Abstract
In vivo imaging techniques can be integrated with classical histochemistry to create an actual histochemistry of water. In particular, Magnetic Resonance Imaging (MRI), an imaging technique primarily used as diagnostic tool in clinical/preclinical research, has excellent anatomical resolution, unlimited penetration depth and intrinsic soft tissue contrast. Thanks to the technological development, MRI is not only capable to provide morphological information but also and more interestingly functional, biophysical and molecular. In this paper we describe the main features of several advanced imaging techniques, such as MRI microscopy, Magnetic Resonance Spectroscopy, functional MRI, Diffusion Tensor Imaging and MRI with contrast agent as a useful support to classical histochemistry.
Key words: Magnetic resonance imaging, magnetic resonance spectroscopy, functional magnetic resonance imaging, diffusion tensor imaging, tissue analysis, contrast agents
Introduction
Technological developments have allowed new approaches to overcome the classical histochemistry issues. In particular, magnetic resonance imaging (MRI) is widely used in the clinical practice as powerful and, non-invasive diagnostic tool; however, it has a substantial relevance also in preclinical and pharmaceutical research.1
MRI is based on the principles of Nuclear Magnetic Resonance (NMR), that measures the absorption of electromagnetic radiation in molecules immersed in a strong magnetic field. These radiations are the cause of nuclear spin transitions in certain atoms (typically 1H or 13C) and the information on the molecular structure are deduced by observing the behavior of atomic nuclei.
At first, MRI was used as a tomographic imaging modality able to produce images of a section of the human body starting from NMR signal. In particular, since the human body is mainly composed by water, MRI exploits the abundance of hydrogen atoms into the body to produce images.
As a morphological technique, MRI conjugates high space resolution ith high soft tissue contrast and sensitivity to alterations of tissue parameters. Moreover, MRI is able to provide not only morphological and structural information, but also functional. More recently it has further evolved toward imaging of molecular events (molecular imaging) that represents a challenge for imaging techniques. Finally, an advanced MRI technique, namely Magnetic Resonance Spectroscopy (MRS) allows in vivo monitoring of metabolites that can be quantified, enabling parametric mapping of their concentration.2
Under certain situations, the potential offered by in vivo imaging appears to be superior to those offered by traditional histology: it is non-invasive, it avoids substances or treatments that strongly interfere with the tissue structure and surrounding environment and it provides the possibility of analyzing hydrated tissues. With this technique it is possible to perform sophisticated studies in living systems, with the opportunity to observe several reactions that take place inside living cells: this gives the opportunity to achieve an actual histochemistry of water, since the hydrogen nuclei contained in the water molecule are able to produce the magnetic resonance signal. Moreover, using appropriated MRI technique, in particular functional MRI (fMRI) and diffusion tensor imaging (DTI) it is also possible to characterize/quantify physical parameters of tissues.
Here, our aim is to describe the main features of advanced in vivo imaging techniques, such as MRI microscopy, MRS, fMRI, DTI and MRI with contrast agent.
MRI microscopy
MRI microscopy is intended as a magnetic resonance technique for imaging of small objects or isolated organs at high spatial resolution (up to about 100 μm). Although it was clear from the beginning3 that the resolution of imaging experiments on small objects (1 mm) could approach microscopic values (around 10 μm), it was only in the second half of 1980s that this field started to develop. Some important applications of MRI microscopy were obtained in the field of material science4 but relevant results have also been obtained in the biological field on living plants, animal cells and tissues.5 The first MRI images of a single cell, a frog ovum with a diameter of 1 mm, was obtained at 400 MHz6 and clearly distinguished between nucleus and cytoplasm. Single cell images have also been obtained on the L7 neuron isolated from the sea hare Aplysia californica5 and researchers have shown that water in nucleus has T2 and diffusion coefficient that are strongly different from those of water contained in cytoplasm,7 suggesting that MRI models of tissue compartmentalization cannot be limited to the consideration of intra and extracellular compartments. MRI microscopy was also performed on cell aggregates, as tumor spheroids4 or isolated tissues and organs for example brain slices imaged at 20×20×300 μm space resolution.8 To date, large projects have been implemented to obtain information on the most widely used laboratory animals by MR microscopy.9 For instance, a study used MRI microscopy as a rapid, non-invasive technique for the phenotyping of mouse mutants.10 As shown in Figure 1 A, B, MRI can offer a number of advantages over traditional histology. For example it is able to produce non-invasive datasets and an accurate calculation of volumes without distortions and consequently the morphometric techniques may be used to allow the identification of novel phenotypes.10 It’s clear that the MRI techniques can not compete with the resolutions of optical techniques but their advantages are: accessibility to MRI parameters at microscopic level, the possibility to follow dynamic events in a meaningful physiological manner and the accessibility to functional and metabolic parameters through localized spectroscopy.11
Figure 1.
