Deluz et al. show that SOX2 and OCT4 remain bound to mitotic chromatin through their respective DNA-binding domains. The absence of SOX2 at the M–G1 transition impairs pluripotency maintenance and abrogates its ability to induce neuroectodermal differentiation but does not affect reprogramming efficiency toward induced pluripotent stem cells.
Keywords: embryonic stem cells, mitotic bookmarking, neuroectoderm, pluripotency
Abstract
Mitotic bookmarking transcription factors remain bound to chromosomes during mitosis and were proposed to regulate phenotypic maintenance of stem and progenitor cells at the mitosis-to-G1 (M–G1) transition. However, mitotic bookmarking remains largely unexplored in most stem cell types, and its functional relevance for cell fate decisions remains unclear. Here we screened for mitotic chromosome binding within the pluripotency network of embryonic stem (ES) cells and show that SOX2 and OCT4 remain bound to mitotic chromatin through their respective DNA-binding domains. Dynamic characterization using photobleaching-based methods and single-molecule imaging revealed quantitatively similar specific DNA interactions, but different nonspecific DNA interactions, of SOX2 and OCT4 with mitotic chromatin. Using ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) to assess the genome-wide distribution of SOX2 on mitotic chromatin, we demonstrate the bookmarking activity of SOX2 on a small set of genes. Finally, we investigated the function of SOX2 mitotic bookmarking in cell fate decisions and show that its absence at the M–G1 transition impairs pluripotency maintenance and abrogates its ability to induce neuroectodermal differentiation but does not affect reprogramming efficiency toward induced pluripotent stem cells. Our study demonstrates the mitotic bookmarking property of SOX2 and reveals its functional importance in pluripotency maintenance and ES cell differentiation.
During mitosis, transcription is globally shut down, and RNA polymerases and most DNA-binding proteins are stripped off the chromosomes (Prescott and Bender 1962; Spencer et al. 2000). However, some transcription factors do not abide by this rule and remain bound to specific genes on mitotic chromosomes. These so-called mitotic bookmarking transcription factors are involved in physiological processes such as phenotypic maintenance (Zaidi et al. 2010, 2014; Kadauke and Blobel 2013; Festuccia et al. 2016) and ribosome biogenesis (Chen et al. 2002; Grob et al. 2014; Lopez-Camacho et al. 2014) as well as pathological events such as oncogenic transformation (Blobel et al. 2009; Pockwinse et al. 2011; Zaidi et al. 2014). However, the contribution of mitotic chromosome retention to these functions remains unknown. Transcription factors with mitotic bookmarking properties are often master regulators of cell fate, and some of them were reported to mediate rapid transcriptional reactivation after mitotic exit (Blobel et al. 2009; Dey et al. 2009; Caravaca et al. 2013). This led to the suggestion that retention on mitotic chromosomes plays a role at the mitosis-to-G1 (M–G1) transition by ensuring proper restoration of the gene expression program and thereby cellular phenotype after cell division (Zaret 2014). This concept is particularly appealing in the context of self-renewing cells such as stem cells, which need to maintain their identity through a large number of cell divisions. However, to date, there is no direct experimental evidence substantiating this hypothesis.
Mammalian embryonic stem (ES) cells are maintained in a pluripotent state by a network of transcription factors (Dunn et al. 2014) in which SOX2 and OCT4 play a central role. Both are strictly required for the maintenance of the pluripotent state (Nichols et al. 1998; Avilion et al. 2003) and act mainly as a heterodimer that binds to a composite DNA motif to activate transcription of genes controlling pluripotency (Remenyi et al. 2003; Cole and Young 2008). Together with KLF4, they also allow reprogramming of terminally differentiated cells to pluripotency (Takahashi and Yamanaka 2006; Nakagawa et al. 2008). SOX2 also plays a role in differentiation by favoring neuroectodermal commitment upon pluripotency exit (Zhao et al. 2004; Thomson et al. 2011). Here we used live-cell imaging and biophysical techniques to demonstrate and characterize the retention of SOX2 and OCT4 on mitotic chromosomes of ES cells and performed ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) to show that SOX2 bookmarks specific genomic loci during mitosis. Using tools allowing us to specifically degrade SOX2 at the M–G1 transition and a new double knock-in reporter cell line to monitor neuroectodermal and mesendodermal differentiation, we dissect the role of SOX2 mitotic bookmarking in pluripotency maintenance, reprogramming, and differentiation.
Results
SOX2 and OCT4 have intrinsic mitotic chromosome-binding (MCB) properties that are independent of the pluripotency context
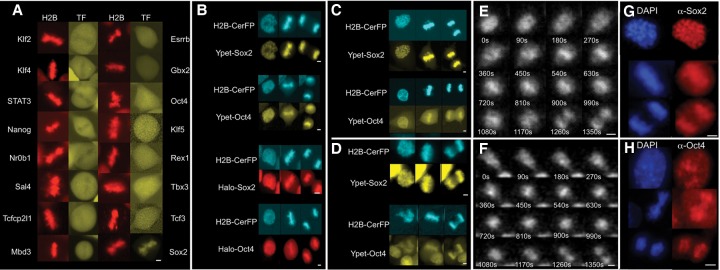
We first aimed to screen for pluripotency transcription factors with the ability to bind mitotic chromosomes. We engineered lentiviral constructs that allow doxycycline (dox)-inducible expression of 16 central pluripotency transcription factors (Dunn et al. 2014), each of which is C-terminally fused to a yellow fluorescent protein (YPet) (Supplemental Methods). We then generated the 16 corresponding mouse ES cell lines that also constitutively expressed H2B-mCherry to identify mitotic cells and a reverse tetracycline transactivator (rtTA3G) to allow for inducible transcription factor-YPet expression and performed live-cell fluorescence imaging 16–24 h after dox treatment. All transcription factors were exclusively or mostly localized in the nucleus, suggesting that the YPet tag did not affect their subcellular localization (Supplemental Fig. S1A). Upon examination of metaphase cells, most transcription factors were either excluded or not clearly enriched on mitotic chromosomes, with the exception of SOX2, which was markedly colocalized with mitotic chromosomes (Fig. 1A). To exclude a widespread effect of YPet in preventing mitotic chromosomal localization, for some of them, we also confirmed the absence of enrichment on mitotic chromosomes by immunofluorescence experiments (Supplemental Fig. S1B) and SNAP tag fusions (Supplemental Fig. S1C).
Figure 1.
SOX2 and OCT4 bind to chromosomes throughout mitosis. (A) Fluorescence snapshots of living ES cells in metaphase expressing H2B-mCherry and dox-inducible fusion proteins to pluripotency transcription factors. (B) Fluorescence snapshots of living E14 cells in prophase (left), metaphase (middle), and anaphase (right) expressing H2B-CerFP and different dox-inducible fusion proteins to SOX2 and OCT4. Cells expressing HaloTag fusions to SOX2 and OCT4 were labeled with HaloTag TMR ligand shortly before imaging. (C,D) Fluorescence snapshots of living NIH-3T3 cells (C) and HEK 293T cells (D) in prophase (left), metaphase (middle), and anaphase (right) expressing H2B-CerFP and YPet-SOX2 or YPet-OCT4. (E,F) Time series from luminescence time-lapse imaging of SOX2-luciferase (SOX2-Luc) (E) and Luc-OCT4 (F) knock-in ES cell lines. (G,H) Immunofluorescence against Sox2 (G) and Oct4 (H) of E14 cells in prophase (top), metaphase (middle), and anaphase (bottom). Bars, 5 µm.
