Abstract
PIWI-interacting RNAs (piRNAs) play essential roles in the defense system against selfish elements in animal germline cells by cooperating with PIWI proteins. A subset of piRNAs is predicted to be generated via the “ping-pong” cascade, which is mainly controlled by two different PIWI proteins. Here we established a cell-based artificial piRNA production system using a silkworm ovarian cultured cell line that is believed to possess a complete piRNA pathway. In addition, we took advantage of a unique silkworm sex-determining one-to-one ping-pong piRNA pair, which enabled us to precisely monitor the behavior of individual artificial piRNAs. With this novel strategy, we successfully generated artificial piRNAs against endogenous protein-coding genes via the expected back-and-forth traveling mechanism. Furthermore, we detected “primary” piRNAs from the upstream region of the artificial “ping-pong” site in the endogenous gene. This artificial piRNA production system experimentally confirms the existence of the “ping-pong” cascade of piRNAs. Also, this system will enable us to identify the factors involved in both, or each, of the “ping” and “pong” cascades and the sequence features that are required for efficient piRNA production.
Keywords: PIWI proteins, piRNAs, ping-pong cycle, silkworm cells, artificial piRNAs
INTRODUCTION
Animals possess a highly tuned defense system against a diverse range of selfish, repeat elements, such as transposons. In animal germline cells, PIWI-clade proteins and 23–30 nucleotide (nt)-long small RNAs that are associated with PIWI proteins (piRNAs) play a major role in this immune system. PIWI proteins possess a slicer activity that is guided by piRNAs; therefore the PIWI–piRNA complex represses transposon activity by cleaving transposon RNAs (Klattenhoff and Theurkauf 2008; Ghildiyal and Zamore 2009). The piRNA production is initiated by the fragmentation of long, single-stranded piRNA precursors that are transcribed from piRNA clusters (Brennecke et al. 2007; Li et al. 2013). This step is mediated by the Zucchini/MitoPLD endoribonuclease (Ipsaro et al. 2012; Nishimasu et al. 2012), and the fragmented RNAs are then incorporated into a PIWI protein with a specific nucleotide preference for uracil (1U) at their 5′-ends (Cora et al. 2014). The PIWI-loaded fragmented RNAs are also cleaved by Zucchini/MitoPLD at a position downstream from the PIWI-protected region (Ipsaro et al. 2012; Nishimasu et al. 2012; Han et al. 2015; Mohn et al. 2015). The 3′-end of the associated RNA is then trimmed by PNLDC1, a homolog of PARN (called Trimmer) in silkworms (Kawaoka et al. 2011a; Izumi et al. 2016) or PARN-1 in worms (Tang et al. 2016), which is coupled with 3′-end modification by the methyltransferase Hen1 (Horwich et al. 2007; Kirino and Mourelatos 2007; Saito et al. 2007; Kamminga et al. 2010). Recently, Zucchini/MitoPLD-dependent cleavage of the precursor RNA was demonstrated to produce additional downstream piRNAs (“phased” piRNAs) by defining their 5′-ends (Han et al. 2015; Homolka et al. 2015; Mohn et al. 2015). These processes are known as the primary pathway.
Bioinformatic analyses of large sets of piRNA sequences suggest a model for the secondary piRNA biogenesis pathway, which is known as the “ping-pong” amplification cycle (Brennecke et al. 2007; Gunawardane et al. 2007; Li et al. 2009a; Malone et al. 2009). In this pathway, PIWI and 1U piRNA complexes cleave their complementary targets across from nts 10 and 11 from the guide piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). The 3′ RNA fragments are in turn incorporated into another subset of PIWI proteins and again processed into mature secondary piRNAs with adenine at the position 10 (10A), which precisely overlap with 1U piRNAs by 10 nt and constitute the “ping-pong” signature (Brennecke et al. 2007; Gunawardane et al. 2007). Next, these secondary 10A piRNAs produce secondary 1U piRNAs by cleaving their complementary target RNAs via cooperation with another PIWI protein. This cleavage-dependent piRNA biogenesis, called the “ping-pong” amplification cycle, is supposed to be broadly conserved among many animals including flies, mice, zebrafish, sponges, and silkworms (Brennecke et al. 2007; Aravin et al. 2008; Grimson et al. 2008; Houwing et al. 2008; Kawaoka et al. 2009).
Integration of an exogenous transgene into the active piRNA clusters is supposed to be required for de novo production of piRNAs with ping-pong signatures (Kawaoka et al. 2012; Muerdter et al. 2012; Yamamoto et al. 2013). Furthermore, the introduction of reporter cassettes containing single or multiple piRNA-dependent cleavage sites, which are predicted to be cleaved by endogenous piRNA–PIWI complexes, results in artificial piRNA production (Xiol et al. 2014; Homolka et al. 2015; Mohn et al. 2015). However, these studies do not provide solid evidence for the presence of the entire piRNA “ping-pong” amplification cycle: (i) Each artificial piRNA was not precisely tracked because artificial piRNAs were produced from the entire reporter cassette including promoter sequences (Kawaoka et al. 2012; Muerdter et al. 2012; Yamamoto et al. 2013; Xiol et al. 2014; Itou et al. 2015; Kuramochi-Miyagawa and Nakano 2015; Shoji and Katsuma 2015); or (ii) artificial piRNAs that were produced from the reporter with the piRNA-dependent cleavage sites were not verified to act in production of partner piRNAs with 10-nt overlaps; therefore, only half of the “ping-pong” cascade was validated (Xiol et al. 2014; Homolka et al. 2015; Mohn et al. 2015; Nishida et al. 2015). Collectively, the piRNA “ping-pong” cascade needs to be verified by solid experimental results.
In 2014, we discovered that the primary determiner of sex in the silkworm Bombyx mori is a female-specific, W chromosome-derived piRNA (Kiuchi et al. 2014). This piRNA, called Feminizer (Fem) piRNA, is loaded into the silkworm PIWI protein Siwi, and the Fem piRNA–Siwi complex cleaves a protein-coding mRNA called Masculinizer (Masc). This cleavage leads to production of Masc-derived piRNA, which is then loaded into another PIWI protein, BmAgo3. Because Masc encodes a protein required for masculinization, cleavage of Masc mRNA by the Fem piRNA–Siwi complex leads to feminization in the silkworm (Kiuchi et al. 2014). Based on the loading preference of Fem and Masc piRNAs to the two silkworm PIWI proteins and the presence of a ping-pong signature between these piRNAs, these sex-determining piRNAs are assumed to be produced by a “ping-pong” cycle (Fig. 1A; Kiuchi et al. 2014). Fem is a repeat element on the silkworm W chromosome, but Fem-derived piRNAs contain almost identical sequences (Kiuchi et al. 2014). In addition, Masc is a single copy, protein-coding gene on the Z chromosome, indicating that unlike most piRNAs, Fem and Masc piRNAs are a unique one-to-one “ping-pong” piRNA pair. Furthermore, this piRNA pair is present in the silkworm ovary-derived BmN-4 cell line (Kiuchi et al. 2014), which has a complete piRNA pathway (Kawaoka et al. 2009). Here, we established an artificial piRNA production system in BmN-4 cells using the Fem–Masc piRNA pair and artificial piRNA precursors for silkworm endogenous protein-coding genes. This system adds experimental evidence for the presence of the piRNA “ping-pong” cascade and will be useful for studying the piRNA “ping-pong” biogenesis cascade.