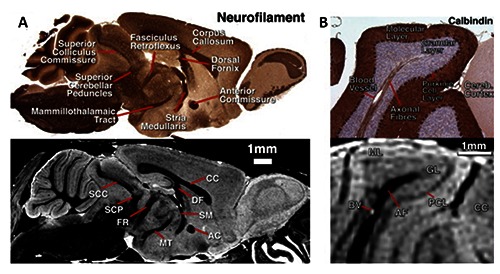
A) Sagittal sections of MRI microscopy and a similar neurofilament histology section in the same brain, showing visible white-matter anatomy. B) Detail through the cerebellum showing the correspondence of structures between histology and MRI microscopy. Adapted from Cleary et al., Neuroimage 2011;56:974-83 with permission.
In vivo magnetic resonance spectroscopy
While MRI maps the spatial distribution of mobile nuclei, the in vivo MRS can be used to obtain further detailed chemical information from localized volumes of interest within the well-defined compartmental structures of the studied system. Therefore, MRS is able to provide spatially localized chemical information at a tissue resolution level: for this reasons it can be considered as a new histochemical technique. It is clear that the intrinsic features of this technique make lipids, carbohydrates and various metabolites mapping possible and, for this reason, MRS has a potential in additional to the classic histochemistry.
Certainly the greatest advantage of this new form of histochemistry is represented by the possibility to carry out localized spectroscopic evaluations. Indeed, by selecting a specific voxel is it possible to analyze the spectrum of the compounds present inside it. This approach appears innovative compared to traditional histochemistry since this analysis can be performed avoiding extraction procedures or tissue processing allowing to limit the artifacts due to these processes. Many metabolites, that are not directly visible with the standard procedures, can be studied by means of localized spectroscopy approaches. Using these approaches it is also possible to obtain selective imaging, identifying peaks linked to a given compound in a specific tissue area. Indeed, MRS allows to compare spectra obtained from normal tissue with spectra derived from pathological tissue. Moreover, metabolic changes arising from pathological conditions and visualized by MRS may not be detected through anatomical images (obtained by conventional MRI) since metabolic changes may precede anatomic changes. Thus, MRS is used for diagnostics, to observe disease progression, monitor therapeutic treatments, and to understand the pathogenesis of diseases.12
The quality of the information obtainable by MRS in vivo, is not only determined by the signal-to-noise ratio (SNR); the separation between peaks is crucial for detection and classification. As the distance between peaks of different chemical species (normally expressed in parts per million, ppm) increases linearly with the resonance frequency (or with the applied magnetic field), proper imaging is required. For example, water (resonance at 4.7 ppm) and fat (resonance of CH2 and CH3 groups at about 1.4 ppm) are separated by 660 Hz at 4.7 T and by 280 Hz at 2 T. In studies on brain, most of the metabolites of interest have resonance frequencies contained in a few ppm (for example lactate 1.35 ppm, N-Acetylaspartate 2.0 ppm, Choline 3.2 ppm), thus high fields are mandatory to resolve the different signal components. A further advantage of high fields for 1H-MRS spectroscopy is that efficient water suppression requires selective excitation of water molecules followed by spoiler gradients; selective excitation of water molecules is easier if the distance between water and metabolites peaks is higher. In biological studies, nuclei of interest are mainly 1H and 31P, the first one mainly used in brain studies and the second one in muscle studies. Several papers reported 1H-MRS studies in different brain pathologies, as stroke, tumors13 and neurological diseases14 (Figure 2 A-C); MR spectra of brain tumors generally report increased lactate and decreased N-Acetylaspartate content,15 indicating neuronal loss. The lipids are the most easily detectable compounds given their high hydrogen content. A clear visualization of areas rich in lipids is permitted by the ability of MRI to differentiate between the hydrogen atoms linked to carbon atoms (of the lipid molecules) and the hydrogen atoms linked to the protons of the water molecules. Adipose tissues are widely represented in the human body and constitute the environment in which a large number of neoplastic lesions develop. Indeed, in tumors there might be lipids signals (Figure 2 D, E) that are absent in healthy tissue.13,16 In this contest, the type of adipose tissue in a particular area and also the composition of lipid accumulation, may represent two important factors in the genesis and progression of cancer. Studies on lipids in cancer revealed that the presence of accumulation of poly-unsaturated fatty acid lipids correlates with the presence of apoptotic cell death.17 The composition of fatty acids in periprostatic tissue is altered in patients with aggressive prostate cancers,18 the characterization of adipose tissue in close proximity to colorectal tumors showed an alteration of monounsaturated fatty acids related to the stage of the tumor.19
Figure 2.
