non-activated carcinogen B(a)P is able to induce a transcriptional induction of DNA repair genes and to compare the transcriptional activation upon B(a)P and BPDE, we used metabolically competent MCF7 breast cancer cells which can convert B(a)P into BPDE. Both B(a)P and BPDE induced the p53 and AP-1 response in MCF7 cells (Supplementary Figure S7C and D). The expression of all known DNA repair genes was analyzed in MCF7 cells upon exposure to BPDE and B(a)P using a qRT-PCR array. The data is provided in Supplementary Table S3. Genes showing >2-fold induction are presented in Figure 8C. Among these genes, p21, DDB2, XPC, XPG and POLH are found to be upregulated by both BPDE and B(a)P. Interestingly, also besides the above mentioned genes, upon B(a)P and BPDE treatment, induction of identical DNA repair genes was observed, indicating that BPDE represents the most important metabolite mediating B(a)P-induced alterations in DNA repair gene expression.
Upregulation of repair and TLS genes in buccal cells in volunteers upon smoking
In order to translate the data to humans exposed to B(a)P, we determined the repair gene expression in individuals following cigarette smoking, using a qRT-PCR assay validated for DDB2, POLH andXPC. We analyzed buccal cells obtained from healthy male volunteers (non-smokers) before and 24 h after smoking of three cigarettes. From 12 volunteers (V1 to V12) included in the study, 10 showed a higher expression level (threshold 2-fold above the control) of DDB2, 8 showed a higher level of XPC and 9 volunteers showed a higher level of POLH following smoking. The induction level was highly variable between the volunteers. Interestingly, two volunteers (V2 and V4) did not respond at all with upregulation of DDB2, XPC and POLH (Figure 8D), pointing to substantial inter-individual differences as to the induction of DNA repair genes in the human population.
DISCUSSION
About 40 years ago, evidence was provided for transcriptional upregulation of DNA repair genes resulting in an SOS or adaptive response in E. coli, protecting the microorganism from cell death (11,12,39). Since not only bacteria, but also multicellular organisms, including humans, are constantly exposed to endogenous and exogenous genotoxic stress, which can induce genomic instability and thus promote carcinogenesis already at low doses (40), it is reasonable to suppose that also eukaryotes respond with the induction of protective functions resulting in an adaptive response. Best analyzed in human cells is the adaptive response induced by IR, which is related to the phenomenon of hormesis (41). Also other DNA damaging insults appear to trigger adaptation. Thus, in our previous work we showed that in human cells exposed to UV-C light an adaptive response can be evoked, which was associated with transcriptional activation of the NER endonucleases XPF and XPG (6–8). Fortunately, humans are under normal conditions not exposed to UV-C light.
To address the question of whether highly relevant chemical environmental carcinogens can stimulate transcription of DNA repair genes and thereby trigger an adaptive response in human cells, we analyzed the expression of DNA repair genes following treatment with the PAH B(a)P and its active metabolite BPDE. B(a)P is one of the most important environmental polycyclic hydrocarbons we are exposed to (42). BPDE binds via the epoxide group to the exocyclic N2-position of guanine in the DNA and thus forms a bulky adduct that can be repaired by NER (43). We observed a strong dose-dependent activation of p53 and AP-1, which resulted in a concerted transcriptional activation of the p53-regulated NER genes DDB2 and XPC and the AP-1 regulated NER genes XPF and XPG. Induction of DDB2 was previously reported in MCF7 and HepG2 cells treated with B(a)P and BPDE, which was p53 dependent (25,44,45). Here we demonstrate that in case of BPDE the concentrations applied are critical for detecting the induction of DNA repair. Thus repair of BPDE adducts, activation of p53 and induction of DNA repair genes was only observed in a non-toxic concentration range. Additional experiments showed that low-dose BPDE treatment not only enhanced the expression of NER genes following a BPDE pre-treatment dose, but also maintained at high level following a subsequent high-dose exposure, which on its own suppressed gene expression. The induction of DDB2, XPC, XPF and XPG appears to be sufficient to maintain the cell's NER capacity in challenged cells, since low-dose BPDE pre-exposure reinforced the removal of BPDE adducts, reduced the persistence of γH2AX foci and induced an adaptive response, protecting cells from killing following a subsequent challenge dose. A BPDE dose, which was below the threshold of activating p53 and AP-1 and high doses, which were toxic on their own, did not elicit an adaptive response. The dose dependence described here is reminiscent of the dose window reported for the radiation-induced adaptive response (41). Of mention, the threshold for inducing an adaptive response by IR is tissue-type dependent and the effect of pre-treatment gradually transits from adverse effects to protection (41,46,47).
