Abstract
Recent advances in next-generation sequencing strategies have led to the discovery of many novel disease genes. We describe here a non-consanguineous family with two affected boys presenting with early onset of severe axonal neuropathy, optic atrophy, intellectual disability, auditory neuropathy and chronic respiratory and gut disturbances. Whole-exome sequencing (WES) was performed on all family members and we identified compound heterozygous variants (c.[760C>A][1528G>C]p.[(Gln254Lys);(Ala510Pro)] in the polyribonucleotide nucleotidyltransferase 1 (PNPT1) gene in both affected individuals. PNPT1 encodes the polynucleotide phosphorylase (PNPase) protein, which is involved in the transport of small RNAs into the mitochondria. These RNAs are involved in the mitochondrial translation machinery, responsible for the synthesis of mitochondrially encoded subunits of the oxidative phosphorylation (OXPHOS) complexes. Both PNPT1 variants are within highly conserved regions and predicted to be damaging. These variants resulted in quaternary defects in the PNPase protein and a clear reduction in protein and mRNA expression of PNPT1 in patient fibroblasts compared with control cells. Protein analysis of the OXPHOS complexes showed a significant reduction in complex I (CI), complex III (CIII) and complex IV (CIV). Enzyme activity of CI and CIV was clearly reduced in patient fibroblasts compared with controls along with a 33% reduction in total mitochondrial protein synthesis. In vitro rescue experiments, using exogenous expression of wild-type PNPT1 in patient fibroblasts, ameliorated the deficiencies in the OXPHOS complex protein expression, supporting the likely pathogenicity of these variants and the importance of WES in efficiently identifying rare genetic disease genes.
Introduction
Advances in next-generation sequencing strategies, particularly whole-exome sequencing (WES), have provided the opportunity to obtain a clearer picture of the genetic basis for many human diseases. WES offers a rapid, efficient and cost-effective sequencing strategy, which has been used with great effect to identify Mendelian disorders.1, 2, 3, 4
Primary mitochondrial respiratory chain disorders may be due to a functional defect of one of the five enzyme complexes involved in oxidative phosphorylation (OXPHOS), or may manifest as multiple OXPHOS defects.5 Because of this, identification of the underlying genetic basis can be particularly challenging given over 200 genes (including nuclear and mitochondrially encoded genes) have been directly implicated in these disorders. More recently, it has been highlighted that a ‘next-generation sequencing’ approach to OXPHOS disease gene identification is a much more effective approach than the traditional approach of sequential Sanger sequencing of candidate disease genes.6
In this study, two brothers from a non-consanguineous family with a severe neurological disorder characterised by profound cognitive impairment and sensory, autonomic, auditory and optic neuropathies, and their unaffected parents underwent WES to identify the causative gene of their presumed Mendelian, disorder. The WES results led to the identification of novel compound heterozygous variants in the polyribonucleotide nucleotidyltransferase 1 (PNPT1) gene in both affected boys, and our functional studies confirmed the pathogenicity of these two variants. Previous reports have shown that variants in the PNPT1 gene cause combined OXPHOS deficiency and a severe non-progressive hypotonia/dystonia/choreoathetosis in one family7 and isolated autosomal-recessive hearing impairment in another.8 Our patients shared features reported in these two families, and in addition had optic atrophy and an auditory neuropathy, and thus the PNPT1 variants observed in these two boys are a likely cause of their clinical phenotype.
Materials and methods
This research was approved by the Human Research Ethics Committee of the Sydney Children’s Hospitals Network, Sydney. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Declaration of Helsinki 1975, as revised in 2000. Informed consent was obtained from all patients included in the study.
WES and bioinformatic analysis
Target sequences of the DNA samples were fragmented to construct the library and the target regions were captured using the Agilent SureSelect Human All Exon 50 Mb Kit (Agilent Technologies, Santa Clara, CA, USA). Libraries were sequenced on the HiSeq 2500 Platform (Illumina Inc., San Diego, CA, USA) with onboard clustering using 100 × 100 bp paired-end reads and TruSeq Rapid PE Cluster Kit-HS and TruSeq Rapid SBS-HS Sequencing Chemistry Kits (Illumina Inc.) following the manufacturer's protocol.