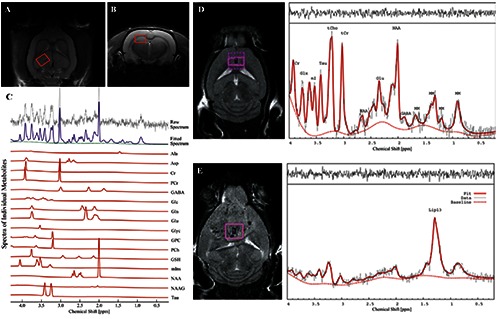
In vivo T2-weighted MR images A) coronal and B) axial showing regions of interest (ROI) for 1H-MRS experiments. Here, the ROI is centered in the right dorsal hippocampal region of the rat brain. C) Quantification of neuro-metabolite signals from in vivo 1H-MRS in the right dorsal hippocampal region of the Stress-induced Sleep Perturbation (SSP) model. D) 1H-MRS voxel location on a T2-weighted MRI and the corresponding 1H-MRS spectra from a control mouse and E) from a mouse with large a glioblastoma. 1HMRS spectra from glioblastoma are characterized by increased lipid signals. tCr, total creatine; Glx, glutamate + glutamine; Ins, myo-Inositol; Tau, Taurine; tCho, total choline; NAA, N-acetyl-aspartate; Glu, glutamate; MM, macromolecules; Lac, Lactate; GABA, gamma-aminobutyric acid. A, B and C panels were adapted from Lee et al., PLoS One 2016;11:e0153346 with permission; D and E panels were adapted from Park et al., PLoS One 2014;9:e94755 with permission.
To date, another important application of 1H-MRS is the measurement of hepatic lipid content. Indeed, lipid content, together with intracellular muscular lipid content, has been correlated with insulin resistance, a predictor for the clinical onset of type 2 diabetes mellitus.20 The assessment of lipid concentration by 1H-MRS is used to develop hepatic lipid-lowering drugs in clinical trial and in preclinical research. Also the measurement of intramyocellular and extramyocellular lipid content is possible by 1H-MRS, and the major advantage is the avoiding of manual cleaning of extramyocellular adipose tissue.20 This technique allows not only a non-invasive approach, but it also removes the quantification errors originated from manual extramyocellular cleaning.
Also carbohydrates are identifiable with localized spectroscopy approaches and MRS has important advantages over conventional approaches in the study of metabolism alteration in several humans disease.21 Glycogen is an important energy reservoir in the human body and it is abundant in the liver and skeletal muscles. Brain glycogen concentration is lower than in other organs,22 making it difficult the study its physiological role and the evaluation of its concentration in vivo. MRS, in particular 13C-MRS, is able to measure glycogen content and turnover in both physiological and pathological contexts in the living brain noninvasively.23,24 Several studies reported that alterations in brain glycogen metabolism are related with brain injury, hypoxia and ischemia.20 Furthermore, MRS allows the assessment of the metabolic profile of many hepatic metabolites in vivo, which cannot be accurately measured using invasive biopsy techniques, due to the instability of these compounds and the invasiveness of biopsies. In addition, the non-invasive features of MRS, combined with the large hepatic blood flow, allows repeated measurements in the same patients, facilitating long-term longitudinal studies with real-time monitoring of metabolic alterations.21
Finally, among macromolecules, proteins are less easily measurable by MRS. In tissues, however protein accumulations, such as in fibrotic tissues, can appear with negative images by virtue of a poor hydration and the scarcity of lipids.
