Abstract
APRIN (PDS5 cohesin associated factor B) interacts with both the cohesin complex and the BRCA2 tumor suppressor. How APRIN influences cohesion and DNA repair processes is not well understood. Here, we show that APRIN is recruited to DNA damage sites. We find that APRIN interacts directly with RAD51, PALB2 and BRCA2. APRIN stimulates RAD51-mediated DNA strand invasion. APRIN also binds DNA with an affinity for D-loop structures and single-strand (ss) DNA. APRIN is a new homologous recombination (HR) mediator as it counteracts the RPA inhibitory effect on RAD51 loading to ssDNA. We show that APRIN strongly improves the annealing of complementary-strand DNA and that it can stimulate this process in synergy with BRCA2. Unlike cohesin constituents, its depletion has no impact on class switch recombination, supporting a specific role for this protein in HR. Furthermore, we show that low APRIN expression levels correlate with a better survival in ovarian cancer patients and that APRIN depletion sensitizes cells to the PARP inhibitor Olaparib in xenografted zebrafish. Our findings establish APRIN as an important and specific actor of HR, with cohesin-independent functions.
INTRODUCTION
Each day, genome integrity is threatened by a multitude of endogenous and exogenous agents than can induce DNA double-strand breaks (DSBs). To neutralize these potentially harmful lesions, cells have implemented diverse mechanisms. One of these, homologous recombination (HR), is an error-free repair pathway that takes place during the S and G2 phases of the cell cycle and allows cells to resolve the damage using the sister chromatid, as a preferred template. Unfortunately, failure to complete HR accurately can lead to cell death or cancer (1–4). Some of the central proteins in this process are RAD51, PALB2 (PArtner and Localizer of BRCA2) and BRCA2 (BReast CAncer 2, early onset). Human BRCA2, a 384 kDa protein, exhibits important Breast Cancer (BRC) repeats controlling the function and localization of RAD51 and other components. Mainly, BRCA2 mediator function is needed to bind single-strand (ss) DNA, to interact directly with RAD51 and to stimulate the recruitment of RAD51 by removing Replication Protein A (RPA) from resected DNA (5,6). RAD51 then searches for homologous sequences (6), through the invasion of the resected DSB ends into the intact sister chromatid (7). This process, called D-loop formation, allows one of the two resected DNA ends to be extended using its homologous sequence. PALB2 (Partner and Localizer of BRCA2) protein, another key HR regulator, can also remove the inhibitory effect of RPA and promote RAD51 activity (8–11).
The cohesin complex, a ring-like structure, is composed of cohesin proteins (SMC1, SMC3, SCC1 and SCC3) and its regulators (PDS5A/B, SA1/2, NIPBL, WAPL, sororin and SCC4) (12). This complex is required for the cohesion between sister chromatids after DNA replication. The function of cohesin in DNA repair is mainly linked to its ability to hold sister chromatids together (13). Nevertheless, cohesin was first discovered as implicated in DNA repair (14). Moreover, the cohesin complex has a dual function following DNA damage: a direct implication in the repair per se and a role in the recruitment of DNA damage checkpoint proteins (15). Cohesin has recently been shown to repress the C-NHEJ and Alt-NHEJ of distant end-breaks in S/G2 phases (16). During G1 phase, cohesin is implicated in the class switch recombination (CSR) by modulating non-homologous end-joining (NHEJ) efficiency (17). Cohesin is also recruited to laser-induced DNA damage (18). Two cohesin complexes differing by one subunit (SA1 or SA2) are present in somatic cells. Cohesin-SA2 is mainly recruited to DNA damage whereas Cohesin-SA1 is implicated in the intra-S checkpoint (19). These observations highlight the versatility of the cohesin complex.
Genetic screens in yeast and fungi have identified several classes of cohesion factors, among them is PDS5 for Precocious Dissociation of Sisters (20). Two PDS5 proteins, PDS5A and PDS5B, are expressed in human cells and interact with cohesin (21). PDS5B (alternatively named APRIN) belongs to the PDS5 family characterized by the presence of HEAT (Huntingtin, Elongation Factor 3 (EF3), Protein Phosphatase 2A (PP2A), and the yeast kinase TOR1) repeats, known to be involved in protein-protein interactions. (22). Cells depleted of PDS5B show defects in metaphasic chromosome morphology (21).
Interestingly, novel roles for APRIN in the DNA damage response were reported. It has been shown that APRIN and other cohesin components associate with BRCA2 in early S-phase. Depletion of APRIN compromises the nuclear localization of RAD51 and BRCA2, disturbs efficient HR and sensitizes cells to DNA damaging agents (23). Moreover, APRIN expression levels are associated with histological grade of breast cancer and the outcome of breast cancer patients treated with DNA-damaging chemotherapy (23). The PDS5–BRCA2 complex has also been found to contribute to meiotic recombination at the nuclear envelope by associating with cohesins and lamins (24). Additionally, PDS5 functions can be regulated by post-translational modifications such as sumoylation, acetylation or phosphorylation (25–27). While APRIN has been clearly linked to both cohesion and HR, its precise role in these processes requires better characterization. The roles of PDS5 proteins in cohesion are highly debated, as different teams propose that PDS5 could have positive and negative involvement in the cohesin complex. The role of APRIN outside of its cohesion regulator activity is also poorly understood. While the conceptual implication of APRIN in HR has been characterized, its precise functions in this mechanism still remain elusive.
Here, we provide new and precise mechanistic insights into the role of APRIN in different steps of HR. We have purified APRIN and analysed its DNA-binding properties. We reveal that APRIN has affinity for D-loop structures and ssDNA. APRIN physically interacts with RAD51, PALB2 and BRCA2. APRIN enhances RAD51 DNA strand-invasion activity and catalyzes the annealing of complementary-strand DNA also in synergy with BRCA2. In contrast, we show that APRIN is not involved in NHEJ repair or CSR. Finally, we find that low APRIN expression levels correlate with a better survival in ovarian cancer patients and that APRIN depletion sensitizes cells to Poly(ADP-Ribose) Polymerase (PARP) inhibitor Olaparib in vivo in zebrafish xenografts. Our findings establish APRIN as an important regulator of HR with functions outside the cohesin complex.
MATERIALS AND METHODS
Cell lines and survival assays
SKOV3 were provided by Anne-Marie Mes-Masson. HEK293T were purchased from ATCC and authenticated. Lentiviruses carrying shRNA constructs (Sigma) were used in order to create cell lines stably expressing shRNA scramble or targeting APRIN or PALB2 mRNA (Pauty, Couturier et al., in preparation). Cell survival was measured using the 3-(4, 5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) Cell Proliferation Assay kit of ATCC bio products with AZD2281 (Olaparib, BPS Bioscience, Inc. (27003)) or ABT-888 (Veliparib, Enzo Life Sciences (ALX-270-444)).
Constructs
FLAG/HIS tagged APRIN was cloned by polymerase chain reaction (PCR) amplification of pCMV SPORT plasmid APRIN-myc-his (gift from Peter Geck) into pFastBAC1-52b-GST using the primers cited in Table 1. pEGFP-APRIN was obtained after the insertion of EcoR1/BamH1 PCR products into pEGFP-C1 (Clontech). APRIN deletion constructs were obtained by mutagenesis using the Q5® Site-Directed Mutagenesis Kit (NEB) and primers cited in Table 1. APRIN fragments were cloned by PCR amplification of pCMV SPORT plasmid APRIN-myc-his (gift from Peter Geck) into pEGFP-C1 using primers cited in Table 1. mCherry-PCNA-19-NLS-4 was purchased from Addgene.
Table 1. Primers used for DNA constructs and their sequences.
| Protein | Primers |
|---|---|
| HIS/FLAG APRIN | JYM 2267, JYM 2268 |
| Oligo names | Sequence |
| JYM2267 | ATTCCATGGATTACAAGGATGACGACGATAAGGCTCATTCAAAGACTAGG |
| JYM2268 | ATTGCGGCCGCTCGCCGTTCCCTTTTAGCACTT |
| Protein | Primers |
| GFP APRIN | JYM 2367B, JYM 2368B |
| Oligo names | Sequence |
| JYM2367B | ATTGAATTCTATGGCTCATTCAAAGACTAGG |
| JYM2368B | ATTGGATCCTCATCGCCGTTCCCTTTTAGC |
| Protein | Primers |
| APRINΔA2 | JYM 2816, JYM 2817 |
| APRINΔA2 | JYM 2820, JYM 2821 |
| APRINΔA5 | JYM 2822, JYM 2823 |
| For these four constructs another PCR step has been done to put back frameshift | |
| APRINΔA5 + NLS | JYM 2824, JYM 2825 |
| Oligo names | Sequence |
| JYM 2816 | ACCAATTTTTTTAATCAGGTTCTGATGCTTGGGAAAACATGCTCCTGCAAGCAGGCTGA |
| JYM 2817 | TCAGCCTGCTTGCAGGAGCATGTTTTCCCAAGCATCAGAACCTGATTAAAAAAATTGGT |
| JYM 2820 | CTAACAACAATATTGCATAGTGATGGAGACATGGAAACTGTAAGCAATGCAAGCAGCAG |
| JYM 2821 | CTGCTGCTTGCATTGCTTACAGTTTCCATGTCTCCATCACTATGCAATATTGTTGTTAG |
| JYM 2822 | ATTGAATTCATGGCTCATTCAAAGACTAGG |
| JYM 2823 | ATTGGATCCTCGTGATGATTTGGTCTGAGA |
| JYM 2824 | TCGAGCTAAGCGCCCCGCCGCCACCAAGAAGGCCGGCCAGGCCAAGAAGAAGAAGCA |
| JYM 2825 | AAGCTGCTTCTTCTTCTTGGCCTGGCCGGCCTTCTTGGTGGCGGCGGGGCGCTTAG |
| JYM 2825 | AAGCTTGCTTCTTCTTCTTGGCCTGGCCGGCCTTCTTGGTGGCGGCGGGGCGCTTAG |
| Fragments | Primers |
| A1 | JYM 2308, JYM 2309 |
| A2 | JYM 2310, JYM 2311 |
| A3 | JYM 2312, JYM 2313 |
| A4 | JYM 2314, JYM 2320 |
| A5 | JYM 2321, JYM 2322 |
| Oligo names | Sequence |
| JYM2308 | ATTGGATCCATGGCTCATTCAAAGACTAGG |
| JYM2309 | ATTGCGGCCGCTTATTTCCCAAGCATCAGAAC |
| JYM2310 | ATTGGATCCATGACATCTATCAGCGATTTG |
| JYM2311 | ATTGCGGCCGCTTATGGACTAACAAGTACTTC |
| JYM2312 | ATTGGATCCATGACATGCTCCTGCAAGCAG |
| JYM2313 | ATTGCGGCCGCTTAGTCTCCATCACTATGCAA |
| JYM2314 | ATTGGATCCATGTTGACAGAACAGGGGAAA |
| JYM2320 | AGCTGCTAGCTTTCTTAATAGAATG |
| JYM2321 | ATTGGATCCATGGAAACTGTAAGCAATGCA |
| JYM2322 | ATTGCGGCCGCTTATCATCGCCGTTCCCTTTT |
Generation of SKOV3 GFP-APRIN cell line
SKOV3 cells were transfected with pEGFPC1-APRIN using X-tremeGENE 9 (Sigma). Clones were selected under a G418 (450 μg/ml) treatment.
