Abstract
A remarkable proportion of factors causing genetic predisposition to breast cancer (BC) are unknown in non-BRCA1/2 families. Exome sequencing was performed for 13 high-risk Finnish hereditary breast and/or ovarian cancer (HBOC) families to detect variants contributing to BC susceptibility. After filtering, 18 candidate variants in DNA damage response (DDR) pathway genes were screened in 129 female HBOC patients, up to 989 female controls, and 31 breast tumours by Sanger sequencing/TaqMan assays. In addition, two variants were further studied in 49 male BC patients and 909 male controls. Second, all variants predicted to affect function in six early-onset BC patients were analysed in detail. Variants in ATM, MYC, PLAU, RAD1, and RRM2B were enriched in female HBOC patients compared with controls (odds ratio 1.16–2.16). A rare nonsynonymous variant in RAD50 was detected in a male BC patient. In addition, a very rare BRCA1 variant was identified in a single high-risk family. None of the variants showed wild-type allele loss in breast tumours. Furthermore, novel variants predicted to affect function were detected in early-onset patients in genes, which target DNA repair and replication, signalling, apoptosis, and cell cycle pathways. Family-specific enrichment of multiple DDR pathway gene defects likely explains BC predisposition in the studied families. These findings provide new information on potential BC-related pathways and an excellent premise for future studies.
Introduction
Breast cancer (BC) is the most common cancer and the leading cause of cancer-related deaths in women in both developed and developing countries. Worldwide, 1.67 million new BC cases are diagnosed each year, and the number of BC-related deaths is 522 000.1 In Finland, BC was diagnosed in 4694 women in 2012, and the annual number of BC diagnoses is predicted to increase (Finnish Cancer Registry). Up to 10% of all BCs are familial.2 Approximately 20–25% of familial BCs are due to germline variants in two high-risk genes, BRCA1 and BRCA2, which are tumour suppressor genes that encode large proteins that act in a common pathway of genome protection and have important roles at different stages in DNA damage response (DDR) and DNA repair.3, 4 Variant in BRCA1/2 confer a high risk of breast and ovarian cancers; the average cumulative cancer risks in a population-based series of unselected breast or ovarian cancer patients have been estimated to be 65% and 39% for BRCA1 variant carriers and 45% and 11% for BRCA2 variant carriers, respectively, by age of 70.5 In the Finnish population, the contribution of BRCA1/2 -variants to the breast/ovarian cancer families and ovarian cancer-only families is ~20% and 26%, respectively.6, 7
In addition to BRCA1/2, rare germline variants in the known high-risk genes TP53, CDH1, STK11, and PTEN predispose to familial cancer syndromes, among which BC is also observed.4 Moreover, genes for which their protein products interact with BRCA1/2 in the DDR pathway are strong candidates for breast and/or ovarian cancer susceptibility, including genes such as ATM, CHEK2, BRIP1, PALB2 that moderately increase the breast/ovarian cancer risk (by two- to fourfold).4 Moreover, large-scale genome-wide association studies have identified common variants in over 70 loci associated with BC, explaining ~14% of the familial risk of the disease.8 Despite the intensive efforts, the familial predisposition remains largely unresolved and the majority of this missing heritability likely comprises of thousands of rare variants, each presenting a minor disease risk.9
In this study, a family-based exome-sequencing approach was used to identify rare variants that contribute to breast/ovarian cancer predisposition in Finnish BRCA1/2 -negative hereditary breast and/or ovarian cancer (HBOC) families, many of which included very early-onset BC patients. The primary aim was to identify variants that are shared between affected family members and that target DDR pathway genes. The secondary aim was to identify novel candidate genes contributing to early-onset BC by exploring genes in any pathway, with a special focus on those that likely has a role in tumorigenesis such as pathways related to the cell cycle, DNA replication, signalling, DNA repair, and apoptosis.
Materials and methods
Study subjects
Genomic DNA samples of 37 individuals from 13 HBOC families were used for whole-exome sequencing. Six of the families belong to a previously characterised cohort of high-risk Finnish BRCA1/2 founder variant-negative HBOC individuals collected from Tampere University Hospital in which index patients have been screened for variants in BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 genes and for copy number variations on a genome-wide scale.10, 11 In one of these families, two CHEK2 variants, c.470T>C and c.1100delC, have been reported.10 Seven other families were selected into this study based on the previously described high-risk hereditary BC criteria.10 The families were recruited from Turku University Hospital, and the index patients had tested negative for BRCA1/2 founder-variants. From each of the 13 families, one to three breast or breast and ovarian cancer-affected individuals were selected for exome sequencing. Corresponding healthy relatives of the affected family members, used as controls, were selected from each family whenever possible. Of the 37 selected individuals, 23 were breast or breast and ovarian cancer-affected females, 1 was a BC-affected male, and 13 were healthy. The clinical characteristics of the 23 affected females and 1 affected male are presented in Supplementary Information. Notably, six of the females were early-onset patients with either BC or bilateral BC diagnosed at an age ≤29 years. Genomic DNA samples of the index patients’ relatives were utilised for candidate variant segregation analyses. Moreover, candidate variants were screened from germline DNA from 129 female HBOC patients from the Tampere and Turku regions, 49 Finnish male BC patients, up to 989 healthy female population controls, and 909 healthy male population controls (more details in Supplementary Information). In addition, variants were analysed from formalin-fixed paraffin-embedded (FFPE) DNA from tumour and normal tissue from 31 Finnish breast or breast and ovarian cancer patients obtained from the Auria Biobank (Turku, Finland; more details in Supplementary Information). All of the exome-sequenced study subjects as well as their relatives have been informed of the analyses and have provided their written consent to use their DNA samples in the study. The Ethical Committees of Tampere and Turku University Hospitals, the National Authority for Medicolegal Affairs, and the Auria Biobank have provided their permission for the research project.
Sample preparation and whole-exome sequencing
Genomic DNA was extracted from peripheral leukocytes by using a Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions. For one patient (id 240010), only FFPE tumour tissue was available. After a pathologist had confirmed the presence of tumour cells, DNA was extracted by using the Arrow DNA kit and NorDiag Arrow automated magnetic bead-based nucleic acid extraction system (DiaSorin, Saluggia (Vercelli), Italy) according to the manufacturer’s instructions at Fimlab Laboratories (Tampere, Finland). Exome capture and sequencing were conducted out by BGI Tech Solutions (Hong Kong, China) Co. Ltd. Exome capture was performed on 3–5 μg of genomic DNA per sample by SureSelect Human All Exon 51M kit (Agilent Technologies, Inc., Santa Clara, CA, USA) followed by 100-bp paired-end sequencing with 50X genome coverage depth per sample on the HiSeq 2000 instrument (Illumina, Inc., San Diego, CA, USA) according to the protocols by Agilent, Illumina, and BGI.