Since OCT4 is a heterodimeric partner of SOX2 and appeared weakly enriched on mitotic chromosomes (Fig. 1A), we generated four stable cell lines using higher lentiviral titers to allow broader expression ranges of N-terminal fusions of YPet or HaloTag to SOX2 and OCT4 under the control of a dox-inducible promoter (TRE3G) as well as constitutive expression of H2B-cerulean (H2B-CerFP) and rtTA3G. We then performed two-color time-lapse fluorescence imaging, revealing colocalization of H2B-CerFP with YPet-tagged and Halo-tagged SOX2 and OCT4 throughout mitosis (Fig. 1B; Supplemental Fig. S2; Supplemental Movies S1, S2). We then investigated MCB of SOX2 and OCT4 in NIH-3T3 and 293T cells, which are not pluripotent and do not express Sox2 and Oct4 endogenously. To do so, we engineered them for inducible expression of YPet-Sox2 or YPet-Oct4 and constitutive expression of H2B-CerFP. We observed that both transcription factors maintained their binding to mitotic chromosomes in these cell lines (Fig. 1C,D), suggesting that this is an inherent property of SOX2 and OCT4 that is independent of the pluripotent context and that they can bind mitotic chromosomes independently of each other. To confirm MCB activity of endogenously expressed SOX2 and OCT4 in living cells, we used the CRISPR/Cas9 technology to generate two heterozygous knock-in ES cell lines in which firefly luciferase (Luc) is fused to the C terminus of SOX2 and the N terminus of OCT4, respectively (Supplemental Methods; Supplemental Fig. S3). This enabled us to observe MCB by luminescence microscopy, allowing highly sensitive measurements that can be used to monitor the expression of endogenous genes in single cells (Suter et al. 2011). Approximately 15 min before cell division, the luminescence signal relocalized to the midplane of the cell perpendicular to the cell division axis, consistent with a colocalization with metaphase chromosomes (Fig. 1E,F; Supplemental Movies S3, S4). Finally, we performed immunofluorescence staining of endogenous Oct4 and Sox2, which also yielded consistent results (Fig. 1G,H).
DNA-binding domain dependency and quantitative differences of SOX2 and OCT4 in MCB
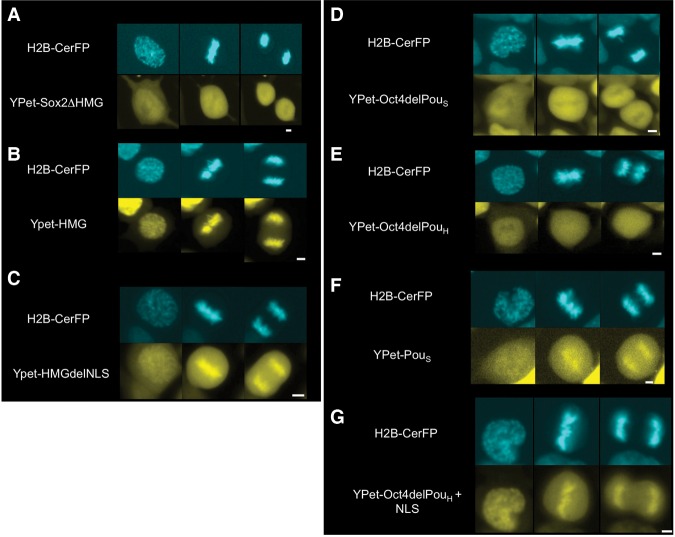
To investigate the role of the DNA-binding domains of SOX2 and OCT4 in MCB, we generated different mutants that were N-terminally fused to YPet and expressed them in ES cells using our dox-inducible system. A deletion mutant of Sox2 devoid of its HMG DNA-binding domain lost its MCB activity (Fig. 2A; Supplemental Fig. S2), whereas the HMG domain alone was sufficient for MCB (Fig. 2B). Interestingly, removal of amino acids 133–135 of the nuclear localization signal (NLS) within the HMG domain reduced its MCB (Fig. 2C). We generated two deletion mutants of Oct4, lacking either the POUS or the POUH DNA-binding domain. Deletion of the POUS domain abrogated MCB (Fig. 2D), in contrast to the POUH domain deletion mutant, which retained a low MCB activity (Fig. 2E; Supplemental Fig. S2). Furthermore, the POUS domain alone displayed MCB (Fig. 2F). This suggests that MCB of OCT4 is mediated mainly by the POUS domain. Interestingly, the substitution of the POUH domain with a NLS (RKRKR) rescued the MCB activity of OCT4 (Fig. 2G). Taken together, these results suggest that the HMG domain of SOX2 and the POUS domain of OCT4 are necessary and sufficient for their interaction with mitotic chromatin and that NLS sequences enhance their MCB.
Figure 2.
MCB of SOX2 and OCT4 is mediated by their HMG and POUS domains, respectively. Fluorescence snapshots of living ES cells expressing H2B-CerFP and dox-inducible YPet fusion to truncated versions of SOX2 and OCT4. (A) SOX2 with a deletion of its HMG DNA-binding domain. (B) HMG DNA-binding domain of SOX2. (C) HMG DNA-binding domain of SOX2 without its NLS. (D) OCT4 with a deletion of its POUS DNA-binding domain. (E) OCT4 with a deletion of its POUH DNA-binding domain. (F) POUS DNA-binding domain of OCT4. (G) OCT4 with a substitution of its POUS DNA-binding domain with a NLS. In each panel, prophase is at the left, metaphase is in the middle, and anaphase is at the right. Bars, 5 µm.
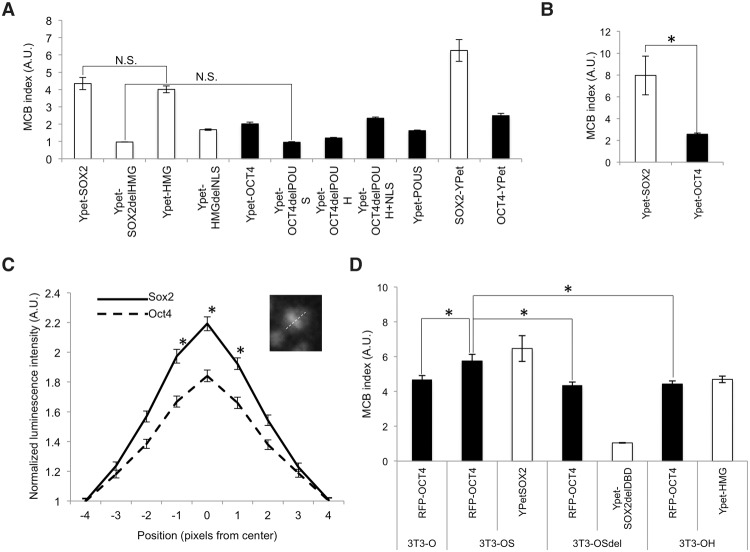
We next quantified the relative MCB strength of SOX2, OCT4, and their truncated versions by calculating the ratio of fluorescent signal on mitotic chromosomes to that in the cytoplasm in wide-field fluorescence microscopy images of cells in metaphase, a quantity that we define as the MCB index. We found a higher MCB index for SOX2 than for OCT4 in ES cells (Fig. 3A), suggesting a stronger MCB activity for SOX2. To verify that the N-terminal fusion to the fluorescent tag did not bias the MCB efficiency of SOX2 and OCT4, we also generated cell lines expressing C-terminal fusions to YPet, which yielded consistent, although slightly higher, MCB indexes for both SOX2 and OCT4 (Fig. 3A). We then asked whether these differences stem from intrinsic properties of SOX2 and OCT4 by measuring their MCB index in NIH-3T3 cells, which also yielded similar results (Fig. 3B). To confirm the differences in MCB of endogenously expressed SOX2 and OCT4, we performed quantitative measurements of their MCB in the Sox2-Luc and Luc-Oct4 knock-in cell lines. Because of the lower spatial resolution of luminescence microscopy and the inability to simultaneously visualize fluorescently tagged H2B, we quantified the MCB properties of SOX2 and OCT4 by measuring their luminescence intensity profile across mitotic cells at metaphase, normalized over the average luminescence level four pixels from the center and on both sides of mitotic chromosomes (Fig. 3C, inset). These measurements confirmed the weaker MCB activity of OCT4 as compared with SOX2 in the endogenous context (Fig. 3C). Next, we investigated the interdependence of SOX2 and OCT4 in the relative strength of their MCB activity, which might be expected, since they can heterodimerize on DNA (Remenyi et al. 2003), and SOX2 was reported to assist DNA binding of OCT4 (Chen et al. 2014). To compare their MCB indexes in the presence or absence of each other, we generated four NIH-3T3 cell lines carrying an inducible construct for the expression of OCT4 fused to TagRFP-T (RFP-OCT4). This cell line (3T3-O) was further transduced to allow inducible expression of YPet-SOX2 (3T3-OS), YPet-SOX2delHMG (3T3-OSdel), or YPet-HMG (3T3-OH). Since coexpression of RFP-OCT4 and YPet-SOX2 decreased cell proliferation and thus the number of cell divisions that we could observe, we performed these experiments at lower doses of dox (20 ng/mL) on 3T3-O, 3T3-OS, 3T3-OSdel, and 3T3-OH cell lines. The MCB index of YPet-SOX2 was decreased only marginally in the presence of RFP-OCT4 (Fig. 3, cf. values from D and B). We observed a similar MCB index of RFP-OCT4 in the 3T3-O, 3T3-OSdel, and 3T3-OH cell lines, but it was significantly increased in the 3T3-OS cell line (Fig. 3D). Thus, SOX2 enhances the mitotic chromatin interaction of OCT4, and the SOX2 HMG domain is required but not sufficient to increase the mitotic chromatin interaction of OCT4.