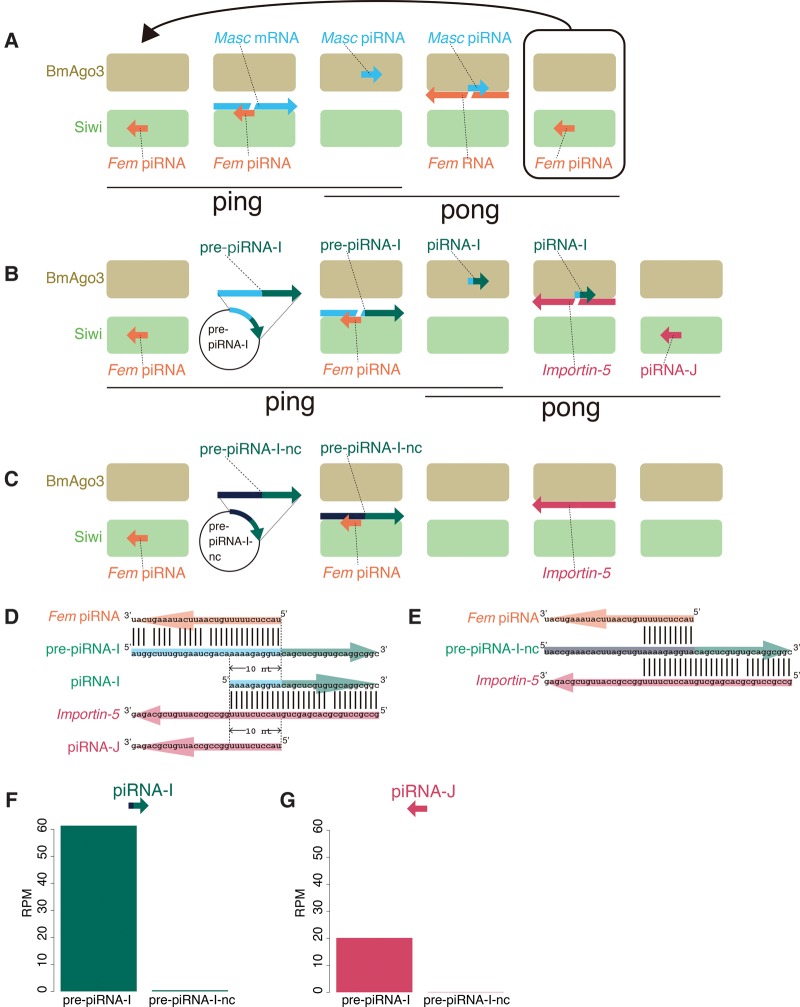
FIGURE 1.
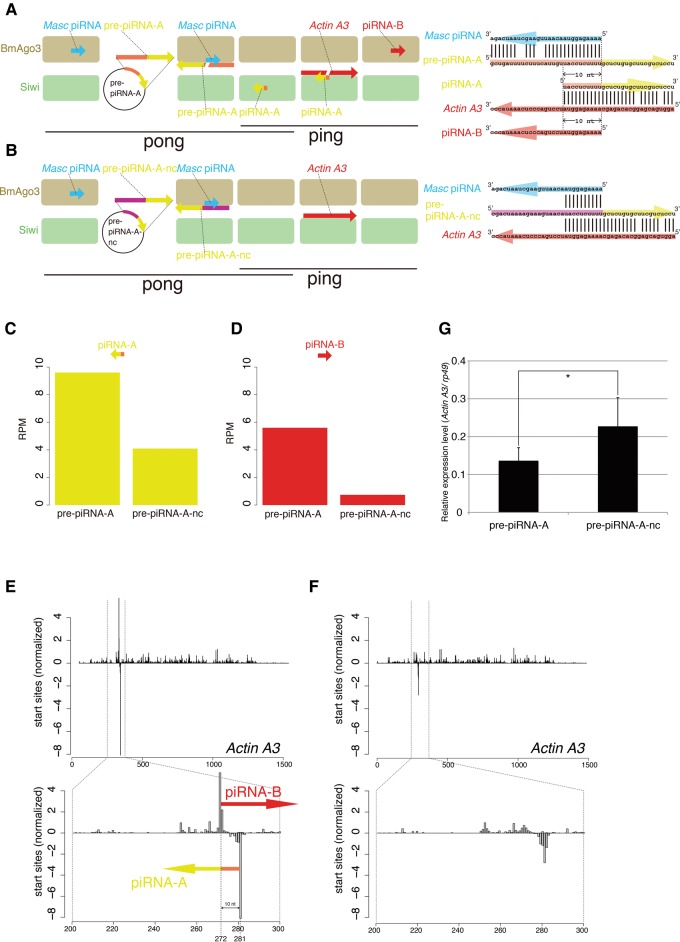
Conceptual diagrams of the artificial piRNA production system using silkworm BmN-4 cells. (A) The silkworm sex determination pathway via a unique one-to-one ping-pong pair, Fem piRNA and Masc piRNA. (B,C) The artificial piRNA production system targeting the silkworm Importin-5 gene. The silkworm ovary-derived cell line BmN-4 was transfected with plasmids expressing pre-piRNA-I (B) or pre-piRNA-I-nc (C). pre-piRNA-I mRNA is cleaved by the Fem piRNA–Siwi complex, whereas pre-piRNA-I-nc mRNA is not cleaved, due to sequence incompatibility. The resulting 3′ pre-piRNA-I fragment is loaded into BmAgo3, producing the piRNA-I–BmAgo3 complex, which recognizes and cleaves silkworm Importin-5 mRNA. The cleaved 3′ fragment is then loaded into another PIWI protein, Siwi, thereby producing the piRNA-J–Siwi complex. (D,E) Sequence design for artificial piRNA precursors. The pre-piRNA-I is designed to be cleaved by the endogenous Fem piRNA–Siwi complex, whereas pre-piRNA-I–nc mRNA contains nontargeted nucleotide sequences. (F,G) Normalized piRNA-I (F) and piRNA-J (G) reads in total piRNA libraries from BmN-4 cells transfected with pre-piRNA-expressing plasmids. Mapping reads against 1811 transposons were used for normalization.
