Abstract
Background: Identifying genes that are essential for mouse embryonic development and survival through term is a powerful and unbiased way to discover possible genetic determinants of human developmental disorders. Characterising the changes in mouse embryos that result from ablation of lethal genes is a necessary first step towards uncovering their role in normal embryonic development and establishing any correlates amongst human congenital abnormalities.
Methods: Here we present results gathered to date in the Deciphering the Mechanisms of Developmental Disorders (DMDD) programme, cataloguing the morphological defects identified from comprehensive imaging of 220 homozygous mutant embryos from 42 lethal and subviable lines, analysed at E14.5.
Results: Virtually all embryos show multiple abnormal phenotypes and amongst the 42 lines these affect most organ systems. Within each mutant line, the phenotypes of individual embryos form distinct but overlapping sets. Subcutaneous edema, malformations of the heart or great vessels, abnormalities in forebrain morphology and the musculature of the eyes are all prevalent phenotypes, as is loss or abnormal size of the hypoglossal nerve.
Conclusions: Overall, the most striking finding is that no matter how profound the malformation, each phenotype shows highly variable penetrance within a mutant line. These findings have challenging implications for efforts to identify human disease correlates.
Keywords: mouse, embryo, phenotype, morphology, high-resolution episcopic microscopy, development, penetrance
Introduction
Animal models have long been used as experimental surrogates for investigating the role of individual genes in human development and disease. The remarkable degree of conservation in gene sequence and role that we now know exists across species confirms the validity of this approach and genetic manipulation in the mouse provides a commonly used way to explore gene function. The most ambitious example of this is the attempt coordinated by the International Mouse Phenotyping Consortium (IMPC) to generate a catalogue of gene function, using a systematic approach to phenotyping of individual gene knockouts (KO) that cover the entire mouse genome. In generating KO lines from about one quarter of the total mouse genome so far, these studies have revealed that around one third of all mammalian genes are essential for life 1– 3, their removal resulting in embryonic or perinatal lethality. The study of such mutant lines provides a unique opportunity to gain a comprehensive overview of the genetic components regulating normal embryo development and, by inference, the identity of genes whose mutation may cause congenital abnormalities or developmental disease.
Deciphering the Mechanisms of Developmental Disorders (DMDD) is a five year, UK-based programme funded by the Wellcome Trust with the goal of studying 240 embryonic lethal KO lines 3. By applying systematic phenotyping methods for homozygous mutant embryos with parallel efforts to identify placental abnormalities and changes in early embryo transcriptome profiles, DMDD offers a foundation for identifying novel genes important for developmental or clinical studies. Here we summarise results to date from detailed examination of homozygous mutant embryos at E14.5 for structural abnormalities.
Materials and methods
Embryos
All embryos were produced by the Wellcome Trust Sanger Institute ( https://www.sanger.ac.uk/mouseportal/) as part of the DMDD project 3. Gene knockout lines produced as part of a systematic programme coordinated by the International Mouse Phenotyping Consortium ( http://www.mousephenotype.org) were designated lethal if no homozygous mutants were present amongst a minimum of 28 pups at P14 and sub-viable if their proportion fell below 13% of total offspring 2. Embryos were harvested from one or more litters at E14.5, fixed in Bouin’s fixative for 24 hours and stored at 4°C in phosphate buffered saline.
Generation of digital volume data
Embryos were initially scored for gross abnormalities under a dissection microscope before preparation for 3D imaging. Briefly, embryos were dehydrated in methanol (10% steps until 90%, followed by 95% and 100%; at least 2 hours each) and embedded in methacrylate resin (JB-4, PolySciences) containing eosin B and acridine orange, as previously described 4– 6. Within each resin block, the embryo was oriented to ensure transverse sectioning along its longitudinal axis. Resin blocks were allowed to polymerise overnight at room temperature, baked at 90°C for 24–48 hours and then subjected to digital volume data generation using high-resolution episcopic microscopy (HREM) 7. HREM data was downsized as appropriate to provide an isotropic voxel size of between 2.5–3 µm, depending on original section thickness.
Data processing and annotation
12 bit raw greyscale image data was adjusted to optimise tissue visualisation using Photoshop 6 (Adobe). Data visualisation and analysis was performed using software packages Amira 5 (ThermoFisher Scientific) and Osirix, versions 6–8 (Pixmeo). Virtual 2D sections were used for phenotype scoring, essentially as described in a recently published protocol 8, each phenotype call being assigned to a 3D point in each embryo image data stack. Abnormalities were classified by using the Mammalian Phenotype (MP) ontology 9 using the most specific MP term that described each defect. 3D volume rendered models were employed for developmental staging from external morphology.
Data analysis
In order to facilitate summarising of detailed phenotype annotation data, two subsets of the MP terms closer to the root of the ontology were chosen to provide structured “high” and “intermediate” level overviews of DMDD phenotype data. These MP ontology slims are shown in Table 4 and Table 5. The MP terms assigned during annotation of the embryos were summarised into the categories defined by the DMDD slims using the Map2Slim algorithm ( https://metacpan.org/pod/distribution/go-perl/scripts/map2slim). All the terms of the DMDD slims that map to terms used to annotate embryo phenotypes are listed in Supplementary Table 1.
MP annotation terms used to describe the phenotypes of each embryo of a line were normalised to remove duplicate terms, and the terms for each embryo were mapped onto the ontology slims. For each line, a set of the unique slim terms observed for the line was generated and lists were produced of all the embryos from the line falling into each of these high or intermediate level categories. This enabled calculation of a penetrance score for each of the broad slim terms, calculated as a ratio of the number of embryos listed for the slim category to the number of homozygous mutant embryos analysed for the line.
To obtain a global view of the phenotypes detected, the frequency of lines showing each of the broad category slim terms were counted across all the lines analysed. In addition, the incidence of embryos scored for every phenotype category described by the slim terms, and the total number of embryos analysed in lines exhibiting each individual phenotype category was counted.
The total number of lines for each slim term that had a penetrance score between 0–0.24, 0.25–0.49, 0.50–0.74 and 0.75–1.00 was recorded. We calculated the cumulative penetrance score for each slim term as the overall sum of the penetrance scores of every line showing this broad category phenotype. In addition, for each of the penetrance intervals listed above, the sum of the penetrance scores was calculated for the lines falling into these categories.
All plots showing analysis of the data were produced using the R software package, version 3.2.1 (2015-06-18) (The R Foundation for Statistical Computing).
Use of animals
The care and use of all mice in this study were in accordance with UK Home Office regulations, UK Animals (Scientific Procedures) Act of 1986 (PPL 80/2485) and were approved by the Wellcome Trust Sanger Institute’s Animal Welfare and Ethical Review Body.
Results
Size of the study
The data for this study comprises 220 homozygous mutant E14.5 embryos analysed by the DMDD programme. All data is presented in Dataset 1 and also available on the DMDD web site ( https://dmdd.org.uk). Embryos were obtained from 42 novel gene knockout lines, 31 classified as lethal and 11 as sub-viable (see Materials and methods; Table 1). This corresponds to an average of approximately 5 embryos for each mutant line, although in practice numbers ranged widely from 1 to 11 as a result of variable breeding efficiency and cost limitations inherent in a large scale screening programme ( Supplementary Figure 1). In total, 722,329 transverse section images obtained from the 220 embryos formed the basis for examining embryo structure and with the addition of digital resection of datasets in coronal and sagittal planes, scoring of phenotypes was based on examination of 1,634,134 images.
Table 1. List of lethal and subviable lines studied.
The gene symbol, Mouse Genome Informatics (MGI) ID for the gene, and allele symbol is listed for each line studied along with the number of homozygous mutant embryos analysed, and the viability status.
| gene_symbol | mgi_id | allele_symbol | viability | # mutant embryos analysed |
|---|---|---|---|---|