Data analysis
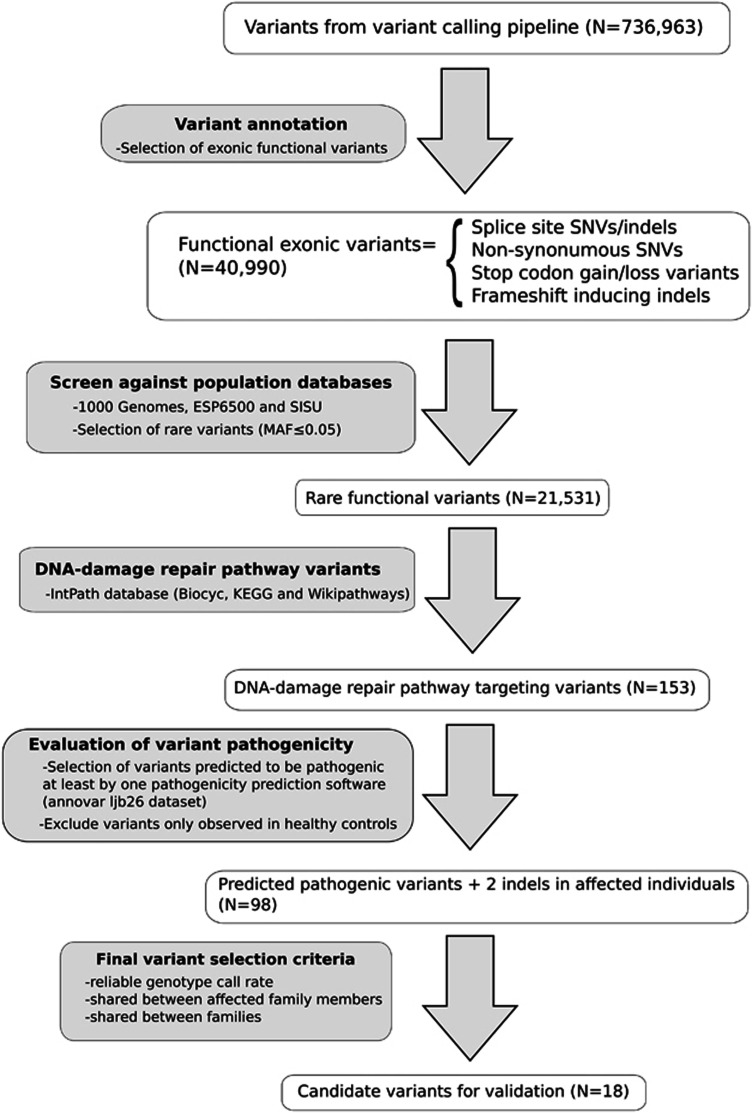
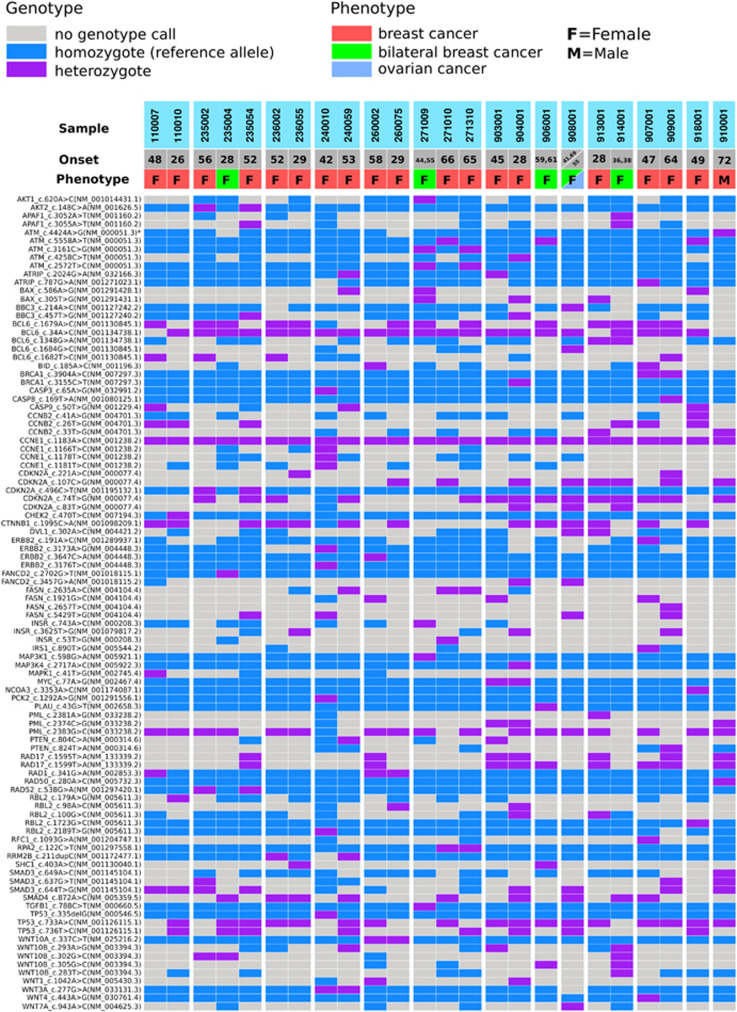
The sequencing data have been deposited at the European Genome-phenome Archive (EGA; www.ega-archive.org) under accession number EGAS00001001835. Exome sequencing detected a large number of variants, and several filtering steps were used to reduce the number of candidate variants for downstream analyses (described in detail in Supplementary Information). The analysis workflow is illustrated in Figure 1. The distribution of variants predicted to affect function in DDR pathway across 24 affected breast or breast and ovarian cancer patients with phenotypic information is presented in Figure 2.
Figure 1.
Data analysis workflow.
Figure 2.
DDR pathway gene variants in 24 breast or breast and ovarian cancer patients.
Variant validation
A total of 18 variants were genotyped by Sanger sequencing or TaqMan SNP genotyping assays from germline DNA from 129 female HBOC patients and up to 989 healthy female controls as well as from DNA from FFPE breast tumour tissue samples from 31 Finnish breast or breast and ovarian cancer patients. The reference sequences of the DDR genes are shown in Figure 2. Two of the variants, RAD50 c.280A>C, and ATM c.4424C>G, which were identified in one male BC patient, were further analysed from the germline DNA of an additional 49 male BC patients and 919 healthy male controls. Either Sanger sequencing or TaqMan assays were used to study segregation of the variants in families in which they were observed by utilising germline DNA samples of index patients’ relatives (Supplementary Information).
