This review by Degrauwe et al. summarizes our current understanding of the functions of IMPs during normal development and focuses on a series of recent observations that have provided new insight into how their physiological functions enable IMPs to play a potentially key role in cancer stem cell maintenance and tumor growth.
Keywords: cancer, IGF2BP, IMP, RNA binding proteins, let-7, stem cells
Abstract
IMPs, also known as insulin-like growth factor 2 (IGF2) messenger RNA (mRNA)-binding proteins (IGF2BPs), are highly conserved oncofetal RNA-binding proteins (RBPs) that regulate RNA processing at several levels, including localization, translation, and stability. Three mammalian IMP paralogs (IMP1–3) have been identified that are expressed in most organs during embryogenesis, where they are believed to play an important role in cell migration, metabolism, and stem cell renewal. Whereas some IMP2 expression is retained in several adult mouse organs, IMP1 and IMP3 are either absent or expressed at very low levels in most tissues after birth. However, all three paralogs can be re-expressed upon malignant transformation and are found in a broad range of cancer types where their expression often correlates with poor prognosis. IMPs appear to resume their physiological functions in malignant cells, which not only contribute to tumor progression but participate in the establishment and maintenance of tumor cell hierarchies. This review summarizes our current understanding of the functions of IMPs during normal development and focuses on a series of recent observations that have provided new insight into how their physiological functions enable IMPs to play a potentially key role in cancer stem cell maintenance and tumor growth.
RNA-binding proteins (RBPs) regulate gene expression by intervening at all stages of messenger RNA (mRNA) metabolism, including transcription, 5′ end capping, precursor mRNA splicing, 3′ end processing, nuclear export, transport, translation, and stability (for review, see Muller-McNicoll and Neugebauer 2013). Individual RBPs may have as many as hundreds to thousands of mRNA targets (Hafner et al. 2010; Anko and Neugebauer 2012; Ascano et al. 2012) and serve as structural components of messenger ribonucleoprotein particles (mRNPs), in which mRNAs are transported from the nucleus to the cytoplasm and whose composition helps determine transcript fate (Muller-McNicoll and Neugebauer 2013). As many RBPs lack enzymatic activity, they can exert some of their regulatory functions by recruiting additional proteins, which may include decay enzymes and translational repressors (Moore and Proudfoot 2009; Schoenberg and Maquat 2012). RBPs thereby not only add to the structural and functional diversity of mRNPs but, more importantly, orchestrate the composition of RNA processing effector complexes to which they confer sequence specificity.
RBPs share post-transcriptional regulation of mRNA stability with microRNAs (miRNAs), 21- to 22-nucleotide (nt) noncoding RNAs (ncRNAs) that mediate post-transcriptional silencing by target mRNA degradation or translational repression (Bartel 2009) and are implicated in the regulation of virtually all biological processes in multicellular organisms (He and Hannon 2004; Ivey and Srivastava 2010). miRNA biogenesis is a multistep process that is not discussed here, and we refer readers to several excellent reviews (Ha and Kim 2014; Lin and Gregory 2015).
MiRNAs and RBPs can exert synergistic or opposing effects on target mRNAs (for review, see van Kouwenhove et al. 2011; Ciafre and Galardi 2013; Ho and Marsden 2014). RBPs can cooperate with miRNAs to effect mRNA destabilization by recruiting miRNA-containing RNA-induced silencing complexes (RISCs), the core of which consists of Argonaute proteins (AGO1–4) (for review, see Ha and Kim 2014), to target mRNAs (Jing et al. 2005; Kim et al. 2009; Glorian et al. 2011) or altering the secondary structure of target mRNAs to render miRNA-binding sites more accessible (Kedde et al. 2010). Conversely, RBPs can compete with miRNAs for binding to or in the vicinity of miRNA recognition binding sites (also referred to as miRNA recognition elements [MREs]) on target mRNAs, including their own, and protect transcripts from miRNA-dependent degradation (Kedde et al. 2007; Kundu et al. 2012; Young et al. 2012; Xue et al. 2013). In some situations, the same RBP can exert a stabilizing or destabilizing effect on different target mRNAs, depending on its interplay with miRNAs that associate with the same transcripts (Srikantan et al. 2012). Determination of mRNA fate by this complex relationship between RBPs and miRNAs affects a broad panel of physiological and pathological events from embryogenesis and differentiation to transformation and tumorigenesis.
Among the ∼600 RBPs annotated in mammalian genomes based on their content of known RBPs and 800 candidate RBPs identified in cell lines using new high-throughput ultraviolet (UV) cross-linking technologies (Baltz et al. 2012; Castello et al. 2012), relatively few have been associated with cancer. RBPs with a recognized role in tumor growth and progression include the TET family (Riggi et al. 2007), the STAR family (Elliott and Rajan 2010), β-catenin (Lee et al. 2007; Kim et al. 2012), LIN28A and LIN28B (Viswanathan et al. 2009; Nguyen et al. 2014), and several RBPs involved in RNA splicing (David and Manley 2010). Recent work from several laboratories has shown that the IMP family of oncofetal RBPs can promote carcinogenesis in part by regulating miRNA activity.
Three mammalian IMP paralogs—IMP1–3, also known as insulin-like growth factor 2 (IGF2) mRNA-binding proteins 1, 2, and 3 (IGF2BP1–3)—have been identified thus far. They belong to the family of zipcode-binding proteins initially proposed to bear the acronym VICKZ based on the first letter of its founding members (Yaniv and Yisraeli 2002), which include Vegetal-1 mRNA-binding protein (Vg1RBP/Vera) in Xenopus laevis; IMP1–3 in mammals; coding region instability determinant (CRD)-binding protein, subsequently shown to be IMP1; K homology (KH) domain-containing protein overexpressed in cancer (KOC), subsequently shown to be IMP3; and zipcode-binding protein 1 (ZBP1) in chickens. ZBP1 was shown to bind a 54-nt sequence element (or zipcode) located in the 3′ untranslated region (UTR) of β-actin mRNA in chicken cells, which resulted in the active transport of β-actin transcripts to regions of polarized cell growth in neurons and fibroblasts (Ross et al. 1997; Bassell et al. 1999; Farina et al. 2003; Huttelmaier et al. 2005). IMP1 (a ZBP1 ortholog) was identified as a protein involved in MYC mRNA stabilization (Doyle et al. 1998). Its mechanism of action was shown to be prevention of MYC degradation by binding to its CRD (hence the initial denomination CRD-binding protein). Similar to ZBP1 in chicken cells, IMP1 was observed to control the subcellular localization of β-actin mRNA in primary mammalian fibroblasts (Farina et al. 2003) and neurons (Tiruchinapalli et al. 2003) by binding to the cis-acting zipcode in the 3′ UTR. Human IMP2, which has diverged phylogenetically from IMP1 and IMP3 and has no known orthologs, was discovered as a result of its binding to IGF2 transcripts (Nielsen et al. 1999). It was also referred to as p62, reflecting the molecular mass of one of its splice variants, and was proposed to be an autoantigen in hepatocellular carcinoma (HCC) (Zhang et al. 1999a). Single-nucleotide polymorphisms (SNPs) have been identified in the second intron of the IMP2 gene that correlate with elevated risk of type 2 diabetes (Christiansen et al. 2009). IMP3, a Vg1-RBP/Vera ortholog initially called KOC, was identified on the basis of its abundance in pancreatic cancer (Mueller-Pillasch et al. 1997). It was subsequently shown to be expressed by a broad range of tumors, and its expression was often found to correlate with poor prognosis (for review, see Lederer et al. 2014).
Physiological expression of IMP family members
Physiological expression of IMPs occurs primarily during development. Mammalian IMPs display a biphasic expression pattern, first appearing in the oocyte and zygote (Nielsen et al. 2001; Yaniv and Yisraeli 2002) and subsequently displaying up-regulation on mouse embryonic day 10.5 (E10.5) to E12.5 (Nielsen et al. 1999; Runge et al. 2000). At mid-gestation, IMPs are expressed in most developing tissues, their expression being highest in neuronal and epithelial cells. Imp1 and Imp3 transcripts are expressed in the forebrain and hindbrain, the snout, the branchial arches, the gut, the tail, the vertebrae, and the skin (Mueller-Pillasch et al. 1999; Mori et al. 2001; Hansen et al. 2004). A similar expression pattern is observed in Xenopus, zebrafish, and Drosophila (Mueller-Pillasch et al. 1999; Zhang et al. 1999b; Nielsen et al. 2000; Adolph et al. 2009). Detailed assessment of expression in mouse brains revealed that, at E10.5, Imp1 is expressed throughout the ventricular zone (VZ) of the entire developing brain with the exception of the floor and roof plates. Between E12.5 and E16.5, Imp1 expression gradually becomes restricted primarily to the dorsomedial telencephalon (DMT), where it continues to be expressed by undifferentiated neural stem/progenitor cells in the VZ and sub-VZ (SVZ) (Nishino et al. 2013). Little or no Imp1 expression is observed in differentiated neurons that accumulate at the cortical plate, and virtually no Imp1 expression remains in the cerebral cortex at birth. However, some Imp1 expression persists in the small and large intestines, kidney, and liver for several days after birth, and low Imp1 expression levels can be detected in the intestines of adult mice (Hansen et al. 2004). In contrast to Imp1, Imp3 mRNA becomes virtually undetectable at birth.
During embryogenesis, Imp2 expression resembles that of Imp1 and Imp3 (Christiansen et al. 2009) and, at E17.5, is observed in the brain—including the neopallial cortex, VZ, and striatum—as well as the nasal cavity, lungs, liver, intestines, and kidney. However, unlike Imp1 and Imp3, Imp2 transcripts are prominent in the perinatal period and in adult mouse tissues, including brain, gut, bone marrow, kidney, lung, muscle, liver, testis, and pancreas (Bell et al. 2013). Thus, IMP2 expression overlaps with that of IMP1 and IMP3 during development but, in contrast to that of its paralogs, persists in several adult organs in mice.
