Abstract
Understanding the functional sequelae of amino-acid replacements is of fundamental importance in medical genetics. Perhaps, the most intuitive way to assess the potential pathogenicity of a given human missense variant is by measuring the degree of evolutionary conservation of the substituted amino-acid residue, a feature that generally serves as a good proxy metric for the functional/structural importance of that residue. However, the presence of putatively compensated variants as the wild-type alleles in orthologous proteins of other mammalian species not only challenges this classical view of amino-acid essentiality but also precludes the accurate evaluation of the functional impact of this type of missense variant using currently available bioinformatic prediction tools. Compensated variants constitute at least 4% of all known missense variants causing human-inherited disease and hence represent an important potential source of error in that they are likely to be disproportionately misclassified as benign variants. The consequent under-reporting of compensated variants is exacerbated in the context of next-generation sequencing where their inappropriate exclusion constitutes an unfortunate natural consequence of the filtering and prioritization of the very large number of variants generated. Here we demonstrate the reduced performance of currently available pathogenicity prediction tools when applied to compensated variants and propose an alternative machine-learning approach to assess likely pathogenicity for this particular type of variant.
It is almost a truism that the degree of evolutionary conservation of an amino-acid residue reflects the structural and/or functional importance of that residue. Human disease-causing variants tend to occur disproportionately in those residues of structural and/or functional importance, and for this reason are subject to negative selection. It is therefore not unreasonable to expect that missense variants, which are disease-causing in human, would also be deleterious in evolutionarily closely related species. However, this expectation has been challenged by the realization that there are numerous examples where human mutant alleles correspond to the wild-type alleles in other mammalian species.1, 2, 3, 4, 5, 6, 7 Such variants have become known as compensated pathogenic deviations (CPDs) following their original designation5 because it is assumed that the apparently benign nature of these missense variants in non-human species is due to the coexistence of other amino-acid substitutions (AASs) that compensate for their otherwise dysfunctional consequences. Among those human mutant residues corresponding to the wild-type residue in mouse5 are an p.(Ala53Thr) (NM_000345.3:c.157G>A) substitution at the α-synuclein (SNCA) locus reported to be associated with familial Parkinson disease;8 the ADA-p.(Arg142Gln) (NM_000022.2:c.425G>A) causing severe combined immunodeficiency;9 and the CFTR-p.(Phe87Leu) (NM_000492.3:c.259T>C) in the cystic fibrosis transmembrane conductance regulator gene underlying cystic fibrosis.10 Another interesting CPD is EIF2B5-p.(Arg113His) (NM_003907.2:c.338G>A), the most common lesion associated with leukoencephalopathy with vanishing white matter,11 which corresponds to the wild-type allele in the genomes of both rat and mouse.4
When the genome sequence of the rhesus macaque was released,3 further examples of CPDs were identified. Among them was the p.(Ile164Thr) (NM_000277.1:c.491T>C) at the phenylalanine hydroxylase12 responsible for the most common human inborn error of metabolism (phenylketonuria) and p.Arg40His (NM_000531.5:c.119G>A) at the X-linked ornithine transcarbamylase (OTC) locus. Although the OTC-p.(Arg40His) replacement leads to the cytosolic degradation of the human enzyme precursor,13 abnormal levels of ammonia were not evident in simian plasma. Moreover, abnormal levels of phenylalanine were not detected in macaque,3 reinforcing the notion that these variants are only deleterious on a human genetic background. Intriguingly, a different OTC variant p.(Thr125Met) associated with fatal hyperammonemia14 was found to correspond to the wild-type allele in chimpanzees.1, 15, 16 Among the CPDs identified through a comparison with the recently reported mountain gorilla genome6 was the NPC1-p.(Asn961Ser) (NM_000271.4:c.2882A>G) that leads to Niemann-Pick disease C.17
Finally, variants associated with ciliopathies at the BBS4 and RPGRIP1L genes associated with Bardet-Biedl and Meckel-Gruber syndromes, respectively, constitute the wild-type alleles in the genomes of several vertebrates.7 The same study reported a de novo variant at the BTG2 locus in which the disease-associated allele corresponded to the wild-type allele in more than 50 vertebrate species.
