Abstract
Oxidative phosphorylation dysfunction has been found in many different disorders. This biochemical pathway depends on mitochondrial protein synthesis. Thus, mutations in components of the mitochondrial translation system can be responsible for some of these pathologies. We identified a new homozygous missense mutation in the mitochondrial translation elongation factor Ts gene in a patient suffering from slowly progressive childhood ataxia and hypertrophic cardiomyopathy. Using cell, biochemical and molecular-genetic protocols, we confirm it as the etiologic factor of this phenotype. Moreover, as an important functional confirmation, we rescued the normal molecular phenotype by expression of the wild-type TSFM cDNA in patient’s fibroblasts. Different TSFM mutations can produce the same or very different clinical phenotypes, going from abortions to moderately severe presentations. On the other hand, the same TSFM mutation can also produce same or different phenotypes within the same range of presentations, therefore suggesting the involvement of unknown factors.
Introduction
The oxidative phosphorylation system (OXPHOS) is a biochemical pathway involved in many key cellular processes. This system includes electron transport chain respiratory complexes I–IV (CI–CIV) and ATP synthase (CV). OXPHOS complexes contain 13 mitochondrial DNA (mtDNA)-encoded polypeptides. Moreover, mtDNA codes for 22 transfer and 2 ribosomal RNAs required for the expression of these polypeptides. Mitochondrial translation also depends on many nuclear DNA-encoded proteins. Mutations in many of these translation components are associated to different pathologic conditions.1
Case Report
The patient is a 23-year-old male born at term from non-consanguineous Galician parents. Ethical approval was obtained from involved Institutional Review Boards. Written informed consents from the patient and the family members were also obtained. He has an elder healthy sister and his mother suffered three abortions prior to her pregnancy. Pregnancy and delivery were uneventful with normal early psychomotor development. Since the age of 6 years, gross- and fine-motor clumsiness, tiptoe gait and frequent falls were observed. Two years later, lack of coordination and reduction of visual acuity with poor school performance were detected. Brain MRI was then normal. A nonobstructive hypertrophic cardiomyopathy without functional repercussion was found at 11 years old. Dystonic movements in hands appeared, and progressively facial and upper-limb dystonia with increasing clumsiness and dysarthric speech emerged at 14 years old. Metabolic screening, lactate in blood and CSF were normal. Brain MRI showed signal intensity mainly on both lenticular nuclei, and to a lesser extent, in the right caudate nucleus (in T2-weighted images). Electromyography was normal. By electroneurography, the lower limbs’ motor conduction velocity was normal and sensory amplitude decreased. Disintegration of somatosensory-evoked potentials of lower extremities was also observed. At present, he has stable hypertrophic cardiomyopathy on medication, normal cognitive function, axial ataxia with dysarthric speech, mild optic atrophy, and frequent dystonic movements in his face, neck and shoulder region.
A muscle biopsy at the age of 16 showed red granular material with Gomori trichrome staining, and measurements of electron transport chain enzyme activities by spectrophotometry showed partial CI and almost complete CIV deficiencies, with increment in CII activity.
