Abstract
The FANCI-FANCD2 (I-D) complex is considered to work with RAD51 to protect the damaged DNA in the stalled replication fork. However, the means by which this DNA protection is accomplished have remained elusive. In the present study, we found that the I-D complex directly binds to RAD51, and stabilizes the RAD51-DNA filament. Unexpectedly, the DNA binding activity of FANCI, but not FANCD2, is explicitly required for the I-D complex-mediated RAD51-DNA filament stabilization. The RAD51 filament stabilized by the I-D complex actually protects the DNA end from nucleolytic degradation by an FA-associated nuclease, FAN1. This DNA end protection is not observed with the RAD51 mutant from FANCR patient cells. These results clearly answer the currently enigmatic question of how RAD51 functions with the I-D complex to prevent genomic instability at the stalled replication fork.
INTRODUCTION
DNA interstrand crosslinks (ICLs), arising from endogenous aldehydes or anti-cancer crosslinking agents, stall replication fork progression (1–3). Fanconi anemia (FA) is an infantile hereditary disorder with severe manifestations, such as bone marrow failure, predisposition to cancers and cellular hypersensitivity to ICL-inducing agents (4–6). Nineteen FA-causal genes have been identified from the FA patient cells (7). Among the FA proteins, FANCI and FANCD2 form the I-D complex, which is monoubiquitinated by the FA core complex, composed of FANCA, -B, -C, -E, -F, -G, -L, -M, FAAP20, FAAP24, FAAP100, MHF1 and MHF2 (8–11). FANCL is a ubiquitin E3 ligase subunit, and interacts with a ubiquitin E2 conjugating enzyme, UBE2T (FANCT) (12–16). The I-D complex preferentially binds to branched DNA structures, such as the stalled replication fork (17), and its DNA binding drastically enhances the FANCL-UBE2T complex-mediated FANCD2 monoubiquitination (18–20). The monoubiquitinated I-D complex is considered to function in recruiting structure-specific nucleases to the damaged DNA site, and to remove the affected bases at the ICL site (21–27).
RAD51 is a eukaryotic recombinase that functions in meiotic homologous recombination and mitotic DNA double strand break (DSB) repair (28,29). During DSB repair, RAD51 promotes homologous pairing between the single-stranded DNA (ssDNA) produced at the DSB site and the homologous double-stranded DNA (dsDNA) (30–32). Recently, a heterozygous RAD51 mutation (RAD51 T131P), in which Thr131 is replaced by Pro, was identified as an FANCR gene in an infant with FA (33). Another missense RAD51 mutation (RAD51 R150Q), in which Arg150 is replaced by Gln, was also reported in bilateral breast cancer patients (34,35). These facts demonstrate that RAD51 plays an important role in the suppression of tumorigenesis.
Emerging evidence has indicated that RAD51 accumulates on ICL-induced stalled replication forks, and protects the stalled replication fork from undesired degradation by exonucleases recruited by the I-D complex (33,36). In fact, defects in RAD51 assembly at the stalled replication fork resulted in excessive nucleolytic degradation of the nascent DNA strand by the exonucleases, including the MRE11, DNA2 and FAN1 nucleases, leading to chromosomal instability (33,37–41). Interestingly, the I-D complex reportedly colocalizes with RAD51 at the nascent DNA region of stalled replication forks, and plays a role in the RAD51-mediated nascent DNA protection (42–45). However, the mechanism by which the I-D complex and RAD51 cooperatively protect genomic DNA in the stalled replication fork has remained elusive.
In the present study, we found that the I-D complex directly binds to RAD51, stabilizes the RAD51-DNA filament and protects the DNA end from nucleolytic degradation by an FA-associated nuclease, FAN1. This work explains how RAD51 and the I-D complex cooperatively function to prevent genomic instability at the stalled replication fork, which is a major target for cancer chemotherapy.
MATERIALS AND METHODS
Plasmids
The DNA fragment encoding chicken RAD51 was amplified by polymerase chain reaction from the DT40 cDNA library, using the following primers: 5′-GGAAT TCCAT ATGGC CATGC AGGTG CAGTT CGAGG C-3′ and 5′-TCCCG CGGAT CCTTA TTCTT TTGCA TCTCC CACTC CATCA G-3′. The amplified DNA fragment was ligated into the NdeI-BamHI sites of the pET15b vector (Novagen). The FANCR mutation, in which Thr131 was replaced by Pro, was introduced in the chicken RAD51 cDNA by using a KOD mutagenesis kit (TOYOBO) with the following primers: 5′-CGGGA AAAAC ACAGT TGTGC CATAC TTTGG-3′ and 5′-GACGA AACTC CCCAA ATAAT TCTGT TATGG AC-3′. The RAD51 F86E mutation, in which Phe86 was replaced by Glu, was introduced into the chicken RAD51 cDNA by using a KOD mutagenesis kit (TOYOBO) with the following primers: 5′-GAAAC CACAG CAACG GAATT CCATC-3′ and 5′-ACCCA TCGGA ACCAG TTTAG CTG-3′.
For the generation of the expression vector for the chicken FAN1 nuclease domain (amino acids 385–1034 of chicken FAN1), the DNA fragment encoding full-length chicken FAN1 was ligated into the NdeI-BamHI sites of the pET15b vector. The cDNA of the FAN1 nuclease domain was amplified by using a KOD mutagenesis kit with the following primers: 5′-CCATA TTACC TCCGA AACTT TTTAA TGGTG TTG-3′ and 5′-GCTGC CGCGC GGCAC CAGGC CGCTG CTG-3′. For the FAN1 nuclease domain mutant, in which Asp977 was replaced by Ala, the amino acid substitution was introduced in the FAN1 cDNA by using a KOD mutagenesis kit with the following primers: 5′-CGCTG GTTGT GTGGA GTACT CACAG CAATC AC-3′ and 5′-CAGGA AGCCC CCCTC TGCAA TGGCG CAGAT CTTTG-3′.
The DNA fragment encoding the chicken FANCD2(Ex7) mutant, in which Lys361, Lys369, Arg399, Lys400, Lys404, Arg407 and Lys974 were replaced by Glu, was generated by the site directed mutagenesis method using a KOD mutagenesis kit with the following primer pairs: 5′-AGCTT GGATT GAAGC TATTG AGAAC AGCAC ATCTG-3′/5′-TCAGA GACAT CTTCC TGGAA TCTTA CAGCT TGC-3′, 5′-GAGGA AGTAT TGGAA AGCAA GATTC GCCTG GGC-3′/5′-AGTTT GTTCT TCGTT CTTGC TGTTC GTAGA ATGG-3′ and 5′-GAACT GGAAC ATGTG CTGAC CCCAG GCTCC AC-3′/5′-CCAGC ACATG TCATC CAGAA GGAAG CAGAG-3′.
Generation of cell line
The construction of the human U2OS cells expressing C-terminally 3xFLAG-tagged FANCD2 will be described elsewhere. In brief, a 3xFLAG-tag sequence was knocked-in at the termination codon of the endogenous FANCD2 gene locus, using TALEN technology. To produce doxycycline-inducible GFP-RAD51 expressing U2OS cells, the cDNA encoding the N-terminally GFP-tagged RAD51 was inserted into the entry vector pENTR (Invitrogen). The resulting vector was transferred into a puromycin-resistant derivative of CSIV-TRE-RfA-UbC-KT (46), using LR Clonase (Invitrogen). U2OS cells were infected with the lentivirus, and were selected in the presence of puromycin (0.5 μg/ml).
Cell culture
U2OS cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). HeLa cells and hTERT-immortalized fibroblasts (1BR3.hTERT) were maintained in DMEM (Low Glucose) supplemented with 10% FBS. FANCD2-deficient fibroblasts stably expressing GFP, GFP-FANCD2 or GFP-FANCD2 K561R, in which the ubiquitination target Lys561 was replaced by Arg, were maintained in α-MEM supplemented with 1 μg/ml puromycin (Sigma-Aldrich) and 15% FBS, as previously described (47). DT40 cells were maintained in RPMI-1640 medium supplemented with 10% fetal calf serum, 1% chicken serum, 2 mM L-glutamine, 50 μM 2-mercaptoethanol, penicillin and streptomycin.
RNA interference and immunoblotting
The FANCD2-specific siRNA (5′-CAGAG UUUGC UUCAC UCUCU AdTdT-3′) and the control siRNA (5′-UCGAA GUAUU CCGCG UACGdT dT-3′) were introduced into immortalized human fibroblasts or U2OS cells using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer's protocol. Whole cellular proteins were prepared 3 days after the siRNA transfection, fractionated by 6 or 10% SDS-PAGE, and immunoblotted with a mouse monoclonal antibody directed against α-tubulin (1:5000; B-5-1-2, Sigma-Aldrich), or a rabbit polyclonal antibody against either FANCD2 (1:5000; NB100-182, Novus) or RAD51 (1:5000; 48). Secondary antibody treatment was performed with a sheep anti-mouse IgG, HPR-linked species-specific F(ab’)2 fragment (1:10 000 for α-tubulin; GE Healthcare) or a donkey anti-rabbit IgG, horseradish peroxidase (HRP)-linked species-specific F(ab’)2 fragment (GE Healthcare; 1:10 000 for RAD51 and FANCD2). Signals were detected by the ECL Prime Western Blotting Detection Reagent (GE Healthcare).