Protein structure and RNA binding
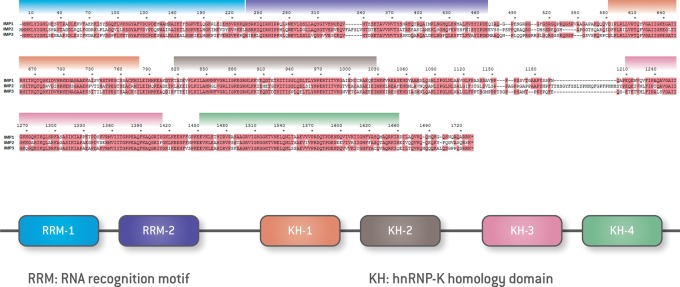
In mammals, the canonical structure of the three IMP proteins is highly similar in terms of domain order and spacing (Fig. 1). The overall amino acid sequence identity between the three proteins is 56% (Bell et al. 2013), with even greater similarity within the domains, consistent with shared functions. IMP1 and IMP3 are the most closely related members of the family, with 73% amino acid sequence identity (Bell et al. 2013). All three proteins contain two RNA recognition motifs (RRMs) in their N-terminal regions and four hnRNP KH domains in their C-terminal regions (Nielsen et al. 1999). In Drosophila, a protein lacking the N-terminal RRM domain but containing four KH domains is proposed to be the Drosophila IMP (dIMP) (Bell et al. 2013).
Figure 1.
Structure of IMP paralogs. (Top) Amino acid sequence alignment and corresponding domain structure of IMP1, IMP2, and IMP3. Sequence similarity among the paralogs is highlighted in red, and colors indicate the boundaries of the domains shown in B. Numbers represent corresponding base numbers of the respective genes. (Bottom) Schematic representation of IMP domains.
All members of the IMP family, irrespective of the organism in which they are expressed, have been shown to bind RNA. In vitro studies have shown that the KH domains are primarily responsible for RNA binding, although the RRMs may contribute to the stabilization of IMP–mRNA complexes (Nielsen et al. 2004; Wachter et al. 2013). However, the architecture of the KH domains allows recognition of only short stretches of RNA with relatively weak binding affinity (Chao et al. 2010) that may be insufficient for appropriate regulation of target mRNA fate. Increased binding affinity and specificity are typically achieved by the presence of multiple copies of these domains within the RBP along with their arrangement in an orientation that optimizes interaction with their partners. Structural analyses of IMP1 KH domains 3 and 4 suggest that they function as a single module by assuming an anti-parallel pseudodimer conformation in which the RNA-binding surfaces are placed on opposite ends of the molecule (Chao et al. 2010). The RBP can therefore recognize its targets through sequence-specific interactions that span two stretches of RNA separated by an appropriate number of nucleotides (Chao et al. 2010). Two low-affinity interactions can thereby generate highly specific RNA binding, suggesting that IMPs may force their associated transcripts to adopt a specific conformation. Based on this notion, a model has been proposed in which binding of KH3 and KH4 to mRNA induces a conformational change in the transcript (Chao et al. 2010), which may create new RNA-binding sites for other factors. The IMP family of RBPs may therefore orchestrate the assembly of higher-order complexes, which may not necessarily require direct interaction with other proteins (Chao et al. 2010). The long half-life of IMP–mRNA complexes in vitro testifies to their stability and supports the notion that IMPs bind their target mRNAs with high affinity (Nielsen et al. 2004).
The mRNA-binding repertoire of IMP family members, which should provide insight into the mechanisms by which they regulate transcript processing and the corresponding cellular functions, is beginning to be elucidated. A genome-wide study in HEK293T cells using photoactivatable ribonucleoside-enhanced UV cross-linking and immunoprecipitation (PAR-CLIP), which provides an almost single-nucleotide resolution of RNA-binding sites, proposed the definition of a putative IMP consensus recognition element as 5′-CAUH-3′ (H = A, U, or C) (Hafner et al. 2010). The candidate recognition element was contained in >75% of the top 1000 clusters for IMP1–3 and identified >100,000 sequence clusters recognized by the IMP family that map to ∼8400 protein-coding transcripts (Hafner et al. 2010). A more recent comparison of IMP RNA-binding specificity using enhanced UV CLIP (eCLIP) in human pluripotent stem cells (hPSCs), which express all three paralogs, revealed significant overlap between IMP1 and IMP2 binding that was not observed between either IMP1 and IMP3 or IMP2 and IMP3 (Conway et al. 2016). IMP1 and IMP2 were found to bind predominantly 3′ UTRs of target genes with enrichment of CA motifs in eCLIP-defined binding sites, whereas IMP3 bound a higher portion of coding regions, which did not correlate with either IMP1 or IMP2 (Conway et al. 2016).
PAR-CLIP applied to a primary proneural glioblastoma (GBM) grown as a glioma stem cell (GSC)-enriched spheroid culture and its nontumorigenic adherent progeny (Degrauwe et al. 2016) revealed that IMP2 preferentially localizes to 3′ UTRs of target mRNAs with high AT content and miRNA-binding sites. IMP2-binding density was enriched on a subset of miRNA-binding sites, suggesting that IMP2 protects these transcripts from miRNA-mediated silencing. Consistent with this notion, transcripts bound by IMP2 on miRNA-binding sites of previously validated miRNA targets constituted the class of mRNAs that was the most strongly silenced upon IMP2 depletion (Degrauwe et al. 2016).
Physiological functions
Several approaches, including reverse genetics in loss-of-function and gain-of-function models, have been used to address the physiological role of IMPs. In Drosophila, aberrant expression of neuronal dIMP resulted in compromised central and peripheral synaptogenesis, suggesting that dIMP is required for appropriate exon guidance, similar to IMP1 in mammalian cells (Boylan et al. 2008). In Xenopus, depletion of Vg1RBP/Vera revealed its requirement for migration of cells from the roof plate of the neural tube and for neural crest migration (Yaniv et al. 2003). These observations are consistent with the well-established function of IMP1 in mRNA localization, as illustrated by IMP1-mediated and neuronal ZBP1-mediated β-actin mRNA transport to the leading edge of migrating fibroblasts (Farina et al. 2003) and to dendrites and growth cones (Zhang et al. 2001; Tiruchinapalli et al. 2003), respectively. Based on these observations, IMP1 has been proposed to enhance neurite outgrowth and axonal guidance. Additional mRNAs whose localization is shown to be regulated by IMP1 include H19 and Tau (Runge et al. 2000; Atlas et al. 2004). The full repertoire of mRNAs subjected to IMP1-mediated transport remains to be determined but is likely to be extensive, as suggested by a study that found 3% of the transcriptomes of HEK293 cells to be represented in IMP1 mRNP granules (Jonson et al. 2007). The most abundant of these transcripts encode proteins implicated in the secretory pathway, ubiquitin-dependent protein degradation, and post-transcriptional control of mRNA processing and translation. The degree to which IMP2 and IMP3 participate in RNA localization remains to be elucidated.
Gene deletion experiments indicate additional physiological functions. IMP1-deficient mice are ∼40% smaller than normal sex-matched littermates, with proportional reduction in the size of most organs, and are subject to significant perinatal mortality, attributed primarily to defects in intestinal development (Hansen et al. 2004). Histological changes in the gut among Imp1−/− mice range from an almost normal appearance to nearly complete loss of villi, decreased mucosal and muscle wall thickness, and modifications of the composition of the intestinal extracellular matrix (Hansen et al. 2004). In surviving mice, the histological modifications become gradually normalized, suggesting that Imp1 deficiency may cause delayed organ maturation (Hansen et al. 2004). Together, these observations raise the possibility that IMP1 may be implicated in the regulation of cell metabolism and/or stem cell maintenance.
Consistent with this notion, recent work suggests that IMP1 regulates changes in stem cell properties in fetal brains (Nishino et al. 2013). Imp1 was found to be expressed by fetal neural stem/progenitor cells in response to Wnt signaling, and the decline in its expression in late fetal development was observed to occur partly as a consequence of let-7 miRNA expression, of which Imp1 is a target (Nishino et al. 2013). Upon Imp1 deletion, fetal neural stem cells (NSCs) became prematurely depleted in the dorsal telencephalon due to accelerated differentiation, resulting in impaired pallial expansion and reduced brain mass. Based on these observations, Imp1 expression by stem/progenitor cells during forebrain development was proposed to regulate the timing of neuronal and glial differentiation, and postnatal loss of its expression was suggested to contribute to the decline in NSC function during adulthood (Nishino et al. 2013). Imp1 therefore appears to be required for the expansion of fetal NSCs but dispensable for self-renewal of adult NSCs. Taken together, these observations suggest that Imp1 may belong to a heterochronic gene network that regulates temporal changes in stem cell properties from fetal development throughout adulthood (Nishino et al. 2013). In addition to regulating NSC expansion, IMP1 may influence other stem cell functions. Thus, in hPSCs, IMP1 has been shown recently to regulate adhesion and promote survival (Conway et al. 2016).
Given that IMP2 is highly expressed in developing brains (with a peak around mid-gestation in mice) (Nielsen et al. 1999) and that its depletion in NSCs favors astrocytic lineage commitment (Fujii et al. 2013), it would seem plausible that, similar to IMP1, IMP2 may participate in NSC/progenitor cell maintenance. Using conditional Imp2 knockout mice crossed to Nestin Cre animals, Imp2 depletion was shown to impair NSC clonogenicity. Consistent with the observations of Fuji et al. (2013), NSCs depleted of Imp2 displayed increased glial and reduced neuronal differentiation (Degrauwe et al. 2016).
The phenotype of mice bearing homozygous Imp2 deletion suggests that IMP2 may play an important role in the control of cell metabolism. Imp2-deficient mice displayed a slightly smaller size than their normal littermates, which corresponded to mildly reduced linear growth and lean mass gain after weaning as well as decreased fat deposition following a high-fat diet with a decrease in adipocyte precursors (Dai et al. 2015). In addition, these mice displayed modestly increased energy expenditure, improved glucose tolerance, resistance to the development of fatty liver, and a longer life span than their wild-type littermates (Dai et al. 2015). IMP2 was discovered to bind uncoupling protein-1 (Ucp1) mRNA along with transcripts of several other mitochondrial proteins and inhibit its translation. The increased energy expenditure of Imp2−/− mice was proposed to be a consequence of increased UCP1 expression and contribute to resistance to obesity and the observed increased life span (Dai et al. 2015). Consistent with the phenotype of Imp2−/− mice, transgenic overexpression of Imp2 in mouse livers resulted in steatosis (Tybl et al. 2011). Interestingly, one of the first SNPs found by a genome-wide association study (GWAS) for type 2 diabetes mellitus (T2DM) is located in intron 2 of IMP2 (Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research 2007; Scott et al. 2007; Zeggini et al. 2007). However, the molecular mechanisms that may implicate IMP2 in T2DM remain to be uncovered.