| 1700067K01Rik | MGI:1920703 | 1700067K01Rik<tm2a(KOMP)Wtsi> | Lethal | 8 |
| 4933434E20Rik | MGI:1914027 | 4933434E20Rik<tm1a(EUCOMM)Wtsi> | Lethal | 6 |
| Adamts3 | MGI:3045353 | Adamts3<tm1b(KOMP)Wtsi> | Lethal | 7 |
| Adcy9 | MGI:108450 | Adcy9<tm1b(EUCOMM)Wtsi> | Subviable | 8 |
| Anks6 | MGI:1922941 | Anks6<tm1b(KOMP)Wtsi> | Lethal | 2 |
| Atp11a | MGI:1354735 | Atp11a<tm1a(KOMP)Wtsi> | Lethal | 5 |
| Brd2 | MGI:99495 | Brd2<em2Wtsi> | Lethal | 5 |
| Camsap3 | MGI:1916947 | Camsap3<tm1a(EUCOMM)Wtsi> | Subviable | 4 |
| Celf4 | MGI:1932407 | Celf4<tm1a(EUCOMM)Wtsi> | Lethal | 5 |
| Chst11 | MGI:1927166 | Chst11<tm1a(KOMP)Wtsi> | Lethal | 10 |
| Chtop | MGI:1913761 | Chtop<tm1a(EUCOMM)Wtsi> | Lethal | 4 |
| Cir1 | MGI:1914185 | Cir1<tm3a(KOMP)Wtsi> | Lethal | 3 |
| Cmip | MGI:1921690 | Cmip<tm1a(EUCOMM)Wtsi> | Lethal | 10 |
| Col4a3bp | MGI:1915268 | Col4a3bp<tm1a(KOMP)Wtsi> | Subviable | 2 |
| Cpt2 | MGI:109176 | Cpt2<tm1b(KOMP)Wtsi> | Subviable | 6 |
| D930028M14Rik | MGI:3687343 | D930028M14Rik<tm1a(EUCOMM)Wtsi> | Lethal | 5 |
| Dbn1 | MGI:1931838 | Dbn1<tm1b(KOMP)Wtsi> | Subviable | 5 |
| Dhx35 | MGI:1918965 | Dhx35<tm1b(EUCOMM)Wtsi> | Lethal | 1 |
| Exoc3l2 | MGI:1921713 | Exoc3l2<tm1b(KOMP)Wtsi> | Lethal | 3 |
| Fam46c | MGI:1921895 | Fam46c<tm1b(KOMP)Wtsi> | Lethal | 7 |
| H13 | MGI:95886 | H13<tm1b(KOMP)Wtsi> | Lethal | 7 |
| Kif1bp | MGI:1919570 | Kif1bp<tm1a(KOMP)Wtsi> | Lethal | 3 |
| Mybphl | MGI:1916003 | Mybphl<tm1b(KOMP)Wtsi> | Subviable | 3 |
| Npat | MGI:107605 | Npat<tm1b(EUCOMM)Wtsi> | Lethal | 1 |
| Nsun2 | MGI:107252 | Nsun2<tm1a(EUCOMM)Wtsi> | Subviable | 6 |
| Nxn | MGI:109331 | Nxn<tm1b(EUCOMM)Wtsi> | Lethal | 3 |
| Otud7b | MGI:2654703 | Otud7b<tm1b(EUCOMM)Wtsi> | Lethal | 1 |
| Pdzk1 | MGI:1928901 | Pdzk1<tm2b(EUCOMM)Wtsi> | Subviable | 9 |
| Polb | MGI:97740 | Polb<tm1a(KOMP)Wtsi> | Lethal | 6 |
| Prrc2b | MGI:1923304 | Prrc2b<tm1a(EUCOMM)Wtsi> | Lethal | 9 |
| Psph | MGI:97788 | Psph<tm1a(EUCOMM)Hmgu> | Lethal | 8 |
| Pth1r | MGI:97801 | Pth1r<tm1a(EUCOMM)Hmgu> | Lethal | 3 |
| Rundc1 | MGI:2144506 | Rundc1<tm1b(EUCOMM)Wtsi> | Subviable | 4 |
| Sh3pxd2a | MGI:1298393 | Sh3pxd2a<tm1b(EUCOMM)Wtsi> | Lethal | 11 |
| Slc25a20 | MGI:1928738 | Slc25a20<tm1a(EUCOMM)Wtsi> | Lethal | 6 |
| Slc5a7 | MGI:1927126 | Slc5a7<tm1a(KOMP)Wtsi> | Lethal | 3 |
| Smg9 | MGI:1919247 | Smg9<tm1b(EUCOMM)Wtsi> | Lethal | 6 |
| Smpd4 | MGI:1924876 | Smpd4<tm2b(KOMP)Wtsi> | Subviable | 3 |
| Ssr2 | MGI:1913506 | Ssr2<tm1b(EUCOMM)Wtsi> | Lethal | 3 |
| Tcf7l2 | MGI:1202879 | Tcf7l2<tm1a(EUCOMM)Wtsi> | Lethal | 5 |
| Traf6 | MGI:108072 | Traf6<tm2a(EUCOMM)Wtsi> | Lethal | 9 |
| Unk | MGI:2442456 | Unk<tm1a(KOMP)Wtsi> | Subviable | 5 |
Incidence of structural abnormalities in homozygous mutant embryos
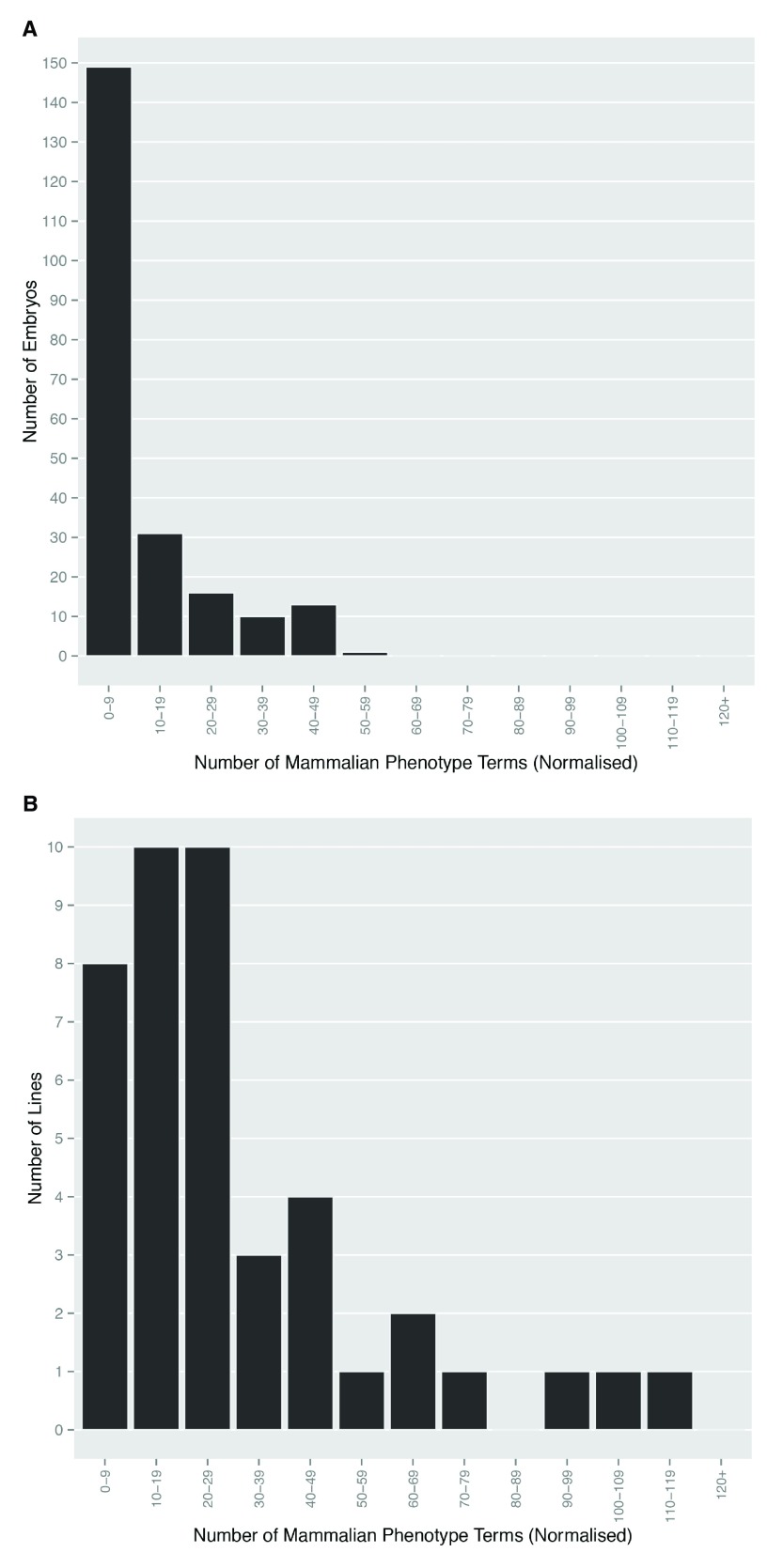
Almost all embryos studied (209/220) showed structural abnormalities that could be identified by a phenotyping procedure previously established and refined from pilot studies 8. The remaining 11 apparently normal embryos were obtained from 9 different lines, each of which yielded several other homozygous mutants bearing detectable morphological abnormalities. We have previously reported that the resolution afforded by 3D datasets obtained by HREM imaging allowed the detection of phenotypic abnormalities spanning in size range from individual nerves and blood vessels to gross organ and tissue malformations 8. In the present study, a total of 398 different MP terms were employed to record a total of 2,939 detected embryo phenotypes ( Dataset 1). Multiple abnormalities were scored in virtually all embryos. Most showed up to 10, but in some embryos as many as 50 phenotypes were recorded ( Figure 1A). Whilst a few phenotypes (for example those affecting different parts of vertebrae or different parts of the vertebral column) were often scored repeatedly within affected embryos, their incidence was insufficient to have a significant impact on the overall distribution of phenotype numbers scored per embryo across the whole study. When analysed by individual mutant line, the incidence of detectable abnormalities is more broadly distributed, with more than half of the 42 lines showing between 10 and 49 different phenotypes ( Figure 1B).
Figure 1. Multiple abnormalities are evident in homozygous mutant embryos.
The Mammalian Phenotype Ontology terms scored for ( A) each embryo, and ( B) each line were normalised to remove duplicate ontology terms. The number of distinct phenotypes scored that fell into categories with a window width of 10 were plotted to show the total number of embryos and lines respectively in each category.
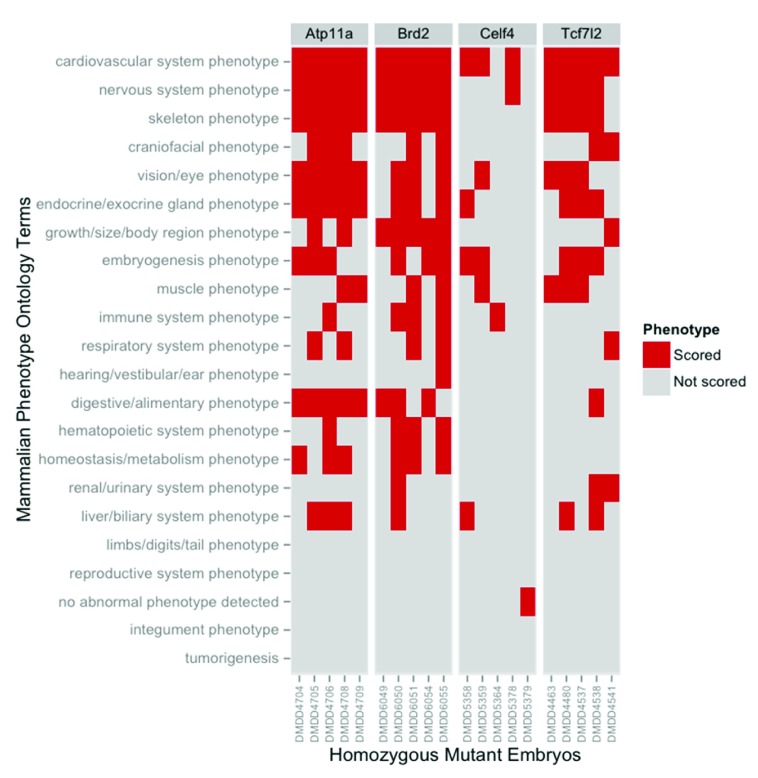
The disparity between phenotype incidence by embryo and phenotype incidence by line is striking and indicates that individual mutant embryos within each line show distinct, but partially overlapping spectra of phenotypes. This is graphically illustrated by data from the four lines shown in Figure 2. The Atp11a allele exemplifies those mutant lines in which a significant proportion of phenotypes were detected across all the embryos scored. (Note that the data is summarised by high level MP ontology slim terms and individual embryos may still differ in the precise phenotypes actually scored. This may reflect differing degrees of severity amongst affected embryos or pleiotropic effects of mutations on individual organ systems). The Brd2 and Tcf712 alleles showed similar, but less pronounced, conservation of phenotype whilst the Celf4 allele is an example of those lines in which there was no detectable consistency to the phenotypes detected across the mutants analysed.
Figure 2. Individual embryos show overlapping but distinct spectra of phenotypes.
The phenotypes annotated for individual embryos were normalised to remove duplicate ontology terms. The distinct terms for each homozygous mutant embryo from four lines were then mapped onto the broad set of ontology categories defined in the high level DMDD slim. The presence or absence of phenotype annotation within each of the high level categories was plotted for each embryo analysed.
Prevalence of individual abnormalities
Table 2 presents the frequency with which individual abnormalities were identified amongst the mutant embryos. Since some phenotypes (such as vertebral abnormalities) are often present multiply in affected embryos, the data is normalised for occurrence by embryo. Interestingly, the most common phenotype detected in this study was subcutaneous edema. This was evident from macroscopic observation of embryos at harvest and confirmed by subsequent HREM imaging ( Figure 3, panels A–C). In total, subcutaneous edema and edema in other body regions (scored with four distinct MP terms) affected one third (72/220) of the embryos and was observed in a little over half (24/42) of the mutant lines. Other prevalent phenotypes included defects affecting the vertebral arches, the ventricular septum of the heart, forebrain morphology and musculature of the developing eyes ( Table 2 and Figure 3). Of particular note is the frequency with which mutant embryos showed abnormalities affecting the architecture or presence of the hypoglossal nerve ( Figure 4, panels A and B). Complete absence of the nerve occurred in 37 embryos, obtained from 12 different mutant lines, with some embryos from a similar number of lines showing abnormal topology or unusual thinness of the nerve (13 and 9 lines respectively). Overall, scored phenotypes affected all the major organ systems at E14.5 ( Figure 5A) and multiple organs or tissues were frequently affected within individual embryos or collectively within a mutant line ( Figure 2 and Supplementary Figure 2 and Supplementary Figure 3). The complete listing of scored phenotypes is presented in Supplementary Table 1, organised according to the MP ontology slims adopted by the DMDD programme (see Table 4 and Table 5), with data ranked according to prevalence in mutant lines.
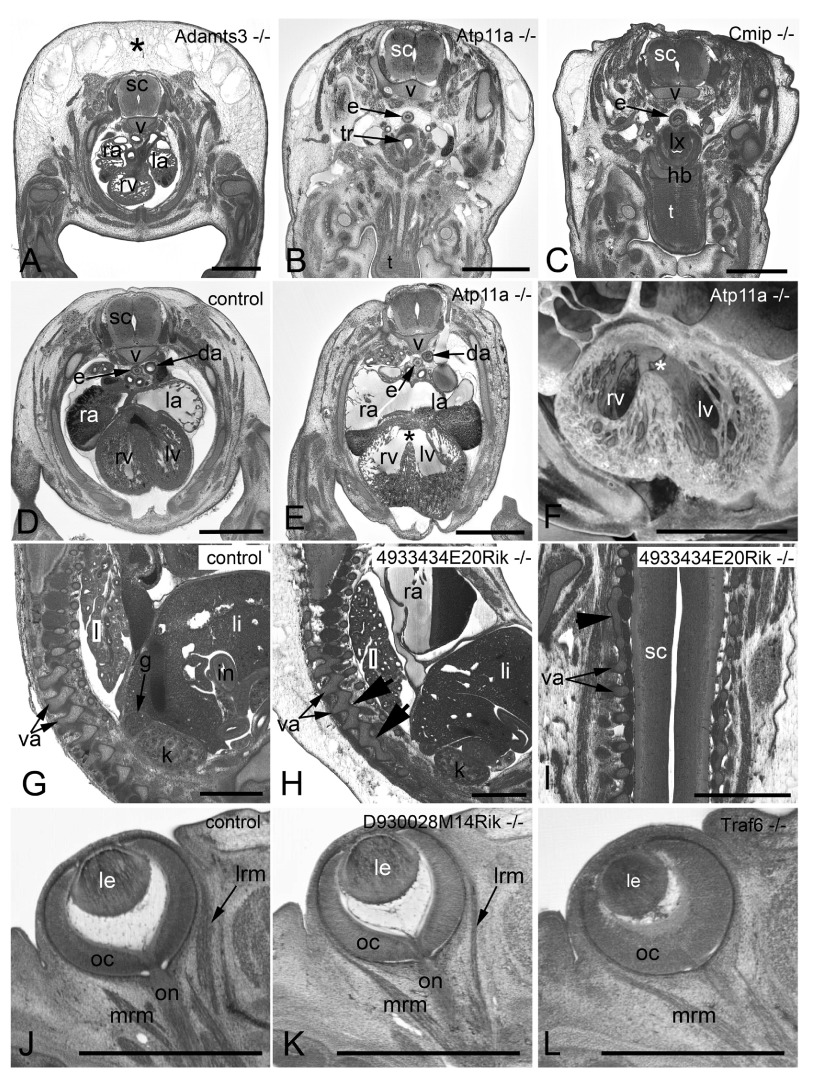
Figure 3. Examples of frequently observed abnormalities.
A– C. Subcutaneous edema. Original HREM sections showing a massive (asterisk) ( A), mild ( B), and unilaterally located subcutaneous edema ( C). Note the shrinkage artefacts in B and C, which complicate post mortem diagnosis. D– F. Perimembraneous septal defect. Normal situation in a control ( D) as appearing in an original HREM section. Defect (asterisk) as appearing in an original HREM section ( E) and a 3D volume model ( F). G– I. Fusion of vertebral arches. Normal situation in a control ( G) as appearing in a sagittal section. Fused articular processes (arrowheads) of subsequent vertebrae in a sagittal ( H) and a coronal section ( I). J– L. Abnormal eye muscle morphology as appearing in original HREM sections. Normal situation in a control ( J). Thinning of the lateral rectus muscle (lrm) ( K). Absence of the lateral rectus muscle (lrm) ( L). da, descending aorta; e, esophagus; g, adrenal gland; hb, hyoid bone; i, intestine; k, kidney; l, lung; la, left atrium; le, lens; li, liver; lrm, lateral rectus muscle; lv, left ventricle; lx, larynx; mrm, medial rectus muscle; oc, optic cup; on, optic nerve; ra, right atrium; rv, right ventricle; sc, spinal chord; t, tongue; tr, trachea; v, body of vertebra; va, arch of vertebra. Scale bars: 1 mm.
