Abstract
G-quadruplexes adopt various folding topologies, but information on their folding pathways remains scarce. Here, we used electrospray mass spectrometry to detect and quantify the specifically bound potassium ions, and circular dichroism to characterize the stacking topology of each ensemble. For human telomeric (hTel) sequences containing the d((GGGTTA)3GGG) core, K+ binding affinity and cooperativity strongly depends on the chosen construct. The shortest sequences bind only one K+ at low KCl concentration, and this 2-quartet G-quadruplex is antiparallel. Flanking bases increase the K+ binding cooperativity. To decipher the folding pathways, we investigated the kinetics of K+ binding to telomeric (hybrid) and c-myc (parallel) G-quadruplexes. G-quadruplexes fold via branched pathways with multiple parallel reactions. Up to six states (one ensemble without K+, two ensembles with 1-K+ and three ensembles with 2-K+) are separated based on their formation rates and ion mobility spectrometry. All G-quadruplexes first form long-lived misfolded structures (off-pathway compared to the most stable structures) containing one K+ and two quartets in an antiparallel stacking arrangement. The results highlight the particular ruggedness of G-quadruplex nucleic acid folding landscapes. Misfolded structures can play important roles for designing artificial G-quadruplex based structures, and for conformational selection by ligands or proteins in a biological context.
INTRODUCTION
G-quadruplex nucleic acid structures can form in important regions of the human genome such as telomeres, genes promoters or transcription start sites (1), and are involved in several cellular regulation pathways such as regulation of gene expression (2–8) and telomere maintenance (9–11). Recently G-quadruplexes have been revealed in human cells using an antibody (12). G-quadruplexes are tetra-helical structures formed by guanine-rich sequences (13). Four guanines, interacting via Hoogsteen hydrogen bonds, form a G-quartet. Structures with at least two stacked G-quartets are called G-quadruplexes. G-quadruplex stabilization is ensured by monovalent cations such as Na+, K+ or NH4+ (14,15), which they intercalate between consecutive G-quartets and are octa-coordinated by the O6 of the guanines (16). G-quadruplexes can adopt different topologies (Figure 1) (17), and this structural polymorphism is exploited in applications such as DNA functionalization and nanodevices (18–20). The topology is defined by which bases are engaged in G-quartets, the orientation of the strands in the stem, the position of the loops connecting the four parts of the stem, and the stacking mode of the bases in the stem (all four characteristics being inter-related) (17). Many parameters influence the adopted topology: the nature of the monovalent cation, the concentration, the ionic strength, the pH, the presence of crowding or dehydration agents, and the sequence.
Figure 1.
Structures and their respective schematic topologies of (A) the hybrid-1 24TTG, (22) (B) the hybrid-2 26TTA, (24) (C) the antiparallel 2-quartet 22GT, (25) (D) the antiparallel in sodium 22AG, (27) (E) the antiparallel 2-quartet 22CTA, (32) and (F) the parallel Pu24.(35) These structures were obtained by NMR in 100 mM K+ except for 22AG whose structure was obtained in 100 mM Na+.
The human telomeric sequence, with the repetitive 3′-end sequence d(TTAGGG)n, is one of the most studied biologically relevant G-quadruplexes. Incidentally, this is also one of the most polymorphic G-quadruplex forming sequences (21). Here, we investigated the intramolecular folding of telomeric sequences with four tracts of guanines, in the presence of K+ cations. All sequences contain the central d((GGGTTA)3GGG) core, but the final topology depends on the flanking bases.
In 100 mM K+ (NMR conditions), a hybrid-1 type G-quadruplex structure was observed for the sequence d(TT(GGGTTA)3GGGA) (abbreviated 24TTG, PDB ID: 2GKU (22), Figure 1A, see Table 1 for abbreviations) and for d(TA(GGGTTA)3GGG) (23TAG, PDB ID: 2JSM (23)), the latter being slightly more polymorphic. The sequence d(TTA(GGGTTA)3GGGTT) (26TTA, PDB ID: 2JPZ (24), Figure 1B) folds into a hybrid-2 type G-quadruplex. The hybrid topologies differ by the order of the loops: in the hybrid-1 the loops are respectively 5′-propeller-lateral-lateral-3′ type. For the hybrid-2 the loops are 5′-lateral-lateral-propeller-3′. The sequence d((GGGTTA)3GGGT)) (22GT, PDB ID: 2KF8 (25), Figure 1C) folds predominantly (∼70%) into another topology, called the hybrid-3. Here, only two quartets are formed. The structure is further stabilized by a triplet of guanines, and the circular dichroism spectrum therefore resembles that of the other hybrid structures (26). 22AG (d(A(GGGTTA)3GGG)), a sequence very frequently used for drug binding assays, folds in an antiparallel structure in sodium (Figure 1D) (27) but is polymorphic in KCl (28–30). The structure of 21G (d((GGGTTA)3GGG)) has not been solved by NMR, most likely because it is also polymorphic. It is the shortest possible human telomeric sequence containing four tracks of guanines, and it is frequently used in FRET (Förster Resonance Energy Transfer) melting assays with the FAM and TAMRA dyes at the 5′ and 3′ ends (31). 21G, with and without the fluorescent tags, have also been included in the present study, as well as the FRET homologue of 26TTA. The modified human telomeric DNA 22CTA (d(A(GGGCTA)3GGG), PDB ID: 2KM3 (32), Figure 1E) was also included. This G-quadruplex structure contains two G-quartets plus a G:C:G:C platform.
Table 1. KD values at 20°C of first and second K+ binding to G-quadruplex forming sequences (results of the fitting of the ESI-MS titration curves using DynaFit, with the errors from the fits).
| Abbreviation | Sequence | G4 structure obtained by NMR in 100 mM K+ | KD1 (×10−6 M) | KD2 (×10−6 M) | KD1/KD2 | KD-Glob (×10−9 M2) |
|---|---|---|---|---|---|---|
| 21G | (GGG TTA)3 GGG | Mixture | 18 ± 1 | 1660 ± 40 | 0.01 | 30 ± 2 |
| 22GT | (GGG TTA)3 GGG T | Hybrid-3 (2-quartet antiparallel) (25) | 22 ± 1 | 1050 ± 30 | 0.02 | 23 ± 2 |
| 22CTA | A (GGG CTA)3 GGG | 2-quartet antiparallel (32) | 271 ± 6 | 6500 ± 600 | 0.04 | 1750 ± 200 |
| 22AG | A (GGG TTA)3 GGG | Mixture | 60 ± 4 | 1170 ± 70 | 0.05 | 70 ± 9 |
| F21T | FAM-(GGG TTA)3 GGG-TAMRA | n.d.a | 177 ± 4 | 700 ± 10 | 0.25 | 123 ± 5 |
| 23TAG | TA (GGG TTA)3 GGG | Hybrid-1 (23) | 310 ± 30 | 170 ± 15 | 2 | 50 ± 10 |
| 26TTA | TTA (GGG TTA)3 GGG TT | Hybrid-2 (24) | 1500 ± 100 | 190 ± 20 | 8 | 280 ± 50 |
| F26T | FAM-TTA (GGG TTA)3 GGG TT-TAMRA | n.d.a | 6500 ± 600 | 560 ± 75 | 12 | 3700 ± 800 |
| 24TTG | TT (GGG TTA)3 GGG A | Hybrid-1 (22) | 270 ± 40 | 10.4 ± 1.6 | 26 | 2.7 ± 0.8 |
| Pu24 | TGAG3TG4AG3TG4A2G2 | Parallel (35) | 550 ± 100 | 3.8 ± 0.9 | 145 | 2.2 ± 0.9 |
aG-quadruplex structure, topology not documented.
KD1 and KD2 are defined in Equations (2) and (3). The sequences are ranked with increasing KD1/KD2 ratio.
Telomeric sequences form intramolecular parallel folds only in the crystal state or in molecular crowing conditions, but this species is not significantly populated in dilute aqueous solution (33,34). To include a parallel quadruplex in our panel, we also studied the sequence Pu24 (d(TGAG3TG4AG3TG4A2G2), PDB ID: 2A5P (35), Figure 1F), a variant of the c-myc oncogene promoter nuclease hypersensitivity element.