IMP2 has also been shown to play a role in muscle cell motility (Boudoukha et al. 2010). IMP2 expression was observed to be elevated in primary myoblasts, myogenic cell lines, and regenerating skeletal muscle (Boudoukha et al. 2010). Its depletion resulted in a change in muscle cell shape and a decrease in motility. Whereas it would seem reasonable to assume that, by analogy to the function of IMP1, the observed changes may be related to alterations in β-actin or other cytoskeletal component mRNA transport, analysis of IMP2-depleted muscle cells revealed alterations in post-translationally modified α-tubulin associated with stable, nondynamic microtubules (Boudoukha et al. 2010).
The effects of IMP3 depletion in the organism remain to be determined, as there are currently no available knockout models.
Mechanisms of action
Evidence thus far indicates that IMP family members regulate a broad range of RNA processing steps, including localization, stability, translation, and possibly nuclear export (Nielsen et al. 2001; Yaniv and Yisraeli 2002; Bell et al. 2013). IMPs are primarily cytoplasmic, but the presence of two nuclear export signals suggests that they may bind at least some of their target RNAs in the nucleus and possibly facilitate their nuclear export (Oleynikov and Singer 2003; Huttelmaier et al. 2005; Pan et al. 2007). In the cytoplasm, IMPs form higher-order mRNP structures, which are large enough to be visible under the light microscope and therefore are referred to as mRNP granules (Mitchell and Parker 2014). IMP mRNP granules are located around the nucleus and in cell protrusions. In neuronal cells, these granules localize to dendrites and growth cones (Zhang et al. 2001; Tiruchinapalli et al. 2003), consistent with the notion that they are destined for local translation. IMP1-containing RNP granules are distributed along microtubules and are reported to travel toward the leading edge of motile cells into the cortical region of lamellipodia at an average of 0.12 µm/sec in an ATP-dependent fashion (Nielsen et al. 2002). IMP1 granules are suggested to constitute a unique mRNP entity distinct from processing bodies (P bodies), stress granules, and neuronal hStaufen granules (Jonson et al. 2007), although possibly related to RNA transport granules, which are prevalent in neurons and oocytes and are suggested to play a role in RNA localization (for review, see Mitchell and Parker 2014). They are reported to be 100–300 nm in diameter and, in addition to IMPs (IMP1 or IMP3), contain 40S ribosomal subunits, shuttling hnRNPs, poly(A)-binding proteins, and mRNAs. They also contain CBP80 and factors belonging to the exon–junction complex but lack eIF4E and eIF4G as well as 60S ribosomal subunits, suggesting that the constituent transcripts are not translated (Jonson et al. 2007; Weidensdorfer et al. 2009). IMP1 may therefore regulate mRNA localization by incorporating its target transcripts into mRNP granules that provide protection from premature degradation and promiscuous translation and unloading them to initiate protein synthesis at appropriate destinations.
Similar to other RBPs, IMP1 regulates the expression of numerous proteins, increasing the levels of some by augmenting the stability of their mRNAs and reducing the levels of others by inhibiting translation (for review, see Bell et al. 2013). Several recent studies have shown that one mechanism by which IMPs promote mRNA stability is by preventing miRNA-mediated silencing (Elcheva et al. 2009; Goswami et al. 2010; Nishino et al. 2013; Busch et al. 2016; Degrauwe et al. 2016). IMP1 was notably observed to promote the expression of the self-renewal factor and let-7 miRNA family target HMGA2 (for review, see Fusco and Fedele 2007), which raised the possibility that IMP1 may counteract let-7 miRNA action during development and thereby contribute to stem cell maintenance (Nishino et al. 2013). let-7 is a highly evolutionarily conserved 13-member family of miRNAs that promotes differentiation and suppresses tumor growth (Pasquinelli et al. 2000). Several let-7 target gene products—including LIN28A, LIN28B, and HMGA2—display both self-renewal and oncogenic functions, and LIN28A/B control let-7-mediated differentiation. LIN28 RBP paralogs, which are expressed in embryonic stem cells (ESCs), progenitor cells, and primordial germ cells (Moss and Tang 2003; West et al. 2009; Viswanathan and Daley 2010; Shyh-Chang and Daley 2013), maintain let-7 expression at low levels by blocking let-7 biogenesis, preventing their transition from the pre-miRNA to the mature miRNA form (Heo et al. 2008, 2009; Shyh-Chang and Daley 2013). LIN28A/B and let-7 thereby form a bistable switch that operates to maintain stemness or induce differentiation (for review, see Thornton and Gregory 2012). In the brain, NSC maintenance and differentiation (Nishino et al. 2008; Yu et al. 2015) as well as glial progenitor cell differentiation to astrocytes (Shenoy et al. 2015) are regulated by let-7 family members and their targets.
Sequestration of miRNA target mRNAs in mRNP granules that are free of Ago/RISC (Weidensdorfer et al. 2009; Busch et al. 2016) is proposed to be one mechanism by which IMP1 protects mRNAs from let-7-mediated silencing (Fig. 2). However, IMP1 has also been observed to disrupt miRNA-183-mediated targeting of the coding region of β-transducin repeat-containing E3 ubiquitin protein ligase (BTRC) transcripts (Elcheva et al. 2009) and miRNA-340-mediated targeting of the 3′ UTR of microphthalmia-induced transcription factor (MITF) mRNA (Goswami et al. 2015). IMP1 thus appears to protect its target mRNAs by at least two mechanisms by preventing miRNA binding to its recognition sites and/or displacing miRNA-containing RISCs and incorporating target mRNAs into mRNP, where they may be stored while awaiting release toward their ultimate fate (Fig. 2). The latter mechanism may help fine-tune the timing of specific transcript translation or decay.
Figure 2.

Mechanisms of mRNA protection from miRNA-mediated silencing by IMPs. Whereas LIN28A/B impair let-7 miRNAs maturation, IMP2 prevents let-7 target gene silencing by direct binding to let-7 MREs on mRNAs. IMP2 is found in P bodies, where it may be in direct competition with AGO2, the catalytic component of the RISC. Although IMP1 and IMP3 may also bind to MREs or in their immediate vicinity, safe-housing transcripts in a RISC-free environment constituted of mRNP granules is currently the best-understood mechanism by which they protect their target mRNAs.
IMP3 has been proposed to function in a way similar to that of IMP1, sequestering LIN28B, HMGA2 transcripts, and mRNAs of several other let-7 target genes into cytoplasmic granules that do not contain RISC/Ago (Fig. 2; Jonson et al. 2014). These IMP3 mRNP granules have been proposed to function as “cytoplasmic safe houses” for a variety of oncogenes and stemness maintenance genes targeted by miRNAs, particularly let-7 family members. Whether IMP3-associated mRNP granules have the same composition as their IMP1-associated counterparts remains to be determined. However, IMP1 and IMP3 have been observed in the same mRNP granules (Jonson et al. 2007), suggesting that the two RBPs share at least a fraction of their mRNPs. IMP3 and let-7 family members are reported to bind 3′ UTRs of target mRNAs in close proximity to each other (Jonson et al. 2014), but the binding appears to occur simultaneously without competition for a common binding site. Sequestration into AGO2/RISC-free mRNP granules may therefore be the principal mechanism of IMP3-mediated mRNA protection from let-7-dependent silencing. In contrast, in pancreatic ductal adenocarcinoma cells (PDACs), IMP3 was reported to compete with miRNAs for common binding sites in target mRNA 3′ UTRs but also promote association of mRNAs with AGO2/RISC, suggesting a function as a bimodal regulator of target mRNA stability (Ennajdaoui et al. 2016). Taken together, these observations suggest that IMP1 and IMP3 may regulate mRNA stability by multiple mechanisms, including sequestration in RISC-free mRNP granules, protection from miRNA-dependent degradation by competing for common binding sites, and enhancement of degradation by recruitment of AGO2/RISC to target mRNAs. Whereas the repertoire of effects that IMP1 and IMP3 may have on target mRNAs is becoming relatively clear, the underlying mechanics require further elucidation. By what mechanism is AGO/RISC excluded from IMP1 and IMP3 mRNP granules? What determines recruitment of AGO/RISC to some IMP3 target transcripts but not others? Furthermore, is there selectivity as to which mRNAs are incorporated into the AGO/RISC-free mRNPs, or are all IMP1 and IMP3 target transcripts included regardless of whether the IMPs have disrupted miRNA-mediated silencing by binding to the same cis-elements or in their immediate vicinity? The availability of genome-wide mRNA-binding repertoires of the IMP family should help address these issues.
IMP2 protects mRNAs from let-7-dependent silencing by binding to the corresponding miRNA-binding sites and preventing miRNA-mediated target transcript degradation (Fig. 2; Degrauwe et al. 2016). In contrast to IMP1 and IMP3, IMP2 was observed in P bodies, the assembly of which is dependent on the pool of nontranslating mRNA and which contain the conserved core of proteins implicated in mRNA decay and translation repression (Decker and Parker 2012; Lui et al. 2014; Anderson et al. 2015) However, mRNA decay itself does not necessarily occur within the bodies, and, in mammalian cells, mRNAs in P bodies can return to translation (Brengues et al. 2005; Anderson and Kedersha 2006; Bhattacharyya et al. 2006). How incorporation into P bodies might influence the fate of IMP2-bound mRNAs is currently unclear. Coimmunoprecipitation experiments uncovered a RNA-dependent association between AGO2 and IMP2 (Degrauwe et al. 2016) whose significance, in light of the maintained expression of let-7 target genes, remains enigmatic. It is conceivable that the interaction has an inhibitory effect on AGO2/RISC, but such a possibility has not been addressed directly.
