Abstract
Multiplex genome engineering in vivo with CRISPR/Cas9 shows great promise as a potential therapeutic approach. The ability to incorporate multiple single guide RNA (sgRNA) cassettes together with Cas9 gene expression in one AAV vector could greatly enhance the efficiency. In a recent Method article, Mefferd and coworkers indicated that small tRNA promoters could be used to drive sgRNA expression to facilitate the construction of a more effective AAV vector. In contrast, we found that when targeting endogenous genomic loci, CRISPR/Cas9 with tRNA promoter-driven sgRNA expression showed much reduced genome editing activity, compared with significant cleavage with U6 promoter-driven sgRNA expression. Though the underlying mechanisms are still under investigation, our study suggests that the CRISPR/Cas9 system with tRNA promoter-driven sgRNA expression needs to be reevaluated before it can be used for therapeutic genome editing.
Keywords: CRISPR/Cas9, genome editing, tRNA promoter
The use of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems for in vivo genome editing has emerged as a potential treatment for human diseases (Cox et al. 2015). The high targeting efficiency of CRISPR/Cas systems opens the possibility for multiplex genome engineering in vivo, which could be effective in treating certain diseases that require targeting of multiple genes or deleting a trunk of DNA sequences. Recent studies have demonstrated that in mouse models of Duchenne muscular dystrophy, mutated Dmd exon 23 can be successfully excised with paired flanking single guide RNAs (sgRNAs) in vivo using the smaller Cas9 ortholog form Staphylococcus aureus (SaCas9) delivered by adeno-associated virus (AAV), resulting in partially restored function of dystrophin protein (Long et al. 2016; Nelson et al. 2016; Tabebordbar et al. 2016). However, the use of two different AAVs for separate delivery of Cas9 and two sgRNAs rendered a relatively low efficiency of successful excision. One approach to increase the efficiency would be to have SaCas9 and two or even more different sgRNAs delivered by one AAV vector (Friedland et al. 2015). As AAV vectors have a DNA packaging limit of ∼5 kb, and the standard AAV–SaCRISPR system almost fills the cargo limitation, it would be helpful to further reduce the total length of the SaCRISPR system in order to make extra space for more sgRNA cassettes and to improve the packaging efficiency.
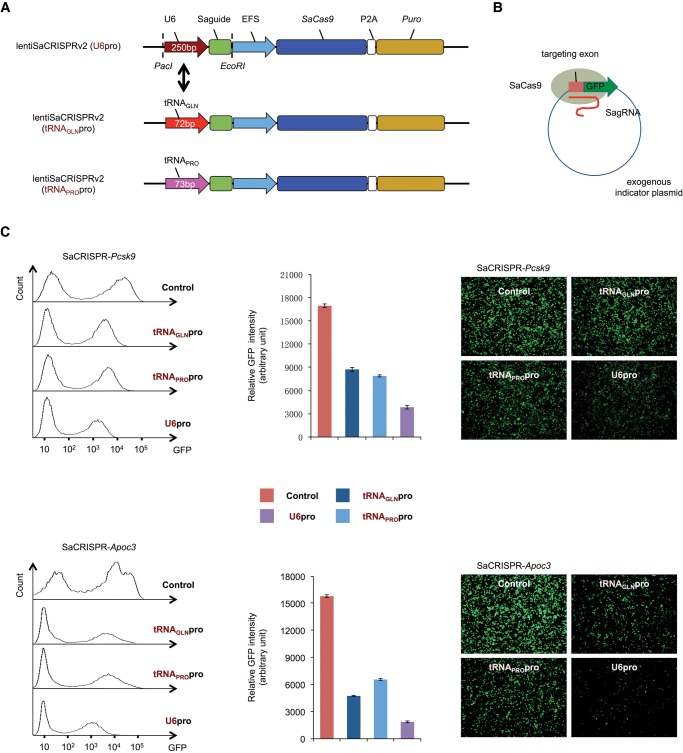
Mefferd et al. (2015) reported recently that expression of sgRNAs can be driven by small tRNA promoters (∼70 bp), making it possible to express two full-length sgRNAs using only ∼350 bp of space, about the size of one sgRNA-expressing cassette with a U6 promoter. We tested the potential of this approach by first comparing side-by-side the genome editing efficiency of CRISPR/Cas9 with U6 promoter- and tRNA promoter-driven sgRNA expression, respectively. For efficient delivery of CRISPR materials to cells in vitro, we reconstructed the original lentiCRISPR v2 for Streptococcus pyogenes (SpCas9) targeting (Sanjana et al. 2014) to make it adaptable for SaCas9 targeting by switching the Cas9 and sgRNA cassettes and named the new vector lentiSaCRISPR v2. We then made three versions of lentiSaCRISPR v2 constructs with different sgRNA promoters—U6pro, tRNAGLNpro, and tRNAPROpro—with the latter two tRNA promoters reported to be able to effectively drive the expression of sgRNAs in SaCRISPR mediated targeting (Fig. 1A; Mefferd et al. 2015). We next made constructs with different lentiSaCRISPR vectors targeting several mouse genes as well as a human gene, using previously screened sgRNA sequences that had shown high on-target efficiency with a U6 promoter.