RESULTS AND DISCUSSION
Design of a “ping-pong”-dependent artificial piRNA production system using silkworm cells
We established an artificial piRNA production system using the silkworm ovary-derived cell line BmN-4, which has a complete “ping-pong” amplification pathway (Kawaoka et al. 2009). We also used the silkworm primary sex determination cascade in which a unique one-to-one ping-pong pair, Fem piRNA and Masc piRNA, function to trigger sexual differentiation in both silkworm embryos and BmN-4 cells (Katsuma et al. 2014; Kiuchi et al. 2014). Fem piRNA preferentially binds to Siwi, a silkworm PIWI protein, whereas Masc piRNA preferentially binds to BmAgo3, another PIWI protein (Fig. 1A). The Fem piRNA–Siwi complex cleaves Masc mRNA, and the resulting 3′ Masc mRNA fragment is loaded into BmAgo3, thereby producing a Masc piRNA–BmAgo3 complex (for simplicity, we refer to this cascade as “ping”) (Fig. 1A). In turn, the Masc piRNA–BmAgo3 complex cleaves Fem RNA, and the cleaved 3′ Fem RNA fragment is then recruited into Siwi, thereby completing the Fem piRNA–Siwi complex (we refer to this cascade as “pong”) (Fig. 1A).
We designed a plasmid expressing the piRNA precursor (pre-piRNA-I) with a perfect 10-nt overlap with Fem piRNA (Fig. 1B,D). Artificial piRNA (piRNA-I) will be produced by cleavage of pre-piRNA-I with the endogenous Fem piRNA–Siwi complex (Fig. 1D). The pre-piRNA-I was designed to contain a sequence that extensively matched the coding region of an endogenously expressed gene, Importin-5. When piRNA-I is successfully produced and loaded into BmAgo3, the piRNA-I–BmAgo3 complex will cleave Importin-5 mRNA, thereby producing Importin-5-derived piRNA (piRNA-J) and piRNA-J–Siwi complex (Fig. 1B,D). If these experimental results are obtained, we have shown both “ping” (piRNA-I production by the Fem piRNA–Siwi complex) and “pong” (piRNA-J production by the piRNA-I–BmAgo3 complex) cascades.
Characterization of artificial piRNAs produced by a ping-pong cascade in silkworm cells
We transfected BmN-4 cells with plasmids expressing pre-piRNA-I or pre-piRNA-I-nc (negative control; designed not to be targeted by the Fem piRNA–Siwi complex, Fig. 1C,E) and generated total piRNA libraries from the transfected cells (Supplemental Fig. S1). By deep sequencing of total piRNA libraries, we found that the peak length distribution was 27–28 nt (Supplemental Fig. S2A,B), which is identical to that observed in naïve BmN-4 cells (Kawaoka et al. 2009; Izumi et al. 2013). We clearly detected piRNA-I in pre-piRNA-I transfected BmN-4 cells but not in pre-piRNA-I-nc transfected cells (Fig. 1F; Supplemental Fig. S3), indicating that pre-piRNA-I is transcribed and cleaved by endogenous Fem piRNA–Siwi complex to produce piRNA-I. With transfection of pre-piRNA-I, we also detected piRNA-J production in BmN-4 cells (Fig. 1G). This indicates the production of the piRNA-I–PIWI complex targeting Importin-5 mRNA (Fig. 1B).
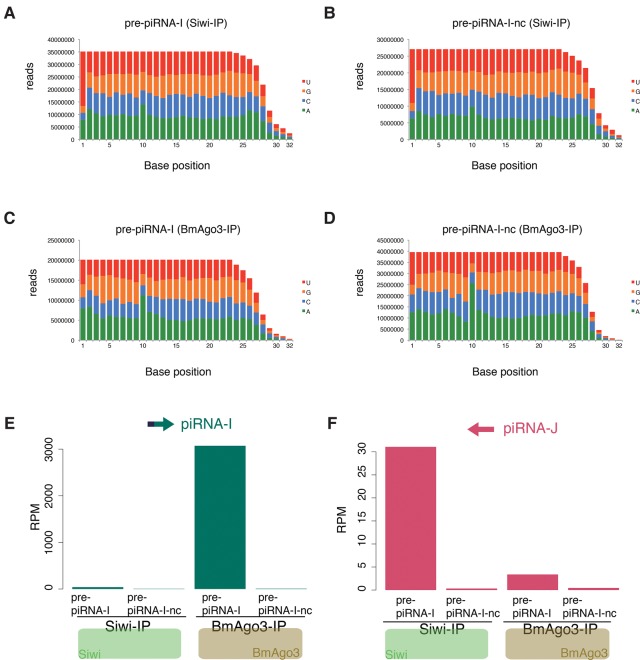
To determine whether correct piRNA loading occurred in this artificial system, we sequenced piRNAs that were immunoprecipitated (IPed) with Siwi antibody or BmAgo3 antibody from BmN-4 cells transfected with plasmids expressing pre-piRNA-I or pre-piRNA-I-nc (Supplemental Fig. S1). Siwi-bound and BmAgo3-bound piRNAs had a peak length distribution of 28 nt and 27 nt, respectively (Supplemental Fig. S2C–F), which is consistent with the results of our previous study (Izumi et al. 2013). Siwi-bound piRNAs showed 1U enrichment, whereas BmAgo3-bound piRNAs lacked this bias (Fig. 2A–D). In contrast, 10A enrichment was observed in BmAgo3-bound piRNAs (Fig. 2C,D). In addition, we found that Fem piRNA preferentially binds to Siwi, whereas Masc piRNA preferentially binds to BmAgo3 in cells transfected with each pre-piRNA expression plasmid (Supplemental Table S1). We counted the piRNA-I and piRNA-J reads in each IPed library. piRNA-I preferentially bound to BmAgo3, whereas piRNA-J preferentially bound to Siwi in pre-piRNA-I transfected cells (Fig. 2E,F; Supplemental Table S2). As observed in total piRNA libraries (Fig. 1F,G), both artificial piRNAs were barely detected in the IPed library from pre-piRNA-I-nc transfected cells (Fig. 2E,F; Supplemental Table S2). These results indicate that artificial piRNAs are correctly loaded into PIWI proteins in this artificial system.
FIGURE 2.
Analyses of Siwi-bound and BmAgo3-bound artificial piRNAs. (A–D) Nucleotide bias in each IPed piRNA library. (E,F) Normalized piRNA-I (E) and piRNA-J (F) reads in anti-Siwi- or anti-BmAgo3-IPed libraries from BmN-4 cells transfected with pre-piRNA-expressing plasmids. Mapping reads against 1811 transposons were used for normalization.