Figure 3.
Differential retention and interdependence of SOX2 and OCT4 for MCB. (A) MCB index of YPet fusions to wild type and mutants of SOX2 and OCT4 in ES cells. All samples were significantly different from each other (P < 0.05) unless specified. (N.S.) P > 0.05. n ≥ 20. (B) MCB index of YPet fusions to SOX2 and OCT4 in NIH-3T3 cells. n ≥ 20 (C) Quantification of luminescence signal across cells in metaphase for SOX2-Luc (solid line) and Luc-OCT4 (dashed line) knock-in cell lines. n ≥ 50. The inset illustrates how the measurements were performed. (D) MCB index in 3T3-O, 3T3-OS, 3T3OSdel, and 3T3OH cell lines upon dox induction with 20 ng/mL. n ≥ 50. (A.U.) Arbitrary units. (*) P < 0.05. Error bars indicate SEM.
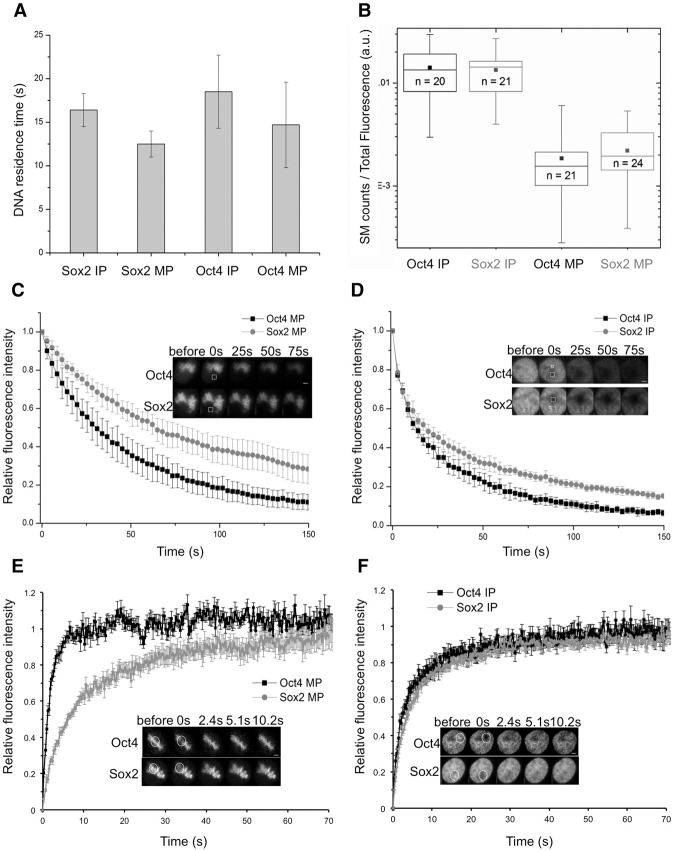
SOX2 and OCT4 display distinct mobility but similar frequencies and residence times of long-lived DNA-binding events on mitotic chromosomes
To determine the residence times of SOX2 and OCT4 on mitotic chromatin, we performed single-molecule live-cell imaging experiments in ES cell lines that allow dox-inducible expression of Halo-SOX2 and Halo-OCT4 that we labeled with the Halo-TMR dye. Cells were treated with 50 ng/mL dox, allowing low Halo-tagged transgene expression levels for accurate identification of single DNA-bound molecules (Gebhardt et al. 2013). We performed measurements on interphase and mitotic cells in the asynchronous population using highly inclined and laminated optical sheet (HILO) microscopy (Tokunaga et al. 2008). To determine residence times on DNA (1/koff), we used a previously described time-lapse imaging strategy (Gebhardt et al. 2013) using imaging parameters that allowed us to measure long-lived specific DNA-binding events. The residence times that we measured in interphase were in close agreement with values described earlier for specific binding of SOX2 and OCT4 to DNA (Chen et al. 2014) and were only slightly shorter on mitotic chromatin; moreover, residence times were similar for both transcription factors (Fig. 4A; Supplemental Fig. S4). We next investigated whether SOX2 and OCT4 have similar relative on rates of DNA binding. As kon = koff/kD and as SOX2 and OCT4 have similar residence times (1/koff), differences in kon should manifest as differences in kD (see the Materials and Methods). Since the equilibrium constant kD is proportional to the ratio of unbound over bound molecules, it scales inversely with the number of binding events divided by the total fluorescence of the cell in a given individual image. The values obtained for this ratio were similar for SOX2 and OCT4 in interphase and mitosis (Fig. 4B), suggesting no major difference in their on rates.
Figure 4.
Dynamics of OCT4 and SOX2 in mitotic and interphase cells. (A) SOX2 and OCT4 DNA residence times measured by time-lapse single-molecule microscopy. Error bars indicate the SE of linear fitting. (B) Relative bound fraction measured as single-molecule count per total cell fluorescence in a single frame (arbitrary units). Boxes represent intervals between the 25th and 75th percentile, and whiskers indicate minimum and maximum values. The number of analyzed cells is indicated inside the boxes. (C,D) Fluorescence loss in photobleaching (FLIP) curves (arbitrary units) for OCT4 and SOX2 in M phase (MP) (C) or interphase (IP) (D). n = 10. Error bars indicate SD. The insets show examples of FLIP time series. (Dashed square) Bleaching area; (solid square) fluorescence recording area. Bars, 2 µm. (E,F) Fluorescence recovery after photobleaching (FRAP) curves (arbitrary units) for OCT4 and SOX2 in M phase (MP) (E) or interphase (IP) (F). n = 10. Error bars indicate SE. The insets show examples of FRAP time series. (Solid circle) Bleaching and fluorescence recording area. Bars, 2 µm.
We next performed fluorescence loss in photobleaching (FLIP) and fluorescence recovery after photobleaching (FRAP), which mainly reflect interactions with nonspecific binding sites (Hager et al. 2009), to measure the mobility of OCT4 and SOX2 in interphase and mitotic cells. YPet-SOX2 and, to a lesser extent, YPet-OCT4 displayed a slower fluorescence loss in mitosis (t1/2 bleach SOX2 = 45.6 sec ± 8.8 sec; t1/2 bleach OCT4 = 29.0 sec ± 8.8 sec) (Fig. 4C) as compared with interphase (t1/2 bleach SOX2 = 13.1 sec ± 3.6 sec; t1/2 bleach OCT4 = 12.9 sec ± 2.2 sec) (Fig. 4D), suggesting that SOX2 and OCT4 are exchanged less between mitotic chromosomes and the mitotic cytoplasm than within the interphase nucleus. In principle, this could be due to a lower mobility of molecules bound to mitotic chromosomes and/or a global retention within the mitotic chromosome environment. FRAP experiments revealed a slower recovery of YPet-SOX2 on mitotic chromosomes (t1/2 recovery = 6.00 sec ± 1.99 sec) (Fig. 4E) as compared with interphase (t1/2 recovery = 3.81 sec ± 1.15 sec) (Fig. 4F), thus confirming the lower mobility of SOX2 in mitotic cells. However, YPet-OCT4 showed the opposite behavior, with a faster recovery on mitotic chromosomes (t1/2 recovery = 1.1 sec ± 0.27 sec) (Fig. 4E) as compared with interphase (t1/2 recovery = 2.83 sec ± 0.91 sec) (Fig. 4F), suggesting that the slower FLIP half-time of mitotic OCT4 is not due to a lower mobility of OCT4 but rather its retention within the mitotic chromosome environment. Since mitotic chromatin consists mainly of highly compacted DNA wrapped around nucleosomes, our results could be explained by the higher affinity of SOX2 for nonspecific sequences on nucleosomal DNA as compared with OCT4 (Soufi et al. 2015).