The structural polymorphism of G-quadruplexes complicates their biophysical studies. The thermodynamically stable folds of telomeric G-quadruplex structures have been well characterized (21) but there is yet no consensus on the folding pathway (36–38). Spectroscopic techniques such as UV-absorbance or UV-melting (39), fluorescence or FRET-melting (40), and circular dichroism (CD) are commonly used. Gibbs free energy of folding can be obtained by assuming a 1-step (2-state) folding (41). However, singular value decomposition (SVD) analysis revealed that thermal folding and unfolding of G-quadruplexes was not 2-state (29,42), and that the isothermal cation-induced folding involves at least three states (37,43). The nature of these states is however still debated. The folding pathway may include a collapse of the unfolded species into hairpin-like structures with G:G base pairs (44–47), which then fold into an antiparallel G-quadruplex (37,48), then interconvert progressively into triplexes intermediates (49–51) before folding properly into a G-quadruplex (37). This model is sequential, and therefore does not consider side-reactions in the mechanism, i.e. ‘parallel’ or ‘branched’ pathways (note that for avoiding confusion between ‘parallel’ G-quadruplex structures and ‘parallel’ reactions in chemical kinetics, we will use the term ‘branched’ instead of ‘parallel’ when talking about mechanisms). Others have proposed branched mechanisms where both the unfolded species, an intermediate (triplex or other) and the G-quadruplex are in equilibrium, with (52) or without intermediates (38,53). Molecular dynamics simulations also suggest that the folding of the human telomeric sequences proceeds through a complex branched mechanism (49,54,55).
A major hurdle in studying the folding mechanisms experimentally is the difficulty in defining the different states (or ensembles) contributing to the mechanism. In the studies cited above, the ensembles were defined based on bulk spectroscopic properties or from single-molecule measurements. In the present study, we used mass spectrometry to separate and quantify the DNA complexes in a different way: with no cation, one potassium ion or two potassium ions specifically bound (56). We studied the equilibrium dissociation constants and the kinetics (on the minute to hour time scale) of K+ binding to several variants of the human telomeric sequences and one c-myc promoter G-quadruplex, at up to 1 mM final KCl concentrations (57). Each ensemble defined based on the potassium binding stoichiometry was then sub-divided based on kinetic behavior, and confirmed by ion mobility spectrometry separation of the mass spectral peaks (IMS-MS). The topology of the ensembles defined are also characterized by CD spectroscopy.
Although we are blind to the details of the early folding events, our experiments allowed to uncover important slow events. While a 1-K+ structure with antiparallel base stacking always forms quickly, hybrid conformations take much longer to form. Branched folding pathways prevail, and the folding energy landscapes are so rugged that misfolded states can linger for several minutes. These results highlight that 2-quartet topologies are worthy of consideration among the biologically relevant ones.
MATERIALS AND METHODS
Materials
Oligonucleotides were purchased in lyophilized and RP-cartridge purified form from Eurogentec (Seraing, Belgium). They were solubilized in nuclease-free grade water from Ambion (Fisher Scientific, Illkirch, France) at approximately 1 mM. The stock concentrations were determined using the Beer-Lambert law, using absorbance at 260 nm on a Uvikon XS, and molar extinction coefficients calculated using the IDT website using the Cavaluzzi-Borer Correction (58). The stock solutions were then diluted to exactly 200 μM. Trimethylammonium acetate (TMAA, Ultra for UPLC, Fluka) and potassium chloride (KCl, >99.999%) were purchased from Sigma-Aldrich (Saint-Quentin Fallavier, France). The pH of the final solutions is 6.9. Table 1 lists the sequences and the abbreviations used in this work.
Electrospray mass spectrometry (ESI-MS)
All equilibrium (ESI-MS) titration experiments were performed in negative ion mode on a LCT Premier mass spectrometer (Waters, Manchester, UK). The titration experiment on the 21G sequence was also repeated on a Bruker Amazon SL mass spectrometer (Bruker, Bremen, Germany). All tuning parameters are given in Supporting Information. Each titration point was obtained from an independent solution (no sequential dilution). The solutions at 10 μM DNA, 100 mM TMAA and the proper amount of KCl were annealed at 85°C for 5 min and cooled at room temperature in the dark for at least one night before measurement in order to avoid any kinetic effect.
The kinetics experiments were carried out on an Agilent 6560 IMS-Q-TOF (Agilent Technologies, Santa Clara, CA, USA) with a dual ESI source in negative ion mode (see supporting information for the tuning parameters). This instrument allows one to monitor the drift time distributions of each m/z range, and this further helps discerning the existence of several ensembles in species of a given stoichiometry. One MS spectrum is accumulated every 1.1 s. The first two minutes are missed or discarded because it is the time lapse to manually mix the solutions and obtain a stable ESI-MS signal and flow rate. The solutions were prepared by mixing the proper volumes of stock solutions (previously annealed in pure water) to reach 10 μM DNA, 100 mM TMAA and the desired KCl concentration. dT6 was also added at 2.5 μM for the mass spectrometry experiments and used as an internal standard to validate that the relative intensities reflect the relative concentrations in solution (see Supporting Information for more detail) (59). The start time of the experiment is the KCl addition (t0). Identical results were obtained whether we annealed the oligonucleotide or not in the TMAA buffer prior to KCl addition.
KD determination and kinetic model evaluation
The concentrations of the species detected by mass spectrometry were obtained after integration of the peaks and subtraction of the nonspecific adducts. We also verified that all complexes have the same response factors. The detailed procedure is reported in the supplementary materials and methods (Supplementary Figures S1–S10). Both KD determination and kinetic model evaluation were carried out using the DynaFit software (4.05.103 BioKin Ltd Watertown, MA, USA) (60). The concentrations of specific complexes of each stoichiometry were fitted as function of the potassium concentration with the chemical Equations (1b) and (2b), correlated to the chemical equilibria (1a) and (2a), respectively.
 |
(1a) |
 |
(1b) |
 |
(2a) |
 |
(2b) |
The DynaFit software works by iteration on the two KD values in order to fit all the concentrations of the three stoichiometries as a function of total KCl added. The KD-Glob is defined by the product of KD1 by KD2.
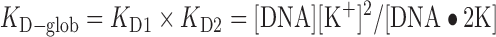 |
(3) |
For the kinetics experiments, we fitted altogether the kinetic data obtained for the 0-K+, specific 1-K+ and 2-K+ ensembles at four different KCl concentrations (100, 200, 500 and 1000 μM), i.e. a total of 12 curves. The chemical reaction models are an input for Dynafit, and the software derives the kinetic differential equations linked with each chemical model, and fits the data. Over 20 different reaction models were tested and ranked using the Akaike information criterion (AIC) (61). The AIC is applicable when the experiments depend on a multitude of parameters, and one is searching for the most useful model to describe the experimental set (62). Several starting initial parameters were tested, and converged to the same results. We also examined manually the fit quality, the errors on the rate constants and their correlation (see results and discussion).
Circular dichroism (CD) spectroscopy
CD is commonly used to characterize the stacking of the guanines in a G-quadruplex, and therefore indirectly gives information on the strand orientations. Parallel G-quadruplexes have only anti-guanines, so anti/anti stacking motifs exist in the resulting topology, and the CD shows a maximum at 260 nm. Antiparallel G-quadruplexes have alternated stacking of the guanines (anti/syn and syn/anti) leading to a minimum in the CD spectrum at 260 nm and a maximum at 290 nm. Hybrid G-quadruplexes have both types of stacking in the same topology, and present two positive peaks at 270 and 290 nm (26). Note that in some cases, 2-quartet structures presenting antiparallel strand orientation can display hybrid-type CD spectra, due to stacking of additional bases on the main two quartets (26).
CD experiments were carried out on solutions prepared exactly as for ESI-MS. Experiments were run on a Jasco J-815 spectrophotometer at 20°C (unless otherwise mentioned)—see supporting information for the instrumental details. CD data were normalized to molar circular-dichroic absoption (Δϵ) based on DNA concentration using Equation (4),
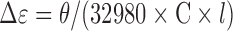 |
(4) |
where θ is the CD ellipticity in milidegrees, C is the DNA concentration in mol/l (in this work: 10−5 mol/l) and l is the pathlength in cm (0.2 cm). The concentrations used for CD are the same as those for MS, to allow direct comparison of the results.
The DynaFit software was used to obtain the molar ellipticity coefficient of each ensemble defined based on ESI-MS, as a function of the wavelength (here, each 5 nm). The software uses the equilibrium or rate constants to calculate the concentration of each ensemble, and then determines the molar response (here the molar ellipticity coefficient) of each ensemble (see supporting information for the data processing details).