Besides NER, TLS is also involved in the bypass of BPDE adducts. Thus POLK is able to bypass B(a)P-adducts of guanine (dG-N2-BPDE) in an error-free manner by inserting dC opposite the lesion (48–50). POLK-defective MEFs were hypersensitive to B(a)P and accumulated mutations upon B(a)P treatment (51). This is conform with a previous report showing that POLK has an impact on the BPDE sensitivity in MEFs (52). POLK has been shown to be regulated by BPDE since a repression of the promoter activity was observed (53). This was not the case in our experiments where POLK was not transcriptionally regulated. Besides POLK, also POLH and POLI have been shown to be involved in bypass of BPDE adducts. POLH inserts A opposite dG-N2-BPDE adducts (48,54,55) and POLI inserts T opposite dA-N6-BPDE adducts (56). The ability to predominantly incorporate A and less frequently T, G or C opposite dG-N2-BPDE adducts correlates with the mutation spectrum of these lesions in mammalian cells, supporting the notion that POLH is involved in generating mutations in humans (54). Nevertheless, TLS performed by human POLH largely stopped opposite or one nucleotide downstream of the lesion (38). In these experiments, extension synthesis performed by human POLK led to an effective error-prone lesion bypass. Another report showed that POLH is partially blocked by the N2-dG and the (-)-trans-N6-dA adducts, but can efficiently bypass the (+)-trans-N6-dA adduct (48).
Our experiments revealed a p53-dependent upregulation of POLH following BPDE in human cells. p53-dependent induction of POLH has been described in human cells upon treatment with IR, camptothecin (57) and DNA interstrand-crosslinking agents (33). We further provide evidence that POLH is involved in protection against BPDE-induced apoptosis and replication arrest. Since POLH can mediate error-prone lesion bypass, it is reasonable to hypothesize that the induction of POLH has an impact on the mutation frequency following BPDE. Our results clearly support this idea. They show that induction of POLH results in a better cellular survival, but at the expense of a higher yield of mutations. Therefore, the adaptive response described here has to be considered as a pro-mutagenic process.
In contrast to POLH, the induction of p21 seems not to be involved in the BPDE-induced adaptive response since its overexpression had no impact on the cytotoxicity of cells treated with toxic challenging doses of BPDE. We cannot exclude, however, that with low BPDE doses p21 also exerts protection by blocking the entry of cells into the S-phase.
It has been shown that short-term exposure to B(a)P induced transient elevation of reactive oxygen species (ROS), leading to extracellular signal-regulated kinase (ERK) pathway activation, cell proliferation and chromosomal DNA damage (58). Therefore, an intriguing question is whether base excision repair (BER) can be activated by B(a)P or BPDE thus contributing to the observed protective effect. However, under our experimental conditions we observed only a very weak and late formation of ROS upon B(a)P exposure (Supplementary Figure S7E) and no ROS formation upon BPDE exposure (data not shown). Therefore, it seems unlikely that oxidative stress is involved in the cytotoxic potential of B(a)P and BPDE.