It has been already mentioned the intrinsic low sensitivity of MRS due to the low concentration of metabolites; a different application of MRS has been explored for which the SNR is also more critical, namely Diffusion weighted MRS (DW-MRS).25 The use of diffusion gradients in imaging as well as in in vitro spectroscopy is well established; diffusion gradients produce a decrease in the signal intensity, more marked for the molecules diffusing faster. A measurement of the signal intensity as a function of the diffusion gradients characteristics (namely the b factor, dependent on gradient duration, intensity and distance between gradient pulses) provides quantitative value for the diffusion coefficients. The technique has been applied to the detection and characterization of brain damage after ischemic insult as well as in the tracking of axonal fibers in brain white matter.26 In DWMRS, diffusion gradients are used in in vivo experiments with the aim of measuring the diffusion coefficient of different metabolites.25 DW-MRS gives information on the size and possibly on the shape of the compartment in which the metabolite is contained and on shape and size of cells, although the interpretation of results is not simple. Several studies have been performed on experimental models of focal cerebral ischemia all reporting a decrease by 25-30% of the apparent diffusion coefficient of brain metabolites (N-Acetylaspartate, Creatine and Choline). In the experimental model of global ischemia, the decrease amounted to 40%.25 These findings are not immediately understandable in the light of cell swelling and increased intracellular water (that occurs in ischemic regions). Increased intracellular tortuosity due to swelling of cell organelles, disaggregation of polyribosomes, increased number of cytoplasmic fibrillary structures could explain this observation.25
The “histo-physic”
Functional MRI (fMRI) and Diffusion Tensor Imaging (DTI) are advanced MRI in vivo techniques allowing the evaluation of parameters that could be defined as physical. DTI provide information about mobility of water and fiber orientation within the sample, while fMRI measures the brain activity by detecting changes associated with blood flow. These approaches have the advantage of being quantitative and they allow a real functional imaging monitoring, for example, the time course of specific parameters during stimulation test. The next chapter describes the main features and applications of fMRI and DTI.
Functional MRI
The term fMRI generally indicates MRI experiments that monitor the neuronal response to a given stimulus. fMRI started in 1990, when Ogawa et al.27 demonstrated that MR images acquired with Gradient Echo sequences can reflect a new kind of contrast dependent on the blood oxygenation level (blood oxygenation level dependent, BOLD). Indeed, BOLD response measures hemodynamic changes (local blood volume and flow or changes in susceptibility) occurring during neuronal activation. An increase in the signal intensity is attributable to a decrease in the local concentration of the intrinsic oxy-hemoglobin contrast agent; such an increase can be due either to an increase in blood flow or in oxygen consumption. In fact deoxy-hemoglobin has paramagnetic properties while oxyhemoglobin does not; deoxy-hemoglobin, contained in blood vessels, disturbs the homogeneity of the static magnetic field. A local decrease in the concentration of deoxy-hemoglobin produces an increase in the signal intensity in T2* weighed sequences.
At first fMRI was used to map brain activity evoked from its sensory, motor, cognitive, and emotional tasks in healthy individuals In the last years this technique has been applied to neurological/neurobehavioral disorders, as epilepsy, multiple sclerosis, Alzheimer’s disease, stroke and cerebral tumors. fMRI has several advantages compared to other functional imaging techniques (e.g., positron emission tomography): it is easy to perform, is reproducible, can be performed with the majority of existing MR scanners and it has superior temporal and spatial resolution.28
In animals, fMRI has been extensively used to detect cerebral activation both in anaesthetized and in awake animals,29,30 but the procedures to analyze these results are quite complicated. Indeed, to analyze fMRI data of animals several steps are required: starting to an Echo Planar Image (EPI, a fastest acquisition MRI method), we have to register our EPI to a common MRI animal brain template, then we overlay the brain atlas to correctly label different brain region and finally we can add the activation maps. All these procedures are shown in Figure 3.
Figure 3.
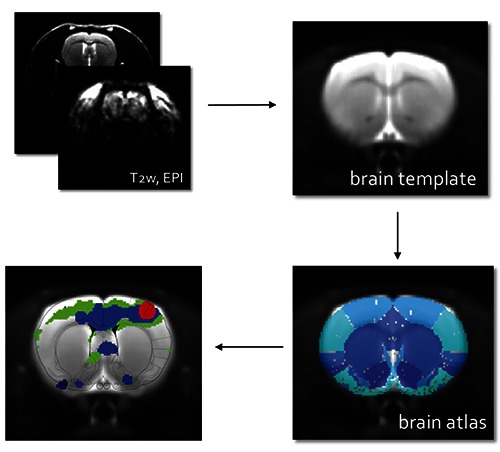
Procedures to analyze fMRI data: acquisition of Echo Planar Images (EPI), registration of EPI to a common MRI brain template, overlapping to brain atlas to correctly label different brain region and addition of the activation maps; the final result is shown in the last panel.