Fastq files generated from the raw sequencing data of the affected boys and their parents were aligned with BWA version 0.5.10-tpx to the 1000 genomes phase 2 (GRCh37) human genome reference. Duplicate molecules were flagged with Picard version 1.74. Local realignment around indels and base call quality score recalibration were performed using GATK version 2.3-9-ge5ebf34. For multisample calling of SNVs and indels, unified Genotyper (GATK) and Reduced BAMs (GATK) were used, respectively. Following this, the called variants were filtered using the Variant Quality Score Recalibration method.
The prioritization strategy of the called variants was designed using the PhenoDB Analysis Tool.9 We prioritized rare functional variants (missense, nonsense, splice site variants and indels) that were heterozygous, homozygous or compound heterozygous in both affected siblings and excluded variants with a minor allele frequency >0.01 in dbSNP 126, 129 and 131 or in the Exome Variant Server (release ESP6500SI-V2) or 1000 Genomes Project. We also excluded all variants found in our in-house controls (CIDRVar 51 Mb). We then generated lists of heterozygous, homozygous and compound heterozygous variants shared by the affected siblings. All the identified variants here were based on the RefSeqGene number NG_033012.1 (NM_033109.4) and NCBI human genome assembly build 37. The data have been submitted to the publically funded databases dbGAP (hhttp://www.ncbi.nlm.nih.gov/gap;IDphs000711.v4.p1) and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/).
OXPHOS enzyme activity assays
Spectrophotometric analysis of OXPHOS enzymes in the skeletal muscle post 600g supernatants and enriched mitochondrial preparations from fibroblasts was performed as described previously.10 The CI and CIV dipstick activity assays were performed using 30 μg of whole-cell lysates from control and patient fibroblasts before and after transfection following the manufacturer’s instructions (MitoSciences, Eugene, OR, USA). The dipsticks were then placed in the MitoSciences MS1000 Dipstick Reader (MitoSciences) for signal intensity quantitation.
Real-time quantitative PCR (qPCR)
Total RNA was extracted from fibroblasts using a protocol combining Trizol (Life Technologies, Carlsbad, CA, USA) and Qiagen RNeasy Kit (Qiagen, Doncaster, VIC, Australia), whereas extraction of blood RNA was performed using the PAXgene Blood RNA Kit (Qiagen), following the manufacturer’s instructions. The concentration and purity of the extracted RNA were determined using the Nanodrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) where the purity was calculated based on the ratio of the absorbance at 260 and 230 nm.
A Rotor-Gene 6000 (Qiagen) was used to perform the qPCR reaction. The qPCR cycling conditions were 95 °C for 12 min; 35 cycles of 95 °C for 10 s, 62 °C for 15 s, 72 °C for 30 s and finally melting at 70–99 °C for 60 s. The housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used as a normalizing gene.
Immunoblotting
Immunoblotting of patient and control fibroblast samples was performed by separating 10 μg protein using 4 to 12% gradient SDS-polyacrylamide gels (Life Technologies, Scoresby, VIC, Australia) and transferred to PVDF membranes (GE Healthcare, Rydalmere, NSW, Australia). Membranes were blocked with 10% (w/v) skim milk prepared in 0.1% PBST overnight on an orbital rotator at 4 °C. Membranes were then probed with human PNPase antibody (1:500 dilution) (Santa Cruz Biotechnology Inc., Dallas, TX, USA) or with anti-OXPHOS (1:500 dilution) (Abcam, Melbourne, VIC, Australia) for 2 h at room temperature, and then reprobed for normalization with anti-GAPDH (Sigma, Sydney, NSW, Australia) or anti-porin (Abcam) (1:1000 dilution) for 1 h.
For BN-PAGE, fibroblast pellets were lysed on ice in Native lysis buffer (Abcam) and then were run according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA).