Immunofluorescence
Immortalized fibroblasts, PD20F cells or U2OS cells were grown on coverslips (Matsunami, 0.17 mm thickness), and were transfected with siRNA, followed by fixation and permeabilization with 0.5% Triton X-100, 3% paraformaldehyde and 2% sucrose in phosphate buffered saline (PBS), for 30 min on ice at 0–25 h after 100 ng/ml mitomycin C (MMC) treatment or at 5 h after 4 mM hydroxyurea (HU) treatment (2–3 days post transfection). For RAD51 staining, the permeabilized cells were treated with rabbit anti-RAD51 (1:2000) and Alexa Fluor 594 goat anti-rabbit IgG (1:500, Molecular Probes). The fluorescence images were obtained using the BZ II viewer application connected to a BZ-9000 fluorescence microscope (KEYENCE) with a Plan Apo λ 40X/NA 0.95 objective lens (Nikon). All captured images were analyzed by the BZ II analyzer software (KEYENCE). To assess the RAD51 foci, the fluorescent intensities of RAD51 in >100 cells were analyzed, and were normalized by each gross area of the nucleus. Statistical differences were determined by Bonferroni's multiple comparison test with the Prism software (GraphPad Software Inc.).
In situ proximity ligation assay (PLA)
PLA experiments were performed according to the manufacturer's protocol, using a DuoLink Kit (Sigma-Aldrich). For the PLA experiments between RAD51 and FLAG-tagged FANCD2, cells were treated with 100 ng/ml MMC or 0.5 mM HU (or not treated) for 24 h, and were fixed with 2% sucrose, 3% bovine serum albumin (BSA) and 0.5% Triton X-100 in PBS, followed by staining with an anti-FLAG tag (anti-DDDDK-tag) mouse Ab (1:500; PM020, MBL) and an anti-RAD51 rabbit Ab (1:500). For the PLA experiments between RAD51 and PCNA, cells were transfected with the control siRNA or the FANCD2 siRNA, and were fixed at 48 h after transfection. The HU treatment (4 mM) was performed for 2.5 h immediately before sample fixation. The cells were then stained with an anti-PCNA mouse Ab (1:500; PC10, Santa Cruz Biotechnology, Inc.) and an anti-RAD51 rabbit Ab (1:500). PLA signals were detected with a fluorescence microscope (BZ-9000), and nuclear PLA signals were analyzed by counting the red signals on DAPI-stained blue areas, using the hybrid cell count software (KEYENCE). In total, >50 cells were analyzed, and were plotted in each column. Statistical differences were determined by the Student's t-test with the Prism software.
Fluorescence recovery after photobleaching (FRAP)
U2OS cells producing GFP-RAD51 were grown in DMEM supplemented with 10% FBS, on glass-bottom dishes, and were transfected with either the control siRNA or the FANCD2 siRNA. After cultivation for 24 h, the cells were treated with doxycycline (1 ng/ml) to induce the GFP-RAD51 production, and were further grown for 24 h in either the presence or absence of MMC (100 ng/ml). FRAP was then performed using a TCS SP5 II confocal microscope (Leica), equipped with an INU incubator system for microscopes (Tokai Hit), at 37°C with a 5% CO2 atmosphere. Two confocal images of a field containing a nucleus were collected, using an HCX PL APO 63x/1.40–0.60 Oil CS objective lens (512 × 512 pixels, zoom 8, scan speed 400 Hz and 2% transmission of 488-nm laser). One-quarter to one-half of each nucleus was bleached using 100% transmission at 488 nm (five iterations), and images were acquired using the original settings at 3 s intervals. The fluorescence intensities of the bleached GFP-RAD51 foci were measured using Image J 1.46r (Rasband, http://rsb.info.nih.gov/ij/). After background subtraction, the intensities of the bleached foci were normalized to the initial intensity before bleaching.
Preparation of recombinant proteins
Human RAD51 was overexpressed in E. coli JM109(DE3) cells, which contain the minor tRNA expression vector (Codon(+)RIL; Stratagene), at 30°C, and was purified by the method described previously (49). Chicken RAD51 was purified by the same method as that for human RAD51. For the purification of the chicken RAD51 T131P mutant, the spermidine precipitation step was omitted. These RAD51 proteins were produced as N-terminally His6-tagged proteins, and the His6 tag was removed by thrombin protease treatment during the purification processes. For the purification of the His6-tagged chicken RAD51 and the His6-tagged chicken RAD51 F86E mutant, the thrombin protease treatment and the spermidine precipitation steps were omitted. The purified His6-tagged RAD51 was dialyzed against 1 l of buffer A, containing 20 mM HEPES-NaOH (pH 7.5), 10% glycerol, 250 mM KCl, 50 mM NaF, 0.25 mM EDTA and 5 mM 2-mercaptoethanol. The His6-tagged chicken RAD51 F86E mutant was further purified by chromatography on a Superdex 200 column (HiLoad 26/60 preparation grade; GE Healthcare) equilibrated with buffer A. In the Ni-NTA agarose (Qiagen) chromatography step for the purification of the His6-tagged chicken RAD51 F86E mutant, the resin was washed with buffer containing 10 mM imidazole, instead of the buffer containing 60 mM imidazole used for the His6-tagged chicken RAD51 purification (wild type).
The FAN1 nuclease domain was overexpressed in E. coli BL21(DE3) cells containing the Codon(+)RIL vector, at 18°C. The cells producing the His6-tagged FAN1 nuclease domain were collected by centrifugation, and were resuspended in buffer B, containing 50 mM Tris-HCl (pH 8.0), 10% glycerol, 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 12 mM imidazole and 5 mM 2-mercaptoethanol. The cells were then disrupted by sonication, and the supernatant was separated from the cell debris by centrifugation at 27 700 × g for 30 min. The supernatant was mixed gently with 3 ml of Ni-NTA agarose resin at 4°C for 1 h, by the batch method. The Ni-NTA agarose resin was packed into an Econo-column (Bio-Rad), and was washed with 150 ml buffer B. The FAN1 nuclease domain was eluted with a 60 ml linear-gradient of 12–400 mM imidazole in buffer B. The peak fractions were pooled, and the sample was immediately dialyzed against 4 l buffer C, containing 20 mM Tris-HCl (pH 8.0), 10% glycerol, 200 mM NaCl and 5 mM 2-mercaptoethanol. The sample was then loaded on a Q Sepharose Fast Flow column (2 ml; GE Healthcare). The unbound fractions were pooled, and were loaded on a Superdex 200 column (HiLoad 16/60 preparation grade; GE Healthcare) equilibrated with buffer C. The purified FAN1 nuclease domain was concentrated, and aliquots were frozen in liquid nitrogen. The FAN1 nuclease domain (D977A) protein was prepared by the same method as that for the wild-type FAN1 nuclease domain.
Chicken FANCI, chicken FANCI(Ex6), chicken FANCD2, human RPA and human DMC1 were purified as described previously (18,50,51). The I-D complex was prepared by mixing FANCI and FANCD2 in a 1:1 stoichiometry. For the purification of FANCD2(Ex7), a Q Sepharose Fast Flow column (3 ml) was employed, instead of a Heparin Sepharose CL-6B column (GE Healthcare). The column was washed with 150 ml buffer C containing 235 mM NaCl, and the proteins were eluted with a 60 ml linear gradient of 235–450 mM NaCl in buffer C. The subsequent purification procedure for FANCD2(Ex7) was the same as that for wild-type FANCD2. The protein concentration was determined by the Bradford method (52), using BSA as the standard protein.
DNA substrates
The 49-mer dsDNA was prepared by annealing oligonucleotide 1, with the sequence 5′-GTCCC AGGCC ATTAC AGATC AATCC TGAGC ATGTT TACCA AGCGC ATTG-3′, and its complementary oligonucleotide, as described previously (53). The 5′-biotinylated 3′-tailed DNA was prepared by annealing the 5′-biotinylated oligonucleotide 3 and the unmodified oligonucleotide 4, with the sequences 5′-GTCCC AGGCC ATTAC AGATC AATCC TGAGC ATGTT TACCA AGCGC ATTGT TTTTT TTTTT TTTTT TTTTT TTTTT TTTTT TTTTT TTTTT TTTTT TTTTT-3′ and 5′-CAATG CGCTT GGTAA ACATG CTCAG GATTG ATCTG TAATG GCCTG GGAC-3′, respectively. For the preparation of the 32P-radiolabeled 3′-tailed DNA, oligonucleotide 4 labeled at the 5′-end was annealed with the unmodified oligonucleotide 3. The 5′-biotinylated replication fork-like DNA was prepared by annealing the 5′-biotinylated oligonucleotide 5 and the unmodified oligonucleotides 6, 7 and 8, with the sequences 5′-GTCCC AGGCC ATTAC AGATC AATCC TGAGC ATGTT TACCA AGCGC ATTGG CCTCG ATCCT ACCAA CCAGA TGACG CGCTG CTACG TGCTA CCGGA AGTCG-3′, 5′-CGACT TCCGG TAGCA CGTAG CAGCG CGTCA TCTGG TTGGT AGGAT CGAGG C-3′, 5′-ATGGC GCAGC GCATC CTGCA GCTGG CGGCC GTTTT TTTTT TTTTT TTTTT TCAAT GCGCT TGGTA AACAT GCTCA GGATT GATCT GTAAT GGCCT GGGAC-3′ and 5′-CGGCC GCCAG CTGCA GGATG CGCTG CGCCA T-3′, respectively. For the preparation of the 32P-radiolabeled replication fork-like DNA, the 5′-end (32P) labeled oligonucleotide 8 was annealed with the unmodified oligonucleotides 5, 6 and 7. All of the oligonucleotides were purchased from Nihon Gene Research Laboratory, as HPLC-purified grade. DNA concentrations are expressed in moles of nucleotides.