The repertoire of mRNAs whose stability is regulated by IMPs may predict, at least in part, the phenotype of IMP paralog-expressing cells. Thus, protection of BCL2 and integrin transcripts, particularly ITGB5, by IMP3 has been reported recently to promote survival and adhesion, respectively, in hPSCs (Conway et al. 2016). Whether these effects are related to protection from let-7 or other miRNAs remains to be determined. Enforced expression of IMP3 in the murine hematopoietic system resulted in increased numbers of hematopoietic stem cells (HSCs), lymphoid-primed multipotent progenitors (LMPPs), and common lymphoid progenitors (CLPs) in the bone marrow, with increased proliferation of HSCs and LMPPs (Palanichamy et al. 2016). Mouse hematopoiesis was skewed toward the myeloid and B-cell lineages accompanied by leukocytosis in the peripheral blood, atypical infiltration of B cells into the thymic medulla, and increased myeloid cell numbers in the spleen (Palanichamy et al. 2016). The observed phenotype was ascribed to stabilization of Myc and Cdk6 mRNAs by IMP3 based on individual nucleotide-resolution CLIP (iCLIP) analysis and reporter assays. However, the mechanism of the stabilization remains to be elucidated, as deletion of single IMP3-binding sites in the 3′ UTRs of Myc and Cdk6 failed to reverse the phenotype of the reporter assays (Palanichamy et al. 2016).
Post-translational modifications of IMPs appear to be essential for their modulation of mRNA fate. Thus, Src-dependent phosphorylation of the linker region between KH domains 2 and 3 of IMP1 is proposed to induce the disassembly of cytoplasmic mRNPs and activate translation of β-actin mRNA (Huttelmaier et al. 2005). Phosphorylation of Vg1RBP/Vera by MAPKs is suggested to modulate the release of Vg1 mRNA from mRNPs localized to the vegetal cortex during meiotic maturation (Git et al. 2009). Dual phosphorylation of IMP2 in the N-terminal linker region between RRM2 and KH1 by mTOR complex 1 in a rapamycin-sensitive manner promotes its association with the leader 3 (L3) 5′ UTR of IGF2 and the translational initiation of the mRNA through eIF4E and 5′ cap-independent internal ribosomal entry, resulting in elevated IGF2 protein synthesis (Dai et al. 2011).
Although most studies thus far have focused on interactions of RBPs with coding transcripts, recent work has uncovered interactions between IMPs and ncRNAs. Thus, interaction of IMP2 with the long ncRNA (lncRNA) MyoD was found to prevent translation of its associated transcripts, N-Ras and C-myc (Gong et al. 2015). A study conducted on mesenchymal GBM showed that IMP2 interacts with the hypoxia-sensitive lncRNA HIF1a-AS2 and facilitates its association with its target transcripts, such as HMGA1 (Mineo et al. 2016). IMPs also appear to have the potential to destabilize lncRNAs, as shown by the ability of IMP1 to recruit the CCR4–NOT deadenylase complex to degrade the liver-specific lncRNA HULC (Hammerle et al. 2013).
Regulation of IMP expression
Most studies on IMP family members have focused on expression patterns during development and mechanisms of action, and comparatively little is known about the regulation of their expression. Nevertheless, there is evidence to suggest that IMP1 expression is subject to regulation by the Wnt pathway (Noubissi et al. 2006; Gu et al. 2008; Nishino et al. 2013). In 293T cells, IMP1 could be induced by β-catenin in a TCF-dependent manner (Noubissi et al. 2006). This was also observed to be the case in human breast cancer cells (Gu et al. 2008), where β-catenin was shown to bind to the IMP1 promoter and transactivate the gene. IMP1 expression in mammalian breast cancer cells correlated with nuclear translocation of endogenous β-catenin, and a highly conserved element (CTTTG-TC) located in the IMP1 promoter was shown to be necessary for β-catenin-mediated regulation of IMP1 gene expression. IMP1 was observed to stabilize β-catenin mRNA, suggesting a positive feedback loop in which β-catenin and IMP1 regulate each other's expression (Gu et al. 2008).
There is also evidence to suggest that IMP1 expression is regulated by c-Myc (Noubissi et al. 2010). The human IMP1 gene contains four consensus c-Myc-binding sequences in its promoter region. Both c-Myc and Max were shown to interact with DNA fragments of the IMP1 promoter containing each of the four sites; mutagenesis and reporter assays revealed that each c-Myc-binding site contributes to the regulation of IMP1 transcription, and a significant increase in IMP1 transcript and protein expression was observed in 293T and HeLa cells engineered to overexpress c-Myc. Thus, c-Myc induces IMP1 transcription by binding to the IMP1 promoter and is in turn stabilized by the IMP1 protein (Bernstein et al. 1992; Prokipcak et al. 1994; Doyle et al. 1998) in a positive feedback loop analogous to that created by IMP1 and β-catenin.
The let-7 target HMGA2, which plays an important role in the maintenance of pluripotency (for review, see Cleynen and Van de Ven 2008), has been shown to regulate IMP2 gene expression (Cleynen et al. 2007). HMGA2 does not have intrinsic transcriptional activation capacity but instead participates in the formation of higher-order nucleoprotein complexes (enhanceosomes) in promoter/enhancer regions of its target genes and is considered to be an “architectural” transcription factor (Cleynen and Van de Ven 2008; Pfannkuche et al. 2009). HMGA2 contains three DNA-binding domains, called “AT hooks,” which recognize AT-rich regions in promoters and enhancers to which they bind in a structure-specific rather than a sequence-specific fashion. IMP2 possesses an AT-rich region in the DNA sequence encoding its first intron, which is required for HMGA2-mediated regulation. Detailed assessment of the sequences surrounding the HMGA2-binding region uncovered a consensus binding site for NF-kB in the immediate vicinity of the AT-rich regulatory region. NF-kB binds to this site and cooperates with HMGA2 to induce IMP2 expression (Cleynen et al. 2007).
Induction of IMP1 and IMP2 expression by c-Myc/β-catenin and HMGA2, respectively, is consistent with both normal and cancer stem cell (CSC) properties as well as the oncofetal expression pattern of IMPs. Although the mechanisms that underlie regulation of IMP expression warrant further exploration, the observations that IMP1 and IMP2 are targets of central players in pluripotency and oncogenesis are consistent with their requirement in normal development and cancer initiation.
Implication in cancer: protection of let-7 target genes and CSC maintenance
IMP family members are re-expressed in a broad range of tumor types, where they appear to resume their physiological functions. By regulating the transport of cytoskeletal protein, adhesion receptor, and secretory protein mRNAs to the leading edge of motile cells and stabilizing mRNAs of numerous oncogenes and pluripotency factors, IMPs may be predicted to participate in several key events that underlie tumor development. Consistent with this notion, transgenic mice engineered to express IMP1 in mammary epithelial cells from the whey acidic protein (WAP) promoter developed mammary tumors in up to 95% of cases, some of which formed metastases (Tessier et al. 2004).
These initial observations were supported by more recent work in colorectal cancer (CRC) patients (Hamilton et al. 2013), where increased IMP1 expression was found to correlate with enhanced metastasis and poor prognosis. Accordingly, overexpression of IMP1 in CRC cell lines promoted their ability to form tumors in immunocompromised mice and disseminate into the bloodstream (Hamilton et al. 2013). Conversely, intestine-specific deletion of Imp1 strongly reduced the number of tumors in the ApcMin/+ model of intestinal tumor development, and human CRC cell line xenografts depleted of IMP1 displayed reduced numbers of circulating tumor cells (CTCs) (Hamilton et al. 2013). IMP1 overexpression decreased E-cadherin expression in CRC cell lines and promoted survival of CRC colonospheres as well as enrichment in the CD44+CD24+ subpopulation associated with tumor-initiating capacity (Hamilton et al. 2013).
Studies on IMP3, which is the IMP family member most widely associated with human cancer (for review, see Lederer et al. 2014), further support a functional implication of IMPs in tumor development. IMP3 expression was first observed in PDACs (Gress et al. 1996) and was associated with up to 97% of invasive tumors (Yantiss et al. 2005). Its expression has been reported in most tumors of the hepato-biliary system, including HCCs (Jeng et al. 2008; Hu et al. 2014), intrahepatic cholangiocarcinoma (Chen et al. 2013b), gallbladder carcinomas (Shi et al. 2013), and bile duct carcinomas (Levy et al. 2010), where it correlates with poor prognosis and invasiveness (Jeng et al. 2008). IMP3 expression in HCC mouse models as well as primary human HCC was observed to be highest in CD133+/CD49f+ cells, which display CSC/tumor-initiating cell (TIC) properties (Chen et al. 2013a), and has been proposed to enhance HCC cell migration and invasiveness by promoting the expression of HMGA2. Along with IMP1, IMP3 is suggested to be among the most highly up-regulated RBPs in HCC (Gutschner et al. 2014).
In breast carcinoma, IMP3 expression was observed to be high in triple-negative tumors (TNBCs). Experiments performed on cell lines suggest that IMP3 expression in these tumors is repressed by estrogen receptor β (ERβ) upon engagement by its ligand, 3βA-diol (Samanta et al. 2012), whereas it is induced by activation of the mitogen-activated protein kinase pathway in response to epidermal growth factor receptor (EGFR) signaling. Based on the discovery that ERβ represses EGFR transcription, ERβ has been proposed to repress IMP3 indirectly as a result of its effect on EGFR (Samanta et al. 2012). This notion is consistent with TNBC biology, where low ERβ levels allow elevated EGFR expression. IMP3 is suggested to contribute to TNBC progression by stimulating invasion and migration of the tumor cells, possibly as a result of stabilizing MMP9 and CD164 (endolyn) transcripts (Samanta et al. 2012). Depletion of IMP3 in TNBC cell lines resulted in increased sensitivity to doxorubicin and mitoxantrone (Samanta et al. 2013). The mechanism responsible for increased resistance to cytotoxic drugs was found to be stabilization of breast cancer resistance protein (BRCP or ABCG2) transcripts responsible for doxorubicin and mitoxantrone efflux. IMP3 was observed to bind BCRP mRNA and thereby regulate protein expression, although it is unclear whether the binding prevents miRNA-mediated silencing. IMP3 has also been suggested to promote stemness in TNBCs based on the observation that it is more highly expressed in TICs than in cells devoid of tumor-initiating capacity (Samanta et al. 2016). Investigation into the possible mechanisms underlying the putative induction or maintenance of stemness by IMP3 in TNBC cells revealed that IMP3 binds to the 5′ UTR of SNAI2 (SLUG) mRNA and regulates its expression (Samanta et al. 2016). Stabilization of SLUG expression results in SOX2 up-regulation, which is implicated in maintaining stemness (Samanta et al. 2016).