Cleavage of the target mRNA by artificial piRNAs in silkworm cells
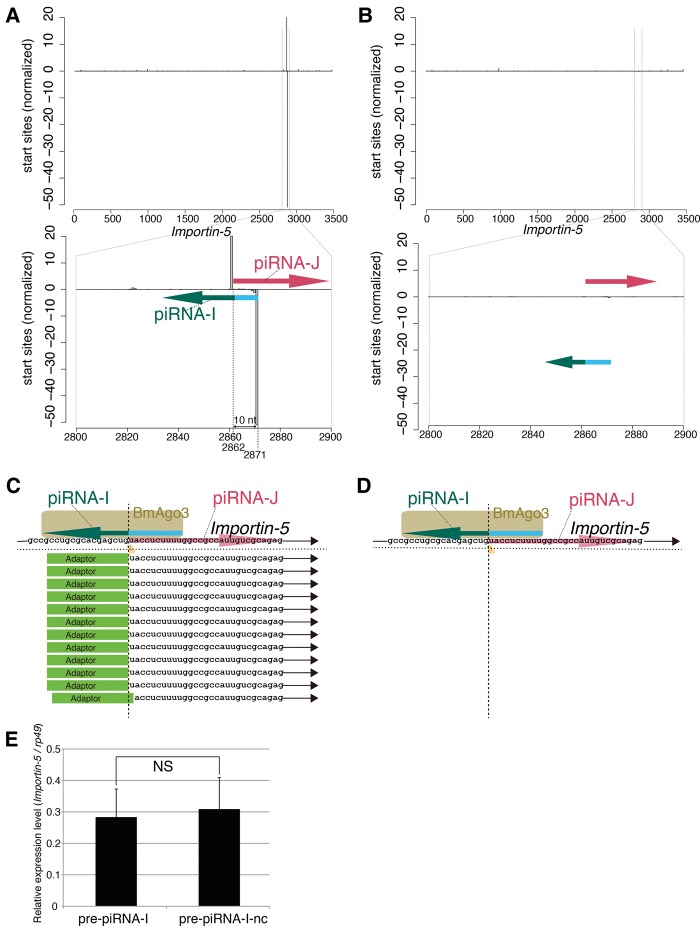
To determine whether an artificial piRNA, piRNA-I, cooperates with BmAgo3 to correctly cleave Importin-5 mRNA, we first mapped total piRNA libraries onto the coding region of Importin-5 to highlight the 5′-end of mapped piRNAs. We observed an obvious ping-pong signature between piRNA-I and piRNA-J in pre-piRNA-I transfected cells (Fig. 3A), but not in pre-piRNA-I-nc transfected cells (Fig. 3B), showing that our artificial piRNA biogenesis system works as designed (Fig. 1B,C). We next performed a modified 5′ rapid amplification of cDNA ends (modified RACE) to determine whether correct cleavage of Importin-5 mRNA occurs. We found that almost all the cloned 5′-ends (11/12) of Importin-5-derived RNA fragments from pre-piRNA-I transfected cells (Fig. 3C) exactly mapped to the predicted piRNA-I cleavage site. In contrast, the cloned fragments from pre-piRNA-I-nc transfected cells did not map to the putative cleavage site of the Importin-5 gene (Fig. 3D), indicating that artificially produced piRNA-I targets and cleaves Importin-5 mRNA with BmAgo3. Finally, we examined the expression level of Importin-5 in BmN-4 cells producing artificial piRNA for Importin-5. We performed RT-qPCR and found that Importin-5 mRNA levels in pre-piRNA-I transfected cells were comparable to those in pre-piRNA-I-nc transfected cells (Fig. 3E). These results indicate that production of a single, gene-specific artificial piRNA has little effect on mRNA levels of the endogenous Importin-5 gene in our BmN-4 system, yet the mRNA is cleaved by the piRNA–PIWI complex.
FIGURE 3.
Artificial piRNA production from the silkworm Importin-5 gene. (A,B) Artificial piRNAs derived from the Importin-5 gene. The start sites of piRNAs mapped to the Importin-5 gene are indicated. (C,D) Identification of the cleavage site of Importin-5 mRNA by the piRNA-I–BmAgo3 complex. Importin-5 mRNA-derived fragments were amplified by a modified RACE method using total RNA from BmN-4 cells transfected with plasmids expressing pre-piRNA-I (C) or pre-piRNA-I-nc (D). The RACE adaptor and cloned 5′-end sequences are indicated. Twelve 5′-ends were determined, with 11 clones showing identical sequences. (E) Expression level of Importin-5. Expression of Importin-5 mRNA was examined by RT-qPCR. Data shown are mean ± standard deviation (n = 5). Data were subjected to Student's t-test (one-sided). NS, not significant (P = 0.3412).
Generation of artificial piRNAs targeting other endogenous genes
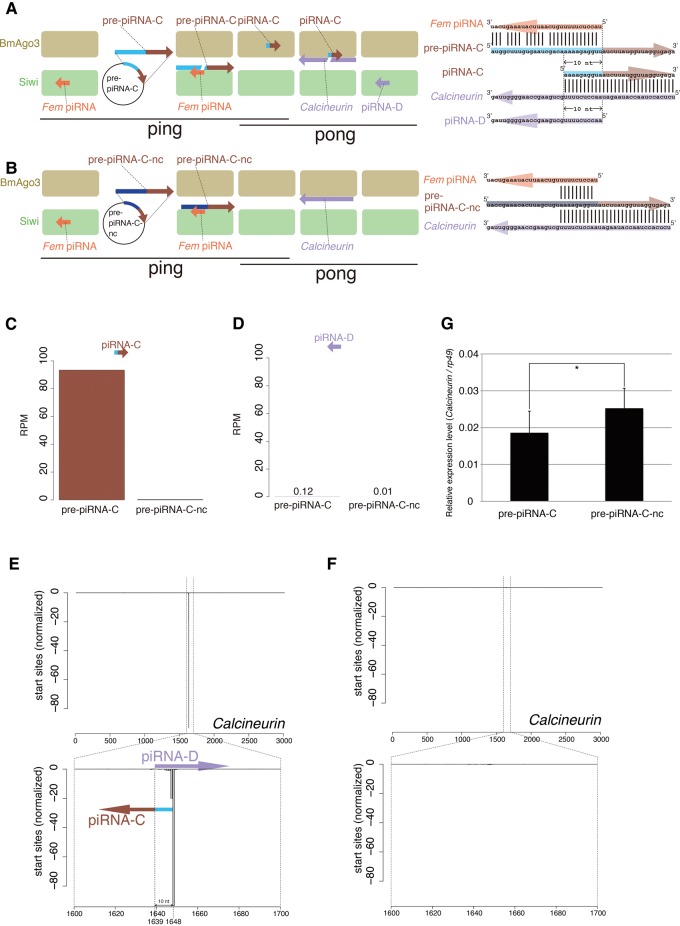
We next asked if artificial piRNAs for endogenous silkworm genes other than Importin-5 are produced in our cell-based system. We designed an artificial piRNA precursor for Calcineurin mRNA. Unlike the piRNA-I for Importin-5, the 5′-end nt of the Fem piRNA sequence is not identical to the corresponding nt of the Calcineurin mRNA sequence (Fig. 4A), which has been shown not to affect the cleavage efficiency by PIWI proteins (Wang et al. 2014). We performed deep sequencing of total piRNA libraries prepared from BmN-4 cells transfected with plasmids expressing pre-piRNA-C and pre-piRNA-C-nc (negative control, Fig. 4B). The length distribution was almost the same as that of naïve BmN-4 cells (Supplemental Fig. S2G,H; Kawaoka et al. 2009; Izumi et al. 2013). We detected production of huge amounts of pre-piRNA-C-derived piRNA (piRNA-C), whereas much less Calcineurin-derived piRNA (piRNA-D) was observed (Fig. 4C,D). The 10-nt overlap between piRNA-C and piRNA-D, i.e., a ping-pong signature, was also detected (Fig. 4E). In addition, both piRNA-C and piRNA-D were barely detected in pre-piRNA-C-nc transfected cells (Fig. 4F). Collectively, similar to the artificial piRNA for Importin-5, Calcineurin mRNA-derived piRNAs can be produced by a “ping-pong” cascade in BmN-4 cells, although the efficiency of piRNA-D production from Calcineurin mRNA is very low.