APRIN HIS-FLAG purification
After infection of Sf9 cells with the indicated baculovirus, spinner packaging was centrifuged and pellets were stored at −80°C. Pellets were then lysed with Lysis buffer (50 mM Tris HCl pH 7.5, 150 mM, 10% glycerol, 0.25% Triton X100, 2 mM ethylenediaminetetraacetic acid (EDTA), 1 mM Dithiothreitol (DTT) and Roche protease inhibitors) and dounced 10 times prior to three rounds of 30-s sonication at 4°C. After another douncing step, the mix was centrifuged 30 min (SS-34 fixed angle rotor at 50 000 g). Supernatants were incubated with M2 flag beads (Affinity gel Sigma) for 3 h at 4°C. After two washing steps with 250 mM NaCl lysis buffer, the mix was incubated in a Mg/ATP buffer (15 mM MgCl2, 5 mM adenosine triphosphate (ATP)) for 30 min at 4°C. Flag peptides (3×) in lysis buffer (500 ng/μl) without EDTA/DTT were used for competitive elutions. Elutions and washes were incubated with TALON beads (Clontech) and P5 buffer (50 mM Na2HPO4, 50 mM NaH2PO4, 500 mM NaCl, 10% glycerol, 0.05% Triton X-100, 5 mM Imidazole) for 1 h at 4°C. After two washes with P5 buffer, the bound proteins were eluted with P500 buffer (50 mM Na2HPO4, 50 mM NaH2PO4, 500 mM NaCl, 10% glycerol, 0.05% Triton X-100, 500 mM Imidazole) for 5 min at 4°C. Sample were dialyzed in a storage buffer (20 mM Tris acetate pH 8.0, 200 mM potassium acetate, 1 mM EDTA, 10% glycerol, 1 mM DTT) and stored at −80°C.
Purification of APRIN glutathione S-transferase fragments
Recombinant APRIN GST fusions (A1 through A5) were purified from BL21 RP (Stratagene), grown at 37°C in LB medium supplemented with 100 μg/ml ampicillin and 25 μg/ml chloramphenicol. At A600 = 0.4, 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the culture and incubated at 28°C for 4 h. The cells were harvested by centrifugation, frozen on dry ice and stored at −80°C. Pellets were lysed with 10 ml of Lysis buffer (phosphate buffered saline (PBS), 1% triton X-100 and protease inhibitors). Cell lysates were sonicated at 4°C and centrifuged 70 min (SS-34 fixed angle rotor at 50 000 g). Supernatants were incubated with 1 ml of Glutathione Sepharose beads 4B (GE Healthcare), 2 h at 4°C. GST was then removed using PreScission Protease (GE Healthcare).
Pull-down assays
GST-pulldown assays using GST alone, or GST-RAD51 (R51T1-R51T4) and APRIN were performed in 500 μl of R51/B2 buffer (20 mM KH2PO4 monobasic pH 7.4, 20 mM KH2PO4 dibasic pH 7.4, 0.5 mM EDTA, 10% glycerol, 0.5% NP40, 1 mM DTT, 200 mM KCl, 1 mg/ml bovine serum albumin (BSA) and protease inhibitors). GST, GST-RAD51 (R51T1-R51T4) were preincubated with dry sepharose beads and R51/B2 buffer for 30 min at room temperature. Then, APRIN was added to the mix and incubated for 1 h at 4°C. Complexes were washed three times with R51/B2 buffer without BSA. Proteins were visualized by Western blotting using the indicated antibodies.
Immunoprecipitation analysis
Purified APRIN and RAD51 were incubated for 15 min at 37°C in IP150 buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% NP40 and protease inhibitors). Flag antibodies were then added to the mix and incubated for 20 min at 4°C. Dry washed Protein A/G Ultralink resin (Thermo Scientific) was added to the mix and incubated 20 min at 4°C. Complexes were washed four times with IP150 buffer and proteins were visualized by Western blotting with the indicated antibodies. Purified APRIN and PALB2 or BRCA2 immunoprecipitations were processed as described above with the appropriate antibodies (Table 2).
Table 2. Antibodies.
| Name | Species | Cat# | Company | WB | IP | IF | Other |
|---|---|---|---|---|---|---|---|
| PDS5B | Rabbit | A300-538A | Bethyl | 1:2500 | 1:5000 | 1:50 | |
| RAD51 | Rabbit | sc-8349 | Santa Cruz | 1:5000 | 1:400 | ||
| PALB2 | Rabbit | A301-246A | Bethyl | 1:2500 | 1:5000 | ||
| HIS | Mouse | 631 212 | Clontech | 1:2500 | |||
| FLAG | Mouse | F3165 | Sigma | 1:5000 | 1:5000 | ||
| pSMC1 pS306 | Rabbit | 4029 | Cell Signaling Technology | 1:100 | |||
| BRCA2 | Mouse | Ab-1 | Calbiochem OP95 | 1:1000 | |||
| DNA-PKCs Ser2056 | Rabbit | ab18192 | ABCAM | 1:500 | |||
| H2AX pS139 | Mouse | 05-636 | Millipore | 1:6000 |
DNA-binding assays
In competition assays, APRIN was added at the indicated concentration to a mix containing 25 nM of 32P-labeled DNA structures (ssDNA, double-strand (ds) DNA, (SA) splayed arms, Holliday junctions (HJ) and D-loop structures) and specific binding buffer (1× APRIN binding buffer (25 mM MOPS, 0,2% tween, 40 mM KCl) and 1 mM MgCl2). The mix was incubated at 37°C for 10 min. Then, complexes were fixed with 0.5% glutaraldehyde during 15 min and reactions were loaded onto a 0.8% Tris/Borate/EDTA (TBE) 1× agarose gel, run at 65 V, dried onto DE81 filter paper and Whatman paper and then visualized by autoradiography. Quantifications were made on a phosphorimager system (Fuji). In DNA binding assays, the same reactions were performed using 50 nM of the indicated oligonucleotide constructs. Sequences of each oligonucleotide are found in Table 3.
Table 3. Primers used for DNA probes and their sequences.
| Structures | Primers |
|---|---|
| ss DNA | JYM 925 |
| ds DNA | JYM 925, JYM 945 |
| SA | JYM 925, JYM 926 |
| D-loop | JYM 1745, JYM 1395, JYM 1396 |
| HJ | JYM,925 JYM 926, JYM 927, JYM 928 |
| Oligo names | Sequence |
| JYM925 | GGGTGAACCTGCAGGTGGGCAAAGATGTCCTAGCAATGTAATCGTCAAGCTTTATGCCGT |
| JYM926 | ACGCTGCCGAATTCTACCAGTGCCAGCGACGGACATCTTTGCCCACCTGCAGGTTCACCC |
| JYM927 | ACGGCATAAAGCTTGACGATTACATTGCTACATGGAGCTGTCTAGAGGATCCGACTATCG |
| JYM928 | CGATAGTCGGATCCTCTAGACAGCTCCATGGTCGCTGGCACTGGTAGAATTCGGCAGCGT |
| JYM945 | ACGGCATAAAGCTTGACGATTACATTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCC |
| JYM1395 | GCCAGGGACGGGGTGAACCTGCAGGTGGGCGGCTGCTCATCGTAGGTTAGTATCGACCTATTGGTAGAATTCGGCAGCGTCATGCGACGGC |
| JYM1396 | GCCGTCGCATGACGCTGCCGAATTCTACCACGCTACTAGGGTGCCTTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCCCGTCCCTGGC |
| JYM 3452 | GGACTCCAGGTCCAGGACCGCTTTTTGCGCGCGCTTTTTGCGGGAGGTCCAGCTGTCCACCTCC |
| JYM 3453 | GGAGGTGGACAGCTGGACCTCCCGCAAAAAGCGCGCGCAAAAAGCGGTCCTGGACCTGGAGTCC |
Complementary single-strand annealing reactions
Reactions (10 μl) contained denatured 32P-labeled dsDNA (50 nM) with APRIN and/or BRCA2 at the indicated concentrations, in reaction buffer (25 mM MOPS pH 7, 60 mM KCl, 0,2% Tween, 1 mM DTT, 1 mM CaCl2). Incubation was performed at 37°C for 15 min. The reaction products were deproteinized by the addition of one-fifth volume of PK buffer (20 mM Tris–Cl pH 7.5 and 2 mg/ml proteinase K) followed by a 30 min incubation period at 37°C. Labeled DNA products were analyzed by electrophoresis (8% polyacrylamide gel electrophoresis gel with TBE1× buffer, run at 150 V for 3 h). The gel was dried and visualized by autoradiography. Quantifications were made on a phosphorimager system.
FAM/BHQ experiments
Reactions were carried out in a 96-well LUMITRAC 200 white in a 100 μl final volume. Fluorescence was monitored with a Cytation 5 Cell Imaging Multi-Mode Reader (BioTek). The wavelengths were chosen according to IDT i.e. 494 nM for excitation and 520 nM for emission.
Reactions with or without APRIN in a reaction buffer (25 mM MOPS pH 7, 20 mM KCl, 0,2% Tween, 2 mM DTT, 1 mM CaCl2) were incubated during 2 min at 25°C during which the basal fluorescence was measured every 6 s. After this timepoint, 20 μl of FAM-oligonucleotide was injected to the reaction and fluorescence was measured during 15 min every 6 s. Then, the BHQ-oligonucleotide was added to the reaction and fluorescence was measured during 20 min every 6 s. Oligonucleotide sequences are found in Table 4.
Table 4. FAM/BHQ oligonucleotide sequences obtained from IDT.
| Sequence | |
|---|---|
| FAM | 5′- 56-FAM/ATAGTTATGGTGAGGACCCCTTTGTTTC-3′ |
| BHQ | 5′- GAAACAAAGGGGTCCTCACCATAACTAT/3BHQ_1/-3′ |
Biotin pull-down assay
Magnetic beads (Roche) bound to a 5′-biotinylated ssDNA polydT 83-mer (corresponding to the final concentration of 1 mM nucleotide) were pre-incubated with a blocking buffer (25 mM Tris–acetate pH 7.5, 40 mM KOAc, 1 mM DTT, 0.02% Igepal and 10 mg/ml BSA) for 20 min at 37°C. The beads were captured with the magnetic particle separator, washed twice with reaction buffer (25 mM Tris–Acetate pH 7.5, 40 mM KOAc, 1 mM DTT, 0.02% Igepal, 1 mg/ml BSA, 5 mM ATP and 0.5 mM MgCl2) and resuspended in the same buffer. hRPA was added and the reaction was incubated for 5 min at 37°C. Then RAD51 was added and the reaction was incubated for 5 min at 37°C. Next, APRIN was added to the reaction for a final volume of 20 μl for 30 min at 37°C. The beads were captured and were washed twice with 500 μl of washing buffer (25 mM Tris–Acetate pH 7.5, 40 mM KOAc, 1 mM DTT, 0.02% Igepal, 5 mM ATP and 0.5 mM MgCl2). Finally, 15 μl of Laemmli buffer was added and beads were heated 5 min at 95°C. The beads were captured again and the supernatants were loaded onto a 8% sodium dodecyl sulphate-polyacrylamide gel electrophoresis Ruby (Invitrogen) and visualized by ultraviolet (UV).
