Abstract
Spliceostatin A (SSA) is a methyl ketal derivative of FR901464, a potent antitumor compound isolated from a culture broth of Pseudomonas sp. no. 2663. These compounds selectively bind to the essential spliceosome component SF3b, a subcomplex of the U2 snRNP, to inhibit pre-mRNA splicing. However, the mechanism of SSA's antitumor activity is unknown. It is noteworthy that SSA causes accumulation of a truncated form of the CDK inhibitor protein p27 translated from CDKN1B pre-mRNA, which is involved in SSA-induced cell-cycle arrest. However, it is still unclear whether pre-mRNAs are uniformly exported from the nucleus following SSA treatment. We performed RNA-seq analysis on nuclear and cytoplasmic fractions of SSA-treated cells. Our statistical analyses showed that intron retention is the major consequence of SSA treatment, and a small number of intron-containing pre-mRNAs leak into the cytoplasm. Using a series of reporter plasmids to investigate the roles of intronic sequences in the pre-mRNA leakage, we showed that the strength of the 5′ splice site affects pre-mRNA leakage. Additionally, we found that the level of pre-mRNA leakage is related to transcript length. These results suggest that the strength of the 5′ splice site and the length of the transcripts are determinants of the pre-mRNA leakage induced by SF3b inhibitors.
Keywords: pre-mRNA splicing, spliceostatin A, RNA-seq, pre-mRNA nuclear retention
INTRODUCTION
Most genes in higher eukaryotes contain introns, which are intervening sequences found between protein-coding regions known as exons. Introns are removed and the exons that flank them are joined together to become mature mRNAs that form templates for protein translation (Dreyfuss et al. 2002). The splicing reaction is carried out by the spliceosome, a macromolecular ribonucleoprotein complex, containing small nuclear RNAs (snRNAs) and more than 200 proteins (Jurica and Moore 2003; Cvitkovic and Jurica 2013).
The spliceosome consists of five major components: U1, U2, U4, U5, and U6 small nuclear ribonucleoprotein particles (snRNPs). To begin, U1 snRNP recognizes the 5′ splice site, which is the 5′ end of the intron. The U2AF complex binds to both the 3′ splice site, which is the 3′ end of the intron, and the polypyrimidine tract, a pyrimidine-rich region just upstream of the 3′ splice site. In addition to these factors, SF1 binds to the branch point sequence (BPS), which is located 18–40 nucleotides (nt) upstream of the 3′ splice site to form complex E. SF1 is replaced by U2 snRNP to form complex A before U4/U6•U5 tri-snRNPs bind to the complex to form complex B. Following subsequent conformational changes, the catalytically active complex named complex C is formed and the splicing reaction can now be carried out (Wahl et al. 2009).
Splicing is an essential cellular function required to maintain the integrity of gene expression. If pre-mRNAs with introns are translated into proteins, truncated and deleterious proteins will be produced since intron sequences harbor a large number of termination codons. Therefore, cells have several mechanisms for preventing pre-mRNA translation. For instance, in the budding yeast Saccharomyces cerevisiae, Mlp1 localizes to the nuclear pore complex to prevent pre-mRNA export from the nucleus (Galy et al. 2004). The retention and splicing complex binds to the SF3b complex, which is a component of U2 snRNP, and has a role in nuclear retention of pre-mRNA (Dziembowski et al. 2004; Wang et al. 2005). SF1 is also involved in nuclear retention of pre-mRNA in yeast (Rutz and Seraphin 2000). In the cytoplasm, nonsense-mediated decay (NMD) is responsible for degrading pre-mRNAs containing premature termination codons (PTCs) that are leaked into the cytoplasm to prevent the production of truncated proteins (Kurosaki and Maquat 2016).
Spliceostatin A (SSA) is a chemically stable, methylated derivative of FR901464, an anticancer bacterial metabolite (Nakajima et al. 1996a,b, 1997). SSA binds to the SF3b complex and inhibits splicing in vivo and in vitro (Kaida et al. 2007). Another SF3b inhibitor, a pladienolide derivative, has been studied clinically as an anticancer agent (Kotake et al. 2007; Eskens et al. 2013). However, it is still unknown how splicing inhibitors induce selective anticancer activity. In addition to inhibiting splicing, SSA has also been shown to affect transcription and pre-mRNA leakage (Kaida et al. 2007; Brody et al. 2011; Kim et al. 2011; Martins et al. 2011; Schmidt et al. 2011; Takemura et al. 2011; Koga et al. 2014, 2015), presumably due to splicing inhibition. Indeed, pre-mRNA of CDKN1B, which encodes the cyclin-dependent kinase inhibitor p27, was shown to leak from the nucleus and be translated into the truncated protein designated as p27* in SSA-treated cells. This finding raised the intriguing possibility that biologically active truncated proteins are involved in the anticancer activity of SF3b inhibitors. However, there is no information regarding the types of pre-mRNAs that leak into the cytoplasm following SSA treatment. In addition, there has been no genome-wide study on splicing patterns that may be changed by SSA.
Here we report the first global analysis of pre-mRNAs in the presence of SSA. SSA treatment caused widespread intron retention but did not significantly alter frequencies of exon skipping, alternative splice site usage, and mutually exclusive exon inclusion. We also found that a limited number of pre-mRNAs leaked into the cytoplasm during SSA treatment. Moreover, the leaked pre-mRNAs are relatively short and associate with weak 5′ splice sites. These results suggest that SSA treatment induces the selective leakage of a particular subset of pre-mRNAs.
RESULTS
SSA treatment causes intron retention
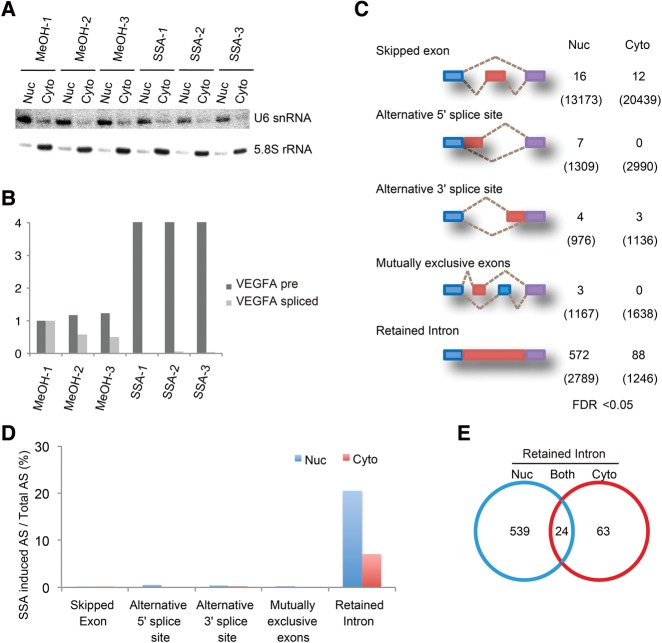
To investigate the global effect of SSA treatment on splicing, we treated HeLa S3 cells with 100 ng mL−1 of SSA for 6 h before fractionation of the cells into cytoplasmic and nuclear fractions. RNA samples were purified from each fraction. Successful fractionation was determined by assessing the distribution of U6 snRNA and 5.8S rRNA (Fig. 1A). To verify that SSA inhibits splicing, we measured the amount of spliced and unspliced forms of VEGFA mRNA. We determined that the level of the unspliced form increased strongly while the spliced form was almost completely eliminated in comparison (Fig. 1B; Furumai et al. 2010). Using these RNA samples, we investigated all splicing events that might occur in both SSA-treated and untreated control cells using RNA-Seq.
