Wada et al. show that FLCN regulates adipose tissue browning via mTOR and the transcription factor TFE3. Adipose-specific deletion of FLCN relieves mTOR-dependent cytoplasmic retention of TFE3, leading to direct induction of the PGC-1 transcriptional coactivators, drivers of mitochondrial biogenesis and the browning program.
Keywords: adipose tissue, beige fat, mitochondria, FLCN, mTOR, TFE3
Abstract
Noncanonical mechanistic target of rapamycin (mTOR) pathways remain poorly understood. Mutations in the tumor suppressor folliculin (FLCN) cause Birt-Hogg-Dubé syndrome, a hamartomatous disease marked by mitochondria-rich kidney tumors. FLCN functionally interacts with mTOR and is expressed in most tissues, but its role in fat has not been explored. We show here that FLCN regulates adipose tissue browning via mTOR and the transcription factor TFE3. Adipose-specific deletion of FLCN relieves mTOR-dependent cytoplasmic retention of TFE3, leading to direct induction of the PGC-1 transcriptional coactivators, drivers of mitochondrial biogenesis and the browning program. Cytoplasmic retention of TFE3 by mTOR is sensitive to ambient amino acids, is independent of growth factor and tuberous sclerosis complex (TSC) signaling, is driven by RagC/D, and is separable from canonical mTOR signaling to S6K. Codeletion of TFE3 in adipose-specific FLCN knockout animals rescues adipose tissue browning, as does codeletion of PGC-1β. Conversely, inducible expression of PGC-1β in white adipose tissue is sufficient to induce beige fat gene expression in vivo. These data thus unveil a novel FLCN–mTOR–TFE3–PGC-1β pathway—separate from the canonical TSC–mTOR–S6K pathway—that regulates browning of adipose tissue.
White adipose tissue (WAT) stores energy, while brown adipose tissue (BAT) dissipates energy via thermogenesis. The activation of mitochondrial biogenesis and a “browning” program in WAT has gained traction recently as a possible approach to combat obesity and diabetes (Harms and Seale 2013). Understanding the mechanisms underlying adipose tissue browning is therefore of great interest.
Mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) is a critical multiprotein signaling hub nucleated around the protein raptor and conserved from yeast to humans that integrates intracellular and extracellular cues to regulate cellular growth and metabolism (Zoncu et al. 2011; Dibble and Manning 2013; Goberdhan et al. 2016). Numerous signals affect mTORC1 function, including amino acids levels, oxygen tension, and the presence of growth factors. At the same time, mTORC1 outputs vary widely from increasing protein synthesis and mRNA synthesis to regulation of lipid synthesis and autophagy. How separate inputs to mTORC1 can segregate to separate outputs (i.e., how functional specificity is achieved) remains an enigmatic aspect of mTOR signaling.
The role of mTOR in adipocyte browning is unclear. Loss of raptor in fat and portions of the brain leads to browning of WAT (Polak et al. 2008), while activation of mTOR in the same tissue distribution via deletion of tuberous sclerosis complex-1 (TSC1) leads to whitening of BAT (Xiang et al. 2015), suggesting that mTOR suppresses adipose browning in vivo. On the other hand, recent work shows that loss of raptor in fat or pharmacological inhibition of mTOR blocks cold-induced browning of WAT (Liu et al. 2016; Tran et al. 2016), demonstrating that mTOR can also promote adipose browning in vivo. A mechanistic reconciliation of these observations is lacking.
Germline heterozygous mutations in folliculin (FLCN) cause Birt-Hogg-Dubé (BHD) syndrome, a hamartomatous disease marked by renal cell tumors with loss of heterozygosity and striking increases in mitochondrial content (Schmidt 2013). In mice, genetic deletion of FLCN in renal cells induces mitochondrial biogenesis via the transcriptional coactivator PGC-1α, although how this occurs is not known (Hasumi et al. 2012). Biochemically, FLCN was identified recently as a GTP-activating protein (GAP) for RagC/D, which, in its GDP-bound state, activates mTORC1, implicating FLCN as a positive regulator of the mTOR pathway (Petit et al. 2013; Tsun et al. 2013). On the other hand, tumors with FLCN deletions typically reveal elevated mTOR activity (Baba et al. 2008), as is also observed following deletion of FLCN in murine hearts (Hasumi et al. 2014), suggesting that, under chronic conditions, counterregulatory signals maintain or even increase mTOR activity in the absence of FLCN. How deletion of FLCN drives PGC-1α and mitochondrial biogenesis and whether the process involves mTOR activation or suppression are not known.
The transcription factor TFE3 is a member of the MiTF gene family that contains four highly homologous transcription factors: MiTF, TFE3, TFEB, and TFEC. MiTF and TFEC have specialized roles in melanocytes and monocytes, respectively (Rehli et al. 1999; Haq et al. 2013; Shoag et al. 2013), while TFE3 and TFEB are more widely expressed, including in adipose tissue. TFEB regulates a broad program of lysosomal biogenesis in numerous cell types (Sardiello et al. 2009; Settembre et al. 2011). The role of TFE3 is less well understood. Conventional TFE3 knockout animals do not show any obvious phenotype (Steingrimsson et al. 2002). In pluripotent stem cells, TFE3 favors the maintenance of a pluripotent state, which, interestingly, is suppressed by FLCN (Betschinger et al. 2013). In renal tumor cells from BHD patients, TFE3 constitutively localizes in the nucleus, and forced FLCN expression retains TFE3 in the cytoplasm (Hong et al. 2010). Finally, we showed recently in myocytes and melanocytes that TFE3 and MiTF directly regulate the expression of PGC-1α and oxidative metabolism, respectively (Haq et al. 2013; Salma et al. 2015).
Taken together, these observations led us to hypothesize that (1) loss of FLCN in adipose tissue would promote mitochondrial biogenesis and browning of WAT; (2) mechanistically, FLCN suppresses mitochondrial biogenesis via mTOR-mediated phosphorylation and cytoplasmic retention of TFE3, leading to reduced PGC-1 expression; (3) FLCN–mTOR–TFE3 signaling in adipose cells would reveal a hub separate from canonical TSC–Rheb–mTOR–S6K signaling; and (4) FLCN–mTOR–TFE3 signaling would thus explain the paradoxical observations on the role of mTOR in adipocyte browning. We tested this hypothesis using adipose-restricted deletion of FLCN in mice.
Results
FLCN deletion in fat leads to browning of WAT
The expression of FLCN protein is comparable in several tissues, including lung, kidney, muscle, and fat (Supplemental Fig. 1A). To test the role of FLCN specifically in adipose tissue, we generated mice with adipose tissue-specific deletion of FLCN (FLCN adipKO) using FLCNlox/lox mice (from Dr. Laura S. Schmidt and Dr. W. Marston Linehan, National Institutes of Health) and adiponectin-Cre mice (from Dr. Evan Rosen, Beth Israel Deaconess Medical Center) (Fig. 1A). FLCN protein expression was strongly reduced in both BAT and inguinal WAT (iWAT) (Fig. 1B, left), and mRNA levels decreased by 90% in BAT and 60% in iWAT (Fig. 1B, right) while remaining intact in other tissues (Supplemental Fig. 1B). Hence, deletion of FLCN in these mice is efficient and specific to adipose tissues.
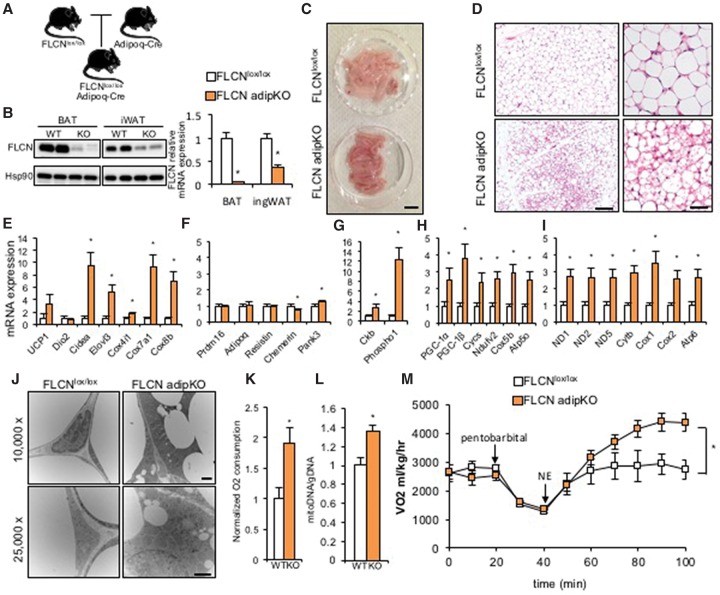
Figure 1.