FIGURE 4.
Artificial piRNA production from the silkworm Calcineurin gene. (A,B) The artificial piRNA production system targeting the silkworm Calcineurin gene. The silkworm ovary-derived cell line BmN-4 is transfected with plasmids expressing pre-piRNA-C (A) or pre-piRNA-C-nc (B). The pre-piRNA-C mRNA is cleaved by the Fem piRNA–Siwi complex, whereas the pre-piRNA-C-nc mRNA is not due to sequence incompatibility. The resulting 3′ pre-piRNA-C fragment is loaded into BmAgo3, thus producing the piRNA-C–BmAgo3 complex, which cleaves silkworm Calcineurin mRNA. The cleaved 3′ fragment is then loaded into another PIWI protein, Siwi, thereby producing the piRNA-D–Siwi complex. (C,D) Normalized piRNA-C (C) and piRNA-D (D) reads in total piRNA libraries from BmN-4 cells transfected with plasmids expressing pre-piRNA-C or pre-piRNA-C-nc. Mapping reads against 1811 transposons were used for normalization. (E,F) Artificial piRNAs derived from the Calcineurin gene. The start sites of piRNAs mapped to the Calcineurin gene are indicated. (G) Expression levels of Calcineurin mRNA. Expression of Calcineurin mRNA was examined by RT-qPCR. Data shown are mean ± standard deviation (n = 5). Data were subjected to Student's t-test (one-sided). (*) P < 0.05 (P = 0.04977).
Finally, we asked whether the artificial “ping-pong” cascade can be recapitulated in BmN-4 cells from the “pong” cascade mediated by cleavage with the Masc piRNA-BmAgo3 complex instead of the Fem piRNA–Siwi complex, which controls the “ping” cascade. We designed an artificial piRNA precursor that can be cleaved by the endogenous Masc piRNA–BmAgo3 complex (Fig. 5A). We constructed a plasmid expressing an artificial piRNA precursor (pre-piRNA-A) for Actin A3 mRNA. We performed deep sequencing of total piRNA libraries from pre-piRNA-A and pre-piRNA-A-nc (negative control that was designed not to be targeted by the Masc piRNA–BmAgo3 complex, Fig. 5B) and found that the length distribution of total piRNAs was normal in both libraries (Supplemental Fig. S2I,J; Kawaoka et al. 2009; Izumi et al. 2013). Additionally, piRNA sequencing revealed production of pre-piRNA-A-derived piRNA (piRNA-A) and Actin-derived piRNA (piRNA-B) in pre-piRNA-A transfected cells (Fig. 5C–E). Production of piRNA-B was rarely observed in pre-piRNA-A-nc transfected cells (Fig. 5D,F). These results demonstrate that the piRNA biogenesis cascade from “pong” to “ping” is recapitulated in our BmN-4-based system, providing additional experimental evidence for the existence of the piRNA “ping-pong” cascade in this piRNA-expressing cell line.
FIGURE 5.
Artificial piRNA production from the silkworm Actin A3 gene. (A,B) The artificial piRNA production system targeting the silkworm Actin A3 gene. BmN-4 cells are transfected with plasmids expressing pre-piRNA-A (A) or pre-piRNA-A-nc (B). The pre-piRNA-A mRNA is cleaved by the Masc piRNA–BmAgo3 complex, whereas pre-piRNA-A-nc mRNA is not. The resulting 3′ pre-piRNA-A fragment is loaded into Siwi, thus producing the piRNA-A–Siwi complex, which targets silkworm Actin A3 mRNA. The cleaved 3′ fragment is then loaded into another PIWI protein, BmAgo3, thereby producing the piRNA-B–BmAgo3 complex. (C,D) Normalized piRNA-A (C) and piRNA-B (D) reads in total piRNA libraries from BmN-4 cells transfected with plasmids expressing pre-piRNA-A or pre-piRNA-A-nc. Mapping reads against 1811 transposons were used for normalization. (E,F) Artificial piRNAs derived from the Actin A3 gene. The start sites of piRNAs mapped to the Actin A3 gene are indicated. (G) Expression levels of Actin A3 mRNA. Expression of Actin A3 mRNA was examined by RT-qPCR. Data shown are mean ± standard deviation (n = 5). Data were subjected to Student's t-test (one-sided). (*) P < 0.05 (P = 0.02046).
In our system, the amount of Importin-5 mRNA was barely affected with transfection of the piRNA precursor (Fig. 3E). On the other hand, Calcineurin and Actin A3 mRNA levels were slightly but significantly reduced by transfecting plasmids expressing piRNA precursors (Figs. 4G, 5G). Because the amount of piRNA-I was comparable to that of piRNA-C (Figs. 1F, 4C), there are other factors that are involved in silencing efficiency by artificial piRNAs. Based on previous studies (Kawaoka et al. 2012; Itou et al. 2015), introduction of long antisense transgenes is more efficient to knockdown endogenous genes by artificial piRNAs compared with expression of specific piRNA precursors that produce a single artificial piRNA. We also observed that the amount of Calcineurin-derived piRNA (piRNA-D) was much less than that of the Importin-5-derived piRNA (piRNA-J), although the level of piRNA-C, which produces piRNA-D by cooperating with BmAgo3, was comparable to that of piRNA-I (Figs. 1F,G, 4C,D). These results suggest that efficiency of piRNA production might be dependent on the target sequence but not the amount of Siwi-loaded artificial piRNAs. Alternatively, Calcineurin mRNA may contain sequences that inhibit efficient production and/or stability of mature piRNAs. Using the Calcineurin mRNA as a model, we are now attempting to identify the sequence properties buried in piRNA precursors that determine the cleavage efficiency or amount of the resulting mature piRNAs.