FIGURE 1.
SSA treatment causes intron retention. (A) Northern blots performed on RNA isolated from nuclear and cytoplasmic fractions of SSA-treated cells (100 ng mL−1, 6 h). MeOH-treated cells were used as an experimental control. Probes against U6 snRNA and 5.8S RNA were used specifically to detect endogenous RNA. (B) RT-PCR analysis of VEGFA pre-mRNA from SSA-treated cells (100 ng mL−1, 6 h). RT-PCR analysis was performed using specific primers against VEGFA pre-mRNA and spliced mRNA. (C) The summary of RNA-seq analysis for alternative splicing. Five representative alternative splicing patterns are depicted. The numbers of statistically significant (FDR < 0.05) splicing pattern changes after SSA treatment are indicated on the right. The numbers of total splicing events are indicated in parenthesis. (D) Ratio of the SSA-induced change in splicing pattern to total alternative splicing events of the data in C. (E) Venn diagram shows the number of introns, the level of which was increased by SSA. The numbers of introns that accumulated only in the nuclear fraction (539 events), only in the cytoplasmic fraction (63 events), and in both (24 events) were all counted.
We classified the alternative splicing events that were observed in both types of cells into five categories (Fig. 1C, numbers in parentheses). We also statistically analyzed the frequency of the splicing pattern changes upon SSA treatment (Fig. 1C,D). Alternative splicing events that occurred after SSA treatment more or less frequently than control were counted as the changed splicing pattern (Fig. 1C, numbers). Although exon skipping was the most frequently observed in total (13,075 cases in the nucleus and 20,439 cases in the cytoplasm), only 16 cases in the nucleus and 12 cases in the cytoplasm out of them were affected by SSA treatment (Fig. 1C,D). In addition, SSA barely affected other alternative splicing patterns, including alternative 5′ splice site selection, alternative 3′ splice site selection, and splicing of mutually exclusive exons. In contrast, SSA treatment affected intron retention most frequently (572 cases out of 2789 cases in the nucleus; 88 cases out of 1246 cases in the cytoplasm) (Fig. 1C,D). Among these, the abundance of 563 introns in the nucleus and 87 introns in the cytoplasm was increased, consistent with the initial finding that SSA is a general splicing inhibitor (Kaida et al. 2007). Hereafter, we refer to “intron-retained pre-mRNAs” as “pre-mRNAs.”
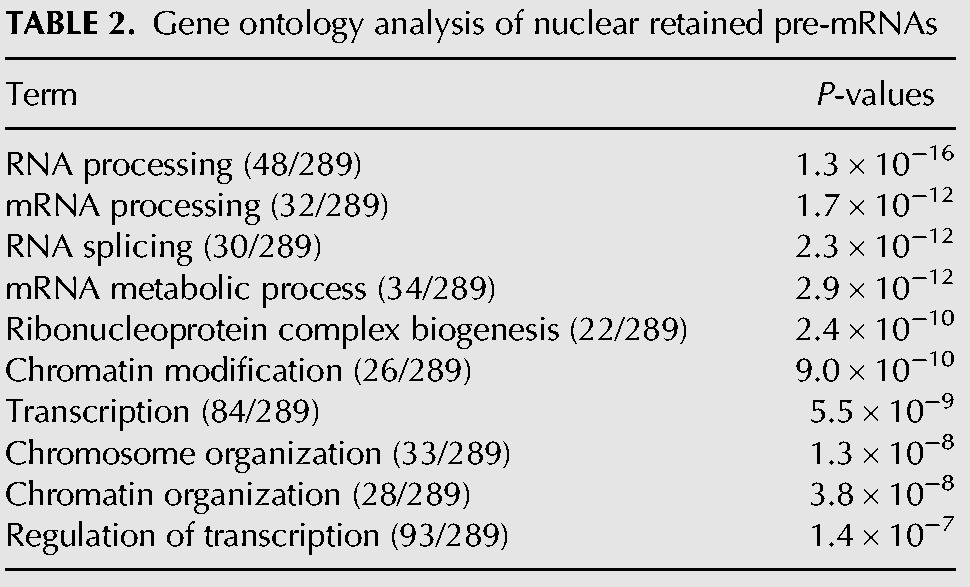
The majority of the introns that were increased by SSA treatment were observed only in the nucleus (Fig. 1E, Nuc). In some cases, however, a remarkable increase in cytoplasmic introns following SSA treatment was observed without showing any increase in nuclear introns (Fig. 1E, Cyto). In addition, intron species that increased in both the fractions upon SSA treatment were categorized as pre-mRNAs increased in both (Fig. 1E, Both). Importantly, only a small number of pre-mRNAs with retained introns, including CDKN1B and NFKBIA as reported previously (Kaida et al. 2007), increased in abundance in the cytoplasm following SSA treatment, suggesting that only a subset of pre-mRNAs leak from the nucleus in SSA-treated cells (Supplemental Table S1). We also performed gene ontology enrichment analysis and found that mRNA species in the Cyto category were different from those in the Nuc category (Tables 1, 2). If the leakage is the consequence of contamination of the Cyto fraction by the Nuc pre-mRNAs during the fractionation steps, similar pre-mRNA species should be enriched in both fractions. Therefore, this result also supports that only a subset of pre-mRNAs leak from the nucleus in SSA-treated cells.
TABLE 1.
Gene ontology analysis of leaked pre-mRNAs
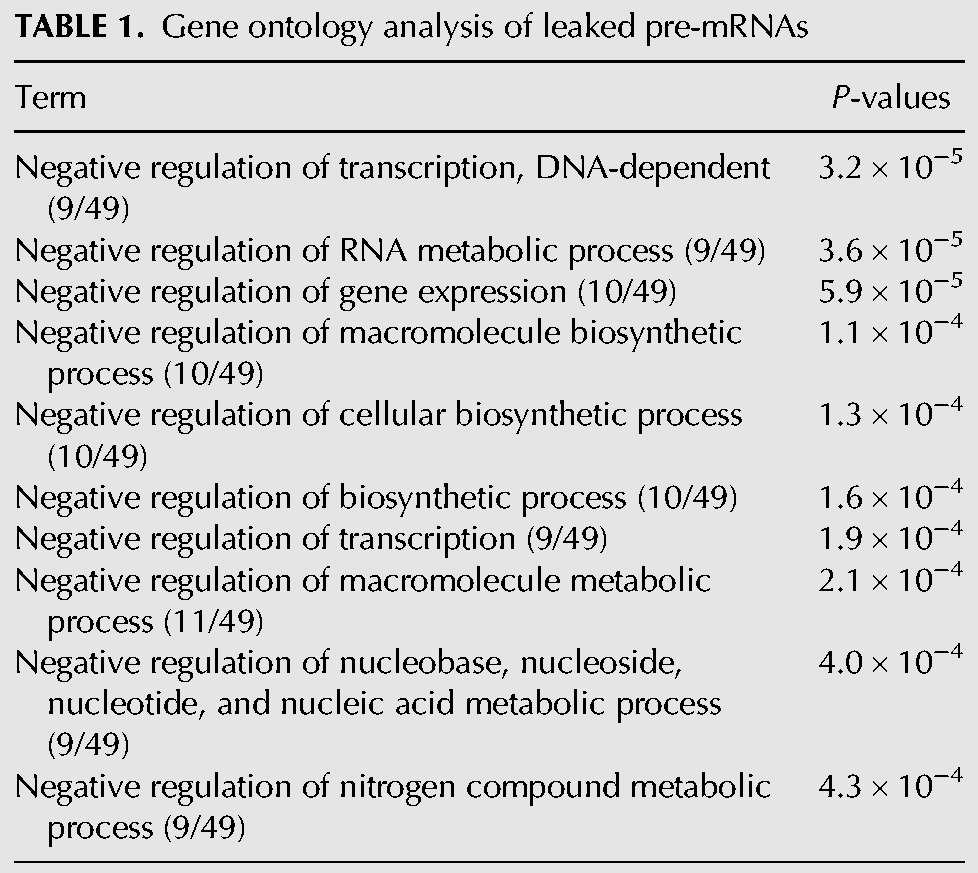
TABLE 2.