At this stage, it is important to mention that these are examples of a more general phenomenon that involves at least 4% of all known missense variants causing human-inherited disease (see Materials and Methods section). In practice, this proportion almost certainly represents a conservative estimate because there is an intrinsic ascertainment bias against the recognition and reporting of CPDs; such variants, by their very nature, are often predicted to be benign. This bias is exacerbated in the context of next-generation sequencing (NGS) where the inappropriate exclusion of CPDs from further consideration constitutes an unfortunate natural consequence of the filtering and prioritization of the very large numbers of candidate variants generated. It follows that we need to take urgent steps both to assess the scale of this problem and to take appropriate remedial action.
It is essential that bioinformatic prediction tools can make accurate and reliable predictions when they are used to assess the functional impact of a putative compensated variant. Some existing tools have already been shown to make incorrect predictions in the case of experimentally validated pathogenic variants.7 Here, we demonstrate, in a larger sample of CPDs, the reduced performance of existing tools to predict the deleterious impact of CPDs when found as disease-associated variants in humans. Further, we present the prototype of a CPD-specific predictor that successfully outperformed currently available tools in terms of its ability to predict the deleterious impact of these variants in humans.
Materials and methods
Detection of CPDs in mammalian species
To identify the amino-acid positions where a deleterious human variant corresponds to the wild-type residue in a non-human mammalian species, herein referred to as a CPD,5 we employed data from the Human Gene Mutation Database (HGMD; http://www.hgmd.org; 52 765 disease-causing missense variants, annotated as DMs; July 2013) to screen 1-to-1 orthologous mammalian protein sequences annotated at the Ensembl Genome Browser (http://www.ensembl.org; release 73).18 Data from a total of 39 mammalian species (35 placental, 3 marsupials and 1 monotreme) were available, and sequences were automatically retrieved and submitted through a series of stringent filters before being considered for subsequent analysis. Whenever more than one sequence matched the gene symbol used in the search (because of redundancy caused by, for example, alternatively spliced isoforms), a pairwise alignment with the human reference sequence was performed and the sequence retained was that which had the highest degree of identity with its human counterpart. In order to avoid highly incomplete sequences, we calculated the pairwise identity to the human sequence and retained only those that were at least 50% identical to the human ortholog. The orthologous sequences that passed these filters were then used in the identification of CPDs by comparing the aligned sequences. For all the cases where a CPD was identified, we applied a strategy similar to that previously documented, 5 namely, screening a flanking window of five amino-acid residues upstream and downstream of the putative CPD site and retaining only those CPDs with no more than four differences with respect to the human sequence within the flanking region. Missense variants at the initiator methionine residue were removed from the analysis. This strategy allowed the identification of 1964 CPDs in a total of 684 protein-coding genes (Supplementary Table S1), a figure that corresponds to ~3.7% of all missense variants analyzed. All the alignments were performed using ClustalO 1.2.0 (www.clustal.org/omega)19 and other tools were developed locally using GNU/Linux-based computers with scripting tools+C programming language.
DM set
A set of 10 211 disease-causing missense variants, which result in an AAS in 2030 genes, was obtained from the HGMD.20 This set of disease-causing variants is representative of data sets that have typically been employed in the training and evaluation of a number of different bioinformatic prediction tools to identify disease-causing AAS. In order to allow an unbiased evaluation of bioinformatic prediction tools, an unseen test set (not used for training) should always be used, otherwise the evaluation represents in-sample error rather than out-of-sample error and hence is likely to be overly optimistic in terms of prediction performance. As some of the prediction tools evaluated here (eg, MetaSVM) either already used, or could have used, HGMD (or other similar overlapping data sets, eg, OMIM) either as training data or in the development of the prediction method, this DM set was selected so as to contain only recently reported (2014 onwards) disease-causing missense variants from HGMD. This data set of variants would therefore not have been available when the various pathogenicity prediction tools being tested were being developed. This set of disease-causing variants is henceforth referred to as the DM set.