Pull-down assays
Purified human and chicken RAD51 were covalently conjugated with Affi-Gel 15 beads (Bio-Rad), according to the manufacturer's protocol. The control beads were prepared by the same method, in the absence of RAD51. For the cell-based pull-down assay, HeLa cell extracts were prepared with lysis buffer, containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 2 mM NaF, 2 mM Na3VO4 and 1x protease inhibitor cocktail (Nacalai Tesque), as previously described (54). DT40 cell extracts were prepared by the same method used for the HeLa cell extract preparation. The human and chicken RAD51 beads (6.4 μg of protein) were gently mixed with the HeLa and DT40 cell extracts (1.5 mg of protein), and were incubated in 20 mM Tris-HCl buffer (pH 7.5), containing 100 mM NaCl, 6.6% glycerol, 0.017% Triton X-100, 0.17% NP-40, 3.3 mM 2-mercaptoethanol, 0.67 mM NaF, 0.67 mM Na3VO4 and 0.67x protease inhibitor cocktail, for 16 h at 4°C. The cell lysates were treated with a nuclease, benzonase (50 unit/ml, Sigma-Aldrich), before the addition of the RAD51 beads. Therefore, the RAD51 binding to the DNA-free FANCI-FANCD2 proteins was detected. After the incubation, the beads were washed 3 times with 1 ml of 20 mM Tris-HCl (pH 7.5) buffer, containing 75 mM NaCl, 10% glycerol, 0.025% Triton X-100 and 5 mM 2-mercaptoethanol. The endogenous FANCI and FANCD2 that copelleted with the RAD51 beads were separated by SDS-PAGE, and were detected by western blotting with a human FANCD2-specific mouse monoclonal antibody (1:250; FI17, Santa Cruz Biotechnology, Inc.) or a rabbit polyclonal antibody against either chicken FANCD2 (1:1000), human FANCI (1:1000; A301-254A, Bethyl Laboratories, Inc.) or chicken FANCI (1:1000). Human RAD51 and chicken RAD51 were detected by Western blotting with a rabbit polyclonal antibody (1:1000).
For the pull-down assay with Ni-NTA beads, chicken RAD51 (5.5 μg) or human DMC1 (5.5 μg) was incubated with either His6-tagged FANCI (3.8 μg), His6-tagged FANCD2 (3.8 μg) or His6-tagged FANCD2 complexed with FANCI (7.6 μg for the I-D complex) for 10 min at 37°C in 100 μl of pull-down buffer, containing 20 mM Tris-HCl (pH 7.5), 10% glycerol, 150 mM NaCl, 0.025% Triton X-100, 5 mM imidazole, 0.2 mM Ethylene diamine tetraacetic acid (EDTA) and 2 mM 2-mercaptoethanol. Ni-NTA agarose beads (4 μl) were added to the reaction mixtures and were gently mixed for 50 min at 23°C. The beads were then washed three times with 1 ml pull-down buffer containing 0.05% Triton X-100. For the pull-down assay with His6-tagged RAD51 proteins, His6-tagged RAD51 (3.5 μg) or His6-tagged RAD51 F86E (2 μg) was gently mixed with Ni-NTA agarose beads (3 μl), for 45 min at 4°C in 55 μl of buffer A. At these protein concentrations, the amounts of RAD51 and RAD51 F86E bound to the Ni-NTA agarose beads were confirmed to be equal. After washing with buffer A (300 μl), the I-D complex (5.5 μg) was added to the beads and was gently mixed in 100 μl of the pull-down buffer containing 50 mM NaF, for 50 min at 23°C. The beads were then washed twice with the pull-down buffer (1 ml) containing 0.05% Triton X-100. The proteins bound to the beads were separated by 5–20%, 10% or 12% SDS-PAGE and were visualized by Coomassie Brilliant Blue staining. The band intensities of the I-D complex, RAD51 and DMC1 were quantitated with an LAS-4000 image analyzer (GE Healthcare), using the MultiGauge ver. 3.2 software (Fujifilm).
For the pull-down assay with magnetic streptavidin beads, chicken RAD51 (3 μM) or human RPA (3 μM) was incubated with a 5′-biotinylated 80-mer poly dT ssDNA (7 μM) conjugated to Dynabeads M-280 Streptavidin (Invitrogen) for 15 min at 37°C, in 20 μl of reaction buffer, containing 35 mM Tris-HCl (pH 8.0), 3% glycerol, 55 mM NaCl, 1 mM AMP-PNP, 2.5 mM MgCl2, 5 mM CaCl2 and 1 mM dithiothreitol. During the incubation period, the reaction mixtures were gently mixed by tapping at 3 min intervals. The beads were then washed once with 20 μl of the reaction buffer, and were incubated with the I-D complex (4 μM), which was preincubated for 15 min at 37°C, in 20 μl of the reaction buffer. The reaction was continued for 30 min at 37°C, with tapping at 3 min intervals. After the 30 min reaction, the beads were washed twice with 20 μl of the reaction buffer, and the proteins bound to the beads were analyzed by 10% SDS-PAGE with Coomassie Brilliant Blue staining. The band intensity of the I-D complex was quantitated with an LAS-4000 image analyzer, using the MultiGauge ver. 3.2 software. The reactions with naked ssDNA beads were performed in the absence of RAD51 and RPA.
RAD51 transfer assay
Chicken RAD51 (5.2 μM) was incubated with a 5′-biotinylated 80-mer poly dT oligo DNA (20.7 μM), a 5′-biotinylated 3′-tailed DNA (20.7 μM) or a 5′-biotinylated replication fork-like DNA (20.7 μM) conjugated to Dynabeads M-280 Streptavidin for 10 min at 37°C, in 11.6 μl of reaction buffer, containing 60 mM Tris-HCl (pH 8.0), 6% glycerol, 108 mM NaCl, 4.3 mM MgCl2, 1.7 mM adenosine triphosphate (ATP) and 1.7 mM dithiothreitol. After the incubation, the I-D complex (2.7 μM), which was preincubated with or without the 49-mer dsDNA (20 μM) for 10 min at 37°C in 6 μl of 13 mM Tris-HCl (pH 8.0) buffer, containing 6.7% glycerol, 133 mM NaCl and 3.3 mM 2-mercaptoethanol, was added to the reaction mixture, followed by an incubation for 10 min at 37°C. For the experiments with the I-D complex-bound 3′-tailed DNA, the I-D complex (94 nM) was preincubated with a 3′-tailed DNA (10.3 μM) conjugated to Dynabeads M-280 Streptavidin, for 10 min at 37°C, in 17 μl of reaction buffer, containing 46 mM Tris-HCl (pH 8.0), 2.4% glycerol, 59 mM NaCl, 2.9 mM MgCl2, 1.2 mM ATP and 1.2 mM dithiothreitol. A 3 μl aliquot of chicken RAD51 (20 μM) was then added to the reaction mixture, which was incubated for 10 min at 37°C. The RAD51 transfer reaction was initiated by the addition of the 49-mer ssDNA oligonucleotide 1 (2.4 μl, final concentration 120 μM), and was continued at 37°C for 1 h. During the reaction, the beads were gently mixed by tapping at 3 min intervals. After the reaction, the beads were washed three times with 20 μl of washing buffer, containing 35 mM Tris-HCl (pH 8.0), 2.5 mM MgCl2, 1 mM ATP, 0.01% NP-40 and 1 mM dithiothreitol and the proteins bound to the beads were analyzed by 10% SDS-PAGE with Coomassie Brilliant Blue staining. The band intensity of the I-D complex was quantitated with an LAS-4000 image analyzer, using the MultiGauge ver. 3.2 software.
Gel-Filtration assay
Gel-filtration analyses were performed with 100 μl of FANCI (5 μM), FANCD2 (5 μM) or the I-D complex (5 μM), as described previously (54,55). The peak fractions at the 7.5-15 ml elution volume were analyzed by 8% SDS-PAGE with Coomassie Brilliant Blue staining.
DNA binding assay
For the experiments with FANCI and FANCD2, the 49-mer dsDNA (5 μM) was incubated with FANCI, FANCD2 or the I-D complex in 10 μl of reaction buffer, containing 20 mM Tris–HCl (pH 8.0), 40 mM NaCl, 2% glycerol, 5 μg/ml BSA and 0.5 mM dithiothreitol, for 10 min at 37°C. For the experiments with the I-D-dsDNA complex, the I-D complex (1 μM) was incubated with or without the 49-mer dsDNA (3.8 μM) for 10 min at 37°C, in 8 μl of 50 mM Tris-HCl (pH 8.0) buffer, containing 4.4% glycerol, 96 mM NaCl, 1.3 mM ATP, 3.1 mM MgCl2 and 1.3 mM dithiothreitol. After the incubation, a 2 μl aliquot of the ssDNA mixture (500 μM oligonucleotide 1 and 15 μM 32P-labeled oligonucleotide 1) was added to the reaction mixture, which was incubated for 5 min at 37°C. For the experiments with RAD51, the 49-mer ssDNA oligonucleotide 1 (5 μM) or the 49-mer dsDNA (5 μM) was incubated with RAD51 in 10 μl of reaction buffer, containing 28 mM HEPES-NaOH (pH 7.5), 60 mM NaCl, 4% glycerol, 2.5 mM MgCl2, 1 mM ATP, 5 μg/ml BSA and 1 mM dithiothreitol, for 10 min at 37°C. The samples were then analyzed by 3.5% or 6% PAGE in 0.2x TBE (18 mM Tris-borate and 0.4 mM EDTA for FANCI, FANCD2 and the I-D complex) or 0.5x TBE (45 mM Tris-borate and 1 mM EDTA for RAD51) buffer. The DNA was visualized by SYBR Gold (Invitrogen) staining. In the control experiments, the samples were deproteinized by adding 2 μl of 1.4% SDS and 8.5 μg/ml proteinase K, followed by an incubation for 5 min at 37°C, before the electrophoresis. The band intensity of the free DNA was quantitated with an LAS-4000 image analyzer or an FLA-7000 laser scanner (Fujifilm), using the MultiGauge ver. 3.2 software.