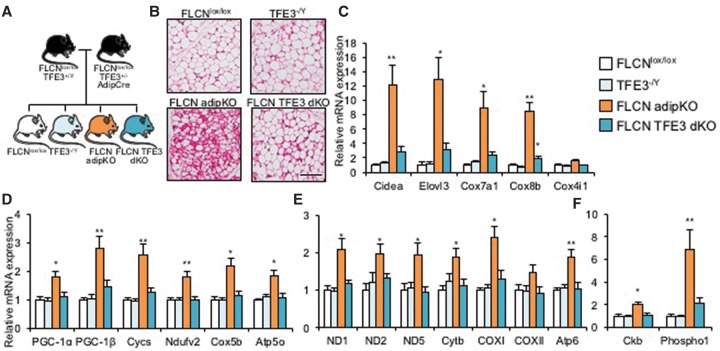
Adipocyte-specific deletion of FLCN induces browning of WAT. (A) Schematic of an animal model for FLCN adipKO. (B, left) Efficiency of FLCN protein deletion in BAT and iWAT. (Right) Relative mRNA expression of FLCN in the indicated fat depots. (C) Gross images of iWAT from FLCNlox/lox and FLCN adipKO mice. Bar, 1 cm. (D) Low-power (left) and high-power (right) images of H&E staining of iWAT from FLCNlox/lox or FLCN adipKO mice. Bars: left, 200 µm; right, 50 µm. (E–I) Relative mRNA expression of the indicated genes in iWAT from FLCNlox/lox or FLCN adipKO mice. (J) Low-power (top) and high-power (bottom) transmission electron microscopy images of iWAT from FLCNlox/lox and FLCN adipKO mice. Bars, 500 nm. (K) Relative O2 consumption of iWAT isolated from FLCNlox/lox and FLCN adipKO mice. (L) Relative mitochondrial DNA content in iWAT from the respective genotypes. (M) Whole-body respiration of FLCNlox/lox and FLCN adipKO animals upon subcutaneous injection of 1 mg/kg norepinephrine (NE) after induction of anesthesia by pentobarbital. (*) P < 0.05 versus FLCNlox/lox. n = 6–8 per group. Two-way analysis of variance (ANOVA) was used for M. Values are represented as mean ± SEM.
iWAT isolated from FLCN adipKO mice was visibly browner in color than iWAT from littermate FLCNlox/lox controls (Fig. 1C). H&E staining of iWAT from FLCN adipKO mice showed numerous multilocular adipocytes, typical histological features of beige adipocytes (Fig. 1D). Well-established brown/beige fat genes, including Cidea, Elovl3, Cox4i1, Cox7a1, and Cox8b, were significantly induced in adipKO WAT (Fig. 1E). Expression of prdm16 and genes thought to be white fat-specific was for the most part unaltered (Fig. 1F). Interestingly, UCP1 was induced less strongly than other brown-specific genes, and Dio2 was not induced (Fig. 1E). Recently, an alternate thermogenic futile cycle involving creatine metabolism was described in beige fat (Kazak et al. 2015). The expression of two key enzymes thought to be critical for this alternate thermogenic pathway, creatine kinase B (Ckb) and phospho1, was strongly induced in FLCN-deficient iWAT compared with controls (Fig. 1G).
In contrast to iWAT, FLCN adipKO BAT was not browner in color than BAT from controls (Supplemental Fig. 1C), and the expression of brown genes was unaltered (Supplemental Fig. 1D). BAT from FLCN adipKO mice appeared lipid-laden, as seen by H&E staining and reflected in increased gross size (Supplemental Fig. 1C), suggesting reduced lipolytic activity in BAT in the context of increased browning of WAT systemically.
Global gene expression studies using RNA sequencing (RNA-seq) of iWAT samples from wild-type and FLCN adipKO followed by gene set enrichment analysis (GSEA) demonstrated that the three gene sets most significantly induced by deletion of FLCN in WAT were oxidative phosphorylation (OXPHOS), fatty acid metabolism, and adipogenesis (Supplemental Fig. 2A). To compare the gene set induced by FLCN deletion with previously established signatures of cold-induced beigeing, we obtained publicly available microarray data sets of iWAT from cold-exposed animals (Xue et al. 2009) and identified 116 genes that were commonly induced after 1 wk and 5 wk of cold exposure. Strikingly, 33 of these cold-inducible genes were also induced in FLCN adipKO (P < 0.00001) (Supplemental Fig. 2B). We confirmed by quantitative PCR (qPCR) that the leading-edge genes from the RNA-seq and cold-inducible genes were highly induced by FLCN deletion (Supplemental Fig. 2C,D). The FLCN adipKO gene signature is thus strongly similar to a cold-inducible gene signature. Together, these data indicate that FLCN actively suppresses in WAT a browning program, perhaps including alternative thermogenic capacities.
FLCN deletion in fat leads to increased mitochondrial content and respiration in iWAT
The expression of nuclear-encoded OXPHOS genes (Cycs, Ndufv5, Cox5b, and Atp5o) was strongly induced twofold to fourfold in iWAT from FLCN adipKO mice compared with controls (Fig. 1H). Mitochondrial genes were similarly induced twofold to threefold (Fig. 1I). OXPHOS protein expression was similarly higher in FLCN adipKO mice (Supplemental Fig. 3A). Transmission electron microscopy (TEM) revealed the presence of extensive networks of mitochondria surrounding small lipid droplets in the adipKO iWAT (Fig. 1J). We next measured the respiratory capacity of isolated minced iWAT using a Clark electrode and found a near doubling of O2 consumption in FLCN-deficient iWAT compared with control (Fig. 1K). Finally, significantly more mitochondrial DNA was found in adipKO iWAT compared with control (Fig. 1L). Protein expression of OXPHOS in BAT was largely unchanged (Supplemental Fig. 3B). Thus, these data demonstrate that loss of FLCN in iWAT leads to marked mitochondrial biogenesis.
Mitochondrial biogenesis and browning of WAT are generally thought to be driven by chronic cold-induced adrenergic signals (Nedergaard and Cannon 2014). Mice under standard laboratory conditions live under relative cold exposure (Nedergaard and Cannon 2014). To test whether relative cold exposure was driving browning of WAT in the FLCN adipKO mice, the animals were housed for 2 wk at thermoneutrality (33°C) (Supplemental Fig. 4A), an exposure that normally reverses any browning seen in WAT (Waldén et al. 2012; Rosenwald et al. 2013). Even at thermoneutrality, however, brown-specific genes and mitochondrial genes remained elevated in both iWAT and eWAT of FLCN adipKO mice (Supplemental Fig. 4B,C), while BAT was not affected (Supplemental Fig. 4D). Hence, the browning pathway suppressed by FLCN in WAT is independent of cold stimulation.
To evaluate the functional consequences of elevated mitochondrial content and browning of adipose tissue, we next analyzed whole-mouse metabolic parameters of the FLCN adipKO animals. Body weight did not differ between FLCN adipKO animals and controls (Supplemental Fig. 5A). The relative amount of fat and lean mass, measured by nuclear magnetic resonance (NMR), also did not differ significantly, although a slight trend was noted toward decreased fat mass in the adipKO animals (Supplemental Fig. 5B). Basal respiratory activity and respiratory exchange ratio (RER) were also not different between the two groups (Supplemental Fig. 5C–E). Thus, at baseline, the induction of browning and mitochondrial biogenesis in FLCN adipKO mice have little impact on systemic metabolism, suggesting that the mitochondria remain coupled in vivo, unlike when the adipose tissue is removed from the mice (Fig. 1K). To test the response of FLCN adipKO animals to adrenergic stimulation, animals were anesthetized and injected subcutaneously with 1 mg/kg norepinephrine followed by measurement of respiratory activity in comprehensive laboratory animal monitoring (CLAM) chambers. FLCN adipKO animals showed dramatically higher respiration than FLCNlox/lox animals upon adrenergic agonist administration, nearly doubling their VO2 (Fig. 1M). To test the response of FLCN adipKO animals to cold, animals were challenged with acute cold exposure (4°C) followed by measurement of rectal temperature, revealing a strong trend (P < 0.07) toward resistance to acute temperature drop (Supplemental Fig. 5F). Collectively, these data thus show that FLCN deletion in fat leads to increased mitochondrial content in WAT independently of cold stimulation as well as markedly higher respiratory capacity both in isolated fat tissue and at the systemic level.
FLCN promotes cytoplasmic retention of TFE3 via mTOR
Several studies have suggested that FLCN inhibits TFE3 transcriptional activity by regulating its subcellular localization (Hong et al. 2010; Betschinger et al. 2013; Martina et al. 2014). To test this possibility in the adipocyte system, we therefore deleted FLCN in immortalized preadipocytes (isolated from iWAT) using two different methods. First, we used the CRISPR/Cas9 system to efficiently target the FLCN locus (Fig. 2A). Immunofluorescence staining of endogenous TFE3 in these cells showed a dramatic nuclear translocalization in the FLCN-deleted cells (Fig. 2B,C). The mRNA expression of GPNMB, a well-established TFE3 target gene (Hong et al. 2010), and PGC-1α was dramatically increased in FLCN-deleted cells compared with control cells (Fig. 2D), demonstrating that nuclear translocation of TFE3 leads to activation of target genes. Interestingly, PGC-1β was also highly increased in these cells, even more so than PGC-1α (Fig. 2D). FLCN deletion in vitro also induced most of the genes identified above by RNA-seq as induced by FLCN deletion in vivo (Supplemental Fig. 6A,B). In a second approach to delete FLCN, preadipocytes isolated from iWAT of FLCNlox/lox mice were immortalized, transduced with tamoxifen-inducible Cre (CreERt2), and treated with vehicle or 4-hydroxytamoxifen (4OHT) for 48 h. 4OHT treatment led to near-complete deletion of FLCN protein (Fig. 2E). As with the CRISPR cells, dramatic translocation of TFE3 to the nucleus was observed in the absence of FLCN (Fig. 2F,G) as well as induction of both PGC-1β and GPNMB (Fig. 2H). 4OHT treatment in wild-type preadipocytes did not alter the gene expression pattern of beige/brown fat genes (Supplemental Fig. 6C). In vivo, immunofluorescence staining of iWAT in FLCN adipKO and FLCNlox/lox mice revealed that TFE3 also localized to the nucleus in the absence of FLCN (Supplemental Fig. 6D). We conclude that FLCN promotes cytoplasmic retention of TFE3 in adipose cells, inhibiting the ability of TFE3 to activate its target genes.