Statistical and bioinformatic analyses
Variants were tested for Hardy–Weinberg equilibrium in controls. The allele frequencies between patients and controls were compared by Fisher’s exact test using PLINK v1.07.12 P-values were two-sided. P<0.05 was considered statistically significant. ESEfinder was used to predict the effects of two ATM variants on exonic splicing enhancer (ESE) elements.13
Variant prioritisation in exome-sequenced early-onset patients
Filtering steps based on the variant function, allele frequency, and predicted effect on function were performed similarly as for all exome-sequenced individuals (described above). Instead of focusing on a certain pathway, gene variants in all pathways were considered. In the case that no pathway information was available, the gene function was obtained from the GeneCards database (http://www.genecards.org/). Catalogue of human somatic mutations in cancer (COSMIC) database search was performed for the variants.14 Four variants were further studied by segregation analysis using Sanger sequencing.
Results
After extensive filtering, a total of 18 variants in DDR pathway genes were selected for further validation and the variants were screened in cohorts of female HBOC patients, male BC patients, and healthy population controls. Observed variant frequencies and their association with breast and/or ovarian cancer are presented in Table 1. All variants were in Hardy–Weinberg equilibrium in the controls. None of the validated variants showed a significant association with breast/ovarian cancer. Five variants, ATM c.5558A>T, MYC c.77A>G, PLAU c.43G>T, RAD1 c.341G>A, and RRM2B c.211dupC, were more commonly detected in female HBOC patients than in female controls (odds ratio (OR) 1.16–2.16), suggesting that these variants may be low-to-moderate risk alleles but further studies are needed to confirm the findings. The rest of the variants were either absent in both patients and controls or more commonly detected in controls.
Table 1. Variant frequencies.
| Carrier frequency | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Females | Males | |||||||||||
| Gene | Nucleotide change | Effect on protein | Rs-number | Frequency in ESP6500 | Affects function by prediction programmea | HBOC cases | Controls | BC cases | Controls | P-value | OR; 95%CI | Mutation loadb |
| AKT2 | c.148C>A | p.(P50T) | rs184042322 | 0.0127 | 3, 5 | 2/127 | 5/280 | — | — | 1 | 0.88; 0.17–4.57 | 3/1 |
| ATM | c.2572T>C | p.(F858L) | rs1800056 | 0.0090 | 1, 4, 5 | 1/129 | 10/975 | — | — | 1 | 0.75; 0.10–5.92 | |
| ATM | c.3161C>G | p.(P1054R) | rs1800057 | 0.0171 | 1, 2, 3, 4, 6 | 1/129 | 14/981 | — | — | 1 | 0.54; 0.07–4.13 | |
| ATM | c.4424A>G | p.(Y1475C) | rs34640941 | 0.0008 | 1, 2, 3 | 0/129 | 1/278 | 0/49 | 0/909 | 1/1c | na | 11/5 |
| ATM | c.5558A>T | p.(D1853V) | rs1801673 | 0.0048 | 1, 3, 4, 6 | 1/129 | 5/989 | — | — | 0.52 | 1.54; 0.18–13.19 | |
| BRCA1 | c.3904A>C | p.(T1302P) | rs80357231 | na | 7 | 0/128 | 0/986 | — | — | 1 | na | 11/6 |
| CDKN2A | c.496C>T | p.(H166Y) | rs181044510 | na | 6 | 3/129 | 7/280 | — | — | 1 | 0.93; 0.24–3.62 | 8/5 |
| MYC | c.77A>G | p.(N26S) | rs4645959 | 0.0289 | 1, 2, 3, 4, 5 | 5/129d | 23/987 | — | — | 0.14 | 2.02; 0.81–5.01 | 2/1 |
| NCOA3 | c.3353A>C | p.(Q1118P) | rs149561356 | 0.0007 | 3, 5 | 0/129 | 7/279 | — | — | 0.10 | na | 11/2 |
| PLAU | c.43G>T | p.(V15L) | rs2227580 | 0.0078 | 7 | 2/129 | 11/984 | — | — | 0.60 | 2.16; 0.30–15.45 | 3/1 |
| RAD1 | c.341G>A | p.(G114D) | rs2308957 | 0.0048 | 1, 2, 3, 4, 6 | 5/129 | 15/464 | — | — | 0.79 | 1.16; 0.42–3.22 | 3/2 |
| RAD50 | c.280A>C | p.(I94L) | rs28903085 | 0.0035 | 3 | 0/129 | 0/187 | 0/49 | 1/909 | 1/1c | na | 1/1 |
| RAD52 | c.538G>A | p.(G180R) | rs7487683 | 0.0246 | 1 | 4/129 | 15/269 | — | — | 0.33 | 0.55; 0.18–1.67 | 1/1 |
| RBL2 | c.1723G>C | p.(E575Q) | rs76818213 | 0.0156 | 1, 2, 3, 4, 5, 6, 7, 8, 9 | 8/129 | 22/261 | — | — | 0.55 | 0.73; 0.32–1.66 | 11/7 |
| RPA2 | c.122C>T | p.(S41F) | rs149249571 | 0.0004 | 6 | 0/129 | 5/467 | — | — | 0.59 | na | 1/1 |
| RRM2B | c.211dupC | p.(R71fs) | — | na | na | 16/128e | 22/247f | — | — | 0.31 | 1.39; 0.73–2.64 | 4/2 |
| WNT3A | c.277G>A | p.(D93N) | rs201274685 | 0.0003 | 1, 2, 3, 4, 6 | 1/129 | 4/468 | — | — | 1 | 0.91; 0.10–8.15 | 2/1 |
| WNT10A | c.337C>T | p.(R113C) | rs141074983 | na | 3 | 1/129 | 10/988 | — | — | 1 | 0.77; 0.10–6.00 | 1/1 |
Abbreviations: BC, breast cancer; CI, confidence interval; HBOC, hereditary breast and/or ovarian cancer; na, not available; OR, odds ratio. Reference sequences: AKT2, NM_001626.5; ATM, NM_000051.3; BRCA1, NM_007297.3; CDKN2A, NM_001195132.1; MYC, NM_002467.4; NCOA3, NM_001174087.1; PLAU, NM_002658.3; RAD1, NM_002853.3; RAD50, NM_005732.3; RAD52, NM_001297420.1; RBL2, NM_005611.3; RPA2, NM_001297558.1; RRM2B, NM_001172477.1; WNT3A, NM_033131.3 and WNT10A, NM_025216.2.
Prediction programmes: 1, Polyphen2_HDIV; 2, Polyphen2_HVAR; 3, MutationTaster; 4, MutationAssessor; 5, LRT; 6, SIFT; 7, FATHMM; 8, RadialSVM; 9, LR.
Total number of identified different variants in a gene/Predicted function-affecting variants identified in a gene.
Females/males.
Homozygous in 1/129 of the HBOC cases.
Homozygous in 1/128 of the HBOC cases.
Homozygous in 2/247 of the female controls.
Clinical features of the female HBOC patients carrying the validated variants are presented in Table 2, with the exception of the RRM2B variant, which was observed with high frequency in HBOC patients. Eighteen variants were also screened from tumour samples of 31 breast or breast and ovarian cancer patients. Observed genotypes are presented in Table 3. No wild-type allele loss was observed for any of these variants.