Gene ontology analysis of nuclear retained pre-mRNAs
SSA causes gene-specific leakage of pre-mRNA
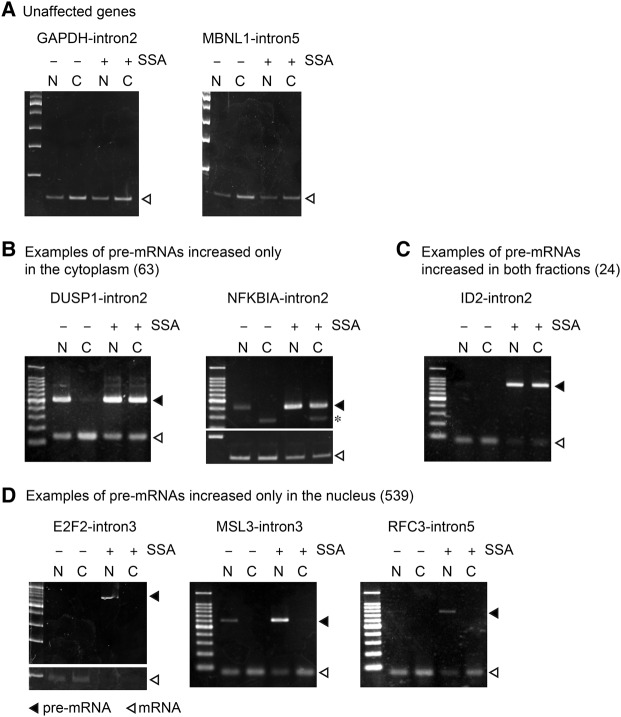
To verify intron retention in SSA-treated cells, we performed RT-PCR to detect both the spliced and unspliced forms of selected genes. We thereby confirmed that some introns that showed no intron retention according to the RNA-seq analysis also did not exhibit intron retention according to RT-PCR in the presence of SSA (Fig. 2A). These pre-mRNAs may be degraded very quickly or their splicing may be resistant to SSA. The pre-mRNAs of several genes were detected even in the nuclei of control cells, presumably due to a time lag between transcription and splicing (Fig. 2B, DUSP1). In addition to intron retention events observed in the nucleus of untreated control cells, we observed a remarkable accumulation of intron-containing pre-mRNAs in SSA-treated cells (Fig. 2B–D). The pre-mRNAs of several genes were localized to both the cytoplasm and nucleus (Fig. 2B,C), while other pre-mRNAs were observed only in the nucleus following SSA treatment (Fig. 2D), suggesting that the leakage of pre-mRNAs from the nucleus caused by SSA occurs in a gene-specific manner.
FIGURE 2.
SSA causes pre-mRNA leakage in a gene-specific manner. (A–D) HeLa S3 cells were treated with SSA (100 ng mL−1, 6 h) and the cells were disrupted and separated into cytoplasmic and nuclear fractions. RNA samples were purified from the fractions. RT-PCR analysis was performed using the RNA samples to detect spliced and unspliced forms of the genes, of which pre-mRNAs were not affected by SSA (A), accumulated only in the cytoplasm (B), accumulated in both fractions (C), and accumulated only in the nucleus (D). Open triangles indicate spliced mRNA, while the filled triangles indicate pre-mRNA. Asterisk indicates nonspecific bands.
We also found that susceptibility to splicing inhibition varies among genes and even among introns within a single pre-mRNA. For example, all the introns in DUSP1 accumulated following SSA treatment, whereas none of the introns in GAPDH were affected by SSA treatment (Supplemental Fig. S1). Intron 1 of NFKBIA was not affected, whereas the other introns of this gene accumulated upon SSA treatment (Supplemental Fig. S1). Presumably, the strength of splicing sites in each intron affects splicing pattern changes after SSA treatment. In addition, we found that the total transcript level (i.e., spliced plus unspliced) of some genes, including NFKBIA, was apparently up-regulated (Fig. 2). This could be caused by transcriptional up-regulation caused, in turn, by the activation of the NFkB pathway by SSA (Khan et al. 2014).
Effects of BPS and splice sites on splicing efficiency and pre-mRNA leakage
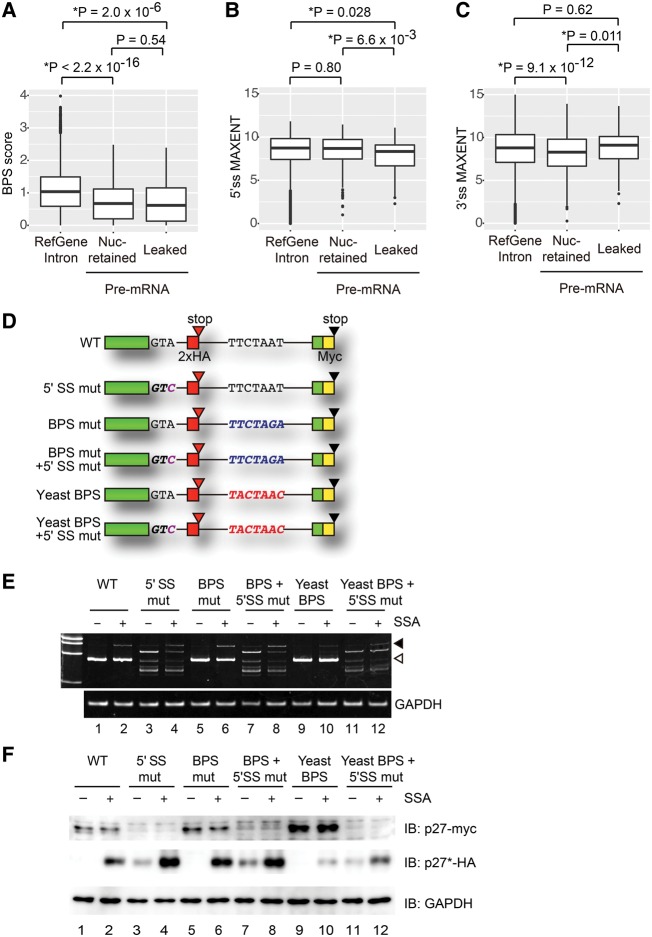
Among the intron-retained species up-regulated in the presence of SSA, most of the pre-mRNAs were localized in the nucleus and only a part of the population was leaked into the cytoplasm (Figs. 1E, 2). To find the significant characteristics differentiating leaked pre-mRNAs and nuclear-retained pre-mRNAs, we separated pre-mRNAs into those increased only in the nucleus (Nuc-retained) and increased only in the cytoplasm, or in both the cytoplasm and the nucleus (Leaked). Next, we first investigated whether the strength of the BPS affects splicing efficiency and pre-mRNA leakage, because SSA binds to the SF3b complex, which affects BPS recognition by the U2 snRNP (Corrionero et al. 2011). We predicted the location of the BPS within the increased introns following SSA treatment (Fig. 1E) before calculating the strength of the predicted BPSs using the SVM-BP program, and compared the values with those of introns in UCSC RefGene (Corvelo et al. 2010).