SOX2 is retained on a small number of genomic loci during mitosis
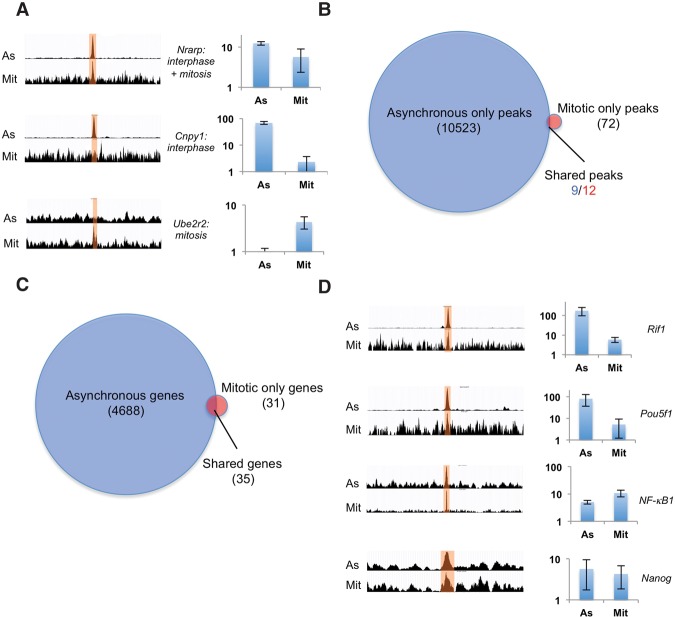
To investigate the genome-wide distribution of SOX2 during mitosis, we performed three independent ChIP-seq experiments on purified mitotic cells and asynchronous cells. Mitotic cells were obtained by 12 h of treatment with 200 ng/mL nocodazole followed by H3S10P labeling and sorting by FACS (Supplemental Fig. S5), as described previously (Kadauke et al. 2012). This resulted in 94.8% mitotic cells (n = 1601) in the sorted population as compared with 3.1% mitotic cells in the asynchronous samples (n = 1029), as assessed by inspection of DAPI staining of cell nuclei (Supplemental Fig. S5). We then performed Western blotting after Sox2 ChIP on mitotic and asynchronous cells, showing that Sox2 was pulled down in mitotic cells, although less efficiently than in asynchronous cells (Supplemental Fig. S5G). We performed ChIP-seq on SOX2 for both mitotic and unsynchronized samples and used model-based analysis of ChIP-seq (MACS2) (Zhang et al. 2008) for peak calling on grouped triplicates from each condition. We included an additional filtering step to remove peaks previously identified as frequent artifacts in high-throughput sequencing data (excessive unstructured anomalous reads mapping) (Supplemental Fig. S6; The ENCODE Project Consortium 2012). High-amplitude peaks called in unsynchronized samples displayed either clear or no enrichment for SOX2 in mitotic samples, as assessed from sequence read visualization and ChIP-qPCR (ChIP combined with quantitative PCR) experiments (Fig. 5A), thus excluding that peaks in mitotic cells are due to contaminating nonmitotic cells, confirming the purity of our mitotic cell preparation. MACS2 analysis yielded 10,523 peaks in asynchronous samples but only 84 peaks in mitotic samples (Fig. 5B). While 35 out of 66 genes bound in mitosis were also bound in unsynchronized samples (Fig. 5C), only a small number of called peaks overlapped between these two data sets (Fig. 5B). Two factors may contribute to the low number of mitotic peaks: (1) the lower pull-down efficiency of SOX2 from mitotic samples, although it is unclear whether this is a general issue in the field, since differences in pull-down efficiency were not measured in other studies on mitotic bookmarking transcription factors, and (2) our stringent peak calling; this is corroborated by the lower number of peaks that we found for GATA1 and FoxA1 when reanalyzing ChIP-seq data from Kadauke et al. (2012) and Caravaca et al. (2013) with our pipeline (Supplemental Fig. S6F,G). This raises the possibility that we might have missed a number of enriched loci, prompting us to perform visual track inspection of mitotic reads in regions where peaks were called only in the asynchronous samples. We indeed observed mitotic read enrichment in these regions and validated visually identified mitotic peaks located close to genes involved in pluripotency regulation (Rif1, Oct4, Nanog, and NFκB1) by ChIP-qPCR (Fig. 5D). Conversely, peak-centered enrichment analysis (Supplemental Fig. S7) and ChIP-qPCR (Supplemental Fig. S8) revealed that peaks called only in mitotic samples were often enriched in the asynchronous samples as well, suggesting a larger overlap of bound loci than indicated by peak calling. While gene ontology analysis did not reveal any significantly enriched gene category in the mitotic samples, peak location analysis revealed a high proportion of mitotic peaks located within and downstream from genes, at the expense of promoters and upstream and intergenic regions (Supplemental Fig. S9). De novo motif analysis (MEME) revealed the known composite motif of Pou5f1::Sox2 as top-ranked in the asynchronous sample (Supplemental Fig. S10), but no motif could be identified from mitotic peaks. While this could be due to the low number of peaks called, it is also possible that DNA sequence-independent properties of mitotic chromatin mediating differential accessibility to DNA mediate SOX2 enrichment at certain genomic locations. Taken together, our data suggest that SOX2 is mostly bound nonspecifically to mitotic chromatin but is significantly enriched in the vicinity of a small number of genes, of which >50% overlap with genes also bound in asynchronous cells.
Figure 5.
ChIP-seq analysis of SOX2 in unsynchronized and mitotic cells. (A) Examples of merged triplicate reads of peaks called in only the asynchronous samples, both the asynchronous and mitotic samples, or only the mitotic samples. For each example, asynchronous reads (As) are shown in the top row, and mitotic reads (Mit) are shown in the bottom row. Bar graphs show ChIP-qPCR results as relative fold enrichment over the input. n = 2. Error bars indicate SE. (B) Number of called peaks in asynchronous versus mitotic samples. The different numbers of shared peaks are due to the occasional occurrence of two peaks in the mitotic samples that match a single peak in the asynchronous sample. (C) Number of genes with SOX2 peaks 20 kb upstream of, 20 kb downstream from, or within genes. (D) Merged triplicate reads of peaks in the vicinity of pluripotency regulators that were not called but were identified as enriched by ChIP-qPCR. Bar graphs show ChIP-qPCR results represented as relative fold enrichment over the input. n = 2. Error bars indicate SE.
Functional interrogation of mitotic bookmarking by degradation of SOX2 at the M–G1 transition
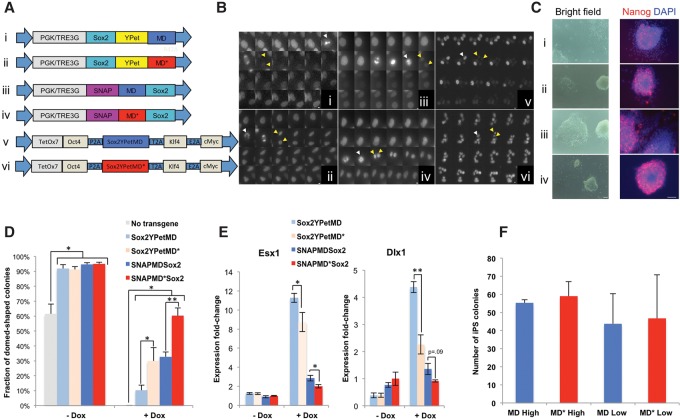
We next asked whether sequestration of SOX2 on mitotic chromosomes and therefore its presence on or in the vicinity of target genes at the M–G1 transition are essential for its functions in cell fate decision making. To do so, we aimed at engineering tools that allowed us to (1) down-regulate SOX2 levels specifically at the M–G1 transition and (2) sort cell populations with defined average SOX2 expression levels to allow for meaningful comparisons with controls that are not degraded during mitosis. We thus fused SOX2 to a mitotic degron (MD) that allows degradation of SOX2 at the metaphase-to-anaphase transition or an inactive mutant thereof carrying a single R42A amino acid substitution that inactivates the destruction box (MD*) (Kadauke et al. 2012). Sox2-MD and Sox2-MD* were also tagged with either YPet or a SNAP tag, allowing us to sort populations according to their tagged SOX2 expression levels (Fig. 6A). To assess the relative levels of these SOX2 fusion proteins in different phases of the cell cycle, we generated four ES cell lines expressing rtTA3G and the four respective TRE3G-driven fusion proteins, which were FACS-sorted 24 h after dox induction using windows spanning the same fluorescence intensities (Supplemental Methods). We then performed time-lapse fluorescence imaging of the YPet fusion and the SNAP-tag fusions labeled with the SiR-SNAP dye (Lukinavicius et al. 2013). While the fluorescence signals for MD-fused SOX2 rapidly decreased during the metaphase-to-anaphase transition followed by an increase of the signal in newly divided cells, the MD*-fused SOX2 did not display observable changes in fluorescence signals during cell division (Fig. 6B; Supplemental Movies S5–S8). We next generated two dox-inducible lentiviral vectors that allow expression of OCT4, KLF4, SOX2-YPet, and cMYC fused to either MD or MD* (OSKM-MD and OSKM-MD*, respectively) and transduced mouse embryonic fibroblasts (MEFs) with either vector together with an expression vector for M2rtTA (Hockemeyer et al. 2008). We also observed a rapid decrease of MD-fused SOX2 that was not observable in the MD*-fused SOX2, but the recovery of the signal was slower than what we observed in ES cells (Fig. 6B).