Primary piRNA production from the upstream region of the artificial piRNA production locus in an endogenous gene
Recent studies in Drosophila and mice revealed that Zucchini/MitoPLD cleaves piRNA precursors and produces the 3′-end of the upstream primary piRNA and downstream “phased” primary piRNAs (Han et al. 2015; Homolka et al. 2015; Mohn et al. 2015). Primary piRNA production from the upstream region of the “ping-pong” loci of the exogenous reporter was also observed in BmN-4 cells transfected with reporter cassettes containing multiple piRNA-dependent cleavage sites (Homolka et al. 2015). We investigated whether primary piRNAs are similarly produced from the artificial piRNA production locus in an endogenous gene Importin-5. Three kinds of 1U primary piRNAs were produced from ∼40-nt upstream of the “ping-pong” site of this gene in BmN-4 cells transfected with pre-piRNA-I but not pre-piRNA-I-nc (Fig. 6A,B). Production of downstream “phased” primary piRNAs was not observed in transfected cells (Fig. 6A,B). We did not clearly detect primary piRNAs from the upstream region of the artificial piRNA production loci in Calcineurin and Actin A3 presumably because of low production of the artificial piRNA from these genes (Figs. 4D, 5D).
FIGURE 6.
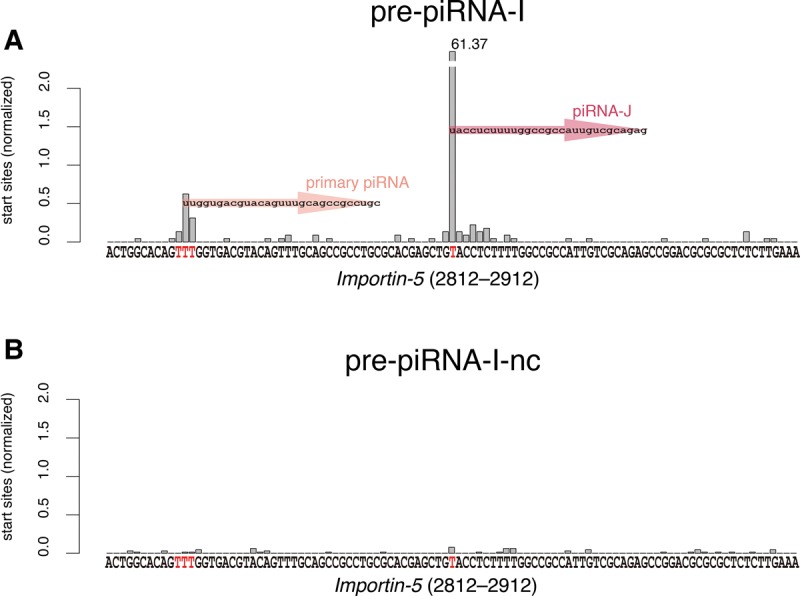
Primary piRNA production from the upstream region of the artificial piRNA production locus in the silkworm Importin-5 gene. Three 1U primary piRNAs were detected at ∼40 nt upstream of the artificial “ping-pong” site of the Importin-5 gene.
Defining the base-pairing required for target sequence recognition by the piRNA–PIWI protein complex
Base-pairing of nts 2–22 at the 5′-end of piRNA target sequence is sufficient for efficient target cleavage by the PIWI protein in mice (Reuter et al. 2011). We aimed to define precisely the base-pairing required for target sequence recognition by the piRNA–PIWI protein complex using our system. We constructed 15 plasmids expressing pre-piRNA-I derivatives, which contained sequential nucleotide mismatches in complementary sequences of Importin-5 (Fig. 7A). We transfected BmN-4 cells with plasmids expressing pre-piRNA-I, pre-piRNA-I-nc, or pre-piRNA-I derivatives, prepared small RNA fractions and performed RT-qPCR for piRNA-J. We found that 17-nt base-pairing of the 5′-end is necessary and 22-nt base-pairing is sufficient for piRNA-J production (Fig. 7B). These results clearly demonstrate that this system is a powerful tool to identify the factors required for piRNA production.
FIGURE 7.
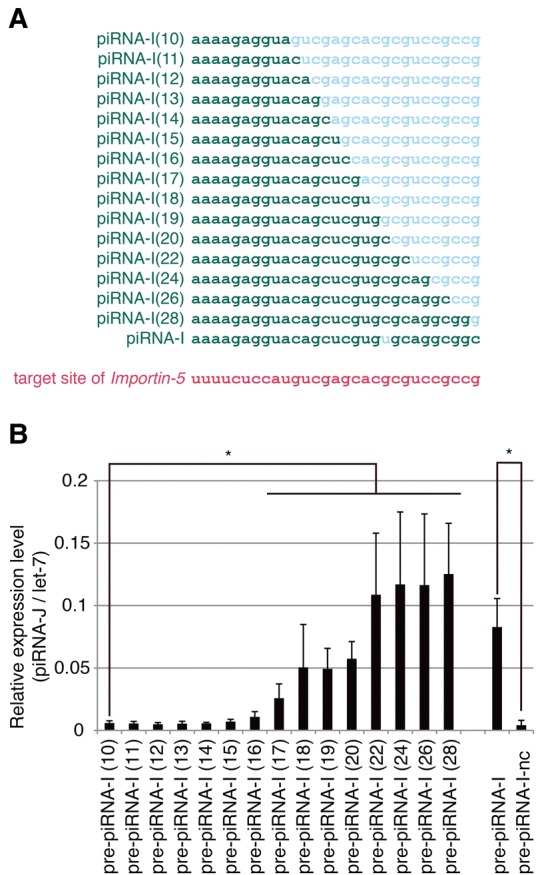
3′ Sequences required for target recognition by the piRNA–PIWI complex. (A) Nucleotide sequences of piRNA-I derivatives possessing identical (green) and different (blue) nucleotides to those of a complementary sequence of Importin-5 (red). (B) Expression levels of piRNA-J in BmN-4 cells transfected with pre-piRNA-I derivatives. The piRNA-J amounts were measured by RT-qPCR and normalized to those of let-7. Data shown are mean ± standard deviation (n = 4). (*) P < 0.05 with the Mann-Whitney U test followed by the Benjamini-Hochberg correction method to control the false discovery rate.
Recently, it was reported that piRNAs target protein-coding genes in spite of low stringent interactions (Vourekas et al. 2016). In our artificial system, we detected cleaved, mature piRNAs by piRNA sequencing or RT-qPCR, whereas Vourekas's study detected the piRNA–mRNA interactions but not their cleavage events. Also, we designed piRNA target sequences with nucleotide mismatches after completely identical sequences (Fig. 7A), which might not be consistent with most of the interactions observed between piRNAs and endogenous targets. To verify whether less stringent interactions between piRNAs and mRNAs are sufficient for piRNA production, further experiments are required by transfecting more plasmids with other types of nucleotide mismatches in pre-piRNA sequences in our artificial system.
