Abstract
Verheij syndrome, also called 8q24.3 microdeletion syndrome, is a rare condition characterized by ante- and postnatal growth retardation, microcephaly, vertebral anomalies, joint laxity/dislocation, developmental delay (DD), cardiac and renal defects and dysmorphic features. Recently, PUF60 (Poly-U Binding Splicing Factor 60 kDa), which encodes a component of the spliceosome, has been discussed as the best candidate gene for the Verheij syndrome phenotype, regarding the cardiac and short stature phenotype. To date, only one patient has been reported with a de novo variant in PUF60 that probably affects function (c.505C>T leading to p.(His169Tyr)) associated with DD, microcephaly, craniofacial and cardiac defects. Additional patients were required to confirm the pathogenesis of this association and further delineate the clinical spectrum. Here we report five patients with de novo heterozygous variants in PUF60 identified using whole exome sequencing. Variants included a splice-site variant (c.24+1G>C), a frameshift variant (p.(Ile136Thrfs*31)), two nonsense variants (p.(Arg448*) and p.(Lys301*)) and a missense change (p.(Val483Ala)). All six patients with a PUF60 variant (the five patients of the present study and the unique reported patient) have the same core facial gestalt as 8q24.3 microdeletions patients, associated with DD. Other findings include feeding difficulties (3/6), cardiac defects (5/6), short stature (5/6), joint laxity and/or dislocation (5/6), vertebral anomalies (3/6), bilateral microphthalmia and irido–retinal coloboma (1/6), bilateral optic nerve hypoplasia (2/6), renal anomalies (2/6) and branchial arch defects (2/6). These results confirm that PUF60 is a major driver for the developmental, craniofacial, skeletal and cardiac phenotypes associated with the 8q24.3 microdeletion.
Introduction
The increasing use of next-generation sequencing technologies, including whole exome sequencing and whole genome sequencing, has led to the massive identification of novel genes with de novo heterozygous variants as a major cause of intellectual disability (ID).1, 2, 3 The PUF60 (Poly-U Binding Splicing Factor 60 kDa) gene encodes a general splicing factor that facilitates 3′ splice-site recognition at the early stages of spliceosome assembly.4, 5 PUF60 has been partly associated with the rare 8q24.3 microdeletion syndrome phenotype, in particular craniofacial and heart manifestations, in six patients and one fetal case. This rare condition, also named Verheij syndrome, is characterized by low birth weight and growth retardation, microcephaly, vertebral anomalies, joint laxity, and/or dislocation, developmental delay (DD), cardiac defects, renal defects and dysmorphic features.6, 7 It can be considered, at least for now, as a contiguous gene syndrome characterized by the existence of apparently unrelated clinical features in the same patient, resulting from a chromosomal rearrangement involving contiguous dosage-sensitive genes. Indeed, in 2013, Dauber et al8 reported on five individuals carrying overlapping 8q24.3 microdeletions that included SCRIB, NRBP2 and PUF60, and demonstrated by using functional experiments on zebrafish that the patients’ phenotype resulted from a combination of endophenotypes induced by the haploinsufficiency of PUF60 (short stature, craniofacial and cardiac anomalies) and SCRIB (coloboma and renal anomalies). They also identified a patient with microcephaly, short stature, DD and heart defects with a de novo c.505C>T variant leading to a p.His169Tyr change in PUF60.8 This is the only patient reported to date with a de novo function-affecting variant in PUF60.
Here, we report five additional patients with similar clinical features and de novo heterozygous variants in PUF60 identified by whole exome sequencing (WES) and confirm the major role of PUF60 in the phenotype of patients with the 8q24.3 microdeletion syndrome.
Subjects and methods
Patients
WES was performed in five unrelated individuals with developmental delay, congenital malformations and distinct facial dysmorphism in order to identify the molecular cause of their conditions, after negative standard genetic investigations and malformation screening. WES was used in a diagnostic approach in patient 1 and international data sharing was initiated to find other patients with very rare de novo PUF60 variants. This allowed us to identify patients 2–5. Written informed consent was obtained from all of the subjects’ parents for WES and for the use of photographs for patients 1, 3, 4 and 5.
Whole exome sequencing
WES was performed for patient 1 at Integragen (Evry, France). Targeted regions were captured using the SureSelect Human All Exon V5 kit (Agilent, Santa Clara, CA, USA) according to the manufacturer’s recommendations. Sequencing of paired-end 76-bp reads was carried out on a HiSeq 2000 (Illumina, San Diego, CA, USA). Reads were aligned to the human genome reference sequence (GRCh37/hg19 build of UCSC Genome Browser; http://genome.ucsc.edu) with the Burrows–Wheeler Aligner (BWA, v.0.6.2). Duplicate paired-end reads were marked with Picard v.1.77 (http://picard.sourceforge.net/). The Genome Analysis Toolkit (GATK) v.2.6–4 was used for base quality score recalibration, indel realignment and variant discovery. Variants were annotated with SeattleSeq SNP Annotation 138 (http://snp.gs.washington.edu/SeattleSeqAnnotation138/). Candidate de novo events were systematically identified by focusing on protein-altering and splice-site variants present at a frequency of <1% in the database of single-nucleotide polymorphisms (dbSNP) (dbSNP 146; http://www.ncbi.nlm.nih.gov/SNP/), absent from 100 local exomes of unaffected individuals and supported by at least four reads in the index subject and absent in both parents (less than two variant reads) at base-pair positions covered by at least four reads in the entire trio. Candidate de novo events were then manually inspected using the Integrative Genomics Viewer (IGV, Broad Institute; http://www.broadinstitute.org/igv/.