Immunoblotting of patient and control muscle samples was performed by extracting muscle protein in solubilisation buffer (125 mM Tris (pH 8.8), 40% (v/v) glycerol, 4% (w/v) SDS, 100 mM DTT, 0.01% bromphenol blue and protease inhibitors) by heating at 94 °C for 3 min before sonication. Equal amounts of protein were loaded onto a 12% Tris-glycine-SDS-polyacrylamide gels (Bio-Rad, Gladesville, NSW, Australia) and transferred to PVDF membranes (GE Healthcare). Membranes were blocked with 10% (w/v) skim milk prepared in 0.1% PBST for 2 h at room temperature. Membranes were then probed with human PNPase antibody (1:500 dilution) (Santa Cruz Biotechnology Inc.) or with anti-human OXPHOS (1:500 dilution) (Abcam), overnight at 4 °C and then reprobed for normalization with anti-GAPDH (1:2000 dilution) (Cell Signaling, Danvers, MA, USA) or anti-porin (Calbiochem, Alexandria, NSW, Australia) (1:1000 dilution). Membranes were developed using ECL detection reagents (Amersham, GE Healthcare LifeSciences, Sydney, NSW, Australia) and exposed to Hyperfilm ECL (Amersham, GE Healthcare LifeSciences).
Mitochondrial protein synthesis assay
Mitochondrial protein synthesis labeling experiments were performed following a previously reported protocol with minor modifications.11 Samples were analyzed on precast 10% Tricine gels (Life Technologies, Scoresby, VIC, Australia).
Cloning and transfection
The pCR2.1-TOPO (Life Technologies, Scoresby, VIC, Australia) vector was used to subclone the wild-type and mutant PNPT1 PCR products before being cloned into the pPM-hPNPT1-His vector (Applied Biological Materials Inc., Richmond, BC, Canada) to express the exogenous wild-type or mutant PNPT1 genes in patient and control fibroblast cells. Fibroblast cells were transfected with Lipofectamine LTX (Life Technologies, Scoresby, VIC, Australia) according to the manufacturer’s instructions. The transfection media was replaced with fresh media after 5 h and the cells were harvested 48 h after transfection.
Statistical analysis
Statistical analyses were carried out using unpaired nonparametric Mann–Whitney U-test (test statistic) and calculated using a two-tailed t-test (GraphPad Prism Software, GraphPad Software Inc., La Jolla, CA, USA). Error bars represent the standard error of the mean (±SEM). A P-value <0.05 was considered to be statistically significant.
Results
Patient clinical summaries
The clinical histories of the two affected brothers are detailed in the Supplementary Material, together with results of the OXPHOS enzymology for S1 (Supplementary Table 1).
WES and Sanger sequencing
Bioinformatic analysis of the WES data identified two candidate genes: PNPT1 and FAM122C (Family With Sequence Similarity 122C). As little is known about the function and role of FAM122C in disease pathogenesis, it seems less likely to be causative and therefore we did not consider it for further studies. The more likely candidate, PNPT1, was pursued. The PNPT1 gene was selected because of its functional relevance to the disease phenotype, which is supported by two previous publications,7, 8 with overlapping phenotypes to those found in our patients. We identified novel heterozygous missense variations in exon 9 (c.[760C>A], p.(Gln254Lys)) and exon 19 (c.[1528G>C], p.(Ala510Pro)) (NG_033012.1) of the PNPT1 gene in both affected boys. Sanger sequencing confirmed the presence of both variants in the affected boys (S1 and S2) and their parents were each heterozygous for one or other of the variants (Supplementary Figure 1A).
In silico analysis predicts that the p.(Gln254Lys) and p.(Ala510Pro) variants have an impact on PNPase protein function
In silico analyses were used to determine the potential effect of these PNPT1 variants on the structure and function of the PNPase protein. Both Gln254 and Ala510 were conserved across in 9 of 14 species analysed (Supplementary Figure 1B). Moreover, in silico tools suggested that the p.(Ala510Pro) variant was likely to have more harmful effects on the function of the protein, whereas the p.(Gln254Lys) variant would be likely to have a milder effect (Supplementary Table 2).
PNPT1 mRNA and PNPase protein expression levels are reduced in patient fibroblasts
To evaluate the effect of these variants on gene expression, mRNA expression of PNPT1 was quantitated using qPCR in fibroblasts from patient S1. Fibroblasts from the other family members were not available for testing. As shown in Figure 1a, mRNA expression of PNPT1 in patient S1 fibroblasts was significantly reduced by 65% compared with a control, indicating that at least one of the variants could affect mRNA expression or its stability. mRNA expression of PNPT1 was measured in blood obtained from all family members; however, no abnormalities were observed (Figure 1a).