The functional activation of the olfactory bulb as a consequence of odors stimuli has been reported31 and investigated also at very high space resolution. Recently, in a fMRI study, Tambalo et al. investigated the functional response to somatosensory stimulation of the forepaw in DA Dark Agouti rats with experimental autoimmune encephalomyelitis (EAE), a widely accepted preclinical model of chronic multiple sclerosis (MS).32 Alterations of the functional response of EAE mice (in the relapsing and chronic phases) were found and this finding was in line with human fMRI studies showing that cortical reorganization occurs in MS patients.33 These results provide additional support to the validity of the DA EAE rat model for fMRI studies and for the development of innovative therapies in EAE/MS research.32 In animals, fMRI can also be performed using exogenous paramagnetic or superparamagnetic contrast agents. When confined in the vascular space, they create a strong inhomogeneity in the static magnetic field and consequently a decrease in signal intensity. The activated regions, where an increase in local blood volume (and contrast agent concentration) occurs, are reflected by MR images as low signal intensity pixel. Particularly relevant are studies performed with ultrasmall superparamagnetic iron oxide particles (USPIO)34-36 that are characterized by a long blood half time (2-3 h). These techniques appear consequently more sensitive compared with BOLD, and less unfavorable at low static magnetic fields (2 T). Such techniques have been widely used in the so called “pharmacological MRI”37 where the effect of stimulatory or inhibitory drugs on brain hemodynamics are studied. For example it has been applied to study the changes in cerebral blood volume induced by administration of cocaine or bicocculine (an antagonist of GABA) in the different parts of the brain.38
Diffusion Tensor Imaging
Another advanced MRI technique that allows the evaluation of some physical characteristics is the Diffusion Tensor Imaging (DTI). DTI can be used to characterize the diffusion of water as a function of spatial location. The two most common diffusion tensor MRI parameters are mean diffusivity (MD) and fractional anisotropy (FA). MD is a measure of the directionally averaged magnitude of diffusion and is related to the integrity of the local brain tissue. FA represents the degree of diffusion anisotropy and reflects the degree of alignment of cellular structure.39 The diffusion of water within the tissues may be altered by changes in the tissue microstructure due to different pathological processes of the central nervous system, including demyelination, axonal damage, inflammation, oedema and ischaemia.
DTI is now becoming widely available in clinical scanners and human clinical studies are actively being performed. On the other hand, high-resolution DTI technology and applications to animal studies for basic neuroscience research have started only recently. In vivo DTI allows to monitor the longitudinal evolution of axonal injuries and the efficacy of interventions in various disease models. Finally, with this technique, we can correlate the findings with histology to investigate disease mechanisms.40
In vivo “staining” (contrast agent)
The basic contrast in the MR images mainly results from regional differences in the intrinsic relaxation times T1 and T2, each of which can be independently chosen to dominate image contrast. In some cases, there is not enough endogenous contrast to characterize tissues or detect blood flow; on these occasions, the imaging capability can be enhanced by the use of contrast agents (CA).41 CA alter the image contrast following intravenous injection. The degree and location of the contrast changes provide substantial diagnostic information. Certain contrast agents are predominantly used to shorten the T1 relaxation time and these are mainly based on low-molecular weight chelates of the gadolinium ion (Gd3+), while the most widely used T2 shortening agents are based on iron oxide (FeO) particles. Depending on their chemical composition, molecular structure and overall size, the in vivo distribution volume and pharmacokinetic properties vary widely between different contrast agents and these largely determine their use in specific diagnostic tests. T1 CA consist mainly of paramagnetic complexes while T2 CA mostly use superparamagnetic ions. Paramagnetic CA contain metal ions that have unpaired electrons and thus a permanent magnetic moment, like Manganese (Mn2+) or Gadolinium (Gd3+). The magnetic moment of the paramagnetic atom interacts with the much smaller magnetic moment of the protons in adjacent water molecules, reducing their longitudinal and transverse relaxation time, resulting in a positive contrast.41 The effect on T1 relaxation time is evident at low concentrations, but at higher concentrations the effect on T2 relaxation time becomes significant, and the signal intensity in T2 weighted acquisitions decreases.42 Paramagnetic metal in their ionic form are cytotoxic, so they must be complexed with chelating compounds, influencing the chemistry of the ions by reducing in vivo toxicity, altering the bio-distribution and affecting the efficiency in shortening T1 and T2. An MRI technique that involves the use of paramagnetic CA is Dynamic contrastenhanced magnetic resonance imaging (DCE-MRI):43-45 it is the acquisition of serial MRI images before, during, and after the administration of an MR contrast agent, as Gd3-DTPA. DCE-MRI gives information about physiological tissue characteristics. Indeed, the concentration of the CA is measured as it passes from the blood vessels to the extracellular space of the tissue and as it goes back to the blood vessels. Many researchers have established the utility of the DCE-MRI in the diagnosis of several kinds of tumors. It appears to provide information in the assessment of carcinomas, according to their stromal content;44 it can successfully demonstrate the nature of a lymphoma and is helpful for making a differential diagnosis from other lesions.