Nuclease protection assay
Chicken RAD51 (0.36 μM) was incubated with 32P-labeled 3′-tailed DNA (1.4 μM) or 32P-labeled replication fork-like DNA (1.4 μM) for 10 min at 37°C, in 7 μl of reaction buffer, containing 26 mM Tris-HCl (pH 8.0), 4.3% glycerol, 71 mM NaCl, 1.4 mM MgCl2, 1.4 mM MnCl2, 1.4 mM ATP, 0.14 mg/ml BSA and 7.1 mM dithiothreitol. The I-D complex (1 μM), which was preincubated with the 49-mer dsDNA (3.8 μM) for 10 min at 37°C in 2 μl of 10 mM Tris-HCl (pH 8.0) buffer, containing 5% glycerol, 100 mM NaCl and 2.5 mM 2-mercaptoethanol, was added, and the mixture was incubated for 10 min at 37°C. The nucleolytic reaction was initiated by the addition of the FAN1 nuclease domain (1 μl, final concentration 0.4 μM), and was continued for 30 min at 37°C. The reaction was stopped by the addition of 2 μl of 1.4% SDS and 8.5 μg/ml proteinase K, followed by an incubation for 15 min at 37°C. After deproteinization, Hi-Di formamide (50 μl, Applied Biosystems) was added to each sample. The samples were boiled for 10 min at 100°C, and were immediately cooled on ice for 5 min. The samples were then fractionated by 12% urea denaturing PAGE in 1x TBE buffer (89 mM Tris-borate and 2 mM EDTA). The gels were exposed to an imaging plate (Fujifilm), and the DNA bands were visualized using an FLA-7000 imaging analyzer. The band intensities were quantitated using the Image J 1.46r software.
RESULTS AND DISCUSSION
FANCD2 is required for stable RAD51 association at replication forks stalled by ICLs
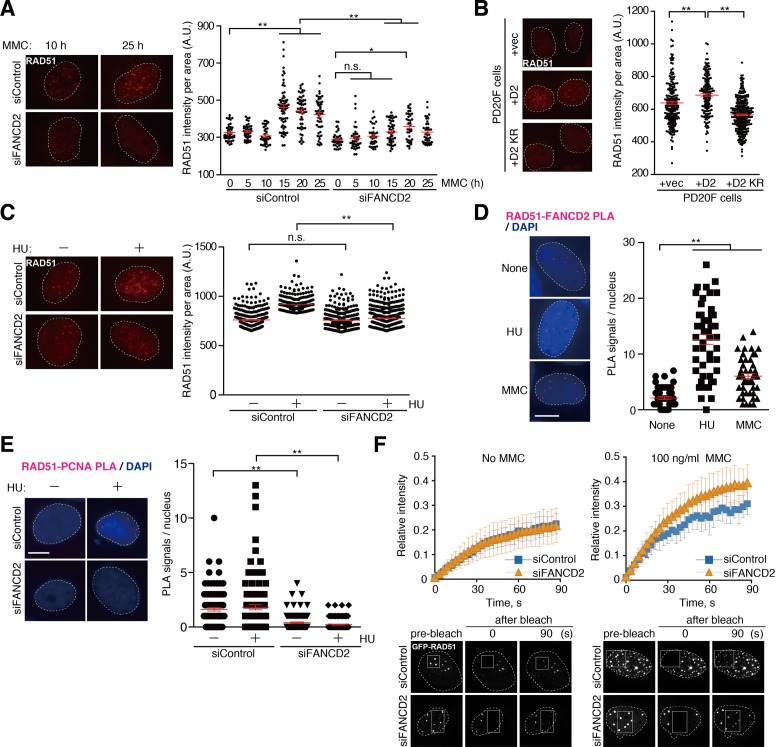
In cells, RAD51-foci intensity was remarkably increased at 15–25 h after treatment with a DNA crosslinker, mitomycin C (MMC), which stalls replication fork progression (Figure 1A, experiments with a control siRNA). We found that the MMC-induced RAD51 foci formation was significantly suppressed at a later time point of MMC treatment in the FANCD2-knockdown cells (Figure 1A, experiments with a FANCD2 siRNA), in which the expression level of FANCD2 was substantially decreased by a FANCD2-specific siRNA to <5% of that in the cells treated with control siRNA (Supplementary Figure S1A). The exogenous production of FANCD2 in FANCD2-deficient PD20F patient cells complemented the MMC-induced RAD51 foci formation (Figure 1B). The monoubiquitination-deficient FANCD2 K561R mutant, which is defective in chromatin targeting (8), did not complement the defective RAD51 recruitment in PD20F cells (Figure 1B). In the PD20F cells, HU, a chemical compound that induces replication fork stalling, promotes the degradation of the stalled replication fork (44). Consistently, in the FANCD2-knockdown cells, the formation of RAD51 foci induced by HU was also suppressed (Figure 1C and Supplementary Figure S1B).
Figure 1.
Stable RAD51 targeting to the stalled replication forks requires FANCD2. (A–C) Replication stress-induced foci formation of RAD51. (A) 1BR3.hTERT fibroblasts transfected with control or FANCD2 siRNA were treated with MMC, and the RAD51 foci were visualized by staining with an anti-RAD51 antibody after incubations for the indicated times. (B) PD20F cells expressing GFP-FANCD2, GFP-FANCD2 K561R (KR) and GFP alone were used for the MMC-induced RAD51 foci formation assay. (C) U2OS cells transfected with control or FANCD2 siRNA were treated with hydroxyurea (HU), and the RAD51 foci intensities were analyzed. The normalized intensities of RAD51 foci are represented as dot plots. Red horizontal lines denote means with standard deviations. Statistical differences were determined by the Student's t-test; *P < 0.01, **P < 0.0001, n.s., not significant. (D and E) In situ proximity ligation assay (PLA). (D) The PLA signals (red) between RAD51 and FLAG-tagged FANCD2 were visualized by staining with anti-RAD51 and anti-FLAG antibodies, in the untreated cells and the cells treated with MMC or HU. (E) U2OS cells transfected with control or FANCD2 siRNA were treated with or without HU, and the PLA signals between RAD51 and PCNA were visualized by staining with anti-RAD51 and anti-PCNA antibodies. The numbers of PLA signals were counted and represented as dot plots. DNA was counterstained with DAPI (blue). Bar: 10 μm. Red horizontal lines denote means with standard deviations. Statistical differences were determined by the Student's t-test; **P < 0.0001. (F) Fluorescence recovery after photobleaching (FRAP) analysis of GFP-RAD51 foci. Two days after the transfection of the control or FANCD2 siRNA, the mobility of GFP-RAD51 was analyzed by bleaching the RAD51 foci, with or without MMC treatment (100 ng/ml, for 24 h). The means of the relative fluorescence intensities with standard deviations (n = 20–22) and representative images are shown. The bleached area is indicated as an open square.
The interaction between RAD51 and FANCD2 in cells was then verified by an in situ PLA. In this assay, the PLA signals are observed as fluorescent foci, if RAD51 and FANCD2 are localized in close proximity in cells (<40 nm) (56). As shown in Figure 1D, detectable amounts of PLA signals were observed in the nuclei of untreated U2OS cells expressing FLAG-tagged FANCD2, while the number of PLA signals was very low in cells treated with either the anti-FLAG or anti-RAD51 antibody alone (Supplementary Figure S1C). In addition, a significant increase in the PLA signal number was observed after the MMC or HU treatment, which stalls replication fork progression (Figure 1D). Importantly, the MMC-induced PLA signals were barely detected with either the anti-FLAG or anti-RAD51 antibody alone, although they were robustly detected in the presence of both anti-FLAG and anti-RAD51 antibodies (Supplementary Figure S1D). We also found that the PLA signals with RAD51 and PCNA were significantly reduced in the FANCD2-knockdown cells after the HU treatment (Figure 1E), indicating that FANCD2 is required for the RAD51 accumulation at the stalled replication forks. Therefore, FANCD2 may directly bind to RAD51, probably on the stalled replication fork in cells.
To assess the stability of RAD51 at the stalled replication forks, we performed the FRAP experiments before and after the MMC treatment, in cells with and without FANCD2. The FRAP measurements were performed for each RAD51 focus. Before the MMC treatment, the RAD51 mobility remained unchanged in the FANCD2-knockdown cells (Figure 1F, left). However, the RAD51 mobility was clearly increased in the FANCD2-knockdown cells after the MMC treatment (Figure 1F, right). These results directly showed that FANCD2 stabilizes the RAD51 bound to stalled replication forks induced by ICLs.