SNP set
As a negative control, a set of 2174 putatively ‘neutral’ common missense SNPs (MAF≥0.4) in 1640 genes from the NHLBI ESP6500 Exomes(http://evs.gs.washington.edu/EVS/) was compiled. This set is henceforth referred to as the SNP set.
Performance evaluation of bioinformatic prediction tools used to identify disease-causing variants
Numerous different prediction methods to identify disease-causing or functional variants have been developed. The four chosen here (SIFT,21 Mutation Assessor,22 PROVEAN23 and MetaSVM24) were selected on the basis that they are commonly used (eg, SIFT) or represent a different approach to the classification problem (eg, MetaSVM, an ensemble approach using population frequency data and the scores from 10 other prediction methods (SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, GERP++, MutationTaster, Mutation Assessor, FATHMM, LRT, SiPhy and PhyloP)). Prediction scores for the CPD, DM and SNP set (where available) were then obtained from dbNSFP (version 3.2c).24 As a means to evaluate performance for each of the four bioinformatic prediction methods, receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC) were calculated for the CPD set versus the SNP set and the DM set versus the SNP set. In this context, an ROC-AUC of 100% would represent a perfect predictor, whereas an AUC of 50% would correspond to a prediction tool making random predictions.
Feature subset ranking
In order to evaluate discriminative features or attributes (commonly used to identify disease-causing variants) in the context of CPDs, an array of features relating to the AAS (eg, solvent accessibility) were derived from SNVBox25 (Supplementary Table S2). Related features were then grouped into seven different subsets (amino-acid-based, exon-based, genomic MSA, protein MSA, protein structure, regional protein composition and annotated functional sites). For more details, please refer to SNVBox.25 The prediction performance of each feature subset (eg, exon-based features) was evaluated using 10-fold cross-validation and a linear support vector machine (SVM)26 for two different data sets (CPD and SNP, DM and SNP) by training the SVM classifier with only the specific feature subset under evaluation. For each feature subset, ROC curves and the AUC were calculated, which allowed us to rank precisely how informative each feature subset was for identifying CPDs and also for identifying disease-causing variants. As a control, a random attribute was generated for each training instance and the performance was measured using the AUC of the ROC.
A machine-learning approach to assess the functional impact of potential CPDs
Current methods to assess the impact of potential disease-causing variants have neither been specifically developed nor evaluated in the context of CPDs. Although they do demonstrate some utility in terms of the functional assessment of CPDs, we set out to develop a novel prototype CPD-specific predictor. Using the features employed in this study (Supplementary Table S1), two different Random forest 27 classification models were built. Random forest can be employed for supervised classification and involves an ensemble of decision trees, which helps to minimize overfitting, making them robust to noise in the data. The first Random forest classification model, termed the 'CPD-trained model', was trained using the 1964 CPD (positive examples) and 2174 SNP variants (negative examples). The second model, termed the 'DM-trained model', was trained using the DM set as positive examples (10 211 instances) and the SNP set as negative examples (2174 instances). As the ‘DM-trained model’ demonstrated a class imbalance with fivefold more positive examples than negative examples, the DM set (majority class) was randomly undersampled (without replacement) to balance the classes. The DM-trained model also excludes any features derived from multiple sequence alignments (MSA). To evaluate both models (‘CPD-trained model’ and ‘DM-trained model’), 10-fold cross-validation was performed and the AUC was then calculated. In the case of 10-fold cross-validation for the ‘DM-trained model’, the positive variants in the ‘test’ fold were derived from the CPD set (therefore, the training fold had disease-causing mutations as positives and common SNPs as negative instances), in order that we evaluated the performance of the model on classifying CPDs (rather than disease-causing mutations in general). Standard benchmarking statistics were employed to evaluate performance such as the true-positive rate (sensitivity), the false-positive rate and the Matthew’s Correlation Coefficient (MCC).28 The MCC was employed as it represents one of the best available measures of prediction quality. It returns a value between −1 and +1; a coefficient of −1 represents the worst possible prediction, 0 a random prediction and +1 a perfect prediction.