Conclusions
Transgenic insertion and expression of reporter cassettes into a specific piRNA cluster or unknown piRNA-generating loci produced piRNAs from both strands of the entire transgene cassette, including the plasmid backbone (Kawaoka et al. 2012; Itou et al. 2015; Kuramochi-Miyagawa and Nakano 2015; Shoji and Katsuma 2015). These artificial piRNAs have “ping-pong” signatures; however, such systems do not completely trace the behavior of each artificial piRNA. In our system, we used a one-to-one ping-pong pair, Fem and Masc piRNAs, and succeeded in producing a predesigned, specific artificial piRNA that individually enabled us to monitor artificial piRNAs. We were also able to separately observe the active “ping” and “pong” cascades (Fig. 8A,B). Using this monitoring system for artificial piRNAs, it might be possible to identify the factors involved in both, or each, of the “ping” and “pong” cascades as well as the sequence properties required for efficient production of “ping-pong” piRNAs and “ping-pong”-triggered primary piRNAs.
FIGURE 8.
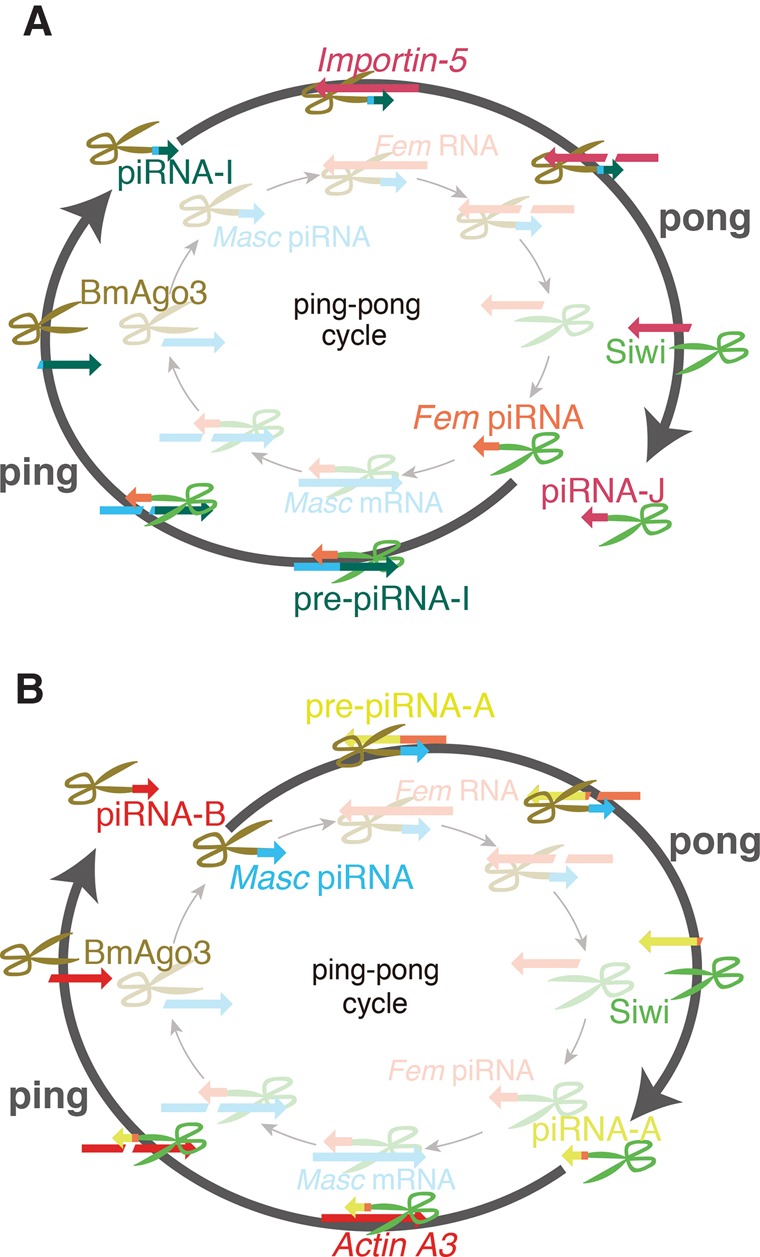
Proof-of-concept evidence for the piRNA “ping-pong” amplification cascade. (A) Artificial piRNA production triggered from the “ping” cascade. Exogenous pre-piRNA-I mRNA is cleaved by the endogenous Fem piRNA–Siwi complex. The resulting 3′ pre-piRNA-I mRNA fragment is loaded into BmAgo3, thus producing the piRNA-I–BmAgo3 complex. The piRNA-I–BmAgo3 complex cleaves the endogenous silkworm Importin-5 mRNA. The 3′ fragment of the cleaved Importin-5 mRNA is loaded into another PIWI protein Siwi, thus producing the piRNA-J–Siwi complex. (B) Artificial piRNA production triggered from the “pong” cascade. Exogenous pre-piRNA-A mRNA is recognized and cleaved by the endogenous Masc piRNA–BmAgo3 complex. The resulting 3′ pre-piRNA-A mRNA fragment is loaded into Siwi, thus producing the piRNA-A–Siwi complex. The piRNA-A–Siwi complex recognizes and cleaves the endogenous silkworm Actin A3 mRNA. The 3′ fragment of the cleaved Actin A3 mRNA is loaded into another PIWI protein BmAgo3, thus producing the piRNA-B–BmAgo3 complex.
MATERIALS AND METHODS
Design and construction of plasmids expressing piRNA precursors
The piRNA precursors for artificial piRNAs were designed as shown in Figures 1, 4, 5, and 7. The oligonucleotides listed in Supplemental Table S3 were annealed and cloned into BamHI and HindIII sites of the pIEX-4 vector (Novagen).
Cell line and transfection
BmN-4 cells were cultured at 27°C in IPL-41 medium (Applichem) supplemented with 10% fetal bovine serum. Transfection of BmN-4 cells with plasmid DNAs was performed using FuGENE HD (Promega). Three days after transfection, cells were harvested and used for total RNA or piRNA preparation.
Preparation of small RNA libraries
Total RNA was prepared from BmN-4 cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Ten micrograms of total RNA was loaded onto 15% denaturing polyacrylamide gels containing 10 M urea, separated by electrophoresis, and then stained with SYBRGold (Invitrogen). Small RNAs were recovered using the ZR small-RNA PAGE Recovery Kit (ZYMO Research). Small RNA libraries were constructed using the small RNA Cloning Kit (TaKaRa). DNA sequencing was performed using the Illumina HiSeq 2500 platform.
Sequence analysis of cloned small RNAs
Illumina HiSeq 2500 sequencing generated small RNA reads of 36 nt in length. The 3′-adaptor sequences were identified and removed, allowing for up to two mismatches. Reads shorter than 23 nt or longer than 32 nt were excluded, thereby obtaining reads of 23–32 nt. Mapping small RNAs to the Bombyx genome (International Silkworm Genome Consortium. 2008) or 1811 Bombyx transposons (Osanai-Futahashi et al. 2008) was performed using bowtie (Langmead et al. 2009). Reads that could be aligned to the genome or transposons up to one mismatch were used to calculate the mapping rate of each library (Supplemental Table S4). Mapping rates against 1811 transposons were used for normalization. Sam files were converted to bam files by SAMtools (Li et al. 2009b) and then to bed files by BEDTools (Quinlan and Hall. 2010). 5′-End positions for each piRNA were obtained from bed files using handmade R programs. Basic information on each piRNA library is provided in Supplemental Tables S1, S2, and S4.