WES was performed in patients 2 and 3 in a trio diagnostic approach (patient and both parents) at the Radboud University Medical Center in Nijmegen, The Netherlands, essentially as described before.9 Exome capture was performed with the Agilent SureSelect Human All Exon v4 enrichment kit (Agilent Technologies). WES was performed on the Illumina HiSeq platform (BGI, Copenhagen, Denmark). Data were analyzed with BWA (read alignment) and GATK (variant calling) software packages. Variants were annotated using an in-house-developed pipeline. Variants were prioritized using an in-house-designed ‘variant interface’ and manual curation. De novo variants were considered the main candidates. Potentially causative variants were confirmed by Sanger sequencing.
WES was performed in patient 4 in a trio research approach at the Charles University and University Hospital Motol in Prague, Czech Republic. Whole exome capture was performed with SeqCap EZ Human Exome Library v3.0 (Roche NimbleGen, Madison, WI, USA). WES was performed on the Illumina HiSeq 1500 platform (Illumina) according to the manufacturer’s recommendations for paired-end 101 bp reads. Sequences aligned using Novoalign (Novocraft, Petaling Jaya, Selangor, Malaysia) were processed according to GATK best practices workflow. Variants were annotated using the GEMINI framework and subjected to several levels of filtering. Common variants (allele frequency >0.01) from public human genetic variation databases (dbSNP, 1000 Genomes, Exome Variant Server (EVS) and the Exome Aggregation Consortium (ExAC)) or from the in-house database were removed.
WES was performed in patient 5 in a trio diagnostic approach (patient and both parents) at the Center of Medical Genetics of the University and University Hospital of Antwerp, Belgium, essentially as described before.10 In brief, exome capture was performed with the SeqCap EZ Human Exome Library enrichment kit (Nimblegen). WES was performed on the Illumina HiSeq platform. Data were analyzed using BWA (read alignment) and GATK (variant calling) software packages. Variants were annotated using an in-house-developed pipeline. Variants were prioritized using our program VariantDB.11 Potentially causative variants were confirmed by Sanger sequencing.
We have submitted data for the variants to the gene variant database at www.LOVD.nl/PUF60 (patient IDs 73735–73739, 78890).
Results
Clinical assessment
Patient 1
This 3-year-old French girl (Figure 1) was the second child of healthy non-consanguineous French parents. There was no family history. The pregnancy was marked by isolated IUGR. She was born at 39 weeks of gestation (WG), she weighed 2675 g (7th centile), had a length of 43 cm (<<3rd centile) and a head circumference of 34 cm (50th centile). At birth, bilateral microphthalmia associated with iris coloboma, a naevus flammeus on the forehead and eyelids, bilateral branchial cleft cysts, bilateral clinodactyly of the fifth digits and valgus deformity of one foot were noted. Several facial features, including asymmetric and posteriorly rotated ears, retrognathism, long philtrum with thin upper lip and anteverted nares, were also noted (Figure 1). She also had a ventricular septal defect and unilateral hip dislocation. Fundus and brain MRI showed bilateral microphthalmia and retinal coloboma involving the optic discs, bilateral optic nerve hypoplasia (Figure 2) and an ectopic posterior pituitary without interruption of the pituitary stalk. Endocrinological tests, abdominal/renal ultrasonography and skeleton X-rays were normal. Standard karyotyping (46,XX) and array-CGH were normal. Molecular analysis of several genes, including RAX, VSX2, OTX2, SOX2 and FOXE3, known to be responsible for microphthalmia, was performed (Sanger sequencing). The results were negative, as was the molecular analysis of a targeted panel of 187 known or candidate genes involved in ocular malformations (Toulouse, France). This panel comprised genes that were already associated with ocular developmental defects (ALDH1A3, ATOH7, BCOR, BMP4, BMP7, c12orf57, CHD7, COX7B, FNBP4, FOXC1, FOXE3, GDF6, GJA8, HCCS, MFRP, OTX2, PAX2, PAX6, PITX2, PITX3, RARB, RAX, SIX6, SMOC1, SOX2, STRA6, TENM3, VAX1 and VSX2) and other candidate genes, including candidates from the retinoic acid or SHH pathways or identified through our previous exome sequencing experiments. Libraries were pooled and sequenced on an Illumina HiSeq 2000 as paired-end 75 bp reads. The bioinformatics analysis of sequencing data was based on the Illumina pipeline (CASAVA1.8.2). Mean coverage of the targeted exons was 250 × and ≥99% of the targeted exons were covered over 25 ×. The proband had developmental delay but was able to sit without support at 11 months. On the last examination at age 3 years, she had just acquired independent walking. Her hearing appeared to be normal. She had poor eye contact because of visual impairment (refraction +6), reacted to light and could follow large moving targets. She had behavioral anomalies with intolerance to frustration and echolalia. She had severe feeding difficulties and could only eat blended food, with frequent vomiting. Her measurements were 83 cm for height (<−3 SD), 10 kg for weight (<−3 SD) and 47.5 cm for OFC (−1.5 SD) with micromelia. Her facial features are summarized in Table 1. Bilateral clinodactyly of the fifth fingers, pectus excavatum and two branchial cleft cysts were also noted (Figure 1).