As expected, accumulated introns in the presence of SSA have relatively weak BPSs compared to RefGene introns (Fig. 3A, RefGene intron vs. pre-mRNA). This observation suggests that the strength of the BPS affects the sensitivity of the splicing machinery to SSA. In contrast, the BPS scores of the nuclear-retained introns were almost the same as those of the leaked ones. Even when the strength of experimentally determined BPS (Mercer et al. 2015) was used for the statistical analysis, there was no correlation between the strength of BPSs and pre-mRNA leakage (Supplemental Fig. S2A). These observations suggest that the strength of the BPS is not always involved in the nuclear retention of pre-mRNA (Fig. 3A). Next, we tested whether the strength of the 5′ and 3′ splice sites is involved in the splicing efficiency and nuclear retention of pre-mRNA upon SSA treatment. Pre-mRNAs that harbor relatively weaker 5′ splice sites were enriched in leaked pre-mRNA in SSA-treated cells, suggesting that the 5′ splice site is involved in pre-mRNA leakage (Fig. 3B). In contrast, 3′ splice sites of leaked pre-mRNA were not weaker but slightly stronger than those of nuclear retained pre-mRNA (Fig. 3C).
FIGURE 3.
Effects of BPS and splice sites on splicing and leakage of pre-mRNA. (A–C) Boxplots of the strength of BPS (BPS scores), 5′ splice site (5′ss MAXENT), and 3′ splice site (3′ss MAXENT). All introns of human RefGenes, nuclear-retained pre-mRNAs that contain introns in the Nuc category in Figure 1E, and leaked pre-mRNAs that contain introns in the Cyto category in Figure 1E were analyzed. (D) Schematic representation of CDKN1B-derived reporters. Each box shows an exon and a line shows an intron. (E) HEK293T cells were transfected with the reporter plasmids, then treated with SSA (100 ng mL−1, 6 h). RNA samples were purified from the cells. RT-PCR analysis was performed for detecting the spliced and unspliced forms of the reporter constructs. Open triangle and filled triangle indicate spliced mRNA and pre-mRNA, respectively. (F) HEK293T cells were transfected with the reporter plasmids and then treated with SSA (100 ng mL−1, 6 h). Total cell extracts were analyzed by immunoblotting. The antibodies used are indicated to the right of the respective blots.
To verify the effect of the BPS and 5′ splice site on splicing efficiency and pre-mRNA leakage, we constructed six reporter CDKN1B plasmids with modified BPSs and/or the 5′ splice site (Fig. 3D; Kaida et al. 2007). The BPS of the CDKN1B reporter gene is TTCTAAT (wild-type; WT). We changed this sequence to either TTCTAGA to weaken the BPS (mutant; BPS mut), or to TACTAAC, which is the same as the BPS in budding yeast (yeast BPS) (Berglund et al. 1997). The yeast BPS is complementary to the sequence of the BPS base-paring site in U2 snRNA and should bind to U2 snRNA more strongly (Berglund et al. 1997). We also introduced a point mutation in the 5′ splice site (GTA to GTC) to abolish the 5′ splice site function (5′ SS mutant; 5′ SS mut). In these reporter constructs, a DNA sequence encoding the 2× HA tag was inserted just upstream of the in-frame stop codon in the intron, while another DNA sequence encoding the Myc tag was inserted just upstream of the canonical stop codon. If the spliced form is translated, the Myc-tagged protein will be observed. In contrast, if the unspliced form leaks into and is translated in the cytoplasm, the HA-tagged protein will be generated.
First, we investigated the splicing efficiency of these reporter constructs in SSA-treated and untreated control cells by RT-PCR. We transfected HEK293T cells with the reporter plasmids before treating the transfected cells 18 h later with 100 ng mL−1 SSA for 6 h. Following SSA treatment for 6 h, the abundance of the unspliced form had increased, suggesting that SSA treatment inhibits the splicing reaction of the reporter gene (Fig. 3E, WT). The spliced form remaining in the SSA-treated cells may be transcribed and spliced before SSA treatment. Since no spliced form of the constructs containing the mutation at the 5′ splice site was observed even in the absence of SSA, the 5′ splice site mutation appeared to completely suppress splicing (Fig. 3E, lanes 3,4,7,8,11,12). Instead, several other bands were observed, suggesting that splicing occurred at cryptic 5′ splice sites (Fig. 3E, lanes 3,4,7,8,11,12). Although the BPS mutation barely affected splicing efficiency compared to WT (Fig. 3E, lanes 1,5), the yeast BPS seemed to be relatively resistant to splicing inhibition by SSA, because a reduced amount of the unspliced form compared to WT was observed in the presence of SSA (Fig. 3E, lanes 2,10) due to strong binding between U2 snRNA and the yeast BPS. Taken together, these results indicate that splicing efficiency in the presence of SSA is affected by the strength of the BPS, which is consistent with the findings of a previous study (Corrionero et al. 2011).
Next, we investigated the levels of proteins produced from pre-mRNAs that leaked into the cytoplasm. Since translation occurs in the cytoplasm, levels of proteins translated from pre-mRNAs should reflect pre-mRNA leakage level. We transfected HEK293T cells with the reporter plasmids and treated them with 100 ng mL−1 of SSA for 6 h. In cells transfected with the WT plasmid, only the Myc-tagged protein, which is translated from spliced mRNA, was observed in the absence of SSA (Fig. 3F, lane 1). Following SSA treatment, however, the HA-tagged protein translated from pre-mRNA was also detected, indicating that SSA treatment causes the leakage of pre-mRNA in addition to splicing inhibition (Fig. 3F, lane 2). After transfection with the 5′ SS mut plasmid, Myc-tagged protein became undetectable, confirming that normal splicing was inhibited by the 5′ splice site mutation (Fig. 3F, lane 3), although alternative splicing events using cryptic 5′ splice sites occurred to some extent (Fig. 3E). SSA treatment greatly increased the amount of HA-tagged protein translated from the 5′ SS mut reporter (Fig. 3F, lanes 3,4), suggesting that SSA allows efficient leakage of pre-mRNA carrying the 5′ splice site mutation. The amount of HA-tagged protein measured was much larger than that from the WT reporter in the presence of SSA (Fig. 3F, lanes 2,4) although the pre-mRNA levels were almost the same (Fig. 3E,F, lanes 2,4), which supports the involvement of the 5′ splice site in the nuclear retention of pre-mRNA.
In cells transfected with the BPS mutant plasmid, the abundance of pre-mRNA and of HA-tagged protein was greatly increased compared to the WT plasmid-transfected cells in the presence of SSA (Fig. 3E,F, lanes 2,6). In contrast, a smaller amount of HA-tagged protein was produced from the yeast BPS plasmid compared to the WT plasmid upon SSA treatment, consistent with less effective inhibition of splicing by SSA (Fig. 3E,F, lanes 2,10). Given the correlation between the amount of pre-mRNA and the amount of HA-tagged protein, these results suggest that the strength of the BPS mainly affects splicing efficiency, which has an impact on the abundance of HA-tagged protein. In the presence of the 5′ splice site mutation together with the yeast BPS, however, much less HA-tagged protein was produced than in the presence of just the 5′ splice site mutation, even though the pre-mRNA levels were almost identical (Fig. 3E,F, lane 3 vs. lane 11 and lane 4 vs. lane 12). This result suggests that BPS affects pre-mRNA leakage in some particular contexts. Taken together, the 5′ splice site seems to play a major role in pre-mRNA retention.