RESULTS
K+ binding equilibria
Mass spectrometry titrations reveal a sequence-dependent K+ binding cooperativity
Thanks to the mass resolution, an independent signal is detected for each potassium binding stoichiometry, allowing non-ambiguous stoichiometry determination. Figure 2A–C shows representative mass spectra (zoom on the 5- charge state) obtained upon KCl titration of three human telomeric sequences, 24TTG, 23TAG and 22GT. Full titration experiments, full-scale mass spectra, zooms on other charge states, and results on other sequences are shown in Supplementary Figures S11–S20. After subtracting the nonspecific adducts contribution (which account for ∼15%, ∼30% or ∼70% of the total signal for charge states 6-, 5- and 4-, respectively, see Supplementary Figure S7), we quantify the concentration of specific K+ complexes of each stoichiometry for each point in the titration (Figure 2D–F, for the other sequences, see Supporting Supplementary Figure S21). After this subtraction, only 1-K+ and 2-K+ specific complexes remain. Interestingly, a 1-K+ complex predominates for the shortest sequences (21G, 22GT and 22AG). Although this was expected for 22GT (∼70% is folded into a 2-quartet form), this was more surprising for 22AG and 21G, usually represented as 3-quartet hybrid G-quadruplexes in K+. The 22CTA sequence presents almost exclusively the 1-K+ stoichiometry, as anticipated based on its 2-quartet structure (32).
Figure 2.
(A–C) Mass spectrometry titrations (20°C) of 10 μM (A) 24TTG, (B) 23TAG and (C) 22GT, in 100 mM TMAA with increasing amounts of KCl (zooms on the 5- charge state, see Supplementary Figures S11-S20 for the full scale mass spectra). (D–F) Concentration of the complexes with 0, 1 or 2 specific potassium ions bound in green, blue and orange, respectively, as a function of total KCl concentration, for (D) 24TTG, (E) 23TAG and (F) 22GT. The error bars are the standard errors on the treatment of the three charge states. The solid lines represent the best fits of the models (Equations 2 and 3).
Figure 2D–F and Supporting Supplementary Figure S21 show the best fits and the experimental data for each sequence, and Table 1 summarizes the KD values. The experiments were successfully reproduced on another mass spectrometer (Supporting Supplementary Figure S22). KD1 is smallest for the 21-mer and 22-mer sequences, and the opposite is found for KD2. This indicates that, although these short sequences contain tracts of three guanines, they predominantly adopt a 1-K+ binding stoichiometry in sub-mM KCl concentrations. The KD-Glob values allow to compare the sequences in terms of their propensity to form high amounts of 2-K+ species at equilibrium, whereas the ratio KD1/KD2 reflects the K+ binding cooperativity. As described previously (63), the 2:1 binding can be considered as positively cooperative if KD1/KD2 > 1/4. Interestingly, all sequences containing more than 22 bases (i.e. more than one flanking base) showed positive cooperativity in K+ binding. 24TTG and Pu24 are the most cooperative. Note that 24TTG is a mutated telomeric sequence, which is particularly reputed for being monomorphic (22). The parallel Pu24 G-quadruplex has an even higher K+ binding cooperativity and in equilibrium conditions almost no 1-K+ complex is formed, whatever the KCl concentration.
The effect of the FRET dyes (FAM and TAMRA) on the KD values was also investigated. For 21G, adding the dyes increases KD1 10-fold and decreases KD2 2.4-fold. Adding the dyes therefore increases the cooperativity by a factor of ∼25. For 26TTA, the same effect is observed, but it is less pronounced as this sequence binds K+ already in a more cooperative manner than 21G. The KD-Glob is also affected by the addition of dyes, reflecting that the affinity for potassium (and hence the relative stability of the folded form) of the labeled oligonucleotides decreases compared to the unlabeled analogues. These results highlight that G-quadruplex folding thermodynamics is modified by fluorophore attachment.
The 1-K+ complexes formed at equilibrium are antiparallel
To deduce the predominant topology of each ensemble distinguished by ESI-MS (the specific 0-K+, 1-K+ and 2-K+ complexes), we monitored the same titration experiments by circular dichroism spectroscopy (Figure 3 and Supporting Supplementary Figure S23). Without KCl in solution the DNA sequences are not folded into G-quadruplexes (green), except for 22GT and 21G whose bands at 295 nm could be indicative of a small fraction of folded G-quadruplex. Because of the high ionic strength (100 mM monovalent cation and anion), it is possible that sequences starting with a 5′-guanine tract form secondary structures not requiring K+ binding, similar to those reported by the Plavec group (44). Here we observed the same CD spectra with and without pre-annealing of the solution.
Figure 3.
Circular dichroism titrations (20°C) of 10 μM (A) 24TTG, (B) 23TAG and (C) 22GT in 100 mM TMAA with increasing amount of KCl (0 in yellow, 25, 50, 75, 100, 200, 500 in gray and 1000 μM KCl in red). (D–F), CD spectra reconstructed for the 0-K+ (green), 1-K+ (blue) and 2-K+ (orange) complexes, for sequences (D) 24TTG (the 1-K+ complex was not abundant enough to reconstruct a CD spectrum), (E) 23TAG and (F) 22GT.
The CD spectra change upon KCl addition to the oligonucleotides, indicating a structural change induced by K+ binding. For 24TTG, the folding into a hybrid G-quadruplex occurs at low KCl concentration, as anticipated based on the MS results. The isoelliptic point in the CD titration is in line with the cooperativity observed by ESI-MS: the 1-K+ species not being significantly populated, the transition is 2-state. For 23TAG and 26TTA, the CD spectrum of hybrid structure(s) is progressively formed, but there is no isoelliptic point, and this indicates the presence of more than two states. This is in line with the ESI-MS titrations that revealed three states, the 1-K+ population being significantly populated at intermediate KCl concentrations (Figure 2).
Interestingly, for 21- and 22-mer sequences (21G, 22GT, 22AG and F21T), the amplitude of the CD signals at 260 nm and 295 nm reaches a maximum at about 0.1 mM KCl (Figure 3C and Supporting Supplementary Figure S23C, D and G) and decreases at higher KCl concentrations. In order to obtain structural information for each ensemble detected and quantified by MS, we deconvoluted the CD spectra into those of the 0-K+, 1-K+ and 2-K+ ensembles, using the abundances deduced from ESI-MS. The generated CD spectra are shown in Figure 3D–F and Supporting Supplementary Figure S24.
For 24TTG and Pu24, the CD spectra for the 1-K+ structures were not deconvoluted because of the low population of 1-K+ ensemble (see for example Figure 3A for 24TTG). Also, given that the titration involves only two main states, the CD titration data can be fitted using Equation (3). The KD-Glob obtained by fitting the CD titration agrees with the one obtained by fitting the ESI-MS titration (2.69 × 10−9 M2 versus 2.70 × 10−9 M2, respectively) (Table 1), and this also validates the quantitative mathematical treatment of the ESI-MS data.
For all the human telomeric sequences in which it is significantly populated, the 1-K+ ensemble has clearly an antiparallel-type CD spectrum. For 22GT, based on the published structure (PDB ID: 2KF8 (25)), we interpret the results as follows. The final structure contains two G-quartets and a triplet of guanines, so the two expected cation binding sites are therefore not equivalent. The highest-affinity binding site is most likely the one in-between the two G-quartets, while the lower-affinity binding site is the one between the G-quartet and the G-triplet. One cation suffices to stabilize a 2-quartet G-quadruplex structure, which is antiparallel. Upon addition of higher amounts of KCl and population of the second binding site, the negative band at 260 nm becomes less negative, due to the triplet stacking, resulting in a hybrid-like CD spectrum (26). Mechanistically, given that no dramatic topology rearrangements is required between the 1-K+ and the 2-K+ structure, the K+ binding events could be sequential.
The 21G and 22AG sequences have similar K+ binding characteristics as 22GT, suggesting potentially similar folding pathways. Interestingly, the sequence d(A(GGGTTA)3GGGT), a close derivative of both 22GT and 22AG, was also shown to form an antiparallel G-quadruplex with two quartets in potassium (64). So, although the 22AG sequence folds into an antiparallel 3-quartet G-quadruplex in NaCl (27), it may form a 2-quartet, antiparallel structure at low KCl concentrations. A very similar CD spectrum has previously been deconvoluted by Bončina et al. for 22AG in presence of a ligand (65). The 22CTA sequence behaves differently from 21G, 22AG and 22GT, in that the 1-K+ stoichiometry is the only one significantly populated (with an antiparallel signature in CD, see Supporting Supplementary Figure S24H). The 22CTA NMR structure does not involve G-triplets, and this could prevent the creation of additional K+ binding sites.