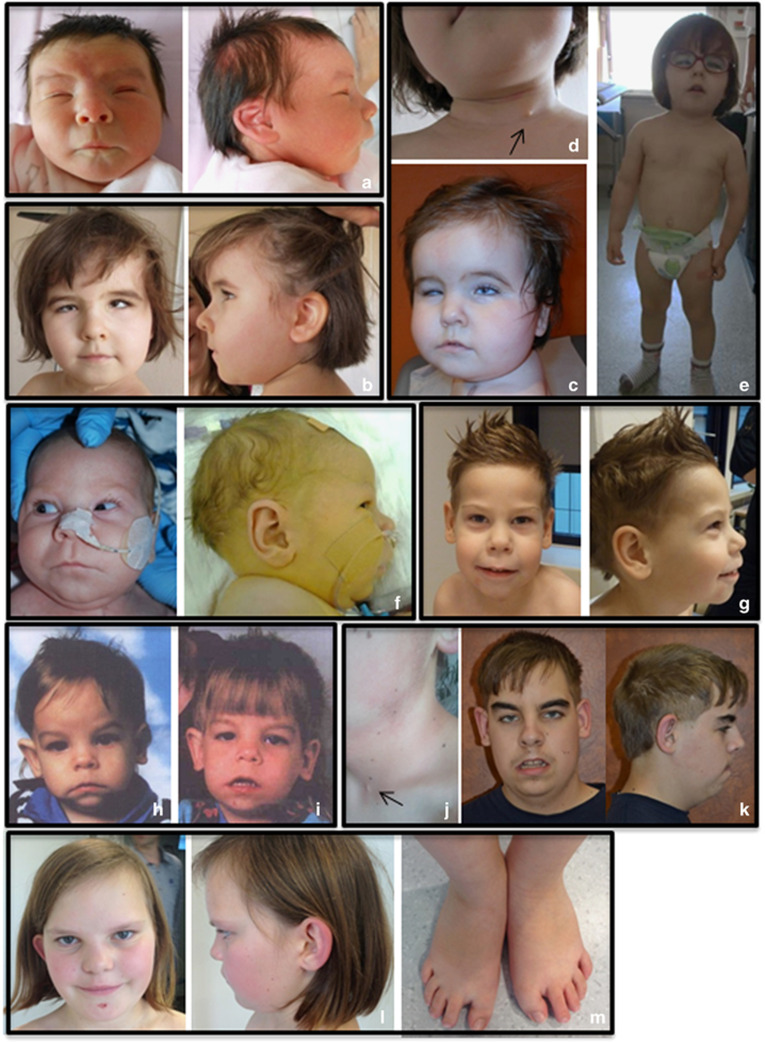
Figure 1.
Facial characteristics of the individuals with PUF60 variants. (a–c) Frontal and lateral views of patient 1 at birth (a) at 3 years and 3 months (b) and at 1 year (c) demonstrating facial dysmorphism including a square face, full cheeks, prominent forehead with bitemporal narrowing, low-set eyebrows, asymmetric and posterior rotated ears, bilateral colobomatous microphtalmia with horizontal nystagmus and convergent strabismus, bilateral epicanthal folds, wide nasal bridge with broad nasal tip and anteverted nares, thin upper lip with a long philtrum, micrognathism and broad short neck. (d) Lateral view of the neck of patient 1 showing a branchial cleft cyst (arrow). (e) Full-body view of patient 1 showing the contrast between the normal head circumference and the low body height and weight with relatively short arms, and a pectus excavatum. (f, g) Frontal and lateral views of patient 3 at birth (f) and at 3 years and 10 months (g) showing an elongated face, bilateral epicanthal folds, short up-slanted palpebral fissures, long eyelashes, anteverted nares, long prominent philtrum, thin upper lip and a short neck. (h–k) Frontal and lateral views of patient 4 at the age of 8 months (h), 2 years (i) and 17 years (k). (j) Lateral view of the neck of patient 4 showing a branchial cleft cyst (arrow). (l, m) Frontal and lateral view of patient 5 at the age of 11 years (l) showing deep set eyes, broad nasal bridge with protruding ears. (m) Flat feet with short halluces and sandal-gap deformity.
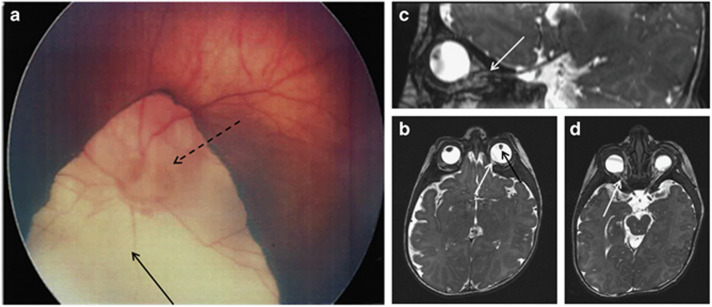
Figure 2.
Fundus and brain MRI results. (a) Fundus view showing the retinal coloboma (full-line arrow) of left eye involving the optic disk (dotted-line arrow) – type IV of the Gopal classification. (b–d) Brain and optic nerve MRI (2 months) showing bilateral microphthalmia (b) associated with irido coloboma (black arrow) and retinal coloboma (white arrow), and the bilaterally thin (hypoplastic) optic nerve hypoplasia predominantly in the intra-orbital portion (c, d) (white arrows).