Figure 4. Other frequently observed abnormalities.
A and B. Abnormal hyopglossal nerve in original HREM sections through the head of Prrc2b -/- ( A) and a Polb -/- ( B) embryo. Note the missing right hypoglossal nerve (arrowhead, inlay) in A and the thinning of both hypoglossal nerves (hn) in B. C– E. Abnormalities that also occur in controls. Persisting craniopharnygeal duct (arrowhead) as appearing in sagittal sections ( C). Split tip of tail featured by volume models ( D) and vesicles (arrowheads) in the lens (le) as appearing in an original HREM section ( E).
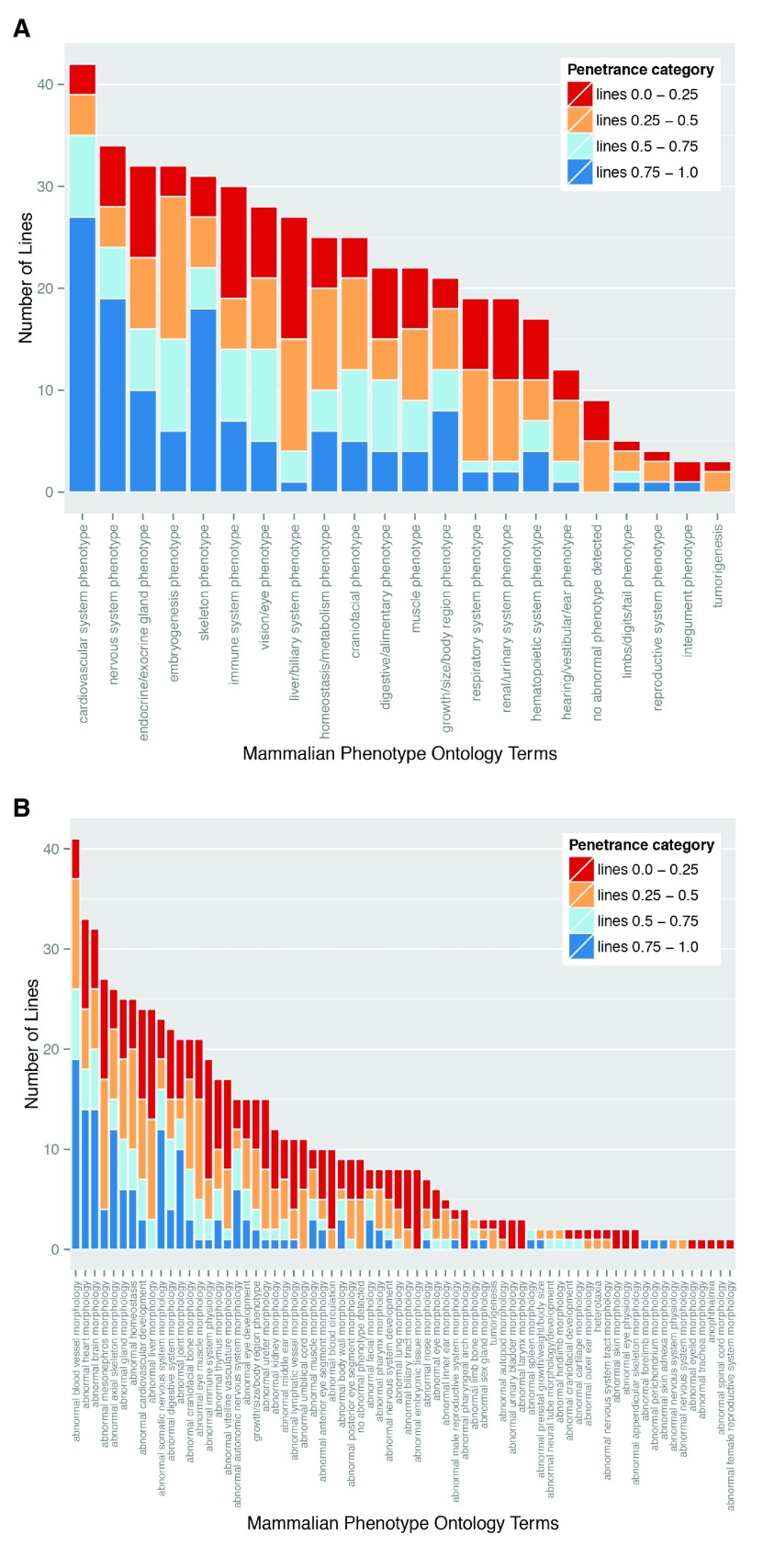
Figure 5. Variable prevalence and penetrance of individual phenotypes.
Data from the global analysis of the frequency of phenotype terms (see Materials and Methods) was plotted to show the number of lines falling into each of the observed phenotype categories. The colours indicate the number of lines falling into each of the distinct penetrance categories. The data was ordered according to line frequency, and subsequently by the numbers seen in the penetrance categories. ( A) shows the phenotype annotations summarised using the high level DMDD ontology slim, ( B) shows the phenotype annotations summarised using the intermediate level DMDD ontology slim.
Table 2. Frequency of phenotypes identified.
The Mammalian Phenotype Ontology terms describing phenotypes observed in each embryo were normalised to remove duplicates and the list then ranked in descending order by frequency of embryos exhibiting each phenotype.
| mp_id | term | frequency |
|---|---|---|
| MP:0013848 | subcutaneous edema | 64 |
| MP:0004613 | fusion of vertebral arches | 61 |
| MP:0010418 | perimembraneous ventricular septal
defect |
49 |
| MP:0000783 | abnormal forebrain morphology | 47 |
| MP:0003686 | abnormal eye muscle morphology | 45 |
| MP:0001015 | small superior cervical ganglion | 45 |
| MP:0010420 | muscular ventricular septal defect | 41 |
| MP:0013835 | absent hypoglossal nerve | 37 |
| MP:0003826 | abnormal Mullerian duct morphology | 33 |
| MP:0014021 | heterochrony | 33 |
| MP:0004269 | abnormal optic cup morphology | 32 |
| MP:0014001 | abnormal vertebral artery topology | 32 |
| MP:0013836 | abnormal hypoglossal nerve topology | 30 |
| MP:0013876 | absent ductus venosus valve | 29 |
| MP:0000284 | double outlet right ventricle | 29 |
| MP:0004666 | absent stapedial artery | 28 |
| MP:0013971 | blood in lymph vessels | 27 |
| MP:0000703 | abnormal thymus morphology | 26 |
| MP:0014000 | anastomosis between internal
carotid artery and basilar artery |
25 |
| MP:0000602 | enlarged liver sinusoidal spaces | 25 |
| MP:0013969 | reduced sympathetic cervical
ganglion size |
25 |
| MP:0008923 | thoracoschisis | 25 |
| MP:0004163 | abnormal adenohypophysis
morphology |
24 |
| MP:0002237 | abnormal nasal cavity morphology | 20 |
| MP:0013986 | abnormal vitelline vein topology | 20 |
| MP:0013967 | abnormal infrahyoid muscle
connection |
18 |
| MP:0004463 | basisphenoid bone foramen | 18 |
| MP:0008128 | abnormal brain internal capsule morphology | 16 |
| MP:0000282 | abnormal interatrial septum
morphology |
16 |
| MP:0004268 | abnormal optic stalk morphology | 16 |
| MP:0013936 | abnormal thymus topology | 16 |
| MP:0014017 | abnormal Wolffian duct connection | 15 |
| MP:0013877 | abnormal ductus venosus valve
morphology |
15 |
| MP:0002239 | abnormal nasal septum morphology | 15 |
| MP:0000497 | abnormal small intestine placement | 15 |
| MP:0000111 | cleft palate | 15 |
| MP:0013859 | abnormal vitelline vein connection | 14 |
| MP:0013826 | absent hypoglossal canal | 14 |
| MP:0013840 | absent segment of posterior cerebral artery | 14 |
| MP:0013875 | trigeminal neuroma | 14 |
| MP:0010496 | abnormal pectinate muscle
morphology |
13 |
| MP:0013834 | thin hypoglossal nerve | 13 |
| MP:0003827 | abnormal Wolffian duct morphology | 12 |
| MP:0013842 | ductus venosus stenosis | 12 |
| MP:0010912 | herniated liver | 12 |
| MP:0013968 | multiple persisting craniopharyngeal
ducts |
12 |
| MP:0011361 | pelvic kidney | 12 |
| MP:0010572 | persistent right dorsal aorta | 12 |
| MP:0002633 | persistent truncus arteriosis | 12 |
| MP:0013931 | abnormal olfactory bulb position | 11 |
| MP:0011683 | dual inferior vena cava | 11 |
| MP:0000914 | exencephaly | 11 |
| MP:0002169 | no abnormal phenotype detected | 11 |
| MP:0000154 | rib fusion | 11 |
| MP:0000161 | scoliosis | 11 |
| MP:0004110 | transposition of great arteries | 11 |
| MP:0012303 | umbilical vein stenosis | 11 |
| MP:0008922 | abnormal cervical rib | 10 |
| MP:0009917 | abnormal hyoid bone body morphology | 10 |
| MP:0009770 | abnormal optic chiasm morphology | 10 |
| MP:0013844 | abnormal perichondrial ossification | 10 |
| MP:0003345 | decreased rib number | 10 |
| MP:0011493 | double ureter | 10 |
| MP:0000445 | short snout | 10 |
| MP:0002951 | small thyroid gland | 10 |
| MP:0013878 | abnormal ductus venosus valve
topology |
9 |
| MP:0000841 | abnormal hindbrain morphology | 9 |
| MP:0010490 | abnormal inferior vena cava valve
morphology |
9 |
| MP:0010853 | abnormal lung position or orientation | 9 |
| MP:0000141 | abnormal vertebral body morphology | 9 |
| MP:0002243 | abnormal vomeronasal organ
morphology |
9 |
| MP:0013970 | absent connection between
subcutaneous lymph vessels and lymph sac |
9 |
| MP:0011667 | double outlet right ventricle with
atrioventricular septal defect |
9 |
| MP:0014019 | embryo cyst | 9 |
| MP:0013977 | symmetric azygos veins | 9 |
| MP:0002092 | abnormal eye morphology | 8 |
| MP:0014023 | abnormal intestine placement | 8 |
| MP:0001303 | abnormal lens morphology | 8 |
| MP:0000632 | abnormal pineal gland morphology | 8 |
| MP:0010602 | abnormal pulmonary valve cusp
morphology |
8 |
| MP:0013985 | abnormal umbilical vein topology | 8 |
| MP:0013965 | abnormally deep median sulcus of
tongue |
8 |
| MP:0010484 | bicuspid aortic valve | 8 |
| MP:0004646 | decreased cervical vertebrae
number |
8 |
| MP:0013915 | abnormal brachial plexus formation | 7 |
| MP:0010436 | abnormal coronary sinus morphology | 7 |
| MP:0000819 | abnormal olfactory bulb morphology | 7 |
| MP:0009570 | abnormal right lung morphology | 7 |
| MP:0003078 | aphakia | 7 |
| MP:0003584 | bifid ureter | 7 |
| MP:0013949 | fusion of axis and occipital bones | 7 |
| MP:0013846 | retropharyngeal edema | 7 |
| MP:0013847 | retropleural edema | 7 |
| MP:0000153 | rib bifurcation | 7 |
| MP:0002191 | abnormal artery morphology | 6 |
| MP:0000079 | abnormal basioccipital bone
morphology |
6 |
| MP:0000788 | abnormal cerebral cortex morphology | 6 |
| MP:0013995 | abnormal external carotid artery
origin |
6 |
| MP:0013845 | abnormal eye muscle topology | 6 |
| MP:0002858 | abnormal posterior semicircular
canal morphology |
6 |
| MP:0000759 | abnormal skeletal muscle morphology | 6 |
| MP:0013871 | abnormal stapedial artery topology | 6 |
| MP:0001146 | abnormal testis morphology | 6 |
| MP:0000681 | abnormal thyroid gland morphology | 6 |
| MP:0004599 | abnormal vertebral arch morphology | 6 |
| MP:0013996 | abnormal vertebral artery origin | 6 |
| MP:0013849 | absent abducens nerve | 6 |
| MP:0000520 | absent kidney | 6 |
| MP:0009725 | absent lens vesicle | 6 |
| MP:0006093 | arteriovenous malformation | 6 |
| MP:0010412 | atrioventricular septal defect | 6 |
| MP:0013932 | fragmented Meckel's cartilage | 6 |
| MP:0000963 | fused dorsal root ganglion | 6 |
| MP:0005157 | holoprosencephaly | 6 |
| MP:0000480 | increased rib number | 6 |
| MP:0013992 | persistent dorsal ophthalmic artery | 6 |
| MP:0013952 | retro-esophageal left subclavian
artery |
6 |
| MP:0004160 | retroesophageal right subclavian
artery |
6 |
| MP:0004158 | right aortic arch | 6 |
| MP:0020301 | short tongue | 6 |
| MP:0002989 | small kidney | 6 |
| MP:0013852 | abnormal Mullerian duct topology | 5 |
| MP:0010595 | abnormal aortic valve cusp morphology | 5 |
| MP:0000297 | abnormal atrioventricular cushion
morphology |
5 |
| MP:0013186 | abnormal basilar artery morphology | 5 |
| MP:0002152 | abnormal brain morphology | 5 |
| MP:0013874 | abnormal ductus venosus topology | 5 |
| MP:0013945 | abnormal elbow joint morphology | 5 |
| MP:0000559 | abnormal femur morphology | 5 |
| MP:0006063 | abnormal inferior vena cava
morphology |
5 |
| MP:0002135 | abnormal kidney morphology | 5 |
| MP:0001879 | abnormal lymphatic vessel
morphology |
5 |
| MP:0005236 | abnormal olfactory nerve morphology | 5 |
| MP:0000150 | abnormal rib morphology | 5 |
| MP:0004539 | absent maxilla | 5 |
| MP:0003451 | absent olfactory bulb | 5 |
| MP:0001014 | absent superior cervical ganglion | 5 |
| MP:0014003 | additional anastomosis between
intracranial vertebral arteries |
5 |
| MP:0012548 | myelocele | 5 |
| MP:0000273 | overriding aortic valve | 5 |
| MP:0000964 | small dorsal root ganglion | 5 |
| MP:0000694 | spleen hypoplasia | 5 |
| MP:0013928 | thin motoric part of trigeminal nerve | 5 |
| MP:0002199 | abnormal brain commissure
morphology |
4 |
| MP:0006065 | abnormal heart position or orientation | 4 |
| MP:0002249 | abnormal larynx morphology | 4 |
| MP:0009820 | abnormal liver vasculature morphology | 4 |
| MP:0005105 | abnormal middle ear ossicle morphology | 4 |
| MP:0004164 | abnormal neurohypophysis morphology | 4 |
| MP:0013994 | abnormal parasellar internal carotid artery branch morphology | 4 |
| MP:0000633 | abnormal pituitary gland morphology | 4 |
| MP:0013980 | abnormal pulmonary artery origin | 4 |
| MP:0011655 | abnormal systemic artery morphology | 4 |
| MP:0011513 | abnormal vertebral artery morphology | 4 |
| MP:0013855 | absent celiac artery | 4 |
| MP:0013833 | absent olfactory nerve | 4 |
| MP:0013362 | absent pineal gland | 4 |
| MP:0014006 | absent posterior communicating artery | 4 |
| MP:0013913 | absent rib-vertebral column attachment | 4 |
| MP:0004846 | absent skeletal muscle | 4 |
| MP:0004603 | absent vertebral arch | 4 |
| MP:0010440 | anomalous pulmonary venous connection | 4 |
| MP:0010530 | cerebral arteriovenous malformation | 4 |
| MP:0010589 | common truncal valve | 4 |
| MP:0003924 | diaphragmatic hernia | 4 |
| MP:0003253 | dilated bile duct | 4 |
| MP:0013879 | duplication of ductus venosus | 4 |
| MP:0008534 | enlarged fourth ventricle | 4 |
| MP:0004612 | fusion of vertebral bodies | 4 |
| MP:0001914 | hemorrhage | 4 |