Activation of DDB2, XPC and POLH was not specific for human fibroblasts. Induction of these genes was also observed in human MCF7 breast cancer cells treated with B(a)P and in primary human bronchial epithelial cells exposed to BPDE. Therefore, it seems to be a general and robust phenomenon, occurring in many (if not all) cell types equipped with wild-type p53. It can therefore be considered as an important part of the DNA damage response (59). The fact that the DNA repair genes induced by BPDE and B(a)P are identical indicates that among the metabolites formed from B(a)P, BPDE seems to be sufficient to activate the DNA damage response. We undertook serious attempts to translate the findings obtained with cultured human cells to humans. Using qPCR, we were able to show that an enhanced expression of DDB2, XPC and POLH occurs in mouth buccal cells of volunteers (non-smokers) 24 h after the consumption of three cigarettes. With this we show for the first time that upregulation of DNA repair genes occurs in humans exposed to relevant (low doses of) carcinogens.
We should note that the adaptive response observed following IR and chemical agents are very likely the result of different mechanisms. For IR, the adaptive response appears to be based on an increased removal of damaged cells by apoptosis, resulting in a decreased frequency of chromosomal aberrations due to elimination of heavily damaged cells (for review see (41,46)). In addition to this, low IR doses have been shown to increase the non-homologous end joining (NHEJ) efficiency and stimulate the repair of chromosomal breaks in human skin fibroblasts (15,60). For chemical agents, adaptive responses were described resulting from induction of free radical detoxification mechanisms and/or DNA repair. Thus, pre-treatment of human lymphocytes with bleomycin induced almost 50% decrease of chromosomal aberrations, while the protective effect of mitomycin C was below 20% (16). For MNNG and N-Nitroso-N-methylurea (MNU) induced adaptive response, enhanced cellular survival (17) and reduced aberration and point mutation frequencies were observed in a pretreatment setting with low doses (18). In rat hepatoma cells, upregulation of O6-methylguanine-DNA methyltransferase (MGMT) and an enhanced repair capacity for O6-methylguanine was shown to be responsible for mutagenic adaptation (61). In this case an error-free repair function was upregulated, diminishing mutations, whereas in the experiments reported here with BPDE an error-prone function, POLH, was induced, reducing cell death and, at the same time, enhancing the frequency of mutations. The killing protection appears to be the result of concomitant upregulation of DDB2, XPC and POLH, reducing overall cell death (colony formation) and the yield of apoptosis (by about 30%). This might be beneficial for the cell population in terms of survival, however, the surviving fraction represents a cell pool harboring mutations, which may increase the risk for carcinogenesis.
Altogether, we show that low-dose exposure of human cells to the highly relevant environmental, food-borne and tobacco smoke derived carcinogen BPDE activates a network of transcriptional alterations of DNA repair and TLS genes, leading to protection against cell death, at the expense of an increased yield of mutations in the survivors. We hypothesize that the induction of either protective (NER) or mutagenic (TLS) DNA damage pathways can impact carcinogenesis. Of note, not all smokers develop cancer, which may lead to the speculation that these individuals may predominantly activate the error-free DNA repair pathway. On the other hand, even passive smokers or irregular smokers can develop cancer indicating these individuals may predominantly activate error-prone repair. Thus, the extent and the balance between the induction of NER and TLS genes may have a great impact on the inter-individual susceptibility to cancer formation in human populations.