Table 1. Phenotype of the patients with a PUF60 variant and the patients with a 8q24.3 deletion comprising PUF60.
| Patients with a PUF60 variant | Patients with a 8q24.3 deletion | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | P4 | P5 | Dauber et al,8 P6 | Verheij et al,6 P1 | Dauber et al,8 P1 | Dauber et al,8 P2 | Dauber et al,8 P3 | Dauber et al,8 P4 | Dauber et al,8 P5 | Wells et al,7 P1 | |
| Gender | F | M | M | M | F | F | M | F | M | F | F | F | F |
| Age | 3 y | 14 y | 7 y | 17 y | 12 y 2 m | 21 y | 4 m | 10 y | 6 y | 17 y | 9 y | 11 y | 28+6 WG |
| Facial features | |||||||||||||
| Square face | + | + | − | + | + | + | + | + | + | + | + | + | + |
| Full cheeks | + | − | + | + | + | − | + | + | + | + | + | + | − |
| Bitemporal narrowing | + | − | + | + | − | ND | + | ND | ND | + | + | + | + |
| Prominent forehead | + | + | + | + | + | + | + | + | + | − | + | + | − |
| Low set eyebrows | + | + | + | + | + | + | + | − | + | − | + | + | + |
| Wide nasal bridge | + | + | − | + | + | − | + | ND | + | − | + | + | + |
| Broad nasal tip | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Anteverted nares | + | + | + | + | − | + | + | + | + | + | + | + | + |
| Long philtrum | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Thin upper lip | + | − | + | + | + | + | + | + | + | + | + | + | + |
| Micro- or retrognathism | + | − | − | + | − | + | + | + | − | − | − | + | + |
| Short neck | + | − | − | + | + | + | + | − | + | + | + | + | + |
| Branchial arch anomalies | Branchial cleft cysts | − | − | Branchial cleft cysts, uvula dimple | − | − | Branchial arch remnant, fistula | Bilateral preauricular pits | − | − | Bilateral preauricular pits | − | − |
| Eye anomalies | |||||||||||||
| Coloboma | + (IRC) | − | − | − | − | − | + (IRC) | + (IC) | + (IRC) | − | − | − | − |
| Other | Bilateral microphtalmia and optic nerve hypoplasia | Strabismus | − | −a | Bilateral optic nerve hypoplasia | − | Hyperopia | ||||||
| Cardiac defects | |||||||||||||
| VSD | + | − | + | − | − | + | + | − | − | + | − | + | − |
| Other | − | − | Truncus arteriosus | Aortic insufficiency | AVSD | Bicuspid aortic valve | − | − | − | Truncus arteriosus | Aortic coarctation | AVSD, hypoplastic aortic arch | |
| Laryngo-tracheo-broncomalacia | − | − | + | − | − | − | + | − | − | − | − | + | − |
| Growth | |||||||||||||
| IUGR | + | − | + | − | − | + | + | + | − | + | + | − | + |
| Short stature | + | − | + | + | + | + | + | + | + | + | + | + | NA |
| Micromelia | + | − | + | − | − | + (asymmetric) | ND | ND | ND | ND | ND | ND | + |
| Microcephaly | − | − | + | − | − | + | + | + | + | − | − | + | − |
| Skeletal anomalies | |||||||||||||
| Joint laxity/dislocation | +b | − | + | + | + | +b | +b | − | + | +c | + | +b | − |
| Distal limb anomalies | + a, b | + a | − | + b, f, i | j, k | + c | + d, e | − | − | − | + | + a, h | + a, f, g |
| Vertebral anomalies | − | − | Fusion of the left facet joints of C5−C6 | Unfused arch of C7, scoliosis | − | Hemivertebrae, scoliosis | − | + | − | Fusion of L5-S1 | − | Hemivertebrae, scoliosis | Mild scoliosis, abnormal third sacral vertebra |
| Feeding difficulties | + | − | + | − | + | − | + | − | − | + | + | + | NA |
| Neurology | |||||||||||||
| Developmental delay | + | + | + | + | + | + | + | − | + | + | + | + | NA |
| Seizures | − | − | − | −d | − | + | + | − | − | − | − | + | − |
| Brain malformation | Posterior pituitary ectopia | ND | − | ND | CC hypoplasia | Enlarged 3rd and lateral ventricles, thin CC | Hypoplastic CC, cerebral atrophy, enlarged ventricles | − | Cerebral atrophy | − | Ischemic encephalomalacia | Volume loss in the left temporal lobe | − |
| Renal anomalies | − | − | − | Hypoplastic kidneys | Pelvic left kidney with unilateral VUR | − | − | Left renal agenesis | − | Left polycystic kidney | Ectopic fused kidneys | Right kidney hypoplasia with renal cyst | − |
| Other | Naevus flammeus on the forehead and eyelids, pectus excavatum | Autistic features | Autistic features, pectus excavatum, scoliosis | Trilobar left lung | Cleft palate, BPD, pes planus, hemifacial microsomia | Coarse hair, CHL | Recurrent infections, hiatus hernia | − | Forehead capillary hemangioma | Cyclic vomiting | Small medial alveolar cleft | ||
| De novo PUF60 variants (NG_033879.1; NM_078480.2) | c.24+1G>C chr8:g. 144911449C>G | c.1342C>T p.(Arg448*) | c.1448T>C p.(Val483Ala) | c.407_410delTCTA p.(Ile136Thrfs*31) | c.901A>T p.(Lys301*) | c.505C>T p.(His169Tyr) | Deletion | Deletion | Deletion | Deletion | Deletion | Deletion | Deletion |
Abbreviations: AVSD, atrioventricular septal defect; BPD, bronchopulmonary dysplasia; CC, corpus callosum; CHL, conductive hearing loss due to middle ear malformation; IC, iris coloboma; IRC, irido–retinal coloboma, IUGR, intrauterine growth retardation; NA, not applicable; ND, not determined, VSD, ventricular septal defect; VUR, vesicoureteral reflux; WG, weeks of gestation.