Figure 6.
The role of SOX2 at the M–G1 transition in pluripotency maintenance and reprogramming to induced pluripotent stem (iPS) cells. (A) Lentiviral constructs used to generate stable SOX2-MD and SOX2-MD* ES cell lines and for reprogramming to iPS cells. (B) Fluorescence time-lapse imaging of dividing ES cells (panels i–iv) or MEFs (panels v,vi) transduced with the corresponding lentiviral vectors (TRE3G promoter for panels i–iv) from A. Consecutive frames are separated by 15 min. Bars: panels i–iv, 5 µm; panels v,vi, 10 µm. White arrows indicate the cell just prior to division, and yellow arrows indicate the two daughter cells. (C) Representative bright-field images (left) and immunofluorescence staining against Nanog (right) of the four 2TS22C sub-cell lines transduced with the corresponding lentiviral vectors from A (PGK promoter) after 1 wk of culture in the presence of dox. (Red) Nanog; (blue) DAPI. Bars, 100 µm. (D) Counting of dome-shaped and flattened colonies after 1 wk of culture with or without dox. n = 3 per condition for the original 2TS22C cell line (no transgene); n = 6 per condition for all other cell lines. (E) RT-qPCR on Esx1 and Dlx1 after 1 wk of culture with or without dox, normalized against values obtained for SNAP-MD*-Sox2 without dox. n = 4 per condition. (F) Counting of iPS cell colonies generated from Oct4-GFP MEFs, as assessed by GFP fluorescence 20 d after transduction. High and low refer to different relative lentiviral titers. n = 3 high; n = 4 low. Error bars indicate SE. (*) P < 0.05; (**) P < 0.01.
We reasoned that there are two equally valid strategies for quantitative comparisons of MD and MD* cell lines: Either MD and MD* should have similar average SOX2 expression levels, at the expense of slightly higher SOX2 levels of the MD cell lines during cell cycle phases in which the MD is inactive, or both cell lines should have similar expression levels in cell cycle phases during which the MD is inactive but with slightly lower average expression levels for the MD cell lines. We were able to generate experimental conditions approximately matching these two strategies by sorting for narrow or large windows of fluorescence intensities, respectively (Supplemental Methods; Supplemental Fig. S11), and implemented these to assess the functional relevance of SOX2 at the M–G1 transition in pluripotency maintenance.
The presence of SOX2 at the M–G1 transition contributes to pluripotency maintenance but is dispensable for reprogramming to induced pluripotent stem (iPS) cells
To investigate the function of SOX2 at the M–G1 transition in pluripotency maintenance, we took advantage of the 2TS22C ES cell line, which allows turning off of Sox2 expression by addition of dox (Masui et al. 2007) and leads to rapid loss of pluripotency. We then generated four sub-cell lines that allow PGK-driven SOX2 fused to either YPet or a SNAP tag in combination with MD or MD* and sorted either narrow (PGK-Sox2-YPet fusion cell lines) or large (PGK-SNAP-Sox2 cell lines) fluorescence windows. We then compared the ability of the different SOX2 fusions to maintain pluripotency in the presence of dox. After 1 wk of dox treatment, we observed more flat colonies with uneven edges expressing lower and more heterogeneous Nanog levels in the MD cell lines when compared with the respective MD* cell lines (Fig. 6C). We then quantified morphological changes by counting the number of flat colonies with uneven edges versus dome-shaped colonies with sharp edges in the four cell lines and the original 2TS22C cell line after 1 wk of culture in the presence or absence of dox. Although both MD-fused and MD*-fused versions of SOX2 partially rescued pluripotency as compared with the original 2TS22C cell line, this effect was significantly stronger in the MD* compared with the respective MD cell lines (Fig. 6D). Using the same culture conditions, we also measured mRNA expression of esx1 and dlx1, two trophectodermal markers that have been shown to be up-regulated upon dox-mediated SOX2 knockdown in the 2TS22C cell line (Masui et al. 2007). We observed higher expression levels of both markers after 1 wk of dox in the MD cell lines as compared with the matched MD* cell lines, thus confirming the higher propensity of ES cells to differentiate in the absence of SOX2 during the M–G1 transition (Fig. 6E). Interestingly, the SNAP-tagged versions of SOX2 were more potent in maintaining pluripotency than the YPet-tagged versions, which could be due to differences in expression levels between SNAP-tagged and YPet-tagged SOX2 cell lines and/or differences in the functionality of SOX2, depending on the tag and/or its location at the N terminus versus the C terminus. Taken together, our results suggest that the M–G1 transition is a key time window for the pluripotency-maintaining function of SOX2.
We next investigated whether the presence of SOX2 at the M–G1 transition is required for reprogramming to iPS cells. To do so, we generated lentiviral particles for the OSKM-MD and OSKM-MD* lentiviral vectors and measured the relative titers of both versions by transducing these vectors together with FuW-M2rtTA in 293T and quantifying YPet expression in the presence of dox by flow cytometry. We then used adjusted relative lentiviral titers of OSKM-MD and OSKM-MD* to transduce Oct4-GFP MEFs (Lengner et al. 2007) using two different concentrations of lentiviral particles differing by a factor of 3. Dox was included in the cell culture medium from day 2–16 after transduction, and reprogrammed colonies were counted on day 20. We did not observe any difference in reprogramming efficiency between OSKM-MD and OSKM-MD*, suggesting that SOX2 expression at the M–G1 transition does not play a role in reprogramming toward iPS cells (Fig. 6F).