In summary, an antiparallel 1-K+ complex, presumably containing two G-quartets, exists at equilibrium, particularly for human telomeric sequences with no or few flanking bases. In such case, a second K+ cation can subsequently bind with lower affinity and without structural change, and a sequential binding pathway is plausible. For sequences containing longer overhangs, the 1-K+ complex is formed only at low KCl concentration and sometimes with low abundance. For those sequences we therefore wanted to determine whether the 1-K+ structure was transiently populated along the folding pathway, and if yes, whether the folding pathways are sequential or branched. To elucidate the pathways, we studied the kinetics of potassium binding to those sequences.
Kinetics of K+ binding
The 1-K+ stoichiometry is transiently populated, even for sequences that bind 2 K+ very cooperatively
On the minute to the hour time scale, we followed the potassium binding stoichiometry by ESI-MS at room temperature (20°C). The recording is started when KCl is added to the solution (t0). After subtraction of the nonspecific adducts, we distinguish and quantify three different stoichiometries (0-K+, 1-K+ and 2-K+) as a function of reaction time.
The least cooperative sequences (21G to F21T in Table 1) form both 1-K+ and 2-K+ ensembles faster than the dead time of our experiment (Supporting Supplementary Figures S25–S29). In contrast, K+ binding to 26TTA, 23TAG and 24TTG was slower (on 20-min to the hour time scale, 26TTA being the fastest, Supporting Supplementary Figures S30-S32). We investigated more in depth the sequences 24TTG, 23TAG and Pu24, because they gave clearer variations of the signals in the KCl concentration range accessible here. Figure 4 shows the temporal evolution of the K+ binding stoichiometries for 24TTG (500 μM KCl) as well as the quantification of the three stoichiometries as a function of time. Those experiments have been quantitatively reproduced on two different MS instruments (Supplementary Figure S33). The transient 1-K+ ensemble is clearly detected even for the very cooperative K+-binding sequence 24TTG (kinetics at other KCl concentrations are shown in supplementary Supplementary Figure S31). This ensemble also appeared for 23TAG and Pu24 (Supplementary Figure S34).
Figure 4.
Kinetics of K+-binding (20°C) monitored by mass spectrometry of 10 μM 24TTG in 100 mM TMAA after adding KCl to reach a final concentration of 500 μM. (A) Mass spectra after addition of KCl at different time (integrated over 1 min accumulation). (B–E) Concentration of the complexes with 0, 1 or 2 potassium in green, blue and orange, respectively, as a function of time after K+ addition. The data were fitted using the models described above each graph.
G-quadruplexes fold via multiply branched pathways
The abundances of 0-K+, 1-K+ and 2-K+ ensembles, measured at four different KCl concentrations (i.e. a total of 12 curves), were fitted globally using over 20 binding models (listed in supporting information Scheme S1). We included some that had been already suggested in the literature. The models, ranked based on the Akaike information criteria—AIC—calculated by DynaFit) are shown in supporting Supplementary Tables S1–S3 for sequences 24TTG, 23TAG and Pu24, respectively. We also visually examined the residuals, and the coefficients of variation (CV) on the fitted rate constants.
The 0-1-2 model, proposed based on UV spectroscopy (66), shows good CV on the rate constants, but very bad residuals when used to fit our data (see Figure 4B and Supplementary Figure S35 for 24TTG), indicating that this model does not properly account for the experimental data. The branched reaction model 0×(1|2|2) with two different 2-K+ complexes is derived from a recent paper from the Schwalbe group (67). The authors demonstrated by NMR that, for 24TTG and 26TTA, the hybrid-2 is formed first before unfolding to form the hybrid-1. We added a 1-K+ ensemble (not mentioned in their study) as another branched reaction. The residuals are lower, but still clearly deviate from zero (Figure 4C and Supplementary Figure S36). We also fitted our data with the model proposed by Chaires (named here 0×((2-1-2)|(2-1-2))) (37), involving first the folding into antiparallel G-quadruplexes, which convert to G-triplexes, before proper folding into hybrid G-quadruplexes. The antiparallel structure was proposed to have three quartets (2-K+), and the triplex ensemble was proposed to bind 1 K+. This model works better, but does convincingly fits of our data even if it contains more species and more adjustable parameters (Supplementary Figure S37). However, if we assume that the antiparallel structures of Chaires’ mechanism have only one K+ ion bound, we reduce the number of species in the mechanism and the fitting becomes more satisfactory (Figure 4D and Supplementary Figure S38, 0×((1-2)|(1-2)) model). The residuals are good, but the CV values are large for most rate constants.
Besides, this is not the only folding mechanism that might fit. Here we have to evaluate models in a framework where the data sets may not cover the whole parameter space of the model: our experiments cover a limited concentration range and time scale, and we separate only three ensembles based on stoichiometry. In the Akaike framework, all tested models are to some extent wrong, but the AIC helps finding the model(s) that maximize predictive accuracy (62). After exploring over 20 binding models, we found that all mechanisms giving satisfactory AIC shared the following characteristics: they have are multiple branches, the two main ones leading respectively to a significantly populated 1-K+ ensemble, and to the final 2-K+ structures. For 24TTG, three ensembles with 2-K+ stoichiometries can be distinguished kinetically: two form slowly on the time scale of the experiment, and one is in fast equilibrium (hence the CVs of its kon and koff are high, and actually only the ratio kon/koff can be reliably determined). The best-ranked model for 24TTG is shown in Figure 4E and is compared with the other best ones in Supplementary Figures S39 and S40.
We performed the same pathway analysis for the kinetics of K+ binding to 23TAG (Supplementary Table S2). The same set of mechanisms with multiple branched reactions were found to fit the best. The most satisfactory fit is obtained with one fewer 2-K+ ensemble (Supplementary Figure S41), using the 0×((1-2)|(1-2)) model. This model is a simplification of the best 0×((1-2)|(1×(2|2))) model, and corresponds to Chaires’ model, as discussed above. The coefficients of variations were high for several rate constants, revealing that two species (1b and 2b) form fast on the time scale of our experiments. The fits using other satisfactory models are shown in supplementary Figures S42–S44.
Finally, in order to test the general applicability of the highly branched pathways, we also studied the folding of the c-myc Pu24 sequence (Supplementary Table S3 and Supplementary Figure S45). The folding of Pu24 is even slower than that of 24TTG (>20 min at 1 mM KCl). Again, we observed clearly a transient 1-K+ complex. We found that the same family of branched mechanisms fits the data. Strikingly, the best-ranked mechanism for the c-myc Pu24 is the same as for the telomeric sequence 24TTG. The values of the rate constants tell us that the 1b and 2b ensembles are in fast equilibrium with the 0 and 1a ensembles, respectively. In summary, highly branched folding pathways with multiple branched reactions is valid for G-quadruplexes of parallel topology as well as for the telomeric sequences, and therefore potentially constitute a tenet of G-quadruplex folding.
Ion mobility spectrometry confirms the presence of up to three distinct ensembles with two potassium cations bound
Separation methods such as size-exclusion chromatography (SEC) are able to separate multimers but not G-quadruplexes of different topologies (68). For this reason, we used ion mobility spectrometry (56,69–71) to separate in the gas phase ions of a given mass-to-charge ratio according to a second dimension linked to their shape. For a given mass and charge, the time tD needed for the ions to drift through the helium-filled cell, in which a uniform electric field is applied, will be influenced by the ion shape: a more extended conformer will be subjected to more friction and will travel more slowly. For G-quadruplex structures, the G-stack core is preserved in the gas phase upon gentle ionization and ion transfer conditions (72). If several solution ensembles give rise to m/z ensembles of significantly different collision cross sections, these will be separated based on their drift time.
A full scale IMS-MS spectrum is shown in the supplementary Figure S46. For each potassium binding stoichiometry, we integrated the ion mobility drift time distribution, at different reaction time intervals following KCl addition (Figure 5A and B for (24TTG)4− and (23TAG)4− respectively at different time points after addition of 500 μM KCl; for the other KCl concentrations and for charge state 5-, see supplementary Figures S47–S49). The drift time distribution reflects the distribution of shapes for each K+ binding stoichiometry. For the 2-K+ complex of 24TTG, we observe very clearly three peaks at short reaction time scale, indicating three conformational ensembles for the 2-K+ stoichiometry. This observation validates a mechanism with three different species having 2-K+, and discards the 0×((1-2)|(1-2)) and 0×(1|1×(2|2)) models.