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Science Foundation (DFG) [CH 665/2-2 to M.C.]; Intramural Research Funding of the University of Mainz (to M.T.T.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Christmann M., Kaina B. Transcriptional regulation of human DNA repair genes following genotoxic stress: trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res. 2013;41:8403–8420. doi: 10.1093/nar/gkt635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith M.L., Ford J.M., Hollander M.C., Bortnick R.A., Amundson S.A., Seo Y.R., Deng C.X., Hanawalt P.C., Fornace A.J., Jr p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol. Cell. Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ford J.M., Hanawalt P.C. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J. Biol. Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 4.Haas S., Kaina B. c-Fos is involved in the cellular defence against the genotoxic effect of UV radiation. Carcinogenesis. 1995;16:985–991. doi: 10.1093/carcin/16.5.985. [DOI] [PubMed] [Google Scholar]
- 5.Kaina B., Haas S., Kappes H. A general role for c-Fos in cellular protection against DNA-damaging carcinogens and cytostatic drugs. Cancer Res. 1997;57:2721–2731. [PubMed] [Google Scholar]
- 6.Christmann M., Tomicic M.T., Aasland D., Kaina B. A role for UV-light-induced c-Fos: stimulation of nucleotide excision repair and protection against sustained JNK activation and apoptosis. Carcinogenesis. 2007;28:183–190. doi: 10.1093/carcin/bgl119. [DOI] [PubMed] [Google Scholar]
- 7.Christmann M., Tomicic M.T., Origer J., Aasland D., Kaina B. c-Fos is required for excision repair of UV-light induced DNA lesions by triggering the re-synthesis of XPF. Nucleic Acids Res. 2006;34:6530–6539. doi: 10.1093/nar/gkl895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomicic M.T., Reischmann P., Rasenberger B., Meise R., Kaina B., Christmann M. Delayed c-Fos activation in human cells triggers XPF induction and an adaptive response to UVC-induced DNA damage and cytotoxicity. Cell. Mol. Life Sci. 2011;68:1785–1798. doi: 10.1007/s00018-010-0546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christmann M., Tomicic M.T., Aasland D., Berdelle N., Kaina B. Three prime exonuclease I (TREX1) is Fos/AP-1 regulated by genotoxic stress and protects against ultraviolet light and benzo(a)pyrene-induced DNA damage. Nucleic Acids Res. 2010;38:6418–6432. doi: 10.1093/nar/gkq455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomicic M.T., Aasland D., Nikolova T., Kaina B., Christmann M. Human three prime exonuclease TREX1 is induced by genotoxic stress and involved in protection of glioma and melanoma cells to anticancer drugs. Biochim. Biophys. Acta. 2013;1833:1832–1843. doi: 10.1016/j.bbamcr.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977;267:281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- 12.Jeggo P., Defais T.M., Samson L., Schendel P. An adaptive response of E. coli to low levels of alkylating agent: comparison with previously characterised DNA repair pathways. Mol. Gen. Genet. 1977;157:1–9. doi: 10.1007/BF00268680. [DOI] [PubMed] [Google Scholar]
- 13.Olsson M., Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J. Biol. Chem. 1980;255:10569–10571. [PubMed] [Google Scholar]
- 14.Foote R.S., Mitra S., Pal B.C. Demethylation of O6-methylguanine in a synthetic DNA polymer by an inducible activity in Escherichia coli. Biochem. Biophys. Res. Commun. 1980;97:654–659. doi: 10.1016/0006-291x(80)90314-9. [DOI] [PubMed] [Google Scholar]