| MP:0003262 | intestinal/bowel diverticulum | 4 |
| MP:0010404 | ostium primum atrial septal defect | 4 |
| MP:0013917 | persistent right 6th pharyngeal arch
artery |
4 |
| MP:0000562 | polydactyly | 4 |
| MP:0001088 | small nodose ganglion | 4 |
| MP:0013827 | thin oculomotor nerve | 4 |
| MP:0013858 | abnormal azygos vein topology | 3 |
| MP:0002928 | abnormal bile duct morphology | 3 |
| MP:0008026 | abnormal brain white matter
morphology |
3 |
| MP:0004607 | abnormal cervical atlas morphology | 3 |
| MP:0000820 | abnormal choroid plexus morphology | 3 |
| MP:0013873 | abnormal ductus venosus morphology | 3 |
| MP:0010439 | abnormal hepatic vein morphology | 3 |
| MP:0000823 | abnormal lateral ventricle morphology | 3 |
| MP:0000598 | abnormal liver morphology | 3 |
| MP:0000897 | abnormal midbrain morphology | 3 |
| MP:0013861 | abnormal pancreas topology | 3 |
| MP:0000613 | abnormal salivary gland morphology | 3 |
| MP:0013943 | abnormal ureter topology | 3 |
| MP:0001100 | abnormal vagus ganglion morphology | 3 |
| MP:0014002 | absent extracranial vertebral artery
segment |
3 |
| MP:0013929 | absent eye muscles | 3 |
| MP:0003722 | absent ureter | 3 |
| MP:0000138 | absent vertebrae | 3 |
| MP:0000640 | adrenal gland hypoplasia | 3 |
| MP:0005262 | coloboma | 3 |
| MP:0010433 | double inlet heart left ventricle | 3 |
| MP:0001785 | edema | 3 |
| MP:0000274 | enlarged heart | 3 |
| MP:0006203 | eye hemorrhage | 3 |
| MP:0005244 | hemopericardium | 3 |
| MP:0013843 | hepatic portal vein stenosis | 3 |
| MP:0011659 | interrupted aortic arch, type b | 3 |
| MP:0013948 | intraembryonal intestine elongation | 3 |
| MP:0013963 | jugular vein stenosis | 3 |
| MP:0000692 | small spleen | 3 |
| MP:0001093 | small trigeminal ganglion | 3 |
| MP:0013828 | thin facial nerve | 3 |
| MP:0004057 | thin myocardium compact layer | 3 |
| MP:0003617 | urinary bladder hypoplasia | 3 |
| MP:0013851 | abnormal Wolffian duct topology | 2 |
| MP:0013857 | abnormal abdominal muscle
morphology |
2 |
| MP:0004113 | abnormal aortic arch morphology | 2 |
| MP:0002747 | abnormal aortic valve morphology | 2 |
| MP:0004181 | abnormal carotid artery
morphology |
2 |
| MP:0013978 | abnormal carotid artery origin | 2 |
| MP:0013975 | abnormal coronary sinus
connection |
2 |
| MP:0002279 | abnormal diaphragm morphology | 2 |
| MP:0013815 | abnormal digastric muscle morphology | 2 |
| MP:0013865 | abnormal dorsal pancreas topology | 2 |
| MP:0000961 | abnormal dorsal root ganglion
morphology |
2 |
| MP:0013950 | abnormal dorsal root ganglion
topology |
2 |
| MP:0006011 | abnormal endolymphatic duct
morphology |
2 |
| MP:0013918 | abnormal endolymphatic sac
topology |
2 |
| MP:0006033 | abnormal external auditory canal
morphology |
2 |
| MP:0000266 | abnormal heart morphology | 2 |
| MP:0003056 | abnormal hyoid bone morphology | 2 |
| MP:0013966 | abnormal infrahyoid muscle
morphology |
2 |
| MP:0000489 | abnormal large intestine morphology | 2 |
| MP:0008986 | abnormal liver parenchyma
morphology |
2 |
| MP:0001175 | abnormal lung morphology | 2 |
| MP:0000458 | abnormal mandible morphology | 2 |
| MP:0003632 | abnormal nervous system
morphology |
2 |
| MP:0001330 | abnormal optic nerve morphology | 2 |
| MP:0002177 | abnormal outer ear morphology | 2 |
| MP:0000492 | abnormal rectum morphology | 2 |
| MP:0002428 | abnormal semicircular canal
morphology |
2 |
| MP:0002746 | abnormal semilunar valve
morphology |
2 |
| MP:0000496 | abnormal small intestine morphology | 2 |
| MP:0005107 | abnormal stapes morphology | 2 |
| MP:0003230 | abnormal umbilical artery
morphology |
2 |
| MP:0002725 | abnormal vein morphology | 2 |
| MP:0009707 | absent external auditory canal | 2 |
| MP:0013987 | absent intrahepatic inferior vena
cava segment |
2 |
| MP:0009771 | absent optic chiasm | 2 |
| MP:0013999 | absent parasellar internal carotid
artery |
2 |
| MP:0013809 | absent pectinate muscle | 2 |
| MP:0004571 | absent vagus nerve | 2 |
| MP:0000140 | absent vertebral pedicles | 2 |
| MP:0003130 | anal atresia | 2 |
| MP:0010463 | aorta stenosis | 2 |
| MP:0004055 | atrium hypoplasia | 2 |
| MP:0010406 | common atrium | 2 |
| MP:0003586 | dilated ureter | 2 |
| MP:0013981 | double lumen aortic arch | 2 |
| MP:0014018 | embryo tumor | 2 |
| MP:0010200 | enlarged lymphatic vessel | 2 |
| MP:0008536 | enlarged third ventricle | 2 |
| MP:0002015 | epithelioid cysts | 2 |
| MP:0004201 | fetal growth retardation | 2 |
| MP:0010977 | fused right lung lobes | 2 |
| MP:0010728 | fusion of atlas and occipital bones | 2 |
| MP:0013982 | inverse situs of great intrathoracic arteries | 2 |
| MP:0010647 | left atrium hypoplasia | 2 |
| MP:0000600 | liver hypoplasia | 2 |
| MP:0000618 | small salivary gland | 2 |
| MP:0001102 | small superior vagus ganglion | 2 |
| MP:0000706 | small thymus | 2 |
| MP:0011249 | abdominal situs inversus | 1 |
| MP:0000639 | abnormal adrenal gland
morphology |
1 |
| MP:0010592 | abnormal atrioventricular septum
morphology |
1 |
| MP:0002745 | abnormal atrioventricular valve
morphology |
1 |
| MP:0001614 | abnormal blood vessel morphology | 1 |
| MP:0000494 | abnormal cecum morphology | 1 |
| MP:0013862 | abnormal cecum position | 1 |
| MP:0010744 | abnormal cervical flexure
morphology |
1 |
| MP:0003048 | abnormal cervical vertebrae
morphology |
1 |
| MP:0009495 | abnormal common bile duct
morphology |
1 |
| MP:0012729 | abnormal common carotid artery
morphology |
1 |
| MP:0013930 | abnormal digastric muscle
connection |
1 |
| MP:0004252 | abnormal direction of heart looping | 1 |
| MP:0014022 | abnormal duodenum topology | 1 |
| MP:0013924 | abnormal dural venous sinus
morphology |
1 |
| MP:0013927 | abnormal facial nerve topology | 1 |
| MP:0006107 | abnormal fetal atrioventricular
canal morphology |
1 |
| MP:0000828 | abnormal fourth ventricle morphology | 1 |
| MP:0005084 | abnormal gallbladder morphology | 1 |
| MP:0003105 | abnormal heart atrium morphology | 1 |
| MP:0003922 | abnormal heart right atrium morphology | 1 |
| MP:0013814 | abnormal hepatic portal vein connection | 1 |
| MP:0013853 | abnormal hepatic portal vein formation | 1 |
| MP:0010668 | abnormal hepatic portal vein
morphology |
1 |
| MP:0013973 | abnormal hepatic vein connection | 1 |
| MP:0005296 | abnormal humerus morphology | 1 |
| MP:0009913 | abnormal hyoid bone greater horn
morphology |
1 |
| MP:0013824 | abnormal hypoglossal canal
morphology |
1 |
| MP:0002859 | abnormal inner ear canal fusion | 1 |
| MP:0009804 | abnormal interventricular foramen
morphology |
1 |
| MP:0000281 | abnormal interventricular septum
morphology |
1 |
| MP:0000477 | abnormal intestine morphology | 1 |
| MP:0013976 | abnormal left vena cava superior
connection |
1 |
| MP:0004881 | abnormal lung size | 1 |
| MP:0013841 | abnormal lymphatic vessel
topology |
1 |
| MP:0003792 | abnormal major salivary gland
morphology |
1 |
| MP:0000455 | abnormal maxilla morphology | 1 |
| MP:0000452 | abnormal mouth morphology | 1 |
| MP:0002108 | abnormal muscle morphology | 1 |
| MP:0004056 | abnormal myocardium compact
layer morphology |
1 |
| MP:0005269 | abnormal occipital bone morphology | 1 |
| MP:0013818 | abnormal oral cavity morphology | 1 |
| MP:0014011 | abnormal ovary tissue architecture | 1 |
| MP:0004509 | abnormal pelvic girdle bone morphology | 1 |
| MP:0002748 | abnormal pulmonary valve morphology | 1 |
| MP:0009571 | abnormal right lung accessory lobe morphology | 1 |
| MP:0009688 | abnormal spinal cord central canal morphology | 1 |
| MP:0008023 | abnormal styloid process morphology | 1 |
| MP:0013979 | abnormal subclavian artery origin | 1 |
| MP:0001011 | abnormal superior cervical ganglion morphology | 1 |
| MP:0000787 | abnormal telencephalon morphology | 1 |
| MP:0005272 | abnormal temporal bone morphology | 1 |
| MP:0000826 | abnormal third ventricle morphology | 1 |
| MP:0002368 | abnormal thymus capsule morphology | 1 |
| MP:0002282 | abnormal trachea morphology | 1 |
| MP:0001065 | abnormal trigeminal nerve morphology | 1 |
| MP:0010667 | abnormal umbilical vein morphology | 1 |
| MP:0000534 | abnormal ureter morphology | 1 |
| MP:0013925 | abnormal vascular plexus formation | 1 |
| MP:0000137 | abnormal vertebrae morphology | 1 |
| MP:0005274 | abnormal viscerocranium morphology | 1 |
| MP:0010666 | abnormal vitelline vein morphology | 1 |
| MP:0014004 | absent basilar artery segment | 1 |
| MP:0008129 | absent brain internal capsule | 1 |
| MP:0013998 | absent canalicular internal carotid artery segment | 1 |
| MP:0008460 | absent dorsal root ganglion | 1 |
| MP:0013880 | absent ductus venosus | 1 |
| MP:0013914 | absent intracranial segment of vertebral artery | 1 |
| MP:0013937 | absent lobe of thyroid gland | 1 |
| MP:0000629 | absent mammary gland | 1 |
| MP:0013926 | absent neurohypophysis | 1 |
| MP:0013988 | absent portal vein segment | 1 |
| MP:0013850 | absent posterior commissure | 1 |
| MP:0000614 | absent salivary gland | 1 |
| MP:0013823 | absent segment of anterior cerebral artery | 1 |
| MP:0000690 | absent spleen | 1 |
| MP:0008386 | absent styloid process | 1 |
| MP:0002728 | absent tibia | 1 |
| MP:0009905 | absent tongue | 1 |
| MP:0001064 | absent trochlear nerve | 1 |
| MP:0013595 | absent vomeronasal organ | 1 |
| MP:0013860 | anastomosis between common carotid and vertebral artery | 1 |
| MP:0014009 | anastomosis between middle cerebral arteries | 1 |
| MP:0001293 | anophthalmia | 1 |
| MP:0003387 | aorta coarctation | 1 |
| MP:0006135 | artery stenosis | 1 |
| MP:0000705 | athymia | 1 |
| MP:0010403 | atrial septal defect | 1 |
| MP:0013935 | basal brain tissue herniation | 1 |
| MP:0010527 | bicuspid pulmonary valve | 1 |
| MP:0011797 | blind ureter | 1 |
| MP:0010607 | common atrioventricular valve | 1 |
| MP:0004686 | decreased length of long bones | 1 |
| MP:0009532 | decreased parotid gland size | 1 |
| MP:0004648 | decreased thoracic vertebrae number | 1 |
| MP:0011965 | decreased total retina thickness | 1 |
| MP:0001247 | dermal cysts | 1 |
| MP:0000825 | dilated lateral ventricles | 1 |
| MP:0009144 | dilated pancreatic duct | 1 |
| MP:0004938 | dilated vasculature | 1 |
| MP:0011380 | enlarged brain ventricles | 1 |
| MP:0013864 | enlarged paraumbilical vein | 1 |
| MP:0003595 | epididymal cyst | 1 |
| MP:0002947 | increased hemangioma incidence | 1 |