Results and discussion
How accurately are CPDs predicted by current bioinformatics tools?
Owing to their occurrence in MSAs, there may well be a tendency for CPDs to evade detection by the predictive tools commonly used to evaluate the functional impact of human missense variants, simply because the mutant residues in question are tolerated in other mammalian species. Most predictive methods rely to some extent on the degree of evolutionary conservation of the mutated residue but, when a mutant residue occurs as the wild-type allele in one or more orthologs, its impact in a human context may not be readily predictable. Thus, for example, using three widely used predictive tools, PolyPhen,29 SIFT21 and Mutation Assessor,30 Jordan and et al.7 failed to predict the deleterious effect of three CPDs (NM_033028.4(BBS4):c.493A>C (p.Asn165His); NM_015272.3(RPGRIP1L):c.2810G>T (p.Arg937Leu); and NM_006763.2(BTG2):c.421G>A (p.Val141Met)) experimentally demonstrated to be disease-associated in human.
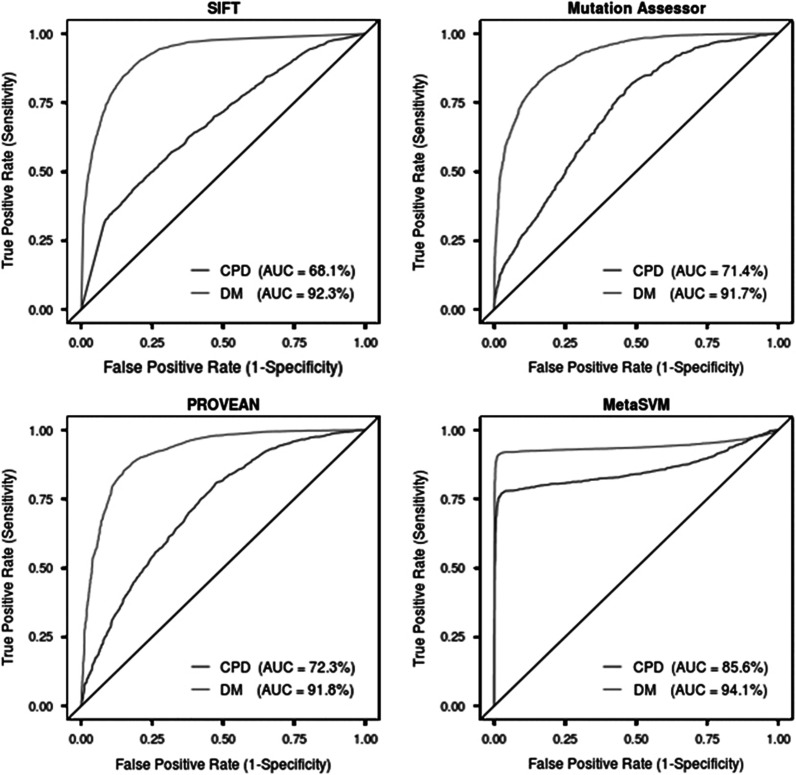
To assess the extent of the ability of existing predictive methods to deal with CPDs, we compared a set of more than 10 200 disease-causing variants from the HGMD with a set of CPDs (Supplementary Table S1) and a set of neutral missense SNPs (MAF≥0.4; Figure 1). The results showed that all four bioinformatic prediction tools tested here exhibited reduced prediction performance (−24.2 to −8.5% AUC) in relation to CPDs as compared with disease-causing variants. The performance (ROC-AUC) of the four tools evaluated in the context of identifying CPDs was as follows: SIFT (68.1% AUC), Mutation Assessor (71.4% AUC), PROVEAN (72.3% AUC) and MetaSVM (85.6%). Of the four tools, SIFT exhibited the largest reduction in prediction performance (−24.2%) for identifying CPDs. MetaSVM, a consensus of scores from 10 different tools (SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, GERP++, MutationTaster, Mutation Assessor, FATHMM, LRT, SiPhy and PhyloP) and the maximum frequency observed in the 1000 Genomes populations, outperformed all other tools in terms of identifying CPDs (AUC=85.6% Figure 1), suggesting that an ensemble approach could prove useful in classifying CPDs.