Superparamagnetic CA are iron oxide nanoparticles (SPION) that exhibit superparamagnetism. By applying an external magnetic field, as is produced by an MRI machine, the magnetic moments of the nanoparticle align in the direction of the applied field. Once the magnetic field is removed, they no longer exhibit any residual magnetic interaction and, as a consequence, the magnetic moments of the nanoparticles return to be randomly oriented (Brownian motion).46 SPION, thanks to their superparamagnetic properties, are effective contrast agents in MRI. When SPION are present in tissue, they disturb the local magnetic field homogeneity and the large susceptibility differences between the iron oxide crystals and nearby protons, leading to a rapid dephasing of surrounding protons, resulting in a decrease in transverse (T2) relaxation times.46 The shorter transverse relaxation time results in a darker image (negative contrast) observed in the vicinity of the SPION; this is referred to as negative contrast. The magnetic properties of SPION can be manipulated by controlling the size of core (magnetite or maghemite), the nature of their biocompatible coatings (hydrodynamic size), the polydispersity and the surface charge. A schematic illustration of nanoparticle’s structure is showed in Figure 4A.
Figure 4.
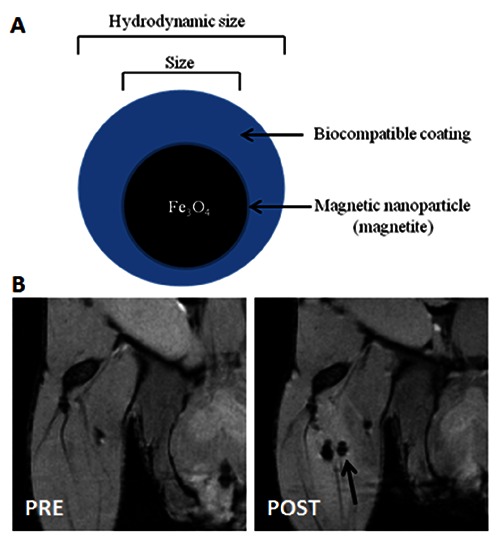
A) Basic schematic diagram of a magnetic nanoparticle’s structure; a magnetic nanoparticle consists of magnetic core and biocompatible coating. B) In vivo MR Images of USPIO-labeled stem cells (2.5x103) PRE and POST intra-muscle injection.
The hydrodynamic and the crystals size are measured by trasmission electron microscopy, X-ray diffraction or dynamic light scattering, while surface charge is measured by zeta potential, which is based on the study of the electrophoretic mobility. The nanoparticles can be classified according to their diameter in: i) ultra-small superparamagnetic iron oxide nanoparticles (USPIO - diameter less than 50 nm); ii) superparamagnetic iron oxide nanoparticles (SPION - d greater than 50 nm but less than 1 μm); and iii) micron-sized particles of iron oxide (MPIO - diameter higher than 1 μm). SPION have been used as MRI contrast agents since 1990.47 Currently, they are clinically used for liver imaging, spleen imaging, tumor imaging, lymph nodes imaging, central nervous system imaging and blood pool agent.48 Due to their safety, biodegradability and efficiency as MRI contrast agent, SPION represent efficient cell labels for cellular imaging. Although not initially developed for cellular imaging, recently they have been successfully adapted for labeling and tracking stem cells and nanovesicles.49 In example, SPION are the most widely used contrast agents for the detection of stem cells in vivo (Figure 4B). In context of stem cells as therapeutic approach for several diseases (i.e., neurodegenerative disease), SPION-labeled stem cells have the potential to increase our knowledge and understanding of post injection stem cell behavior and migration processes.50,51 This studies supports the idea that SPION-based MRI could be applicable as non-invasive imaging approach for cells tracking, leading to a better understanding of physiological and pathological mechanisms related to cells migration.