The observations in CRC, HCC, and TNBCs all point toward a role for IMP1 and IMP3 in promoting or preserving tumor cell subpopulations with stem cell features and thereby contributing to the establishment of tumor cell hierarchies that are responsible for heterogeneity in a broad range of tumors. Tumor cell heterogeneity, which is a hallmark of solid tumors, can arise by several mechanisms (for review, see Meacham and Morrison 2013). The best established among these, which is referred to as clonal evolution, relies on intrinsic differences among cancer cells resulting from stochastic genetic (Nowell 1976) and epigenetic (Baylin and Jones 2011) changes that are subject to selection within tumors. In some malignancies, however, nongenetic determinants related to developmental pathways play a central role in tumor heterogeneity. In these cancers, a small population of tumorigenic cells has the unique capacity to generate nontumorigenic progeny, which results in a hierarchical organization that is a caricature of normal tissue architecture. Hematopoietic malignancies as well as a subset of solid tumors have been shown to possess subpopulations of cells endowed with stem cell properties that include expression of ESC genes and asymmetric division (Lobo et al. 2007; Kreso and Dick 2014). These cells also display tumor-initiating capacity and are generally believed to represent the tumor repopulating force, preserving their own numbers through self-renewal and generating more differentiated progeny that comprises the bulk of the tumor. Together, these properties functionally define CSCs.
Evidence from several laboratories provides a candidate mechanism that underlies the association between IMP expression and CSC-like phenotypes. All three IMP paralogs have been shown to protect let-7 target genes from silencing. They therefore fulfill a function similar to that of LIN28A/B, albeit by distinct mechanisms, and may cooperate with LIN28A/B to induce or maintain stemness in tumor cells (Fig. 3). Similar to IMPs, LIN28 paralogs are expressed in a broad range of tumor types, including many pediatric malignancies (Carmel-Gross et al. 2016), and predominantly in aggressive, poorly differentiated tumor cell subpopulations (Viswanathan et al. 2009; Viswanathan and Daley 2010; Shyh-Chang and Daley 2013). LIN28 expression can be induced by numerous mediators, one of which is NF-kB, the master regulator of the inflammatory response. NF-kB induces IL-6, which promotes LIN28B expression and inhibition of let-7 maturation (Iliopoulos et al. 2009), leading to increased expression of several oncogenes and stemness maintenance genes that may be relevant to the development of a cell hierarchy.
Figure 3.
Schematic representation of the principal known functions of IMPs that may contribute to CSC properties and maintenance. All three IMP paralogs as well as LIN28A/B block let-7 activity, albeit by different mechanisms, and their transcripts are also let-7 targets. Poorly differentiated tumor cells that display self-renewal and tumor-initiating properties express elevated levels of LIN28 paralogs, usually in conjunction with IMP family members, although some such cells express either LIN28 or IMP paralogs. Low let-7 levels facilitate expression of LIN28, IMPs, and HMGA2. In addition, LIN28 and IMPs stabilize each others’ expression as well as that of HMGA2; HMGA2 in turn promotes expression of IMP2. The main functions of IMPs recognized thus far that are associated with their elevated expression in tumor cells include maintenance of stemness or pluripotency; transport of cytoskeletal and adhesion receptor mRNAs to the cell periphery, which promote migration and invasiveness; enhancement of IGF2 translation; and transport of several nuclear-derived mRNAs that encode various components of the respiratory chain to mitochondria, promoting oxidative phosphorylation (OXPHOS).
IMP family members are commonly re-expressed in tumor cells that contain one or both of the LIN28 paralogs (Busch et al. 2016; our unpublished observations). In ovarian cancer cell lines, LIN28B and IMP1 appear to display synergistic protection of let-7 target transcripts, including HMGA2, as well as mutual protection, as they are both let-7 targets (Busch et al. 2016). Depletion of both IMP1 and LIN28B in these cells decreases their clonogenicity, migration, and tumor-initiating capacity (Busch et al. 2016). Similar to its action in embryonic cells, the mechanism by which IMP1 prevents let-7 target gene silencing in cancer cells appears to be by sequestration of its bound transcripts into mRNP granules devoid of RISC/Ago (Fig. 2). IMP1 may therefore promote CSC maintenance at least in part by preventing degradation of stemness and oncogene transcripts, including LIN28B and HMGA2.
IMP3 may function in a way similar to that of IMP1, sequestering LIN28 and HMGA2 mRNA into cytoplasmic granules that do not contain RISC/Ago (Fig. 2; Jonson et al. 2014). However, IMP3 may further promote CSC development and maintenance by destabilizing mRNAs that encode proteins implicated in differentiation and/or tumor suppression based on the discovery that IMP3 can recruit AGO/RISC to some of its mRNA targets (Ennajdaoui et al. 2016).
IMP2 has been less extensively studied in cancer, one notable exception being GBM, where our group assessed IMP2 mRNA binding by RIP-ChIP (Janiszewska et al. 2012) and, more recently, PAR-CLIP (Degrauwe et al. 2016). IMP2 expression was shown to be more elevated in GSCs than in the tumor bulk, suggesting a possible functional implication in GSC emergence and/or maintenance. In GSCs, the expression of both let-7 family members and their canonical target transcripts, including IMP2 itself, was observed in the absence of LIN28 paralogs, suggesting that IMP2 expression alone may suffice to protect let-7 target genes and maintain stemness. Consistent with this notion, analysis of IMP2 binding to mRNA revealed that IMP2 preferentially localizes to 3′ UTRs and binds to a subset of MREs, including let-7 target sites (Fig. 2), resulting in protection of the corresponding transcripts from RISC/AGO-mediated silencing (Degrauwe et al. 2016).
As expected, LIN28B and IMP2 were observed to engage in the same functional pathway in GSCs. Thus, loss of clonogenicity and tumor-initiating capacity by GSCs depleted of IMP2 was rescued by enforced LIN28B expression, which led to a robust decrease in mature let-7 levels and restoration of let-7 target gene expression. Depletion of IMP2 alone, which does not affect let-7 biogenesis, did not significantly alter mature let-7 expression.
Cooperation between LIN28 and dIMP has recently been highlighted in neuroblast (NB)-derived tumors in Drosophila melanogaster (Narbonne-Reveau et al. 2016). Together with the transcription factor Chinmo, dIMP and LIN28 form the core of an early oncogenic module that controls Drosophila NB (dNB) mitotic activity and that can be co-opted by susceptible cells to initiate malignant growth during early larval development. Whereas Chinmo behaves as an oncogene whose overexpression in dNBs is sufficient to cause dNB amplification and initiate tumor growth, dIMP appears to be required to sustain tumor growth beyond developmental stages at least in part by maintaining Chinmo expression. LIN28, which is positively regulated by Chinmo in the tumors, appears to be dispensable for tumor growth but may promote self-renewal of Chinmo+/dIMP+ NBs and thereby increase tumor growth potential. Coexpression of Chinmo, dIMP, and LIN28 occurs in a small subset of dNBs, which sustain unlimited tumor growth. Whether this subpopulation of cells represents CSCs or a transient amplifying population of progenitors derived from a distinct set of CSCs remains to be determined. The model, however, may be relevant to highly aggressive pediatric neural tumors that are often initiated during early development (Narbonne-Reveau et al. 2016).
Implication of IMP family members in functions relevant to cancer other than stem cell maintenance
Several IMP functions that are not directly related to maintenance of self-renewal and pluripotency may contribute to IMP promotion of tumor growth and progression. Some of these may be related to interference with miRNAs other than let-7 family members, whereas others may be unrelated to miRNA activity. The transport of cytoskeletal and secretory protein mRNAs as well as stabilization of cell adhesion receptor transcripts, including CD44 and ITGB5, may be relevant to tumor progression. IMP1 and IMP3 have been shown to enhance formation of invadopodia (Vainer et al. 2008), and enforced expression of IMP1 promotes invasiveness of tumor cells in vitro (for review, see Fig. 3; Bell et al. 2013). Consistent with these observations, expression of IMPs was reported in metastasizing CRCs and was observed to be particularly high at the invasive edge (Vainer et al. 2008). However, in another in vitro study, depletion of IMP1 in the breast cancer cell lines T47D and MDA-MB231 enhanced migration, whereas enforced expression of ZBP1 had the opposite effect (Gu et al. 2008). In addition to β-actin mRNA, IMP1 was observed to bind and transport mRNAs encoding E-cadherin, α-actinin, and the Arp2/3 complex, which are implicated in the stabilization of cell–cell contacts and focal adhesions. Based on these findings, it would appear that IMP1 may help maintain cell adhesion and polarity and possibly diminish or even abolish chemotactic responsiveness (Lapidus et al. 2007). These seemingly contradictory observations may reflect the breadth of IMP1 regulation of mRNA stability, with the capability of affecting mRNAs that encode proteins implicated in opposing functions, including cell polarization, lamellipodia formation, and migration (Lapidus et al. 2007; Vainer et al. 2008). Cell context-specific events, such as the presence of other RBPs and miRNA expression repertoires that modulate IMP1 activity, may help determine whether IMP1 preferentially promotes polarity or migration. More recently, the loss of IMP3 was reported to reduce PDAC invasiveness (Ennajdaoui et al. 2016). Numerous IMP3 targets related to invasiveness and implicated in focal adhesions, adherens junctions, actin cytoskeleton, and cell migration were identified by iCLIP in PDAC cells. Which of these are stabilized by protection against miRNA-mediated decay or destabilized as a result of AGO/RISC recruitment remains to be determined (Ennajdaoui et al. 2016).
An important IMP function that may be implicated in cancer development is the regulation of cell metabolism. This appears to be a function preferentially associated with IMP2, although it does not exclude participation of other IMPs whose relationship to metabolism has not been explored to the same depth. IMP2 was observed to bind numerous nuclear-encoded mRNAs associated with mitochondrial function (Vainer et al. 2008; Janiszewska et al. 2012), and it is possible that IMP2 helps localize the transcripts to mitochondria in a fashion analogous to IMP1- and IMP3-mediated transport of cytoskeletal and adhesion protein transcripts to the leading edge of motile cells. Consistent with this notion, IMP2 was found to bind mitochondria and regulate respiratory complex formation, favoring oxidative phosphorylation (OXPHOS) (Janiszewska et al. 2012). Regulation of OXPHOS by IMP2 may provide an important contribution to GSC maintenance (Fig. 3), as these cells appear to rely predominantly on OXPHOS for their energy requirements. It will be important to determine whether OXPHOS is the mechanism of choice for energy provision by CSCs of other tumor types.