| MP:0001634 | internal hemorrhage | 1 |
| MP:0011974 | intestinal stenosis | 1 |
| MP:0001916 | intracerebral hemorrhage | 1 |
| MP:0003178 | left pulmonary isomerism | 1 |
| MP:0013953 | left sided brachiocephalic trunk | 1 |
| MP:0003327 | liver cysts | 1 |
| MP:0003888 | liver hemorrhage | 1 |
| MP:0000162 | lordosis | 1 |
| MP:0010854 | lung situs inversus | 1 |
| MP:0005287 | narrow eye opening | 1 |
| MP:0004442 | occipital bone foramen | 1 |
| MP:0000565 | oligodactyly | 1 |
| MP:0006221 | optic nerve hypoplasia | 1 |
| MP:0013933 | short Meckel's cartilage | 1 |
| MP:0002766 | situs inversus | 1 |
| MP:0002768 | small adrenal glands | 1 |
| MP:0001306 | small lens | 1 |
| MP:0013923 | small prevertebral sympathetic ganglia | 1 |
| MP:0006254 | thin cerebral cortex | 1 |
| MP:0013829 | thin splanchnic nerve | 1 |
| MP:0013832 | thin vagus nerve | 1 |
| MP:0003499 | thyroid hypoplasia | 1 |
| MP:0009904 | tongue hypoplasia | 1 |
| MP:0011697 | vacuolated lens | 1 |
| MP:0013831 | vagus nerve compression | 1 |
| MP:0004609 | vertebral fusion | 1 |
Individual phenotypes show highly variable penetrance
Perhaps the most striking finding of the DMDD study is the almost complete absence of any fully penetrant abnormalities. Amongst lines for which more than a single embryo was analysed, only three phenotypes showed 100% penetrance: abnormal perichondrial ossification (1 line; 10 mutant embryos), small nodose ganglion (1 line; 4 embryos) and small trigeminal ganglion (1 line, 3 embryos). Furthermore, most defects showed surprisingly low penetrance. A penetrance greater than 75% within the line was only found for 7% of detected phenotypes. In contrast, over half (55%) of the scored abnormalities had a penetrance of 25% or less. ( Table 3). This is graphically illustrated in Figure 5A, in which the scored phenotypes are clustered according to high level MP ontology terms (broadly reflecting distinct organ systems, tissues or body regions) and the prevalence of each in the 42 mutant lines categorised by penetrance. All phenotypes show a broad range of penetrance, about half showing roughly symmetrical distribution of penetrance, with similar numbers of lines both above and below 50%. Interestingly, it is possible also to distinguish several phenotypes where penetrance is noticeably skewed. Abnormalities affecting the cardiovascular system, nervous system and skeleton all affected a relatively large number of lines and each showed a striking bias towards higher penetrance values. A second group of abnormalities encompassing liver/biliary, respiratory, renal and hearing systems showed a converse bias to penetrance values below 50% ( Figure 5A).
Table 3. Variability in phenotype penetrance.
Every distinct phenotype scored in each line was listed along with its penetrance (i.e. the number of embryos showing the phenotype divided by the total number of embryos analysed for that line). Scored phenotypes were then ranked by penetrance value to obtain the proportions falling within the four ranges shown. (Note that all data from the lines Otud7b,Npat and Dhx35 were removed from the analysis, since in each case, these were obtained from examination of a single embryo).
| Penetrance range | Phenotypes scored | % |
|---|---|---|
| <25% | 673 | 55.21% |
| 26–50% | 343 | 28.14% |
| 51–75% | 118 | 9.68% |
| >75% | 85 | 6.97% |
Table 4. High level MP ontology slim used by DMDD.
A list of the Mammalian Phenotype Ontology IDs and names of terms selected as the high level ontology slim.
| MP:0002169 | no abnormal phenotype detected |
| MP:0005375 | adipose tissue phenotype |
| MP:0005386 | behavior/neurological phenotype |
| MP:0005385 | cardiovascular system phenotype |
| MP:0005384 | cellular phenotype |
| MP:0005382 | craniofacial phenotype |
| MP:0005381 | digestive/alimentary phenotype |
| MP:0005380 | embryogenesis phenotype |
| MP:0005379 | endocrine/exocrine gland phenotype |
| MP:0005378 | growth/size/body region phenotype |
| MP:0005377 | hearing/vestibular/ear phenotype |
| MP:0005397 | hematopoietic system phenotype |
| MP:0005376 | homeostasis/metabolism phenotype |
| MP:0005387 | immune system phenotype |
| MP:0010771 | integument phenotype |
| MP:0005371 | limbs/digits/tail phenotype |
| MP:0005370 | liver/biliary system phenotype |
| MP:0010768 | mortality/aging |
| MP:0005369 | muscle phenotype |
| MP:0003631 | nervous system phenotype |
| MP:0001186 | pigmentation phenotype |
| MP:0005367 | renal/urinary system phenotype |
| MP:0005389 | reproductive system phenotype |
| MP:0005388 | respiratory system phenotype |
| MP:0005390 | skeleton phenotype |
| MP:0005394 | taste/olfaction phenotype |
| MP:0002006 | tumorigenesis |
| MP:0005391 | vision/eye phenotype |
Table 5. Intermediate level MP ontology slim used by DMDD.
A list of the Mammalian Phenotype Ontology IDs and names of terms selected as the intermediate level ontology slim.
| MP:0000001 | mammalian phenotype |
| MP:0002873 | normal phenotype |
| MP:0002169 | no abnormal phenotype detected |
| MP:0005375 | adipose tissue phenotype |
| MP:0000003 | abnormal adipose tissue morphology |
| MP:0005666 | abnormal adipose tissue physiology |
| MP:0004924 | abnormal behavior |
| MP:0020222 | abnormal alertness |
| MP:0011275 | abnormal behavioral response to light |
| MP:0009745 | abnormal behavioral response to xenobiotic |
| MP:0001502 | abnormal circadian rhythm |
| MP:0002069 | abnormal consumption behavior |
| MP:0002572 | abnormal emotion/affect behavior |
| MP:0001440 | abnormal grooming behavior |
| MP:0010698 | abnormal impulsive behavior control |
| MP:0002063 | abnormal learning/memory/conditioning |
| MP:0002066 | abnormal motor capabilities/coordination/movement |
| MP:0002067 | abnormal sensory capabilities/reflexes/nociception |
| MP:0011396 | abnormal sleep behavior |
| MP:0002557 | abnormal social/conspecific interaction |
| MP:0001529 | abnormal vocalization |
| MP:0002822 | catalepsy |
| MP:0002899 | fatigue |
| MP:0002064 | seizures |
| MP:0002127 | abnormal cardiovascular system morphology |
| MP:0001614 | abnormal blood vessel morphology |
| MP:0002925 | abnormal cardiovascular development |
| MP:0000266 | abnormal heart morphology |
| MP:0003279 | aneurysm |
| MP:0013332 | peliosis |
| MP:0001544 | abnormal cardiovascular system physiology |
| MP:0002128 | abnormal blood circulation |
| MP:0010695 | abnormal blood pressure regulation |
| MP:0000249 | abnormal blood vessel physiology |
| MP:0004039 | abnormal cardiac cell glucose uptake |
| MP:0002972 | abnormal cardiac muscle contractility |
| MP:0004084 | abnormal cardiac muscle relaxation |
| MP:0011926 | abnormal cardiac valve physiology |
| MP:0011390 | abnormal fetal cardiomyocyte physiology |
| MP:0011925 | abnormal heart echocardiography feature |
| MP:0008775 | abnormal heart ventricle pressure |
| MP:0004085 | abnormal heartbeat |
| MP:0003137 | abnormal impulse conducting system conduction |
| MP:0020095 | abnormal mean heart rate adaptation |
| MP:0004215 | abnormal myocardial fiber physiology |
| MP:0003547 | abnormal pulmonary pressure |
| MP:0020092 | abnormal susceptibility to aortic cartilaginous metaplasia |
| MP:0020098 | abnormal susceptibility to diet-induced aortic fatty streak lesions |
| MP:0000230 | abnormal systemic arterial blood pressure |
| MP:0004484 | altered response of heart to induced stress |
| MP:0000343 | altered response to myocardial infarction |
| MP:0005330 | cardiomyopathy |
| MP:0006138 | congestive heart failure |
| MP:0001853 | heart inflammation |
| MP:0003328 | portal hypertension |
| MP:0005384 | cellular phenotype |
| MP:0000358 | abnormal cell morphology |
| MP:0005621 | abnormal cell physiology |
| MP:0013258 | abnormal extracellular matrix morphology |
| MP:0003121 | genetic imprinting |
| MP:0005382 | craniofacial phenotype |
| MP:0000428 | abnormal craniofacial morphology |
| MP:0002116 | abnormal craniofacial bone morphology |
| MP:0003935 | abnormal craniofacial development |
| MP:0003743 | abnormal facial morphology |
| MP:0011495 | abnormal head shape |
| MP:0002177 | abnormal outer ear morphology |
| MP:0005381 | digestive/alimentary phenotype |
| MP:0000462 | abnormal digestive system morphology |
| MP:0001663 | abnormal digestive system physiology |
| MP:0005380 | embryogenesis phenotype |
| MP:0001672 | abnormal embryogenesis/ development |
| MP:0002084 | abnormal developmental patterning |
| MP:0001697 | abnormal embryo size |
| MP:0002085 | abnormal embryonic tissue morphology |
| MP:0008926 | abnormal anterior definitive endoderm morphology |
| MP:0013230 | abnormal cervical sinus morphology |
| MP:0003085 | abnormal egg cylinder morphology |
| MP:0010115 | abnormal embryonic cloaca morphology |
| MP:3000001 | abnormal gastrula morphology |
| MP:0011411 | abnormal gonadal ridge morphology |
| MP:0011257 | abnormal head fold morphology |
| MP:0011260 | abnormal head mesenchyme morphology |
| MP:0012187 | abnormal intraembryonic coelom morphology |
| MP:0005650 | abnormal limb bud morphology |
| MP:0006301 | abnormal mesenchyme morphology |
| MP:0008487 | abnormal mesonephros morphology |
| MP:0011256 | abnormal neural fold morphology |
| MP:0005657 | abnormal neural plate morphology |
| MP:0002151 | abnormal neural tube morphology/development |
| MP:0002825 | abnormal notochord morphology |
| MP:0002884 | abnormal pharyngeal arch morphology |
| MP:0013231 | abnormal pharyngeal groove morphology |
| MP:0013232 | abnormal pharyngeal membrane morphology |
| MP:0006031 | abnormal pharyngeal pouch morphology |
| MP:0012496 | abnormal pleuropericardial membrane morphology |
| MP:0002399 | abnormal pluripotent precursor cell morphology/development |
| MP:0013217 | abnormal posterior definitive endoderm morphology |
| MP:0003885 | abnormal rostral-caudal body axis extension |
| MP:0012252 | abnormal septum transversum morphology |
| MP:0001688 | abnormal somite development |
| MP:0002861 | abnormal tail bud morphology |
| MP:0011258 | abnormal tail fold morphology |
| MP:0001674 | abnormal triploblastic development |
| MP:0011835 | abnormal urogenital fold morphology |
| MP:0011853 | abnormal urorectal septum morphology |
| MP:0003988 | disorganized embryonic tissue |
| MP:0013241 | embryo tissue necrosis |