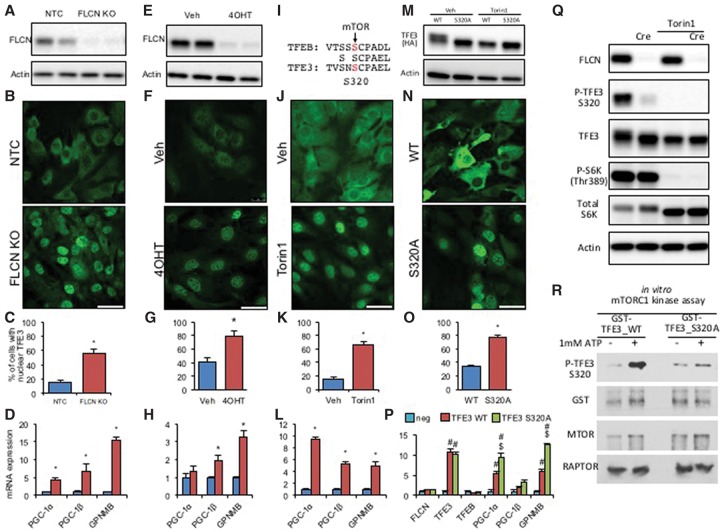
Figure 2.
FLCN deletion and mTOR inhibition promote TFE3 nuclear localization to induce TFE3 target genes in preadipocytes. (A) Western blot of FLCN in nontarget control (control) or FLCN guide RNA (gRNA) plus CRISPR/Cas9-expressing stromal vascular fraction (SVF) cells. (B) Immunofluorescence staining of TFE3 in control or FLCN-deleted SVF cells. (C) Quantification results of TFE3 nuclear localization shown in B. (D) Relative mRNA expression of TFE3 target genes in the control or FLCN-deleted SVF cell line. (E) Western blot of FLCN in vehicle (veh) or 4OHT-treated (10 μM for 48 h) CreERt2-expressing SVF cells isolated from FLCNlox/lox iWAT. (F–H) As in B–D but with FLCNlox/lox CreERt2 cells treated with vehicle versus 4OHT. (I) Schematic of the mTOR phosphorylation site in TFEB and its conservation in TFE3. (J–L) As in B–D but with cells treated with vehicle versus 250 nM mTOR inhibitor Torin1 for 1 h (J,K) or 4 h (L). (M) Western blot of stably expressed HA-tagged wild-type TFE3 or mTOR site-mutated TFE3 (S320A). Cells were treated with vehicle (veh) or Torin1 for 2 h. (N–P) As in B–D but with cells expressing TFE3 wild type or TFE3 S320A. (Q) Western blot of FLCN, phospho-TFE3 (Ser320), TFE3, phospho-S6K (Thr 389), and total S6K in FLCNlox/lox cells infected with adenovirus harboring Cre versus GFP (mock) and 250 nM Torin1 for 2 h versus vehicle. (R) In vitro mTORC1 kinase assay of TFE3: GST-fused TFE3 wild-type and S320A mutant were incubated with purified mTORC1, and phosphorylated Ser320 was detected by anti-pTFE3 (S320) antibody. (*) P < 0.05 by Student's t-test; (#) P < 0.05 versus mock; ($) P < 0.05 versus TFE3 wild type. n = 3 or 4 per group. Values are represented as mean ± SEM. Bars, 50 µm.
TFE3 is homologous to TFEB, which was reported to be phosphorylated at Ser211 by mTOR (Martina et al. 2012; Settembre et al. 2012). The Ser211 residue (S320 in mouse TFE3; S321 in human TFE3) and the adjacent sequences are largely conserved between TFEB and TFE3 (Fig. 2I). We thus reasoned that mTOR would phosphorylate TFE3 at this site, leading to sequestration of TFE3 in the cytoplasm (Martina et al. 2014). Consistent with this notion, Torin1, a potent mTOR inhibitor, completely relocalized TFE3 to the nucleus (Fig. 2J,K), leading to strong induction of TFE3 target genes PGC-1α, PGC-1β, and GPNMB (Fig. 2L). We next introduced an alanine substitution at the putative mTOR phosphorylation site of TFE3 (S320A) (Fig. 2M). Exogenous expression of this S320A mutant led to almost entirely nuclear localization of TFE3 (Fig. 2N,O) and significantly higher expression of PGC-1α, PGC-1β, and GPNMB than with wild-type TFE3 (Fig. 2P).
Interestingly, whereas exogenously expressed wild-type TFE3 reveals two distinct bands by SDS-PAGE and Western blotting, the S320A mutant reveals only the faster-migrating band (Fig. 2M). Torin1 treatment of wild-type TFE3 similarly removed the presence of the slower-migrating band (Fig. 2M), as did treatment of cell extracts with calf intestinal phosphatase (CIP) (Supplemental Fig. 6E), implying that Ser320 is phosphorylated by mTOR. To test this notion directly, we used an antibody that specifically recognizes TFE3 phosphorylated at the Ser320 position (Martina et al. 2016). Western blotting with this antibody revealed a strong signal in preadipocyte cells, indicating the presence of TFE3 phosphorylated at Ser320 (Fig. 2Q). Strikingly, Ser320 phosphorylation was nearly abolished by either FLCN deletion or Torin1 treatment (Fig. 2Q). Finally, we performed an in vitro kinase assay to test whether mTORC1 directly phosphorylates TFE3 at Ser320. A TFE3 fragment containing the amino acid sequence surrounding S320 was fused with GST and expressed in bacteria. Purified GST-TFE3 protein was then incubated with purified mTORC1 complex, and the phospho-S320 TFE3 antibody was used to detect S320 phosphorylation. We observed that, in the presence of ATP, mTORC1 robustly phosphorylates the S320 residue of TFE3. Moreover, the phosphorylation is entirely blocked by alanine substitution of the serine residue (Fig. 2R). Collectively, those results demonstrate that FLCN regulates the phosphorylation status of TFE3 at Ser320, that loss of FLCN leads to constitutive nuclear localization of TFE3 via loss of Ser320 phosphorylation, and that mTORC1 directly phosphorylates TFE3 at Ser320.
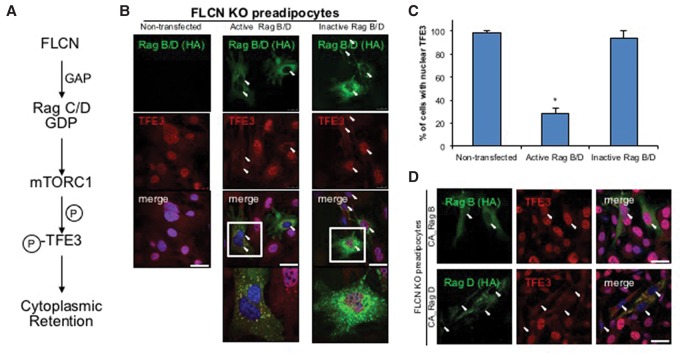
Finally, to formally test whether TFE3 nuclear localization in response to FLCN deletion requires the inhibition of mTOR (Fig. 3A), we asked whether TFE3 nuclear localization could be reversed by forced activation of mTORC1. To this end, we overexpressed constitutively active RagB and RagD in FLCN-deficient preadipocytes, in which TFE3 is constitutively nuclear. Most of the cells transfected with active RagB and RagD retained TFE3 in the cytoplasm (Fig. 3B,C). In contrast, inactive RagB/D failed to sequester TFE3 to the cytoplasm (Fig. 3B [right panels], C). On higher magnification (Fig. 3B, bottom panels), TFE3 colocalized with active RagB/D in cytoplasmic punctae (Fig. 3B, left) but not with inactive RagB/D (Fig. 3B, right). Transfection of cells with either constitutively active RagB or RagD alone revealed that RagD, but not RagB, could promote TFE3 sequestration to the cytoplasm (Fig. 3D). Together, these data demonstrate that FLCN acts via RagC/D to activate mTOR phosphorylation of TFE3 at Ser320, leading to its cytoplasmic retention and inactivation.
Figure 3.