Results
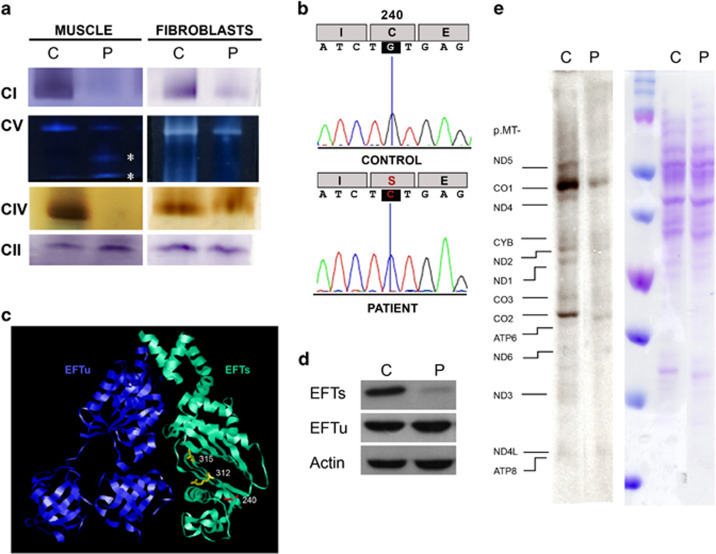
To confirm this biochemical defect, we analyzed the BN-PAGE in-gel activity in muscle and skin fibroblasts.2 An almost complete absence of muscle CI, CIV and CV in-gel activity was observed (Figure 1a). Moreover, CV sub-complexes were also detected. A moderate increase in CII in-gel activity corroborated the spectrophotometric result described in the case report. Although less pronounced than muscle fibers, cultured fibroblasts also showed a defect in CI, CIV and CV in-gel activities (Figure 1a).
Figure 1.
OXPHOS characteristics. (a) OXPHOS complex (CI, CII, CIV and CV) in-gel activities from muscle and fibroblasts of patient (P) and control (C). Asterisks indicate CV sub-complexes. (b) TSFM cDNA sequence obtained by Sanger method. (c) Structural modeling of EFTu–EFTs dimer. The locations of EFTs amino acids that have been previously associated with missense pathologic mutations are showed in yellow color, and the new one, here reported, is shown in red color. (d) EFTs and EFTu immunoblots. (e) Mitochondrial translation products. Gels show loading controls (right) and electrophoretic patterns of mitochondrial translation products (left) from patient (P) and control (C) fibroblasts. Gels of this figure have been cropped to improve the clarity and conciseness of the presentation. See Supplementary Figure S3 for uncropped gel images.
This deficiency in several OXPHOS complexes with mtDNA-encoded subunits suggested a defect on the organelle genome. DNA of all available family members was isolated from peripheral white blood cells and from the patient muscle. However, mtDNA depletion and mutations were ruled out. Simultaneous sequencing in MiSeq platform (Illumina, San Diego, CA, USA) of the coding regions of 150 nuclear genes for mitochondrial proteins previously associated with OXPHOS defects was performed after an in-solution hybridization enrichment method with a custom Sure Select XT kit (Agilent, Santa Clara, CA, USA). A heterozygous variant [c.385C>T (p.(Pro129Ser))] in the NDUFS1 gene (NG_009248.1; NM_005006.6; NP_004997.4) was observed. We also found a c.719G>C homozygous transversion in TSFM gene (OMIM *604723; NG_016971.1; GRCh38 Chr12:57,796,324; NM_005726.5) exon 6 (Figure 1b). This exon, numbered like in Ahola et al,3 was also amplified and resequenced using the Sanger method in both parents and a healthy sister, and the change was found heterozygous. This variant (http://databases.lovd.nl/shared/individuals/00063251) has been previously reported only once in SNPdb (http://www.ncbi.nlm.nih.gov/projects/SNP/rs750799705, 11 September 2015) in a Galician heterozygous individual (http://www.ciberer.es/bier/CSVS/).
The c.719G>C variant is predicted to change a cysteine to serine at amino acid position 240 of the EFTs protein (NP_005717.3), that is conserved in 99 (63.9%) out of 155 animal species analyzed (GenBank, 11 September 2015; Supplementary Figure S1). The three-dimensional structure of the bovine elongation factor Tu (EFTu)–EFTs dimer (PDB 1XB2)4 was obtained with the RasMol 2.6 program (http://www.rasmol.org). The cysteine 240 is located in EFTs subdomain C β5 strand from the core domain. This subdomain interacts with the domain III of mitochondrial translation EFTu. However, the (p.(Cys240Ser)) substitution is positioned in the other side of the β-sheet and it does not affect the contact surface with EFTu (Figure 1c). Western blots for these proteins were performed using anti-TSFM (EFTs) (1:1000, AV38672, Sigma, St Louis, MO, USA), anti-TUFM (EFTu) (1:1000, SAB1406560, Sigma) and anti-actin (1:2000, A2066, Sigma) as primary antibodies.5 This mutation provokes an important decrease in the EFTs levels, but the EFTu amount is not affected (Figure 1d). A mitochondrial protein synthesis analysis6 also showed an important decrease in the levels of mtDNA-encoded polypeptides (Figure 1e).