NHEJ in vitro reporter assays
As previously described (28), a linearized, 32P-end labeled pBluescript was incubated for the indicated times with a nuclear extract derived from HeLa cells treated with a scrambled siRNA or an siRNA directed against APRIN (siRNA sequences are found in Table 5).
Table 5. siRNA and shRNA sequences.
| Sequence | |
|---|---|
| shAPRIN | CCGGCCAGAGTATGTTGTTCCATATCTCGAGATATGGAACAACATACTCTGGTTTTT |
| siAPRIN | GAACAAUCAAUAGAUGGA |
| si53BP1 | ON-TARGETplus Human TP53BP1 siRNA Dharmacon L-003548 |
Cell fractionation
3 × 106 cells were washed with PBS and resuspended in 200 μl of solution A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 10 mM NaF, 1 mM Na3VO4 and protease inhibitors). Triton X-100 was added to a final concentration of 0.1%, and the cells were left on ice for 5 min. Cytoplasmic proteins (S1) were separated from nuclei (P2) by low-speed centrifugation (1400 g for 4 min).
Isolated nuclei were washed three times with solution A and lysed in 200 μl of solution B (3 mM EDTA, 0.2 mM Ethylene glycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetic acid (EGTA), 1 mM DTT and protease inhibitors). After a 30 min incubation at 4°C, soluble nuclear proteins (S2) were separated from chromatin (P2) by centrifugation (1700 g for 4 min). Isolated chromatin was washed three times with solution B and separated by centrifugation (1700 g for 4 min). Finally, chromatin was resuspended in 200 μl of Laemmli buffer and sheared by sonication. Proteins were visualized by Western blotting using the indicated antibodies.
Immunofluorescence
For DNA-PKcs foci, U2OS cells were treated with neocarzinostatin (Sigma - #9014-02-2) 100 ng/ml during 1 h. Cells were washed two times with PBS and fixed with 4% PFA and cold methanol. Cells were then permeabilized with Triton-X (Sigma) and quenched with Sodium borohydride. After blocking in a BSA/Goat Serum mix, cells were incubated with DNA-PKcs Ser2056 antiboody for 2 h. After washing, cells were incubated with secondary antibody for 1 h at room temperature. After three more washes, cells were mounted in with the ProLong® Gold Antifade Mountant with DAPI (Life Technologies Inc., #P-36931).
Laser-induced DNA damage and time-lapse microscopy analysis
SKOV3 cells stably expressing GFP-APRIN and HeLa cells transiently expressing GFP-APRIN or deletion mutants were grown in 35-mm fluorodishes (World Precision Instruments, Inc.) and micro-irradiated along a track in cell nuclei using a 405 nm UV-laser coupled into a Leica TCS SP5 II confocal microscope driven by Leica LAS AF software. The settings were as follows: laser power output 100%, format 512 × 512 pixels, scan speed 100 Hz, scan iterations 5, mode bidirectional, zoom 2×. In the case of HeLa cells, the GFP-APRIN constructs (1 μg) were transfected overnight with Effectene transfection reagent (Qiagen) prior to microirradition. Cells were then fixed at the indicated time points and processed for immunofluorescence according to Cell Signaling Technology general protocol with antibodies to SMC1 pS360, RAD51 and H2AX pS139. Secondary antibodies Alexa Fluor 568 (1:1000, Invitrogen, #A-11004) was used to detect γ-H2AX and Alexa 647 (1:1000, Invitrogen, #A-21244) for p-SMC1 and RAD51 staining. Coverslips were mounted onto slides with ProLong® Gold Antifade Mountant with DAPI (PLife Technologies Inc., #P-36931). Images were acquired at a magnification of 63× using a Leica CTR 6000 microscope followed by background subtraction and deconvolution with the Volocity software v 6.0 (PerkinElmer).
For time-lapse microscopy, 250 000 cells from the indicated HeLa cell lines were seeded onto 35-mm fluorodishes and transfected 20 h later with 1 μg mCherry-PCNA and GFP-APRIN or YFP-PALB2-WT with Effectene transfection reagent (Qiagen). The next day, cells were micro-irradiated with the point-bleach mode using the confocal system and settings described above. The recruitment of GFP-APRIN or YFP-PALB2 to laser-induced DNA damage sites was then monitored over time by imaging cells every 2 min for 1 h. The fluorescence intensity of the indicated DNA constructs at DNA damage sites relative to an unirradiated area was quantified and plotted over time. Data show the mean relative fluorescence intensity ± S.E. of ∼50 cells per condition from at least three independent experiments.
Generation of CRISPR/Cas APRIN deletion clones in CH12F3-2 cells
Guide RNA sequences were designed using CRISPR Design Tool (http://tools.genome-engineering.org). Two strategies with two gRNA designed to produce a genomic deletion were used, either targeting exon 3 (for a 54 bp deletion) or exon 4 and exon 6, producing a 964 bp DNA deletion (See Table 6 for sequences). Each gRNA sequence and their complement were ligated in pX458 (pSpCas9(BB)-2A-GFP, Addgene #48138). Cells were cotransfected with the two guides in pX458 by electroporation using nucleofector kit V (Amaxa) following the manufacturer's protocol. Single GFP positive cells were sorted in 96-well plates 24 h post-transfection. DNA from the resulting clones was extracted with DirectPCR (cell) (Viagen) + Proteinase K (Biobasic) following the manufacturer's protocol. APRIN knock-out clones were identified by PCR using primers listed in Table 7. Knock-out clones were confirmed by western blot using rabbit anti-PDS5B antibody.
Table 6. Guide RNAs used for CRISPR-Cas deletion of APRIN exons.
| Sequence | |
|---|---|
| Exon 3 gRNA | 5′- AGTTTATCGGGAGACGTGTA-3′ |
| Exon 3 gRNA | 5′- TGATAAAGATGTTCGTTTAC-3′ |
| Exon 4 gRNA | 5′-GTTTATAACAAGGCAACTAA-3′ |
| Exon 6 gRNA | 5′-TGAAGGTGATACCGTATCTC-3′ |
Table 7. PCR primers to validate CRISPR-Cas deletions.
| Sequence | |
|---|---|
| Exon 3 deletion forward | 5′-GACCAGGACTCTGAAGAGGAAAA-3′ |
| Exon 3 deletion reverse | 5′-GTCCAGGCAATACAACAGAA-3′ |
| Exon 4 to 6 deletion forward | 5′-TCTTCCATGTTTACAAAGTTGGTG-3′ |
| Exon 4 to 6 deletion reverse | 5′-CATTTCACATACCTTATGGGCAGG-3′ |
Monitoring class switch recombination
IgM to IgA switching was assayed in CH12F3-2 cells activated for 3 days with 1 ng/ml TGF-β1, 10 ng/ml IL-4 and 1 μg/ml agonist anti-CD40 (BD). IgA expression was measured by flow cytometry using anti-mouse IgA-PE antibody in which stimulation with anti-CD40, TGF-β1 and IL-4 induces AID and isotype switching to IgA.
Annexin staining
0.5 × 106 cells were resuspended in 100 μl Annexin V binding buffer (BD Biosciences) and stained with 3ul APC-Annexin V (BD Biosciences) for 15 min at room temperature. A total of 400 μl Annexin V binding buffer was then added with 2 μg/ml Propidium Iodide (Bioshop). Annexin V positive cells were acquired on a FACSCalibur (BD Biosciences).
Zebrafish husbandry, embryo collection and embryo staging
Use of zebrafish in this study was approved by the Dalhousie University Animal Care Committee (protocol no. 11–129). Zebrafish were maintained according to standard protocols (29). Embryos were collected and grown in E3 embryo media (5 mM NaCl, 0.17 mM KCl, 0.4 mM CaCl2 and 0.16 mM MgSO4, pH 7.5, supplemented with 1 × 10-5% Methylene Blue [v/v]) at 28.5°C. The casper (roy orbison-/-; nacre-/) double mutant zebrafish line was employed to avoid any autofluorescence that might impede direct visualization of fluorescent leukemia cells (30). Embryos were developmentally staged according to standard protocol (29).
Xenotransplantation of APRIN depleted cells into Zebrafish embryos, dissociation and immunofluorescence
Using a protocol adapted from Haldi et al. (31), mutant casper zebrafish embryos at 48 h post fertilization (hpf) were anaesthetized with 0.090 mg/ml Tricaine (Sigma-Aldrich) and used for cell transplantation. Briefly, CM-DiI-labeled HeLa cells were re-suspended in Fetal Bovine Serum (FBS) and loaded into a pulled glass micropipette and ∼50 cells were delivered, as a single injection, into the yolk sac of each embryo (Injection conditions: 0.1 seconds, 2.5 psi) using a PLI-100A Pico-Liter Injector (Warner instruments) while under observation using a Zeiss SteREO Discovery.V8 microscope. Following injection, embryos were allowed to recover at 28°C for 1 h before transfer to 35°C where they remained for the experiment.
At 48 h post injection (hpi), only embryos with a uniform fluorescent cell mass at the site of injection were used for proliferation studies. Embryo xenotransplanted with human cancer cells were then maintained in groups of 15–20 and drugs were added directly to the fish water, at indicated concentrations and embryos were incubated for 48 h until 144hpf. Embryos treated with vehicle (0.1% Dimethyl sulfoxide (DMSO)) served as a negative control for drug efficacy (32). For the fluorescence imaging, a filter with excitation/emission wavelengths of 550/605 nm was used and all embryos were photographed under the same settings.
To determine rate of cellular proliferation in vivo positively injected embryos were placed into groups of 15–20 embryos and euthanized by a Tricaine overdose at the time intervals: 48hpi (0hpt) and 96hpi (48hpt) and dissociated into a single cell suspension using the protocol outlined by Corkery et al. (32). The dissociations were analyzed using the inverted Axio Observer Z1 microscope to determine the number of fluorescent cells per embryo, and cellular proliferation was represented as a fold change of the number of fluorescent cells present at 48hpi.
Statistical Analysis were performed with GraphPad Prism 5.0. For parameter comparisons between groups, an unpaired two-tailed Student's t-test or one-way ANOVA followed by Dunnett's multiple comparison test were used when appropriate. P-values of <0.05 were considered significant. Results are reported as mean ± SEM.
Patients and tissue specimens
Tumor samples were obtained from patients who underwent surgery for ovarian cancer at the Department of Gynecologic Oncology Centre Hospitalier de l'Université de Montréal (CHUM). A gynecologist-oncologist established disease stage as defined by the Federation International of Gynecology and Obstetrics (FIGO). An independent pathologist reviewed tumor histopathology and grade. Tissue selection criteria for this study were based on independent confirmation of high grade serous (HGS) histopathology in samples from chemotherapy-naive patients. Samples were collected between 1992 and 2012. Patient disease free survival was calculated from the diagnostic date until the first progression or last follow-up. Clinical data on progression-free interval were defined according to computed tomographic imaging alone or in combination with blood CA125 levels. Patient overall survival was defined as the time from surgery to death from ovarian cancer or last follow-up. Patients known to be still alive at time of analysis were censored at time of their last follow-up.