Distal limb anomalies: a: clinodactyly of the fifth digit; b: valgus deformity of the feet; c: absent right thumb, hypoplastic metacarpal and scaphoid bones; d: bilateral post-axial polydactyly; e: oligodactyly and fusion of the third and fourth metatarsal of the left foot; f: brachymesophalangy of the fifth digit; g: rocker-bottom feet; h: Bilateral transverse palmar creases; i: overlying proximal thumb; j: brachydactyly, k: flat feet with short halluces and sandal gap deformity.
No focused ophthalmological examinations have been performed since the age of 3 years.
Hip dislocation.
Elbow subluxation with malformation of the great sigmoid cavity.
EEG showed epileptiform discharges with absence of seizures.
Patient 2
The second patient, a 14-year-old boy, was the fourth child of healthy, non-consanguineous Dutch parents. He was born after an uneventful pregnancy. His growth was normal. Developmental delay was noted from the age of 1 year. At the age of 6 years, his IQ was 46 and an autism spectrum disorder was diagnosed. The family history was negative for ID or congenital malformations. Distinct dysmorphic features were noted: exotropia, a square face with a prominent forehead, low-set eyebrows, a wide nasal bridge with broad nasal tip and anteverted nares, a long philtrum and bilateral clinodactyly of the fifth digits (Figure 1 and Table 1). The parents did not permit a brain MRI. FMR1 testing and metabolic screening were negative. Array-CGH revealed two numerical variants inherited from the parents: hg19 chr11:g.(133072607_133078519)_(133691682_133693512)dup; chr13:g.(48465866_48466014)_(48540617_48540958)del (ISCN 2016 nomenclature: arr[GRCh37] 11q25(133078519_133691683)x3 pat, 13q14.2(48466014_48540618)x1 mat). The maternal deletion at 13q14.2 (75 kb) only includes the SUCLA2 gene that is associated with autosomal recessive mitochondrial depletion syndrome type 5. SUCLA2 gene was therefore sequenced in the child, but no variants were detected on the other allele. The paternal duplication at 11q25 (600 Kb) includes OPCML gene that has uniquely been involved in ovarian and colorectal cancer to date. Hence, the absence of potential causative gene, notably syndromic ID gene, in both rearrangements, combined to the fact that they were inherited from unaffected parents, led us to consider those as not likely to contribute to the patient’s phenotype. In addition, no affected patients have been reported in the literature or in the DECIPHER (https://decipher.sanger.ac.uk/) database with such chromosomal aberrations.
Patient 3
This 7-year-old Dutch boy was the first child of healthy, non-consanguineous Dutch parents. The family history was unremarkable as was the pregnancy. He was born at 38 WG, weighed 2570 g (−2.1 SD) and his height was 49.5 cm (−0.7 SD). He was microcephalic (head circumference 32 cm (−3.2 SD) at age 3 weeks). Soon after birth, he was diagnosed with a heart defect, including a truncus arteriosus and a ventricular septal defect, for which he had multiple surgical interventions. He developed an inspiratory stridor and was diagnosed with a cricoid stenosis at the age of 2 years, probably because of multiple intubations. He also had difficulties swallowing and was fed by a percutaneous gastric tube from age 2 years. His facial features included an elongated face, short up-slanted palpebral fissures, bilateral epicanthal folds, long eyelashes, anteverted nares, long prominent philtrum, thin upper lip, short neck and micromelia (Figure 1 and Table 1). Ocular coloboma were not detected. A cervical spine CT-scan showed fusion of the left facet-joints of C5-C6. The brain MRI was normal. There were no renal malformations.
On follow-up, he showed DD and growth retardation (height −2 SD, weight −2.5 SD and head circumference <−2.5 SD). At the age of 4 years and 5 months, his verbal IQ was normal (100) but his performance IQ was much lower (67). His motor skills showed a delay of 1 year. At the last evaluation (6 years and 10 months), he attended a special school, because of his developmental delay and behavioral problems.
A 105K oligo-array (Agilent) was normal and molecular analysis of the NIPBL and SMC1A genes (Cornelia de Lange syndrome) was negative.
Patient 4
Patient 4 is a 17-year-old Czech boy, the first child of healthy non-consanguineous parents. The delivery was induced at 36 WG. His birth weight was 2470 g, length was 48 cm and head circumference was 33 cm. He needed intensive care because of respiratory distress syndrome during the first 3 weeks of life. Abdominal and hip ultrasound showed hypoplastic kidneys with left pyelectasis and hip dysplasia, respectively. His psychomotor development was delayed from the beginning (sitting without support at the age of 9 months, crawling at the age of 11 months and first steps from the age of 18 months). Eye examinations performed when he was a toddler were normal and no further examinations were done. Speech development was also delayed, with babbling until 4 years of age. At the last examination, speech was slow with signs of dyslalia. At the age of 4 years, moderate aortic insufficiency was revealed, associated with asymmetric coarctation and a dysplastic aortic valve. At 17 years of age, the height was 164.5 cm (−2.1 SD), weight was 59.6 kg (−0.9 SD) and the head circumference was 54 cm (<−1.6 SD). He had bifrontal narrowing, fused, thick and arched eyebrows, large dysplastic ears, a wide nasal bridge and long nose with a broad nasal tip and anteverted nares, a long smooth philtrum, thin upper lip and short neck. A branchial cleft cyst was noted on the neck as well as a fistula above the uvula. Skeletal features included slight shortening of the trunk, significant flattening of the chest, pectus excavatum, scoliosis, valgus deformity of the feet, long fingers and toes, overlying proximal thumbs, short fifth fingers with clinodactyly on the right digit, bilateral cervical ribs and an unfused arch of vertebra C7. He has moderate ID and autism, hyperactivity and transitional tic disorder. The EEG showed epileptiform discharges with the absence of seizures. SNP array analysis (Illumina HumanCytoSNP-12 BeadChips) was performed and was normal. Brain MRI has not yet been performed.