Figure 1.
Performance evaluation of four different bioinformatic tools (SIFT, Mutation Assessor, PROVEAN and MetaSVM) used to identify disease-causing variants. Each bioinformatic tool was evaluated on CPDs (lower line) and DMs (upper line). ROC curves and the AUC were calculated for each data set (CPD and DM) with a different prediction tool. An AUC of 100% would represent a perfect predictor, whereas an AUC of 50% would represent a prediction tool making only random predictions (denoted by the black diagonal line). Note: MetaSVM is an ensemble prediction method based on 10 other prediction tools.
Evaluation of features commonly used to identify disease-causing AAS
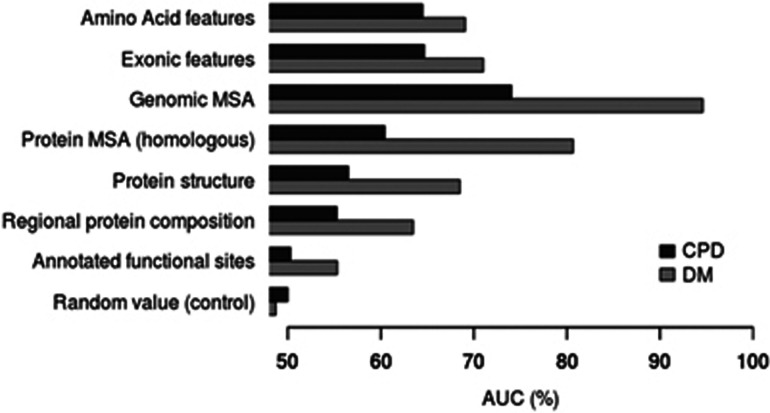
In order to perform an evaluation in the context of CPDs, the discriminatory power of groups of related features (eg, structural features; Supplementary Table S2) commonly used to distinguish disease-causing missense variants from neutral polymorphic missense variants was computed using 10-fold cross-validation with a linear SVM (Table 1 and Figure 2). Each feature subset was evaluated in the contexts both of identifying CPDs and disease-causing mutations, in order to compare and contrast how informative the different features are for the two different classes of disease-causing mutations (CPD set versus DM set). The most informative feature subsets discriminating disease-causing variants from common putatively neutral polymorphisms (DM versus SNP) were derived from MSA (94.6% AUC for genomic MSA and 80.7% AUC for protein MSA; Figure 2—DM set). These MSA-derived features (genomic and protein MSA) also demonstrated the largest reduction in performance (−20% in AUC; Table 1) when used to discriminate between the CPDs and the SNP set (as compared with the DM versus the SNP set; Figure 2—CPD set). MSA-derived features include information pertaining to evolutionary conservation (genomic or protein level). As these features proved to be the most informative (of all the features studied) in terms of discriminating between disease-causing mutations and putatively neutral SNPs, it is likely that the lower level of evolutionary conservation was responsible for the reduced performance in discriminating between CPDs and the SNP set. As all feature subsets demonstrated reduced performance in classifying CPDs as compared with disease-causing mutations, it is apparent that CPDs cannot simply be regarded as disease-causing mutations that happen to occur in evolutionarily unconserved residues; rather, CPDs must differ from standard disease-causing mutations in terms of other features such as the immediate protein context and the structural location of the affected residue within the body of the protein molecule.
Table 1. Evaluation of feature subsets (ROC-AUC, 10-fold cross-validation with linear support vector machine classifier) to discriminate between CPDs versus SNPs and disease-causing mutations versus common SNPs (DM versus SNPs).
| Feature subset | CPD-AUC (%) | DM-AUC (%) | Performance reduction for CPD (CPD-AUC versus DM-AUC) |
|---|---|---|---|
| Genomic MSA | 74.0 | 94.6 | −20.6 |
| Protein MSA (homologous) | 60.4 | 80.7 | −20.2 |
| Local protein structure | 56.5 | 68.5 | −12.0 |
| Regional protein composition | 55.3 | 63.5 | −8.2 |
| Exonic features | 64.7 | 71.0 | −6.4 |
| Annotated functional sites | 50.3 | 55.3 | −5.0 |
| Amino-acid features | 64.5 | 69.1 | −4.6 |
| Random value (control) | 50.0 | 48.7 | 1.3 |
Features ranked by performance reduction between CPD set and DM set.