An interesting observation is that the effect of IMPs on tumor progression may differ according to whether they are expressed in tumor cells or the tumor stroma. Thus, loss of IMP1 expression in the stroma has been reported recently to promote tumor growth in the azoxymethan (AOM)/dextran sodium sulphate (DSS) model of colitis-associated cancer (Hamilton et al. 2015). IMP1-floxed mice were crossed with DermoCre animals, resulting in IMP1 deletion in mesoderm-derived tissues, including fibroblasts, macrophages, and endothelial cells as of E9.5. Loss of IMP1 in stromal cells did not appear to affect intestinal homeostasis or the constitution of the intestinal stroma itself. However, mice lacking IMP1 in their stromal cells developed a greater number of as well as larger tumors in response to AOM/DSS than their wild-type counterparts (Hamilton et al. 2015). The inflammation provoked by AOM/DSS was more severe in mice lacking IMP1 in their stromal cells, and inflammatory cell infiltrates in the tumors were denser with stronger evidence of a wound healing-type response. Candidate proposed factors that may underlie the increased inflammation and tissue remodeling include increased secretion of several proinflammatory cytokines as well as Gremlin 1 (GREM1) and hepatocyte growth factor (HGF). HGF has been implicated in CRC pathogenesis (Rasola et al. 2007) and is an important paracrine signaling component in diverse cancer types. Pro-HGF and active HGF were observed to be increased in fibroblasts upon IMP1 depletion, but the transcript level was unaffected, suggesting regulation at the post-transcriptional/translational level (Hamilton et al. 2015). The mechanisms by which GREM1 and HGF are induced in response to inflammatory stimuli in the context of IMP1 depletion remain to be elucidated, but the observations suggest that expression of IMP1 in the tumor microenvironment may play a restrictive role in tumor growth.
IMP and LIN28 paralogs share multiple functions
LIN28 and IMP paralogs display intriguingly overlapping functions at the pathophysiological level, albeit by distinct molecular mechanisms that rely on different effector complexes. A case in point is protection of let-7 target genes, which appears to be a central function of both LIN28 and IMP. Whereas IMPs sequester let-7 target transcripts in RISC/Ago-free mRNP granules or disrupt let-7 binding to their recognition sites, LIN28 paralogs selectively inhibit let-7 biogenesis by binding to the terminal loop or pre-element (preE) of pre-let-7 and pri-let-7 and recruiting Zcchc11, a terminal uridylyl transferase (TUTase; also known as TUT4). Zcchc11/TUT4 catalyzes the addition of an oligo-uridine 3′ tail, which is a signal for pre-let-7 degradation (Thornton and Gregory 2012). By protecting let-7 target genes, both RBPs contribute directly to CSC establishment and maintenance. Both sets of paralogs are also let-7 targets and share many common target mRNAs. Moreover, they display several additional functional similarities that may be relevant to CSC biology, some but probably not all of which are a consequence of let-7 blockade.
LIN28A/B and IMPs display a temporally and spatially similar expression pattern during normal development (Christiansen et al. 2009; Balzer et al. 2010) and share the ability to enhance translation of IGF-2 (Polesskaya et al. 2007; Dai et al. 2011; Yang et al. 2015). They can block glial differentiation (Balzer et al. 2010; Fujii et al. 2013) and are implicated in the control of cellular bioenergetics (Janiszewska et al. 2012; Dai et al. 2015), and LIN28 is involved in the regulation of glucose metabolism (Zhu et al. 2011) and in coordinating growth through let-7-dependent regulation of numerous metabolic genes (Zhu et al. 2011; Shyh-Chang and Daley 2013). LIN28 can also enhance OXPHOS during wound healing through let-7-independent mechanisms, and inhibition of OXPHOS reduces its ability to promote tissue repair (Shyh-Chang et al. 2013). These functions appear to be analogous to those of IMP2-dependent promotion of OXPHOS in GSCs, although the mechanisms and effector complexes used may be distinct, as IMP2 regulation of respiratory complex function requires physical interaction with mitochondria (Janiszewska et al. 2012). Although the mechanisms by which IMP2 and LIN28 regulate cellular bioenergetics require further elucidation, this particular property shared by the two sets of paralogs may provide an important contribution to tumor heterogeneity.
Are IMPs a part of a targetable CSC module?
There is compelling evidence to suggest that by regulating let-7 biogenesis and mRNA targeting, respectively, the LIN28 paralogs and the IMP family of RBPs provide key players in determining the balance between pluripotency maintenance and differentiation in cancer cell subpopulations. LIN28A and LIN28B function upstream of IMPs to regulate let-7 biogenesis, and, in their presence, mature let-7 expression remains low, allowing cells to maintain plasticity. In the absence of LIN28, IMPs may provide the primary protection of let-7 target genes from silencing by either sequestration into RISC/Ago-free compartments (IMP1 and IMP3) or binding let-7 miRNA-binding sites in 3′ UTRs (IMP2), as observed in GSCs. Expression of both LIN28 and IMPs may therefore appear redundant unless IMPs provide a necessary fail-safe mechanism for let-7 target gene preservation or complementary functions required for CSC function. One possibility is that, in the presence of LIN28, IMPs may have a limited effect on let-7 target gene maintenance but may tilt cellular bioenergetics toward OXPHOS, which may be the preferred mode of ATP generation in CSCs of at least some cancer types and preserve relevant target genes of other miRNAs to generate a proinvasive phenotype and contribute to survival.
Implication of IMPs in CSC generation/maintenance does not appear to be confined to any single tumor but may be a general requirement for CSC maintenance irrespective of tumor type. This notion is supported by experiments using TNBC, CRC, and HCC cell lines, which have shown that IMP1/IMP3 play a role in promoting a stem cell phenotype that includes resistance to cytotoxic drugs and increased metastatic proclivity. In ovarian cancer, IMP1 appears to play the major role in promoting tumorigenesis along with LIN28B, whereas, in GBM, IMP2, possibly with some participation of IMP3, ensures GSC maintenance in the absence of LIN28 paralogs. Based on the notion that expression of LIN28A/B and IMPs in cancer cells recapitulates developmental events, the expression levels and patterns of LIN28 and IMP paralogs may reflect those of the corresponding embryonic cells and provide clues as to the cell of origin of at least some tumors. Careful assessment of expression of all LIN28 and IMP paralogs in tumor cell subpopulations, which is currently largely lacking, is therefore warranted. From a functional standpoint, it will be important to determine the relative contribution of the different IMP paralogs to CSC establishment and maintenance in different cancer types and whether the presence of more than one paralog serves merely as a backup or whether each paralog fulfills distinct functions, possibly by targeting or protecting different mRNAs from degradation by distinct sets of miRNAs. Furthermore, different IMP paralogs may be subject to distinct influences by other RBPs expressed in a cell context-specific fashion, resulting in enhanced, suppressed, or modified functions. Similarly, their functional relationship to LIN28 paralogs remains to be clarified. In other words, do IMPs and LIN28A/B have similar or distinct effects on self-renewal versus tumor-initiating capacity? In the presence of LIN28 and a low let-7 expression level, which IMP functions other than let-7 target gene protection contribute to the fine-tuning of the CSC phenotype? How might they synergize with analogous LIN28 functions to promote CSC/TIC properties?
Viewed from a different perspective, a central and perhaps essential event in CSC maintenance in diverse cancer types appears to be keeping let-7 family member expression and function in check, as is the case in ESC/progenitor cell maintenance during normal development. The fact that control of let-7 expression relies on at least three distinct mechanisms—biogenesis blockade by the LIN28 paralogs, protection of target mRNAs by IMP family members, and sequestration of mature let-7 forms by H19 and possibly other transcripts, which remove let-7 by providing a sponge effect (Kallen et al. 2013)—testifies to the importance of let-7 in regulating the switch between stemness and differentiation. Reactivation of such a central developmental switch underscores the similarity between cellular hierarchies in normally developing tissues and cancer and suggests that IMP family members along with LIN28 paralogs may provide core components of a CSC determination and maintenance module. If so, they or their regulatory circuits may become prime therapeutic targets whose neutralization may be effective in blunting the driving force of a broad range of malignancies, particularly solid tumors whose treatment remains a major challenge.
Acknowledgments
Work on IMPs was supported by the Swiss National Foundation (FNS) grant 310030_150024 and a Swiss Institute for Experimental Cancer Research (ISREC) grant.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.287540.116.