Table 2. Clinical features of the patients with the validated variants.
| Variant | Family id | Cancer (age at dg) | Histology, grade | Receptor status | Family history of BC/OC (age at dg if known) |
|---|---|---|---|---|---|
| AKT2 c.148C>A | 131 | Bil. BC (54) | Intraductal and Ductal, 2 | na | Bil. BC (46), BC (48) |
| AKT2 c.148C>A | 244 | BC (45) | Ductal, 2 | ER+, PR+, HER2− | Bil. BC (<45), 2 × BC (<35, 46) |
| ATM c.2572T>C ATM c.3161C>G | TuFamBC10 | BC (49) | Ductal, 3 | ER+, PR+, HER2+ | Bil. BC (43) |
| ATM c.5558A>T | TuFamBC5 | BC (32) | Ductal, 1 | ER+, PR+, HER2− | 3 × BC (44, 49, 70) |
| CDKN2A c.496C>T RAD1 c.341G>A | 122 | BC (25) | Ductal, 2 | ER−, PR−, HER2+ | — |
| CDKN2A c.496C>T RBL2 c.1723G>C | 104 | BC (50), OC (41) | BC: Ductal, 2; OC: na | ER+,PR+, HER2− | — |
| CDKN2A c.496C>T MYC c.77A>G | 213 | BC (68) | Lobular, na | ER+, PR−, HER2– | 3 × BC (47, 50, 62) |
| MYC c.77A>Ga | 256 | BC (62) | Ductal, 3 | ER−, PR−, HER2− | 3 × BC (48, 60, 67) |
| MYC c.77A>G | 208 | BC (50) | Ductal, 2 | ER+, PR+, HER2− | 3 × BC |
| MYC c.77A>G | TuFamBC3 | BC (28) | Ductal, 3 | ER−, PR–, HER2− | BC (45) |
| MYC c.77A>G | TuFam43 | BC (49), OC (77) | BC: na; OC: endometrioid, 2 | Breast: na; ovarian: ER−, PR+, HER2 na | 3 × BC (62, 69, 69) |
| PLAU c.43G>T | TuFam47 | BC (55) | Mucinous, 2 | ER+, PR+, HER2− | 5 × BC (45, 51, 55, 57, 81) |
| PLAU c.43G>T | 258 | BC (57) | Lobular, 2 | ER+, PR+, HER2− | 2 × BC (53, 53) |
| RAD1 c.341G>A | TuFam50 | BC (51) | Lobular, 2 | ER+, PR+, HER2− | 2 × BC (46, 64) |
| RAD1 c.341G>A | TuFamBC27 | BC (50, 60) | Ductal and Tubular, 1; | na | 4 × BC (35, 40, 45, 45) |
| RAD1 c.341G>A RAD52 c.538G>A | 111 | BC (45) | Ductal, 2 | ER+, PR+, HER2− | BC, OC |
| RAD1 c.341G>A RBL2 c.1723G>C | 218 | BC (55) | Intraductal, na | na | BC (36) |
| RAD52 c.538G>A | TuFam28 | BC (43) | Lobular, 2 | ER+, PR+, HER2− | 2 × BC (48, -) |
| RAD52 c.538G>A | 229 | Bil. BC (68) | na | na | Bil. BC (50, 70) |
| RAD52 c.538G>A | TuFamBC16 | BC (74) | Ductal, 2 | ER+, PR+, HER2− | Bil. BC (57), BC (80) |
| RBL2 c.1723G>C | TuFam48 | BC (42) | Ductal, 2 | ER+, PR+, HER2− | BC (38) |
| RBL2 c.1723G>C | TuFamBC4 | BC (32) | Ductal, 3 | ER−, PR+, HER2− | 2 × BC (early onset, 60) |
| RBL2 c.1723G>C | TuFam69 | BC (25) | Ductal, 3 | ER−, PR−, HER2− | 2 × BC (50, 70) |
| RBL2 c.1723G>C | TuFamBC1 | BC (38) | Ductal, 3 | ER+, PR+, HER2+ | Bil. BC (61), 2 × BC (36, 59) |
| RBL2 c.1723G>C | 115 | BC (59) | Lobular, 2 | ER+, PR+, HER2– | 5 × BC |
| RBL2 c.1723G>C | TuFamBC9 | Bil. BC (44) | Ductal, na | ER+, PR+, HER2− | BC (58) |
| WNT3A c.277G>A | TuFam58 | BC (67) | Ductal, 3 | ER+, PR+, HER2− | 2 × BC (30, 58) |
| WNT10A c.337C>T | TuFam65 | BC (48) | Ductal, 2 | ER+, PR+, HER2− | 3 × BC (39, -, -) |
Abbreviations: BC, bilateral breast cancer; DCIS, ductal carcinoma in situ; na, not available; OC, ovarian cancer.
homozygous.
Table 3. Observed genotypes in breast tumour samples.
| Variant | Observed genotypes |
|---|---|
| AKT2 c.148C>A | CC=21; CA=0; AA=0 |
| ATM c.2572T>C | TT=30; TC=1; CC=0 |
| ATM c.3161C>G | CC=29; CG=2; GG=0 |
| ATM c.4424A>G | AA=31; AG=0; GG=0 |
| ATM c.5558A>T | AA=31; AT=0; TT=0 |
| BRCA1 c.3904A>C | AA=31; AC=0; CC=0 |
| CDKN2A c.496C>T | CC=31; CT=0; TT=0 |
| MYC c.77A>G | AA=31; AG=0; GG=0 |
| NCOA3 c.3353A>C | AA=29; AC=2; CC=0 |
| PLAU c.43G>T | GG=28; GT=3; TT=0 |
| RAD1 c.341G>A | GG=30; GA=1; AA=0 |
| RAD50 c.280A>C | AA=30; AC=1; CC=0 |
| RAD52 c.538G>A | GG=30; GA=0; AA=1a |
| RBL2 c.1723G>C | GG=29; GC=2; CC=0 |
| RPA2 c.122C>T | CC=31; CT=0; TT=0 |
| RRM2B c.211dupC | – |
| WNT3A c.277G>A | GG=31; GA=0; AA=0 |
| WNT10A c.337C>T | CC=31; CT=0; TT=0 |
Genotype AA was also observed in the normal tissue.
Based on available DNA samples, the variants were further studied in families in which they were observed. The PLAU c.43G>T variant was detected in one bilateral BC patient (onset at 59 and 61 years of age) by exome sequencing (Figure 2; patient 906001). The patient had three BC-affected sisters (diagnosed at 49, 50, and 56 years of age) but no DNA samples from these relatives were available. However, two out of three patients’ healthy daughters (current ages 41 and 34 years) were confirmed to carry the variant. In addition, the PLAU variant was also detected in a BC-affected female relative of the patient with the mucinous type of BC observed in the validation experiments (Table 2). The female relative had received a diagnosis of grade 1 ductal carcinoma in situ at 51 years of age.