Figure 5.
(A and B) Arrival time distribution in function of the time for the 4- charge state for the 2-K+ stoichiometry at 500 μM KCl from green (3 min) to red (24h) through gray, respectively for 10 μM (A) 24TTG and (B) 23TAG. On each panel, arrival time distributions were area-normalized for visualization of the relative abundances. (C and D) Simulation of the concentrations of the species obtained using the best models at 500 μM KCl: (C) 0×((1-2)|(1×(2|2))) for 24TTG and (D) 0×((1-2)|(1-2))) for 23TAG. (E and F) Deconvoluted CD spectra for the ensembles defined by the kinetics model, respectively for (E) 24TTG and (F) 23TAG.
Using the kinetic parameters obtained from the fitting we simulated the fraction of each of the ensembles as a function of time (see Figure 5C for the best model 0×((1-2)|(1×(2|2))) on 24TTG, and Supplementary Figures S38–S40 for the other models). The three complexes with 2-K+ appear at different time scales. One is formed fastest then disappears (2c), one forms slowest (2a) and the last one forms with intermediate speed (2b). Consistent results between simulated relative abundances and ion mobility peak abundances are obtained at the other KCl concentrations for the mechanisms.
For 23TAG, the simpler 0×((1-2)|(1-2)) model is compatible with the IMS data: only two ensembles are distinguished by ion mobility on the 2-K+ stoichiometry (Figure 5B Supplementary Figure and S49). For the 1-K+ stoichiometry, we observe only one broad mobility peak (Supplementary Figures S47–S49). This is also consistent with the abundances predicted by the best mechanisms: indeed, only one of the two 1-K+ species (1b) is significantly populated. For Pu24, ion mobility separation gives only one 2-K+ ensemble (not shown). Note however that, although several ion mobility peaks indicate several populations initially in solution, the reverse is not true: several populations can end up in structures of very similar collision cross sections, not separable by ion mobility spectrometry.
Circular dichroism characterization of the transient species
To obtain more structural information on the sub-ensembles, we investigated their stacking arrangements by CD spectroscopy. The binding/folding kinetics were monitored by CD and correlated with the abundances predicted from ESI-MS (Supplementary Figures S50-58). Using a global fitting with the best models, we generated the CD spectra for each kinetically distinguishable species that is significantly populated (i.e. species 0, 1b, 2a, 2b and 2c for 24TTG, see Figure 5E and Supplementary Figure S38-40, bottom panel). In practice, the lowly populated 1a species was discarded by setting its Δϵ to zero at all wavelengths. Similarly, we extracted the CD spectra of four species for 23TAG (Figure 5F and Supplementary Figure S41), and five for Pu24 (Supplementary Figure S45).
For all telomeric sequences, the 0-K+ species’ CD is the one at the starting point of the kinetic. Two species with 2-K+ (2a and 2b) present CD spectra typical of hybrid G-quadruplex stacking (maxima at both 295 and 265 nm). They however differ around 245 nm. There is currently no interpretation of the 245-nm CD signal in terms of structure, and this subject may be worthy of further investigation. For 24TTG, the third 2-K+ species (2c), is never very abundant over time, so the molar ellipticity coefficients are entailed with larger errors and the reconstruction of its CD spectrum gave different results depending on the kinetic model. Using the model 0×((1-2)|(1×(2|2))), a negative band at around 270–280 nm is obtained, which is reminiscent of the recently published CD spectrum of a left handed G-quadruplex (73) (even though less intense in our case). Using the model 0×(1|2|(1×(2|2))) (Supplementary Figure S40), however, two positive peaks between 250 and 290 nm are obtained, reminiscent of the CD spectrum reported for the Giardia telomeric sequence d(TAGGG)4, which folds into an antiparallel 2-quartet form (26,74). Both structural hypotheses for these off-pathway 2-K+ folds are worthy of future investigation by higher-resolution techniques or by molecular modeling.
The other major off-pathway structure of 24TTG, i.e. the 1-K+ ensemble ‘1b’, robustly presents an antiparallel-type CD spectrum, independently of the fitting model. For 23TAG as well, species ‘1b’ shows a negative band around 260–270 nm and a positive band around 290 nm (Supplementary Figure S41). This highlights again the prevalence of 1-K+/2-quartet antiparallel structures in the folding mechanism, which is off-pathway as compared to the most stable 2-K+ species. The deconvolution of the CD for Pu24 gave intriguing results as well (Supplementary Figure S45). Both off-pathway species (1b and 2c) display positive CD signals at 260 and 290 nm. The spectrum of 1b is again very similar to the CD spectrum reported for the 2-quartet antiparallel Giardia telomeric sequence d(TAGGG)4. The CD spectrum of 2c, which forms sequentially from 1b, is similar to that of 1b but with a higher intensity for the positive peak at 260 nm, and this could be due to an additional homo-stacking on the ‘Giardia-like’ antiparallel 2-quartet structure.
Finally, to check if the proposed mechanisms also hold for higher KCl concentrations and temperatures, we monitored the folding of 24TTG and Pu24 by CD at 37°C with 1 and 25 mM KCl (Supplementary Figures S55 and S58). For 24TTG, the folding remained slow even at 25 mM KCl, as also shown by Chaires’ group (37). The spectrum recorded <0.8 min after the addition of KCl (highlighted in black in Supplementary Figure S55) presents a significant peak at 295 nm, typical of antiparallel arrangements. At 37°C, the folding of Pu24 is faster (Supplementary Figure S58), yet the short time scale spectrum presents a band at 298 nm as also monitored in the previous conditions. The folding kinetics monitored at 298 nm showed that this peak persists for about 5 min. These experiments confirm that off-pathway antiparallel structures linger for minutes, even at higher KCl concentration and physiological temperature.
DISCUSSION
In summary, our results suggest a general G-quadruplex folding landscape in which 1-K+ complexes, having presumably antiparallel 2-quartet structures, form fast, but are off-pathway as compared to the final states. These antiparallel 2-quartet structures can therefore be considered as ‘misfolded’ structures.
The 1-K+ antiparallel G-quadruplex structures
Potassium ions can bind with very different cooperativity to different telomeric sequences. The shortest sequences have the lowest K+ binding cooperativity, and the 1-K+ structure predominates at low KCl concentration. This was unexpected for sequences such as 21G and 22AG, but for sequences 22GT and 22CTA this observation is in line their 2-quartet structure (hence presumably one preferred inter-quartet K+ binding site). The Chaires group measured indirectly the K+/oligonucleotide stoichiometry by measuring the K+ free concentration with a fluorescent probe, and deduced that a wide range of stoichiometries can be formed (75). By MS, we observe directly the complexes with only one or two specific potassium ions, the rest being nonspecific adducts that also form in non-G-quadruplex sequences. The Chaires group also observed a complete folding at 2–3 mM KCl, concentration at which the K+ binding stoichiometry is 2:1 (K:DNA) (43), and this fully agrees with our results. Higher stoichiometries described previously could therefore come from nonspecific interaction with the negatively charged backbone (14). Single molecule FRET imaging also revealed two folded ensembles for 21G (d((GGGTTA)3GGG)) (52), or 23TAG (d(TA(GGGTTA)3)) in which a low FRET ensemble is mainly present at low KCl concentration (76). In both studies, those ensembles were associated to parallel and antiparallel/hybrid structures. Based on our results, we would propose the ‘antiparallel’ species to be the 1-K+/2-quartet antiparallel G-quadruplex and the ‘parallel’ ones to be the folded hybrid G-quadruplexes.
Using differential scanning calorimetry (DSC), the Vesnaver group proposed a folding mechanism for 22AG in which both a folded, an unfolded and an intermediate are in equilibrium (53). The authors also estimated the difference in number of cations bound to these three states, and found 1.5 ± 0.1 cations for the intermediate and 3.2 ± 0.2 for the final folded structure. The numbers of cations bound, extracted from fitting, do not match the integer numbers detected directly by mass spectrometry, but the difference could again reflect the extra nonspecific adducts. The intermediate is often considered in the literature as a triplex, but in light of our results we rather propose the major intermediate ensemble to be antiparallel structures with one K+ and two G-quartets, and the final form to be hybrid structures with 2-K+ and, depending on the sequences, either three G-quartets or two G-quartets and one G-triplet.