Epithelial serous ovarian tumor tissue microarray (TMA)
A gynecologic pathologist reviewed all cases. The grade and type of ovarian carcinoma were identified and areas of interest were marked on slides. Two cores of 0.6 mm for each tissue sample were arrayed onto one recipient paraffin block. This tissue array was composed of cores from 213 HGS tumors (two cores per patient sample) and was built on two recipient blocks. The tumor tissue microarray (TMA) was then sectioned, stained with hematoxylin-eosin and subjected to another review of tissue pathology to verify the presence of tumor material in each core. Four patients were removed from analysis after pathology review because they did not match with the inclusion criteria.
Immunofluorescence studies on TMA
We produced an epithelial mask using several specific epithelial antigens to distinguish stromal and epithelial components within tissue. To ensure coverage of all epithelial cells we used a cocktail of CK18, CK19 and CK7 that were all labeled to emit in the orange channel (AF546). APRIN was labeled to emit in the red channel (Cy5) and slides were also stained with DAPI (blue) to identify nuclei. This allowed us to distinguish specific staining of APRIN in the nucleus and cytoplasm of epithelial cells. Specifically, antigen retrieval was carried out with Cell Conditioning 1 using the Bench Mark XT automated stainer (Ventana Medical System Inc.). Primary antibody against APRIN was diluted 1:50 in PBS and incubated at 37°C for 60 min. The following steps were manually done on the bench (while previous steps were performed using the BenchMark XT automated stainer (Ventana Medical System Inc.)) under conditions to protect slides from light. Secondary fluorescent antibody against APRIN was incubated for 45 min at room temperature (RT): anti-mouse Cy5 (, Life Technologies Inc., #A10524) diluted 1:250 in 1× PBS. TMA slides were then blocked for 60 min with Mouse-On-Mouse blocking reagent (one drop in 250 μl PBS, MKB-2213, Vector Laboratories, CA, USA). Slides were subsequently incubated for 90 min at RT with a mix of anti-CK18, anti-CK19 and anti-CK7 (1:100 in 1× PBS) and then 45 min at RT with secondary fluorescent anti-mouse AF546 (Life Technologies Inc., #A10036). Finally, slides were incubated 15 min at RT with a 0.1% (w/v) solution of Sudan Black in 70% ethanol to quench tissue auto-fluorescence. Slides were mounted with the ProLong® Gold Antifade Mountant with DAPI (Life Technologies Inc., P-36931). Between each step, except after the blocking steps, TMA slides were washed twice with 1× PBS. Slides were stored at 4°C and scanned the next day. A negative control TMA slide was done in parallel and incubated with 1× PBS in place of all primary antibodies.
Staining quantification of TMA
The staining and scoring were performed blinded to the study end-point. The tissue sections were scanned with a 20 × 0.75 NA objective with a resolution of 0.3225 mm with the Olympus microscope and VS110 slide scanner linked to an OlyVia® image viewer software (xvViewer.exe). Fluorescent staining was then quantified by the VisiomorphDP software (Visiopharm, Denmark) allowing for automated image analysis. The staining via CK18/CK19/CK7 markers was used to limit the analysis to epithelial areas as the region of interest #1 (ROI #1), while DAPI served to circumscribe nuclei compartment. Cores were manually reviewed and surrounding tissues with necrosis or inflammatory zones were removed from the analysis. In each TMA core, APRIN fluorescence was measured and a mean fluorescence intensity (MFI) score was calculated in the whole epithelial cell compartment as well as in nuclei of epithelial cells more specifically. Continuous MFI values were then collected by the VisiomorphDP software and transferred to Excel files to define epithelial and nuclear APRIN variable expression. The positive and negative signals were based on designated threshold values (mean intensities + standard deviation) and used to calculate the frequency of positive nuclei per core. The average MFI of both tumor cores from the same patient was used for analysis.
Statistical analysis in TMA experiments
All statistical analyses were performed using SPSS software version 16.0 (SPSS Inc., Chicago, IL, USA) where p = 0.05 were deemed significant. APRIN expression thresholds of 1400 were determined by the receiver operating characteristic (ROC) curve. Survival curves (overall survival and disease free survival) were plotted by the Kaplan-Meier estimator, compared using the log rank test and analyzed for significant differences. The Spearman's correlation analysis (two-tailed) was used to obtain correlation with clinical data and biomarker.
RESULTS
APRIN is recruited to laser-induced DNA damage sites
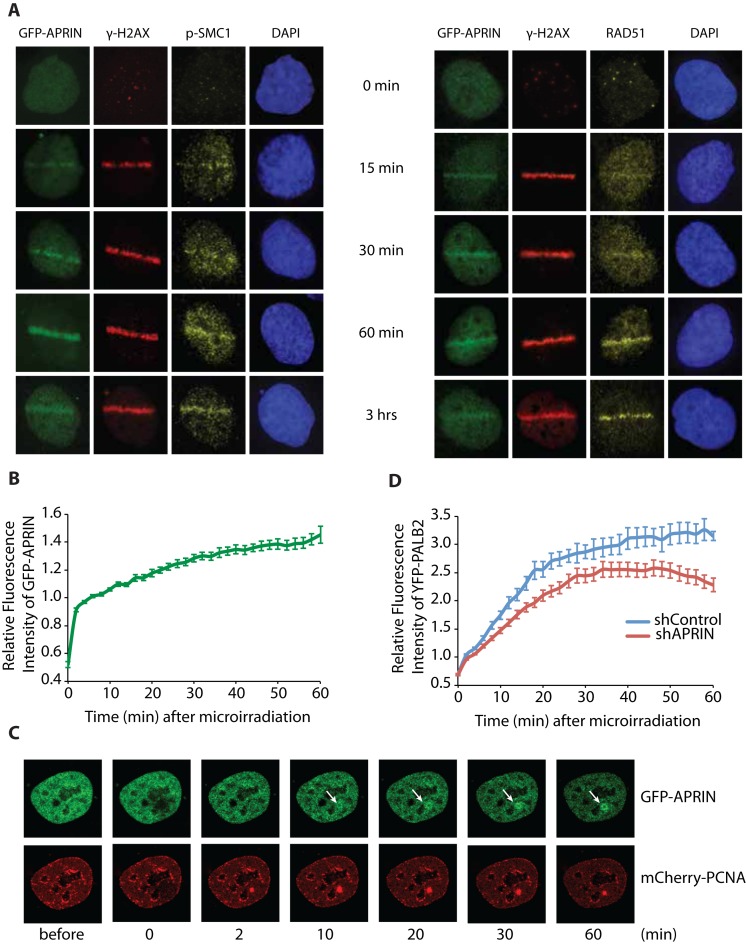
Brough et al. have shown that APRIN was present at, or near DNA damage sites using chromatin immunoprecipitations (23). In order to get more insight into the dynamics of this recruitment, we studied the localization of APRIN after DNA damage and compared it to that of RAD51 and p-SMC1, a member of the cohesin complex with a role in DNA repair (33). Thus, we explored the recruitment of APRIN after a laser-induced damage in a GFP-APRIN SKOV3 stable cell line. Using immunofluorescence time course analysis of recruitment after DNA damage, we found that APRIN is recruited to the damage sites 15 min after release from laser-track. We compared this recruitment to that of p-SMC1 and showed similar recruitment (within 15 min after the damage) (Figure 1A). We showed that APRIN was recruited before RAD51 protein, known to be a marker of homology-directed DNA repair. Moreover, we found that APRIN presents, together with RAD51, a peak accumulation at 60 min post-damage (Figure 1A). These results are in correlation with a previous study showing a prominent recruitment of RAD51 60 min after DNA damage (34).
Figure 1.
APRIN is recruited to laser-induced DNA damage sites and its depletion impacts the recruitment of PALB2 to DNA damage sites. (A) Representative images of laser-irradiated SKOV3 GFP-APRIN (green) cells and subjected to immunofluorescence procedures for detection of p-SMC1 (yellow, left) or RAD51 (yellow, right) and γ-H2AX (red), at different times post-DNA damage (indicated at the top of each panel). DAPI (blue) represents the nucleus. (B) Quantitative evaluation and representative live-cell images (C) of APRIN recruitment kinetics in S-phase HeLa cells co-transfected with GFP-APRIN and mCherry-PCNA after laser microirradiation. (D) YFP-PALB2 recruitment to laser-induced DSBs is diminished in shAPRIN cells.
To examine the functional relevance of APRIN recruitment, we used quantitative microscopy on live cells. Briefly, we generated localized DNA damage in a specific region within the nucleus and followed the recruitment of GFP-APRIN by taking pictures every 2 min during 1 h. Interestingly, we show that S/G2 phase cells (as detected by mCherry-PCNA distribution) exhibited 1.4-fold greater median APRIN recruitment to laser-induced DNA damage (Figure 1B and C).
We monitored subsequently if the depletion of APRIN could have an impact on PALB2 or the opposite. APRIN depletion leads to a reduction, but not impairment, of PALB2 recruitment to laser-induced DSBs suggesting a functional relationship between these proteins (Figure 1D). ShRNA knockdown of PALB2 did not affect APRIN recruitment (Supplementary Figure S1A). We found that APRIN and RAD51 colocalized at laser-induced DNA damage (Supplementary Figure S1B). These results suggest that APRIN is recruited to DNA damage sites and possibly function in HR with RAD51 and PALB2.
APRIN interacts directly with RAD51, PALB2 and BRCA2
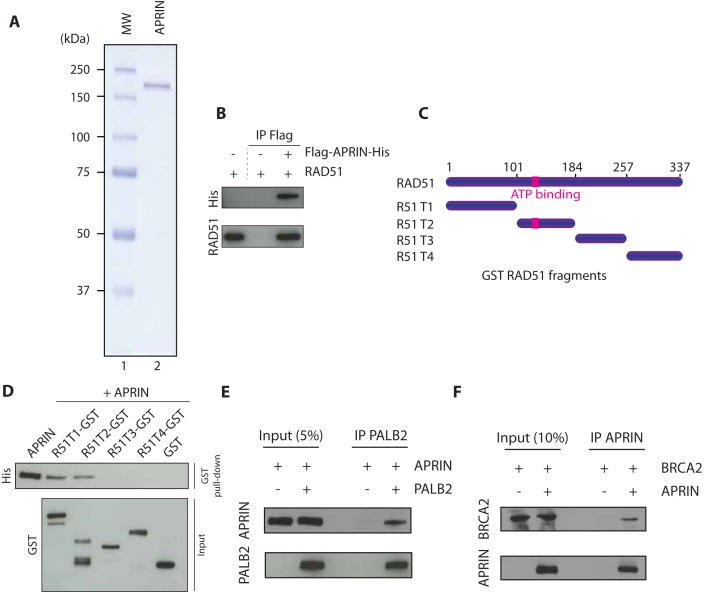
Using immunoprecipitation analysis from cell extracts, it has been previously shown that APRIN interacts in vivo with BRCA2, PALB2 and RAD51 (23). Having established that APRIN influences PALB2 recruitment and co-localized with RAD51, we scrutinized if APRIN could interact directly with HR proteins using purified components. To do so, we purified APRIN to near homogeneity by double affinity (Figure 2A and Supplementary Figure S2A), from Spodoptera frugiperda insect cells (Sf9). The protein was pure as judged by Coomassie blue and silver staining (Supplementary Figure S2B). Using co-immunoprecipitation assays of purified proteins, we observed that human RAD51 and APRIN bind directly with each other (Figure 2B). APRIN interacts on two separate regions of RAD51 (amino acids 1–100, and 101–184, the latter bearing the RAD51 ATPase domain) (Figure 2C and D). Furthermore, using the same assays we showed that APRIN interacted directly with full-length proteins PALB2 (Figure 2E) and BRCA2 (Figure 2F). These results identify APRIN as a new component of the HR machinery.