Patient 5
This female proband was born at 41 WG as the first child of non-consanguineous Belgian parents. Birth weight was 3560 g for a length of 50 cm. At birth, she was diagnosed with atrioventricular septal defect that was surgically corrected at age 1 month. Lung imaging revealed a trilobar left lung. She was also diagnosed with a left pelvic kidney associated with unilateral vesicoureteral reflux. Over time, left kidney function decreased significantly. During the first months of life, she had severe feeding problems and was fed by gastric tube. She developed postnatal growth retardation. At the age of 11 years and 2 months, she had a skeletal age of 9 years and 2 months. At the age of 3 years, she underwent tonsillectomy because of extreme snoring and obstructive apnea. She attended a regular school until the age of 7 years but is now in special education. MRI of the brain showed hypoplasia of the corpus callosum. The ophthalmological evaluation revealed bilateral optic hypoplasia.
At the clinical examination at age 12 years and 2 months, occipitofrontal circumference was 53.2 cm (0 SD), length 131 cm (−2.5 SD) and weight 33.3 kg (−1 SD). Craniofacial characteristics included deep set eyes, a broad nasal bridge and protruding ears. Both hands showed brachydactyly. She had flat feet with short halluces and sandal-gap deformity. Skin was normal.
Exome sequencing
Exome sequencing from a blood-derived DNA sample from patient 1 generated over 2.96 Gigabases of mappable sequence with a median sequencing depth provided for RefSeq coding exons and splice junctions of 85.8 ×. Depth of coverage of RefSeq coding bases and splice junctions was 94.2 × for at least 10 reads and 91 × for at least 20 reads calculated with the Genome Analysis Toolkit Depth of Coverage tool. After filtering, among the 16 622 non-synonymous splice-site and indel variants, 368 rare variants remained. Seventeen candidate variations of OMIM genes were found. Among these, after studying their expression and the disorders they were involved in, and comparisons with our patient’s phenotype, only three OMIM gene variants were retained. These candidate variants were validated and tested in parental DNA samples. A PAX6 missense change (NM_000280.4:c.518C>T, p.(Thr173Met)) and a PACS1 missense change (NM_018026.3:c.2197C>T, p.(Arg733Trp)) were found to be inherited from an asymptomatic parent, and a de novo PUF60 NM_078480.2:c.24+1G>C (chr8:g.144911449C>G) variant was found. Taking into account the familial segregation and reverse phenotyping in favor of Verheij syndrome,12 the PUF60 variant was considered the one affecting gene function and thus the likely cause of the phenotype (Table 2). Indeed, exon 1 skipping is expected as the wild-type donor site is predicted to be broken (Human Splicing Finder score of −31.86: most probably affecting splicing; http://www.umd.be/HSF3/HSF.html).
Table 2. Pathogenicity of each variant evaluated by different databases.
| Variant (hg19) | Amino-acid change | dbSNP | Frequency in ExAC | Grantham | Polyphen | GERP | CADD | MutationTaster | SIFT | |
|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | chr8:g.144911449C>G NG_033879.1; NM_078480.2:c.24+1G>C | NA | NA | 0 | NA | NA | 4.740 | 21.400 | 1 | NA |
| Patient 2 | chr8:g.144899118G>A NG_033879.1; NM_078480.2:c.1342C>T | p.(Arg448*) | NA | 0 | NA | NA | 5.280 | 36.000 | 1 | NA |
| Patient 3 | chr8:g.144898922A>G NG_033879.1; NM_078480.2:c.1448T>C | p.(Val483Ala) | NA | 0 | 64 | 0.989 | 5.270 | 24.100 | 0.999 | 0.001 |
| Patient 4 | chr8:g.144900643_144900646delTAGA NG_033879.1; NM_078480.2:c.407_410delTCTA | p.(Ile136Thrfs*31) | NA | 0 | NA | NA | 5.180 | NA | 1 | NA |
| Patient 5 | chr8:g.144899869T>A NG_033879.1; NM_07848.2:c.901A>T | p.(Lys301*) | NA | 0 | NA | NA | 3.930 | 33.000 | 1 | NA |
Trio WES and data analysis of patients 2 and 3 and their parents did not reveal any function-affecting variants of genes known to be associated with ID. Exome-wide analysis revealed a de novo PUF60 nonsense variant (NM_078480.2:c.1342C>T; p.(Arg448*)) in patient 2, and a de novo missense variant in PUF60 (NM_078480.2; c.1448T>C; p.(Val483Ala)) in patient 3 (Table 1 and Figure 3) that were considered of interest given the patients’ phenotypes. The nonsense variant p. Arg448* in exon 11 leads to protein truncation at position 448 with loss of RRM3 domain (462–549) (Uniprot). These variant were confirmed by Sanger sequencing. Both PUF60 variants identified were absent from all of the public genetic variant databases used (dbSNP142; Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP); ExAC, accessed in February 2016). Genomic positions were conserved (GERP 5.270) and bioinformatics tools supported the deleterious effect of the de novo missense change (PolyPhen-2 0.989, PhastCons 0,998, Grantham 64, CADD 24,1, Mutation Taster 0.99).