References
- Adolph SK, DeLotto R, Nielsen FC, Christiansen J. 2009. Embryonic expression of Drosophila IMP in the developing CNS and PNS. Gene Expr Patterns 9: 138–143. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. 2006. RNA granules. J Cell Biol 172: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, Ivanov P. 2015. Stress granules, P-bodies and cancer. Biochim Biophys Acta 1849: 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anko ML, Neugebauer KM. 2012. RNA–protein interactions in vivo: global gets specific. Trends Biochem Sci 37: 255–262. [DOI] [PubMed] [Google Scholar]
- Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. 2012. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA 3: 159–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas R, Behar L, Elliott E, Ginzburg I. 2004. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J Neurochem 89: 613–626. [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. [DOI] [PubMed] [Google Scholar]
- Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. 2010. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development 137: 891–900. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Oleynikov Y, Singer RH. 1999. The travels of mRNAs through all cells large and small. FASEB J 13: 447–454. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2011. A decade of exploring the cancer epigenome: biological and translational implications. Nat Rev Cancer 11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JL, Wachter K, Muhleck B, Pazaitis N, Kohn M, Lederer M, Huttelmaier S. 2013. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci 70: 2657–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein PL, Herrick DJ, Prokipcak RD, Ross J. 1992. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev 6: 642–654. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124. [DOI] [PubMed] [Google Scholar]
- Boudoukha S, Cuvellier S, Polesskaya A. 2010. Role of the RNA-binding protein IMP-2 in muscle cell motility. Mol Cell Biol 30: 5710–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan KL, Mische S, Li M, Marques G, Morin X, Chia W, Hays TS. 2008. Motility screen identifies Drosophila IGF-II mRNA-binding protein–zipcode-binding protein acting in oogenesis and synaptogenesis. PLoS Genet 4: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch B, Bley N, Muller S, Glass M, Misiak D, Lederer M, Vetter M, Strauss HG, Thomssen C, Huttelmaier S. 2016. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Res 44: 3845–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmel-Gross I, Bollag N, Armon L, Urbach A. 2016. LIN28: a stem cell factor with a key role in pediatric tumor formation. Stem Cells Dev 25: 367–377. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. [DOI] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. 2010. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev 24: 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Tsukamoto H, Liu JC, Kashiwabara C, Feldman D, Sher L, Dooley S, French SW, Mishra L, Petrovic L, et al. 2013a. Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J Clin Invest 123: 2832–2849. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen YL, Jeng YM, Hsu HC, Lai HS, Lee PH, Lai PL, Yuan RH. 2013b. Expression of insulin-like growth factor II mRNA-binding protein 3 predicts early recurrence and poor prognosis in intrahepatic cholangiocarcinoma. Int J Surg 11: 85–91. [DOI] [PubMed] [Google Scholar]
- Christiansen J, Kolte AM, Hansen T, Nielsen FC. 2009. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J Mol Endocrinol 43: 187–195. [DOI] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S. 2013. microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol 10: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I, Van de Ven WJ. 2008. The HMGA proteins: a myriad of functions (Review). Int J Oncol 32: 289–305. [PubMed] [Google Scholar]
- Cleynen I, Brants JR, Peeters K, Deckers R, Debiec-Rychter M, Sciot R, Van de Ven WJ, Petit MM. 2007. HMGA2 regulates transcription of the Imp2 gene via an intronic regulatory element in cooperation with nuclear factor-κB. Mol Cancer Res 5: 363–372. [DOI] [PubMed] [Google Scholar]
- Conway AE, Van Nostrand EL, Pratt GA, Aigner S, Wilbert ML, Sundararaman B, Freese P, Lambert NJ, Sathe S, Liang TY, et al. 2016. Enhanced CLIP uncovers IMP protein-RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Rep 15: 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai N, Rapley J, Angel M, Yanik MF, Blower MD, Avruch J. 2011. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes Dev 25: 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai N, Zhao L, Wrighting D, Kramer D, Majithia A, Wang Y, Cracan V, Borges-Rivera D, Mootha VK, Nahrendorf M, et al. 2015. IGF2BP2/IMP2-Deficient mice resist obesity through enhanced translation of Ucp1 mRNA and Other mRNAs encoding mitochondrial proteins. Cell Metab 21: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL. 2010. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 24: 2343–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. 2012. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4: a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrauwe N, Schlumpf TB, Janiszewska M, Martin P, Cauderay A, Provero P, Riggi N, Suva ML, Paro R, Stamenkovic I. 2016. The RNA binding protein IMP2 preserves glioblastoma stem cells by preventing let-7 target gene silencing. Cell Rep 15: 1634–1647. [DOI] [PubMed] [Google Scholar]
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. 2007. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336. [DOI] [PubMed] [Google Scholar]
- Doyle GA, Betz NA, Leeds PF, Fleisig AJ, Prokipcak RD, Ross J. 1998. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res 26: 5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. 2009. CRD-BP protects the coding region of βTrCP1 mRNA from miR-183-mediated degradation. Mol Cell 35: 240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DJ, Rajan P. 2010. The role of the RNA-binding protein Sam68 in mammary tumourigenesis. J Pathol 222: 223–226. [DOI] [PubMed] [Google Scholar]
- Ennajdaoui H, Howard JM, Sterne-Weiler T, Jahanbani F, Coyne DJ, Uren PJ, Dargyte M, Katzman S, Draper JM, Wallace A, et al. 2016. IGF2BP3 modulates the interaction of invasion-associated transcripts with RISC. Cell Rep 15: 1876–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Kishi Y, Gotoh Y. 2013. IMP2 regulates differentiation potentials of mouse neocortical neural precursor cells. Genes Cells 18: 79–89. [DOI] [PubMed] [Google Scholar]
- Fusco A, Fedele M. 2007. Roles of HMGA proteins in cancer. Nat Rev Cancer 7: 899–910. [DOI] [PubMed] [Google Scholar]
- Git A, Allison R, Perdiguero E, Nebreda AR, Houliston E, Standart N. 2009. Vg1RBP phosphorylation by Erk2 MAP kinase correlates with the cortical release of Vg1 mRNA during meiotic maturation of Xenopus oocytes. RNA 15: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorian V, Maillot G, Poles S, Iacovoni JS, Favre G, Vagner S. 2011. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ 18: 1692–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, Glass DJ. 2015. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell 34: 181–191. [DOI] [PubMed] [Google Scholar]
- Goswami S, Tarapore RS, Teslaa JJ, Grinblat Y, Setaluri V, Spiegelman VS. 2010. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein. J Biol Chem 285: 20532–20540. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Goswami S, Tarapore RS, Poenitzsch Strong AM, TeSlaa JJ, Grinblat Y, Setaluri V, Spiegelman VS. 2015. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor (MITF) mRNA is inhibited by coding region determinant-binding protein (CRD-BP). J Biol Chem 290: 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gress TM, Muller-Pillasch F, Geng M, Zimmerhackl F, Zehetner G, Friess H, Buchler M, Adler G, Lehrach H. 1996. A pancreatic cancer-specific expression profile. Oncogene 13: 1819–1830. [PubMed] [Google Scholar]
- Gu W, Wells AL, Pan F, Singer RH. 2008. Feedback regulation between zipcode binding protein 1 and β-catenin mRNAs in breast cancer cells. Mol Cell Biol 28: 4963–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Pazaitis N, Bley N, Fiskin E, Uckelmann H, Heim A, Grobeta M, Hofmann N, Geffers R, et al. 2014. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology 59: 1900–1911. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton KE, Noubissi FK, Katti PS, Hahn CM, Davey SR, Lundsmith ET, Klein-Szanto AJ, Rhim AD, Spiegelman VS, Rustgi AK. 2013. IMP1 promotes tumor growth, dissemination and a tumor-initiating cell phenotype in colorectal cancer cell xenografts. Carcinogenesis 34: 2647–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton KE, Chatterji P, Lundsmith ET, Andres SF, Giroux V, Hicks PD, Noubissi FK, Spiegelman VS, Rustgi AK. 2015. Loss of stromal IMP1 promotes a tumorigenic microenvironment in the colon. Mol Cancer Res 13: 1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T, Breuhahn K, et al. 2013. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology 58: 1703–1712. [DOI] [PubMed] [Google Scholar]
- Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC. 2004. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol 24: 4448–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708. [DOI] [PubMed] [Google Scholar]
- Ho JJ, Marsden PA. 2014. Competition and collaboration between RNA-binding proteins and microRNAs. Wiley Interdiscip Rev RNA 5: 69–86. [DOI] [PubMed] [Google Scholar]
- Hu S, Wu X, Zhou B, Xu Z, Qin J, Lu H, Lv L, Gao Y, Deng L, Yin J, et al. 2014. IMP3 combined with CD44s, a novel predictor for prognosis of patients with hepatocellular carcinoma. J Cancer Res Clin Oncol 140: 883–893. [DOI] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. 2005. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. 2009. An epigenetic switch involving NF-κB, Lin28, Let-7 microRNA, and IL6 links inflammation to cell transformation. Cell 139: 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Srivastava D. 2010. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 7: 36–41. [DOI] [PubMed] [Google Scholar]
- Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, Radovanovic I, Rheinbay E, Provero P, Stamenkovic I. 2012. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev 26: 1926–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng YM, Chang CC, Hu FC, Chou HY, Kao HL, Wang TH, Hsu HC. 2008. RNA-binding protein insulin-like growth factor II mRNA-binding protein 3 expression promotes tumor invasion and predicts early recurrence and poor prognosis in hepatocellular carcinoma. Hepatology 48: 1118–1127. [DOI] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120: 623–634. [DOI] [PubMed] [Google Scholar]
- Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC. 2007. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics 6: 798–811. [DOI] [PubMed] [Google Scholar]
- Jonson L, Christiansen J, Hansen TV, Vikesa J, Yamamoto Y, Nielsen FC. 2014. IMP3 RNP safe houses prevent miRNA-directed HMGA2 mRNA decay in cancer and development. Cell Rep 7: 539–551. [DOI] [PubMed] [Google Scholar]
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, et al. 2013. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 52: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. 2007. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131: 1273–1286. [DOI] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. 2010. A Pumilio-induced RNA structure switch in p27–3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12: 1014–1020. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. 2009. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 23: 1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Kwak H, Lee HK, Hyun S, Jeong S. 2012. β-Catenin recognizes a specific RNA motif in the cyclooxygenase-2 mRNA 3′-UTR and interacts with HuR in colon cancer cells. Nucleic Acids Res 40: 6863–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, Dick JE. 2014. Evolution of the cancer stem cell model. Cell Stem Cell 14: 275–291. [DOI] [PubMed] [Google Scholar]
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. 2012. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res 40: 5088–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus K, Wyckoff J, Mouneimne G, Lorenz M, Soon L, Condeelis JS, Singer RH. 2007. ZBP1 enhances cell polarity and reduces chemotaxis. J Cell Sci 120: 3173–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer M, Bley N, Schleifer C, Huttelmaier S. 2014. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol 29: 3–12. [DOI] [PubMed] [Google Scholar]