Short transcripts are prone to leak from the nucleus
To further characterize pre-mRNAs that leaked into the cytoplasm in the presence of SSA, we statistically compared the transcript length, intron length, and exon number of leaked pre-mRNAs as well as nuclear retained pre-mRNAs (Fig. 4A–C). The average transcript length of leaked pre-mRNAs is significantly shorter than that of nuclear retained pre-mRNAs, suggesting that shorter transcripts tend to leak from the nucleus (Fig. 4A). The average length of accumulated introns in the presence of SSA was significantly shorter than that of RefGene (Fig. 4B). However, the intron length did not significantly affect pre-mRNA leakage (Fig. 4B). Furthermore, we found that transcripts with a smaller number of exons are more likely to be exported from the nucleus (Fig. 4C). Since gene length is usually reflected by the exon number, these results collectively suggest that short pre-mRNAs tend to leak from the nucleus.
FIGURE 4.
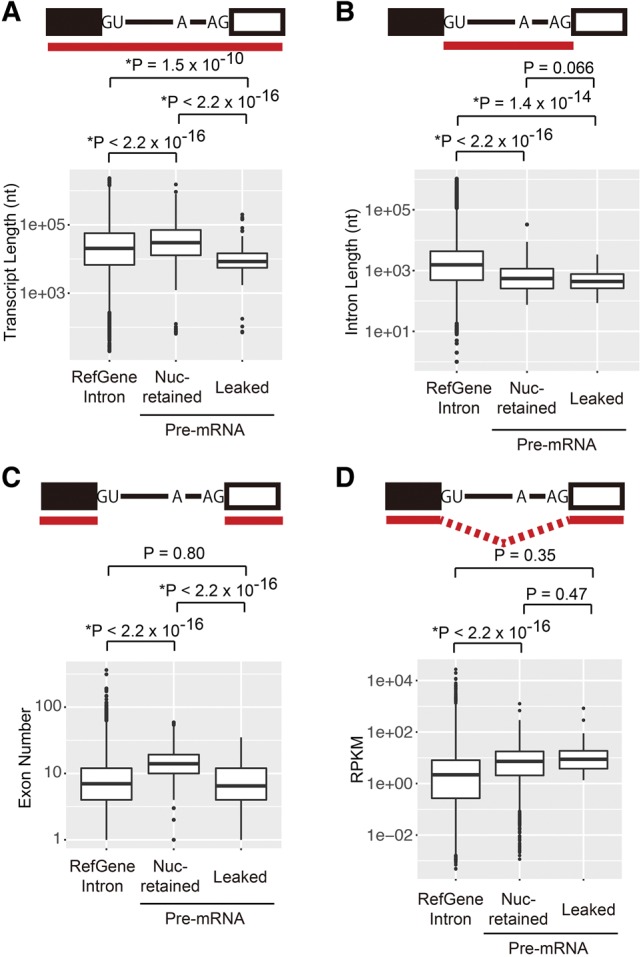
Short pre-mRNAs are prone to leak from the nucleus. (A) Boxplots of the transcript length of human RefGenes, nuclear retained pre-mRNAs, and leaked pre-mRNAs. (B) Boxplots of the intron length of human RefGenes, nuclear retained pre-mRNAs, and leaked pre-mRNAs. (C) Boxplots of the exon number of human RefGenes, nuclear retained pre-mRNAs, and leaked pre-mRNAs. (D) Boxplots of the RPKM of human RefGenes, nuclear retained pre-mRNAs, and leaked pre-mRNAs.
Next, we tested whether relatively abundant pre-mRNAs leak from the nucleus. To this end, we calculated the reads per kilobase of exon per million mapped sequence reads (RPKM) that reflect abundance of pre-mRNAs. We found no significant difference in the RPKM values between leaked and nuclear retained pre-mRNAs (Fig. 4D), suggesting that the abundance of pre-mRNA does not affect the leakage of pre-mRNA. Taken together, the length of pre-mRNA is involved in the pre-mRNA leakage induced by SSA.
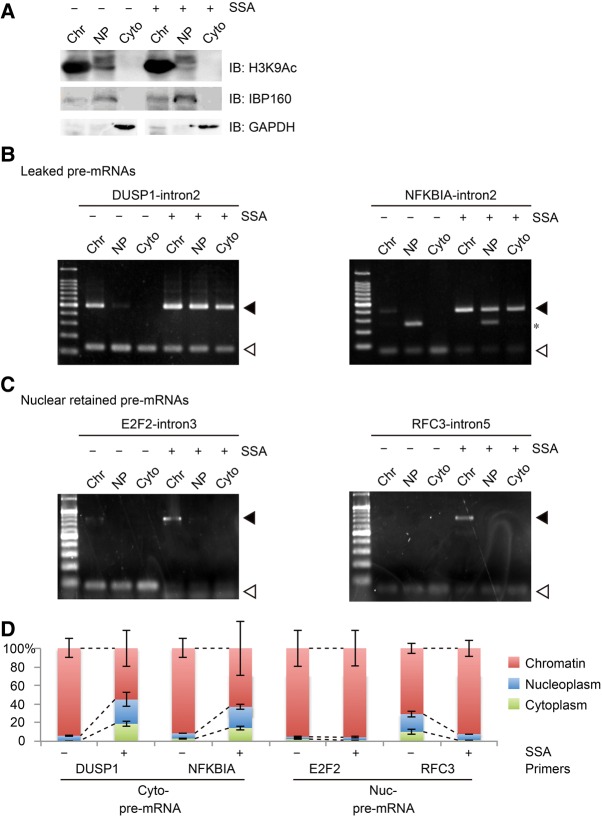
Gene-specific anchoring of pre-mRNA to the chromatin region
To understand the molecular mechanisms underlying the leakage of pre-mRNA, we further separated the nuclear fraction into nucleoplasm and chromatin-bound fraction and analyzed the distribution of pre-mRNAs in those fractions. Successful fractionation was verified by detecting histone H3K9Ac, IBP160, and GAPDH as chromatin, nucleoplasmic, and cytoplasmic markers, respectively (Fig. 5A). Without SSA treatment, pre-mRNAs were mainly observed in the chromatin fraction, presumably because most pre-mRNAs are subjected to cotranscriptional splicing, in which introns are excised before the pre-mRNA dissociates from chromatin (Fig. 5B,C). Following SSA treatment, DUSP1 and NFKBIA pre-mRNAs, which leaked from the nucleus, were observed in all three fractions, confirming that pre-mRNAs detected in the cytoplasm were released from chromatin (Figs. 2B, 5B). In contrast, E2F2 and RFC3 pre-mRNAs, which had been detected only in the nucleus (Fig. 2D), were also present only in the chromatin fraction but not in the nucleoplasm of cells treated with SSA, which suggests that nuclear-retained pre-mRNAs are tethered by chromatin (Fig. 5C). Essentially the same results were obtained by semiquantitative analysis using qRT-PCR (Fig. 5D). These results demonstrate that relatively long pre-mRNAs that are bound to chromatin during splicing are resistant to SSA-induced leakage into the cytoplasm.