The MYC c.77A>G variant was detected by exome sequencing in one family in two BC-affected relatives who received BC diagnoses at 28 and 45 years, respectively (Figure 2; patients 903001 and 904001). In genotyping, the variant was observed in homozygous form in one (0.8%, 1/129) BC patient but in none of the 987 controls. The patient with the homozygous MYC variant had high-grade triple-negative ductal carcinoma diagnosed at 61 years of age and had a family history of three other BC cases (mother, sister, sister’s daughter; Table 2). Moreover, the BC-affected sister had received a colon cancer diagnosis at 66 years of age. In addition, throat and stomach cancers were reported in the index’s grandparents, but the cancers were not confirmed. Further analyses identified the MYC variant in its homozygous form in the index patient’s healthy sister (current age 81 years) and in its heterozygous form in the index patient’s healthy daughter as well as in three other healthy relatives (Supplementary Information). Unfortunately, no DNA samples from BC-affected relatives were available.
The BRCA1 c.3904A>C variant was identified in two BC-affected females, a mother and daughter, in a single family by exome sequencing (Figure 2; patients 907001 and 909001). In addition, the BRCA1 variant was detected in affected daughter’s healthy daughter who is currently 22 years of age. Moreover, three other BC cases have been reported in the maternal side of the index patient’s family, but no DNA samples were available from these individuals.
The ATM c.5558A>T variant was detected in three BC-affected females from three different families by exome sequencing (Figure 2; patients 271010, 906001, and 918001) and in 1 out of 129 (0.8%) female HBOC patients by genotyping (Table 1). Interestingly, a known variant that affects correct splicing, c.5557G>A, was unexpectedly detected in the adjacent nucleotide position in the validation experiments. Preliminary predictions using ESEfinder indicated that the c.5558A>T variant introduces a new exonic splicing enhancer site in both the presence and the absence of the c.5557G>A variant. Two of the exome-sequenced patients, 271010 and 906001, had both of the variants and received a diagnosis of lobular BC at the age of 66 years and bilateral ductal BC at the ages of 59 and 61 years, respectively. A total of 0.2% (2/989) of the healthy females was found to have both of the variants. The patients carrying only the c.5558A>T variant had ductal high-grade triple-negative BC diagnosed at 49 years of age (Figure 2; patient 918001) and an early-onset BC (Table 2).
The RAD50 c.280A>C and ATM c.4424A>G variants were detected in one male BC patient by exome sequencing (Figure 2; patient 910001). The male BC patient had BC diagnosed at the age of 72 and a family history of one female BC case (daughter, diagnosed at the age of 28). Unfortunately, the daughter’s DNA sample was not available for further analyses. The RAD50 variant was also observed in 1 out of the 909 (0.1%) male controls but was absent in the cohorts of female HBOC patients and male BC patients as well as in female controls, whereas the ATM variant was absent in a cohort of female HBOC patients, male BC patients and in 909 male controls but was observed in 0.4% (1/278) of the female controls (Table 1).
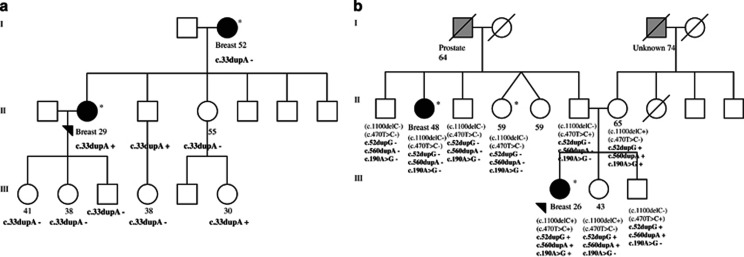
The sequencing data for the six early-onset patients was re-analysed and all rare variants predicted to affect function that were present only in early-onset patients (compared with exome-sequenced patients diagnosed >40 years) in any possible pathways were considered as good candidates for the disease susceptibility. A list of 56 candidate variants (50 nonsynonymous SNVs, 3 stopgains, and 3 frameshift indels), in genes that have a role in important cellular functions such the cell cycle, proliferation, apoptosis, adhesion, different signalling pathways and the DDR is presented in Table 4. Based on available DNA samples, four variants were further studied in families in which they were observed. Segregation of three frameshift altering variants, BNIPL c.33dupA, MAGEF1 c.52dupG, and EDN3 c.560dupA, and one nonsynonymous variant, APEX1 c.190A>G, is presented in Figure 3.
Table 4. Candidate variants in early-onset breast cancer patients.
| Gene | Variant | N in cases (sample id) | Pathway or function | Cosmic db search tissue distribution |
|---|---|---|---|---|
| AKAP13 | c.571G>A, p.(G191R) | 1 (235004) | G Protein signalling | — |
| AKAP8 | c.1513A>G, p.(N505D) | 1 (236055) | G Protein signalling | — |
| APEX1 | c.190A>G, p.(I64V)a | 1 (110001) | TSH signalling, base excision repair | — |
| BIRC6 | c.2675A>G, p.(E892G) | 1 (110001) | Ubiquitin-mediated proteolysis, apoptosis modulation and signalling | — |
| BNIPL | c.33dupA, p.(T11fs)a | 1 (236055) | Interacts with BCL2, promotes cell death | — |
| BRCA1 | c.3155C>T, p.(P1052L) | 1 (904001) | Ubiquitin-mediated proteolysis, DNA damage response | — |
| CDC45 | c.326A>G, p.(E109G) | 1 (236055) | DNA replication, cell cycle | — |
| CDKN2B | c.56C>A, p.(A19D) | 1 (236055) | Cell cycle, TGF beta signalling, pathways in cancer | Hematopoietic and lymphoid |
| CHEK2 | c.470T>C, p.(I157T) | 1 (110010) | DNA damage response, p53 signalling, cell cycle | — |
| CINP | c.159C>G, p.(N53K) | 2 (260075, 913001) | DNA replication, checkpoint signalling | — |