Figure 2.
APRIN physically interacts to RAD51, PALB2 and BRCA2. (A) SDS-PAGE gel of purified His/Flag APRIN stained with Coomassie blue. Lane 1, Molecular weight markers; Lane 2, APRIN purified protein (250 ng). (B) Co-immunoprecipitation of purified APRIN and RAD51 using antibodies against histidine tag (His), followed by western blotting with anti-His antibody. (C) Scheme of the RAD51 deletion variants fused to GST. (D) GST alone or GST-tagged RAD51 fragments purified from Escherichia coli were incubated with APRIN. Sepharose beads were used to immunoprecipitate complexes. (E) Co-immunoprecipitation of purified APRIN (500 ng) and PALB2 (500 ng) using antibodies against PALB2, followed by western blotting with anti-APRIN or anti-PALB2 antibodies. (F) Co-immunoprecipitation of purified APRIN (500 ng) and BRCA2 (100 ng) using antibodies against APRIN, followed by western blotting with APRIN or BRCA2 antibodies.
Purified APRIN binds different DNA intermediates of HR
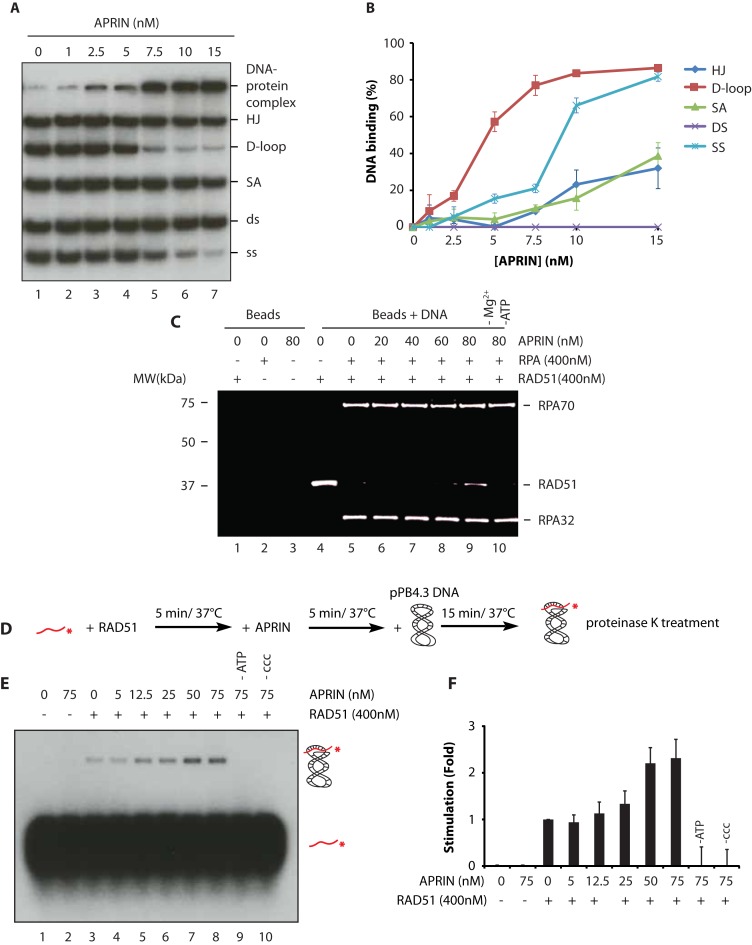
One of the characteristics of HR mediators is their ability to bind DNA with specific affinity for ssDNA. APRIN bears AT hook domains known to be involved in DNA binding (Supplementary Figure S2C) (22) and is a chromatin-associated protein (Supplementary Figure S2D). These observations suggested that APRIN could also be implicated in DNA repair via its likely interaction with DNA. Using DNA electrophoretic mobility shift assays (EMSA), we showed that APRIN can indeed bind diverse HR intermediates, such as ssDNA, SA, HJ or displacement-loop (D-loop), but surprisingly not dsDNA (Supplementary Figure S3). We then analyzed the relative affinity of APRIN for the different DNA structures using competition assays. Thus, we mixed all five DNA substrates in the same reaction and quantified APRIN–DNA interactions. At 5 nM of APRIN, 60% of D-loop was bound, followed by ssDNA and splayed arms (15–20%), compared with 1–15% for all other DNA substrates (Figure 3A and B). Hence, we showed that APRIN binds preferentially D-loop structures over ssDNA, SA and HJ. Together, these results reveal the ability of APRIN to bind different DNA substrates, with a strong preferential affinity for D-loop structures.
Figure 3.
APRIN binds DNA with preference for single-strand (ss) DNA and D-loop structures and enhances RAD51 mediated D-loop formation. (A) Competition electrophoretic mobility shift assays (EMSAs) were performed with purified APRIN and a mix containing equal quantities of ssDNA, double-strand (ds) DNA, splayed arms (SA), Holliday junctions (HJ), Displacement-loop (D-loop) on 8% acrylamide gel. (B) Quantification with Phosphor Imager of the EMSAs from four independent experiments. Error bars represent the corresponding standard error. (C) RPA displacement assay. RPA bound to a ssDNA oligonucleotide prevents RAD51 assembly (lane 5) whereas addition of APRIN (20–80 nM) stimulates RAD51 filament formation in presence of RPA (lanes 6–9). (D) Scheme of the different steps of D-loop formation. (E) D-loop formation mediated by APRIN alone (lane 2), RAD51 alone (lane 3) and a titration of APRIN (lanes 4–8). (F) Phosphor Imager quantification of the results from three independent experiments. Error bars represent the corresponding standard error.
To further characterize the DNA binding domains of APRIN, we purified to near homogeneity five protein fragments (named A1 to A5) from bacteria, spanning APRIN coding sequence (Supplementary Figure S4A and B). Using EMSA, we found that 50–100 nM of A2 and A4 could bind ssDNA whereas A1, A3 and the C-terminal region (A5) do not bind DNA (Supplementary Figure S4C). To understand the function of these domains in the recruitment of APRIN to DNA damage sites, we created different versions of APRIN lacking the A2, A4 or A5 regions (Supplementary Figure S4D). Due to the presence of a NLS on the C-terminal domain of APRIN, we also complemented the A5-deleted mutant with the NLS from nucleoplasmin. Laser track experiments showed that although all mutants were still recruited to DNA damage sites, the mutants lacking A2 or A4 fragments were less efficiently recruited (Supplementary Figure S4E). These results suggest that the A2 and A4 regions promote APRIN recruitment to DNA damage sites.
APRIN is a new HR mediator that stimulates RAD51-mediated D-loop formation
APRIN is important for efficient HR (23). Since we have shown that APRIN is recruited to damage sites, interacts directly with RAD51 and binds DNA, we tested whether it could be a mediator of HR by its ability to counteract the RPA inhibitory effect on RAD51. Indeed, prior to strand invasion, the ssDNA binding protein RPA must be displaced from the ss tail of the newly resected DSB to allow RAD51 filament formation. Such function is accomplished by BRCA2 or PALB2 recombination mediators to promote the presynaptic filament assembly (10).
The addition of 400 nM totally blocks the association of RAD51 with ssDNA-coupled beads. In the presence of RPA, RAD51 loading on ssDNA coupled beads was strongly inhibited (Figure 3C, compare lanes 4 and 5). When APRIN was added after RAD51 and RRA, RAD51 gained access to the ssDNA (Figure 3F, compare lane 9). These results suggest that APRIN, as PALB2 and BRCA2, is a mediator of HR.
Having establish APRIN as a new HR mediator, we analyzed whether APRIN could enhance RAD51-mediated D-loop formation. We performed D-loop assays, as previously described (35), using labeled 63 nt probes in presence of saturating concentration of RAD51 (300 nM) and a super-coiled plasmid (pPB4.3) (Figure 3D). No product of D-loop was observed with APRIN alone (Figure 3E, lane 2). We then quantified strand invasion in presence of increasing concentration of APRIN and RAD51 (Figure 3D–E). APRIN stimulated RAD51-mediated D-loop formation by 2-fold. These results suggest that APRIN is a mediator of HR with a role in RAD51 D-loop formation.
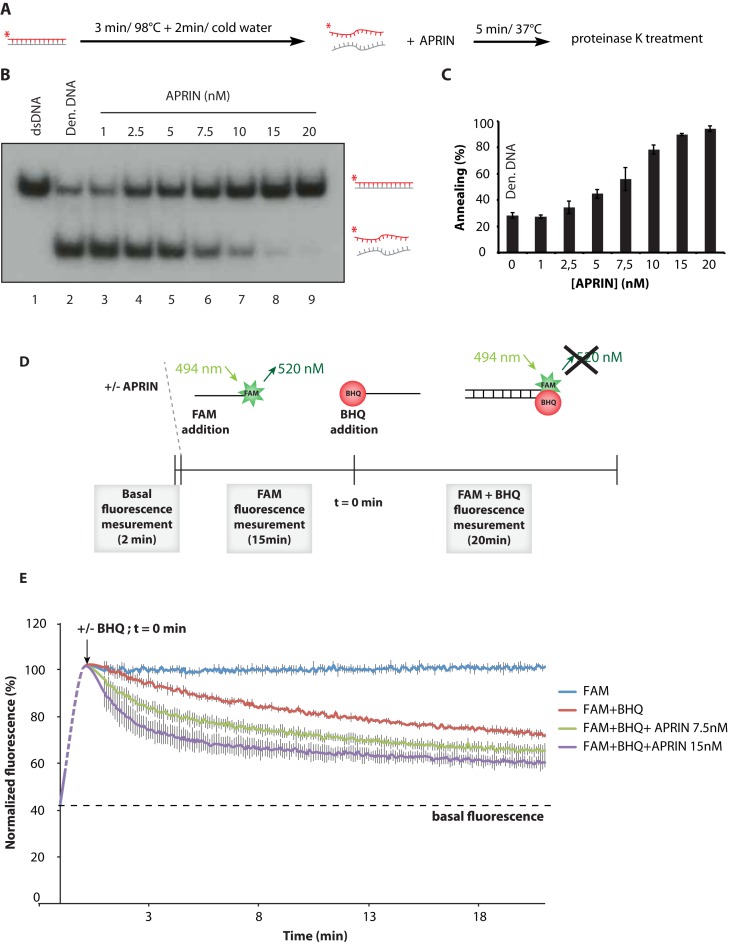
APRIN enhances the annealing of complementary single-strands
The high affinity of APRIN for ssDNA and its implication in RAD51 D-loop formation led us to investigate next the role of APRIN in complementary-strand annealing. In this assay, we denatured a 100 bp dsDNA fragment. The product was cooled down and APRIN was added or not to the reaction (Figure 4A). While low levels of spontaneous strand annealing are observed in the control reaction alone (28%, Figure 4B, lane 2), APRIN greatly enhanced the formation of dsDNA (∼100% for 20 nM of APRIN, Figure 4B, lane 9 and Figure 3C), indicating that it promotes annealing of complementary ssDNA. To confirm this result, we designed a system implicating two-labeled complementary DNA probes. One of the two oligonucleotides was labeled with a fluorescent fluorescein (FAM) moiety at the 5′ terminus and a complementary oligonucleotide bore a Black Hole Quencher (BHQ). Briefly, at 494 nm the FAM is excited and emits light at 592 nM. When the BHQ is sufficiently near to the FAM, it blocks this phenomenon leading to an absence of emission (Figure 3D). Having established a condition where auto-quenching was low, we added APRIN to the reaction. We found that APRIN efficiently accelerates the annealing of complementary DNA strands (Figure 3E).