Figure 2.
Evaluation of different groups of commonly used features (feature subset ranking) for identifying disease-causing AAS. Each feature subset is then evaluated (10-fold cross-validation using a linear support vector machine) in the context of (i) discriminating between CPDs and common polymorphisms (CPD versus SNP set), (ii) discriminating between disease-causing variants (DMs) and common polymorphisms (DM versus SNP set). The AUC of the ROC curve was then calculated for each feature subset; features employed in each subset are shown in Supplementary Table S1. As a control, a random feature was computed for each training example and the AUC of the ROC calculated.
A machine-learning approach to evaluate potential CPDs
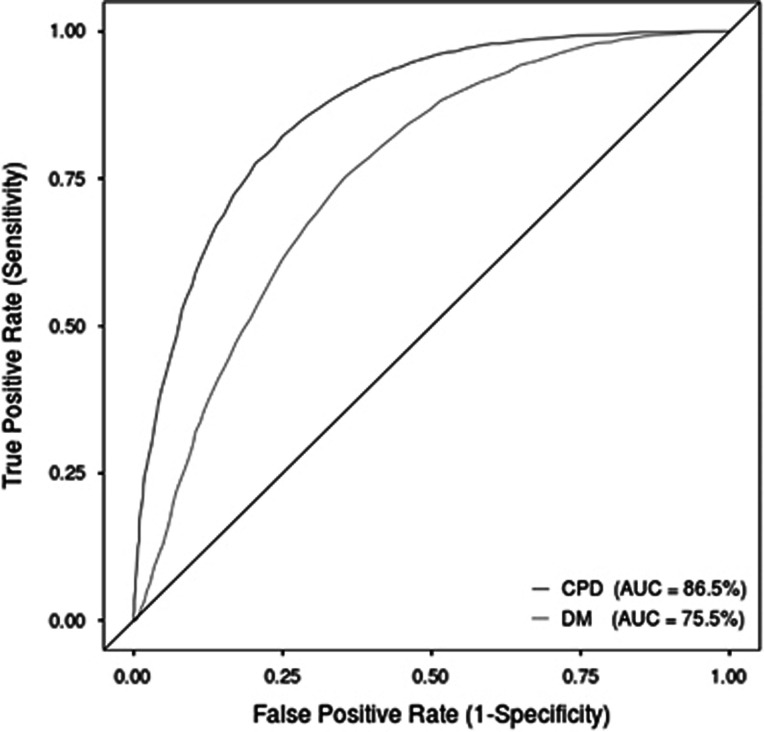
We next prototyped two different machine-learning approaches to assess the pathogenicity of potential CPDs using the features indicated in Supplementary Table S2 (see Materials and Methods section). The CPD-trained model (AUC=86.21%) outperformed the DM-trained model (with no MSA features) by over 10% based on the AUC (Table 2 and Figure 3). Both models shared similar false-positive rates with the DM-trained model exhibiting reduced sensitivity (−23.2%) as compared with the CPD-trained model. In the context of identifying CPDs, the CPD model prototyped here outperformed all existing bioinformatic prediction methods evaluated in this study, indicating utility in developing a specific CPD predictor. The work performed here also demonstrates that bioinformatic prediction methods (eg, MetaSVM; 85.6% AUC) not specifically developed for predicting the pathogenicity of CPDs can still be useful in this context. Future avenues to explore would include the further development of the prototype and the evaluation of an ensemble model combining a general model such as MetaSVM with a CPD-specific model. It may also be worthwhile trying to identify novel CPD-specific features, which could prove useful in classifying CPDs.