| COL11A2 | c.32T>A, p.(L11H) | 1 (110010) | ECM-receptor interaction, focal adhesion | — |
| COL4A6 | c.3481A>G, p.(I1161V) | 1 (110010) | ECM-receptor interaction, pathways in cancer, focal adhesion | — |
| COL6A2 | c.679G>A, p.(D227N) | 1 (260075) | ECM-receptor interaction, focal adhesion | — |
| DENND2D | c.46C>T, p.(R16*) | 1 (904001) | Promotes the exchange of GDP to GTP | — |
| DHH | c.25C>G, p.(P9A) | 1 (260075) | Hedgehog signalling | — |
| DTX4 | c.1243C>T, p.(R415C) | 1 (235004) | Notch signalling | — |
| EDN3 | c.560dupA, p.(E187fs)a | 1 (110010) | Variety of cellular roles including proliferation, migration, differentiation | — |
| EFCAB13 | c.1009A>T, p.(K337*) | 1 (260075) | Calcium ion binding | — |
| EXO1 | c.836A>G, p.(N279S) | 1 (913001) | Mismatch repair | — |
| FANCD2 | c.2702G>T, p.(G901V) | 1 (235004) | DNA damage response | — |
| FBXW8 | c.1409C>T, p.(T470M) | 1 (913001) | Ubiquitin-mediated proteolysis | — |
| FOCAD | c.5047G>A, p.(A1683T) | 2 (110010, 904001) | Tumour suppressor in glioma and colorectal cancer | — |
| LAMA1 | c.2186G>A, p.(R729H) | 1 (913001) | ECM-receptor interaction, pathways in cancer, focal adhesion | Ovary |
| LAMA5 | c.5035C>T, p.(R1679W) | 2 (236005, 260075) | ECM-receptor interaction, pathways in cancer, focal adhesion | Pancreas and biliary tract |
| LAMA5 | c.3062C>T, p.(A1021V) | 1 (913001) | ECM-receptor interaction, pathways in cancer, focal adhesion | — |
| LAMA5 | c.7367G>A, p.(R2456H) | 1 (235004) | ECM-receptor interaction, pathways in cancer, focal adhesion | — |
| LAMA5 | c.6413G>T, p.(S2138I) | 1 (913001) | ECM-receptor interaction, pathways in cancer, focal adhesion | — |
| LAMB1 | c.2869G>A, p.(D957N) | 1 (260075) | ECM-receptor interaction, pathways in cancer, focal adhesion | — |
| LAMB2 | c.1306G>A, p.(G436S) | 1 (235004) | ECM-receptor interaction, pathways in cancer, focal adhesion | — |
| LAMC3 | c.1687C>T, p.(R563W) | 1 (235004) | ECM-receptor interaction, pathways in cancer, focal adhesion | Central nervous system |
| LIG1 | c.841G>A, p.(V281M) | 1 (904001) | DNA replication, mismatch repair, base and nucleotide excision repair | — |
| LRP2 | c.6850A>G, p.(T2284A) | 1 (913001) | Hedgehog signalling | — |
| LRP2 | c.5107C>T, p.(P1703S) | 1 (235004) | Hedgehog signalling | — |
| MAD1L1 | c.175C>T, p.(R59C) | 1 (913001) | Cell cycle, progesterone-mediated oocyte maturation | — |
| MAGEF1 | c.52dupG, p.(E18fs)a | 1 (110010) | Enhancer of ubiquitin ligase activity | Large intestine |
| MAP3K4 | c.2717A>C, p.(H906P) | 1 (904001) | DNA damage response, MAPK signalling | — |
| MBD4 | c.1073T>C, p.(I358T) | 1 (236005) | Base excision repair | — |
| NEIL3 | c.516G>C, p.(Q172H) | 1 (110010) | Base excision repair | — |
| NLRP4 | c.1912G>A, p.(G638R) | 1 (904001) | NOD signalling | — |
| NUMBL | c.1347T>G, p.(F449L) | 1 (235004) | Notch signalling | — |
| PLD1 | c.1192C>T, p.(R398C) | 2 (236055, 904001) | Glycerophospholipid metabolism, pathways in cancer | — |
| PRDM1 | c.1739C>T, p.(P580L) | 1 (235004) | NOD signalling | — |
| RASGRP3 | c.844G>A, p.(G282S) | 1 (236055) | Integrated cancer, MAPK signalling | — |
| RBL2 | c.98A>C, p.(D33A) | 2 (260075, 904001) | DNA damage response, TGF beta signalling, cell cycle | — |
| RBL2 | c.100G>C, p.(A34P) | 2 (904001, 913001) | DNA damage response, TGF beta signalling, cell cycle | — |
| RBL2 | c.179A>G, p.(E60G) | 1 (110010) | DNA damage response, TGF beta signalling, cell cycle | — |
| RET | c.2876G>A, p.(R959Q) | 1 (235004) | Pathways in cancer | Large intestine |
| RICTOR | c.3221A>G, p.(D1074G) | 1 (235004) | TOR signalling, mTOR signalling | — |
| S1PR5 | c.953T>A, p.(L318Q) | 2 (235004, 913001) | Signal transduction of S1P receptor | — |
| SOX17 | c.83G>T, p.(G28V) | 1 (235004) | Wnt signalling | — |
| TAB3 | c.743C>T, p.(T248M) | 1 (236055) | NOD-like receptor signalling | Large intestine |
| TICRR | c.1993C>T, p.(R665*) | 1 (260075) | DNA replication | — |
| TNC | c.1642G>A, p.(V548M) | 1 (110010) | ECM-receptor interaction, focal adhesion | — |
| TNC | c.2977G>C, p.(V993L) | 1 (913001) | ECM-receptor interaction, focal adhesion | — |
| UBE2Q1 | c.727A>C, p.(N243H) | 1 (235004) | Ubiquitin-mediated proteolysis | — |
| UBE3A | c.532G>A, p.(A178T) | 1 (913001) | Ubiquitin-mediated proteolysis | — |
Reference sequences: AKAP8:NM_005858.3; AKAP13:NM_006738.5; APEX1:NM_001244249.1; BIRC6:NM_016252.3; BNIPL:NM_138278.3; BRCA1:NM_007297.3; CDC45:NM_001178011.2; CDKN2B:NM_004936.3; CHEK2:NM_007194.3; CINP:NM_032630.2; COL4A6:NM_001287759.1; COL6A2:NM_001849.3; COL11A2:NM_001163771.1; DENND2D:NM_024901.4; DHH:NM_021044.2; DTX4:NM_015177.1; EDN3:NM_207032.2; EFCAB13:NM_001195192.1; EPSTI1:NM_001002264.2; EXO1:NM_003686.4; FANCD2:NM_001018115.2; FBXW8:NM_012174.1; FOCAD:NM_017794.4; LAMA1:NM_005559.3; LAMA5:NM_005560.4; LAMB1:NM_002291.2; LAMB2:NM_002292.3; LAMC3:NM_006059.3; LIG1:NM_001289064.1; LRP2:NM_004525.2; MAD1L1:NM_001013836.1; MAGEF1:NM_022149.4; MAP3K4:NM_005922.3; MBD4:NM_001276270.1; NEIL3:NM_018248.2; NLRP4:NM_134444.4; NUMBL:NM_001289979.1; PLD1:NM_001130081.2; PRDM1:NM_182907.2; RASGRP3:NM_015376.2; RBL2:NM_005611.3; RET:NM_020630.4; RICTOR:NM_001285439.1; SOX17:NM_022454.3; S1PR5:NM_001166215.1; TAB3:NM_152787.4; TICRR:NM_152259.3; TNC:NM_002160.3; UBE2Q1:NM_017582.6; UBE3A:NM_130838.1.
Segregation was further studied in the family.