- Lee HK, Kwak HY, Hur J, Kim IA, Yang JS, Park MW, Yu J, Jeong S. 2007. β-Catenin regulates multiple steps of RNA metabolism as revealed by the RNA aptamer in colon cancer cells. Cancer Res 67: 9315–9321. [DOI] [PubMed] [Google Scholar]
- Levy M, Lin F, Xu H, Dhall D, Spaulding BO, Wang HL. 2010. S100P, von Hippel-Lindau gene product, and IMP3 serve as a useful immunohistochemical panel in the diagnosis of adenocarcinoma on endoscopic bile duct biopsy. Hum Pathol 41: 1210–1219. [DOI] [PubMed] [Google Scholar]
- Lin S, Gregory RI. 2015. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15: 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo NA, Shimono Y, Qian D, Clarke MF. 2007. The biology of cancer stem cells. Annu Rev Cell Dev Biol 23: 675–699. [DOI] [PubMed] [Google Scholar]
- Lui J, Castelli LM, Pizzinga M, Simpson CE, Hoyle NP, Bailey KL, Campbell SG, Ashe MP. 2014. Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep 9: 944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. 2013. Tumour heterogeneity and cancer cell plasticity. Nature 501: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo M, Ricklefs F, Rooj AK, Lyons SM, Ivanov P, Ansari KI, Nakano I, Chiocca EA, Godlewski J, Bronisz A. 2016. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep 15: 2500–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Parker R. 2014. Principles and properties of eukaryotic mRNPs. Mol Cell 54: 547–558. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700. [DOI] [PubMed] [Google Scholar]
- Mori H, Sakakibara S, Imai T, Nakamura Y, Iijima T, Suzuki A, Yuasa Y, Takeda M, Okano H. 2001. Expression of mouse igf2 mRNA-binding protein 3 and its implications for the developing central nervous system. J Neurosci Res 64: 132–143. [DOI] [PubMed] [Google Scholar]
- Moss EG, Tang L. 2003. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol 258: 432–442. [DOI] [PubMed] [Google Scholar]
- Mueller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, Varga G, Friess H, Buchler M, Beger HG, et al. 1997. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene 14: 2729–2733. [DOI] [PubMed] [Google Scholar]
- Mueller-Pillasch F, Pohl B, Wilda M, Lacher U, Beil M, Wallrapp C, Hameister H, Knochel W, Adler G, Gress TM. 1999. Expression of the highly conserved RNA binding protein KOC in embryogenesis. Mech Dev 88: 95–99. [DOI] [PubMed] [Google Scholar]
- Muller-McNicoll M, Neugebauer KM. 2013. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 14: 275–287. [DOI] [PubMed] [Google Scholar]
- Narbonne-Reveau K, Lanet E, Dillard C, Foppolo S, Chen CH, Parrinello H, Rialle S, Sokol NS, Maurange C. 2016. Neural stem cell-encoded temporal patterning delineates an early window of malignant susceptibility in Drosophila. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LH, Robinton DA, Seligson MT, Wu L, Li L, Rakheja D, Comerford SA, Ramezani S, Sun X, Parikh MS, et al. 2014. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 26: 248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 19: 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Cilius Nielsen F, Kragh Jakobsen R, Christiansen J. 2000. The biphasic expression of IMP/Vg1-RBP is conserved between vertebrates and Drosophila. Mech Dev 96: 129–132. [DOI] [PubMed] [Google Scholar]
- Nielsen FC, Nielsen J, Christiansen J. 2001. A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand J Clin Lab Invest Suppl 234: 93–99. [PubMed] [Google Scholar]
- Nielsen FC, Nielsen J, Kristensen MA, Koch G, Christiansen J. 2002. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J Cell Sci 115: 2087–2097. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Kristensen MA, Willemoes M, Nielsen FC, Christiansen J. 2004. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Res 32: 4368–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ. 2008. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 135: 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J, Kim S, Zhu Y, Zhu H, Morrison SJ. 2013. A network of heterochronic genes including Imp1 regulates temporal changes in stem cell properties. Elife 2: e00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubissi FK, Elcheva I, Bhatia N, Shakoori A, Ougolkov A, Liu J, Minamoto T, Ross J, Fuchs SY, Spiegelman VS. 2006. CRD-BP mediates stabilization of βTrCP1 and c-myc mRNA in response to β-catenin signalling. Nature 441: 898–901. [DOI] [PubMed] [Google Scholar]
- Noubissi FK, Nikiforov MA, Colburn N, Spiegelman VS. 2010. Transcriptional regulation of CRD-BP by c-myc: implications for c-myc functions. Genes Cancer 1: 1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. 1976. The clonal evolution of tumor cell populations. Science 194: 23–28. [DOI] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. 2003. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol 13: 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanichamy JK, Tran TM, Howard JM, Contreras JR, Fernando TR, Sterne-Weiler T, Katzman S, Toloue M, Yan W, Basso G, et al. 2016. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Invest 126: 1495–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Huttelmaier S, Singer RH, Gu W. 2007. ZBP2 facilitates binding of ZBP1 to β-actin mRNA during transcription. Mol Cell Biol 27: 8340–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89. [DOI] [PubMed] [Google Scholar]
- Pfannkuche K, Summer H, Li O, Hescheler J, Droge P. 2009. The high mobility group protein HMGA2: a co-regulator of chromatin structure and pluripotency in stem cells? Stem Cell Rev 5: 224–230. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. 2007. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev 21: 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokipcak RD, Herrick DJ, Ross J. 1994. Purification and properties of a protein that binds to the C-terminal coding region of human c-myc mRNA. J Biol Chem 269: 9261–9269. [PubMed] [Google Scholar]
- Rasola A, Fassetta M, De Bacco F, D'Alessandro L, Gramaglia D, Di Renzo MF, Comoglio PM. 2007. A positive feedback loop between hepatocyte growth factor receptor and β-catenin sustains colorectal cancer cell invasive growth. Oncogene 26: 1078–1087. [DOI] [PubMed] [Google Scholar]
- Riggi N, Cironi L, Suva ML, Stamenkovic I. 2007. Sarcomas: genetics, signalling, and cellular origins. Part 1: the fellowship of TET. J Pathol 213: 4–20. [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge S, Nielsen FC, Nielsen J, Lykke-Andersen J, Wewer UM, Christiansen J. 2000. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J Biol Chem 275: 29562–29569. [DOI] [PubMed] [Google Scholar]
- Samanta S, Sharma VM, Khan A, Mercurio AM. 2012. Regulation of IMP3 by EGFR signaling and repression by ERβ: implications for triple-negative breast cancer. Oncogene 31: 4689–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S, Pursell B, Mercurio AM. 2013. IMP3 protein promotes chemoresistance in breast cancer cells by regulating breast cancer resistance protein (ABCG2) expression. J Biol Chem 288: 12569–12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S, Sun H, Goel HL, Pursell B, Chang C, Khan A, Greiner DL, Cao S, Lim E, Shultz LD, et al. 2016. IMP3 promotes stem-like properties in triple-negative breast cancer by regulating SLUG. Oncogene 35: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE. 2012. Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A, Danial M, Blelloch RH. 2015. Let-7 and miR-125 cooperate to prime progenitors for astrogliogenesis. EMBO J 34: 1180–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Liu H, Wang HL, Prichard JW, Lin F. 2013. Diagnostic utility of von Hippel-Lindau gene product, maspin, IMP3, and S100P in adenocarcinoma of the gallbladder. Hum Pathol 44: 503–511. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ. 2013. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12: 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ. 2013. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 155: 778–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Tominaga K, Gorospe M. 2012. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci 13: 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier CR, Doyle GA, Clark BA, Pitot HC, Ross J. 2004. Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res 64: 209–214. [DOI] [PubMed] [Google Scholar]
- Thornton JE, Gregory RI. 2012. How does Lin28 let-7 control development and disease? Trends Cell Biol 22: 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. 2003. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and β-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci 23: 3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tybl E, Shi FD, Kessler SM, Tierling S, Walter J, Bohle RM, Wieland S, Zhang J, Tan EM, Kiemer AK. 2011. Overexpression of the IGF2-mRNA binding protein p62 in transgenic mice induces a steatotic phenotype. J Hepatol 54: 994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainer G, Vainer-Mosse E, Pikarsky A, Shenoy SM, Oberman F, Yeffet A, Singer RH, Pikarsky E, Yisraeli JK. 2008. A role for VICKZ proteins in the progression of colorectal carcinomas: regulating lamellipodia formation. J Pathol 215: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R. 2011. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11: 644–656. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ. 2010. Lin28: a microRNA regulator with a macro role. Cell 140: 445–449. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. 2009. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 41: 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter K, Kohn M, Stohr N, Huttelmaier S. 2013. Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Biol Chem 394: 1077–1090. [DOI] [PubMed] [Google Scholar]
- Weidensdorfer D, Stohr N, Baude A, Lederer M, Kohn M, Schierhorn A, Buchmeier S, Wahle E, Huttelmaier S. 2009. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15: 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al. 2009. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460: 909–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. 2013. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152: 82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Yang SL, Herrlinger S, Liang C, Dzieciatkowska M, Hansen KC, Desai R, Nagy A, Niswander L, Moss EG, et al. 2015. Lin28 promotes the proliferative capacity of neural progenitor cells in brain development. Development 142: 1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K, Yisraeli JK. 2002. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene 287: 49–54. [DOI] [PubMed] [Google Scholar]
- Yaniv K, Fainsod A, Kalcheim C, Yisraeli JK. 2003. The RNA-binding protein Vg1 RBP is required for cell migration during early neural development. Development 130: 5649–5661. [DOI] [PubMed] [Google Scholar]
- Yantiss RK, Woda BA, Fanger GR, Kalos M, Whalen GF, Tada H, Andersen DK, Rock KL, Dresser K. 2005. KOC (K homology domain containing protein overexpressed in cancer): a novel molecular marker that distinguishes between benign and malignant lesions of the pancreas. Am J Surg Pathol 29: 188–195. [DOI] [PubMed] [Google Scholar]
- Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA. 2012. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol Cancer Res 10: 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KR, Shin JH, Kim JJ, Koog MG, Lee JY, Choi SW, Kim HS, Seo Y, Lee S, Shin TH, et al. 2015. Rapid and efficient direct conversion of human adult somatic cells into neural stem cells by HMGA2/let-7b. Cell Rep 10: 441–452. [DOI] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, et al. 2007. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316: 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Chan EK, Peng XX, Tan EM. 1999a. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J Exp Med 189: 1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yaniv K, Oberman F, Wolke U, Git A, Fromer M, Taylor WL, Meyer D, Standart N, Raz E, et al. 1999b. Vg1 RBP intracellular distribution and evolutionarily conserved expression at multiple stages during development. Mech Dev 88: 101–106. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. 2001. Neurotrophin-induced transport of a β-actin mRNP complex increases β-actin levels and stimulates growth cone motility. Neuron 31: 261–275. [DOI] [PubMed] [Google Scholar]
- Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. 2011. The Lin28/let-7 axis regulates glucose metabolism. Cell 147: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]