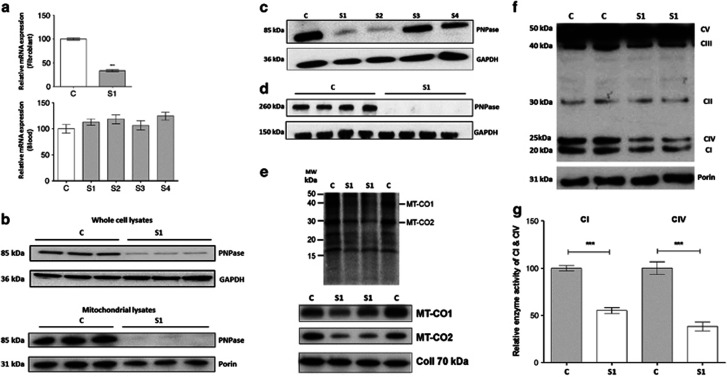
Figure 1.
Assay of PNPT1 mRNA expression levels, protein levels, mitochondrial protein synthesis and respiratory chain enzyme activity in the fibroblasts and blood of control and patient. (a) Relative mRNA expression of PNPT1 in mRNA extracted from fibroblasts and blood of proband (S1), affected brother (S2), unaffected father (S3) and unaffected mother (S4). The expression level was normalized relative to GAPDH. (b) The PNPase protein level in subject S1 and control fibroblasts using whole-cell lysates and mitochondrial lysates. GAPDH was used as a loading control. (c) PNPase protein levels in control and all family members using protein extracts from white blood cells. GAPDH was used as a loading control. (d) Non-denaturing gel electrophoresis analysis was performed to examine the assembly of PNPase in S1 and control fibroblasts. GAPDH was used as a loading control. (e) Analysis of mitochondrial translation in fibroblasts from S1 and two controls using a [35S]methionine pulse-chase labeling experiment. Phosphorimage in the upper panel and corresponding immunoblot in the lower panel. (f) Protein levels of OXPHOS complex subunits (CI NDUFB8, CII 30 kDa, CIII Core 2, CIV MT-CO2, CVα) in subject S1 and control fibroblasts using OXPHOS cocktail antibodies. Porin was used as a loading control. (g) CI and CIV enzyme activity dipstick assay in S1 and control fibroblasts.
To assess whether the PNPase protein levels were reduced, western blotting was conducted using whole-cell lysates from patient S1 fibroblasts. In agreement with the mRNA levels, PNPase was found to be reduced by ~80% compared with the control (Figure 1b). The PNPase protein is located predominantly in the mitochondria, and thus it was hypothesized that using mitochondrial lysates would show an even greater reduction in the protein expression level. As expected, we observed a greater reduction in the PNPase level in patient S1 using mitochondrial lysates (Figure 1b).
PNPase protein levels were evaluated in all family members using protein extracted from white blood cells. Western blot analysis of whole-cell lysates showed an obvious reduction in PNPase in both affected boys (S1 and S2) with an ~80% reduction compared with the control. The parental samples (S3 and S4) showed levels comparable to the control (Figure 1c). Owing to the lack of sample availability, PNPase protein levels could not be analyzed in white blood cell mitochondrial lysates.
PNPase protein levels were also measured in whole-cell lysates from a muscle biopsy taken from S1 and were compared with those of two aged-matched controls (C1 and C2). Normal levels of the PNPase protein was observed in S1 (Supplementary Figure 6A).
PNPase complex assembly is altered in the affected sibling S1
The human PNPase protein assembles into a trimeric complex and therefore non-denaturing gel electrophoresis was performed to determine whether the identified variants had an effect on the assembly of PNPase into its functional trimeric state. The PNPase complex was found to be totally absent in patient S1 fibroblast samples, suggesting that these variants severely impair the assembly of the complex (Figure 1d).
Mitochondrial protein translation is impaired in the affected sibling S1
Mitochondrial protein synthesis was investigated using [35S]methionine pulse-chase labelling. As shown in Figure 1e (upper panel), the phosphorimage of the 35S gel revealed a marked reduction in the synthesis of the mtDNA-encoded proteins MT-CO1 (mitochondrially encoded cytochrome c oxidase I) and MT-CO2 (mitochondrially encoded cytochrome c oxidase II) in fibroblasts from patient S1 compared with the control, where the total incorporation of [35S]methionine into mitochondrial extracts of patient S1 fibroblasts was reduced by 33% compared with the control (Supplementary Figure 2). This reduction in the mtDNA-encoded, CIV subunits (MTCO1, MT-CO2) was confirmed by wWestern blot, whereas the nuclear-encoded subunit of CII showed no reduction (Figure 1e; lower panel). These results indicate that the PNPT1 p.Gln254Lys and p.Ala510Pro variants together result in a defect in mitochondrial translation.