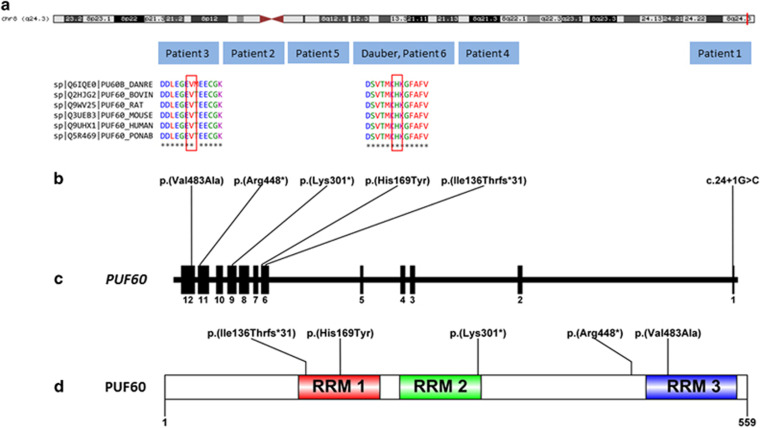
Figure 3.
Genetic location of the PUF60 variants identified to date. (a) Schematic human chromosome 8. The vertical red line indicates the position of the PUF60 gene at 8q24.3. (b) Variants identified in PUF60 by WES in the five patients of the present study and in the patient reported by dauber et al8, and conserved amino acid sequences among mammals. (c) A focus on the PUF60 locus with a schematic representation of exons (vertical black bars). (d) Distribution of amino-acid changes in PUF60. RRM, RNA recognition motif. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Trio WES and data analysis of patient 4 revealed a private heterozygous de novo deletion of four nucleotides in the PUF60 gene (NM_078480.2:c.407_410delTCTA, p.I136Tfs*31). Thanks to reverse phenotyping, the patient’s phenotype was deemed to correspond to the Verheij syndrome. This variant was also absent from all of the public genetic variant databases used (dbSNP142; Exome Variant Server, NHLBI GO ESP; ExAC, accessed in June 2016). The genomic position was conserved (GERP 4.28).
Trio WES and data analysis of patient 5 and her parents revealed a single de novo variant in known ID genes from a predefined panel, namely a nonsense mutation in the PUF60 gene: NM_07848.2: c.901C>T leading to p.Lys301*. Taking into account the familial segregation, the nature of the identified variant and reverse phenotyping in favor of Verheij syndrome, the PUF60 variant was considered to be the likely cause of the phenotype. De novo occurrence of the PUF60 variant was confirmed by Sanger sequencing.
Discussion
Here, we report five de novo PUF60 variants in five distinct new sporadic patients sharing common congenital anomalies associated with ID. This raises the total number of patient with these common phenotypes harboring PUF60 de novo variants to six.
Before this study, only one patient carrying a de novo variant in PUF60 had been reported by Dauber et al8 (patient 6). This female patient shared overlapping clinical features with patients harboring the previously described 8q24.3 deletion. These features included developmental delay, ante- and postnatal growth retardation, microcephaly, skeletal anomalies including hemivertebrae, scoliosis, distal limb anomalies and bilateral hip dislocation, craniofacial and cardiac defects (Table 1). The authors’ findings suggested that the patient’s variant (c.505C>T variant leading to a p.His169Tyr) was a loss-of-function allele that perturbed the splicing patterns of PUF60 itself. They showed, in an elegant manner using morpholino experiments in zebrafish, that PUF60 and SCRIB (a gene contiguous to PUF60) could be considered the main drivers of the 8q24.3 deletion-associated phenotype, SCRIB for coloboma and renal defects and PUF60 for the craniofacial, short stature and cardiac phenotype.8
Our five additional patients carrying a de novo heterozygous variant in PUF60 display obvious overlapping features with the first patient and with the individuals harboring the 8q24.3 microdeletion, as summarized in Table 1. Indeed, all six patients (present study and patient 6 from the study of Dauber et al8) carrying a PUF60 variant have the same core facial gestalt (Figure 1 and Table 1).6, 7, 8 The other overlapping clinical features were DD (6/6), cardiac defect (5/6), short stature (5/6), vertebral anomalies (3/6), moderate distal limb anomalies (5/6) and joint laxity/dislocation (5/6), suggesting that PUF60 variants that affect function are likely to be associated with this part of the phenotype observed in these patients (Table 1), namely the developmental, craniofacial, skeletal and cardiac phenotype. These findings are consistent with the significant and reproducible reduction of body length observed in the puf60 morphants by Dauber et al,8 and the demonstration that PUF60 loss of function drove a cardiac defect phenotype in 34% of puf60 MO-injected zebra fish embryos (Table 1).8 Other variable clinical features such as severe feeding difficulties (3/6), bilateral optic nerve hypoplasia (2/6) and branchial arch defects (2/6), the latter having been described in 3/7 8q24 deletion patients, were also observed in these patients.6, 7, 8 Taken together, these observations emphasize the major role of PUF60 in the phenotype of Verheij syndrome and contribute to the phenotypical delineation of this condition.