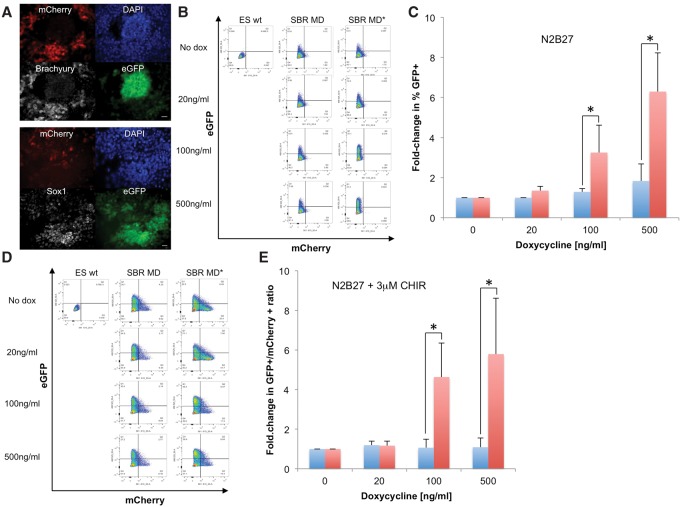
SOX2 is required at the M–G1 transition to drive neuroectodermal differentiation
SOX2 has been shown to favor neuroectodermal over mesendodermal differentiation (Thomson et al. 2011) and accelerate neuroectodermal commitment of ES cells upon overexpression (Zhao et al. 2004). To study the role of mitotic SOX2 in neuroectodermal differentiation, we generated a double-reporter cell line to facilitate quantitative assessment of neuroectodermal and mesendodermal commitment. We used CRISPR–Cas9 to knock in a P2A-eGFP cassette downstream from the endogenous Sox1-coding sequence and a P2A-mCherry cassette downstream from the endogenous Brachyury-coding sequence (Supplemental Methods; Supplemental Fig. S12), allowing expression of eGFP in neuroectodermal cells and mCherry in mesendodermal cells (Ying et al. 2003; Kubo et al. 2004). We refer to this cell line as SBR (Sox1/Brachyury reporter). We confirmed the specificity of our reporters by differentiating the SBR cell line for 4 d and inspecting the colocalization of SOX1 and BRACHYURY immunoreactivity with eGFP and mCherry expression, respectively (Fig. 7A). The SBR cell line was then used to generate two sub-cell lines expressing rtTA3G and either SNAP-MD-Sox2 or SNAP-MD*-Sox2. Since we intended to modulate average expression levels in both cell lines using different doses of dox, we used a single sorting strategy using a unique, narrow fluorescence intensity window using the SiR-SNAP dye after overnight incubation with 100 ng/mL dox (Supplemental Fig. S11). We then compared the potency of MD-Sox2 and MD*-Sox2 in enhancing neuroectodermal differentiation by culturing cells in N2B27 for 3 d in the presence of various doses of dox followed by flow cytometry to measure the fraction of eGFP+ cells. Whereas the SBR SNAP-MD*-Sox2 cell line displayed dose-dependent increased neuroectodermal commitment, differentiation of the SBR SNAP-MD-Sox2 cell line did not change upon dox addition (Fig. 7B,C). We then compared the potency of MD-Sox2 and MD*-Sox2 in favoring neuroectodermal commitment over mesendodermal commitment by treating cells in N2B27 medium for 2 d followed by 2 d in N2B27 with 3 µM CHIR, which enhances mesendodermal commitment (Thomson et al. 2011). We observed a dox dose-dependent increase in the eGFP+/mCherry+ ratio in the SBR SNAP-MD*-Sox2 cell line but not in the SBR SNAP-MD-Sox2 cell line (Fig. 7D,E), suggesting that mitotic degradation of SOX2 suppresses its ability to favor neuroectodermal over mesendodermal commitment (Fig. 7D,E). We thus conclude that the presence of SOX2 at the M–G1 transition is required for its ability to enhance neuroectodermal differentiation.
Figure 7.
SOX2 is required at the M–G1 transition to promote neuroectodermal differentiation. (A) Immunofluorescence analysis of BRACHYURY (top) and SOX1 (bottom) expression in the SBR cell line after 4 d of differentiation in N2B27. Bars, 20 µm. (B) Flow cytometry of the SBR cell line expressing dox-inducible SNAP-MD-Sox2 (SBR MD) or SNAP-MD*-Sox2 (SBR MD*) after 3 d of differentiation in N2B27 with different doses of dox. (ES wt) Negative control. (C) Quantification of six independent flow cytometry experiments as shown in B, expressed as fold change in the fraction of eGFP+ cells and normalized over the values obtained without dox. (*) P < 0.05. (Blue bars) SNAP-MD-Sox2; (red bars) SNAP-MD*-Sox2. (D) Flow cytometry of the SBR cell line expressing dox-inducible SNAP-MD-Sox2 or SNAP-MD*-Sox2 after 4 d of differentiation with different doses of dox in N2B27 with the addition of 3 µM CHIR after 48 h to induce mesendodermal differentiation. (ES wt) Negative control. (E) Quantification of seven independent flow cytometry experiments as shown in D, expressed as fold change in the eGFP+/mCherry+ ratio and normalized over the value of this ratio without dox. (Blue bars) SNAP-MD-Sox2; (red bars) SNAP-MD*-Sox2. (*) P < 0.05.
Discussion
Here we provide evidence of MCB of SOX2 and OCT4 and mitotic bookmarking by SOX2. Our results are in contrast to recent immunofluorescence-based experiments reporting that OCT4 does not bind to mitotic chromosomes (Galonska et al. 2014). However, several studies reported unclear (Caravaca et al. 2013) or false negative results (Chen et al. 2002; Blobel et al. 2009; Mishra et al. 2009; Wu et al. 2015) when using immunofluorescence to assess MCB, which was suggested to be due to limited accessibility of epitopes to antibodies within the dense mitotic environment (Chen et al. 2002; Kadauke and Blobel 2013; Wu et al. 2015) but also formaldehyde fixation artifacts (Pallier et al. 2003; Kumar et al. 2008). Similar to previous reports on other transcription factors binding to mitotic chromosomes (Pallier et al. 2003; Kumar et al. 2008; Lerner et al. 2016), we were unable to observe mitotic chromosome localization of SOX2 and OCT4 after formaldehyde fixation (data not shown). However, using a mixture of 95% methanol and 5% acetic acid previously shown to preserve the localization of transcription factors bound to mitotic chromosomes (Kumar et al. 2008), we could observe MCB of endogenous SOX2 and OCT4 by immunofluorescence. Furthermore, here we used three unrelated tags to assess MCB of both overexpressed and endogenously expressed SOX2 and OCT4 in living cells and showed that their DNA-binding domains are necessary and sufficient for MCB. Interestingly, we observed a significant contribution of NLS sequences to the colocalization of SOX2 and OCT4 with mitotic chromosomes, in line with the reported sequence-independent electrostatic interactions of NLS with DNA (Simeoni et al. 2003; Mann et al. 2008).
While mitotic bookmarking by sequence-specific transcription factors was reported >10 years ago (Xing et al. 2005), its biological function remains unclear. A prevailing idea in the field is that mitotic bookmarking could serve as a mechanism to maintain cell identity (Kadauke et al. 2012; Zaidi et al. 2014; Zaret 2014). This hypothesis was fueled by experimental evidence of mitotic bookmarking as a mechanism for rapid gene reactivation in G1 (Blobel et al. 2009; Dey et al. 2009; Kadauke et al. 2012) and thus potentially contributing to robust maintenance of cell type-specific gene expression programs after cell division. However, recent studies have challenged the role of binding to specific DNA sites during mitosis for rapid transcriptional reactivation of targeted genes (Caravaca et al. 2013; Hsiung et al. 2016), and, most importantly, the functional relevance of mitotic bookmarking in cell fate decision making has not been demonstrated so far. Our study reveals a key time window for the action of SOX2 on cell fate decisions, which temporally overlaps with its mitotic bookmarking activity and the re-establishment of transcriptional activity in newly divided cells (Hsiung et al. 2016). Interestingly, the functional importance of SOX2 at the M–G1 transition differed across the cell fate decisions that we studied. The impaired but not null pluripotency maintenance that we observed upon mitotic degradation of SOX2 could be due to incomplete SOX2 degradation at the M–G1 transition, but it is also likely that SOX2 expression in other cell cycle phases plays a role in pluripotency maintenance. Surprisingly, we did not observe any effect of mitotic SOX2 degradation on its ability to reprogram MEFs to iPS cells. This is unlikely to be due to differences in temporal pattern of SOX2 degradation in MEFs, since its increase in G1 was slower than in ES cells (Fig. 6B), but we cannot exclude that the residual levels of SOX2 at the M–G1 transition were sufficient for its maximal potency in mediating reprogramming. The most striking impact on cell fate decisions that we observed was the abrogation of the neuroectodermal differentiation-promoting activity of SOX2 when absent at the M–G1 transition. In ES cells, the decisions to commit to the mesendodermal versus neuroectodermal lineages take place in early and late G1, respectively (Pauklin and Vallier 2013), and are regulated notably by the cell cycle machinery (Pauklin et al. 2016). In our experiments with dox-inducible SOX2-MD, expression levels increase rapidly after cell division, and thus the absence of neuroectodermal fate enhancement is unlikely to result from insufficient expression at the end of the G1 phase but rather suggests that SOX2 acts during the M–G1 transition to enhance neuroectodermal commitment.