Immunoprecipitation of Siwi-bound and BmAgo3-bound piRNAs
Immunoprecipitation of Siwi-bound and BmAgo3-bound piRNAs was performed using anti-Siwi and anti-BmAgo3 antibodies (Kawaoka et al. 2009), respectively. IPed piRNAs were isolated using TRIzol reagent, electrophoresed and stained with SYBRGold. Small RNA libraries were constructed as described above. Basic information on each IPed piRNA library is provided in Supplemental Tables S1, S2, and S4.
Modified RACE
5′-Ends of mRNA-derived RNA fragments were determined by a modified RACE procedure (Watanabe et al. 2011), with the primers listed in Supplemental Table S3.
RT-qPCR
Total RNA was subjected to reverse transcription with avian myeloblastosis virus reverse transcriptase and oligo(dT) primer (TaKaRa). RT-qPCR was performed using the KAPA SYBR FAST qPCR Kit (Kapa Biosystems) and the specific primers listed in Supplemental Table S3.
Small RNA fractions were enriched with the aid of a mirVana miRNA Isolation Kit (Ambion) according to the manufacturer's instruction. Reverse transcription and following qPCR for piRNA-J and let-7 were performed as described previously (Kawaoka et al. 2011b) with the specific primers listed in Supplemental Table S3.
DATA DEPOSITION
Deep sequencing data obtained in this study are available under the accession number DRA004111 (DDBJ).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yukihide Tomari and Takashi Kiuchi for critical reading of the manuscript and Munetaka Kawamoto for clerical assistance. K.S. is a recipient of a fellowship from the Japan Society for the Promotion of Science (15J00469). This work was supported by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (grant number 26034A) to S.K., Grant-in-Aid for Scientific Research (A) (grant number 15H02482) to S.K. and T.S., and Grant-in-Aid for Scientific Research (B) (grant number 16KT0064) to S.K. and Y.S.
Author contributions: S.K. and K.S. conceived and designed the experiments. K.S. performed all of the molecular biological experiments and bioinformatic analyses. Y.S. and S.S. performed deep sequencing. K.S., Y.S., S.S., T.S., and S.K. performed data analysis. All of the authors discussed the data and helped with manuscript preparation. S.K. and K.S. wrote the manuscript with intellectual input from all authors. S.K. supervised the project.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.058875.116.
REFERENCES
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Cora E, Pandey RR, Xiol J, Taylor J, Sachidanandam R, McCarthy AA, Pillai RS. 2014. The MID-PIWI module of Piwi proteins specifies nucleotide- and strand-biases of piRNAs. RNA 20: 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka D, Pandey RR, Goriaux C, Brasset E, Vaury C, Sachidanandam R, Fauvarque MO, Pillai RS. 2015. PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep 12: 418–428. [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Silkworm Genome Consortium. 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol 38: 1036–1045. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou D, Shiromoto Y, Yukiho SY, Ishii C, Nishimura T, Ogonuki N, Ogura A, Hasuwa H, Fujihara Y, Kuramochi-Miyagawa S, et al. 2015. Induction of DNA methylation by artificial piRNA production in male germ cells. Curr Biol 25: 901–906. [DOI] [PubMed] [Google Scholar]
- Izumi N, Kawaoka S, Yasuhara S, Suzuki Y, Sugano S, Katsuma S, Tomari Y. 2013. Hsp90 facilitates accurate loading of precursor piRNAs into PIWI proteins. RNA 19: 896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Shoji K, Sakaguchi Y, Honda S, Kirino Y, Suzuki T, Katsuma S, Tomari Y. 2016. Identification and functional analysis of the pre-piRNA 3′ Trimmer in silkworms. Cell 164: 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF. 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 29: 3688–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuma S, Kawamoto M, Kiuchi T. 2014. Guardian small RNAs and sex determination. RNA Biol 11: 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S. 2009. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 15: 1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 2011a. 3′ End formation of PIWI-interacting RNAs in vitro. Mol Cell 43: 1015–1022. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Kadota K, Arai Y, Suzuki Y, Fujii T, Abe H, Yasukochi Y, Mita K, Sugano S, Shimizu K, et al. 2011b. The silkworm W chromosome is a source of female-enriched piRNAs. RNA 12: 2144–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Mitsutake H, Kiuchi T, Kobayashi M, Yoshikawa M, Suzuki Y, Sugano S, Shimada T, Kobayashi J, Tomari Y, et al. 2012. A role for transcription from a piRNA cluster in de novo piRNA production. RNA 18: 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. 2007. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13: 1397–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, Ishihara G, Kawaoka S, Sugano S, Shimada T, et al. 2014. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509: 633–636. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. 2008. Biogenesis and germline functions of piRNAs. Development 135: 3–9. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Nakano T. 2015. Reply to Shoji and Katsuma. Curr Biol 25: R710. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009a. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009b. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Moore MJ, Zamore PD. 2013. Defining piRNA primary transcripts. Cell Cycle 12: 1657–1658. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J. 2015. piRNA-guider slicing specifies transcripts for Zucchini-dependent phased piRNA biogenesis. Science 348: 812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA. 2012. Production of artificial piRNAs in flies and mice. RNA 18: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Iwasaki YW, Murota Y, Nagao A, Mannen T, Kato Y, Siomi H, Siomi MC. 2015. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep 10: 193–203. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491: 284–287. [DOI] [PubMed] [Google Scholar]
- Osanai-Futahashi M, Suetsugu Y, Mita K, Fujiwara H. 2008. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem Mol Biol 38: 1046–1057. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267. [DOI] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji K, Katsuma S. 2015. Is the expression of sense and antisense transgenes really sufficient for artificial piRNA production? Curr Biol 25: R708–R710. [DOI] [PubMed] [Google Scholar]
- Tang W, Tu S, Lee H-C, Weng Z, Mello CC. 2016. The RNase PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell 164: 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Alexiou P, Vrettos N, Maragkakis M, Mourelatos Z. 2016. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 531: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yoshikawa M, Han BW, Izumi N, Tomari Y, Weng Z, Zamore PD. 2014. The initial uridine of primary piRNAs does not create the tenth adenine that is the hallmark of secondary piRNAs. Mol Cell 56: 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. 2011. Role of piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332: 848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Couté Y, Conn S, Kadlec J, Sachidanandam R, et al. 2014. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157: 1698–1711. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Watanabe T, Hoki Y, Shirane K, Li Y, Ichiiyanagi K, Kuramochi-Miyagawa S, Toyoda A, Fujiyama A, Oginuma M, et al. 2013. Targeted gene silencing in mouse germ cells by insertion of a homologous DNA into a piRNA generating locus. Genome Res 23: 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.