The constitutively active Rag complex rescues TFE3 cytoplasmic retention in the absence of FLCN. (A) Schematic of mTORC1 activation by FLCN and subsequent phosphorylation of TFE3 by mTORC1. (B) Immunofluorescence staining of TFE3. Constitutively active or inactive RagB and RagD (HA-tagged) were overexpressed in FLCN-deficient preadipocytes. (Green) RagB/D (HA); (red) TFE3; (blue) DAPI. (C) Quantification result of B. (D) Constitutively active (CA) RagB or RagD was individually overexpressed in FLCN knockout preadipocytes. (Green) RagB or RagD (HA); (red) TFE3; (blue) DAPI. Arrowheads indicate nuclei of RagB/D transfected cells. (*) P < 0.05 versus nontransfected control. Values are represented as mean ± SEM. Bars, 50 µm.
Cytoplasmic retention of TFE3 and phosphorylation of S6K are separable arms of mTOR signaling
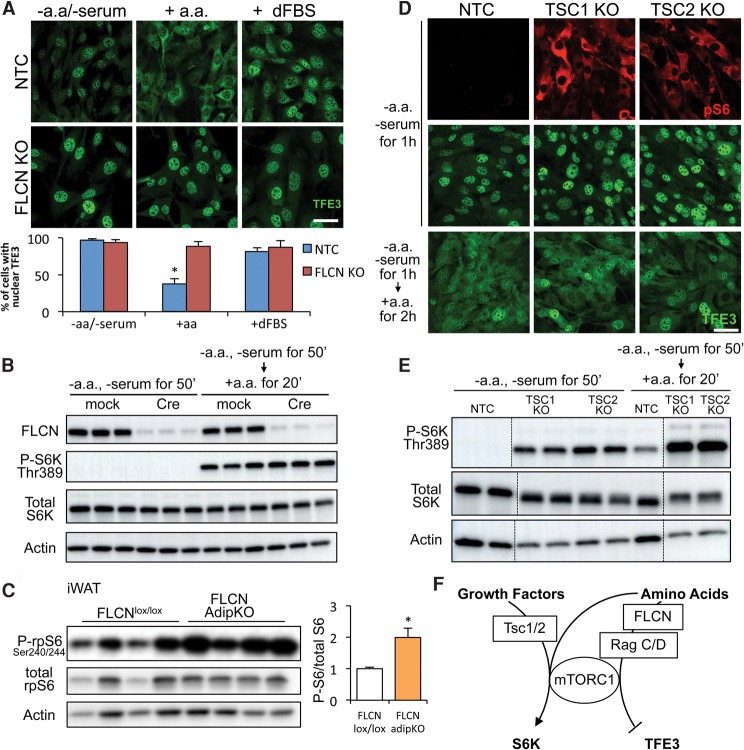
The cytoplasmic retention of TFE3 by mTOR in preadipocytes is sensitive to amino acids but not to the presence of growth factor-rich FBS dialyzed to remove amino acids (dFBS) (Fig. 4A). In the absence of FLCN, TFE3 is nuclear even in the presence of amino acids (Fig. 4A), consistent with the reported role of FLCN as a transducer of amino acid sensing (Petit et al. 2013; Tsun et al. 2013). Surprisingly, however, amino acids induced the phosphorylation of S6K even in cells lacking FLCN (Fig. 4B). Similarly, phosphorylation of rpS6, the canonical S6K substrate, was intact in adipose tissue from FLCN adipKO animals (Fig. 4C). Thus, signaling from mTOR to one of its canonical substrates remains intact in the absence of FLCN despite the complete abrogation of mTOR signaling to TFE3. To further test the notion that mTOR–S6K and mTOR–TFE3 signaling pathways are separable, we evaluated the phosphorylation of rpS6 and cellular localization of TFE3 in cells lacking TSC1 or TSC2. The absence of either TSC renders mTOR activity constitutive (Tee et al. 2002), as reflected by strong phosphorylation of rpS6 (Fig. 4D) and S6K (Fig. 4E) even in the absence of amino acids or serum. Strikingly, however, TFE3 remained nuclear in these cells (Fig. 4D). Only upon addition of amino acids did TFE3 again become cytoplasmic (Fig. 4D), and cytoplasmic retention depended on FLCN (Supplemental Fig. 7A,B). These data demonstrate that the FLCN–mTOR–TFE3 pathway described here represents a branch of mTOR signaling that is separable from the canonical TSC–mTOR–S6K branch (Fig. 4F).
Figure 4.
The FLCN–mTORC1–TFE3 pathway is separable from the canonical mTOR–S6K pathway. (A, top) Immunofluorescence staining of TFE3 in control (NTC) or FLCN knockout cells. Cells were starved in amino acid-free DMEM for 1 h or starved and restimulated with amino acids or 10% dFBS for 2 h, as indicated. (Bottom) The graph shows quantification of the percentage of cells with nuclear TFE3. (*) P < 0.05 versus NTC − amino acid/− serum. (B) Western blot to phospho-S6K Thr389. Preadipocytes isolated from FLCNlox/lox mice were infected with adenovirus harboring GFP (mock) or Cre to knock out FLCN. Cells were then starved or starved and restimulated with amino acids, as indicated. Total S6K and actin were used as loading controls. (C) Western blot to phospho-rpS6 Ser240/244 in iWAT depots isolated from FLCNlox/lox or FLCN adipKO mice. Animals were fasted overnight and refed ad lib for 4 h. Quantification of phospho-rpS6 is shown at the right, normalized to total rpS6. (*) P < 0.05 by Student's t-test. (D) Immunofluorescence staining of phospho-S6 (red) and TFE3 (green) in control (NTC) cells and cells lacking TSC1 or TSC2. Cells were starved or starved and restimulated with amino acids as indicated. TSC1 or TSC2 genes were deleted by the CRISPR/Cas9 system. (E) Western blot to phospho-S6K. Control and TSC1 or TSC2 knockout preadipocytes were starved or starved and restimulated with amino acids, as indicated (dashed lines indicate separation within the same gel). (F) Diagram of separable mTORC1 hubs. Values are represented as mean ± SEM. Bars, 50 μm.
Codeletion of TFE3 with FLCN reverses white fat browning in vivo
We next tested whether TFE3 was the dominant transcription factor downstream from FLCN in the regulation of mitochondrial biogenesis and browning in adipose tissue. To this end, we took advantage of the fact that TFE3 is X-linked and crossed the adipKO mice with female TFE3+/− mice, generating male mice lacking both FLCN and TFE3 in fat (FLCN TFE3 double knockout) and littermate controls (Fig. 5A). By H&E staining of iWAT, the appearance of multilocular adipocytes was much less frequent in FLCN TFE3 double knockout than in FLCN adipKO mice (Fig. 5B). mRNA expression of brown/beige fat genes induced in FLCN adipKO mice was nearly completely normalized by codeletion of TFE3 (Fig. 5C). The same normalization was true for nuclear-encoded mitochondrial genes and both PGC-1 coactivators (Fig. 5D), mitochondria-encoded genes (Fig. 5E), and genes of the creatine futile cycle (Fig. 5F). The induction of mitochondrial protein expression in adipKO iWAT was similarly reversed by TFE3 codeletion (Supplemental Fig. 8A,B). The expression of the top genes identified above by RNA-seq as inducible by FLCN deletion in vivo was also entirely normalized by codeletion of TFE3 (Supplemental Fig. 2C,D), further supporting the model that TFE3 is the dominant factor downstream from FLCN. These data thus provide an epistatic demonstration in vivo that FLCN in adipose tissue regulates browning and mitochondrial biogenesis via the TFE3 transcription factor.
Figure 5.
Codeletion of TFE3 with FLCN reverses the browning of white fat. (A) Breeding scheme to obtain FLCN/TFE3 double-deficient male mice. (B) H&E staining of iWAT from animals of the noted genotypes. Bar, 100 µm. (C–F) relative mRNA expression from brown/beige fat-specific genes (C), nuclear encoded mitochondrial genes (D), mitochondria-encoded mitochondrial genes (E), and creatine futile cycle genes (F). (*) P < 0.05; (**) P < 0.01 versus FLCNlox/lox. n = 8 per group. Values are represented as mean ± SEM.
PGC-1β is a direct target of TFE3 and is necessary and sufficient for browning of white fat in the absence of FLCN
We noted that PGC-1β was significantly induced by FLCN deletion in vivo, even more so than PGC-1α (Figs. 1,5). Moreover, the induction of PGC-1β by FLCN deletion was dependent on TFE3 (Fig. 5D), suggesting that TFE3 may directly regulate PGC-1β transcription. Interestingly, TFE3 ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) experiments in pluripotent stem cells incidentally identified PGC-1β as a possible TFE3 target (Betschinger et al. 2013). We found that the putative TFE3-binding region is located in the first intron of PGC-1β, a frequent site of regulatory regions. The binding region is highly conserved among mammals and contains a number of canonical E-box-binding sites (Supplemental Fig. 9A). Moreover, the region is highly sensitive to DNase cleavage and has a high content of H3K4 methylations, both reliable indicators of the presence of enhancers (Supplemental Fig. 9A). Using ChIP-qPCR, we found that TFE3 binds efficiently to this region in the adipocyte lineage (Supplemental Fig. 9B). Moreover, TFE3 binding to the PGC-1β enhancer was enriched upon FLCN deletion (Supplemental Fig. 9B), consistent with the translocation of TFE3 to the nucleus in this context. TFE3 thus binds directly to the PGC-1β gene in the adipocyte lineage.