The I-D complex binds to RAD51
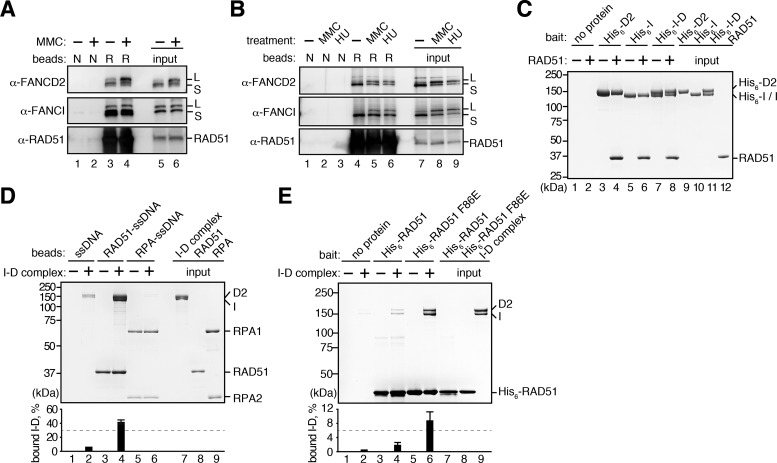
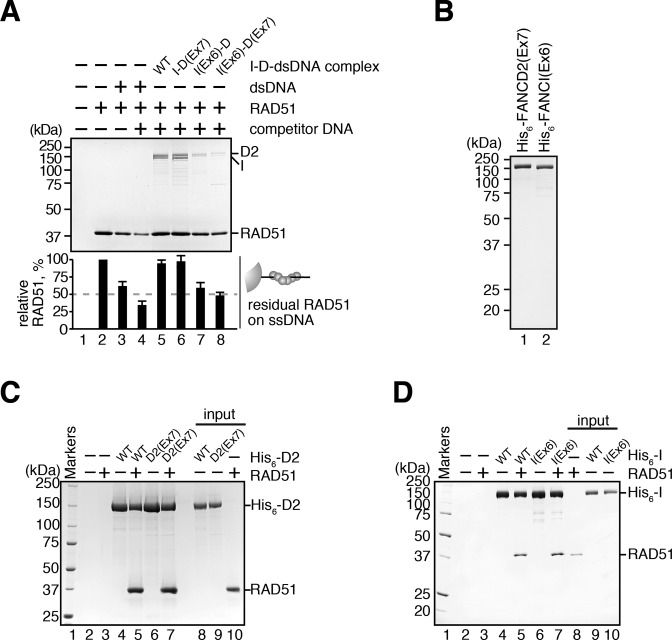
FANCD2 reportedly recruits CtIP to the stalled replication fork (47). CtIP is known to mediate the DNA end resection, which is prerequisite for subsequent RAD51 loading. This suggested the possibility that FANCD2 may indirectly recruit RAD51 through the CtIP function. To eliminate this possibility, we tested whether FANCD2 binds to RAD51. To do so, we prepared RAD51-conjugated beads, and then performed the pull-down assay with human cell extracts. Endogenous FANCD2 copelleted with RAD51 beads, together with FANCI, indicating that the endogenous I-D complex binds to RAD51 (Figure 2A). The MMC treatment stimulated the FANCD2 monoubiquitination and both the ubiquitinated and non-ubiquitinated forms of FANCI and FANCD2 efficiently bound to RAD51 (Figure 2A). This RAD51-FANCI-FANCD2 interaction was also detected in pull-down assays with chicken DT40 cell extracts (Figure 2B). These pull-down assays were performed with cell lysates treated with a nuclease, benzonase, thus suggesting that RAD51 binds to the DNA-free I-D complex. We then tested the direct interaction between RAD51 and the I-D complex, using purified chicken RAD51, FANCI and FANCD2 (Supplementary Figure S2A and B). We found that RAD51 was efficiently captured by the Ni-NTA agarose beads bound to His6-tagged FANCI, FANCD2 or the I-D complex (Figure 2C). These results indicated that both subunits of the I-D complex directly bind to RAD51. The I-D complex also bound to DMC1, a meiosis-specific RAD51 isoform, but its affinity was quite low, as compared to that of RAD51 (Supplementary Figure S2C and D).
Figure 2.
FANCD2 and FANCI interact with RAD51. (A) Pull-down assay with the HeLa cell extracts. The beads without RAD51 (N) or with RAD51 (R) were incubated with the nuclease-treated cell extracts, and the proteins bound to the beads were detected by Western blotting. The cell extracts were prepared from untreated and MMC-treated HeLa cells. (B) Pull-down assay with the DT40 cell extracts. The nuclease-treated cell extracts were prepared from untreated, MMC-treated and HU-treated DT40 cells. (C) Pull-down assay with Ni-NTA beads. RAD51 bound to His6-tagged FANCI, FANCD2 and the I-D complex was copelleted with the Ni-NTA beads, and the proteins were analyzed by SDS-PAGE. (D) Pull-down assay with single-stranded DNA (ssDNA) beads. The beads conjugated with ssDNA, the RAD51-ssDNA complex or the RPA-ssDNA complex were incubated with the I-D complex. The I-D complex that copelleted with the beads was analyzed by SDS-PAGE. The amounts of the I-D complex in the bound fractions were quantitated, and the mean percentages of three independent experiments are indicated with the standard deviations. (E) Pull-down assay with His6-tagged RAD51 proteins. The I-D complex was copelleted with His6-RAD51 or His6-RAD51 F86E bound to Ni-NTA beads, and the proteins were analyzed by SDS-PAGE with Coomassie Brilliant Blue staining. The amounts of the I-D complex in the bound fractions were quantitated, and the mean percentages of three independent experiments are indicated with the standard deviations.
RAD51 binds to ssDNA, and forms the RAD51-ssDNA filament in the presence of ATP (57–60). We then tested whether the I-D complex binds to the RAD51-ssDNA filament. We assembled the RAD51-ssDNA filament with poly dT ssDNA conjugated to magnetic beads (ssDNA beads) in the presence of a non-hydrolyzable ATP analog, AMP-PNP, which allows stable RAD51-filament formation on ssDNA. The purified I-D complex copelleted with the ssDNA beads, because the I-D complex itself binds to ssDNA (17) (Figure 2D, lane 2). Interestingly, however, a substantial amount of the I-D complex was captured with the RAD51-ssDNA filament, as compared to the experiments without RAD51 (Figure 2D, lane 4). In contrast, the I-D complex did not copellet with RPA-coated ssDNA, which is formed at stalled forks before RAD51 loading (36) (Figure 2D, lane 6, and Supplementary Figure S2E). These results indicated that the I-D complex specifically binds to the RAD51-ssDNA complex, but not the RPA-ssDNA complex, probably by its RAD51-binding activity.
Intriguingly, we found that the I-D complex bound more efficiently to the RAD51 F86E mutant, which is reportedly defective in the polymer formation (61), than the wild-type RAD51 (Figure 2E and Supplementary Figure S2F). In the RAD51 F86E mutant, the RAD51–RAD51 interface, which is buried by the polymer formation, is exposed to the solvent. This finding suggested that the I-D complex may bind to the buried RAD51 surface in the RAD51 polymer. Such RAD51 surface may be accessible at the end of the RAD51 polymer. Therefore, the I-D complex may bind to the end of the RAD51 filament, formed on the stalled replication fork (See Figure 7).
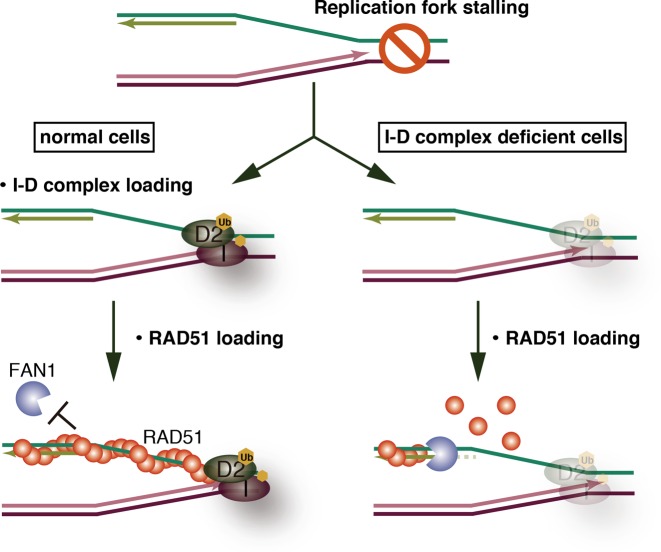
Figure 7.
Model for cooperative function of the ID complex and RAD51 in replication fork protection. The I-D complex preferentially binds to the branched DNA structure at the stalled replication fork. RAD51 is subsequently assembled on the ssDNA region, and forms the nucleoprotein filament covering the ssDNA-dsDNA junction. The I-D complex, which directly binds to RAD51, prevents RAD51 from dissociating from the end of the nucleoprotein filament. Consequently, the RAD51 nucleoprotein filament is stabilized, and the ssDNA-dsDNA junction may not be accessible to exonucleases. In the I-D complex-deficient cells, the spontaneous disassembly of RAD51 may allow an exonuclease to access the 5′-end of the newly synthesized lagging strand and induce undesired replication fork degradation, leading to chromosomal aberrations.