FIGURE 5.
Nuclear-retained pre-mRNAs are localized in the chromatin fraction. (A) Validation of cellular fractionation into chromatin, nucleoplasm, and cytoplasmic fractions. H3K9Ac, IBP160, and GAPDH were used as markers for chromatin, nucleoplasm, and cytoplasmic fractionation, respectively. (B,C) HeLa cells were treated with SSA (100 ng mL−1, 6 h) before they were disrupted and separated into chromatin, nucleoplasmic, and cytoplasmic fractions. RNA samples were purified from the fractions. RT-PCR analysis was performed using RNA samples to detect the spliced and unspliced forms of the leaked pre-mRNA (B) and nuclear retained pre-mRNA (C). Open triangles indicate the spliced mRNA, while the filled triangles indicate the pre-mRNA. Asterisk indicates a nonspecific band. (D) The amount of pre-mRNA in the three fractions from control and SSA-treated cells was measured by quantitative RT-PCR using the same sample as B and C.
DISCUSSION
Nuclear retention of pre-mRNA is one of the key mechanisms for ensuring the integrity of gene expression, because the export and/or leakage of pre-mRNA, which is followed by its translation in the cytoplasm, results in the production of nonfunctional and deleterious proteins. Such proteins may cause cell death or several pathogenic alterations, including cancer (Eskens et al. 2013). It is therefore important to understand the molecular mechanisms underlying the nuclear retention of pre-mRNA. In this study, we performed global sequence analysis and found that intron retention is the primary consequence of splicing inhibition by SSA. This conclusion is also supported by our reanalysis of the RNA-seq data presented in a recent report by Kfir et al. (2015), which showed that SF3B1 knockdown causes intron retention most frequently (Supplemental Fig. S2). Although the accumulation of a large number of pre-mRNA species was triggered by SSA treatment, only a small subset of pre-mRNAs, including CDKN1B and NFKBIA, leaked specifically into the cytoplasm upon SSA treatment. Therefore, the pre-mRNAs that leaked into the cytoplasm following SSA treatment appear to have specific structural or functional features to bypass the nuclear retention mechanism. We found two features that affect the retention of pre-mRNA: the strength of the 5′ splice site and the transcript length.
We found that the majority of pre-mRNAs were retained in the nucleus even in the presence of SSA that binds and inhibits SF3b, the subcomplex that recognizes the BPS. A series of assays using the reporter plasmids and the statistical analysis in this study revealed that the BPS influences splicing efficiency, but its role in the nuclear retention of pre-mRNA is marginal, which is consistent with a previous report showing that U2 snRNA and SF3b155, components of U2 snRNP, are not required for pre-mRNA nuclear retention in mammals (Takemura et al. 2011). However, mutations in the BPS and even its deletion caused elevated pre-mRNA leakage in yeast (Legrain and Rosbash 1989; Rain and Legrain 1997). It seems possible that the yeast BPSs are well conserved and can bind tightly to U2 snRNA, whereas mammalian BPSs are more diverse in their sequence, and their ability to bind to U2 snRNA is not as strong. Consistent with this, we also showed that replacement of the BPS in the plasmids containing the 5′ splice site mutation with yeast BPS inhibited pre-mRNA leakage (Fig. 3). Therefore, the mammalian BPS and U2 snRNP may not play a critical role in nuclear retention. These observations suggest that the BPS is not the primary factor in determining SSA-induced pre-mRNA leakage, although the strength of the BPS appears to correlate with the amount of leaked pre-mRNA. It seems likely that modulating splicing efficiency by changing BPS activity affects the level of pre-mRNA leakage in mammals. However, we cannot rule out the possibility that the strength of the BPS regulates pre-mRNA leakage in some specific context, so careful investigations should be carried out to understand the effect of BPS on pre-mRNA leakage in mammals.
Nuclear-retained pre-mRNAs accumulated in the presence of SSA were significantly longer in size than leaked pre-mRNAs (Fig. 4). This observation suggests that short pre-mRNAs can escape the nuclear retention machinery more efficiently. However, it seems possible that the reduced abundance of long pre-mRNAs in the cytoplasm is due to rapid degradation of long pre-mRNAs by NMD in the cytoplasm because long pre-mRNAs are more likely to harbor PTCs that trigger NMD (Kurosaki and Maquat 2016). In fact, most of the accumulated introns were located within the coding region (517 introns out of 539 in the nuclear-retained fraction, and 70 introns out of 87 in the leaked fraction). Such introns have the potential to generate PTCs. To test this possibility, we treated HeLa cells with cycloheximide (CHX), an NMD inhibitor, together with SSA, and subsequently compared pre-mRNA levels between CHX-treated and control cells. E2F2 and RFC3 pre-mRNAs, which were detected only in the nucleus (Fig. 2), were not up-regulated after NMD inhibition by CHX (Supplemental Fig. S3), although SNHG1, a well-known NMD target (Tycowski et al. 1996), was up-regulated following NMD inhibition (Supplemental Fig. S3). These results suggest that the tendency of short pre-mRNAs to leak from the nucleus is not due to NMD-mediated cytoplasmic degradation of long pre-mRNAs such as E2F2 and RFC3.
The nuclear-retained pre-mRNAs were detected mainly in the chromatin fraction (Fig. 5), suggesting that unknown nuclear retention factor(s) associated with chromatin tethers pre-mRNAs in the nucleus. Because U1 snRNP and U2AF65, which prevent pre-mRNA leakage from the nucleus (Takemura et al. 2011), can recognize pre-mRNA cotranscriptionally at the chromatin region (Bentley 2014), these proteins are potential candidates for nuclear retention factors that impede the SSA-induced nuclear export of pre-mRNAs. Long transcripts tend to have more introns, meaning that the transcripts have more 5′ splice sites and polypyrimidine tracts, which form binding sites for U1 snRNP and U2AF65, respectively. Even after splicing inhibition by SSA treatment, U1 snRNP and U2AF65 continued to bind to pre-mRNA (Roybal and Jurica 2010; Corrionero et al. 2011). Furthermore, the 5′ splice site was reported to be important for nuclear retention in both yeast and mammals (Fig. 3; Legrain and Rosbash 1989; Rain and Legrain 1997). These observations support the idea that these factors are responsible for nuclear retention of long pre-mRNAs. Consistent with this, short transcripts usually have a smaller number of 5′ splice sites and polypyrimidine tracts, which may allow short transcripts to escape from the nuclear retention machinery. Indeed, we found that pre-mRNAs that leaked into the cytoplasm are relatively short and contain a smaller number of exons, further supporting a correlation between the pre-mRNA length and their nuclear retention. However, it is also possible that factors other than U1 snRNP and U2AF65 are involved in nuclear retention. Therefore, it is necessary to determine the role of U1 snRNP and U2AF65 in pre-mRNA nuclear retention following SSA treatment and to explore other factors such as chromatin-associated proteins for nuclear retention in future studies.