Increasing the number of flanking bases decreases the population of the 1-K+ complex to the profit of the 2-K+ one. Vorlìčková's group reported that if flanking bases are added the CD signature becomes less antiparallel-like and more hybrid-like (77). Here, we bring the crucial information that the antiparallel structures actually contain only one specifically bound potassium ion.
The sequence-dependent K+ binding cooperativity
Past reports based on DSC also showed that the ΔG° of folding of human telomeric G-quadruplexes could vary by up to ∼7 kcal/mol from one human telomeric sequence to another (78). The large differences can actually be correlated to the influence of a single nucleotide on the K+ binding constants and binding cooperativity. Here, we show that the K+ binding constants (and therefore the ΔG° values) are highly influenced even by a single flanking base or by flanking fluorophores.
In traditional spectroscopic methods, the binding cooperativity can be assessed by the Hill coefficient (79). For the short variant 21G (d((GGGTTA)3GGG)) (66), a Hill coefficient of 0.9 ± 0.1 was obtained by the Balasubramanian group. This indicates a non-cooperative binding of K+, in agreement with our results on the shortest sequences. The authors also reported a positive cooperativity with a Hill coefficient of 1.8 ± 0.1 for TERRA, a parallel RNA G-quadruplex. Our experiments on Pu24 show that the K+ binding to this parallel G-quadruplex is also very cooperative. The Chaires group also commented on potassium (and sodium) binding cooperativity, and showed that adding nucleotides increases the binding cooperativity (80). Our results are in line with previous studies, but the ability to quantitatively determine the individual KD values for the first and second potassium binding event (Equations 2 and 3, Table 1) refines the interpretation of equilibrium binding data obtained from calorimetric or spectroscopic studies. Similar effects have been reported when terminal thymine residues are added to synthetic sequences (81), or when fluorescent probes increase the K+ binding cooperativity and modify the folding thermodynamics (31).
A generalized G-quadruplex folding pathway
Several G-quadruplex folding pathways have been proposed previously, including intermediates such as hairpins and triplexes. Thanks to the ability to separate populations based on the K+ binding stoichiometry and on distinct kinetic ensembles, we can propose the generalized folding landscape shown in Figure 6. The kinetic data revealed a complex mechanism with several branched reactions resulting in several ‘branches’ in the folding landscape. At least five groups of structures are involved for 23TAG and six for 24TTG: one unfolded ensemble, two ensembles with only one potassium cation and two or three ensembles with two cations.
Figure 6.
Proposed folding pathway. The number of cations is shown by the number of green circles, and the quartet guanine relative orientations are shown by arrows. Example topologies compatible with each ensemble are shown in gray (anti bases in light gray, syn bases in dark gray).
The sequences fold into the 1-K+ antiparallel G-quadruplex (1b), then grab one more cation (2c) to form a 22GT-like structure (or a Giardia-like structure). The reason this pathway is the fastest may lie in probability: there are many more ways to form a misaligned (or ‘misfolded’) 2-quartet quadruplex than ways to form a perfectly aligned 3-quartet structure, so although the latter may be enthalpically favorable, the former may be entropically favorable. Future temperature-dependent experiments or calorimetric experiments will help in validating this hypothesis. Importantly, the 1-K+ antiparallel ensemble is significantly populated at the beginning of the reaction and persists on time scales up to 20 min at 20°C and 1 mM KCl (even though it is not a thermodynamically stable final state for all sequences longer than 22-nt). It is still detected at 37°C and 25 mM KCl. These structures later rearrange into the proper hybrid-1 (2a) and/or -2 (2b), but to do so they must first completely turn back to unfolded forms without specifically bound cations. This turning back is complete for sequences 23TAG and 24TTG. Previous kinetic studies indicated that the folding of 24TTG is about five times slower than for 22AG when initiated with 25 mM KCl addition (80) and we observe the same trend. The turning back may however not always be complete, and some sequences may permanently have part of their population sampling the ‘misfolded’ pathway. Chaires also observed that the folding of 22AG is faster than sequences with flanking bases (37). It is possible that the lack of flanking bases lowers the steric hindrance, stabilizing the ‘2c’ complex and therefore stopping the folding in this state. The folding landscape proposed therefore also holds for those short sequences, which preferentially take the fastest branch (0-1b-2c).
Previous works have been devoted to elucidating the folding pathways of G-quadruplexes (82), and multi-pathway mechanisms were also predicted structurally by the extensive modeling work of the Sponer group (49,55). Hairpins and triplexes are commonly invoked reaction intermediates. Where would these intermediates fit within our classification according to the number of specific cations bound to each state? Mashimo et al. (83) proposes a collapse from the unfolded species to a hairpin ensemble, and the Plavec group has demonstrated cation-independent formation of GG base pairs (44). Hairpins would therefore belong to our 0-K+ ensemble. The G-triplex is also a commonly invoked intermediate (37,38,53). Our experiments could neither directly confirm nor dismiss any triplex structure. However, taking into account that molecular modeling shows significant cation affinity for the triplets (49), and that the control sequences used to subtract nonspecific adducts contained fewer than three G-tracts, a G-triplex structure is a good candidate for our low-abundance 1-K+ intermediate 1a. Based on existing data (67), we attributed the firstly folded 2-K+ hybrid G-quadruplex to the hybrid-2, whereas the hybrid-1 is formed at longer time scales. The rate constants then suggest that intermediate 1a is structurally closer to the hybrid-2 than to the hybrid-1 structure. Molecular modeling shows that hairpins and G-triplexes are stable only when their strands are antiparallel (49,54). We therefore propose that for telomeric sequences, the 1a intermediate would have its 5′-end folded in an antiparallel manner. The fact that 1a is not abundantly populated means that if its formation is the rate-limiting step en route to folding into perfect G-quadruplex structures.
Interestingly, the folding landscape proposed here holds also for the Pu24 sequence (the only difference being that the final 2-K+ structures are parallel). A branched mechanism is needed to fit the data, and an antiparallel 2-quartet structure is consistently a prominent off-pathway species. The mechanism is also valid for the recently studied synthetic sequences d((G3T2)3G3) and d(T(G3T2)3G3T) (81), which were presented as artificial structural switches that respond to cation presence and cation concentration.
CONCLUSION
The detection and quantification of the 1-K+ and 2-K+ ensembles, made possible by ESI-MS, provide novel insight into the folding pathway of DNA G-quadruplexes. Unlike the sequential folding pathway that is typically considered, we demonstrated that misfolded species may be formed, and significantly influence the global folding kinetics. In that respect, nucleic acids (and especially G-quadruplex folding) differs from proteins in that their folding energy landscape is roughened by numerous hydrogen bonds and ionic interactions, which create high barriers between ensembles. As a result, misfolded structure are not just a mere frustration along the folding pathway, but can persist on the second to the minute time scale.
The importance of 2-quartet misfolded structures should therefore not be ignored. For example, 2-quartet structures have been found in the context of longer, more biologically-relevant sequences (84). In the oligonucleotide sequences studied here, often used for biophysical studies, the 1-K+ structures are not negligible. Our results also highlight the critical effect of choosing the exact sequence (terminal bases or mutations) and of labeling the sequences, as those modifications can drive the folding into different pathways. This aspect should be taken into account for future studies, and in comparing published results obtained on different sequence variants.
Admittedly the potassium concentration and temperature of our experimental studies are not the physiologically relevant ones. Changing the concentrations will change the relative abundances of the different ensembles in the population, yet the folding pathway is an inherent property of the system. Besides, even if 2-quartet ‘misfolded’ quadruplexes are a minor part of the population, conformational selection processes may still fish them out. The implications of the proposed folding pathway are therefore also important in the field of G-quadruplex ligands design. Indeed, the characterization of ligand binding to G-quadruplexes should not be limited to binding affinity, but should also consider ligand-induced changes in the target conformation. It was recently shown that the binding of some of the most potent ligands known to date (360A, Phen-DC3 and pyridostatin) was accompanied by the ejection of one cation (85,86), suggesting that the ligands preferentially bind to 2-quartet antiparallel structures. Species that are off-pathway in the folding of model oligonucleotides in vitro can become the predominant species in another environment. One should therefore not overlook that ligands and/or proteins may displace the equilibrium toward the ‘misfolded’ 1-K+ antiparallel structures. Better understanding of the folding pathways, including minor branches, may help revealing new structural targets and new strategies for inter-G-quadruplex selectivity.