Figure 4.
APRIN is proficient in ss annealing. (A) Scheme of the different steps of the homologous strand-annealing assay. (B) APRIN promotes ss annealing. Lane 1, purified 60-bp duplex DNA (dsDNA). Reactions contained denatured 60-bp duplex DNA in buffer with no protein (lane 2; den. DNA: denatured DNA) or with purified APRIN (1–20 nM), lanes 3–9. (C) Phosphor Imager quantification of the annealing from three independent experiments. Error bars represent the corresponding standard error. (D) Scheme of the FAM/BHQ annealing experiment. In brief, APRIN was added to the reaction and the basal fluorescence was analyzed during 2 min. Then, 5′-FAM (6-carboxyfluorescein amidite), a fluorescent dye for labeling oligonucleotides, was added to the reaction for 15 min. The FAM is reactive, and has an absorbance maximum of 494 nm with an emission maximum of 520 nm. Lastly, BHQ (Black Hole Quencher) homologous oligonucleotide was added to the reaction after which the fluorescence was measured every 6 s with a Cytation5 BioteK plate reader. (E) Quantification of the results from three independent experiments. The maximum of fluorescence at t = 0 min corresponds to the mean fluorescence during the 15 min of the reaction containing APRIN and the oligonucleotide. Error bars represent the corresponding standard error.
APRIN enhances complementary-strand annealing in synergy with BRCA2
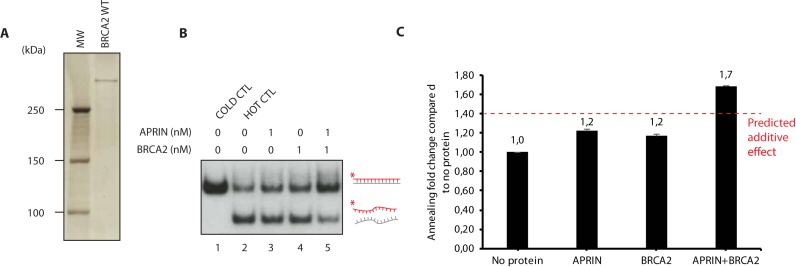
Given that BRCA2 and APRIN interact with each other ((23) and Figure 2F) and that APRIN has a strong DNA annealing activity (Figure 2A–E), we analysed the impact of this interaction on complementary-strand annealing in presence of BRCA2. It is important to note that APRIN ss annealing activity is more robust than PALB2 (Supplementary Figure S5A and B). We purified human BRCA2 to homogeneity (36) (Figure 5A) and performed further strand annealing experiments. When using purified APRIN and BRCA2 proteins independently and at very low concentration (1 and 0.1 nM respectively), the annealing was enhanced significatively (1.2-fold more compared to no protein) (Figure 5B and C). Interestingly, at the same concentrations, APRIN and BRCA2 cooperate to increase annealing to 1.7-fold, the stimulation being stronger than the predicted additive effects (Figure 5B). These results suggest that BRCA2 and APRIN have synergistic roles in the synaptic step of HR, which implicates complementary strand annealing.
Figure 5.
APRIN enhances the homologous-strand annealing in synergy with BRCA2. (A) Silver-stained SDS page of human BRCA2. (B) APRIN and BRCA2 annealing assays. Lane 1, purified 60-bp duplex DNA. Reactions contained denatured 60-bp duplex DNA in buffer with no protein (lane 2; den. DNA: denatured DNA) or with purified APRIN alone (1 nM, lane 3), BRCA2 alone (1 nM, lane 4) or a mixture of APRIN and BRCA2 (lane 5). (C) Phosphor Imager quantification of the result from three independent experiments. Error bars represent the corresponding standard error.
Unlike cohesins, APRIN does not play a role in NHEJ or CSR
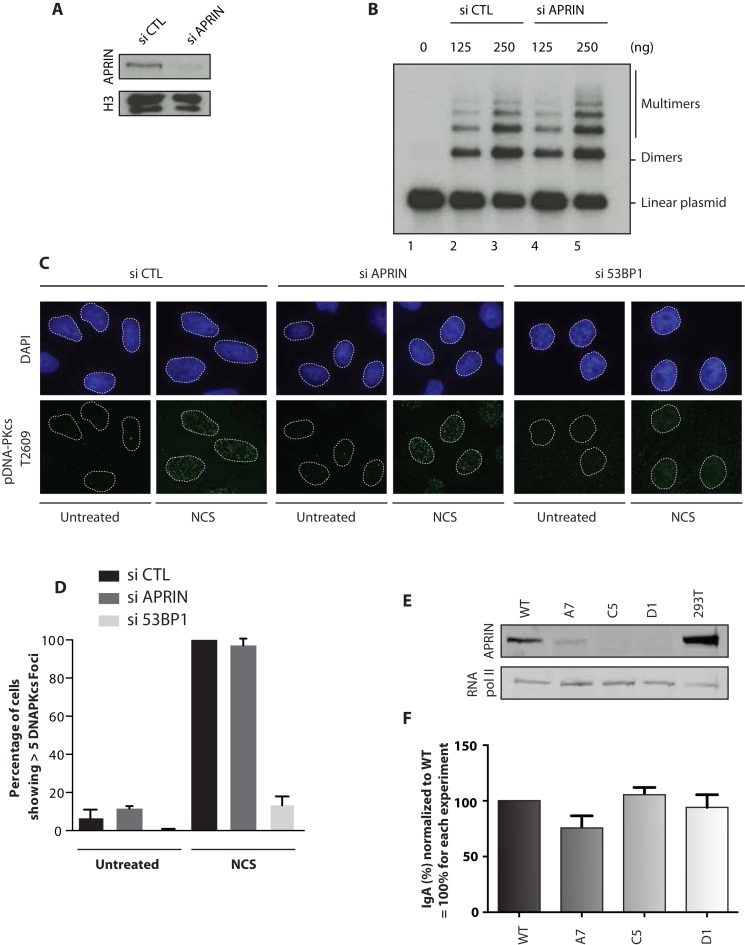
It has been shown that cohesins are implicated in NHEJ during CSR (17). Since APRIN is a cohesin binding protein (21), we reasoned that investigating whether APRIN plays a role in NHEJ and CSR would indicate whether APRIN has cohesin-dependent roles. We first used a cell-free NHEJ assay (28) that measures the ligation of a 32P-labeled linearized plasmid following incubation with nuclear extracts derived from siRNA control or siRNA APRIN transfected 293T cells (Figure 6A). The knockdown of APRIN in 293T cells did not have any impact on NHEJ kinetics in vitro compared to the control condition (Figure 6B). To confirm this result, we investigated whether APRIN affected DNA-PKcs phosphorylation at Ser2056 in vivo, which is correlated with active NHEJ (37). As expected, DNA-PKcs autophosphorylation level induced by neocarzinostatin treatment was not affected by APRIN depletion as opposed to 53BP1, a key player in the promotion of NHEJ and inhibition of HR (38), whose knock-down caused reduced DNA-PKcs Ser2056 foci formation (Figure 6C and D).
Figure 6.
APRIN is not implicated in non-homologous end-joining (NHEJ) nor switch class recombination (CSR). (A) Depletion in 293T using siRNA control (si CTL) or siRNA against APRIN (si APRIN). (B) A linearized, 32P-end labeled pBluescript was incubated for 30 min with a nuclear extract (125 or 250 ng) derived from 293T cells treated with si CTL (lanes 2–3) or si APRIN (lanes 4–5). (C) Immunofluorescence staining of phosphorylated DNA-PKcs in untreated or neocarzinostatin (NCS) treated cells at 100 ng/ml for 1 h. (D) Quantification of the number of foci from three independent experiments. Error bars represent the corresponding standard error. (E) Mouse B-cell line CH12F3 were depleted from APRIN using the CRISPR-Cas9 method. A7 clone represents a deletion in exon 3. C5 and D1 clones represent deletion of exon 5 and a part of exon 6. (F) Quantification of the switching between IgM and IgA. After stimulation with TGFB1, IL-4 and anti-CD40, cells were labeled with anti-IgA and analyzed using FACS. Error bars represent the corresponding standard error.
We then studied the impact of APRIN depletion on the CSR. To do so, we quantified the switch between IgM and IgA in murine B lymphoma CH12F3 cells. Using the CRISPR-Cas9 method, we eliminated APRIN in CH12F3 cells following two strategies: one consisting in the deletion of exon 3 and the other in the deletion of exon 5 and parts of exon 4 and 6 of APRIN. We selected a single clone of the first strategy (A7) and two from the second (C5 and D1) (Figure 6E). The depletion of APRIN in CH12F3 did not have any impact on the cell viability (Supplementary Figure S6A and B). After TGFB1, IL-4 and anti-CD40 stimulation, cells were labeled with an antibody against IgA and analyzed by FACs. We found that APRIN depletion does not have any impact on CSR in CH12F3 cells (Figure 6F), supporting an exclusive implication of APRIN in HR.