| MP:0008932 | abnormal embryonic tissue physiology |
| MP:0003890 | abnormal embryonic-extraembryonic boundary morphology |
| MP:0002086 | abnormal extraembryonic tissue morphology |
| MP:0001726 | abnormal allantois morphology |
| MP:0005029 | abnormal amnion morphology |
| MP:0011199 | abnormal amniotic cavity morphology |
| MP:0002836 | abnormal chorion morphology |
| MP:0011202 | abnormal ectoplacental cavity morphology |
| MP:0003396 | abnormal embryonic hematopoiesis |
| MP:0011200 | abnormal extraembryonic coelom morphology |
| MP:0010736 | abnormal extraembryonic ectoderm morphology |
| MP:0001724 | abnormal extraembryonic endoderm formation |
| MP:0006323 | abnormal extraembryonic mesoderm development |
| MP:0011203 | abnormal parietal yolk sac morphology |
| MP:0001711 | abnormal placenta morphology |
| MP:0011197 | abnormal proamniotic cavity morphology |
| MP:0001725 | abnormal umbilical cord morphology |
| MP:0011201 | abnormal visceral yolk sac cavity morphology |
| MP:0001718 | abnormal visceral yolk sac morphology |
| MP:0003229 | abnormal vitelline vasculature morphology |
| MP:0002582 | disorganized extraembryonic tissue |
| MP:0004264 | abnormal extraembryonic tissue physiology |
| MP:0004966 | abnormal inner cell mass proliferation |
| MP:0009781 | abnormal preimplantation embryo development |
| MP:0011186 | abnormal visceral endoderm morphology |
| MP:0012028 | abnormal visceral endoderm physiology |
| MP:0001730 | embryonic growth arrest |
| MP:0003984 | embryonic growth retardation |
| MP:0005379 | endocrine/exocrine gland phenotype |
| MP:0002163 | abnormal gland morphology |
| MP:0002164 | abnormal gland physiology |
| MP:0005378 | growth/size/body region phenotype |
| MP:0009701 | abnormal birth body size |
| MP:0005451 | abnormal body composition |
| MP:0003385 | abnormal body wall morphology |
| MP:0004134 | abnormal chest morphology |
| MP:0000432 | abnormal head morphology |
| MP:0012719 | abnormal neck morphology |
| MP:0002089 | abnormal postnatal growth/weight/body size |
| MP:0004196 | abnormal prenatal growth/weight/body size |
| MP:0001270 | distended abdomen |
| MP:0004133 | heterotaxia |
| MP:0013328 | visceromegaly |
| MP:0005377 | hearing/vestibular/ear phenotype |
| MP:0002102 | abnormal ear morphology |
| MP:0003938 | abnormal ear development |
| MP:0000026 | abnormal inner ear morphology |
| MP:0000049 | abnormal middle ear morphology |
| MP:0002177 | abnormal outer ear morphology |
| MP:0003878 | abnormal ear physiology |
| MP:0005397 | hematopoietic system phenotype |
| MP:0002396 | abnormal hematopoietic system morphology/development |
| MP:0002429 | abnormal blood cell morphology/development |
| MP:0002398 | abnormal bone marrow cell morphology/development |
| MP:0004808 | abnormal hematopoietic stem cell morphology |
| MP:0000689 | abnormal spleen morphology |
| MP:0000703 | abnormal thymus morphology |
| MP:0001545 | abnormal hematopoietic system physiology |
| MP:0005376 | homeostasis/metabolism phenotype |
| MP:0001764 | abnormal homeostasis |
| MP:0005266 | abnormal metabolism |
| MP:0008872 | abnormal physiological response to xenobiotic |
| MP:0005164 | abnormal response to injury |
| MP:0000604 | amyloidosis |
| MP:0013027 | wounding |
| MP:0005387 | immune system phenotype |
| MP:0000685 | abnormal immune system morphology |
| MP:0000716 | abnormal immune system cell morphology |
| MP:0002722 | abnormal immune system organ morphology |
| MP:0001879 | abnormal lymphatic vessel morphology |
| MP:0001790 | abnormal immune system physiology |
| MP:0010771 | integument phenotype |
| MP:0010678 | abnormal skin adnexa morphology |
| MP:0010680 | abnormal skin adnexa physiology |
| MP:0002060 | abnormal skin morphology |
| MP:0005501 | abnormal skin physiology |
| MP:0001968 | abnormal touch/ nociception |
| MP:0005371 | limbs/digits/tail phenotype |
| MP:0002109 | abnormal limb morphology |
| MP:0000572 | abnormal autopod morphology |
| MP:0000550 | abnormal forelimb morphology |
| MP:0000556 | abnormal hindlimb morphology |
| MP:0002115 | abnormal limb bone morphology |
| MP:0006279 | abnormal limb development |
| MP:0012000 | abnormal limb position |
| MP:0000549 | absent limbs |
| MP:0008985 | hemimelia |
| MP:0013069 | limb wound |
| MP:0000548 | long limbs |
| MP:0013133 | pale limbs |
| MP:0000547 | short limbs |
| MP:0020288 | supernumerary limbs |
| MP:0002111 | abnormal tail morphology |
| MP:0005370 | liver/biliary system phenotype |
| MP:0002138 | abnormal hepatobiliary system morphology |
| MP:0005083 | abnormal biliary tract morphology |
| MP:0003943 | abnormal hepatobiliary system development |
| MP:0000598 | abnormal liver morphology |
| MP:0010040 | abnormal oval cell morphology |
| MP:0002139 | abnormal hepatobiliary system physiology |
| MP:0010768 | mortality/aging |
| MP:0005369 | muscle phenotype |
| MP:0002108 | abnormal muscle morphology |
| MP:0002106 | abnormal muscle physiology |
| MP:0003631 | nervous system phenotype |
| MP:0003632 | abnormal nervous system morphology |
| MP:0002751 | abnormal autonomic nervous system morphology |
| MP:0002152 | abnormal brain morphology |
| MP:0002653 | abnormal ependyma morphology |
| MP:0003634 | abnormal glial cell morphology |
| MP:0002184 | abnormal innervation |
| MP:0005623 | abnormal meninges morphology |
| MP:0003861 | abnormal nervous system development |
| MP:0000778 | abnormal nervous system tract morphology |
| MP:0002882 | abnormal neuron morphology |
| MP:0002752 | abnormal somatic nervous system morphology |
| MP:0000955 | abnormal spinal cord morphology |
| MP:0008493 | alpha-synuclein inclusion body |
| MP:0003329 | amyloid beta deposits |
| MP:0012260 | encephalomeningocele |
| MP:0002229 | neurodegeneration |
| MP:0003012 | no phenotypic analysis |
| MP:0005395 | other phenotype |
| MP:0001186 | pigmentation phenotype |
| MP:0005367 | renal/urinary system phenotype |
| MP:0000516 | abnormal renal/urinary system morphology |
| MP:0011782 | abnormal internal urethral orifice morphology |
| MP:0002135 | abnormal kidney morphology |
| MP:0005187 | abnormal penis morphology |
| MP:0000534 | abnormal ureter morphology |
| MP:0011487 | abnormal ureteropelvic junction morphology |
| MP:0011488 | abnormal ureterovesical junction morphology |
| MP:0000537 | abnormal urethra morphology |
| MP:0000538 | abnormal urinary bladder morphology |
| MP:0003942 | abnormal urinary system development |
| MP:0003630 | abnormal urothelium morphology |
| MP:0003129 | persistent cloaca |
| MP:0005360 | urolithiasis |
| MP:0005502 | abnormal renal/urinary system physiology |
| MP:0003633 | abnormal nervous system physiology |
| MP:0005389 | reproductive system phenotype |
| MP:0002160 | abnormal reproductive system morphology |
| MP:0001119 | abnormal female reproductive system morphology |
| MP:0001929 | abnormal gametogenesis |
| MP:0005149 | abnormal gubernaculum morphology |
| MP:0003673 | abnormal inguinal canal morphology |
| MP:0001145 | abnormal male reproductive system morphology |
| MP:0003315 | abnormal perineum morphology |
| MP:0003936 | abnormal reproductive system development |
| MP:0002210 | abnormal sex determination |
| MP:0000653 | abnormal sex gland morphology |
| MP:0013055 | genital wound |
| MP:0001919 | abnormal reproductive system physiology |
| MP:0005388 | respiratory system phenotype |
| MP:0002132 | abnormal respiratory system morphology |
| MP:0002249 | abnormal larynx morphology |
| MP:0001175 | abnormal lung morphology |
| MP:0002233 | abnormal nose morphology |
| MP:0002240 | abnormal paranasal sinus morphology |
| MP:0002234 | abnormal pharynx morphology |
| MP:0010820 | abnormal pleura morphology |
| MP:0012684 | abnormal pleural cavity morphology |
| MP:0010942 | abnormal respiratory epithelium morphology |
| MP:0003115 | abnormal respiratory system development |
| MP:0002282 | abnormal trachea morphology |
| MP:0002133 | abnormal respiratory system physiology |
| MP:0005390 | skeleton phenotype |
| MP:0005508 | abnormal skeleton morphology |
| MP:0009250 | abnormal appendicular skeleton morphology |
| MP:0002114 | abnormal axial skeleton morphology |
| MP:0003795 | abnormal bone structure |
| MP:0000163 | abnormal cartilage morphology |
| MP:0011849 | abnormal clitoral bone morphology |
| MP:0002932 | abnormal joint morphology |
| MP:0005504 | abnormal ligament morphology |
| MP:0006322 | abnormal perichondrium morphology |
| MP:0002113 | abnormal skeleton development |
| MP:0005503 | abnormal tendon morphology |
| MP:0000566 | synostosis |
| MP:0001533 | abnormal skeleton physiology |
| MP:0005394 | taste/olfaction phenotype |
| MP:0005500 | abnormal gustatory system morphology |
| MP:0001002 | abnormal taste bud morphology |
| MP:0001985 | abnormal gustatory system physiology |
| MP:0005499 | abnormal olfactory system morphology |
| MP:0006292 | abnormal nasal placode morphology |
| MP:0008789 | abnormal olfactory epithelium morphology |
| MP:0012067 | abnormal olfactory gland morphology |
| MP:0001983 | abnormal olfactory system physiology |
| MP:0002006 | tumorigenesis |
| MP:0005391 | vision/eye phenotype |
| MP:0002092 | abnormal eye morphology |
| MP:0005193 | abnormal anterior eye segment morphology |
| MP:0001286 | abnormal eye development |
| MP:0001299 | abnormal eye distance/ position |
| MP:0003686 | abnormal eye muscle morphology |
| MP:0001324 | abnormal eye pigmentation |
| MP:0002697 | abnormal eye size |
| MP:0001340 | abnormal eyelid morphology |
| MP:0008968 | abnormal lacrimal apparatus morphology |
| MP:0010030 | abnormal orbit morphology |
| MP:0005195 | abnormal posterior eye segment morphology |
| MP:0002698 | abnormal sclera morphology |
| MP:0005197 | abnormal uvea morphology |
| MP:0001293 | anophthalmia |
| MP:0006209 | calcified intraocular region |
| MP:0013146 | eye lesions |
| MP:0009859 | eye opacity |
| MP:0013170 | eye swellings |
| MP:0006225 | ocular rupture |
| MP:0001788 | periorbital edema |
| MP:0005254 | strabismus |
| MP:0005253 | abnormal eye physiology |
Table 6. New MP terms derived from embryo phenotyping.
A list of the Mammalian Phenotype Ontology IDs along with their corresponding term name. These have been added to the ontology to allow annotation of abnormalities observed in the embryos which could not be adequately described by existing terms.