Supplementary Material
Acknowledgments
The authors acknowledge Miho Takematsu for technical assistance and Frédéric Rosu for technical support and reading the manuscript. We thank the Structural Biophysical Chemistry platform of the Institut Européen de Chimie et Biologie (CNRS UMS 3033/Inserm US001/Univ. Bordeaux) for access to the mass spectrometers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Inserm [ATIP-Avenir Grant no. R12086GS]; Conseil Régional Aquitaine [20121304005]; EU Seventh Framework Programme [FP7-PEOPLE-2012-CIG-333611]. Funding for open access charge: FP7 Marie Curie Career Integration Grant ‘BIOPHYMS’.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maizels N., Gray L.T. The G4 Genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. An RNA G-quadruplex in the 5’ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahoon L.A., Seifert H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugaut A., Balasubramanian S. 5’-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogoi S., Xodo L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D., Guo K., Rusche J.J., Hurley L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Patel D.J. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry. 1992;31:8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- 10.Mergny J.-L., Riou J.-F., Mailliet P., Teulade-Fichou M.-P., Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzo A., Salvati E., Porru M., D'Angelo C., Stevens M.F., D'Incalci M., Leonetti C., Gilson E., Zupi G., Biroccio A. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res. 2009;37:5353–5364. doi: 10.1093/nar/gkp582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 14.Wong A., Wu G. Selective binding of monovalent cations to the stacking G-quartet structure formed by guanosine 5‘-monophosphate: a solid-state NMR study. J. Am. Chem. Soc. 2003;125:13895–13905. doi: 10.1021/ja0302174. [DOI] [PubMed] [Google Scholar]
- 15.Largy E., Mergny J.-L., Gabelica V. Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. In: Sigel A, Sigel H, Sigel RKO, editors. The Alkali Metal Ions: Their Role for Life. Springer; 2016. pp. 203–258. [DOI] [PubMed] [Google Scholar]
- 16.Parkinson G.N. Fundamentals of quadruplex structures. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge: Royal Society of Chemistry; 2006. pp. 1–30. [Google Scholar]
- 17.Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chemistry. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- 18.Davis J.T. G-quartets 40 years later: from 5’-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 19.Tintoré M., Eritja R., Fábrega C. DNA nanoarchitectures: steps towards biological applications. ChemBioChem. 2014;15:1374–1390. doi: 10.1002/cbic.201402014. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J., Amrane S., Korkut D.N., Bourdoncle A., He H.-Z., Ma D.-L., Mergny J.-L. Combination of i-motif and G-quadruplex structures within the same strand: formation and application. Angew. Chemie Int. Ed. 2013;52:7742–7746. doi: 10.1002/anie.201301278. [DOI] [PubMed] [Google Scholar]
- 21.Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 22.Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phan A.T., Kuryavyi V., Luu K.N., Patel D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai J., Carver M., Punchihewa C., Jones R.A., Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim K.W., Amrane S., Bouaziz S., Xu W., Mu Y., Patel D.J., Luu K.N., Phan A.T. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karsisiotis A.I., Hessari N.M., Novellino E., Spada G.P., Randazzo A., Webba da Silva M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. Engl. 2011;50:10645–10648. doi: 10.1002/anie.201105193. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d(AG3(T2AG3)3) G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 28.Gray R.D., Petraccone L., Trent J.O., Chaires J.B. Characterization of a K+-induced conformational switch in a human telomeric DNA oligonucleotide using 2-aminopurine fluorescence. Biochemistry. 2010;49:179–194. doi: 10.1021/bi901357r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buscaglia R., Gray R.D., Chaires J.B. Thermodynamic characterization of human telomere quadruplex unfolding. Biopolymers. 2013;99:1006–1018. doi: 10.1002/bip.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller M.C., Buscaglia R., Chaires J.B., Lane A.N., Trent J.O. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3’ sequence of d(AG3(TTAG3)3) J. Am. Chem. Soc. 2010;132:17105–17107. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- 31.De Rache A., Mergny J.-L. Assessment of selectivity of G-quadruplex ligands via an optimised FRET melting assay. Biochimie. 2015;115:194–202. doi: 10.1016/j.biochi.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Lim K.W., Alberti P., Guédin A., Lacroix L., Riou J.F., Royle N.J., Mergny J.L., Phan A.T. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G·C·G·C tetrad. Nucleic Acids Res. 2009;37:6239–6248. doi: 10.1093/nar/gkp630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansel R., Lohr F., Foldynova-Trantirkova S., Bamberg E., Trantirek L., Dotsch V. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011;39:5768–5775. doi: 10.1093/nar/gkr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., Correia J.J., Wang L., Trent J.O., Chaires J.B. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phan A.T., Kuryavyi V., Gaw H.Y., Patel D.J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabelica V. A pilgrim's guide to G-quadruplex nucleic acid folding. Biochimie. 2014;105:1–3. doi: 10.1016/j.biochi.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Gray R.D., Trent J.O., Chaires J.B. Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 2014;426:1629–50. doi: 10.1016/j.jmb.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koirala D., Ghimire C., Bohrer C., Sannohe Y., Sugiyama H., Mao H. Long-loop G-quadruplexes are misfolded population minorities with fast transition kinetics in human telomeric sequences. J. Am. Chem. Soc. 2013;135:2235–2241. doi: 10.1021/ja309668t. [DOI] [PubMed] [Google Scholar]
- 39.Mergny J.-L., Lacroix L. Current Protocols in Nucleic Acid Chemistry. Hoboken: John Wiley & Sons, Inc; 2009. UV Melting of G-Quadruplexes; pp. 17.1.1–17.1.15. [DOI] [PubMed] [Google Scholar]
- 40.De Cian A., Guittat L., Kaiser M., Saccà B., Amrane S., Bourdoncle A., Alberti P., Teulade-Fichou M.-P., Lacroix L., Mergny J.-L. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007;42:183–195. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 41.Mergny J., Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;537:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 42.Gray R.D., Buscaglia R., Chaires J.B. Populated intermediates in the thermal unfolding of the human telomeric quadruplex. J. Am. Chem. Soc. 2012;134:16834–16844. doi: 10.1021/ja307543z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray R.D., Chaires J.B. Isothermal folding of G-quadruplexes. Methods. 2012;57:47–55. doi: 10.1016/j.ymeth.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ceru S., Sket P., Prislan I., Lah J., Plavec J. A new pathway of DNA G-quadruplex formation. Angew. Chem. Int. Ed. Engl. 2014;53:4881–4884. doi: 10.1002/anie.201400531. [DOI] [PubMed] [Google Scholar]
- 45.Mashimo T., Sugiyama H. Folding pathways of human telomeric hybrid G-quadruplex structure. Nucleic Acids Symp. Ser. (Oxf). 2007 doi: 10.1093/nass/nrm120. doi:10.1093/nass/nrm120. [DOI] [PubMed] [Google Scholar]
- 46.Mashimo T., Sannohe Y., Yagi H., Sugiyama H. Folding pathways of hybrid-1 and hybrid-2 G-quadruplex structures. Nucleic Acids Symp. Ser. (Oxf). 2008 doi: 10.1093/nass/nrn208. doi:10.1093/nass/nrn208. [DOI] [PubMed] [Google Scholar]
- 47.Rajendran A., Endo M., Hidaka K., Sugiyama H. Direct and single-molecule visualization of the solution-state structures of G-hairpin and G-triplex intermediates. Angew. Chem. Int. Ed. 2014;53:4107–4112. doi: 10.1002/anie.201308903. [DOI] [PubMed] [Google Scholar]