FIGURE 1.
Study of editing efficiency of CRISPR/Cas9 with tRNA and U6 promoter-driven sgRNAs by using exogenous indicator plasmids. (A) LentiSaCRISPR v2 was modified from lentiCRISPR v2 by switching the respected Cas9 and sgRNA cassettes. tRNAGLN(GGTTCCATGGTGTAATGGTTAGCACTCTGGACTCTGAATCCAGCGATCCGAGTTCAAATCTCGGTGGAACCT) and tRNAPRO(TGGCTCGTTGGTCTAGGGGTATGATTCTCGCTTCGGGTGCGAGAGGTCCCGGGTTCAAATCCCGGACGAGCCC) promoter-driven sgRNA cassettes were synthesized and inserted between PacI and EcoRI cutting sites. (B) A schematic diagram of an editing efficiency study by using exogenous indicator plasmids. (C) gRNAs targeting endogenous mouse Pcsk9 and Apoc3 genes were cloned into a lentiSaCRISPR v2 plasmid. An extra G was added for Pcsk9 and Apoc3 targeting when the U6 promoter was used. Indicator plasmids were constructed by inserting sequences of targeted exons to the pEGFP-N1 plasmid between two restriction enzyme sites at the MCS region and 5′ to the GFP gene. Different combinations of lentiSaCRISPR and indicator plasmids were cotransfected to HEK 293T cells at a mass ratio of 3:1 as indicated, and cells were harvested for FACS analysis and GFP imaging after 72 h. Three independent experiments were carried out, and representative images from one experiment are shown. Average of GFP mean intensity in FACS analysis from all three experiments with SEM is indicated. The gRNA sequences are listed in Supplemental Table 1.
We initially used indicator plasmids for comparison of editing efficiency with different promoter-driven sgRNAs (Fig. 1B). Indicator plasmids were constructed with targeting sequences of each gene—exon1 of the Pcsk9 gene and exon 3 of the Apoc3 gene—inserted 3′ to a translation initiation codon and 5′ to the green fluorescent protein (GFP) cassette. Different lentiSaCRISPR constructs were then generated, carrying sgRNAs targeting the mouse Apoc3 and Pcsk9 genes with either a tRNA promoter or a U6 promoter, respectively. Indicator plasmids were next cotransfected into HEK 293T cells with different lentiSaCRISPR constructs. Cells were harvested 72 h post-transfection, and GFP intensity was determined either by FACS analysis or by imaging (Fig. 1C). We observed remarkably decreased GFP intensity (>75%) with the lentiSaCRISPR construct using a U6 promoter, indicating effective editing in targeting sequences that generates frameshift mutations and consequently causes destruction of GFP expression. We also observed a significant decrease of GFP intensity with lentiSaCRISPR constructs using tRNA promoters (>50%), although the editing was less effective compared to lentiSaCRISPR constructs using a U6 promoter.