It has been reported that some of the splicing inhibitors that target SF3b show strong antitumor activity with low toxicity (Mizui et al. 2004; Sakai et al. 2004a,b; Sato et al. 2014). Hence, a derivative of the splicing inhibitor pladienolide B has been studied clinically (Eskens et al. 2013). However, it is highly probable that general splicing inhibition will cause severe toxicity. The mechanism of how SF3b inhibitors induce selective killing of cancer cells is still an open question. The selective leakage and translation of pre-mRNAs may be one such mechanism for the selective antitumor activity of SSA and pladienolide derivatives because only a small subset of pre-mRNAs are exported to the cytoplasm, where some functional proteins may be translated. Indeed, p27*, a stable and active form of the CDK inhibitor p27, was translated from CDKN1B pre-mRNA, which was shown to be responsible for arresting the cell cycle following SSA treatment (Kaida et al. 2007). However, the accumulation of an active isoform such as p27* may be an exceptional case, as most pre-mRNA translation generates short truncated proteins due to the presence of termination codons in intron sequences. Such truncated proteins should be inactive or may even inhibit the native protein by serving as dominant negative forms. It is also possible that such truncated proteins gain novel activity to regulate oncogenic signals in cancer cells. Another possible mechanism would be gene-specific down-regulation of gene expression. VEGFA, which is important for angiogenesis, is a candidate for such down-regulation (Furumai et al. 2010). SSA treatment down-regulates VEGFA expression and inhibits angiogenesis (Furumai et al. 2010). SSA treatment causes defective recruitment of RNA polymerase II to the VEGFA gene (Furumai et al. 2010) and also causes inhibition of transcription elongation in a gene-specific manner (Koga et al. 2014, 2015). In addition, some genes important for cell-cycle progression, including those that encode cyclin A2 and the Aurora kinases, are also down-regulated in SSA-treated cells (Corrionero et al. 2011). Further studies focusing on the dynamics and function of pre-mRNA accumulated in the SSA-treated cells should be conducted to identify the mechanism that underlies the selective killing of cancer cells.
MATERIALS AND METHODS
Cell culture and transfection
HeLa S3 cell culture was performed as previously described (Kaida et al. 2007). Lipofectamine2000 (Life Technologies) was used for transfection according to the manufacturer's protocol.
Northern blotting analysis
RNA samples were separated by denaturing polyacrylamide gel electrophoresis (PAGE) before being transferred to Hybond N+ membranes (GE Healthcare). 32P-labeled RNA probes were transcribed using the mirVana miRNA Probe Construction Kit (Life Technologies). The probe for U6 snRNA was previously described (Yoshimoto et al. 2009). The sequence of the 5.8S probe was GGACACATTGATCATCGACACTTCGTTTTCCTGTCTC. Hybridization with oligonucleotide RNA probes was performed in PERFECTHYB PLUS (Sigma-Aldrich) as previously described (Yoshimoto et al. 2009).
RNA-seq library preparation
The RNA-seq library was prepared as previously reported (Kawaji et al. 2014). HeLa cells were harvested, and nuclear and cytoplasmic RNAs were extracted from them using PARIS kits (Life Technologies). Five micrograms of purified RNA were treated with the RiboZero rRNA removal kit (Human, Mouse, Rat) (Epicentre) to eliminate rRNA. Following rRNA depletion, an RNA-seq library was prepared with the RNA-seq library preparation method starting from the RNA fragmentation step. The library was sequenced on the HiSeq2000 (Illumina) platform. Sequence reads were deposited on NCBI under accession number GSE72156.
Data analysis
The sequence reads we obtained were filtered using TagDust (Lassmann et al. 2009) before they were mapped to the human hg19 genome using Tophat2 (Kim et al. 2013). The statistics of eliminating artifacts and mapping are shown in Supplemental Table S2. Transcript assembly based on mapped sequence reads was performed using Cufflinks (Trapnell et al. 2012). Alternative splicing isoforms were analyzed by rMATS (Shen et al. 2014) using Cufflink GTF files as input. Significant splicing events were defined if the shown false discovery rate (FDR) was less than 0.05. Splice site strength was calculated using MAXENT (Yeo and Burge 2004). The BP score was calculated using SVM-BP (Corvelo et al. 2010). Only the best SVM-scored branch point that resided in the last 100-nt window of the intron was used for further calculation. The strength of BPS was also calculated using experimentally verified branch point data sets (Mercer et al. 2015) and the RNAcofold program of Vienna RNA package (Lorenz et al. 2011). Intronic RPKM (reads per kilobase of intron per million mapped sequence reads) values were calculated using the RSeQC package (Khan et al. 2014). Data visualization was performed using the R/Bioconductor package (http://www.bioconductor.org). For statistical analysis, a Wilcoxon rank sum test was used to calculate a P-value.
RT-PCR analysis
Extracted RNAs were reverse transcribed using a PrimeScript Reverse Transcriptase kit (Takara). Obtained cDNAs were amplified using Ex Taq PCR enzyme (Takara) for 23–30 cycles and analyzed using either a 2.5% agarose gel or an 8% polyacrylamide gel.
Primers used for RT-PCR were as follows:
- DUSP1:
-
TGCAGTACCCCACTCTACGA (fwd),
GAGACGTTGATCAAGGCAGTG (rev)
- ID2:
-
TGTCAAATGACAGCAAAGCAC (fwd),
GTTGTTGTTGTGCAAAGAATAAAAG (rev)
- NFKBIA:
-
TCCTCAACTTCCAGAACAACC (fwd),
TCAGCAATTTCTGGCTGGT (rev)
- E2F2:
-
GCGCATCTATGACATCACCA (fwd),
TCAAACATTCCCCTGCCTA (rev)
- MSL3:
-
TGATCCATTTTAATGGTTGGAA (fwd),
CATCGGTATCACGAAGCACA (rev)
- RFC3:
-
AGTAGGTGCTTGGCGGTTC (fwd),
CACAGTAGATAACACGTGGCAAA (rev)
- GAPDH:
-
AGCCACATCGCTCAGACAC (fwd),
GCCCAATACGACCAAATCC (rev)
- MBNL1:
-
TGCCACAAGTGTTCCCTTC (fwd),
TGTTCGGCAGATATTATGGGTA (rev)
Primers specific for pre-mRNAs are as follows:
- DUSP1-pre:
-
TGCAGTACCCCACTCTACGA (fwd),
CTATATTCTCCCTGGCACTACTC (rev)
- NFKBIA-pre:
-
TCCTCAACTTCCAGAACAACC (fwd),
GGATCTGGGGTGACTCTGC (rev)
- E2F2-pre:
-
GCGCATCTATGACATCACCA (fwd),
CCACTGGCCGCCCAGGCA (rev)
- RFC3-pre:
-
AGTAGGTGCTTGGCGGTTC (fwd),
TACAATATTCAGAGAATCTGAACTG (rev)
The primers used to specifically amplify the p27 int-HA reporter-derived mRNA are as follows:
GAGCTCTCTGGCTAACTAGAGAACCCACTG (fwd)
GCTGGCAACTAGAAGGCACAGTCGAGGCTG (rev)
Primers specific for VEGFA mRNA have been described previously (Furumai et al. 2010).
Chromatin fractionation
Chromatin, nucleoplasmic, and cytoplasmic fractions were isolated as described previously (Pandya-Jones and Black 2009). Fractionated RNAs were extracted using the Nucleospin RNA kit (Machery-Nagel), and treatment with DNase I-containing buffer was used to completely digest genomic DNA.
Plasmid construction
The p27 int-HA plasmid was previously described (Kaida et al. 2007). To construct the p27 int-HA mutants, PCR-based mutagenesis was performed using the following primers:
For yeast BP: CCAGCCATTGTTTTTACTAACAAAGATTGTGTGTTC (fwd), GAACACACAATCTTTGTTAGTAAAAACAATGGCTGG (rev).