PNPT1 variants affect the mitochondrial respiratory chain complexes and the enzyme activity of CI and CIV
Protein expression of the OXPHOS complexes was investigated in fibroblasts from patient S1 using western blot analysis. By using an anti-OXPHOS cocktail (containing five different antibodies to detect a subunit of each of the different OXPHOS complexes, which are labile if the complex is not correctly assembled), a marked reduction in CI, CIII and CIV was revealed, indicating combined OXPHOS deficiencies in this patient (Figure 1f). Relative expression levels were quantitated by calculating the ratio of expression of each OXPHOS protein relative to the expression of porin in each sample. Protein expression of CI, CIII and CIV in S1 fibroblasts showed a reduction by 26%, 22% and 48%, respectively, compared with the control (Supplementary Figure 3). The other complexes, CII and CV, did not show a significant reduction in the relative protein expression levels compared with the control fibroblasts (Supplementary Figure 3). In addition, the enzyme activities of CI and CIV were determined in patient S1 fibroblasts using dipstick assays, and showed a significant reduction in CI and CIV activity, by 45% and 62%, respectively, compared with the control (Figure 1g).
Protein expression of the OXPHOS complexes were also measured in whole-cell lysates of the muscle biopsies and, in accordance with the PNPase expression, no differences in the levels of the OXPHOS complexes were observed in S1 compared with the two controls (Supplementary Figure 6b).
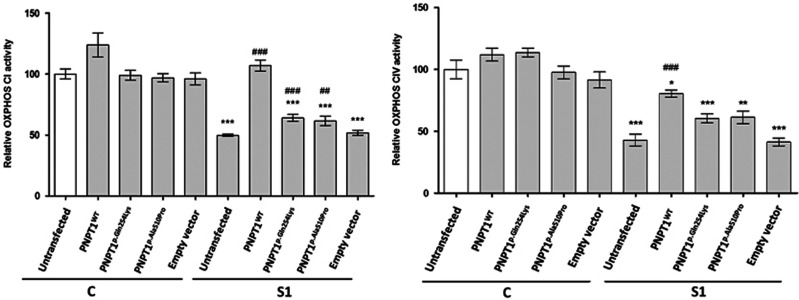
Exogenous expression of PNPT1 rescues CI and CIV enzyme activity
For the rescue experiments, patient S1 and control fibroblasts were transfected with PNPT1wt, PNPT1p.Gln254Lys, PNPT1p.Ala510Pro and empty vector constructs separately. Transfection with PNPT1wt in S1 cells increased CI enzyme activity levels by 115% compared with the untransfected S1 cells (P<0.001), whereas S1 cells expressing PNPT1p.Gln254Lys and PNPT1p.Ala510Pro showed only a partial increase in the activity of CI by 29% (P<0.001) and 25% (P<0.01), respectively, compared with the untransfected cells. S1 fibroblasts transfected with the empty vector did not show any rescue of CI enzyme activity (Figure 2). In contrast, the control fibroblasts transfected with PNPT1wt, PNPT1p.Gln254Lys, PNPT1p.Ala510Pro or the empty vector showed no significant increase in CI enzyme activity (Figure 2).
Figure 2.
Rescue experiments for CI and CIV enzyme activity. Fibroblasts from S1 and a control were transfected with PNPT1wt (wild-type), PNPT1p.Gln254Lys (p.(Gln254Lys)), PNPT1p.Ala510Pro (p.(Ala510Pro)) and empty vector constructs. Untransfected cells from the control and S1 were included. Data are means±SEM from three independent transfection experiments, each analysed in triplicate (n=9). *P<0.05, **P<0.01 and ***P<0.001 significantly different from untransfected control cells. ##P<0.01 and ###P<0.001 are significantly different from untransfected S1 cells.