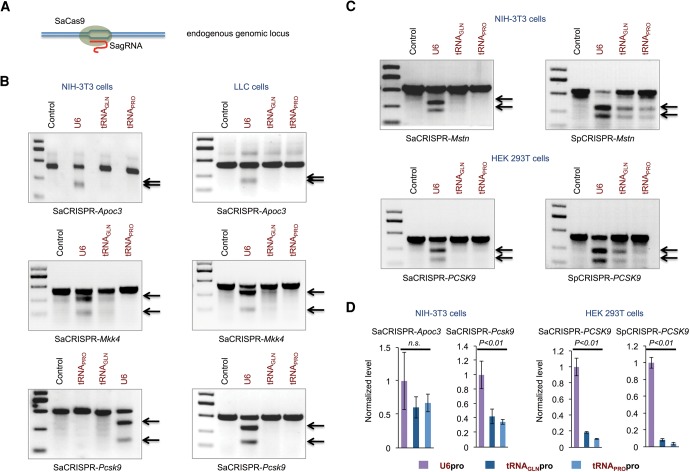
Since exogenous expression levels of indicator plasmids may interfere with studies of editing efficiency, we reasoned that using Surveyor nuclease assays to evaluate the editing efficiency at endogenous genomic sites targeted by the same sgRNA driven with either the U6 promoter or tRNA promoters would be the fairest comparison (Fig. 2A). For effective delivery of CRISPR materials to a mouse cell line, we made lentiviruses with different lentiSaCRISPR vectors targeting the mouse Apoc3, Mkk4, and Pcsk9 genes. We then infected mouse NIH-3T3 cells with lentiviruses, followed by puromycin selection to enrich for successfully transduced cells. Four days post virus infection, cells were harvested for genomic DNA extraction and Surveyor assays. We found that cells infected with U6 promoter-driven sgRNA viruses had substantial mutagenesis (∼50% mutagenesis) at the on-target sites of each gene, but we failed to detect significant editing with tRNA promoter-driven sgRNA viruses in all three targeting loci tested (Fig. 2B). To exclude the possibility that this was a result of different tRNA promoter activity or tRNA processing in NIH-3T3 cells, we repeated the same experiment in the mouse Lewis lung carcinoma (LLC) cell line. Similar results were obtained showing that lentiSaCRISPR constructs with tRNA promoter-driven sgRNA displayed much reduced editing activity compared to constructs with U6 promoter-driven sgRNA (Fig. 2B).
FIGURE 2.
Study of genome editing efficiency of CRISPR/Cas9 with tRNA and U6 promoter-driven sgRNAs by directly accessing mutagenesis rate at the endogenous genomic loci. (A) A schematic diagram of editing efficiency study by directly accessing the endogenous genomic loci. (B) Viral vectors targeting endogenous mouse Apoc3, Mkk4, and Pcsk9 genes were produced in HEK 293T cells with different lentiSaCRISPR constructs, and mouse NIH-3T3 cells and LLC cells were infected and selected with puromycin to enrich for cells transduced with viruses. Cells were then harvested for genomic DNA extraction and Surveyor assays for editing efficiency studies. (C) Viral vectors targeting endogenous mouse Mstn gene and human PCSK9 gene were produced with both lentiSaCRISPR and lentiSpCRISPR constructs, and mouse NIH-3T3 cells (for Mstn targeting) and human HEK 293T cells (for PCSK9 targeting) were infected, selected with puromycin and harvested for Surveyor assays. The gRNA sequences as well as sequences of primers used in Surveyor assays in B and C are listed in Supplemental Table 1. Arrows show the cleavage products resulting from the Surveyor assays; the intensity of the cleavage product bands relative to the uncleaved product band corresponds to the mutagenesis rate. (D) Cells infected with viruses as indicated were collected and subjected to small-RNA extraction. Poly(A) tails were added to RNA before reverse transcription was performed. The expression level of sgRNAs was evaluated with quantitative PCR using SYBR green. 18S RNA level was used as an internal control. The sequences of primers for qRT-PCR were listed in Supplemental Table 1. All data are represented as means with SEM. One-way ANOVA was used for statistical analysis.
We then determined whether this is a specific phenomenon to SaCas9. We screened for two targeting sites—one in the mouse Mstn gene (PAM=TGGAG), the other in the human PCSK9 gene (PAM=AGGAG)—that are targetable by both SaCas9 and SpCas9. We also reengineered the lentiSpCRISPR v2 vector and made three versions of lentiSpCRISPR v2 constructs with different sgRNA promoters—U6pro, tRNAGLNpro, and tRNAPROpro. We next made lentiviruses with different versions of lentiSaCRISPR and lentiSpCRISPR vectors targeting both genes, respectively. NIH-3T3 cells (for the Mstn gene) and HEK 293T cells (for the PCSK9 gene) were then infected by different viruses as indicated, then puromycin-selected and assayed for editing activities. We found that with the same gRNA targeting sequences, lentiSpCRISPR constructs displayed higher editing efficiency than lentiSaCRISPR constructs at both targeting sites (Fig. 2C). Similar to targeting with SaCas9, editing efficiency was also significantly reduced with tRNA promoter-driven sgRNA compared to U6 promoter-driven sgRNA with SpCas9 (Fig. 2C).