For BPmut: GCCATTGTTTTTTCTAGAAAAGATTGTGTGTTC (fwd), GAACACACAATCTTTTCTAGAAAAAACAATGGC (rev).
For the 5′ mutant: CGACCTGCAACCGACGGTCATGACCCTTTCCCAACCA (fwd), TGGTTGGGAAAGGGTCATGACCGTCGGTTGCAGGTCG (rev).
Immunoblotting
Immunoblotting was carried out as described previously (Yoshimoto et al. 2009). The antibodies used in this study were as follows: anti-GAPDH (1:500 dilution; Medical and Biological Laboratories [MBL]), anti-IBP160 (Hirose et al. 2006) (1:1000), anti-Lysine 9 acetylated Histone H3 (1:500 dilution; Abcam), anti-α-tubulin (B-5-1-2) antibody (1:500 dilution; Sigma-Aldrich), anti-Myc tag antibody (1:500 dilution; Santa Cruz Biotechnology), anti-HA tag antibody (1:1000 dilution; MBL), peroxidase-conjugated Affinipure goat anti-mouse IgG + IgM (H + L) (Jackson ImmunoResearch), and peroxidase-conjugated Affinipure goat anti-rabbit IgG (H + L) (1:10,000 dilution; Jackson ImmunoResearch).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by Grants-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (M.Y.), the Japan Science and Technology Corporation, the CREST Research Project (M.Y.), and the Hori Information Science Promotion Foundation (R.Y.). We also thank Dr. Yuko Kimura and Dr. Shinichi Nakagawa for discussions and careful reading of the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.058065.116.
REFERENCES
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787. [DOI] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. 2011. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol 9: e1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrionero A, Minana B, Valcarcel J. 2011. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev 25: 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvelo A, Hallegger M, Smith CW, Eyras E. 2010. Genome-wide association between branch point properties and alternative splicing. PLoS Comput Biol 6: e1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitkovic I, Jurica MS. 2013. Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res 41(Database issue): D132–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205. [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J 23: 4847–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskens FA, Ramos FJ, Burger H, O'Brien JP, Piera A, de Jonge MJ, Mizui Y, Wiemer EA, Carreras MJ, Baselga J, et al. 2013. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin Cancer Res 19: 6296–6304. [DOI] [PubMed] [Google Scholar]
- Furumai R, Uchida K, Komi Y, Yoneyama M, Ishigami K, Watanabe H, Kojima S, Yoshida M. 2010. Spliceostatin A blocks angiogenesis by inhibiting global gene expression including VEGF. Cancer Sci 101: 2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73. [DOI] [PubMed] [Google Scholar]
- Hirose T, Ideue T, Nagai M, Hagiwara M, Shu MD, Steitz JA. 2006. A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol Cell 23: 673–684. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12: 5–14. [DOI] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3: 576–583. [DOI] [PubMed] [Google Scholar]
- Kawaji H, Lizio M, Itoh M, Kanamori-Katayama M, Kaiho A, Nishiyori-Sueki H, Shin JW, Kojima-Ishiyama M, Kawano M, Murata M, et al. 2014. Comparison of CAGE and RNA-seq transcriptome profiling using clonally amplified and single-molecule next-generation sequencing. Genome Res 24: 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfir N, Lev-Maor G, Glaich O, Alajem A, Datta A, Sze SK, Meshorer E, Ast G. 2015. SF3B1 association with chromatin determines splicing outcomes. Cell Rep 11: 618–629. [DOI] [PubMed] [Google Scholar]
- Khan K, Schneider-Poetsch T, Ishfaq M, Ito A, Yoshimoto R, Mukaida N, Yoshida M. 2014. Splicing inhibition induces gene expression through canonical NF-κB pathway and extracellular signal-related kinase activation. FEBS Lett 588: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL. 2011. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci 108: 13564–13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, Satoh T, Takasaki I, Kawamura Y, Yoshida M, Kaida D. 2014. U2 snRNP is required for expression of the 3′ end of genes. PLoS One 9: e98015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, Hayashi M, Kaida D. 2015. Splicing inhibition decreases phosphorylation level of Ser2 in Pol II CTD. Nucleic Acids Res 43: 8258–8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. 2007. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3: 570–575. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. 2016. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129: 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Hayashizaki Y, Daub CO. 2009. TagDust—a program to eliminate artifacts from next generation sequencing data. Bioinformatics 25: 2839–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Rosbash M. 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M. 2011. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat Struct Mol Biol 18: 1115–1123. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Clark MB, Andersen SB, Brunck ME, Haerty W, Crawford J, Taft RJ, Nielsen LK, Dinger ME, Mattick JS. 2015. Genome-wide discovery of human splicing branchpoints. Genome Res 25: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizui Y, Sakai T, Iwata M, Uenaka T, Okamoto K, Shimizu H, Yamori T, Yoshimatsu K, Asada M. 2004. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J Antibiot (Tokyo) 57: 188–196. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Hori Y, Terano H, Okuhara M, Manda T, Matsumoto S, Shimomura K. 1996a. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J Antibiot (Tokyo) 49: 1204–1211. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Sato B, Fujita T, Takase S, Terano H, Okuhara M. 1996b. New antitumor substances, FR901463, FR901464 and FR901465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J Antibiot (Tokyo) 49: 1196–1203. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Takase S, Terano H, Tanaka H. 1997. New antitumor substances, FR901463, FR901464 and FR901465. III. Structures of FR901463, FR901464 and FR901465. J Antibiot (Tokyo) 50: 96–99. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. 2009. Co-transcriptional splicing of constitutive and alternative exons. RNA 15: 1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, Legrain P. 1997. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J 16: 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal GA, Jurica MS. 2010. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res 38: 6664–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz B, Seraphin B. 2000. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J 19: 1873–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Asai N, Okuda A, Kawamura N, Mizui Y. 2004a. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. II. Physico-chemical properties and structure elucidation. J Antibiot (Tokyo) 57: 180–187. [DOI] [PubMed] [Google Scholar]
- Sakai T, Sameshima T, Matsufuji M, Kawamura N, Dobashi K, Mizui Y. 2004b. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. I. Taxonomy, fermentation, isolation and screening. J Antibiot (Tokyo) 57: 173–179. [DOI] [PubMed] [Google Scholar]
- Sato M, Muguruma N, Nakagawa T, Okamoto K, Kimura T, Kitamura S, Yano H, Sannomiya K, Goji T, Miyamoto H, et al. 2014. High antitumor activity of pladienolide B and its derivative in gastric cancer. Cancer Sci 105: 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Basyuk E, Robert MC, Yoshida M, Villemin JP, Auboeuf D, Aitken S, Bertrand E. 2011. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J Cell Biol 193: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci 111: E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura R, Takeiwa T, Taniguchi I, McCloskey A, Ohno M. 2011. Multiple factors in the early splicing complex are involved in the nuclear retention of pre-mRNAs in mammalian cells. Genes Cells 16: 1035–1049. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Steitz JA. 1996. A mammalian gene with introns instead of exons generating stable RNA products. Nature 379: 464–466. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wang Q, He J, Lynn B, Rymond BC. 2005. Interactions of the yeast SF3b splicing factor. Mol Cell Biol 25: 10745–10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394. [DOI] [PubMed] [Google Scholar]
- Yoshimoto R, Kataoka N, Okawa K, Ohno M. 2009. Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res 37: 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.