- 48.Su D.G.T., Fang H., Gross M.L., Taylor J.-S.A. Photocrosslinking of human telomeric G-quadruplex loops by anti cyclobutane thymine dimer formation. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12861–12866. doi: 10.1073/pnas.0902386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stadlbauer P., Trantírek L., Cheatham T.E., Koča J., Sponer J. Triplex intermediates in folding of human telomeric quadruplexes probed by microsecond-scale molecular dynamics simulations. Biochimie. 2014;105:22–35. doi: 10.1016/j.biochi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Limongelli V., De Tito S., Cerofolini L., Fragai M., Pagano B., Trotta R., Cosconati S., Marinelli L., Novellino E., Bertini I., et al. The G-triplex DNA. Angew. Chem. Int. Ed. Engl. 2013;52:2269–2273. doi: 10.1002/anie.201206522. [DOI] [PubMed] [Google Scholar]
- 51.Koirala D., Mashimo T., Sannohe Y., Yu Z., Mao H., Sugiyama H. Intramolecular folding in three tandem guanine repeats of human telomeric DNA. Chem. Commun. (Camb). 2012;48:2006–2008. doi: 10.1039/c2cc16752b. [DOI] [PubMed] [Google Scholar]
- 52.Lee J.Y., Okumus B., Kim D.S., Ha T. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bončina M., Lah J., Prislan I., Vesnaver G. Energetic basis of human telomeric DNA folding into G-quadruplex structures. J. Am. Chem. Soc. 2012;134:9657–9663. doi: 10.1021/ja300605n. [DOI] [PubMed] [Google Scholar]
- 54.Stadlbauer P., Kührová P., Banáš P., Koča J., Bussi G., Trantírek L., Otyepka M., Šponer J. Hairpins participating in folding of human telomeric sequence quadruplexes studied by standard and T-REMD simulations. Nucleic Acids Res. 2015;43:9626–9644. doi: 10.1093/nar/gkv994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stadlbauer P., Krepl M., Cheatham T.E., Koca J., Sponer J. Structural dynamics of possible late-stage intermediates in folding of quadruplex DNA studied by molecular simulations. Nucleic Acids Res. 2013;41:7128–7143. doi: 10.1093/nar/gkt412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferreira R., Marchand A., Gabelica V. Mass spectrometry and ion mobility spectrometry of G-quadruplexes. A study of solvent effects on dimer formation and structural transitions in the telomeric DNA sequence d(TAGGGTTAGGGT) Methods. 2012;57:56–63. doi: 10.1016/j.ymeth.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 57.Marchand A., Gabelica V. Native electrospray mass spectrometry of DNA G-quadruplexes in potassium solution. J. Am. Soc. Mass Spectrom. 2014;25:1146–1154. doi: 10.1007/s13361-014-0890-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cavaluzzi M.J., Borer P.N. Revised UV extinction coefficients for nucleoside-5’-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gabelica V., Rosu F., De Pauw E. A simple method to determine electrospray response factors of noncovalent complexes. Anal. Chem. 2009;81:6708–6715. doi: 10.1021/ac900785m. [DOI] [PubMed] [Google Scholar]
- 60.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 61.Akaike H. Information theory and an extension of the maximum likelihood principle. Int. Symp. Inf. Theory. 1973 doi:10.1016/j.econlet.2011.12.027. [Google Scholar]
- 62.Aho K., Derryberry D., Peterson T. Model selection for ecologists: the worldviews of AIC and BIC. Ecology. 2014;95:631–636. doi: 10.1890/13-1452.1. [DOI] [PubMed] [Google Scholar]
- 63.Rosu F., De Pauw E., Gabelica V. Electrospray mass spectrometry to study drug-nucleic acids interactions. Biochimie. 2008;90:1074–1087. doi: 10.1016/j.biochi.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Z., Dai J., Veliath E., Jones R.A., Yang D. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010;38:1009–1021. doi: 10.1093/nar/gkp1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bončina M., Hamon F., Islam B., Teulade-Fichou M.-P., Vesnaver G., Haider S., Lah J. Dominant driving forces in human telomere quadruplex binding-induced structural alterations. Biophys. J. 2015;108:2903–2911. doi: 10.1016/j.bpj.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang A.Y.Q., Balasubramanian S. The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J. Am. Chem. Soc. 2012;134:19297–19308. doi: 10.1021/ja309851t. [DOI] [PubMed] [Google Scholar]
- 67.Bessi I., Jonker H.R.A., Richter C., Schwalbe H. Involvement of long-lived intermediate states in the complex folding pathway of the human telomeric G-quadruplex. Angew. Chem. 2015;127:8564–8568. doi: 10.1002/anie.201502286. [DOI] [PubMed] [Google Scholar]
- 68.Largy E., Mergny J.-L. Shape matters: size-exclusion HPLC for the study of nucleic acid structural polymorphism. Nucleic Acids Res. 2014;42:e149. doi: 10.1093/nar/gku751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bohrer B.C., Merenbloom S.I., Koeniger S.L., Hilderbrand A.E., Clemmer D.E. Biomolecule analysis by ion mobility spectrometry. Annu. Rev. Anal. Chem. 2008;1:293–327. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gabelica V., Baker E.S., Teulade-Fichou M.-P., De Pauw E., Bowers M.T. Stabilization and structure of telomeric and c-myc region intramolecular G-quadruplexes: the role of central cations and small planar ligands. J. Am. Chem. Soc. 2007;129:895–904. doi: 10.1021/ja065989p. [DOI] [PubMed] [Google Scholar]
- 71.Shi L., Holliday A.E., Shi H., Zhu F., Ewing M.A., Russell D.H., Clemmer D.E. Characterizing intermediates along the transition from polyproline I to polyproline II using ion mobility spectrometry-mass spectrometry. J. Am. Chem. Soc. 2014;136:12702–12711. doi: 10.1021/ja505899g. [DOI] [PubMed] [Google Scholar]
- 72.Balthasart F., Plavec J., Gabelica V. Ammonium ion binding to DNA G-quadruplexes: do electrospray mass spectra faithfully reflect the solution-phase species. J. Am. Soc. Mass Spectrom. 2013;24:1–8. doi: 10.1007/s13361-013-0649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chung W.J., Heddi B., Schmitt E., Lim K.W., Mechulam Y., Phan A.T. Structure of a left-handed DNA G-quadruplex. Proc. Natl. Acad. Sci. U.S.A. 2015;112:2729–2733. doi: 10.1073/pnas.1418718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu L., Lim K.W., Bouaziz S., Phan A.T. Giardia telomeric sequence d(TAGGG) 4 forms two intramolecular G-quadruplexes in K + solution: effect of loop length and sequence on the folding topology. J. Am. Chem. Soc. 2009;131:16824–16831. doi: 10.1021/ja905611c. [DOI] [PubMed] [Google Scholar]
- 75.Gray R.D., Chaires J.B. Linkage of cation binding and folding in human telomeric quadruplex DNA. Biophys. Chem. 2011;159:205–209. doi: 10.1016/j.bpc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Long X., Stone M.D. Kinetic partitioning modulates human telomere DNA G-quadruplex structural polymorphism. PLoS One. 2013;8:e83420. doi: 10.1371/journal.pone.0083420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Renciuk D., Kejnovská I., Skoláková P., Bednárová K., Motlová J., Vorlícková M. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009;37:6625–6634. doi: 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lane A.N., Chaires J.B., Gray R.D., Trent J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weiss J.N. The Hill equation revisited: uses and misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- 80.Gray R.D., Chaires J.B. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Largy E., Marchand A., Amrane S., Gabelica V., Mergny J.-L. Quadruplex turncoats: cation-dependent folding and stability of quadruplex-DNA double switches. J. Am. Chem. Soc. 2016;138:2780–2792. doi: 10.1021/jacs.5b13130. [DOI] [PubMed] [Google Scholar]
- 82.Chaires J.B. Human telomeric G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J. 2010;277:1098–1106. doi: 10.1111/j.1742-4658.2009.07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mashimo T., Yagi H., Sannohe Y., Rajendran A., Sugiyama H. Folding pathways of human telomeric type-1 and type-2 G-Quadruplex structures. J. Am. Chem. Soc. 2010;132:14910–14918. doi: 10.1021/ja105806u. [DOI] [PubMed] [Google Scholar]
- 84.Hänsel R., Löhr F., Trantirek L., Dötsch V. High-resolution insight into G-overhang architecture. J. Am. Chem. Soc. 2013;135:2816–2824. doi: 10.1021/ja312403b. [DOI] [PubMed] [Google Scholar]
- 85.Marchand A., Granzhan A., Iida K., Tsushima Y., Ma Y., Nagasawa K., Teulade-Fichou M.-P., Gabelica V. Ligand-induced conformational changes with cation ejection upon binding to human telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2015;137:750–756. doi: 10.1021/ja5099403. [DOI] [PubMed] [Google Scholar]
- 86.Bončina M., Podlipnik Č., Piantanida I., Eilmes J., Teulade-Fichou M.-P., Vesnaver G., Lah J. Thermodynamic fingerprints of ligand binding to human telomeric G-quadruplexes. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1167. doi:10.1093/nar/gkv1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