Figure 3.
(a) Segregation of the BNIPL c.33dupA in pedigree 236. (b) Segregation of the EDN3 c.560dupA, MAGEF1 c.52dupG, and APEX1 c.190A>G in pedigree 110. Two CHEK2 variants, c.470T>C and c.1100delC (presented in parenthesis), have been described previously in family 110.10 Exome-sequenced samples are marked with an asterisque. Plus indicates heterozygous variant and minus indicates no variant. Females are marked with circles, and males are marked with squares. The index patient is marked with an arrow. Breast cancer is marked with a black circle and with the age at diagnosis. Other cancers are indicated with grey squares. The current ages of healthy females are presented. Deceased individuals are indicated with a slash. Generations are presented in Roman numerals.
Discussion
In the current study, the aim was to identify rare variants that would contribute to HBOC susceptibility in Finnish BRCA1/2-negative families. An exome sequencing approach was used to analyse a total of 13 high-risk HBOC families, 6 of which were involved in our previous analyses. A total of 18 DDR pathway gene variants were considered the most potential ones for validation analyses after filtering. However, none of the validated variants showed a significant association with breast/ovarian cancer upon screening the variants in cohorts of female HBOC patients, male BC patients, and healthy controls; this finding might be explained by the rarity of the variant and the limited number of the analysed patients. Variants in ATM, MYC, PLAU, RAD1, and RRM2B were enriched in female HBOC patients compared with controls, suggesting that these variants may be low-to-moderate risk alleles but additional analyses of larger sample set are needed to further validate the findings.
ATM is a protein kinase that has a central role as an activator of the DDR cascade after DNA double-strand breaks. Heterozygous ATM variants have been shown to increase the BC risk by approximately twofold.15 Here, an interesting observation was that several ATM variants were enriched, particularly in one family (Figure 2; patients 271009, 271010, and 271310). A polygenic risk model for BC has been suggested; according to this model, several low-to-moderate risk alleles can act multiplicatively on the cancer risk.16 Moreover, an additional ATM variant, c.5557G>A, was identified in the validation experiments. The c.5557G>A is a common variant that has been associated with bilateral BC in the Finnish population.17 In line with this, one of the patients with the c.5557G>A had bilateral BC. Moreover, the c.5557G>A variant has been reported to have an effect on splicing.18 Similar findings was observed here for the c.5558A>T variant, which was predicted to create a new splicing site. Splicing variants have been reported to be common in ATM;18 therefore, additional analyses are warranted.
MYC is a well-known oncogene that has a central role in growth control, differentiation, and apoptosis, and its abnormal expression is associated with many human cancers including tumours of epithelial origin.19 An interesting finding here was that a rare homozygous form of the MYC variant was observed in one BC patient who was diagnosed with an aggressive BC at the age of 61 years and who had familial aggregation of breast and other cancers. However, further analysis identified the homozygous variant in the index patient’s healthy sister (current age 81 years), indicating a likely neutral role of the homozygous variant.
The PLAU gene encodes for the urokinase-type plasminogen activator protease (uPA), which has a crucial role in the process of cancer metastasis.20 Here, the PLAU c.43G>T variant was detected in three BC patients who were diagnosed at an older age. However, an interesting observation was that one of the BC patients had been diagnosed with the mucinous type of the disease. Mucinous breast carcinoma is a rare histological subtype that accounts for ~2% of all BCs and is related to favourable outcomes.21 Here, the mucinous subtype was very rare, as it was observed only one HBOC patient (1/129, 0.8%). Clearly, further analyses in a larger sample set are warranted to investigate whether the PLAU variant relates to the mucinous subtype; such finding might hold clinical value. Moreover, further evidence of the enrichment of the PLAU variant in BC patients was obtained by also identifying the germline variant in its heterozygous form in 9.7% (3/31) of breast tumour samples
RAD1 encodes a component of the heterotrimeric cell cycle checkpoint complex that participates in cell cycle checkpoint activation and DNA repair, and RRM2B encodes ribonucleotide reductase, which is directly involved in the p53 checkpoint for the repair of damaged DNA.22, 23 Both RAD1 and RRM2B variants were frequent in genotyped female HBOC patients as well as in female controls, indicating that they are either neutral or low-risk common variants. Because both of the variants are frequent, larger sample sets would be needed to show their possible contribution to breast/ovarian cancer risk.
Interestingly, one BRCA1 variant, c.3904A>T, which was detected in a single exome-sequenced family, was absent both in genotyped female HBOC patients and healthy female controls indicating that this family-specific variant is extremely rare and likely deleterious; this variant indeed warrants further investigations because it might have clinical relevance in genetic counselling of high-risk HBOC families. Moreover, the RAD50 c.280A>C variant, which was observed in a male BC patient by exome sequencing, was in fact absent in both female HBOC patients and healthy controls and was extremely rare (0.1%, 1/909) in male controls, indicating that it might contribute to BC susceptibility, particularly in males. Another variant detected in a male BC patient by exome sequencing, ATM c.4424A>G, was absent both in male BC patients and controls and present in one female control (0.4%, 1/278); these findings suggest that it is a very rare variant that is not specific for male BC.
In addition, rare DDR pathway gene variants were observed in single individuals in the studied families (Figure 2); these variants may modify the cancer risk in the family and explain the phenotypic heterogeneity. Especially, variants in AKT1, ATRIP, BRCA1, CASP3, CASP8, ERBB2, FANCD2, MAP3K1, MAP3K4, PCK2, TGFB1, TP53, and WNT4 are of great interest. Many of these genes have been highlighted as significantly mutated genes in BC by integrated molecular analyses.24 Supported by previous findings, the variants in these genes can be considered as good candidates for BC susceptibility and warrant further investigations.
Notably, although exome sequencing is a useful tool for new variant identification, the causal variants may remain undetected owing to technical challenges. Here, exome sequencing failed to detect the CHEK2 c.1100delC variant in a patient 110010 (Figure 2), who is known to carry the two CHEK2 variants c.470T>C and c.1100delC based on previous analyses.10 The c.1100delC variant is located in a gene region that shows homology with other chromosomal regions; this might be the reason why it was not identified by exome sequencing.
Deeper analyses on six early-onset patients revealed defects in interesting candidate genes in pathways related to cell cycle, proliferation, apoptosis, adhesion, and DDR, among other signalling pathways thus providing a fruitful starting point for later studies. For instance, frameshift altering variants were detected in the BNIPL, EDN3, and MAGEF1 genes. BNIPL is an apoptosis-associated protein that interacts with BCL2 and promotes the invasion and metastasis of human hepatocellular carcinoma cells.25 Segregation analysis confirmed that the BNIPL frameshift variant (c.33dupA) was not detected in index patient’s healthy daughters, sister, and brother’s daughter (Figure 3a), which supports that the variant is likely deleterious. Instead, the variant was observed in index patient’s brother and sister’s healthy daughter who is currently 30 years of age. Because index’s healthy sister did not carry the variant her daughter’s variant is of paternal origin.