For CIV, transfection with PNPT1wt increased the enzyme activity levels in S1 fibroblast by 84% compared with the untransfected S1 cells (P<0.001), whereas S1 cells transfected with PNPT1p.Gln254Lys, PNPT1p.Ala510Pro or the empty vector showed no significant increase in the activity of CIV, compared with the untransfected cells (Figure 2). Control fibroblasts transfected with PNPT1wt, PNPT1p.Gln254Lys, PNPT1p.Ala510Pro and empty vector did not show a significant change in CIV enzyme activity (Figure 2).
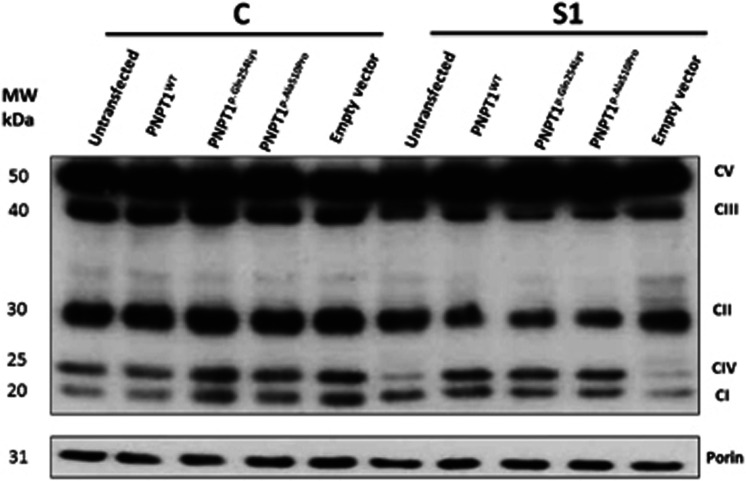
Exogenous expression of PNPT1 rescues CI, CIII and CIV protein expression
Rescue experiments were performed to determine whether the protein levels of CI, CIII and CIV could be restored in fibroblasts from patient S1 and a control fibroblast line transfected with PNPT1wt, PNPT1p.Gln254Lys, PNPT1p.Ala510Pro and empty vector constructs using an anti-OXPHOS cocktail (Figure 3), once satisfactory transfection efficiencies was demonstrated (Supplementary Figure 4). Rescue of protein expression of CI, CIII and CIV in S1 fibroblasts transfected with PNPT1wt was observed by 45%, 20% and 36%, respectively, compared with the untransfected S1 fibroblasts. S1 cells expressing PNPT1p.Gln254Lys also showed rescue in the protein expression of CI, CIII and CIV by 22%, 7% and 19%, respectively, compared with the levels in the untransfected cells (Supplementary Data). Patient S1 fibroblasts transfected with PNPT1p.Ala510Pro increased CI, CIII and CIV protein expression by 24%, 11% and 21%, respectively, compared with the levels seen in untransfected cells (Supplementary Figure 5). Protein expression of CII and CV in S1 fibroblasts transfected with PNPT1wt, PNPT1p.Gln254Lys and PNPT1p.Ala510Pro remained unchanged. S1 fibroblasts transfected with the empty vector did not show a significant change in the protein expression of all five complexes (Figure 3 and Supplementary Figure 5). Control fibroblasts transfected with PNPT1wt, PNPT1p.Gln254Lys, PNPT1p.Ala510Pro and empty vector did not show a significant increase in the expression of all complexes.
Figure 3.
Rescue experiments for CI, CII, CIII, CIV and CIV protein expression. Fibroblasts from S1 and a normal control were transfected with PNPT1wt (wild-type), PNPT1p.Gln254Lys (p.(Gln254Lys)), PNPT1p.Ala510Pro (p.(Ala510Pro)) and empty vector constructs. Porin was used as a loading control. This is a representative figure, and experiments were performed independently six times (n=6).
Discussion
Here, we report two affected brothers who presented with severe multisystem disease. Although the clinical picture was suggestive of a mitochondrial respiratory chain disorder, blood and CSF lactate, and muscle respiratory chain enzyme assays were all normal. Using a WES approach, we identified two novel compound heterozygous missense variations in the PNPT1 gene. Variants in PNPT1 have previously been reported to result in combined OXPHOS deficiency (MIM 614932) and autosomal recessive deafness (MIM 614934).7, 8 Our patients had severe multisystem disease, with profound cognitive impairment, growth failure, sensory and autonomic neuropathies, optic atrophy and an auditory neuropathy, as well as respiratory and gut abnormalities. The differences in clinical severity are not immediately obvious, but may be a reflection of differences in functional severity of the respiratory chain enzyme activities. These results enabled the parents of these two boys to proceed to pregnancy planning using preimplantation genetic diagnosis, which led to the birth of a girl, heterozygous for the c.760C>A variant, who is now aged 4 months and is clinically normal.