Criticisms in the use of MRI histochemistry
Disadvantages are in the higher cost of magnets and electronics. In general, the cost of scanners increases proportionally to the generated static magnetic field beause: for example, the cost of a scanner operating at 7 T is about 33% higher than the same instrument operating at 4.7 T.
Among the limitations of the method there is the possibility to analyze a limited number of elements. Indeed, MRI can only be performed on isotopes with a nuclear spin resulting “not null” and with a sufficiently high natural abundance to be detected. The most exploited element for diagnostic purposes is hydrogen, whose resonance signal allows to highlight the contrast between the different types of tissues, discriminating with precision, and to obtain information on a large number of organic compounds. However, it should also be mentioned the possibility of analyzing carbon, phosphorus and fluoride.
Despite the limitations illustrated above, advanced MRI techniques, in particular MRS and fMRI, can represent a complement and a support to conventional histochemical techniques.
References
- 1.Medhi B, Misra S, Avti PK, Kumar P, Kumar H, Singh B. Role of neuroimaging in drug development. Rev Neurosci 2014;25:663-73. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Figueiras R, Baleato-Gonzalez S, Padhani AR, Oleaga L, Vilanova JC, Luna A, et al. Proton magnetic resonance spectroscopy in oncology: the fingerprints of cancer? Diagn Interv Radiol 2016;22:75-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mansfield P. MRI imaging in biomedicine. Advances in magnetic resonance. Academic Press, New York: 1982. [Google Scholar]
- 4.Callaghan PC. Principles of nuclear magnetic resonance microscopy. Clarendon Press, Oxford: 1993. [Google Scholar]
- 5.Blackband SJ, Buckley DL, Bui JD, Phillips MI. NMR microscopy—beginnings and new directions. Magn Reson Mater Phy 1999;9:112-6. [DOI] [PubMed] [Google Scholar]
- 6.Aguayo JB, Blackband SJ, Schoeniger J, Mattingly MA, Hintermann M. Nuclear magnetic resonance imaging of a single cell. Nature 1986;322:190-1. [DOI] [PubMed] [Google Scholar]
- 7.Bowtell RW, Peters A, Sharp JC, Mansfield P, Hsu EW, Aiken N, et al. NMR microscopy of single neurons using spin echo and line narrowed 2DFT imaging. Magn Reson Med 1995;33:790-4. [DOI] [PubMed] [Google Scholar]
- 8.Blackband SJ, Bui JD, Buckley DL, Zelles T, Plant HD, Inglis BA, et al. MR microscopy of perfused brain slices. Magn Reson Med 1997;38:1012-5. [DOI] [PubMed] [Google Scholar]
- 9.Johnson GA, Cofer GP, Gewalt SL, Hedlund LW. Morphologic phenotyping with MR microscopy: the visible mouse. Radiology 2002;222:789-93. [DOI] [PubMed] [Google Scholar]
- 10.Cleary JO, Wiseman FK, Norris FC, Price AN, Choy M, Tybulewicz VL, et al. Structural correlates of active-staining following magnetic resonance microscopy in the mouse brain. Neuroimage 2011;56: 974-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lunati E, Farace P, Nicolato E, Righetti C, Marzola P, Sbarbati A, et al. Polyunsaturated fatty acids mapping by (1)H MR-chemical shift imaging. Magn Reson Med 2001;46:879-83. [DOI] [PubMed] [Google Scholar]
- 12.Ratai EM, Gilberto Gonzalez R. Clinical magnetic resonance spectroscopy of the central nervous system. Handb Clin Neurol 2016;135:93-116. [DOI] [PubMed] [Google Scholar]
- 13.Park JH, Lee H, Makaryus R, Yu M, Smith SD, Sayed K, et al. Metabolic profiling of dividing cells in live rodent brain by proton magnetic resonance spectroscopy (1HMRS) and LCModel analysis. PLoS One 2014;9:e94755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee DW, Chung S, Yoo HJ, Kim SJ, Woo CW, Kim ST, et al. Neurochemical changes associated with stress-induced sleep disturbance in rats: in vivo and in vitro measurements. PLoS One 2016;11:e0153346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillies RJ, Morse DL. In vivo magnetic resonance spectroscopy in cancer. Annu Rev Biomed Eng 2005;7:287-326. [DOI] [PubMed] [Google Scholar]