The M and G1 phases of the cell cycle were shown to be a window of opportunity for cellular differentiation and reprogramming (Sela et al. 2012; Coronado et al. 2013; Pauklin and Vallier 2013; Halley-Stott et al. 2014). However, the molecular mechanisms underlying the sensitivity of the M–G1 transition to the action of transcription factors on cell fate decisions remain unclear. Despite the clear enrichment of SOX2 on mitotic chromosomes observed by microscopy, we found a surprisingly small number of genes specifically bound during mitosis. In addition to the technical reasons invoked above, this might be due to mostly nonspecific DNA binding of SOX2 and/or DNA-independent binding of SOX2 to mitotic chromosomes. In contrast to GATA1 (Kadauke et al. 2012), RBPJ (Lake et al. 2014), or MLL (Blobel et al. 2009) but similar to FOXA1 (Caravaca et al. 2013), we did not find enrichment of a particular class of genes bound by SOX2 during mitosis. The functional relevance of the sites specifically bound on mitotic chromosomes remains unclear, and there appears to be no quantitative difference in early G1 transcriptional activity of genes bound by FOXA1 or GATA1 in mitosis compared with genes bound in interphase only (Caravaca et al. 2013; Hsiung et al. 2016). Interestingly, nonspecific binding to mitotic chromosomes was also shown to regulate transcriptional reactivation in G1 (Caravaca et al. 2013). This raises the possibility that the high local concentration of mitotically bound transcription factors could mediate robust and fast occupancy of specific DNA sites in early G1, which could serve as a mechanism to ensure primacy against competing transcription factors in regulating target genes.
The role of mitotic bookmarking in stem cell regulatory networks remains largely unexplored. We found that OCT4 and potentially several other pluripotency transcription factors (Fig. 1A) are also partially retained on mitotic chromosomes, suggesting that mitotic bookmarking could be widespread within the pluripotency network. The evidence that we present here on the functional role of mitotic bookmarking by SOX2 paves the way for future studies to identify and assess the functional relevance of other mitotic bookmarking factors in stem cell maintenance and differentiation.
Materials and methods
Live fluorescence microscopy
Live fluorescence imaging was performed at the biomolecular screening facility of the Swiss Federal Institute of Technology on an IN Cell Analyzer 2200 apparatus (GE healthcare) with controlled atmosphere (5% CO2) and temperature (37°C) for long-term imaging and a 20× magnification objective. One day before imaging, cells were seeded in black-walled 96-well plates either uncoated (NIH-3T3 and 293T cells) or coated with E-Cadherin (R&D Systems, catalog no. 748-EC-050) as described (Nagaoka et al. 2006), and transgene expression was induced with dox (100 ng/mL unless specified otherwise). To reduce background fluorescence, the cell culture medium was exchanged before imaging for Fluorobrite DMEM (Life Technologies) supplemented with the respective additives and dox concentrations for the different cell lines. Images were acquired for 16–24 h every 5 min using the following fluorescence channels: CFP channel for CerFP, YFP channel for YPet, TexasRed channel for TagRFP-T, Cy3 channel for the Halo-TMR dye, and Cy5 channel for the SiR-SNAP dye.
Immunofluorescence
E14 ES cells were fixed for 20 min with a chilled mix of 95% methanol/5% acetic acid that was maintained at −20°C, permeabilized with chilled PBS-Triton for 15 min, and blocked with PBS and 1% BSA for 30–60 min. Samples were incubated with primary antibodies rabbit anti-Sox2 (1:200; Life Technologies) and rabbit anti-Oct4 (1:500; Abcam, ab19857) in PBS and 1% BSA overnight at 4°C. Samples were washed twice in PBS and then incubated with Alexa 555 anti-rabbit (1:1000; Life Technologies, catalog no. 481400) in PBS and 1% BSA for 45–60 min followed by three washes with 0.1% PBS-Tween, incubation with 2 ng/mL DAPI, three washes with PBS and 0.1% Tween, and two washes with PBS.
MCB index calculation
We performed background subtraction on time-lapse data using Fiji software and manually measured mean fluorescence intensities of the SOX2 and OCT4 fusion proteins on metaphase chromatin divided by the intensity of a nearby area inside the cell.
Residence time measurements
To extract DNA residence times from raw data, we first extracted molecular localizations in each frame using Octane software. Peak detection thresholds were set individually for each sample set to account for variations in image background level. We identified molecule trajectories as the group of localizations in adjacent frames belonging to the same bound molecule using the following parameters: differences in position <160 nm (to avoid selection of moving nonbound molecules) and temporal distances less than two frames (to allow for missed localizations and fluorophore blinking). For each molecule trajectory, the length was assumed to be equal to the time that the molecule stays bound while unbleached. We fitted the distribution of trajectory lengths to an exponential curve to obtain an effective binding time for each gap time. Finally, we combined effective binding times obtained from imaging with different gaps to disentangle bleaching rate from DNA off rates, allowing us to obtain DNA residence times. This analysis is described in more detail in Gebhardt et al. (2013).
FLIP and FRAP
FLIP and FRAP experiments were performed on a Zeiss LSM 700 confocal microscope equipped with a Plan-Apochromat 63×/1.40 oil DIC M27 objective (Zeiss) with controlled temperature and CO2. Since a 488-nm laser (Zeiss) was used for imaging and bleaching of the sample regions, we used ES cell lines that allowed inducible YPet-SOX2 or YPet-OCT4 expression but were devoid of H2B-CerFP expression to avoid potential cross-talk between the two channels. Twenty-four hours before imaging, transgene expression was induced with 100 ng/mL dox. To reduce background fluorescence, the cell culture medium was exchanged before imaging for Fluorobrite DMEM (Life Technologies) supplemented as described in the Supplemental Material for ES cells and with dox.
FLIP
In mitotic cells, we measured time traces of average fluorescence from a square region in the area containing condensed chromosomes while continuously bleaching a region in the cytoplasm. In interphase cells, both bleaching and measurement regions were inside the nucleus. For both mitotic and interphase cells, the size of the bleaching area was 4 µm2, the size of the measurement area was 1 µm2, and the distance between the bleaching and measurement regions was ∼3 µm.
FRAP
In mitotic cells, we pulse-bleached a round region in the area containing condensed chromosomes. We then measured time traces of average recovered fluorescence in the bleached region using 1.5% and 3% laser power for bleaching for Sox2 and Oct4, respectively. In interphase cells, the bleached and measured region was inside the nucleus. For both mitotic and interphase cells, the size of the bleaching and measurement area was 1.5 µm2. Images were collected using a recording zoom of 2× at 0.3-sec intervals, including five prebleached images.
Chip-seq analysis
Reads for each individual sample/replicate were mapped to the mouse genome (mm10 assembly) using the mapping module of HTSstation (based on Bowtie2) (David et al. 2014) with the option “discard PCR duplicates.” Triplicates were then combined to give a total of 1.53 × 108 mapped reads for the asynchronous samples, 8.3 × 107 mapped reads for the corresponding inputs, 1.2 × 108 mapped reads for the mitotic samples, and 1 × 108 mapped reads for the corresponding inputs. To avoid any potential bias due to unbalanced samples, ChIP samples were down-sampled preliminarily to the peak calling procedure. SOX2 peaks in the asynchronous and mitotic samples were identified with MACS2 (version 2.1) (Feng et al. 2012) from combined triplicates versus respective inputs. Since most reads fell into regions of nonspecific binding in the mitotic samples, this made peak identification for this ChIP more difficult, and, similar to Caravaca et al. (2013), we used a more lenient q-value threshold to define mitotic peaks. Therefore, a q-value threshold of 0.01 was used for the asynchronous samples, and a q-value threshold of 0.05 was used for the mitotic samples. Peaks overlapping any region found in the ENCODE blacklist (converted from mm9 to mm10 with LiftOver) (The ENCODE Project Consortium 2012) were excluded. BioScript (http://bioscript.epfl.ch) was used to annotate peaks to mm10 genes (operation Annotate) and quantify signals in regions surrounding peak summits (operation plotFeature). Motifs overrepresented in peaks were searched in sequences of 300 base pairs (centered on MACS2 summits), using MEME-ChIP (Machanick and Bailey 2011) with default parameters and using asynchronous sequences as background.
Statistical tests
For Figure 3 (all panels), a two-tailed Student's t-test with unequal variance was used. For Figure 6, D–F, a two-tailed paired Student's t-test was used. For Figure 7, C and E, a Shapiro-Wilk test determined that the samples were nonnormally distributed; thus, a Wilcoxon-signed rank test was used to determine the statistical significance of the data.