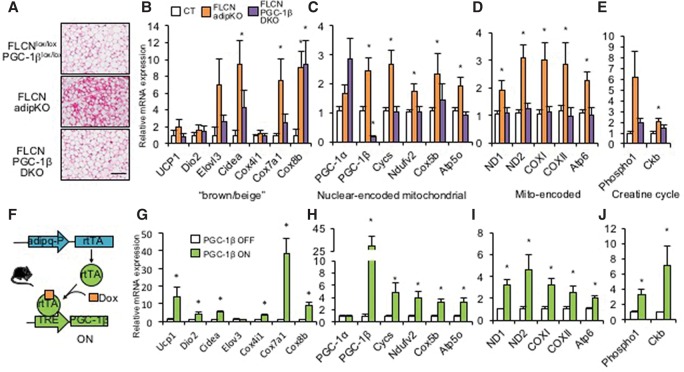
To test in vivo whether PGC-1β is required downstream from FLCN and TFE3 in the regulation of mitochondrial biogenesis and browning in adipose tissue, we used PGC-1β floxed mice (gift of Dr. Daniel Kelly) (Lai et al. 2008) to generate adipose tissue-specific FLCN and PGC-1β double-deficient animals. By H&E staining of iWAT, the appearance of multilocular adipocytes seen in FLCN adipKO mice was absent in double-knockout mice (Fig. 6A). The mRNA expression of most brown/beige fat genes induced in FLCN adipKO mice was largely normalized by codeletion of PGC-1β (Fig. 6B). Similar normalization was seen for nuclear-encoded mitochondrial genes, mitochondria-encoded genes, genes of the creatine futile cycle, and mitochondrial respiratory chain protein expression despite an apparent compensatory increase in PGC-1α expression (Fig. 6C–E; Supplemental Fig. 10A). FLCN in adipose tissue thus regulates browning and mitochondrial biogenesis via PGC-1β.
Figure 6.
PGC-1β is required for white fat browning upon FLCN deletion and is sufficient to induce browning gene expression in WAT in vivo. (A) H&E staining of iWAT from animals of the indicated genotypes. Bar, 100 µm. (B–E) Relative mRNA expression in iWAT from brown/beige fat-specific genes (B), nuclear-encoded mitochondrial genes (C), mitochondria-encoded mitochondrial genes (D), and creatine futile cycle genes (E). (*) P < 0.05 versus FLCNlox/lox, PGC-1βlox/lox. (F) Schematic of adipose tissue-specific doxycycline-inducible PGC-1β expression. (G–J) Relative mRNA expression as in B–E, respectively, in PGC-1β off versus PGC-1β on (2 wk) mice. (*) P < 0.05 versus PGC-1β off. n = 6–8 per group. Values are represented as mean ± SEM.
To test whether PGC-1β is sufficient to induce browning of white fat, we generated mice with tetracycline-inducible PGC-1β overexpression by crossing mice with PGC-1β under control of a tetracycline-responsive element (TRE-PGC-1β, a gift from Dr. Daniel Kelly) (Martin et al. 2014) with adiponectin-rtTA (adipo-rtTA, a gift from Dr. Phillip Scherer) (Wang et al. 2010) mice (Fig. 6F) and fed the mice doxycycline chow for 2 wk to induce PGC-1β expression. Transient PGC-1β overexpression strongly induced expression of brown/beige fat genes, nuclear-encoded mitochondrial genes, mitochondrial-encoded genes, and genes of the creatine futile cycle (Fig. 6G–J) as well as respiratory chain proteins (Supplemental Fig. 10B). Interestingly, multilocular adipocytes were not seen after transient induction of PGC-1β (Supplemental Fig. 10C), suggesting that induction for >2 wk is required to see these histological changes. The TFE3 target gene GPNMB was not induced by PGC-1β, consistent with the idea that PGC-1β acts downstream from TFE3. The majority of leading-edge genes identified above from the RNA-seq data as induced by FLCN deletion or cold exposure were also induced by PGC-1β in vivo (Supplemental Fig. 10D,E). Interestingly, the correlation was strongest for cold-inducible genes, suggesting that PGC-1β more specifically regulates the cold-induced subprogram of FLCN deletion.
Collectively, these data demonstrate that TFE3 directly regulates PGC-1β gene expression by binding to a conserved enhancer region in the first intron of PGC-1β and that PGC-1β is necessary and at least in part sufficient to mediate the activation of the browning/mitochondrial gene programs that occurs upon FLCN deletion.
Discussion
We describe here a novel regulatory axis of adipocyte browning. At the core of this axis is FLCN, a tumor suppressor protein originally identified as mutated in the hereditary hamartomatous BHD syndrome, a hallmark of which is the appearance of mitochondria-rich kidney tumors (Schmidt 2013). Mechanistically, we show in cell culture and with conclusive epistatic experiments in vivo that FLCN suppresses mitochondrial biogenesis via mTOR-mediated suppression of TFE3 and PGC-1β.
Separable mTOR signaling branches
Our data indicate the existence of two separable mTOR-dependent signaling pathways in adipose cells: an FLCN–mTORC1–TFE3 branch and a TSC1/2–mTORC1–S6K–S6 branch. Consistent with separable mTORC1 branches, phosphorylation of the canonical mTORC1 target S6K is preserved in both cells and adipose tissue that lack FLCN (Fig. 4B,C), while the mTOR-mediated cytoplasmic retention of TFE3 is completely lost. The same is true in cells lacking either TSC1 or TSC2 and exposed to low ambient amino acids: S6K and S6 phosphorylation is high, while TFE3 remains nuclear. Separate branches of mTOR signaling have been postulated and ascribed to different mTOR “hubs” potentially segregated in subcellular compartments (Goberdhan et al. 2016). Our data provide mechanistic evidence of such a separate branch. In the context of adipocytes, the FLCN–mTORC1–TFE3 branch responds to amino acid presence. It is possible that this process is physically separate from TSC–mTOR signaling; for example, occurring at the lysosome versus the cytoplasm. Alternatively, an FLCN–mTORC1-containing complex may be uniquely capable of phosphorylating TFE3 and not S6K.
Reconciling the role of mTOR in adipocyte browning
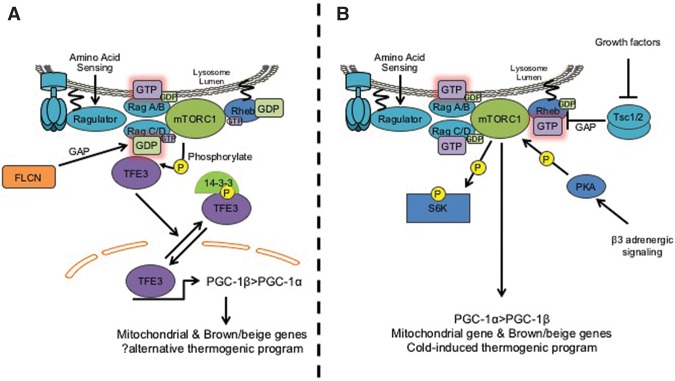
The existence of separable mTOR pathways in adipocytes helps to explain conflicting results in the literature on the role of mTOR in adipocyte browning. Loss of raptor and mTORC1 activity in fat leads to browning of WAT (Polak et al. 2008; Tran et al. 2016), while converse activation of mTOR via deletion of TSC1 leads to whitening of BAT (Xiang et al. 2015), thus suggesting that mTOR suppresses adipose browning. In contrast, however, two reports now show that loss of raptor in fat or pharmacological inhibition of mTOR dramatically blocks cold-induced browning of WAT and that mTOR is a direct target of adrenergic signaling and protein kinase A (PKA) (Liu et al. 2016; Tran et al. 2016). Thus, mTOR appears to have paradoxical both positive and negative roles in adipose browning in vivo. Here we provide a mechanistic explanation for this paradox: mTOR signaling in adipose tissue has at least two branches, one of which (the FLCN–mTOR–TFE3 branch) suppresses browning (Fig. 7A), while the other (the adrenergic mTOR branch) promotes browning (Fig. 7B). In this light, it becomes clear how loss of raptor both blocks cold-induced browning (i.e., loss of the adrenergic mTOR branch) and simultaneously increases basal levels of browning (i.e., loss of the FLCN–mTOR–TFE3 branch) (Fig. 7B), thus reconciling conflicting reports in the literature.
Figure 7.
Proposed models of two separable mTORC1 pathways. (A) Amino acid sensing in the presence of FLCN leads to activation of RagC/D, leading to recruitment of TFE3 to the Rag complex and subsequent phosphorylation by mTORC1. Phosphorylated TFE3 is bound by 14-3-3 and sequestered in the cytoplasm. In the absence of FLCN, TFE3 translocates to the nucleus and activates PGC-1β > PGC-1α and mitochondrial and brown/beige fat genes. (B) The canonical mTORC1–S6K pathway induced by amino acid or growth factor stimulation does not require FLCN, and this pathway is separable from TFE3 phosphorylation. Growth factors modulate mTORC1 activity via repression of TSC1/2, while β3 adrenergic signaling modulates mTORC1 activity via PKA-mediated phosphorylation of mTOR. The PKA–mTORC1 branch induces the classic thermogenic browning program and also leads to S6K phosphorylation but not cytoplasmic sequestration of TFE3.