Table 2. Performance benchmarks for the identification of CPDs based on the two machine-learning models prototyped in this study.
| Data set | TPR (%) | FPR (%) | MCC | AUC of ROC |
|---|---|---|---|---|
| CPD-trained model | 78.05 | 21.75 | 0.56 | 86.21 |
| DM-trained model (no MSA features) | 54.84 | 21.98 | 0.34 | 75.10 |
Abbreviations: FPR, false-positive rate; MCC, Matthew’s Correlation Coefficient; MSA, multiple sequence alignments; TPR, true-positive rate.
The first model, termed the 'CPD-trained model', was trained using the sets of CPDs and common SNPs employed in this study (CPD and SNP sets). The second model, the 'DM-trained model', was trained using disease-causing mutations and common SNPs (DM and SNP sets) but excludes any features derived from MSA. The Random Forest machine-learning algorithm was employed, and evaluation was performed using a variation of 10-fold cross-validation, whereby the positive evaluation set in each fold comprised unseen examples from the CPD set for both models (DM-trained model and CPD-trained model). An MCC of −1 represents the worst possible prediction, 0 a random prediction and +1 a perfect prediction.
Figure 3.
Performance evaluation for the identification of CPDs by means of a ROC curve using 10-fold cross-validation of the two Random Forest prediction models prototyped in this study. The first model, termed the 'CPD-trained model', was trained using the CPD and SNP sets employed in this study (red line). The second model, the 'DM-trained model', which excludes any features derived from MSAs, was trained using disease-causing variants (DM and SNP sets) but in each ‘test’ fold the positive variants were derived from the CPD set (blue line). The AUC was then calculated for each model (CPD-trained model and DM-trained model). An AUC of 100% would represent a perfect predictor, whereas an AUC of 50% would correspond to a prediction tool making random predictions (represented by the diagonal line). The full colour version of this figure is available at European Journal of Human Genetics online.
In conclusion, the scale of the CPD phenomenon is such that a significant proportion of disease-associated human variants (a minimum of 3.7%) are found as the wild-type allele in at least one of the other mammalian species analyzed here. In terms of evaluating the pathological impact of such variants, traditional approaches suffer from the serious drawback of relying upon the evolutionary conservation score between homologous proteins irrespective of the influence of genetic variation at other amino-acid positions. Here we demonstrate the poor performance of established mutation prediction tools to assess the pathological significance of CPDs and show that the development of new tools, which result in increased prediction accuracy, is possible. The in silico assessment of pathogenesis for novel CPDs identified by whole-exome/genome NGS studies currently requires a different protocol from that employed for the bulk of non-compensated variants. We propose a two-stage analysis, whereby whole-exome/genome data should first be screened for potential CPDs (using the strategy employed in this study (and in others)5 to identify missense variants where the mutant amino acid represents the wild-type amino acid in another mammalian species). A method such as the novel CPD prediction tool prototyped here could then be applied to identify any high confidence CPD candidates for further analysis. This work represents the first step toward the development of a new algorithm for identifying CPDs. The prototype algorithm presented here remains to be extensively evaluated but nevertheless promises to form the backbone of a CPD-specific protocol that could help to avoid the misclassification of a sizable proportion of pathological missense variants as benign.