Depletion of APRIN leads to the sensitization of cells to PARP inhibition Olaparib and Veliparib
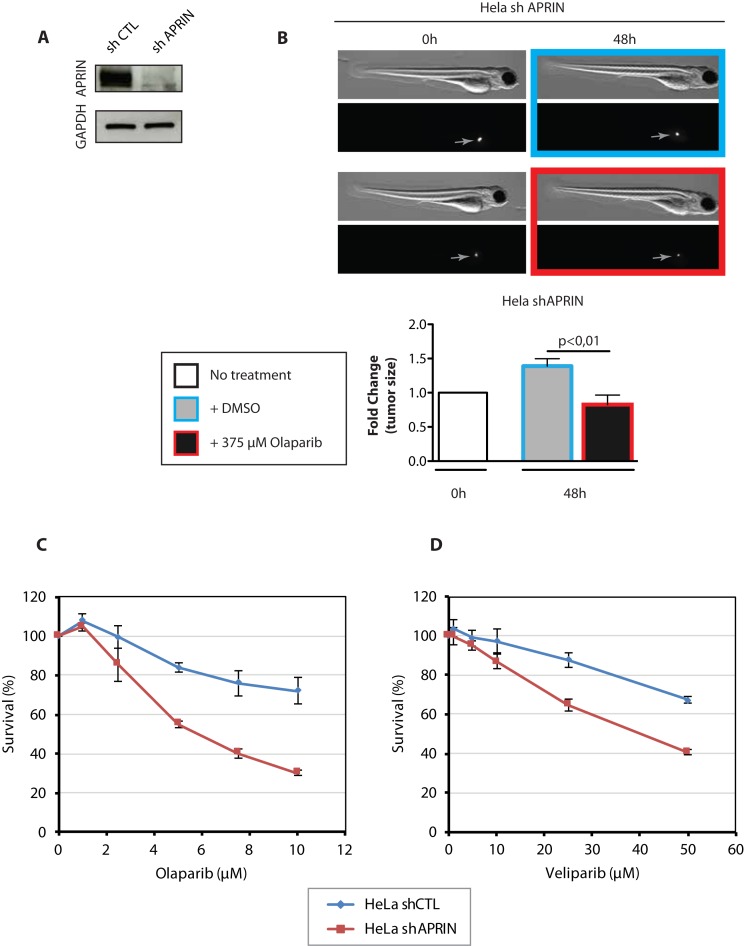
It has been previously shown that APRIN depletion leads to the sensitization of cells to PARP inhibitors or other DNA damaging agents (23). Thus, we wanted to examine this concept in a living animal. Hence, we created HeLa cell line stably expressing APRIN shRNA (shAPRIN) (Figure 7A). These cells were CM-DiI-labeled and implanted in 48 h post-fertilization zebrafish embryos. Forty-eight hours post-injection, embryos with a uniform mass (indicated by a gray arrow in Figure 7B) were treated with the PARP inhibitor Olaparib or vehicle (DMSO) for an additional 48 h, after which tumor size was quantified ex vivo by counting tumor cell number following dissociation of engrafted embryos (Figure 7B). We found that APRIN-depleted cells showed sensitivity to PARP inhibition Olaparib, leading to a decreased tumor size compared to vehicle treated tumors (Figure 7B). We then tested whether HeLa cells depleted for APRIN by shRNA were sensitive to the PARP inhibitor Olaparib as previously described (23). Using MTT assays, we found that the depletion of APRIN sensitized cells to Olaparib (Figure 7C). We also showed that these cells were sensitive to Veliparib, another PARP inhibitor (Figure 7D).
Figure 7.
APRIN-depleted HeLa cells are sensitive to PARP inhibitor treatment. (A) Western blot showing the depletion of APRIN using APRIN shRNA (shAPRIN) or control shRNA (shCTL). (B) Representative bright field and fluorescent images of zebrafish (top) along with the bar graphs showing cellular proliferation in vivo (bottom). Uniform fluorescent mass is indicated by the gray arrow. An unpaired two-tailed Student's t-test or one-way ANOVA followed by Dunnett's multiple comparison test were used when appropriate. P-values of <0.05 were considered significant. Results are reported as mean ± SEM. (C) Survival curves obtained from MTT assays on shCTL or shAPRIN HeLa cells in presence of PARP inhibitor Olaparib at the indicated concentration. (D) Survival curves obtained from MTT assay on shCTL or shAPRIN HeLa cells in presence of PARP inhibitor Veliparib. Error bars represent the corresponding Standard Error. Survival experiments have been performed for 5 days.
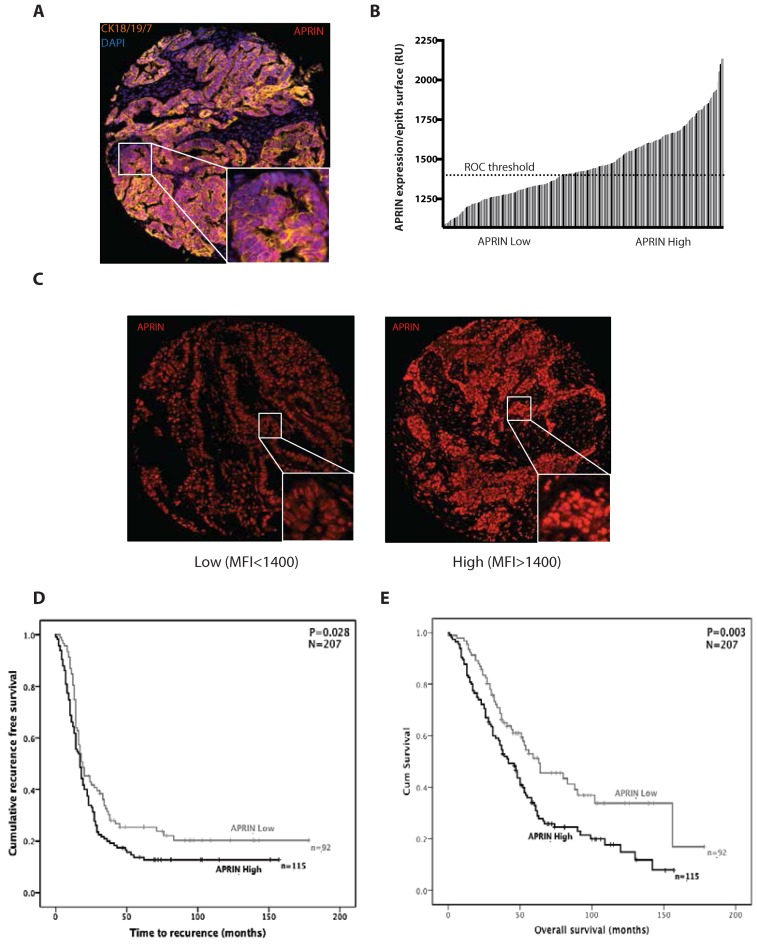
Low levels of APRIN correlate with better survival in high grade serous (HGS) epithelial ovarian cancer patients
It has been suggested that APRIN is a potential tumor suppressor since it is downregulated or suppressed in many cancers (39). Moreover, Brough et al. showed that APRIN levels are modulated in accordance to the breast cancer grade of patients. Since APRIN interacts with BRCA2 whose mutations can lead to ovarian cancers, we investigated the expression of APRIN on a TMA containing 207 cases of HGS ovarian cancer cases (Table 8). To specifically assess the impact of APRIN expression on ovarian tumor cells, we performed immunofluorescence double-staining for APRIN and epithelial cytokeratin 7/18/19 (Figure 8A) and determined the intensity of APRIN expression per epithelial surface unit (Figure 8B). Two groups were created based on a ROC curve analysis (ROC curve is most suitable to determine the optimal cut off values (43)): (i) samples with a MFI of <1400 were considered as ‘low’ APRIN expression; (ii) samples with a MFI over 1400, were considered as ‘High’ APRIN expression (Figure 8C). High levels of APRIN (Figure 8D and E) in tumor cells were associated with a significantly shorter time of recurrence (mean ± SD: 53.96 ± 7.04 months for low APRIN versus 35.39 ± 4.5 months for high APRIN; p = 0.028) (Figure 8D) and decreased overall survival (mean ± SD: 86.25 ± 7.9 months for low APRIN versus 56.9 ± 4.6 months for high APRIN; p = 0.003) (Figure 8E). APRIN expression in tumor cells was also associated with a worse overall survival and correlate with the residual disease as determined by Spearman's correlation analysis (Supplementary Table SI).
Table 8. Clinicopathological characteristics of the HGS EOC cores present on the TMA (n = 207).
| Median (range) or n (%) | ||
|---|---|---|
| Age (years) | 61 (34–89) | |
| FIGO stage | I | 13 (6) |
| II | 23 (11) | |
| III | 147 (71) | |
| IV | 25 (12) | |
| Debulking | Optimal | 96 (46) |
| Non optimal | 91 (44) | |
| unknown | 21 (10) | |
| Chemotherapy | platinum-based | 191 (92) |
| taxane alone | 2 (1) | |
| unknown | 15 (7) | |
| Follow-up time (months) | 36 (1–156) | |
| Dis. Progression time (months) | 14 (0–76) |
Figure 8.
Low APRIN expression levels are correlated with better survival of ovarian cancer patients. (A) Co-expression of APRIN (red) and epithelial cytokeratin-7/18/19 (orange) were analyzed by immunofluorescence. (B) APRIN protein expression in tumor cells was quantified by immunofluorescence, and measured for 207 cases of HGS ovarian cancer on TMA. Expression per epithelial surface unit was calculated as the integrated intensity of APRIN in the cytokeratin compartment divided by the number of pixels present in the cytokeratin compartment (bars represent individual cases). (C) Representative images of low and high immunofluorescence staining against APRIN in TMA of specimens from ovarian cancer patients. (D and E) Subjects with low or high APRIN expression (as determined by ROC analysis) were plotted to determine the association between APRIN expression in tumor cells with the time to recurrence (D) or overall survival (E). A receiver operating characteristic (ROC) curve, is a graphical plot that illustrates the performance of binary classifier system as its discrimination threshold is varied. ROC curve is most suitable to determine the optimal cut off values (43). Significance (P) was determined by log-rank test. n, number of patients in low or high APRIN expression group. N, total number of patients. MFI: mean fluorescence intensity; RU: relative units.
DISCUSSION
In this manuscript, we investigated the activity and interacting partners of APRIN, uncovering novel functions for this protein in HR. Brough et al., suggested that APRIN could be an interface between cohesin and HR proteins. We propose that APRIN could have a direct role in HR and possibly in different steps of the pathway.
Brough et al. showed that APRIN colocalizes occasionnally with RAD51 (23). In this study, we have found that APRIN is recruited to DNA damage using laser-induced DSBs. We showed that APRIN interacts directly with RAD51, PALB2 and BRCA2. It has been previously shown that this complex exists in vivo (23). We hence propose that APRIN could be a new component of this complex. We showed that APRIN enhances RAD51-mediated D-loop formation. Using in vitro experiments, we showed that APRIN interacts strongly with DNA, with a marked preference for the D-loop and ssDNA structures mimicking different steps of HR. Moreover, whereas APRIN is unable to stimulate D-loop formation on its own, we showed that it independently stimulates complementary-strand annealing, an activity that is enhanced with the presence of the BRCA2 protein. We believe that this role could explain why the interaction between BRCA2 and APRIN has been found to be S-phase specific (23).
We found that APRIN is specifically implicated in HR, as it is not involved in NHEJ or CSR. As known, APRIN is a cohesion-interacting factor with roles in cohesion regulation. Previous studies have shown that cohesins are specifically recruited to laser-induced DNA damage (18). Moreover, Kong et al. proposed that the presence of a sister chromatid and/or an additional factor(s) contributes to the cell cycle-specific cohesin-SA2/NIPBL recruitment (19). The fact that APRIN interacts directly with PALB2 and RAD51 makes it a candidate for the specific recruitment of the cohesin complex to DSB during S/G2 phases. We showed that APRIN is recruited to laser-induced DNA damage sites at the same time as p-SMC1. This recruitment of APRIN is dependent on the A2 and A4 domains. This is in accordance with recent findings showing that APRIN interacts with SMC1 through two distinct domains (40) which are located on the A2 and A4 fragments. Hence, we postulate that APRIN recruitment to DNA damage sites would be dependent on A2, A4 domains and cohesin. This recruitment is not dependent on PALB2.