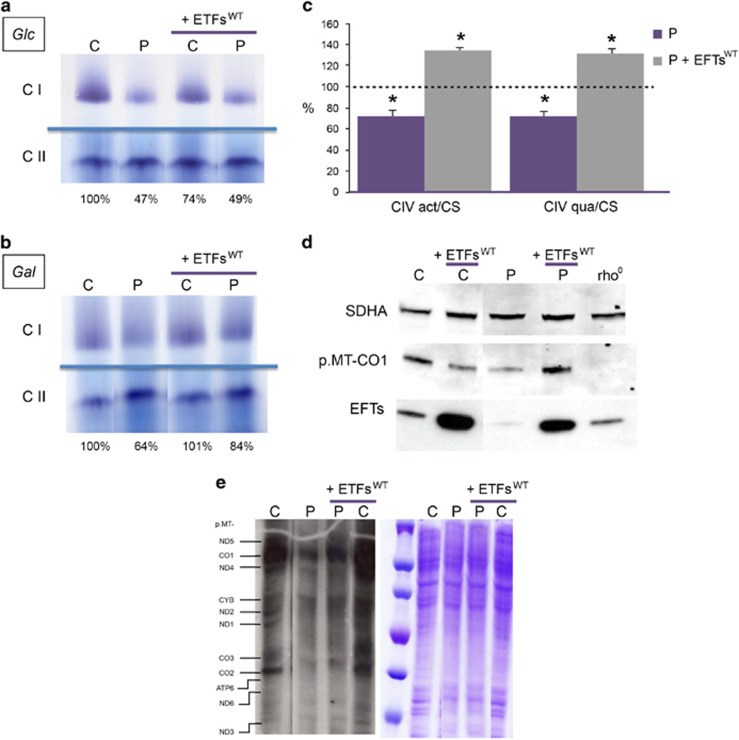
To confirm that this TSFM mutation was responsible of the phenotype, we tried to recover the mitochondrial translation capacity by transfecting mutant fibroblasts with the human wild-type TSFM cDNA (Supplementary Figure S2). These cell lines were grown in high-glucose (25 mm) DMEM supplemented with fetal bovine serum 10%. CI and CII in-gel activities were determined (Figure 2a). Compared with wild-type fibroblasts, CI/CII ratios were decreased in mutant fibroblast, both in transfected and untransfected cells. To make cells more dependent on OXPHOS function,7, 8 fibroblasts were grown in DMEM-no glucose supplemented with galactose (5 mm) and fetal bovine serum 10%. In galactose medium, CI/CII ratios were also decreased in mutant cells, but they were partially recovered in transfected cells (Figure 2b). EFTs transfection does not increase CI activity in transfected control fibroblasts grown in high-glucose or galactose medium. Because this transfection does not have effect on mutant fibroblasts grown in glucose medium, the partial recovery of CI activity in transfected mutant fibroblasts grown in galactose medium seems to be a specific effect. In high-glucose, wild-type and mutant cells obtain energy from glycolysis. In galactose, CI activity will not largely change in control cells because they are able to obtain energy in an efficient way. On the contrary, mutant cells will not fulfill energy requirements, and mitochondrial signals will be generated to increase mitochondrial biogenesis and CI activity. The in-gel activity technique is only semiquantitative. When a quantitative method (ELISA) for CIV determination was applied,6 a significant decrease in CIV activity and quantity was observed in mutant fibroblasts (Figure 2c). Interestingly, the wild-type EFTs overexpression in mutant fibroblasts recovers CIV activity and quantity (Figure 2c), and p.MT-CO1 levels (Figure 2d), a CIV subunit involved in CIV assembly and activity. Moreover, and taking into consideration the loading control, the levels of other mtDNA-encoded polypeptides also seem partially recovered (Figure 2e).