- 15.Klammer H., Kadhim M., Iliakis G. Evidence of an adaptive response targeting DNA nonhomologous end joining and its transmission to bystander cells. Cancer Res. 2010;70:8498–8506. doi: 10.1158/0008-5472.CAN-10-1181. [DOI] [PubMed] [Google Scholar]
- 16.Schlade-Bartusiak K., Stembalska-Kozlowska A., Bernady M., Kudyba M., Sasiadek M. Analysis of adaptive response to bleomycin and mitomycin C. Mutat. Res. 2002;513:75–81. doi: 10.1016/s1383-5718(01)00288-1. [DOI] [PubMed] [Google Scholar]
- 17.Samson L., Schwartz J.L. Evidence for an adaptive DNA repair pathway in CHO and human skin fibroblast cell lines. Nature. 1980;287:861–863. doi: 10.1038/287861a0. [DOI] [PubMed] [Google Scholar]
- 18.Kaina B. Enhanced survival and reduced mutation and aberration frequencies induced in V79 chinese hamster cells pre-exposed to low levels of methylating agents. Mutat. Res. 1982;93:195–211. doi: 10.1016/0027-5107(82)90135-x. [DOI] [PubMed] [Google Scholar]
- 19.Nikolova T., Huttner E. Adaptive and synergistic effects of a low-dose ENU pretreatment on the frequency of chromosomal aberrations induced by a challenge dose of ENU in human peripheral blood lymphocytes in vitro. Mutat. Res. 1996;357:131–141. doi: 10.1016/0027-5107(96)00094-2. [DOI] [PubMed] [Google Scholar]
- 20.Doll R., Peto R., Boreham J., Sutherland I. Mortality in relation to smoking: 50 years' observations on male British doctors. BMJ. 2004;328:1519. doi: 10.1136/bmj.38142.554479.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Group I.W. IARC monographs on the evaluation of carcinogenic risks to humans: tobacco smoke and involuntary smoking. IARCPress. 2012;100E [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips D.H., Venitt S. DNA and protein adducts in human tissues resulting from exposure to tobacco smoke. Int. J. Cancer. 2012;131:2733–2753. doi: 10.1002/ijc.27827. [DOI] [PubMed] [Google Scholar]
- 23.Slaga T.J., Bracken W.J., Gleason G., Levin W., Yagi H., Jerina D.M., Conney A.H. Marked differences in the skin tumor-initiating activities of the optical enantiomers of the diastereomeric benzo(a)pyrene 7,8-diol-9,10-epoxides. Cancer Res. 1979;39:67–71. [PubMed] [Google Scholar]
- 24.Phillips D.H., Hewer A., Seidel A., Steinbrecher T., Schrode R., Oesch F., Glatt H. Relationship between mutagenicity and DNA adduct formation in mammalian cells for fjord- and bay-region diol-epoxides of polycyclic aromatic hydrocarbons. Chem. Biol. Interact. 1991;80:177–186. doi: 10.1016/0009-2797(91)90023-z. [DOI] [PubMed] [Google Scholar]
- 25.Hockley S.L., Arlt V.M., Brewer D., Giddings I., Phillips D.H. Time- and concentration-dependent changes in gene expression induced by benzo(a)pyrene in two human cell lines, MCF-7 and HepG2. BMC Genomics. 2006;7:260. doi: 10.1186/1471-2164-7-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dreij K., Seidel A., Jernstrom B. Differential removal of DNA adducts derived from anti-diol epoxides of dibenzo(a,l)pyrene and benzo(a)pyrene in human cells. Chem. Res. Toxicol. 2005;18:655–664. doi: 10.1021/tx0497090. [DOI] [PubMed] [Google Scholar]
- 27.Talaska G., Jaeger M., Reilman R., Collins T., Warshawsky D. Chronic, topical exposure to benzo(a)pyrene induces relatively high steady-state levels of DNA adducts in target tissues and alters kinetics of adduct loss. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7789–7793. doi: 10.1073/pnas.93.15.7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess M.T., Gunz D., Luneva N., Geacintov N.E., Naegeli H. Base pair conformation-dependent excision of benzo(a)pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol. Cell. Biol. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platt K., Oesch F. Efficient synthesis of non-K-region trans-dihydro diols of polycyclic aromatic hydrocarbons from o-quinones and catechols. J. Org. Chem. 1983;48:265–268. [Google Scholar]