The observation that RAD51 protein is recruited after the cohesin reinforce the idea that APRIN could serve as a coordinating platform, proposed by Brough et al., with an important regulatory role in HR. In fact, APRIN can stimulate RAD51-mediated D-loop formation. It can also strongly stimulate annealing of complementary-strand DNA. The latter could help cohesin to trap homologous sister chromatids but our finding that APRIN annealing activity is enhanced by BRCA2 argues against this hypothesis. Nevertheless, the cooperation with BRCA2 further implicates APRIN in the mechanisms of HR. This is in accordance with the observations that APRIN depletion leads to a decrease in RAD51 foci (23) or to a decrease in the recruitment of PALB2 to DNA damage site (this study).
It has previously been shown that the knockdown of cohesin or its regulatory subunits results in impaired CSR and increased usage of microhomology-based end joining (17). Nevertheless, depletion of APRIN does not lead to an impaired CSR. Moreover, APRIN is not implicated in NHEJ. Thus, our data reinforce the implication of APRIN exclusively in HR and suggest a model in which APRIN functions in various steps of the mechanism (Supplementary Figure S7). APRIN might also be tightly regulated within this model. Recently, Elia et al. have shown that under IR and UV damage APRIN is phosphorylated on its C-terminal part by ATM/ATR (41). We believe that this modification could be important to modulate the activity of APRIN during HR and to contribute to its cell cycle-dependent activity.
APRIN has been characterized as a candidate tumor suppressor protein (42). Recent studies have shown that APRIN expression levels are associated with histological grade in breast cancer and the outcome of breast cancer patients treated with DNA-damaging chemotherapy. Here, we examined APRIN expression status in ovarian cancers and showed that low levels of APRIN expression correlate with a better survival of patients. In zebrafish, APRIN depletion leads to sensitization of cells to PARP inhibition in our xenograft experiments, similar to that reported for other chemical agents (such as HU, MMC, Olaparib) (23), (Olaparib and Veliparib) (this study). Our work therefore helps to understand how APRIN is directly and specifically implicated in HR and identifies APRIN as a potential prognostic factor in ovarian cancer and as a potential target for treatment.
Supplementary Material
Acknowledgments
We are grateful to Dr Isabelle Brodeur for critical reading the manuscript. We thank Masson lab members for suggestions. All authors have contributed to the analysis and interpretation of results as well as to the revision of the manuscript. We thank Peter Geck for the gift of the APRIN cDNA.
Disclaimer: The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Luc Bélanger Scholarship (to A.M.C.); Michèle St-Pierre—Institut du cancer de Montréal Research Fund (to H.F.); Nova Scotia Health Research Foundation (NSHRF) Scholar Award (to V.L.B.); Beatrice Hunter Cancer Research Institute (BHCRI) Trainee Award with funds provided by the CIBC, as part of The Terry Fox Foundation Strategic Health Research Training Program in Cancer Research at CIHR (to V.L.B.); HOG Scholarship (Oncology Axis, CHU de Québec) (to J.N.); Ovarian Cancer Canada Trainee Travel Award (to J.P.); FRQS Chercheur National Investigator and FRQS chair in genome stability (to J.Y.M.); BHCRI (to G.D., J.N.B.); Canada Research Chair tier 2 (to J.M.D.N.); Cancer Research Society (CRS); Canadian Cancer Society Research Institute (CCSRI) [702806](to J.Y.M.); Collaborative Health Research Project (CHRP) Operating Grant (to G.D., J.N.B.); Natural Sciences and Engineering Research Council (NSERC); Canadian Institutes of Health Research (CIHR); CIHR Grant [MOP-130535 to J.M.D.N.]. Funding for open access charge: CHIR. Tumor banking was supported by the Banque de tissus et données of the Réseau de recherche sur le cancer of the Fond de recherche du Québec—Santé (FRQS), associated with the Canadian Tumor Repository Network (CTRNet). H.F., A-M.M-M. and D.M.P. are researchers of the Centre de recherche du CHUM which receives support from the FRQS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Prakash R., Zhang Y., Feng W., Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velic D., Couturier A.M., Ferreira M.T., Rodrigue A., Poirier G.G., Fleury F., Masson J.Y. DNA damage signalling and repair inhibitors: the long-sought-after achilles' heel of cancer. Biomolecules. 2015;5:3204–3259. doi: 10.3390/biom5043204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma P., Greenberg R.A. Noncanonical views of homology-directed DNA repair. Genes Dev. 2016;30:1138–1154. doi: 10.1101/gad.280545.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen R.B., Carreira A., Kowalczykowski S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung P., Krejci L., Van Komen S., Sehorn M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 7.West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 8.Dray E., Etchin J., Wiese C., Saro D., Williams G.J., Hammel M., Yu X., Galkin V.E., Liu D., Tsai M.S., et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buisson R., Dion-Côté A.-M., Coulombe Y., Launay H., Cai H., Stasiak A.Z., Stasiak A., Xia B., Masson J.-Y. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buisson R., Masson J.Y. PALB2 self-interaction controls homologous recombination. Nucleic Acids Res. 2012;40:10312–10323. doi: 10.1093/nar/gks807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauty J., Rodrigue A., Couturier A., Buisson R., Masson J.Y. Exploring the roles of PALB2 at the crossroads of DNA repair and cancer. Biochem. J. 2014;460:331–342. doi: 10.1042/BJ20140208. [DOI] [PubMed] [Google Scholar]
- 12.Losada A. Cohesin in cancer: chromosome segregation and beyond. Nat. Rev. Cancer. 2014;14:389–393. doi: 10.1038/nrc3743. [DOI] [PubMed] [Google Scholar]
- 13.Dorsett D., Ström L. The ancient and evolving roles of cohesin in DNA repair and gene expression. Curr. Biol. 2012;22:R240–R250. doi: 10.1016/j.cub.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birkenbihl R.P., Subramani S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jessberger R. Cohesin's dual role in the DNA damage response: repair and checkpoint activation. EMBO J. 2009;28:2491–2493. doi: 10.1038/emboj.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelot C., Guirouilh-Barbat J., Le Guen T., Dardillac E., Chailleux C., Canitrot Y., Lopez B.S. The cohesin complex prevents the end joining of distant DNA double-strand ends. Mol. Cell. 2016;61:15–26. doi: 10.1016/j.molcel.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Thomas-Claudepierre A.-S., Schiavo E., Heyer V., Fournier M., Page A., Robert I., Reina-San-Martin B. The cohesin complex regulates immunoglobulin class switch recombination. J. Exp. Med. 2013;210:2495–2502. doi: 10.1084/jem.20130166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.S., Krasieva T.B., LaMorte V., Taylor A.M., Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 2002;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- 19.Kong X., Ball A.R., Jr, Pham H.X., Zeng W., Chen H.Y., Schmiesing J.A., Kim J.S., Berns M., Yokomori K. Distinct functions of human cohesin-SA1 and cohesin-SA2 in double-strand break repair. Mol. Cell. Biol. 2014;34:685–698. doi: 10.1128/MCB.01503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlmann F. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 2003;13:R104–R114. doi: 10.1016/s0960-9822(03)00039-3. [DOI] [PubMed] [Google Scholar]
- 21.Losada A., Yokochi T., Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 22.Maffini M., Denes V., Sonnenschein C., Soto A., Geck P. APRIN is a unique Pds5 paralog with features of a chromatin regulator in hormonal differentiation. J. Steroid Biochem. Mol. Biol. 2008;108:32–43. doi: 10.1016/j.jsbmb.2007.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brough R., Bajrami I., Vatcheva R., Natrajan R., Reis-Filho J.S., Lord C.J., Ashworth A. APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 2012;31:1160–1176. doi: 10.1038/emboj.2011.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusch T. Brca2–Pds5 complexes mobilize persistent meiotic recombination sites to the nuclear envelope. J. Cell Sci. 2015;128:717–727. doi: 10.1242/jcs.159988. [DOI] [PubMed] [Google Scholar]
- 25.Sumara I., Vorlaufer E., Gieffers C., Peters B.H., Peters J.M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stead K., Aguilar C., Hartman T., Drexel M., Meluh P., Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J. Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanov D., Schleiffer A., Eisenhaber F., Mechtler K., Haering C.H., Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- 28.Krietsch J., Caron M.C., Gagne J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., Masson J.Y. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012;40:10287–10301. doi: 10.1093/nar/gks798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 30.White R.M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E., et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haldi M., Ton C., Seng W.L., McGrath P. Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis. 2006;9:139–151. doi: 10.1007/s10456-006-9040-2. [DOI] [PubMed] [Google Scholar]
- 32.Corkery D.P., Dellaire G., Berman J.N. Leukaemia xenotransplantation in zebrafish–chemotherapy response assay in vivo. Br. J. Haematol. 2011;153:786–789. doi: 10.1111/j.1365-2141.2011.08661.x. [DOI] [PubMed] [Google Scholar]
- 33.Sjögren C., Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 34.Coker H., Brockdorff N. SMCHD1 accumulates at DNA damage sites and facilitates the repair of DNA double-strand breaks. J. Cell Sci. 2014;127:1869–1874. doi: 10.1242/jcs.140020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genois M.-M., Mukherjee A., Ubeda J.-M., Buisson R., Paquet E., Roy G., Plourde M., Coulombe Y., Ouellette M., Masson J.-Y. Interactions between BRCA2 and RAD51 for promoting homologous recombination in Leishmania infantum. Nucleic Acids Res. 2012;40:6570–6584. doi: 10.1093/nar/gks306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buisson R., Niraj J., Pauty J., Maity R., Zhao W., Coulombe Y., Sung P., Masson J.Y. Breast cancer proteins PALB2 and BRCA2 stimulate polymerase eta in recombination-associated DNA synthesis at blocked replication forks. Cell Rep. 2014;6:553–564. doi: 10.1016/j.celrep.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis A.J., Chen B.P., Chen D.J. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Rep. 2014;17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bunting S.F., Callen E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim M.S., An C.H., Yoo N.J., Lee S.H. Frameshift mutations of chromosome cohesion–related genes SGOL1 and PDS5B in gastric and colorectal cancers with high microsatellite instability. Hum. Pathol. 2013;44:2234–2240. doi: 10.1016/j.humpath.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 40.Ouyang Z., Zheng G., Tomchick D.R., Luo X., Yu H. Structural basis and IP6 requirement for Pds5-dependent cohesin dynamics. Mol. Cell. 2016;62:248–259. doi: 10.1016/j.molcel.2016.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elia A.E.H., Boardman A.P., Wang D.C., Huttlin E.L., Everley R.A., Dephoure N., Zhou C., Koren I., Gygi S.P., Elledge S.J. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol. Cell. 2015;59:867–881. doi: 10.1016/j.molcel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denes V., Pilichowska M., Makarovskiy A., Carpinito G., Geck P. Loss of a cohesin-linked suppressor APRIN (Pds5b) disrupts stem cell programs in embryonal carcinoma: an emerging cohesin role in tumor suppression. Oncogene. 2010;29:3446–3452. doi: 10.1038/onc.2010.100. [DOI] [PubMed] [Google Scholar]
- 43.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J. Intern. Med. 2013;4:627–635. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.