The I-D complex bound to DNA stabilizes the RAD51–DNA filament
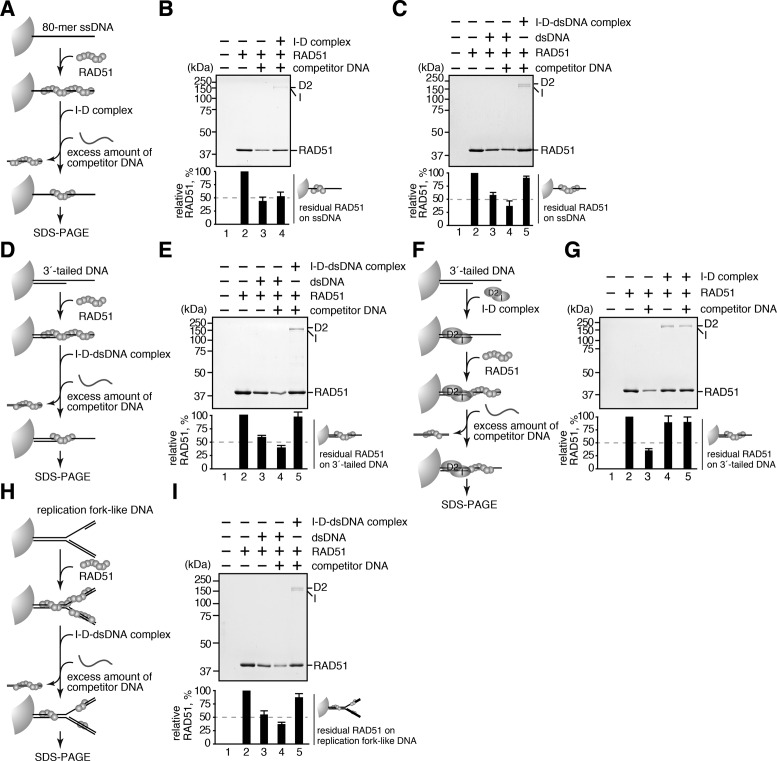
To evaluate the effect of the I-D complex binding to the RAD51-DNA complex, we performed the RAD51 transfer assay (Figure 3A). In this assay, the RAD51–ssDNA filament was assembled on the ssDNA beads in the presence of ATP. A competitor ssDNA (10-fold molar excess) was then added, which stripped the RAD51 from the RAD51–ssDNA beads. The residual RAD51 on the ssDNA beads was finally estimated by SDS-PAGE (Figure 3B). Under the conditions used here, about 60% of the RAD51 was disassembled from the RAD51–ssDNA beads in the presence of the competitor DNA (Figure 3B, lanes 2 and 3). The RAD51 transfer was not detected in the presence of AMP–PNP that stabilizes the RAD51 filament on the ssDNA (Supplementary Figures S3A and B). The I-D complex slightly increased the amount of RAD51 retained on the ssDNA beads (Figure 3B, lane 4). In this assay, the I-D complex bound to the RAD51-ssDNA filament was co-pelleted (Figure 3B, lane 4). This indicates that the I-D complex alone does not affect the stability of the RAD51–ssDNA complex. However, unexpectedly, the I-D complex bound to a 49-mer double-stranded DNA (I-D-dsDNA complex) significantly stabilized the RAD51–ssDNA complex (Figure 3C, lane 5), although the dsDNA alone used for the I-D-dsDNA complex stripped the RAD51 bound to the ssDNA beads (Figure 3C, lane 3). The I-D-dsDNA complex did not bind to the free ssDNA (Supplementary Figure S3C), eliminating the possibility that the I-D-dsDNA complex bound and sequestered the competitor ssDNA. Since the RAD51 filament is assembled on the ssDNA–dsDNA junction by BRCA2 (62), we hypothesized that the I-D complex may stabilize the RAD51 nucleoprotein filament formed on the ssDNA–dsDNA junction (Figure 3D). Consistent with this idea, the I-D-dsDNA complex robustly enhanced the stability of the RAD51 filament formed on the 3′-tailed DNA (Figure 3E). These results indicated that the I-D complex bound to dsDNA stabilizes the RAD51-DNA complex.
Figure 3.
The I-D complex stabilizes the RAD51-DNA nucleoprotein filament. (A) Schematic diagram of the RAD51 transfer assay with ssDNA beads. (B and C) RAD51 transfer assay with ssDNA beads in the presence of (B) the I-D complex or the (C) I-D-dsDNA complex. The RAD51 retained on the ssDNA beads was analyzed by SDS-PAGE and quantitated. The mean percentages of three independent experiments are indicated as bars with standard deviations. (D) Schematic diagram of the RAD51 transfer assay with 3′-tailed DNA beads. (E) RAD51 transfer assay with 3′-tailed DNA beads in the presence of the I-D-dsDNA complex. The RAD51 retained on the 3′-tailed DNA beads was analyzed, as in panel C. (F) Schematic diagram of the RAD51 transfer assay with the I-D complex-bound 3′-tailed DNA beads. (G) RAD51 transfer assay with the I-D complex-bound 3′-tailed DNA beads. The RAD51 retained on the 3′-tailed DNA beads was analyzed, as in panel C. (H) Schematic diagram of the RAD51 transfer assay with replication fork-like DNA beads. (I) RAD51 transfer assay with replication fork-like DNA beads in the presence of the I-D-dsDNA complex. The RAD51 retained on the replication fork-like DNA beads was analyzed, as in panel C.
We then tested whether the I-D complex pre-bound to the 3′-tailed DNA stabilizes the RAD51–ssDNA filament in cis. In this experiment, the I-D-3′-tailed DNA complex was formed in an I-D: 3′-tailed DNA = 1.4: 1 molar ratio (in molecules). We found that the I-D complex pre-assembled on the 3′-tailed DNA also stabilizes the RAD51–DNA filament (Figure 3F and G). This may explain how the I-D complex protects stalled replication forks from degradation, via the RAD51-filament stabilization (44).
Finally, we performed the RAD51 transfer assay with the replication fork-like DNA. As shown in Figure 3H and I, the I-D-dsDNA complex efficiently stabilized the RAD51 complex formed on the replication fork-like DNA.
The DNA binding activity of FANCI, but not FANCD2, in the I-D complex is required for the RAD51-DNA filament stabilization
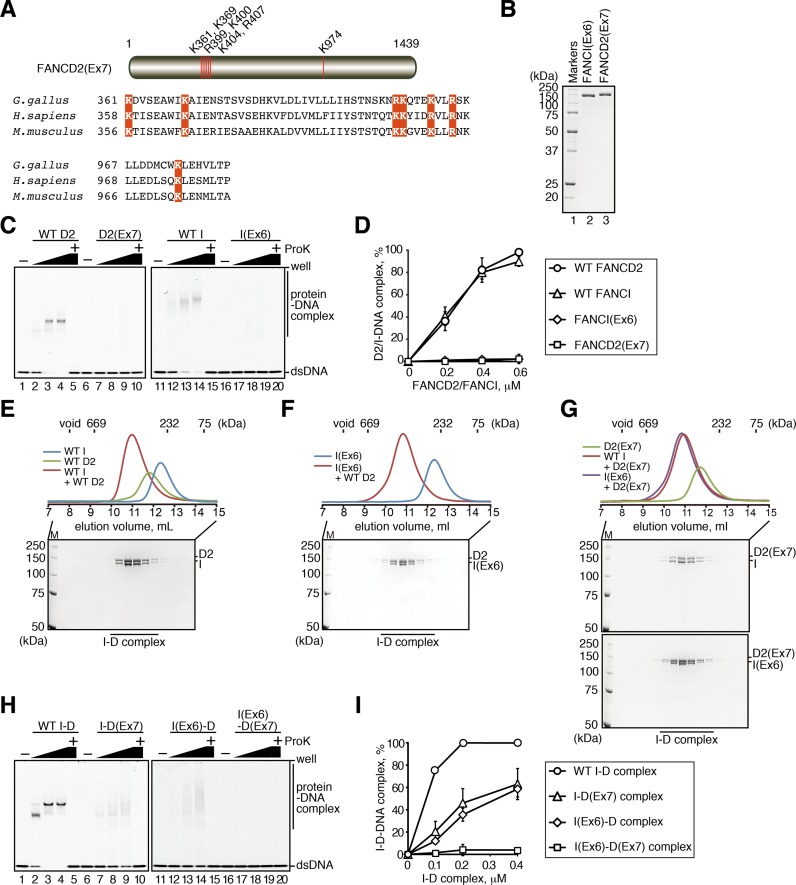
We next determined whether the DNA-binding activity of the I-D complex is essential for the RAD51–ssDNA stabilization. To do so, we purified the FANCI and FANCD2 mutants, FANCI(Ex6) and FANCD2(Ex7), which are defective in the DNA-binding activity (Figure 4B). The DNA-binding deficient FANCI(Ex6) mutant was established previously (18). We then designed the FANCD2(Ex7) mutant (Figure 4A). As shown in Figure 4C and D, both FANCI(Ex6) and FANCD2(Ex7) were completely defective in DNA binding. In contrast, FANCI(Ex6) and FANCD2(Ex7) retained the normal I-D complex formation activity (Figure 4E–G).
Figure 4.
Preparation of FANCI and FANCD2 mutants. (A) Schematic representation of the chicken FANCD2(Ex7) mutant. The basic amino acids Lys361, Lys369, Arg399, Lys400, Lys404, Arg407 and Lys974, located near the predicted DNA-binding surface of FANCD2 (64), were replaced by Glu. Alignment of the amino-acid sequences of the Homo sapiens, Mus musculus and Gallus gallus FANCD2 proteins, with the residues mutated in this study colored red. (B) The purified FANCI(Ex6) and FANCD2(Ex7) mutants. FANCI(Ex6) and FANCD2(Ex7) were analyzed by 12% SDS-PAGE with Coomassie Brilliant Blue staining. (C) DNA binding assay with FANCI(Ex6) and FANCD2(Ex7). The 49-mer dsDNA was incubated with increasing amounts (0, 0.2, 0.4 and 0.6 μM) of FANCD2, FANCD2(Ex7), FANCI or FANCI(Ex6) and the samples were analyzed by PAGE with SYBR Gold staining. Lanes 5, 10, 15 and 20 indicate control experiments, in which the samples were deproteinized before electrophoresis. (D) Graphic representation of the experiments shown in panel C. The intensity of the free DNA band was quantitated, and the amounts of DNA bound to the proteins were estimated. The mean percentages of three independent experiments are plotted against the protein concentration, with standard deviations. (E–G) Gel filtration analysis of the I-D complex formation. FANCI, FANCD2 and the mixture of FANCI and FANCD2 were fractionated on the Superdex 200 gel filtration column. The peak fractions (elution volume 7.5–15 ml) were analyzed by 8% SDS-PAGE with Coomassie Brilliant Blue staining. Experiments with (E) FANCI and FANCD2, (F) FANCI(Ex6) and FANCD2, (G, top) FANCI and FANCD2(Ex7) and (G, bottom) FANCI(Ex6) and FANCD2(Ex7). The void volume of the Superdex 200 column and the elution volumes of thyroglobulin (669 kDa), catalase (232 kDa) and conalbumin (75 kDa) are indicated on the gel filtration profiles. (H) DNA binding assay with the I-D complex mutants. Experiments were performed by the same method as in panel C. The concentrations of the I-D, I-D(Ex7), I(Ex6)-D2 and I(Ex6)-D2(Ex7) complexes were 0, 0.1, 0.2 and 0.4 μM. (I) Graphic representation of the experiments shown in panel H. The amounts of DNA bound to the proteins were analyzed, as in panel D.