Both branches of mTOR signaling in WAT terminate in part on the PGC-1 coactivators. Interestingly, the FLCN–mTOR–TFE3 branch appears to favor PGC-1β, while the adrenergic mTOR branch appears to favor PGC-1α. Loss of PGC-1β alone reverses the induction of browning by FLCN deletion in vivo, and, strikingly, PGC-1β alone is sufficient to activate a browning program in mice (Fig. 6F–J). In this context, it is also interesting to note that UCP-1 is poorly induced by FLCN deletion and that, in contrast, genes of the recently described alternative creatine futile cycle in beige adipocytes (Kazak et al. 2015) are robustly induced. Indeed, consistent with the relatively poor induction of UCP1 at baseline and the observation that PGC-1β induces more coupled mitochondria than does PGC-1α (St-Pierre et al. 2003), we found no difference in body weight or basal respiratory activity in FLCN adipKO animals, suggesting that their browned WATs are relatively coupled in the resting state. It thus may be that the FLCN–mTOR–TFE3–PGC-1β and adrenergic mTOR–S6K–PGC-1α pathways differ in the quality of tissue browning achieved.
TFE3 as a hub for adipose tissue browning
A recent study (Yan et al. 2016) also reported on the phenotype of adipose-specific deletion of FLCN using the same genetic model of adiponectin-Cre to delete FLCN in adipose cells. The study reported that FLCN deletion in fat leads to browning of adipose tissue, cold resistance, obesity resistance upon high-fat diet challenge, and improved glucose metabolism. Mechanistically, the investigators provided evidence in mouse embryonic fibroblasts (MEFs) in favor of an FLCN–AMPK pathway that directly or indirectly activates a PGC-1α/ERRα complex. Here, we provide conclusive evidence—in context-relevant cells and intact animals—for an FLCN–mTOR–TFE3–PGC-1 pathway. Both pathways may contribute to adipose browning and may in fact overlap. The mTOR–TFE3 mechanism that we describe is likely dominant because we demonstrate conclusively in vivo with mouse epistatic experiments the absolute requirement of TFE3 (and PGC-1β) for adipose browning in the context of FLCN deletion (Figs. 5,6). It is possible that the FLCN–AMPK pathway proposed by Yan et al. (2016) superimposes on this pathway by modulating mTOR/TFE3. A recent report (Young et al. 2016) showed in embryoid bodies that AMPK promotes the activity of TFEB, a molecule highly homologous to TFE3, and that this regulation of TFEB by AMPK occurs at least partly through mTOR. It is therefore possible that AMPK also activates TFE3 in adipose tissue via the same mTOR mechanism. Taken together, TFE3 therefore could be the key integrator of mTOR and AMPK signals downstream from FLCN (Supplemental Fig. 11).
We found in cell culture and epistatic experiments in vivo that TFE3 is a key transcription factor downstream from FLCN and mTOR in the regulation of the WAT browning/beigeing program and that TFE3 binds to a highly conserved, highly active chromatin region within the promoter region of PGC-1β. Interestingly, in skeletal muscle cells, TFE3 appears to preferentially induce PGC-1α (Salma et al. 2015), and PGC-1α deletion rescues the mitochondrial biogenesis induced by FLCN deletion in skeletal muscle (Hasumi et al. 2012). PGC-1α and PGC-1β may therefore have different levels of dominance downstream from the FLCN–TFE3 axis in different contexts. Compensatory induction of PGC-1α expression was observed upon PGC-1β deletion in FLCN knockout animals (Fig. 6C) but was not sufficient to reverse the phenotype conferred by PGC-1β deletion. Nevertheless, it remains likely that PGC-1α and PGC-1β have at least partially overlapping roles in adipose tissue, as elsewhere (Rowe et al. 2013). It is also noteworthy that our data indicate that the FLCN–TFE3 axis and PGC-1β govern overlapping but not identical gene programs in vivo and that this overlap was most pronounced in the mitochondrial program and cold-inducible genes, implying that PGC-1β regulates a specific branch of the FLCN–TFE3 program. MiTF/TFE family members also have redundant functionality (Steingrimsson et al. 2002), but our data indicate that, with respect to tissue browning downstream from FLCN, TFE3 dominates. Consistent with this finding, TFE3 gene expression in adipose tissue is higher than other members of the family. It remains possible, however, that heterodimers of TFE3 and TFEB (Fisher et al. 1991) or other MiTF/TFE members work together, and therefore deletion of either of them can be critical. Experiments with codeletion of TFEB and FLCN will help to answer this question.
In conclusion, we show that an FLCN–TFE3 axis drives browning of WAT, FLCN regulates subcellular localization of TFE3 via mTOR-mediated phosphorylation, the FLCN–mTOR–TFE3 axis acts independently of cold stimulation and the canonical mTOR–S6K branch of mTOR signaling, TFE3 directly regulates PGC-1β gene expression, and PGC-1β is necessary and sufficient for beige/mitochondrial gene induction in the absence of FLCN. FLCN, mTOR, and TFE3 thus form a key nexus that likely integrates metabolic cues to coordinate mitochondrial biogenesis and beigeing of WAT.
Materials and methods
Animals
Male animals (12–15 wk old) were used. All procedures of animal experiments were approved by Beth Israel Deaconess Medical Center and University of Pennsylvania Animal Care and Use Committee. FLCN floxed animals (Baba et al. 2008) were crossed with Adipoq-Cre mice (from Dr. Evan Rosen) to obtain FLCN adipKO. PGC-1β floxed animals (Lai et al. 2008) or TFE3-deficient animals (Steingrimsson et al. 2002) were bred to FLCN adipKO to obtain FLCN, PGC-1β adipose tissue-specific double-deficient animals and FLCN TFE3 double-knockout animals, respectively. The Tet-responsive PGC-1β driver (TRE-PGC-1β; a gift of Dr. Kelly, Birnham Institute) (Martin et al. 2014) was crossed with adipoq-rtTA (a gift of Dr. Philipp Scherer, University of Texas Southwestern) (Wang et al. 2010) to generate adipose tissue-specific PGC-1β inducers. All animal experiments were littermate-controlled and were performed on C57B6 and FVB or 129 mixed strains. Mice were housed in 12-h light and dark cycles and fed with standard rodent diets or special diets if specified.
Antibodies
FLCN antibody was a gift from Dr. Laura Schmidt, and phospho-TFE3 (Ser320) antibody has been described (Martina et al. 2016). Other antibodies used were as follows: TFE3 (Sigma, HPA023881), total OXPHOS rodent WB antibody cocktail (Abcam, ab110413), FLCN (Abcam, ab124885), HA tag (Cell Signaling Technology, 2367), β-actin (Cell Signaling Technology, 4970), phospho-p70S6K (Thr389) (Cell Signaling Technology, 9234), total p70S6K (Cell Signaling Technology, 2708), phospho-S6 ribosomal protein (Ser240/244) (Cell Signaling Technology, 2215), total S6 ribosomal protein (Cell Signaling Technology, 2217), and HSP90 (Cell Signaling Technology, 4877).
Gene expression analysis
mRNA expression was quantified by RT-qPCR. Total RNA was extracted by using Trizol (Thermo Fisher Scientific) for tissue samples or TurboCapture mRNA kit (Qiagen) for cells. qPCR was performed by using the SYBR Green method (iQ SYBR Green super mix), and values were normalized to HPRT, 36B4, and TBP expression by the ΔΔCt method. Primer sequences are in Supplemental Table 1.
In vitro mTORC1 kinase assay
BL21 Escherichia coli was transformed with pGEX-4T-1 encoding GST-TFE3 wild type or GST-TFE3 S320A, and GST-fused protein was purified by GST pull-down method. Amino acid sequences surrounding S320 were as follows (the serine residue or substituted alanine residue is underlined): TFE3 wild-type (TPAITVSNSCPAELPNIK) and TFE3 S320A (TPAITVSNACPAELPNIK). The transformed bacteria were grown at 28°C in 2× YTA medium containing 100 μg/mL ampicillin until A600 of 0.6–0.8 followed by 100 nM IPTG treatment for 2 h to induce protein expression. The cells were harvested by centrifugation at 6000g for 15 min at 4°C and resuspended in ice-cold PBS. The cells were lysed with sonicator followed by incubation with 1% Triton X-100 for 30 min at 4°C. Cell lysates were cleared by centrifugation at 12,000g for 10 min at 4°C and incubated with a 50% slurry of glutathione Sepharose 4B (GE Healthcare Life Sciences) for 2 h at 4°C. After rinsing three times with PBS, proteins were eluted with 1 mL of elution buffer (50 mM Tris-HCl, 10 mM reduced glutathione at pH 8.0).
Experimental details of in vitro kinase assay were described elsewhere (Yu et al. 2011). Briefly, 6xHIS-tagged raptor was transfected in Expi293 cells (Invitrogen), and lysates were run on a nickel column and eluted with 300 mM imidazole. Purified mTORC1 was concentrated, washed, and stored in 50 mM Tris (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 0.2% CHAPS, 50% glycerol, and 1 mM DTT.