Acknowledgments
IPATIMUP integrates the i3S Research Unit, which is partially supported by FCT, the Portuguese Foundation for Science and Technology. This work is funded by FEDER funds through the Operational Programme for Competitiveness Factors—COMPETE and National Funds through the FCT-Foundation for Science and Technology, under the projects ‘PEst-C/SAU/LA0003/2013’. DNC and MM gratefully acknowledge financial support from Qiagen Inc. through a License Agreement with Cardiff University.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Azevedo L, Carneiro J, van Asch B, Moleirinho A, Pereira F, Amorim A: Epistatic interactions modulate the evolution of mammalian mitochondrial respiratory complex components. BMC Genomics 2009; 10: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Zhang J: Why are some human disease-associated mutations fixed in mice? Trends Genet 2003; 19: 678–681. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG et al: Evolutionary and biomedical insights from the rhesus macaque genome. Science 2007; 316: 222–234. [DOI] [PubMed] [Google Scholar]
- Huang H, Winter EE, Wang H et al: Evolutionary conservation and selection of human disease gene orthologs in the rat and mouse genomes. Genome Biol 2004; 5: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS, Sunyaev S, Kondrashov FA: Dobzhansky-Muller incompatibilities in protein evolution. Proc Natl Acad Sci USA 2002; 99: 14878–14883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Prado-Martinez J, Sudmant PH et al: Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 2015; 348: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DM, Frangakis SG, Golzio C et al: Identification of cis-suppression of human disease mutations by comparative genomics. Nature 2015; 524: 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E et al: Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science 1997; 276: 2045–2047. [DOI] [PubMed] [Google Scholar]
- Santisteban I, Arredondo-Vega FX, Kelly S et al: Three new adenosine deaminase mutations that define a splicing enhancer and cause severe and partial phenotypes: implications for evolution of a CpG hotspot and expression of a transduced ADA cDNA. Hum Mol Genet 1995; 4: 2081–2087. [DOI] [PubMed] [Google Scholar]
- Bienvenu T, Petitpretz P, Beldjord C, Kaplan JC: A missense mutation (F87L) in exon 3 of the cystic fibrosis transmembrane conductance regulator gene. Hum Mutat 1994; 3: 395–396. [DOI] [PubMed] [Google Scholar]
- Gascon-Bayarri J, Campdelacreu J, Sanchez-Castaneda C et al: Leukoencephalopathy with vanishing white matter presenting with presenile dementia. J Neurol Neurosurg Psychiatry 2009; 80: 810–811. [DOI] [PubMed] [Google Scholar]
- Bénit P, Rey F, Melle D, Munnich A, Rey J: Five novel missense mutations of the phenylalanine hydroxylase gene in phenylketonuria. Hum Mutat 1994; 4: 229–231. [DOI] [PubMed] [Google Scholar]
- Mavinakere M, Morizono H, Shi D, Allewell NM, Tuchman M: The clinically variable R40H mutant ornithine carbamoyltransferase shows cytosolic degradation of the precursor protein in CHO cells. J Inherit Metab Dis 2001; 24: 614–622. [DOI] [PubMed] [Google Scholar]
- Gilbert-Dussardier B, Segues B, Rozet JM et al: Partial duplication [dup. TCAC (178)] and novel point mutations (T125M, G188R, A209V, and H302L) of the ornithine transcarbamylase gene in congenital hyperammonemia. Hum Mutat 1996; 8: 74–76. [DOI] [PubMed] [Google Scholar]
- Azevedo L, Suriano G, van Asch B, Harding RM, Amorim A: Epistatic interactions: how strong in disease and evolution? Trends Genet 2006; 22: 581–585. [DOI] [PubMed] [Google Scholar]
- Consortium TCSaA: Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 2005; 437: 69–87. [DOI] [PubMed] [Google Scholar]
- Dvorakova L, Sikora J, Hrebicek M et al: Subclinical course of adult visceral Niemann–Pick type C1 disease. A rare or underdiagnosed disorder? J Inherit Metab Dis 2006; 29: 591–591. [DOI] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR et al: Ensembl 2013. Nucleic Acids Res 2012; 41: D48–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D et al: Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7: 539–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN: The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2014; 133: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S: SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 2003; 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C: Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol 2007; 8: R232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Chan AP: PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015; 31: 2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wu C, Li C, Boerwinkle E: dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat 2016; 37: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WC, Kim D, Carter H, Diekhans M, Ryan MC, Karchin R: CHASM and SNVBox: toolkit for detecting biologically important single nucleotide mutations in cancer. Bioinformatics 2011; 27: 2147–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik VN, Vapnik V: Statistical Learning Theory Vol 1. Wiley: New York, NY, USA, 1998. [Google Scholar]
- Breiman L: Random Forests. Mach Learn 2001; 45: 5–32. [Google Scholar]
- Matthews BW: Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta 1975; 405: 442–451. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C: Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 2011; 39: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.