Interestingly, one of the patients of the present study with a PUF60 de novo variant had bilateral microphthalmia and irido–retinal coloboma (Table 1 and Figure 2). The presence of coloboma, as well as renal defects, observed in patients with the 8q24.3 deletion has been attributed to the SCRIB deletion, as 53% and 32% of the zebrafish embryos injected with a splice-blocking morpholino against scrib showed, respectively, coloboma and gross edema with abnormal diffusion of dextran, reflecting the loss of ability to maintain fluid homeostasis.8 In addition, SCRIB plays a key role in planar cell polarity and is involved in the formation of the pronephros and eye.13 SCRIB seems to remain a good candidate gene for kidney anomalies, as only two of the six patients with a PUF60 variant had a renal phenotype. However, the presence of a coloboma in one patient from this report prompted us to reconsider the conclusion that the coloboma observed in (3/7) patients with Verheij syndrome was induced by SCRIB haploinsufficiency alone (Table 1).
We observed several distal limb anomalies in both groups of variant or deletion carriers, including clinodactyly of the fifth digit, valgus deformity of feet, absent right thumb, hypoplastic metacarpal and scaphoid bones, bilateral post-axial polydactyly, oligodactyly and fusion of the third and fourth metatarsal of the left foot, brachymesophalangy of the fifth digit and rocker bottom feet. None of these were recurrent except for clinodactyly of the fifth digit, which was present in 4/11 patients, although this feature is not very specific. It is well known that PUF60 encodes a splicing factor that interacts directly with SF3B4, a component of the U2 pre-mRNA spliceosomal complex involved in Nager syndrome, suggesting that PUF60 and SF3B4 haploinsufficiency-associated phenotypes are partially due to a defect of spliceosome function.8, 14 Interestingly, the patient carrying a PUF60 variant reported by Dauber et al8 (patient 6) harbors clinical signs belonging to the Nager syndrome spectrum, including hemifacial microsomia with micro-retrognathism and preaxial upper-limb anomalies with an absent right thumb and hypoplastic metacarpal and scaphoid bones (Table 1).8
In conclusion, these results highlight the power of WES as an unbiased NGS-based strategy for identifying new genes causing very rare multisystemic conditions, 15 and the importance of data sharing to identify their underlying clinical expression that is otherwise hampered by the paucity of reported cases. This strategy allowed us to report autosomal dominant PUF60 variants in five unreported patients with overlapping features of 8q24.3 microdeletion syndrome and, thereby, to confirm the involvement of PUF60 in the Verheij syndrome phenotype. Further screening of PUF60 in other patients will provide important information about the frequency of PUF60 variants and may help to further define the phenotype associated with these variants.
Acknowledgments
We thank the patients and their families involved in the study, the University of Burgundy Centre de Calcul (CCuB) for technical support and management of the informatics platform. The Regional Council of Burgundy and the Centre Hospitalo-Universitaire de Dijon supported this work (PARI 2014). Grant supports were NF-CZ11-PDP-3-003-2014, NT/14200 and 00064203. We thank Philip Bastable from the ‘Pôle de Recherche’ of Dijon University Hospital for a helpful review of this article.
Footnotes
The authors declare no conflict of interest.
References
- Rauch A, Wieczorek D, Graf E et al: Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–1682. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT et al: Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347. [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, Lücke S, Green MR.: U2AF homology motifs: protein recognition in the RRM world. Genes Dev 2004; 18: 1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings ML, Allemand E, Duelli DM, Myers MP, Krainer AR.: Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF(65). PLoS One 2007; 2: e538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheij JB, de Munnik SA, Dijkhuizen T et al: An 8.35 Mb overlapping interstitial deletion of 8q24 in two patients with coloboma, congenital heart defect, limb abnormalities, psychomotor retardation and convulsions. Eur J Med Genet 2009; 52: 353–357. [DOI] [PubMed] [Google Scholar]
- Wells C, Spaggiari E, Malan V et al: First fetal case of the 8q24.3 contiguous genes syndrome. Am J Med Genet 2016; 170: 239–242. [DOI] [PubMed] [Google Scholar]
- Dauber A, Golzio C, Guenot C et al: SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am J Hum Genet 2013; 93: 798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Feenstra I, Gilissen C et al: A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat 2013; 34: 1721–1726. [DOI] [PubMed] [Google Scholar]
- Helsmoortel C, Vandeweyer G, Ordoukhanian P, Van Nieuwerburgh F, Van der Aa N, Kooy RF.: Challenges and opportunities in the investigation of unexplained intellectual disability using family-based whole-exome sequencing. Clin Genet 2015; 88: 140–148. [DOI] [PubMed] [Google Scholar]
- Vandeweyer G, Van Laer L, Loeys B, Van den Bulcke T, Kooy RF.: VariantDB: a flexible annotation and filtering portal for next generation sequencing data. Genome Med 2014; 6: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennekam RC, Biesecker LG.: Next-generation sequencing demands next-generation phenotyping. Hum Mutat 2012; 33: 884–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouloudaki K, Puetz M, Simons M et al: Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc Natl Acad Sci USA 2009; 106: 8579–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier FP, Caluseriu O, Ng S et al: Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet 2012; 90: 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycott KM, Dyment DA, Sawyer SL, Vanstone MR, Beaulieu CL: Identification of genes for childhood heritable diseases. Annu Rev Med 2014; 65: 19–31. [DOI] [PubMed] [Google Scholar]