EDN3 belong to a family of endothelins, which are vasoactive peptides, important signalling molecules that function in fundamental cellular processes such as proliferation, migration, and differentiation. Altered endothelin signalling has a role in carcinogenesis, and the loss of EDN3 expression has been reported in human BC.26 MAGEF1 belongs to a superfamily of MAGE proteins, called cancer–testis antigens, that mediate normal cellular functions and are ubiquitously expressed in both normal and tumour tissue.27 Altered expression of MAGE proteins has been implicated in BC.28 Segregation analysis of the EDN3 (c.560dupA), the MAGEF1 (c.52dupG), and the APEX1 (c.190A>G) variants in the same family showed incomplete segregation (Figure 3b). Because index’s mother is healthy, it is likely that combination of these and other variants explain the index’s early onset. Interestingly, in this family, two deleterious CHEK2 variants, c.470T>C and c.1100delC have been reported previously.10 It can be seen in Figure 3b that the truncating CHEK2 c.1100delC variant is not segregating with the disease in the family, indicating its moderate character. It would be interesting to study further if the EDN3, MAGEF1, and APEX1 variants could be modifiers of the CHEK2 variants. Intriguingly, germline defects in APEX1 have been reported to be associated with BC susceptibility in other population.29 Moreover, variants that induce premature stop codons were identified in the DENND2D, EFCAB13, and TICRR genes. Of these genes for instance, EFCAB13 is a very interesting candidate gene, because it functions in calcium ion binding, and it has been associated with familial prostate cancer in Finland by a recent study.30
In conclusion, family-specific enrichment of multiple DDR pathway gene defects likely explains a proportion of BC predisposition in high-risk HBOC families. Moreover, interesting candidate genes targeting pathways involved in DNA replication, apoptosis, and the cell cycle, among other signalling pathways, were identified in early-onset BC patients, thus providing novel information on potential BC-related pathways and an excellent premise for future studies.
Acknowledgments
We would like to thank all participating patients and their families. This work was supported by the Finnish Cancer Organisations, the Sigrid Juselius Foundation, and the Academy of Finland (251074) for JS. The Finnish Doctoral Programme in Biomedicine and Biotechnology, the Orion-Farmos Research Foundation, the Emil Aaltonen Foundation, the Ida Montini Foundation, and the Finnish Cancer Organisations have supported KM.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Ferlay J, Soerjomataram I, Dikshit R et al: Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- Claus EB, Schildkraut JM, Thompson WD, Risch NJ: The genetic attributable risk of breast and ovarian cancer. Cancer 1996; 77: 2318–2324. [DOI] [PubMed] [Google Scholar]
- Roy R, Chun J, Powell SN: BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 2011; 12: 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M: Genetic susceptibility to breast cancer. Mol Oncol 2010; 4: 174–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou A, Pharoah PD, Narod S et al: Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 2003; 72: 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartikainen JM, Kataja V, Pirskanen M et al: Screening for BRCA1 and BRCA2 mutations in Eastern Finnish breast/ovarian cancer families. Clin Genet 2007; 72: 311–320. [DOI] [PubMed] [Google Scholar]
- Sarantaus L, Auranen A, Nevanlinna H: BRCA1 and BRCA2 mutations among Finnish ovarian carcinoma families. Int J Oncol 2001; 18: 831–835. [DOI] [PubMed] [Google Scholar]
- Michailidou K, Beesley J, Lindstrom S et al: Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet 2015; 47: 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Kim YC, Snyder C et al: Family-specific, novel, deleterious germline variants provide a rich resource to identify genetic predispositions for BRCAx familial breast cancer. BMC Cancer 2014; 14: 470–2407-14-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusisto KM, Bebel A, Vihinen M, Schleutker J, Sallinen SL: Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals. Breast Cancer Res 2011; 13: R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusisto KM, Akinrinade O, Vihinen M, Kankuri-Tammilehto M, Laasanen S, Schleutker J: Copy number variation analysis in familial BRCA1/2-negative Finnish breast and ovarian Cancer. PLoS One 2013; 8: e71802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR: ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 2003; 31: 3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P et al: COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015; 43: D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Rahman N: ATM and breast cancer susceptibility. Oncogene 2006; 25: 5906–5911. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Pharoah PD, McMullan G et al: A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer 2002; 86: 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen K, Rapakko K, Karppinen SM, Erkko H, Nieminen P, Winqvist R: Association of common ATM polymorphism with bilateral breast cancer. Int J Cancer 2005; 116: 69–72. [DOI] [PubMed] [Google Scholar]
- Thorstenson YR, Roxas A, Kroiss R et al: Contributions of ATM mutations to familial breast and ovarian cancer. Cancer Res 2003; 63: 3325–3333. [PubMed] [Google Scholar]
- Hoffman B, Liebermann DA: Apoptotic signaling by c-MYC. Oncogene 2008; 27: 6462–6472. [DOI] [PubMed] [Google Scholar]
- Moquet-Torcy G, Tolza C, Piechaczyk M, Jariel-Encontre I: Transcriptional complexity and roles of Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer. Nucleic Acids Res 2014; 42: 11011–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Geyer FC, Reis-Filho JS: Histological types of breast cancer: how special are they? Mol Oncol 2010; 4: 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T: Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex. J Biol Chem 2009; 284: 20457–20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Arakawa H, Yamaguchi T et al: A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 2000; 404: 42–49. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network: Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Qin W, Li J et al: BNIPL-2 promotes the invasion and metastasis of human hepatocellular carcinoma cells. Oncol Rep 2007; 17: 605–610. [PubMed] [Google Scholar]
- Wiesmann F, Veeck J, Galm O et al: Frequent loss of endothelin-3 (EDN3) expression due to epigenetic inactivation in human breast cancer. Breast Cancer Res 2009; 11: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone B, Schummer M, Paley PJ et al: MAGE-F1, a novel ubiquitously expressed member of the MAGE superfamily. Gene 2001; 267: 173–182. [DOI] [PubMed] [Google Scholar]
- Du Q, Zhang Y, Tian XX, Li Y, Fang WG: MAGE-D1 inhibits proliferation, migration and invasion of human breast cancer cells. Oncol Rep 2009; 22: 659–665. [DOI] [PubMed] [Google Scholar]
- Ali K, Mahjabeen I, Sabir M et al: Germline variations of apurinic/apyrimidinic endonuclease 1 (APEX1) detected in female breast cancer patients. Asian Pac J Cancer Prev 2014; 15: 7589–7595. [DOI] [PubMed] [Google Scholar]
- Laitinen VH, Rantapero T, Fischer D et al: Fine-mapping the 2q37 and 17q11.2-q22 loci for novel genes and sequence variants associated with a genetic predisposition to prostate cancer. Int J Cancer 2015; 136: 2316–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.