The PNPT1 gene encodes the mitochondrial polynucleotide phosphorylase (PNPase) protein, which has 28 exons and is located at chromosome 2p16.1. The PNPase protein consists of five domains, including two RNase PH-like 3′–5′ exoribonucleases domains, two RNA-binding domains, KH and S1, and an α-helical domain.12, 13, 14 The N terminus of this protein has a mitochondrial targeting sequence that is required for subcellular localization to mitochondria.15 PNPase localizes predominantly to the mitochondrial intermembrane space, and assembles into a homotrimeric complex,7, 15 but has also been found to be located in the mitochondrial matrix because of its interaction with the matrix-localised RNA helicase hSUV3, and in the cytoplasm, where it is involved in the degradation of microRNA and mRNA,16 as well as regulation of the importation of cytosolic RNAs into the mitochondrial matrix.14, 17 Moreover, PNPase has an in vivo role in mitochondrial morphology and respiration through unknown mechanisms,18 maintenance of mitochondrial homeostasis and the ability to protect cells from oxidative stress.19
The evolutionary comparison, together with the in silico analysis, predicts that the p.(Gln254Lys) and p.(Ala510Pro) variants in PNPT1 are most likely to be pathogenic, despite that the Gln254 and Ala510 residues are not located in any of the PNPase functional domains and prompted us to make further investigations. Indeed, our studies on the effect of these variants showed that these patients have reduced expression levels of PNPT1 at the message and protein level in their fibroblasts and reduced protein levels in their white blood cells.
The trimerization of PNPase is essential for the RNA processing and transportation function of PNPase protein, and so the ability of PNPase to self-assemble was investigated by non-denaturing BN-PAGE. We identified a marked effect of the identified PNPT1 variants on PNPase trimerization, in agreement with an earlier report,8 where the PNPase complex was totally absent, suggesting that these variants result in an unstable monomer, with the consequent detrimental effect on assembly of the complex. Considering the key role of PNPase for the import of small RNAs,8 we investigated the potential effect on mitochondrial translation by studying the mitochondrial protein synthesis. Impairment of mitochondrial translation was confirmed with a 33% reduction in the mitochondrial protein synthesis found in patient fibroblasts compared with the control.
Analysis of the OXPHOS complexes in patient fibroblasts showed combined OXPHOS protein expression deficiency with reductions in CI, CIII and CIV, together with a marked reduction in CI and CIV enzyme activity. It is interesting to note that traditional spectrophotometric enzyme assays were normal for all complexes in the muscle and fibroblasts from this individual. Normal results were also reported for the skeletal muscle and fibroblast OXPHOS enzymes in a previous patient with PNPT1 mutations,8 and these observations reflect tissue specificity and assay methods. In the fibroblasts, the dipstick enzyme assays and western blotting analyses were performed on whole-cell lysates, whereas the spectrophotometric enzyme assays were performed on enriched mitochondrial preparations. We have seen a similar pattern of normal spectrophotometric enzyme activities but decreased amounts of OXPHOS subunits or dipstick enzyme assays in fibroblasts from other patients with mtDNA translation defects caused by mutations in genes including GFM1, TSFM and NARS2. This pattern likely reflects preferential loss of the most damaged mitochondria in mitochondrial preparations compared with whole-cell samples. Our data emphasise the value of WES even in patients with apparently normal results of OXPHOS respiratory chain enzymology, and the need to consider the most appropriate methods for functional validation of putative pathogenic mutations in OXPHOS genes.
Our studies show that PNPT1 variations identified in the affected proband S1 result in OXPHOS deficiency and are therefore likely to be pathogenic based on our rescue experiments where overexpression of the wild-type PNPT1 (PNPT1wt) in the patient fibroblasts led to a significant rescue of the enzyme activity levels of CI and CIV and in the protein expression levels of CI, CIII and CIV. Interestingly, the fibroblasts from S1-expressing constructs carrying the variants, p.(Gln254Lys) or p.(Ala510Pro), also resulted in an increase in CI and CIV enzyme activity levels and of CI, CIII and CIV protein levels. This evidence of partial correction is probably a consequence of the overexpression of the mutant proteins, with resultant partial restoration of protein function. We can surmise that these variants affect mitochondrial protein translation and therefore caused OXPHOS deficiency, particularly involving the components of the OXPHOS complexes that are mitochondrially translated.