Furthermore, we evaluated the expression level of sgRNAs in cells infected with different viruses using qRT-PCR. As shown in Figure 2D, we found that the expression levels of sgRNAs in cells infected with either lentiSaCRISPR viruses (targeting the mouse Pcsk9 gene and the human PCSK9 gene) or lentiSpCRISPR viruses (targeting the human PCSK9 gene) were all significantly lower with tRNA promoters compared to U6 promoters. The expression level of sgRNA targeting the mouse Apoc3 gene also displayed a trend of reduction with tRNA promoters compared to U6 promoters, although statistically not significant. To further exclude the possibility that the difference in sgRNA expression or editing activity was a result of variability in delivered copy number of different CRISPR constructs, genomic DNA from cells was analyzed for CRISPR copy number by qRT-PCR. We found that there were no significant differences in copy number between different groups treated with lentiSaCRISPRs targeting either the Apoc3 or Pcsk9 gene with indicated sgRNA promoters (Supplemental Fig. 1). Altogether, these data indicated that the lower editing efficiency with tRNA promoter-driven sgRNA compared to U6 promoter-driven sgRNA might be a result of the reduced expression level of sgRNAs with tRNA promoters.
In summary, with this side-to-side comparison targeting the same sequences at either exogenous plasmids or endogenous genomic sites with different sgRNA promoters, we found significantly lower targeting efficiency with tRNA promoter-driven sgRNA expression compared to the U6 promoter. This result is in contrast to the conclusions reported by Mefferd et al. (2015), the robust production of sgRNAs driven by tRNA promoters as well as comparable editing efficiency between CRISPR/Cas9 with tRNA promoter- and U6 promoter-expressed sgRNAs. We note two major differences between our experimental systems that might be able to explain the contradictory results. (i) Different methodologies to study CRISPR activities. In the Mefferd et al. (2015) study, genome-editing activities with different promoter-driven sgRNAs were only studied with exogenous cotransfected indicator plasmids instead of directly assessing the endogenous genomic loci. As is shown in our study, while significant editing activity (>50%) was detected with indicator plasmids, editing activity was barely notable with Surveyor assays when targeting the endogenous genomic locus. It is probable that editing activity is overestimated with exogenous plasmids, because there are significantly more copies and easier access to the targeted locus when editing exogenous plasmids in comparison to endogenous genomic loci. This could potentially minimize the differences in apparent editing activity between CRISPR/Cas9 using tRNA and U6 promoters. (ii) Different targeting sequences were used. Mefferd et al. (2015) noted in their study that targeting sequences with high GC content could inhibit tRNase Z processing, suggesting that expression efficiency of sgRNAs driven by tRNA promoters could be sequence-dependent. Among the sgRNA sequences tested in our systems, one is GC rich (Pcsk9, GC content = 71%), with the others having normal GC content (Apoc3, GC content = 59%; Mkk4, GC content = 38%, Mstn, GC content = 55%; PCSK9, GC content = 65%). It is possible that other features of sgRNA sequences could also have an influence on tRNase Z processing, which will need further investigation.
Due to the limited number of sgRNA sequences as well as tRNA promoters we have tested in this study, it may be premature to conclude that CRISPR targeting does not work well with tRNA promoter-driven sgRNA expression. Nonetheless, our study raises the concern that tRNA promoters may not work as consistently as the U6 promoter. The efficiency of sgRNA expression driven by tRNA promoters may be sequence-dependent, and screening for activity at specific target endogenous genomic loci will be necessary before tRNA promoters can be used for therapeutic genome editing.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kiran Musunuru (University of Pennsylvania) for helpful comments, suggestions, and critical reading of this manuscript and Haiwei Zhang from Haibing Zhang's laboratory (Institute for Nutritional Sciences, SIBS) for generously sharing the pEGFP-N1 plasmid. This work was supported by the Shanghai Institutes for Biological Sciences Fellowship (Y5Y1X41491) (Y.Z.), the National Natural Science Foundation of China (81500614 to Y.Z.), the Hundred Talents Program of the Chinese Academy of Sciences (Q.D.), the National Youth 1000 Talents Program (Q.D.), the Shanghai Pujiang Program (15PJ1409200 to Q.D.), and the National Natural Science Foundation of China (31670829 to Q.D.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.057596.116.
REFERENCES
- Cox DB, Platt RJ, Zhang F. 2015. Therapeutic genome editing: prospects and challenges. Nat Med 21: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, et al. 2015. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol 16: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. 2016. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351: 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefferd AL, Kornepati AV, Bogerd HP, Kennedy EM, Cullen BR. 2015. Expression of CRISPR/Cas single guide RNAs using small tRNA promoters. RNA 21: 1683–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, et al. 2016. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351: 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. 2016. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.