We then tested the DNA binding activities of the I-D complexes, I(Ex6)-D, I-D(Ex7) and I(Ex6)-D(Ex7). We found that I(Ex6)-D and I-D(Ex7) still retained the DNA binding activity, although it was less than half of that of the ID complex (Figure 4H and I). As expected, I(Ex6)-D(Ex7) was completely defective in the DNA binding activity (Figure 4H and I).
Intriguingly, we found that the I(Ex6)-D complex incubated with dsDNA was significantly defective in the RAD51–ssDNA complex stabilization (Figure 5A, lane 7). Similarly, the I(Ex6)-D(Ex7) complex incubated with dsDNA was also defective in this activity (Figure 5A, lane 8). In sharp contrast, the I-D(Ex7) complex incubated with dsDNA was completely proficient in the stabilization of the RAD51–ssDNA complex (Figure 5A, lane 6). These results indicated that the DNA-binding of FANCI, but not FANCD2, is responsible for the I-D complex-mediated stabilization of the RAD51–ssDNA complex. In this assay, the RAD51–ssDNA beads reproducibly captured smaller amounts of the I-D complexes containing the FANCI(Ex6) mutant (Figure 5A, lanes 7 and 8), although equal amounts of the input I-D complexes were used. Neither FANCI(Ex6) nor FANCD2(Ex7) exhibited any obvious defect in the RAD51-binding activity (Figure 5B–D). The low amount of the I(Ex6)-D complex detected in this assay may be responsible for the low amount of RAD51 retained on the ssDNA beads.
Figure 5.
The DNA binding activity of FANCI is required for the I-D complex-mediated stabilization of the RAD51-DNA filament. (A) The RAD51 transfer assay with ssDNA beads in the presence of the I-D, I-D(Ex7), I(Ex6)-D or I(Ex6)-D(Ex7) complex. The RAD51 retained on the ssDNA beads was analyzed, as in Figure 3C. (B) The purified His6-tagged FANCI(Ex6) and FANCD2(Ex7) mutants. His6-FANCD2(Ex7) and His6-FANCI(Ex6) were analyzed by 12% SDS-PAGE with Coomassie Brilliant Blue staining. (C) Pull-down assay with His6-FANCD2(Ex7). RAD51 bound to His6-tagged FANCD2 or His6-tagged FANCD2(Ex7) was copelleted by the Ni-NTA beads, and the proteins were analyzed by 10% SDS-PAGE with Coomassie Brilliant Blue staining. (D) Pull-down assay with His6-FANCI(Ex6). Experiments were performed by the same method as in panel C, except with 12% SDS-PAGE with Coomassie Brilliant Blue staining.
The RAD51–DNA filament stabilized by the I-D complex protects the DNA end
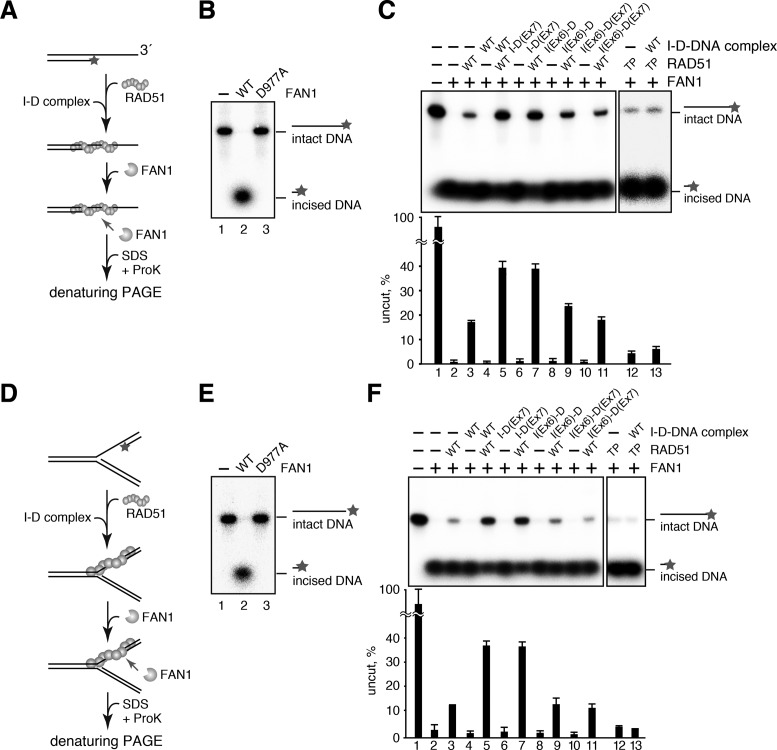
The I-D complex and RAD51 reportedly play essential roles for preventing improper nascent DNA resection at a stalled replication fork (44). The 5′-3′ exonucleases, such as FAN1 and DNA2, have been implicated in such destructive DNA degradation (33,40,41). We then assessed whether the RAD51-filament stabilization by the I-D-dsDNA complex functions to protect the 5′-DNA end at the ssDNA–dsDNA junction. To do so, we performed the nuclease protection assay (Figure 6A). In this assay, the RAD51 nucleoprotein filament was assembled on a 3′-tailed DNA, which contained an ssDNA–dsDNA junction, with ATP and then incubated with the FAN1 nuclease domain (Supplementary Figure S4A). The 5′-end of the shorter strand, located at the ssDNA–dsDNA junction, was labeled by 32P. Consistent with a previous report (63), the labeled 5′-end at the DNA junction was completely nibbled by FAN1, but not by the nuclease-deficient FAN1 D977A mutant (Figure 6B, lanes 2 and 3). The FAN1-mediated DNA nibbling was weakly protected in the presence of RAD51 (Figure 6C, lanes 2 and 3). However, the 5′-DNA end protection by RAD51 was remarkably stimulated by the I-D-dsDNA complex (Figure 6C, lane 5), although the I-D-dsDNA complex alone did not inhibit the FAN1 nuclease activity (Figure 6C, lane 4). The DNA-binding activity of FANCI in the I-D complex was strictly required for the DNA protection by RAD51, as revealed by the I-D complexes containing the DNA-binding deficient FANCI(Ex6), but not FANCD2(Ex7) (Figure 6C, lanes 7, 9 and 11). FANCR-patient cells containing the RAD51 T131P mutation reportedly exhibit excessive nucleolytic degradation of the stalled replication fork, leading to chromosomal aberrations (33). Consistently, we found that the RAD51 T131P mutant was defective in protecting the 5′-DNA end of the tailed DNA from FAN1, even in the presence of the I-D-dsDNA complex (Figure 6C, lanes 12 and 13, and Supplementary Figure S4B–D). We repeated the nuclease protection assay using a replication fork-like DNA substrate, and confirmed that the results are perfectly consistent with those from the experiments with the 3′-overhang DNA (Figure 6D–F).
Figure 6.
The I-D complex protects a DNA end from undesired nucleolytic processing by RAD51-DNA filament stabilization. (A) Schematic diagram of the nuclease protection assay with the 3′-tailed DNA. (B) The 3′-tailed DNA, labeled at the 5′-end of the short strand with 32P, was incubated with the FAN1 nuclease domain or the FAN1 D977A mutant, and the resulting DNA fragments were analyzed by denaturing PAGE. (C) Nuclease protection assay with the 3′-tailed DNA. RAD51 or RAD51 T131P (TP), which contained a mutation found in a FANCR patient, was assembled on the 3′-tailed DNA in the presence of the I-D, I-D(Ex7), I(Ex6)-D or I(Ex6)-D(Ex7) complex that was preincubated with dsDNA. After an incubation with FAN1, the resulting DNA fragments were analyzed by denaturing PAGE. Band intensities of undigested 32P-labeled strand DNA were quantitated, and mean percentages of three independent experiments are indicated as bars with standard deviations. (D) Schematic diagram of the nuclease protection assay with the replication fork-like DNA. (E) The replication fork-like DNA, with the 5′-end of the shortest strand labeled with 32P, was incubated with the FAN1 nuclease domain or the FAN1 D977A mutant, and the resulting DNA fragments were analyzed, as in panel B. (F) Nuclease protection assay with the replication fork-like DNA. Experiments were performed as in panel C. Stars denote 32P at a 5′-DNA end.
Collectively, the I-D complex-mediated stabilization of the RAD51 nucleoprotein filament may function to prevent destructive DNA degradation by exonucleases during replication and at stalled replication forks. Since the I-D complex preferentially binds to branched DNA (17), it may prevent RAD51 dissociation from the end of the RAD51–DNA filament at the stalled replication fork (Figure 7). In addition, the binding of the I-D complex to the end of the RAD51 filament was also suggested by the enhanced I-D complex binding to the RAD51 F86E mutant, which is defective in the RAD51 polymer formation (Figure 2E). The stable RAD51–DNA filament covering the ssDNA-dsDNA junction may substantially protect its 5′-end from undesired nucleolytic degradation by exonucleases.