| MP:0013809 | absent pectinate muscle |
| MP:0013810 | absent brachiocephalic trunk |
| MP:0013812 | enlarged orbital veins |
| MP:0013813 | dilated hepatic portal vein |
| MP:0013814 | abnormal hepatic portal vein connection |
| MP:0013816 | absent digastric muscle |
| MP:0013817 | absent nasal cavity |
| MP:0013818 | abnormal oral cavity morphology |
| MP:0013819 | abnormal acromioclavicular joint morphology |
| MP:0013820 | absent optic cup |
| MP:0013823 | absent segment of anterior cerebral artery |
| MP:0013825 | small hypoglossal canal |
| MP:0013826 | absent hypoglossal canal |
| MP:0013827 | thin oculomotor nerve |
| MP:0013828 | thin facial nerve |
| MP:0013829 | thin splanchnic nerve |
| MP:0013830 | abnormal intrathoracic topology of vagus nerve |
| MP:0013831 | vagus nerve compression |
| MP:0013832 | thin vagus nerve |
| MP:0013833 | absent olfactory nerve |
| MP:0013834 | thin hypoglossal nerve |
| MP:0013835 | absent hypoglossal nerve |
| MP:0013836 | abnormal hypoglossal nerve topology |
| MP:0013837 | abnormal vagus nerve topology |
| MP:0013838 | small caudate nucleus |
| MP:0013840 | absent segment of posterior cerebral artery |
| MP:0013841 | abnormal lymphatic vessel topology |
| MP:0013842 | ductus venosus stenosis |
| MP:0013843 | hepatic portal vein stenosis |
| MP:0013844 | abnormal perichondrial ossification |
| MP:0013845 | abnormal eye muscle topology |
| MP:0013846 | retropharyngeal edema |
| MP:0013847 | retropleural edema |
| MP:0013848 | subcutaneous edema |
| MP:0013849 | absent abducens nerve |
| MP:0013850 | absent posterior commissure |
| MP:0013851 | abnormal Wolffian duct topology |
| MP:0013852 | abnormal Mullerian duct topology |
| MP:0013853 | abnormal hepatic portal vein formation |
| MP:0013855 | absent celiac artery |
| MP:0013857 | abnormal abdominal muscle morphology |
| MP:0013858 | abnormal azygos vein topology |
| MP:0013859 | abnormal vitelline vein connection |
| MP:0013860 | anastomosis between common carotid and vertebral artery |
| MP:0013861 | abnormal pancreas topology |
| MP:0013862 | abnormal cecum position |
| MP:0013864 | enlarged paraumbilical vein |
| MP:0013865 | abnormal dorsal pancreas topology |
| MP:0013868 | abnormal ventral pancreas topology |
| MP:0013869 | vascular diverticulum |
| MP:0013870 | absent proximal internal carotid artery segment |
| MP:0013871 | abnormal stapedial artery topology |
| MP:0013873 | abnormal ductus venosus morphology |
| MP:0013874 | abnormal ductus venosus topology |
| MP:0013875 | trigeminal neuroma |
| MP:0013876 | absent ductus venosus valve |
| MP:0013877 | abnormal ductus venosus valve morphology |
| MP:0013878 | abnormal ductus venosus valve topology |
| MP:0013879 | duplication of ductus venosus |
| MP:0013880 | absent ductus venosus |
| MP:0013913 | absent rib-vertebral column attachment |
| MP:0013914 | absent intracranial segment of vertebral artery |
| MP:0013915 | abnormal brachial plexus formation |
| MP:0013916 | decreased intestine length |
| MP:0013917 | persistent right 6th pharyngeal arch artery |
| MP:0013918 | abnormal endolymphatic sac topology |
| MP:0013923 | small prevertebral sympathetic ganglia |
| MP:0013924 | abnormal dural venous sinus morphology |
| MP:0013925 | abnormal vascular plexus formation |
| MP:0013926 | absent neurohypophysis |
| MP:0013927 | abnormal facial nerve topology |
| MP:0013928 | thin motoric part of trigeminal nerve |
| MP:0013929 | absent eye muscles |
| MP:0013930 | abnormal digastric muscle connection |
| MP:0013931 | abnormal olfactory bulb position |
| MP:0013932 | fragmented Meckel's cartilage |
| MP:0013933 | short Meckel's cartilage |
| MP:0013934 | supratentorial ventricles enlargement |
| MP:0013935 | basal brain tissue herniation |
| MP:0013936 | abnormal thymus topology |
| MP:0013937 | absent lobe of thyroid gland |
| MP:0013938 | abnormal esophagus topology |
| MP:0013943 | abnormal ureter topology |
| MP:0013944 | persistent cloacal membrane |
| MP:0013945 | abnormal elbow joint morphology |
| MP:0013946 | abnormal perirectal tissue morphology |
| MP:0013947 | abnormal paraaortic body morphology |
| MP:0013948 | intraembryonal intestine elongation |
| MP:0013949 | fusion of axis and occipital bones |
| MP:0013950 | abnormal dorsal root ganglion topology |
| MP:0013951 | abnormal descending aorta topology |
| MP:0013952 | retro-esophageal left subclavian artery |
| MP:0013953 | left sided brachiocephalic trunk |
| MP:0013963 | jugular vein stenosis |
| MP:0013964 | absent tongue muscles |
| MP:0013965 | abnormally deep median sulcus of tongue |
| MP:0013967 | abnormal infrahyoid muscle connection |
| MP:0013968 | multiple persisting craniopharyngeal ducts |
| MP:0013969 | reduced sympathetic cervical ganglion size |
| MP:0013970 | absent connection between subcutaneous lymph vessels and lymph sac |
| MP:0013971 | blood in lymph vessels |
| MP:0013972 | occipital vertebra |
| MP:0013973 | abnormal hepatic vein connection |
| MP:0013974 | abnormal coronary vein connection |
| MP:0013975 | abnormal coronary sinus connection |
| MP:0013976 | abnormal left vena cava superior connection |
| MP:0013977 | symmetric azygos veins |
| MP:0013978 | abnormal carotid artery origin |
| MP:0013979 | abnormal subclavian artery origin |
| MP:0013980 | abnormal pulmonary artery origin |
| MP:0013981 | double lumen aortic arch |
| MP:0013982 | inverse situs of great intrathoracic arteries |
| MP:0013984 | abnormal superior mesenterial vein connection |
| MP:0013985 | abnormal umbilical vein topology |
| MP:0013986 | abnormal vitelline vein topology |
| MP:0013987 | absent intrahepatic inferior vena cava segment |
| MP:0013988 | absent portal vein segment |
| MP:0013989 | symmetric hepatic veins |
| MP:0013991 | abnormal common iliac artery origin |
| MP:0013992 | persistent dorsal ophthalmic artery |
| MP:0013993 | anastomosis between basilar artery and common carotid artery |
| MP:0013994 | abnormal parasellar internal carotid artery branch morphology |
| MP:0013995 | abnormal external carotid artery origin |
| MP:0013996 | abnormal vertebral artery origin |
| MP:0013997 | abnormal internal carotid artery topology |
| MP:0013998 | absent canalicular internal carotid artery segment |
| MP:0013999 | absent parasellar internal carotid artery |
| MP:0014000 | anastomosis between internal carotid artery and basilar artery |
| MP:0014001 | abnormal vertebral artery topology |
| MP:0014002 | absent extracranial vertebral artery segment |
| MP:0014003 | additional anastomosis between intracranial vertebral arteries |
| MP:0014004 | absent basilar artery segment |
| MP:0014006 | absent posterior communicating artery |
| MP:0014008 | absent labyrinthine artery |
| MP:0014009 | anastomosis between middle cerebral arteries |
| MP:0014011 | abnormal ovary tissue architecture |
| MP:0014017 | abnormal Wolffian duct connection |
| MP:0014018 | embryo tumor |
| MP:0014019 | embryo cyst |
| MP:0014020 | intramural bleeding in blood vessel wall |
| MP:0014021 | heterochrony |
| MP:0014022 | abnormal duodenum topology |
When grouped into such high level MP ontology terms, the most common group of abnormalities are those affecting the cardiovascular system, examples of which affect embryos in every single mutant line studied. Almost as prevalent are nervous system phenotypes, which are detected in 80% of the lines studied. Re-plotting the data summarised by intermediate level MP term slim provides a more detailed view of the prevalence and variability in penetrance of phenotypes ( Figure 5B). At this level of resolution, for example, cardiovascular defects are subdivided into two broad categories; those encompassing abnormalities in blood vessel morphology or topology (“abnormal blood vessel morphology” and most phenotypes within “abnormal cardiovascular development”) and those affecting the heart and its great vessels (“abnormal heart morphology”). Viewed in this way, it is clear that detection of cardiovascular defects in all lines examined results from the presence of phenotypes in the vasculature. These range from relatively major defects such as absence of the ductus venosus, interrupted aortic arch or arterial stenosis, to more minor alterations in vascular topology in different regions of the embryo. Cardiac abnormalities nevertheless remain prevalent, affecting almost two thirds (27/42) of the mutant lines. These encompass malformations in all regions of the four-chambered heart and its great vessels, including both atrial and ventricular septal defects, atrioventricular septal defects, common arterial trunk, double outlet right ventricle, transposition of the great arteries, bicuspid aortic valve, common truncal valve and abnormally thin myocardium. After blood vessel and cardiac abnormalities, the third most prevalent group of phenotypes detected were those affecting brain morphology ( Figure 5B), most commonly the forebrain ( Figure 6 and Supplementary Table 1).
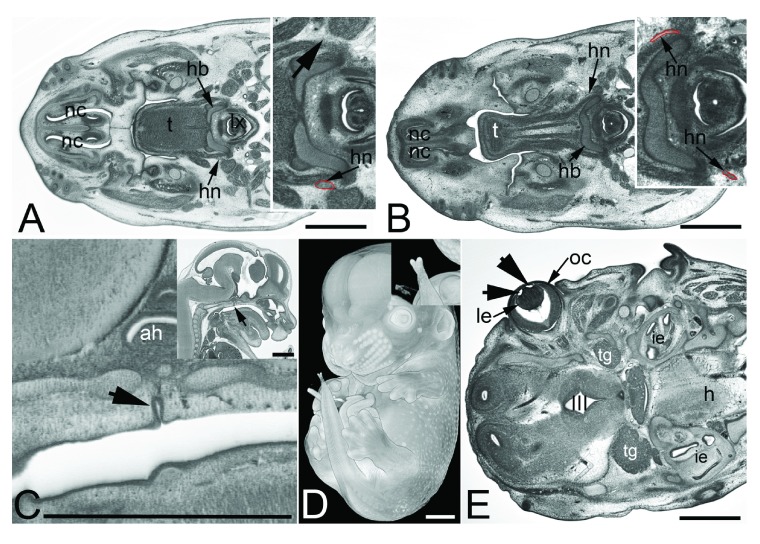
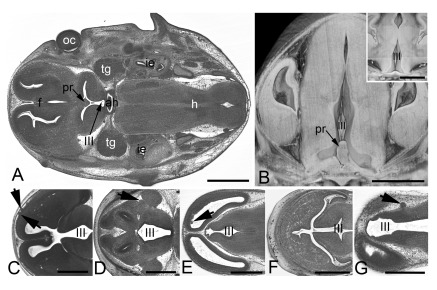
Figure 6. Abnormal brain morphology phenotypes.
A and B. Tissue protrusion (pr) into the 3rd ventricle (III) in an original HREM-section ( A) and a volume model ( B). Inlay in B shows normal situation in a control. C. Irregular tissue protrusions (arrowheads) on the brain surface in a 4933434E20Rik -/- embryo. D. Abnormal tissue (arrowhead) at the cortex near the lateral sulcus in a Polb -/- embryo. E. Abnormal frontal wall of the lateral ventricles in a H13 -/- embryo. F. Abnormal morphology and tissue architecture (arrowhead) of the frontal forebrain in a Chtop -/- embryo. G. Abnormal morphology of the wall of the 3rd ventricle and protrusions (arrowhead) on the surface of the diencephalon in a Brd2 -/- embryo. ah, adenohypophysis; f, forebrain; h, hindbrain; ie, inner ear; oc, optic cup; pr, tissue protrusion; tg, trigeminal ganglion; III, 3rd ventricle; Scale bars 1 mm
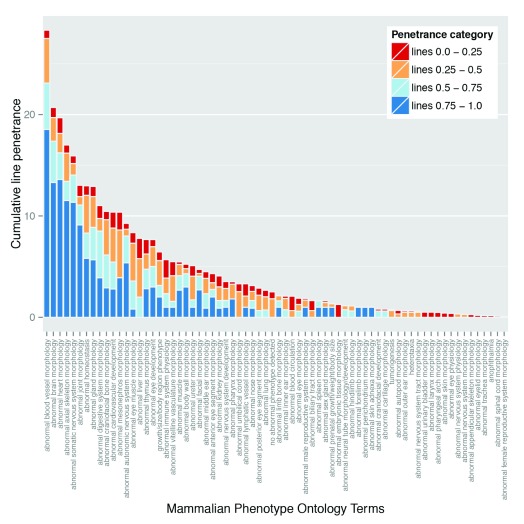
In order to assess the relative significance of each phenotype in the context of variable penetrance, we re-examined their ranking distribution after weighting each phenotype according to its individual prevalence. This provides a plot of cumulative line penetrance for each of the 70 intermediate level MP term slim ( Figure 7). Whilst abnormalities in blood vessel morphology and structure of the heart remain amongst the most prevalent phenotypes, weighting by penetrance has a significant impact on the ranking of other phenotypes. Notably, the relative ranking of “abnormal brain morphology” and “abnormal somatic nervous system morphology” is increased, with both now lying in the five most prevalent abnormalities scored. This change is largely driven by the relatively high prevalence associated with abnormalities in forebrain morphology and hypoglossal nerve structure or presence, respectively.
Figure 7. Cumulative penetrance of individual phenotypes.
Data from the global analysis of the frequency of phenotype terms (see Materials and Methods) was plotted to show the cumulative penetrance score for each of the phenotype categories observed (i.e. the overall sum of the penetrance scores recorded for the lines showing the phenotype). The Mammalian Phenotype Ontology terms assigned during embryo phenotyping were summarised using the intermediate level DMDD ontology slim, and the data was ordered according to the cumulative penetrance score. The colours indicate the contribution of lines falling into each of the distinct penetrance categories to the cumulative penetrance score.
Phenotyping embryos required new MP terms
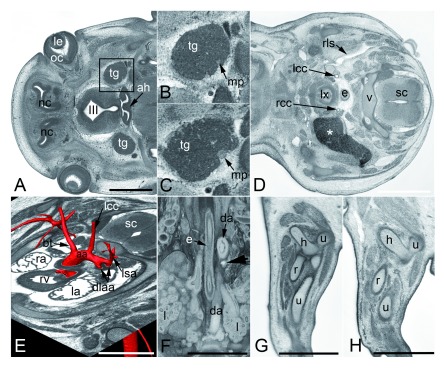
Adoption of a formal, standardised ontology for scoring abnormalities provides an essential framework for analysing the data and facilitating structured search enquiries. However, during the course of the DMDD programme and its pilot study 8, it became clear that additional terms were required in order to adequately describe abnormalities in embryo, as opposed to adult structures. A further outcome of the DMDD study has therefore been the creation of 142 new MP terms to accommodate the range of abnormalities we have observed ( Table 6). These include, for example, thin motoric part of the trigeminal nerve (MP:0013928; http://www.ontobee.org/ontology/MP?iri=http://purl.obolibrary.org/obo/MP_0013928), blood in lymph vessels (MP:0013971; http://www.ontobee.org/ontology/MP?iri=http://purl.obolibrary.org/obo/MP_0013971), double lumen aortic arch (MP:0013981; http://www.ontobee.org/ontology/MP?iri=http://purl.obolibrary.org/obo/MP_0013981), abnormal elbow joint morphology (MP:0013945; http://www.ontobee.org/ontology/MP?iri=http://purl.obolibrary.org/obo/MP_0013945), and intramural bleeding in blood vessel wall (MP:0014020; http://www.ontobee.org/ontology/MP?iri=http://purl.obolibrary.org/obo/MP_0014020)( Figure 8).