Figure 2.
Wild-type EFTs overexpression (+EFTswt) in patient fibroblasts recovers OXPHOS parameters. CI and CII in-gel activities from control (C) and patient (P) fibroblasts grew in high-glucose (25 mm) (a) and galactose (5 mm) medium (b). The control CI/CII ratios are considered 100%. (c) CIV activity/citrate synthase activity (CIV act/CS) and CIV quantity/CS activity (CIV qua/CS) ratios. Dot line represents mean values of untransfected and transfected control fibroblasts. Bars indicate the mean values (and SDs, n=3) of untransfected (P) and transfected (P+EFTsWT) mutant fibroblasts. Asterisks denote statistically significant P-values (mutant vs control) (unpaired t-test) (d) p.MT-CO1 immunoblot. An immunoblot for the nDNA-encoded CII subunit, SDHA, is also shown as a loading control. EFTs immunoblots confirm the overexpression of this translation factor in the transfected cells. Rho0 indicates cells with no mtDNA and, therefore, without the mtDNA-encoded p.MT-CO1. (e) Mitochondrial translation products. Gels show loading controls (right) and electrophoretic patterns of mitochondrial translation products (left) from patient (P) and control (C) fibroblasts. Gels of this figure have been cropped to improve the clarity and conciseness of the presentation. See Supplementary Figure S3 for uncropped gel images.
Discussion
Massive sequencing approaches have largely increased the possibility of false diagnoses, that is, the consideration of a non-disease-causing variant as a pathogenic mutation. These errors would make difficult the identification of the real disease causations and would have a huge impact on patients’ care, on other family members who test negative or positive for the false-positive variants and on future family planning.9 Therefore, performing functional analysis on relevant cells and validating the effect of variants by restoring the phenotype after complementing the genetic deficiency are fundamental approaches to define pathogenicity. In our case, the deficiency in activity of several respiratory complexes with mtDNA-encoded subunits, the low p.MT-CO1 levels, the altered mitochondrial protein synthesis, the EFTs absence in patient fibroblasts and the correction of the biochemical defect by overexpressing the wild-type EFTs confirm the EFTs mutation as the etiologic factor explaining the phenotype of this patient.
Two other previously described EFTs pathologic missense mutations c.997C>T (p.(Arg312Trp)) and c.944G>A (p.(Cys315Tyr)) also located in subdomain C, but in the β6 strand, affect the β-sandwich core, especially the contact surface for interaction with EFTu (Figure 1c), and the levels of both factors are decreased (Supplementary Table S1).3, 10 Seven patients from five different pedigrees harbored, in homozygosity, a EFTs (p.(Arg312Trp)) amino acid substitution. Despite pulse labeling of mitochondrial translation products only revealed a moderate reduction in mitochondrial protein synthesis, all of these patients died of encephalomyopathy, hypertrophic cardiomyopathy or liver failure before the age of 2 months.10, 11, 12 Surprisingly, the patient here reported, presenting ataxia and hypertrophic cardiomyopathy, was alive at the age of 23 years. His mutation also decreased the EFTs levels, but the EFTu levels were normal. However, mitochondrial translation was greatly decreased. Very interestingly, it has recently been described an individual with low EFTs but normal EFTu levels and a clear reduction in mitochondrial translation (Supplementary Table S1).3 This patient harbored compound heterozygous EFTs mutations c.[856C>T][106+4A>G]. He presented ataxia, optical and peripheral neuropathy, and was also alive at the age of 21 years, but he did not show cardiomyopathy.3 In a different pedigree, two sisters harbored (p.(Cys315Tyr))/(p.(Gln286Ter)) mutations, suffered from hypertrophic cardiomyopathy and survived for >15 years. These patients had low levels of both EFTs and EFTu, but mitochondrial translation was partially decreased in one of them and unaffected in the other one.3 Thus, there is no a clear correlation between EFTs/EFTu levels and mitochondrial protein synthesis. Apparently, a large decrease in EFTu levels must provoke a compensatory response that, in same degree, corrects the mitochondrial translation defect. This defect is not corrected when EFTu levels are not highly decreased. Although differences in EFTu levels or mitochondrial translation are not related with the affected organs and do not seem to be the reason for an extended survival, low EFTs and EFTu levels appear to be associated with an early age of onset.