Supplementary Material
Acknowledgments
We thank Andrea Alber for providing the SNAP-tagged Nanog, Esrrb, and Klf4 ES cell lines. We thank Luigi Bozzo, Thierry Laroche, and José Artacho from the Swiss Federal Institute of Technology (EPFL) Bioimaging and Optics Core Facility (EPFL-BIOP); Valérie Glutz, Loïc Tauzin, Miguel Garcia, and André Mozes at the EPFL Flow Cytometry Core Facility (EPFL-FCCF); and Fabien Kuttler at the EPFL Biomolecular Screening Facility (EPFL-BSF) for assistance in imaging. We thank Michael Imbeault for advice on ChIP-seq experiments, Massimiliano Caiazzo and Matthias Lütolf for providing Oct4-GFP MEFs and for advice on reprogramming, and Didier Trono and Pierre Gönczy for insightful comments on the manuscript. Work from the laboratory of D.M.S. was supported by the Carigest Foundation, the Swiss National Science Foundation (grant no. PP00P3_144828), and the Novartis Foundation for Biomedical Research (grant no. 15A018). Work from the laboratory of S.M. was supported by the Swiss National Science Foundation (grant no. CR33I2_149850).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.289256.116.
References
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. 2009. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell 36: 970–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. 2013. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev 27: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Hinkley CS, Henry RW, Huang S. 2002. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell 13: 276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. 2014. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156: 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Young RA. 2008. Mapping key features of transcriptional regulatory circuitry in embryonic stem cells. Cold Spring Harb Symp Quant Biol 73: 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado D, Godet M, Bourillot PY, Tapponnier Y, Bernat A, Petit M, Afanassieff M, Markossian S, Malashicheva A, Iacone R, et al. 2013. A short G1 phase is an intrinsic determinant of naive embryonic stem cell pluripotency. Stem Cell Res 10: 118–131. [DOI] [PubMed] [Google Scholar]
- David FP, Delafontaine J, Carat S, Ross FJ, Lefebvre G, Jarosz Y, Sinclair L, Noordermeer D, Rougemont J, Leleu M. 2014. HTSstation: a Web application and open-access libraries for high-throughput sequencing data analysis. PLoS One 9: e85879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. 2009. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell 20: 4899–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. 2014. Defining an essential transcription factor program for naive pluripotency. Science 344: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N, Dubois A, Vandormael-Pournin S, Gallego Tejeda E, Mouren A, Bessonard S, Mueller F, Proux C, Cohen-Tannoudji M, Navarro P. 2016. Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat Cell Biol 18: 1139–1148. [DOI] [PubMed] [Google Scholar]
- Galonska C, Smith ZD, Meissner A. 2014. In vivo and in vitro dynamics of undifferentiated embryonic cell transcription factor 1. Stem Cell Rep 2: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. 2013. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods 10: 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob A, Colleran C, McStay B. 2014. Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev 28: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. 2009. Transcription dynamics. Mol Cell 35: 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley-Stott RP, Jullien J, Pasque V, Gurdon J. 2014. Mitosis gives a brief window of opportunity for a change in gene transcription. PLoS Biol 12: e1001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. 2008. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3: 346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung CC, Bartman CR, Huang P, Ginart P, Stonestrom AJ, Keller CA, Face C, Jahn KS, Evans P, Sankaranarayanan L, et al. 2016. A hyperactive transcriptional state marks genome reactivation at the mitosis-G1 transition. Genes Dev 30: 1423–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Blobel GA. 2013. Mitotic bookmarking by transcription factors. Epigenetics Chromatin 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. 2012. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ, Keller G. 2004. Development of definitive endoderm from embryonic stem cells in culture. Development 131: 1651–1662. [DOI] [PubMed] [Google Scholar]
- Kumar S, Chaturvedi NK, Kumar S, Tyagi RK. 2008. Agonist-mediated docking of androgen receptor onto the mitotic chromatin platform discriminates intrinsic mode of action of prostate cancer drugs. Biochim Biophys Acta 1783: 59–73. [DOI] [PubMed] [Google Scholar]
- Lake RJ, Tsai PF, Choi I, Won KJ, Fan HY. 2014. RBPJ, the major transcriptional effector of Notch signaling, remains associated with chromatin throughout mitosis, suggesting a role in mitotic bookmarking. PLoS Genet 10: e1004204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R. 2007. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell 1: 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner J, Bagattin A, Verdeguer F, Makinistoglu MP, Garbay S, Felix T, Heidet L, Pontoglio M. 2016. Human mutations affect the epigenetic/bookmarking function of HNF1B. Nucleic Acids Res 44: 8097–8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Camacho C, van Wijnen AJ, Lian JB, Stein JL, Stein GS. 2014. CBFβ and the leukemogenic fusion protein CBFβ-SMMHC associate with mitotic chromosomes to epigenetically regulate ribosomal genes. J Cell Biochem 115: 2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavicius G, Umezawa K, Olivier N, Honigmann A, Yang G, Plass T, Mueller V, Reymond L, Correa IR Jr, Luo ZG, et al. 2013. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat Chem 5: 132–139. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann A, Thakur G, Shukla V, Ganguli M. 2008. Peptides in DNA delivery: current insights and future directions. Drug Discov Today 13: 152–160. [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, et al. 2007. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 9: 625–635. [DOI] [PubMed] [Google Scholar]
- Mishra BP, Ansari KI, Mandal SS. 2009. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. FEBS J 276: 1629–1640. [DOI] [PubMed] [Google Scholar]
- Nagaoka M, Koshimizu U, Yuasa S, Hattori F, Chen H, Tanaka T, Okabe M, Fukuda K, Akaike T. 2006. E-cadherin-coated plates maintain pluripotent ES cells without colony formation. PLoS One 1: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. 2008. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26: 101–106. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391. [DOI] [PubMed] [Google Scholar]
- Pallier C, Scaffidi P, Chopineau-Proust S, Agresti A, Nordmann P, Bianchi ME, Marechal V. 2003. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol Biol Cell 14: 3414–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S, Vallier L. 2013. The cell-cycle state of stem cells determines cell fate propensity. Cell 155: 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S, Madrigal P, Bertero A, Vallier L. 2016. Initiation of stem cell differentiation involves cell cycle-dependent regulation of developmental genes by Cyclin D. Genes Dev 30: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pockwinse SM, Kota KP, Quaresma AJ, Imbalzano AN, Lian JB, van Wijnen AJ, Stein JL, Stein GS, Nickerson JA. 2011. Live cell imaging of the cancer-related transcription factor RUNX2 during mitotic progression. J Cell Physiol 226: 1383–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM, Bender MA. 1962. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res 26: 260–268. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. 2003. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev 17: 2048–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela Y, Molotski N, Golan S, Itskovitz-Eldor J, Soen Y. 2012. Human embryonic stem cells exhibit increased propensity to differentiate during the G1 phase prior to phosphorylation of retinoblastoma protein. Stem Cells 30: 1097–1108. [DOI] [PubMed] [Google Scholar]
- Simeoni F, Morris MC, Heitz F, Divita G. 2003. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res 31: 2717–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161: 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CA, Kruhlak MJ, Jenkins HL, Sun X, Bazett-Jones DP. 2000. Mitotic transcription repression in vivo in the absence of nucleosomal chromatin condensation. J Cell Biol 150: 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. 2011. Mammalian genes are transcribed with widely different bursting kinetics. Science 332: 472–474. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. 2011. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145: 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. 2008. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods 5: 159–161. [DOI] [PubMed] [Google Scholar]
- Wu TF, Yao YL, Lai IL, Lai CC, Lin PL, Yang WM. 2015. Loading of PAX3 to mitotic chromosomes is mediated by arginine methylation and associated with Waardenburg syndrome. J Biol Chem 290: 20556–20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK, Sarge KD. 2005. Mechanism of hsp70i gene bookmarking. Science 307: 421–423. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21: 183–186. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. 2010. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet 11: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Grandy RA, Lopez-Camacho C, Montecino M, van Wijnen AJ, Lian JB, Stein JL, Stein GS. 2014. Bookmarking target genes in mitosis: a shared epigenetic trait of phenotypic transcription factors and oncogenes? Cancer Res 74: 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. 2014. Genome reactivation after the silence in mitosis: recapitulating mechanisms of development? Dev Cell 29: 132–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Nichols J, Smith AG, Li M. 2004. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol Cell Neurosci 27: 332–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.