Figure 8. Examples of new MP phenotypes.
A– C. “Thin motoric part of trigeminal nerve”. Original HREM sections through the head of a Polb -/- embryo ( A, B) and a control ( C). Box in A indicates section displayed in B. D. “Blood in lymph vessels”, as appearing in an original HREM section through the neck of a 1700067K01Rik -/- embryo. Note the blood filled left lymph sac (asterisk). Use the right sided lymph sac (rls) as a control. E. Double lumen aortic arch. Surface model of the great intrathoracic arteries on top of an original HREM section of a Pdzk1 -/- embryo. (Compare with 16). F. “Intramural bleeding in blood vessel wall” (arrowhead) in the descending aorta (da) of an Akap9 -/- embryo from the DMDD pilot study 8. Coronal section through a volume model. G– H. “Abnormal elbow joint morphology” Sagittal sections. Normal situation in a control ( G). Fusion of humerus (h) und ulna material (u) in an Atp11a -/- embryo. aa, aortic arch; ah, adenohypophysis; bt, brachiocephalic trunk; da, descending aorta; dlaa, double lumen aortic arch; e, esophagus; h, humerus; l, lung; la, left atrium; lcc, left common carotid artery; le, lens; lsa, left subclavian artery; lx, larynx; mp, motoric part of trigeminal nerve; nc, nasal cavity; oc, optic cup; r, radius; ra, right atrium; rcc, right common carotid artery; rv, right ventricle; sc, spinal chord; tg, trigeminal ganglion; u, ulna; v, vertrebral body; III, 3rd ventricle; Scale bars 1 mm.
Discussion
Since approximately one third of gene knockouts in the mouse prove to be embryonic or perinatal lethal 1– 3, further study of such lines offers a unique opportunity to better understand the genetic regulation of embryo development and identify genetic determinants of congenital abnormalities. The data accumulated during three years of the DMDD programme provide the first opportunity to study in detail the identity, range and prevalence of morphological abnormalities in such mutants and offer a window on the opportunities (and pitfalls) such systematic studies present.
The current analysis is restricted to a single developmental stage (E14.5) when most organ systems of the embryo have developed their definitive fetal appearance and the body plan is broadly similar to that of the adult mouse. Whilst this provides obvious practical advantages for a systematic, high throughput phenotyping programme, it is of course an arbitrary choice with respect to the time course of individual gene function and the consequences of gene ablation. Indeed, about 60% of the lethal lines entering the DMDD pipeline fail to provide homozygous mutant offspring by E14.5, with half of those causing lethality prior to E9.5 [see also 2]. The data here therefore comes from a subset of lethal lines. Furthermore, phenotypes observed at a single time point most likely combine more immediate consequences of individual gene loss with more distant or secondary consequences. Teasing out the role of regulative or compensatory changes from primary effects of gene loss is likely to be difficult. Despite these caveats, there are, nevertheless, several striking findings that emerge from detailed phenotype analysis.
Our finding that some manifestation of edema (generally subcutaneous) is the most common phenotype could indicate an unappreciated complexity in the genetic controls regulating fluid balance or tissue integrity of vascular or lymphatic components. Edema may also represent a common outcome for a wide range of pathophysiological perturbations, as has been proposed for the association of non-immune hydrops fetalis with human fetal loss 10, 11. The prevalence of cardiovascular defects is also consistent with the well established finding that cardiac abnormalities are the most common congenital defect in human newborns 12. Some caution is necessary in considering the mouse data, since as we have shown, a significant proportion of cardiovascular phenotypes comprise apparently minor alterations in blood vessel topology, the impact of which on normal development remains unclear. However, in addition to these, the lines we have studied show a range of severe abnormalities in cardiac structure that are both relatively prevalent and mirror the range of congenital abnormalities seen in humans. Despite the largely random selection of genes studied in screens such as DMDD, their identification as embryonic lethal therefore provides a dramatic enrichment for potential cardiac developmental disease alleles. Phenotypes affecting neural tissue also prove to be relatively prevalent in mutant embryos. We are limited in the present analysis to identifying a subset of neural deficits readily identified from HREM imaging. This restricts identifiable phenotypes to relatively gross alterations in brain and neural tube morphology, or changes affecting major nerves. Amongst the latter, the frequency with which abnormalities affecting the hypoglossal nerve have been detected is perhaps not so surprising, since these (like abnormalities detected in the motoric portion of the trigeminal nerve) may compromise suckling and lead to perinatal lethality.
The multiplicity of phenotypes frequently detected in individual mutant embryos is not unexpected, given the nature of a single time point screening procedure combined with the likely pleiotropic effects of individual gene loss. However, the most striking and surprising finding to emerge from the DMDD phenotype data is that virtually all phenotypes are incompletely (and frequently poorly) penetrant, despite the use of the isogenic C57BL/6N mouse strain. Combined with the observation of overlapping but distinct spectra of phenotypes between individual embryos from a single line, these findings are challenging to understand, and at a minimum point towards unknown stochastic components affecting the etiology of each phenotype or the compensatory responses they elicit 2. They also demonstrate that efforts to identify linkage between mouse embryo phenotypes and human developmental disease are likely to require sophisticated bioinformatic analysis beyond the obvious issues raised by species differences in anatomy and physiology.
It is also worth noting the several practical lessons which have become evident through the course of DMDD studies and which may be of value for similar embryo phenotyping programmes. The most pressing of these is basing phenotype detection on comparison of each mutant embryo with an appropriately staged normal counterpart 13. Embryos harvested at E14.5 vary markedly in their developmental progress and many tissues and organs are actively remodelled during this period. This is most obvious for the topology of the intestine, the position of the palatal shelves and the interventricular communication between left and right sides of the heart. Only with precise developmental staging is accurate phenotyping of these features possible (Geyer, Reissig, Rose, Wilson, Prin, Szumska, Ramirez-Solis, Tudor, White, Mohun and Weninger, unpublished report, ).
Whilst the precise range and detail of phenotypes that can be scored will necessarily be dictated by the nature of the imaging modality and the method of phenotype identification (compare, for example, 14, 15 with the manual annotation used in the present study), a common challenge is the development of protocols to minimise occurrence or subsequent scoring of apparent abnormalities that are more likely artefacts of sample preparation or processing. These can range from the more obvious ruptures of the embryo skin or damaged external features during dissection, to tissue shrinkage or swelling (causing organ deformation) as a result of dehydration, fixation or embedding. In addition to such artefacts, our study has demonstrated that a number of apparent abnormalities are common to both homozygous mutant embryos and wild-type controls from the C57BL/6N mouse strain. These include splitting of the tail tip, persistence of the craniopharyngeal duct with associated fenestration of head bones and the presence of vesicles in the lens of the eye ( Figure 4, panels C-E). A systematic appraisal of such phenotypes is currently in preparation.
Finally, the power of phenotypic screens such as DMDD to inform our understanding of developmental disease rests heavily on the detail with which abnormalities are scored. However, the very complexity we have seen this generates makes it all the more urgent to distinguish phenotypes not just through the nature of the morphological abnormality, but through its capacity, individually or in concert with others, to compromise subsequent fetal survival.
Data availability
Dataset 1 Zenodo: All phenotypes from lethal and sub-viable lines scored by DMDD to date (Nov 2016), 10.5281/zenodo.163506 17
The cumulative list of all scored phenotypes analysed in this study is presented in Dataset 1. The intermediate and high level slims of the MP ontology used in the analysis are presented in Table 4 and Table 5. All data used in this study is also available from the DMDD web site ( https://dmdd.org.uk) where phenotype annotations are available in tabular format by embryo and by line. In addition, they are identified at their appropriate locations within each 3D dataset of embryo images, which can be viewed in all three orthogonal section planes.
Acknowledgements
We are grateful for the contributions made by past and present members of the DMDD consortium and the support of their institutions, without which the DMDD programme would not be possible.
Funding Statement
This work was supported by the Wellcome Trust [100160], [FC001157], [FC001117]; Cancer Research UK [FC001157], [FC001117]; and the UK Medical Research Council [FC001157], [FC001117].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 1 approved, 2 approved with reservations]
Supplementary material
Supplementary Figure 1: Embryo numbers analysed for each mutant line.
The number of annotated embryos scored for each of the 42 lines was used to plot the variation in numbers of embryos analysed per line.
Supplementary Figure 2: Distribution of phenotypes amongst DMDD mutant lines (high level MP ontology slim).
Data from the global analysis of the frequency of phenotype terms (see Materials and Methods) is plotted to show the penetrance of phenotypes scored for each line, indicated by a colour gradient from light yellow (no penetrance) to dark red (100% penetrance). Each line is labelled after the symbol of the gene disrupted (see also Table 1), and the number of homozygous mutant embryos analysed for each line is shown. Phenotype annotations are summarised using the high level DMDD ontology slim.
Supplementary Figure 3: Distribution of phenotypes amongst DMDD mutant lines (intermediate level MP ontology slim).
As in Supplementary Figure 2, except that phenotype annotations are summarised using the intermediate level DMDD ontology slim.
Supplementary Table 1: Embryo phenotypes, organised by frequency.
The data from the global analysis of the frequency of phenotype terms (see Materials and methods) is presented in a structured fashion showing the relationship between Mammalian Phenotype Ontology terms included in the DMDD high level ontology slim, the DMDD intermediate level ontology slim, and the original annotation terms. The first three columns list the ID, term and frequency of DMDD high level ontology slim terms, columns 4–6 list the ID, term and frequency of the intermediate level ontology slim terms that cluster under the high level term listed in column 1, and for each intermediate level term the annotation phenotype terms are shown in order of frequency in columns 7–9. Column 10 lists the lines in which the phenotype listed in columns 7 was observed. The table rows are ordered according to the frequency of the high level ontology terms, intermediate ontology terms and original annotation terms.
References
- 1. Adams D, Baldock R, Bhattacharya S, et al. : Bloomsbury report on mouse embryo phenotyping: recommendations from the IMPC workshop on embryonic lethal screening. Dis Model Mech. 2013;6(3):571–9. 10.1242/dmm.011833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dickinson ME, Flenniken AM, Ji X, et al. : High-throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508–514. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mohun T, Adams DJ, Baldock R, et al. : Deciphering the Mechanisms of Developmental Disorders (DMDD): a new programme for phenotyping embryonic lethal mice. Dis Model Mech. 2013;6(3):562–6. 10.1242/dmm.011957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mohun TJ, Weninger WJ: Imaging heart development using high-resolution episcopic microscopy. Curr Opin Genet Dev. 2011;21(5):573–8. 10.1016/j.gde.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mohun TJ, Weninger WJ: Embedding embryos for high-resolution episcopic microscopy (HREM). Cold Spring Harb Protoc. 2012;2012(6):678–80. 10.1101/pdb.prot069583 [DOI] [PubMed] [Google Scholar]
- 6. Weninger WJ, Geyer SH, Mohun TJ, et al. : High-resolution episcopic microscopy: a rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat Embryol (Berl). 2006;211(3):213–21. 10.1007/s00429-005-0073-x [DOI] [PubMed] [Google Scholar]
- 7. Mohun TJ, Weninger WJ: Generation of volume data by episcopic three-dimensional imaging of embryos. Cold Spring Harb Protoc. 2012;2012(6):681–2. 10.1101/pdb.prot069591 [DOI] [PubMed] [Google Scholar]
- 8. Weninger WJ, Geyer SH, Martineau A, et al. : Phenotyping structural abnormalities in mouse embryos using high-resolution episcopic microscopy. Dis Model Mech. 2014;7(10):1143–52. 10.1242/dmm.016337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith CL, Eppig JT: The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm Genome. 2012;23(9–10):653–68. 10.1007/s00335-012-9421-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boycott KM, Vanstone MR, Bulman DE, et al. : Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14(10):681–91. 10.1038/nrg3555 [DOI] [PubMed] [Google Scholar]
- 11. Shamseldin HE, Tulbah M, Kurdi W, et al. : Identification of embryonic lethal genes in humans by autozygosity mapping and exome sequencing in consanguineous families. Genome Biol. 2015;16(1):116. 10.1186/s13059-015-0681-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffman JI, Kaplan S: The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39(12):1890–900. 10.1016/S0735-1097(02)01886-7 [DOI] [PubMed] [Google Scholar]
- 13. Wong MD, van Eede MC, Spring S, et al. : 4D atlas of the mouse embryo for precise morphological staging. Development. 2015;142(20):3583–91. 10.1242/dev.125872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wong MD, Dorr AE, Walls JR, et al. : A novel 3D mouse embryo atlas based on micro-CT. Development. 2012;139(17):3248–56. 10.1242/dev.082016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wong MD, Maezawa Y, Lerch JP, et al. : Automated pipeline for anatomical phenotyping of mouse embryos using micro-CT. Development. 2014;141(12):2533–41. 10.1242/dev.107722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geyer SH, Weninger WJ: Some mice feature 5th pharyngeal arch arteries and double-lumen aortic arch malformations. Cells Tissues Organs. 2012;196(1):90–8. 10.1159/000330789 [DOI] [PubMed] [Google Scholar]
- 17. Wilson R: Table 4: All pheotypes from lethal and subviable lines scored by DMDD to date (Nove 2016) [Data set]. Zenodo. 2016. Data Source