Kinase assays were performed for 1 h at 37°C in 50 mM HEPES (pH 7.5), 0.01% Tween20, 1 mM EGTA, 10 mM MgCl2, 2 mM DTT, and 100 nM staurosporine (general kinase inhibitor that does not inhibit mTOR) (Tee and Proud 2001) with or without 1 mM ATP. Kinase assays were stopped in Laemmli sample loading buffer supplemented with 10 mM EDTA and heated for 5 min to 95°C.
Cell isolation from fat tissues and cell culture
iWAT depots isolated from FLCNlox/lox animals were finely minced by scissors and digested by collagenase D (Roche) and dispase II (Sigma). Digested tissue was strained by a 100-µm nylon cell strainer and centrifuged to collect stromal vascular fraction (SVF). SVF was resuspended in DMEM/F12 (Life Technologies) containing 10% FBS and plated on gelatin-coated cell culture dishes. The following day, attached cells were rinsed with PBS and infected with retroviral vector harboring SV40 TLA. Immortalized cells were selected by cell passage. Adipogenic capacity of immortalized cells was confirmed by standard adipogenesis cell culture method. Human embryonic kidney 293T (HEK293T) cells were cultured in DMEM containing 10% FBS.
Plasmids, transfection, and virus vector production
pRK5-HA GST RagB 54L (plasmid 19302), pRK5-HA GST RagB 99L (plasmid 19303), pRK5-HA GST RagD 121L (plasmid 19309), and pRK5-HA GST RagD 77L (plasmid 19308) were purchased from Addgene (Sancak et al. 2008). Plasmid transfection was performed by using Lipofectamine 3000 (Life Technologies). The TFE3 construct (mouse TFE3, HA-tagged) was a gift from Dr. David Fisher and Dr. Nancy Salma. TFE3 cDNA was transferred to pLenti CMV Puro DEST (a gift from Eric Campeau) (Addgene, plasmid 17452). Lenti transfer plasmid, psPAX2 (a gift from Didier Trono) (Addgene, plasmid 12260), and pMD2.G (a gift from Didier Trono) (Addgene, plasmid 12259) were cotransfected to HEK293T cells by Fugene HD (Promega). Seventy-two hours later, supernatant was harvested, and cell debris was removed by a low-protein-binding 0.45-µm syringe filter. Virus particles containing supernatant were stored at −80°C until use. MSCV CreERT2 puro was a gift from Dr. Tyler Jacks (Addgene, plasmid 22776). pBABE-neo largeTcDNA was a gift from Dr. Bob Weinberg (Addgene, plasmid 1780). Retrovirus particles were generated by cotransfection of retroviral transfer vector, envelope vector, and packaging vector into 293T cells. The virus-harvesting method was the same as with lentivirus vector. For lenti or retroviral infection, the spinfection method was used. Cells were plated on six-well multiwell plates, and 2 mL of virus containing supernatant and 2 mL of fresh medium were added per well. Polybrene was added to medium with a final concentration of 8 µg/mL. A six-well plate was centrifuged at 1000g for 1.5 h (spinfection), and then the medium was replaced with fresh medium. Drug selection with the appropriate marker was started 24–48 h after the spinfection.
Gene editing by the CRISPR/Cas9 system
The protocol distributed by the Zhang laboratory at Massachusetts Institute of Technology was used. lentiCRISPR version 2 was a gift from Dr. Feng Zhang (Addgene, plasmid 52961). Guide RNAs (gRNAs) specific to genes of interests were designed by using a Web-based algorithm from the Zhang laboratory (Optimized CRISPR Design, http://crispr.mit.edu). Briefly, gRNA was designed after the start codon as close as possible, and the top one or two candidates were picked up and validated by Western blot. The gRNA sequence was as follows (the pam sequence is excluded): mouse nontarget control (5′-ATTGTTCGACCGTCTACGGG-3′), mouse flcn (5′-GAGCCCTTGGTGAGTCCATA-3′), mouse tsc1 (5′-GCTCCCCAATGTTGGCTAAC-3′), and mouse tsc2 (5′-TTGATGCAATGTATTCGTCA-3′).
Western blot analysis
Tissue or cell samples were lysed by RIPA buffer containing phosphatase inhibitor (PhosSTOP, Roche) and proteinase inhibitor (Complete miniproteinase inhibitor cocktail, Roche). Samples were sonicated followed by centrifugation to remove insoluble debris or lipid. Protein concentration was quantified by BCA protein assay kit (Thermo Fisher Scientific), and the same amount of protein (10–20 µg) was loaded to 4%–20% gradient Tris-glycine polyacrylamide gel (Bio-Rad) and electrophoresed (SDS-PAGE). After the SDS-PAGE, protein samples were transferred to PVDF membrane (Millipore). Membranes were blocked by 5% skim milk for 1 h and then incubated with the respective primary antibodies. Appropriate HRP-conjugated secondary antibodies were selected according to the host species of primary antibodies, and images were taken by using a digital imager (GE Healthcare Life Sciences, ImageQuant LAS 4000). The result was quantified by using ImageJ.
Physiology assay protocols
Body composition of animals was analyzed by NMR at the Animal Physiology Core, Division of Endocrinology, Beth Israel Deaconess Medical Center. Mice were individually housed in cages, and their metabolic physiology (VO2, VCO2, and RER) was monitored by comprehensive laboratory animal monitoring system (CLAMS) at the Animal Physiology Core, Division of Endocrinology, Beth Israel Deaconess Medical Center.
Respiration assay of isolated adipose tissues
Isolated iWAT (20–25 mg) was weighed, minced by curved scissors, and resuspended in warm respiration buffer (2% bovine serum albumin [BSA], 0.45% glucose, 10 mM sodium pyruvate in PBS). Next, resuspended tissue was applied to the Clark electrode chamber (Strathkelvin Instruments) to measure O2 consumption. Values were normalized to tissue weight.
Histology analysis and immunofluorescence staining of cells
Isolated tissues were fixed in 3.7% formaldehyde for 24–48 h followed by dehydration steps. Tissues were embedded in paraffin and sectioned, and then H&E staining was performed. Stained sections were examined under light microscope, and images were captured. Cells were plated on the glass slips coated with gelatin and fibronectin. Twenty-four hours to 48 h after the plating, cells were serum- and nutrient-starved in Earle's balanced salt solution for 1 h followed by 1 h of serum stimulation with 10% FBS in DMEM/F12 medium (Life Technologies). Torin1 was added to the medium in a 250 nM final concentration. Cells were fixed with 3.7% formaldehyde in PBS and permeabilized with 0.3% Triton X/PBS (PBST). Blocking and primary antibody incubation were performed in PBST containing 5% normal goat serum and 1% BSA overnight followed by secondary antibody incubation. Pictures of the cells were taken via confocal microscopy.
In vivo respiration assay
Animals were individually housed in metabolic chambers, and basal respiratory activity was measured in the Mouse Phenotyping Core at the University of Pennsylvania (directed by Dr. Rexford Ahima). Next, animals were sedated by intraperitoneal injection of pentobarbital followed by subcutaneous injection of 1 mg of norepinephrine per kilogram of bodyweight (with or without norepinephrine plus bitartrate salt from Sigma Aldrich). Respiration was measured up to 2 h after norepinephrine injection.
RNA-seq analysis
FastQ files were aligned against the mouse reference genome (mm9) using the STAR aligner (Dobin et al. 2013). STAR was used to also compute the gene counts for individual genes using the Ensembl (version 67) gene annotations. Gene counts, represented as counts per million (CPM), were first nominalized using the TMM method in the edgeR R package, and genes with 25% of samples with a CPM <1 were deemed as low-expressed and removed from further analysis. Data were transformed using the VOOM function from the Limma package (Law et al. 2014). Differential gene expression was performed using a linear model with the Limma package. GSEA was performed using the CAMERA gene set test procedure (Wu and Smyth 2012) with the Hallmark gene set from the Molecular Signature Database. The data set can be found in the Gene Expression Omnibus (GEO; accession number GSE89762).
Transcriptome comparisons
Raw microarray data of iWAT from cold-exposed animals was obtained from the Gene Omnibus Expression (GSE1343) (Xue et al. 2009), and a differential gene expression analysis was performed using the R packages Oligo and Limma. Gene sets of the 1-wk/4°C and 5-wk/4°C comparison were created from the up-regulated genes (P < 0.05 and log fold change >1.5). To determine whether the intersection of the 170 up-regulated FLCN adipKO genes with 1-wk/4°C and 5-wk/4°C was significant, an empirical P-value was obtained from the permutation test. Briefly, a random sample of 170 genes was obtained from all of the expressed genes (n) and compared with up-regulated genes in 1-wk/4°C and 5-wk/4°C 100,000 times to create a distribution of random overlaps. The empirical P is the probability of obtaining 33 genes in a random distribution.