- 30.Yagi H., Thakker D.R., Hernandez O., Koreeda M., Jerina D.M. Synthesis and reactions of the highly mutagenic 7,8-diol 9,10-epoxides of the carcinogen benzo(a)pyrene. J. Am. Chem. Soc. 1977;99:1604–1611. doi: 10.1021/ja00447a053. [DOI] [PubMed] [Google Scholar]
- 31.Nicoletti I., Migliorati G., Pagliacci M.C., Grignani F., Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 32.Tomicic M.T., Friedrichs C., Christmann M., Wutzler P., Thust R., Kaina B. Apoptosis induced by (E)-5-(2-bromovinyl)-2′-deoxyuridine in varicella zoster virus thymidine kinase-expressing cells is driven by activation of c-Jun/activator protein-1 and Fas ligand/caspase-8. Mol. Pharmacol. 2003;63:439–449. doi: 10.1124/mol.63.2.439. [DOI] [PubMed] [Google Scholar]
- 33.Tomicic M.T., Aasland D., Naumann S.C., Meise R., Barckhausen C., Kaina B., Christmann M. Translesion polymerase eta is upregulated by cancer therapeutics and confers anticancer drug resistance. Cancer Res. 2014;74:5585–5596. doi: 10.1158/0008-5472.CAN-14-0953. [DOI] [PubMed] [Google Scholar]
- 34.Hanelt S., Helbig R., Hartmann A., Lang M., Seidel A., Speit G. A comparative investigation of DNA adducts, DNA strand breaks and gene mutations induced by benzo(a)pyrene and (+/-)-anti-benzo(a)pyrene-7,8-diol 9,10-oxide in cultured human cells. Mutat. Res. 1997;390:179–188. doi: 10.1016/s0165-1218(97)00019-0. [DOI] [PubMed] [Google Scholar]
- 35.Christmann M., Kaina B. Nuclear translocation of mismatch repair proteins MSH2 and MSH6 as a response of cells to alkylating agents. J. Biol. Chem. 2000;275:36256–36262. doi: 10.1074/jbc.M005377200. [DOI] [PubMed] [Google Scholar]
- 36.Christmann M., Tomicic M.T., Kaina B. Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutS{alpha} mismatch-binding activity. Nucleic Acids Res. 2002;30:1959–1966. doi: 10.1093/nar/30.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichwan S.J., Yamada S., Sumrejkanchanakij P., Ibrahim-Auerkari E., Eto K., Ikeda M.A. Defect in serine 46 phosphorylation of p53 contributes to acquisition of p53 resistance in oral squamous cell carcinoma cells. Oncogene. 2006;25:1216–1224. doi: 10.1038/sj.onc.1209158. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Wu X., Guo D., Rechkoblit O., Geacintov N.E., Wang Z. Two-step error-prone bypass of the (+)- and (-)-trans-anti-BPDE-N2-dG adducts by human DNA polymerases eta and kappa. Mutat. Res. 2002;510:23–35. doi: 10.1016/s0027-5107(02)00249-x. [DOI] [PubMed] [Google Scholar]
- 39.Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 1975;5A:355–367. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- 40.Langie S.A., Koppen G., Desaulniers D., Al-Mulla F., Al-Temaimi R., Amedei A., Azqueta A., Bisson W.H., Brown D.G., Brunborg G., et al. Causes of genome instability: the effect of low dose chemical exposures in modern society. Carcinogenesis. 2015;36(Suppl. 1):S61–S88. doi: 10.1093/carcin/bgv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchel R.E. The dose window for radiation-induced protective adaptive responses. Dose Response. 2010;8:192–208. doi: 10.2203/dose-response.09-039.Mitchel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips D.H. Polycyclic aromatic hydrocarbons in the diet. Mutat. Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 43.Gunz D., Hess M.T., Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 44.Hockley S.L., Arlt V.M., Brewer D., Te Poele R., Workman P., Giddings I., Phillips D.H. AHR- and DNA-damage-mediated gene expression responses induced by Benzo(a)pyrene in human cell lines. Chem. Res. Toxicol. 2007;20:1797–1810. doi: 10.1021/tx700252n. [DOI] [PubMed] [Google Scholar]
- 45.Hockley S.L., Arlt V.M., Jahnke G., Hartwig A., Giddings I., Phillips D.H. Identification through microarray gene expression analysis of cellular responses to benzo(a)pyrene and its diol-epoxide that are dependent or independent of p53. Carcinogenesis. 2008;29:202–210. doi: 10.1093/carcin/bgm227. [DOI] [PubMed] [Google Scholar]