In conclusion, we report here novel compound heterozygous variants in two affected siblings from a non-consanguineous family that led to structural and functional defects of the mitochondrial respiratory chain, and which are most likely to have caused the complex multisystem phenotype in these boys. This study clearly highlights the important role of PNPase in mitochondrial translation and the generation of a functional OXPHOS system. This study also shows the efficacy of WES to identify disease-causing genes in rare genetic disorders.
Acknowledgments
This work was supported by a scholarship to A Alodaib provided by the Academic and Training Affairs at King Faisal Specialist Hospital and Research Center and the Ministry of Higher Education (Riyadh, Saudi Arabia). This research was supported by Australian NHMRC Grant 1026891 (to JC), an NHMRC Principal Research Fellowship (to DRT) and an NHGRI Grant 1U54HG006542. Finally, we also gratefully acknowledge donations to JC by the Crane and Perkins families.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Ng SB, Turner EH, Robertson PD et al: Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009; 461: 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan M, Chong JX, Uricchio L et al: Exome sequencing reveals a novel mutation for autosomal recessive non-syndromic mental retardation in the TECR gene on chromosome 19p13. Hum Mol Genet 2011; 20: 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Arts HH, Hoischen A et al: Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am J Hum Genet 2010; 87: 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C et al: Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 2010; 42: 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes MJ, Riley LG, Christodoulou J: Mitochondrial respiratory chain disorders in childhood: insights into diagnosis and management in the new era of genomic medicine. Biochim Biophys Acta 2014; 1840: 1368–1379. [DOI] [PubMed] [Google Scholar]
- Lake NJ, Compton AG, Rahman S, Thorburn DR: Leigh syndrome: One disorder, more than 75 monogenic causes. Ann Neurol 2016; 79: 190–203. [DOI] [PubMed] [Google Scholar]
- von Ameln S, Wang G, Boulouiz R et al: A mutation in PNPT1, encoding mitochondrial-RNA-import protein PNPase, causes hereditary hearing loss. Am J Hum Genet 2012; 91: 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedrenne V, Gowher A, De Lonlay P et al: Mutation in PNPT1, which encodes a polyribonucleotide nucleotidyltransferase, impairs RNA import into mitochondria and causes respiratory-chain deficiency. Am J Hum Genet 2012; 91: 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A: New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat 2015; 36: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Thorburn DR: Biochemical analyses of the electron transport chain complexes by spectrophotometry. Methods Mol Biol 2012; 837: 49–62. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Lazarou M, Ryan MT: Analysis of respiratory chain complex assembly with radiolabeled nuclear- and mitochondrial-encoded subunits. Methods Enzymol 2009; 456: 321–339. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Fisher PB: Polynucleotide phosphorylase: an evolutionary conserved gene with an expanding repertoire of functions. Pharmacol Ther 2006; 112: 243–263. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Park ES, Emdad L, Randolph A, Valerie K, Fisher PB: Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35 mediating cellular senescence. Mol Cell Biol 2005; 25: 7333–7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokhi UK, Bacolod MD, Dasgupta S et al: Identification of genes potentially regulated by human polynucleotide phosphorylase (hPNPaseold-35 using melanoma as a model. PLoS One 2013; 8: e76284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwowarski J, Grzechnik P, Dziembowski A, Dmochowska A, Minczuk M, Stepien PP: Human polynucleotide phosphorylase, hPNPase, is localized in mitochondria. J Mol Biol 2003; 329: 853–857. [DOI] [PubMed] [Google Scholar]
- Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ: Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res 2013; 41: 1223–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Bhutia SK, Sokhi UK et al: Human polynucleotide phosphorylase (hPNPaseold-35: an evolutionary conserved gene with an expanding repertoire of RNA degradation functions. Oncogene 2011; 30: 1733–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Chen HW, Oktay Y et al: PNPASE regulates RNA import into mitochondria. Cell 2010; 142: 456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li Z: Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochem Biophys Res Commun 2008; 372: 288–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.