Statistical analysis
Values are presented as mean ± SEM. Student's t-test (two-tailed unpaired t-test) was performed for single comparison. For the comparison of the groups, more than three one-way analyses of variance (ANOVAs) were performed, followed by a t-test (Bonferroni correction). For statistical analysis of in vivo respiration assay, two-way ANOVA was used. P < 0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Bridget Gosis for technical support and critical discussion; Dr. Antonio Davila Jr. for expert advice; Dr. Eleftheria Maratos-Flier in Beth Israel Deaconess Medical Center/Harvard Medical School for CLAMS and DEXA analyses; Dr. Rexford Ahima, Dr. Frederick Anokye-Danso, and the Diabetes Research Center (DRC) Mouse Phenotyping Core at the University of Pennsylvania for in vivo respiration assay (P30-DK19525); Andrea Stout and Jasmine Zhao from the Cell and Developmental Biology Microscopy Core at the University of Pennsylvania for confocal microscopy imaging; Raymond Meade from the Electron Microscopy Resource laboratory at the Perelman School of Medicine, University of Pennsylvania, for TEM imaging; and Lan Cheng from the Histology and Gene Expression Core at the University of Pennsylvania Cardiovascular Institute for histology sample processing. S.W. is supported by an American Diabetes Association Postdoctoral Fellowship Award (1-16-PDF-117) and was supported by the Toyobo Biotechnology Foundation. M.N. is supported by a University of Pennsylvania Cell and Molecular Biology training grant (T32 GM-07229). G.L. is supported by the LAM Foundation, the Tuberous Sclerosis Alliance, and the National Research Foundation of Korea. J.R. was supported by grants from the National Institutes of Health [T32GM007592] and a Mentored Research Training Grant from the Foundation for Anesthesia Education and Research. J.A.M. and R.P. were supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute of the National Institutes of Health. J.B. is supported by the National Institutes of Health (GM51405 and HL121266). J.A.B. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (DK098656) and National Institute on Aging (AG043483). Z.A. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (DK107667) and National Heart, Lung, and Blood Institute (HL094499). S.W. designed and conducted the experiments, analyzed/interpreted the data, and wrote the manuscript. C.J. and M.N. conducted experiments and analyzed data. Y.H.I., G.L., and J.A.B conducted the in vitro kinase assay and gave intellectual input on mTOR signaling. A.B. and M.M. performed bioinformatics analysis on RNA-seq data. J.L. and A.H. generated critical reagents. J.A.M. and R.P. generated the phospho-TFE3 (Ser320)-specific antibody and gave intellectual input on TFE3 biology. G.C.R., J.R., J.A.B., and P.S. assisted in critical discussion and study design. Z.A. designed the study, interpreted the data, and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.287953.116.
References
- Baba M, Furihata M, Hong S-B, Tessarollo L, Haines DC, Southon E, Patel V, Igarashi P, Alvord WG, Leighty R, et al. 2008. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 and Akt–mTOR activation, cell hyperproliferation, and polycystic kidneys. J Natl Cancer Inst 100: 140–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. 2013. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153: 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. 2013. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol 15: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DE, Carr CS, Parent LA, Sharp PA. 1991. TFEB has DNA-binding and oligomerization properties of a unique helix–loop–helix/leucine-zipper family. Genes Dev 5: 2342–2352. [DOI] [PubMed] [Google Scholar]
- Goberdhan DCI, Wilson C, Harris AL. 2016. Amino acid sensing by mTORC1: intracellular transporters mark the spot. Cell Metab 23: 580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, et al. 2013. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 23: 302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M, Seale P. 2013. Brown and beige fat: development, function and therapeutic potential. Nat Med 19: 1252–1263. [DOI] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hasumi Y, Huang Y, Oh H, Hughes RM, Klein ME, Takikita S, Nagashima K, Schmidt LS, et al. 2012. Regulation of mitochondrial oxidative metabolism by tumor suppressor FLCN. J Natl Cancer Inst 104: 1750–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi Y, Baba M, Hasumi H, Huang Y, Lang M, Reindorf R, Oh H-B, Sciarretta S, Nagashima K, Haines DC, et al. 2014. Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum Mol Genet 23: 5706–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-B, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM. 2010. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One 5: e15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, Vetrivelan R, Lu GZ, Laznik-Bogoslavski D, Hasenfuss SC, et al. 2015. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. 2008. Transcriptional coactivators PGC-lα and PGC-lβ control overlapping programs required for perinatal maturation of the heart. Genes Dev 22: 1948–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Bordicchia M, Zhang C, Fang H, Wei W, Li J, Guilherme A, Guntur K, Czech MP, Collins S. 2016. Activation of mTORC1 is essential for β-adrenergic stimulation of adipose browning. J Clin Invest 126: 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin OJ, Lai L, Soundarapandian MM, Leone TC, Zorzano A, Keller MP, Attie AD, Muoio DM, Kelly DP. 2014. A role for peroxisome proliferator-activated receptor γ coactivator-1 in the control of mitochondrial dynamics during postnatal cardiac growth. Circ Res 114: 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, Puertollano R. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7: ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Brady OA, Puertollano R. 2016. TFEB and TFE3 are novel components of the integrated stress response. EMBO J 35: 479–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J, Cannon B. 2014. The browning of white adipose tissue: some burning issues. Cell Metab 20: 396–407. [DOI] [PubMed] [Google Scholar]
- Petit CS, Roczniak-Ferguson A, Ferguson SM. 2013. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 202: 1107–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Cybulski N, Feige JN, Auwerx J, Rüegg MA, Hall MN. 2008. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab 8: 399–410. [DOI] [PubMed] [Google Scholar]
- Rehli M, Lichanska A, Cassady AI, Ostrowski MC, Hume DA. 1999. TFEC is a macrophage-restricted member of the microphthalmia-TFE subfamily of basic helix–loop–helix leucine zipper transcription factors. J Immunol 162: 1559–1565. [PubMed] [Google Scholar]
- Rosenwald M, Perdikari A, Rülicke T, Wolfrum C. 2013. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol 15: 659–667. [DOI] [PubMed] [Google Scholar]
- Rowe GC, Patten IS, Zsengeller ZK, El-Khoury R, Okutsu M, Bampoh S, Koulisis N, Farrell C, Hirshman MF, Yan Z, et al. 2013. Disconnecting mitochondrial content from respiratory chain capacity in PGC-1-deficient skeletal muscle. Cell Rep 3: 1449–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salma N, Song JS, Arany Z, Fisher DE. 2015. Transcription factor Tfe3 directly regulates Pgc-1α in muscle. J Cell Physiol 230: 2330–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. 2009. A gene network regulating lysosomal biogenesis and function. Science 325: 473–477. [DOI] [PubMed] [Google Scholar]
- Schmidt LS. 2013. Birt-Hogg-Dubé syndrome: from gene discovery to molecularly targeted therapies. Fam Cancer 12: 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoag J, Haq R, Zhang M, Liu L, Rowe GC, Jiang A, Koulisis N, Farrel C, Amos CI, Wei Q, et al. 2013. PGC-1 coactivators regulate MITF and the tanning response. Mol Cell 49: 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA. 2002. Mitf and Tfe3, two members of the Mitf–Tfe family of bHLH-zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci 99: 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. 2003. Bioenergetic analysis of peroxisome proliferator-activated receptor γ coactivators 1α and 1β (PGC-1α and PGC-1β) in muscle cells. J Biol Chem 278: 26597–26603. [DOI] [PubMed] [Google Scholar]
- Tee AR, Proud CG. 2001. Staurosporine inhibits phosphorylation of translational regulators linked to mTOR. Cell Death Differ 8: 841–849. [DOI] [PubMed] [Google Scholar]
- Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. 2002. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci 99: 13571–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran CM, Mukherjee S, Ye L, Frederick DW, Kissig M, Davis JG, Lamming DW, Seale P, Baur JA. 2016. Rapamycin blocks induction of the thermogenic program in white adipose tissue. Diabetes 65: db150502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini D. 2013. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 52: 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldén TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. 2012. Recruited vs. nonrecruited molecular signatures of brown, ‘brite,’ and white adipose tissues. Am J Physiol Endocrinol Metab 302: E19–E31. [DOI] [PubMed] [Google Scholar]
- Wang ZV, Deng Y, Wang QA, Sun K, Scherer PE. 2010. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology 151: 2933–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Smyth GK. 2012. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res 40: e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Lan H, Tang H, Yuan F, Xu Y, Zhao J, Li Y, Zhang W. 2015. Tuberous sclerosis complex 1-mechanistic target of rapamycin complex 1 signaling determines brown-to-white adipocyte phenotypic switch. Diabetes 64: 519–528. [DOI] [PubMed] [Google Scholar]
- Xue Y, Petrovic N, Cao R, Larsson O, Lim S, Chen S, Feldmann HM, Liang Z, Zhu Z, Nedergaard J, et al. 2009. Hypoxia-independent angiogenesis in adipose tissues during cold acclimation. Cell Metab 9: 99–109. [DOI] [PubMed] [Google Scholar]
- Yan M, Audet-walsh É, Manteghi S, Dufour CR, Walker B, Baba M, St-pierre J, Giguère V, Pause A. 2016. Chronic AMPK activation via loss of FLCN induces functional beige adipose tissue through PGC-1 α/ERRα. Genes Dev 30: 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NP, Kamireddy A, Van Nostrand JL, Eichner LJ, Shokhirev MN, Dayn Y, Shaw RJ. 2016. AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes Dev 30: 535–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon S-O, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. 2011. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332: 1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.