Supplementary Material
Acknowledgments
The authors thank Dr Makoto Nakanishi and Dr Yoshikazu Johmura (University of Tokyo, Tokyo, Japan) for generously providing the puromycin-resistant derivative of CSIV-TRE-RfA-UbC-KT.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS KAKENHI [JP25116002 to H.K., JP25250023 to H.K., JP24310042 to M.T., JP23114010 to M.T. and JP26830128 to K.S., in part]; Waseda University Grant for Special Research Projects [2015B-319 to K.S.]; JSPS Research Fellowship for Young Scientists [to D.T.]; H.K. is a research fellow of the Waseda Research Institute of Science and Engineering, and is also supported by Waseda University. Funding for open access charge: Waseda University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Deans A.J., West S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langevin F., Crossan G.P., Rosado I.V., Arends M.J., Patel K.J. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 3.Pontel L.B., Rosado I.V., Burgos-Barragan G., Garaycoechea J.I., Yu R., Arends M.J., Chandrasekaran G., Broecker V., Wei W., Liu L, et al. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol. Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auerbach A.D. Fanconi anemia and its diagnosis. Mutat. Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H., D'Andrea A.D. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kottemann M.C., Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceccaldi R., Sarangi P., D'Andrea AD. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D'Andrea A.D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 9.Sims A.E., Spiteri E., Sims R.J., Arita A.G., Lach F.P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2007;14:564–597. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 10.Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D'Andrea A.D., et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longerich S., Li J., Xiong Y., Sung P., Kupfer GM. Stress and DNA repair biology of the Fanconi anemia pathway. Blood. 2014;124:2812–2819. doi: 10.1182/blood-2014-04-526293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meetei A.R., de Winter J.P., Medhurst A.L., Wallisch M., Waisfisz Q., van de Vrugt H.J., Oostra A.B., Yan Z., Ling C., Bishop C.E., et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 13.Alpi A.F., Pace P.E., Babu M.M., Patel K.J. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Hira A., Yoshida K., Sato K., Okuno Y., Shiraishi Y., Chiba K., Tanaka H., Miyano S., Shimamoto A., Tahara H., et al. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am. J. Hum. Genet. 2015;96:1001–1007. doi: 10.1016/j.ajhg.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rickman K.A., Lach F.P., Abhyankar A., Donovan F.X., Sanborn E.M., Kennedy J.A., Sougnez C, Gabriel S.B., Elemento O., Chandrasekharappa S.C., et al. Deficiency of UBE2T, the E2 ubiquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T subtype of Fanconi anemia. Cell Rep. 2015;12:35–41. doi: 10.1016/j.celrep.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Virts E.L., Jankowska A., Mackay C., Glaas M.F., Wiek C., Kelich S.L., Lottmann N., Kennedy F.M., Marchal C., Lehnert E., et al. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum. Mol. Genet. 2015;24:5093–5108. doi: 10.1093/hmg/ddv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan F., El Hokayem J., Zhou W., Zhang Y. FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J. Biol. Chem. 2009;284:24443–24452. doi: 10.1074/jbc.M109.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato K., Toda K., Ishiai M., Takata M., Kurumizaka H. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res. 2012;40:4553–4561. doi: 10.1093/nar/gks053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longerich S., Kwon Y., Tsai M.S., Hlaing A.S., Kupfer G.M., Sung P. Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA. Nucleic Acids Res. 2014;42:5657–5670. doi: 10.1093/nar/gku198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajendra E., Oestergaard V.H., Langevin F., Wang M., Dornan G.L., Patel K.J., Passmore L.A. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol. Cell. 2014;54:858–869. doi: 10.1016/j.molcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knipscheer P., Räschle M., Smogorzewska A., Enoiu M., Ho T.V., Schärer O.D., Elledge S.J., Walter J.C. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kratz K., Schöpf B., Kaden S., Sendoel A., Eberhard R., Lademann C., Cannavó E., Sartori A.A., Hengartner M.O., Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 23.Liu T., Ghosal G., Yuan J., Chen J., Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 24.MacKay C., Déclais A.C., Lundin C., Agostinho A., Deans A.J., MacArtney T.J., Hofmann K., Gartner A., West S.C., Helleday T., et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smogorzewska A., Desetty R., Saito T.T., Schlabach M., Lach F.P., Sowa M.E., Clark A.B., Kunkel T.A., Harper J.W., Colaiácovo M.P., et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto K.N., Kobayashi S., Tsuda M., Kurumizaka H., Takata M., Kono K., Jiricny J., Takeda S., Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein Douwel D., Boonen R.A., Long D.T., Szypowska A.A., Räschle M., Walter J.C., Knipscheer P. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 29.Sung P., Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 30.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 31.Sung P., Robberson D.L. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 32.Baumann P., Benson F.E., West S.C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 33.Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K., et al. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato M., Yano K., Matsuo F., Saito H., Katagiri T., Kurumizaka H., Yoshimoto M., Kasumi F., Akiyama F., Sakamoto G., et al. Identification of Rad51 alteration in patients with bilateral breast cancer. J. Hum. Genet. 2000;45:133–137. doi: 10.1007/s100380050199. [DOI] [PubMed] [Google Scholar]
- 35.Ishida T., Takizawa Y., Sakane I., Kurumizaka H. Altered DNA binding by the human Rad51-R150Q mutant found in breast cancer patients. Biol. Pharm. Bull. 2007;30:1374–1378. doi: 10.1248/bpb.30.1374. [DOI] [PubMed] [Google Scholar]
- 36.Long D.T., Räschle M., Joukov V., Walter J.C. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hashimoto Y., Chaudhuri A.R., Lopes M., Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaudhury I., Stroik D.R., Sobeck A. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol. Cell Biol. 2014;34:3939–3954. doi: 10.1128/MCB.00457-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higgs M.R., Reynolds J.J., Winczura A., Blackford A.N., Borel V., Miller E.S., Zlatanou A., Nieminuszczy J., Ryan E.L., Davies N.J., et al. BOD1L is required to suppress deleterious resection of stressed replication forks. Mol. Cell. 2015;59:462–477. doi: 10.1016/j.molcel.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 42.Taniguchi T., Garcia-Higuera I., Andreassen P.R., Gregory R.C., Grompe M., D'Andrea A.D. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 43.Hussain S., Wilson J.B., Medhurst A.L., Hejna J., Witt E., Ananth S., Davies A., Masson J.Y., Moses R., West S.C., et al. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum. Mol. Genet. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 44.Schlacher K., Wu H., Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lossaint G., Larroque M., Ribeyre C., Bec N., Larroque C., Décaillet C., Gari K., Constantinou A. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol. Cell. 2013;51:678–690. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 46.Kurita R., Suda N., Sudo K., Miharada K., Hiroyama T., Miyoshi H., Tani K., Nakamura Y. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8:e59890. doi: 10.1371/journal.pone.0059890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unno J., Itaya A., Taoka M., Sato K., Tomida J., Sakai W., Sugasawa K., Ishiai M., Ikura T., Isobe T., et al. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 2014;7:1039–1047. doi: 10.1016/j.celrep.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Kim J.S., Krasieva T.B., Kurumizaka H., Chen D.J., Taylor A.M., Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishida T., Takizawa Y., Sakane I., Kurumizaka H. The Lys313 residue of the human Rad51 protein negatively regulates the strand-exchange activity. Genes Cells. 2008;13:91–103. doi: 10.1111/j.1365-2443.2007.01143.x. [DOI] [PubMed] [Google Scholar]
- 50.Henricksen L.A., Umbricht C.B., Wold M.S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 51.Hikiba J., Hirota K., Kagawa W., Ikawa S., Kinebuchi T., Sakane I., Takizawa Y., Yokoyama S., Mandon-Pépin B., Nicolas A., et al. Structural and functional analyses of the DMC1-M200V polymorphism found in the human population. Nucleic Acids Res. 2008;36:4181–4190. doi: 10.1093/nar/gkn362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 53.Yokoyama H., Kurumizaka H., Ikawa S., Yokoyama S., Shibata T. Holliday junction binding activity of the human Rad51B protein. J. Biol. Chem. 2003;278:2767–2772. doi: 10.1074/jbc.M210899200. [DOI] [PubMed] [Google Scholar]
- 54.Sato K., Ishiai M., Takata M., Kurumizaka H. Defective FANCI binding by a Fanconi anemia-related FANCD2 mutant. PLoS One. 2014;9:e114752. doi: 10.1371/journal.pone.0114752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato K., Ishiai M., Toda K., Furukoshi S., Osakabe A., Tachiwana H., Takizawa Y., Kagawa W., Kitao H., Dohmae N., et al. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J. 2012;31:3524–3536. doi: 10.1038/emboj.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 57.Ogawa T., Yu X., Shinohara A., Egelman E.H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 58.Benson F.E., Stasiak A., West S.C. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sung P., Stratton S.A. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- 60.Chi P., Van Komen S., Sehorn M.G., Sigurdsson S., Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst). 2006;5:381–390. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 61.Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 62.Yang H., Li Q., Fan J., Holloman W.K., Pavletich N.P. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 63.Wang R., Persky N.S., Yoo B., Ouerfelli O., Smogorzewska A., Elledge S.J., Pavletich N.P. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science. 2014;346:1127–1130. doi: 10.1126/science.1258973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joo W., Xu G., Persky N.S., Smogorzewska A., Rudge D.G., Buzovetsky O., Elledge S.J., Pavletich N.P. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011;333:312–316. doi: 10.1126/science.1205805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.