More interestingly, it has been proposed that TSFM mutations can provoke early developmental lethality.3 Thus, the three miscarriages reported in this family could also be owing to this TSFM mutation found in the index case, although we did not have the opportunity to study these cases. Other factors would be responsible for the marked differences between these four cases.
In summary, all data presented in this manuscript confirm that the TSFM mutation here reported is the etiologic factor of the disease.
Acknowledgments
We thank the patient and his family for participating in this study. We also thank Santiago Morales for his assistance with the figures and María Victoria Sebastián for her help in the laboratory. This work was supported by grants from Instituto de Salud Carlos III (FIS-PI13/02177, PI14/00005, PI14/00028 and PI14/00070); Departamento de Ciencia, Tecnología y Universidad del Gobierno de Aragón (Grupos Consolidados B33) and FEDER Funding Program from the European Union; and Asociación de Enfermos de Patología Mitocondrial (AEPMI). The CIBERER is an initiative of the ISCIII.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Boczonadi V, Horvath R: Mitochondria: impaired mitochondrial translation in human disease. Int J Biochem Cell Biol 2014; 48: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Karas M, Schagger H: High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 2007; 6: 1215–1225. [DOI] [PubMed] [Google Scholar]
- Ahola S, Isohanni P, Euro L et al: Mitochondrial EFTs defects in juvenile-onset Leigh disease, ataxia, neuropathy, and optic atrophy. Neurology 2014; 83: 743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen MG, Navratil T, Spremulli LL, Nyborg J: Crystal structure of the bovine mitochondrial elongation factor Tu.Ts complex. J Biol Chem 2005; 280: 5071–5081. [DOI] [PubMed] [Google Scholar]
- Lopez-Gallardo E, Solano A, Herrero-Martin MD et al: NARP syndrome in a patient harbouring an insertion in the MT-ATP6 gene that results in a truncated protein. J Med Genet 2009; 46: 64–67. [DOI] [PubMed] [Google Scholar]
- Gomez-Duran A, Pacheu-Grau D, Lopez-Gallardo E et al: Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet 2010; 19: 3343–3353. [DOI] [PubMed] [Google Scholar]
- Weber K, Ridderskamp D, Alfert M, Hoyer S, Wiesner RJ: Cultivation in glucose-deprived medium stimulates mitochondrial biogenesis and oxidative metabolism in HepG2 hepatoma cells. Biol Chem 2002; 383: 283–290. [DOI] [PubMed] [Google Scholar]
- Valente L, Tiranti V, Marsano RM et al: Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EFTu. Am J Hum Genet 2007; 80: 44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shen Y: When a "disease-causing mutation" is not a pathogenic variant. Clin Chem 2014; 60: 711–713. [DOI] [PubMed] [Google Scholar]
- Smeitink JA, Elpeleg O, Antonicka H et al: Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am J Hum Genet 2006; 79: 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Compton AG, Hershman SG et al: Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med 2012; 4: 118ra10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedrenne V, Galmiche L, Chretien D, de Lonlay P, Munnich A, Rotig A: Mutation in the mitochondrial translation elongation factor EFTs results in severe infantile liver failure. J Hepatol 2012; 56: 294–297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.