- 46.Mitchel R.E. Low doses of radiation are protective in vitro and in vivo: evolutionary origins. Dose Response. 2006;4:75–90. doi: 10.2203/dose-response.04-002.Mitchel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchel R.E., Burchart P., Wyatt H. A lower dose threshold for the in vivo protective adaptive response to radiation. Tumorigenesis in chronically exposed normal and Trp53 heterozygous C57BL/6 mice. Radiat. Res. 2008;170:765–775. doi: 10.1667/RR1414.1. [DOI] [PubMed] [Google Scholar]
- 48.Rechkoblit O., Zhang Y., Guo D., Wang Z., Amin S., Krzeminsky J., Louneva N., Geacintov N.E. trans-Lesion synthesis past bulky benzo(a)pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki N., Ohashi E., Kolbanovskiy A., Geacintov N.E., Grollman A.P., Ohmori H., Shibutani S. Translesion synthesis by human DNA polymerase kappa on a DNA template containing a single stereoisomer of dG-(+)- or dG-(-)-anti-N(2)-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y., Yuan F., Wu X., Wang M., Rechkoblit O., Taylor J.S., Geacintov N.E., Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogi T., Shinkai Y., Tanaka K., Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo(a)pyrene. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bi X., Slater D.M., Ohmori H., Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo(a)pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J. Biol. Chem. 2005;280:22343–22355. doi: 10.1074/jbc.M501562200. [DOI] [PubMed] [Google Scholar]
- 53.Zhu H., Fan Y., Shen J., Qi H., Shao J. Characterization of human DNA polymerase kappa promoter in response to benzo(a)pyrene diol epoxide. Environ. Toxicol. Pharmacol. 2012;33:205–211. doi: 10.1016/j.etap.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y., Yuan F., Wu X., Rechkoblit O., Taylor J.S., Geacintov N.E., Wang Z. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 2000;28:4717–4724. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiapperino D., Kroth H., Kramarczuk I.H., Sayer J.M., Masutani C., Hanaoka F., Jerina D.M., Cheh A.M. Preferential misincorporation of purine nucleotides by human DNA polymerase eta opposite benzo(a)pyrene 7,8-diol 9,10-epoxide deoxyguanosine adducts. J. Biol. Chem. 2002;277:11765–11771. doi: 10.1074/jbc.M112139200. [DOI] [PubMed] [Google Scholar]
- 56.Frank E.G., Sayer J.M., Kroth H., Ohashi E., Ohmori H., Jerina D.M., Woodgate R. Translesion replication of benzo(a)pyrene and benzo(c)phenanthrene diol epoxide adducts of deoxyadenosine and deoxyguanosine by human DNA polymerase iota. Nucleic Acids Res. 2002;30:5284–5292. doi: 10.1093/nar/gkf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu G., Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol. Cell. Biol. 2006;26:1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rathore K., Choudhary S., Odoi A., Wang H.C. Green tea catechin intervention of reactive oxygen species-mediated ERK pathway activation and chronically induced breast cell carcinogenesis. Carcinogenesis. 2012;33:174–183. doi: 10.1093/carcin/bgr244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roos W.P., Thomas A.D., Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 60.Azzam E.I., Raaphorst G.P., Mitchel R.E. Radiation-induced adaptive response for protection against micronucleus formation and neoplastic transformation in C3H 10T1/2 mouse embryo cells. Radiat. Res. 1994;138:S28–S31. [PubMed] [Google Scholar]
- 61.Fritz G., Tano K., Mitra S., Kaina B. Inducibility of the DNA repair gene encoding O6-methylguanine-DNA methyltransferase in mammalian cells by DNA-damaging treatments. Mol. Cell. Biol. 1991;11:4660–4668. doi: 10.1128/mcb.11.9.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.