- 16.Hakumaki JM, Poptani H, Puumalainen AM, Loimas S, Paljarvi LA, Yla-Herttuala S, et al. Quantitative 1H nuclear magnetic resonance diffusion spectroscopy of BT4C rat glioma during thymidine kinase-mediated gene therapy in vivo: identification of apoptotic response. Cancer Res 1998;58):3791-9. [PubMed] [Google Scholar]
- 17.Hakumaki JM, Poptani H, Sandmair AM, Yla-Herttuala S, Kauppinen RA. 1H MRS detects polyunsaturated fatty acid accumulation during gene therapy of glioma: implications for the in vivo detection of apoptosis. Nat Med 1999;5):1323-7. [DOI] [PubMed] [Google Scholar]
- 18.Iordanescu G, Brendler C, Crawford SE, Wyrwicz AM, Venkatasubramanian PN, Doll JA. MRS measured fatty acid composition of periprostatic adipose tissue correlates with pathological measures of prostate cancer aggressiveness. J Magn Reson Imaging 2015;42:651-7. [DOI] [PubMed] [Google Scholar]
- 19.Mosconi E, Minicozzi A, Marzola P, Cordiano C, Sbarbati A. (1) H-MR spectroscopy characterization of the adipose tissue associated with colorectal tumor. J Magn Reson Imaging 2014;39:469-74. [DOI] [PubMed] [Google Scholar]
- 20.Hwang JH, Choi CS. Use of in vivo magnetic resonance spectroscopy for studying metabolic diseases. Exp Mol Med 2015;47:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dagnelie PC, Leij-Halfwerk S. Magnetic resonance spectroscopy to study hepatic metabolism in diffuse liver diseases, diabetes and cancer. World J Gastroenterol 2010;16:1577-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roden M. Non-invasive studies of glycogen metabolism in human skeletal muscle using nuclear magnetic resonance spectroscopy. Curr Opin Clin Nutr Metab Care 2001;4:261-6. [DOI] [PubMed] [Google Scholar]
- 23.Cho YD, Choi GH, Lee SP, Kim JK. (1)HMRS metabolic patterns for distinguishing between meningiomas and other brain tumors. Magn Reson Imaging 2003;21:663-72. [DOI] [PubMed] [Google Scholar]
- 24.Oz G, Kumar A, Rao JP, Kodl CT, Chow L, Eberly LE, et al. Human brain glycogen metabolism during and after hypoglycemia. Diabetes 2009;58:1978-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolay K, Braun KP, Graaf RA, Dijkhuizen RM, Kruiskamp MJ. Diffusion NMR spectroscopy. NMR Biomed 2001;14:94-111. [DOI] [PubMed] [Google Scholar]
- 26.Mori S, van Zijl PC. Fiber tracking: principles and strategies - a technical review. NMR Biomed 2002;15:468-80. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. P Natl Acad Sci USA 1990;87:9868-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bobholz JA, Rao SM, Saykin AJ, Pliskin N. Clinical use of functional magnetic resonance imaging: reflections on the new CPT codes. Neuropsychol Rev 2007;17:189-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benveniste H, Blackband SJ. Translational neuroscience and magnetic-resonance microscopy. Lancet Neurol 2006;5:536-44. [DOI] [PubMed] [Google Scholar]
- 30.Driehuys B, Nouls J, Badea A, Bucholz E, Ghaghada K, Petiet A, et al. Small animal imaging with magnetic resonance microscopy. ILAR J 2008;49:35-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu F, Kida I, Hyder F, Shulman RG. Assessment and discrimination of odor stimuli in rat olfactory bulb by dynamic functional MRI. P Natl Acad Sci USA 2000;97:10601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tambalo S, Peruzzotti-Jametti L, Rigolio R, Fiorini S, Bontempi P, Mallucci G, et al. Functional magnetic resonance imaging of rats with experimental autoimmune encephalomyelitis reveals brain cortex remodeling. J Neurosci 2015;35:10088-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filippi M, Rocca MA. Functional MR imaging in multiple sclerosis. Neuroimaging Clin N Am 2009;19:59-70. [DOI] [PubMed] [Google Scholar]
- 34.Mandeville JB, Marota JJ, Kosofsky BE, Keltner JR, Weissleder R, Rosen BR, et al. Dynamic functional imaging of relative cerebral blood volume during rat forepaw stimulation. Magn Reson Med 1998;39:615-24. [DOI] [PubMed] [Google Scholar]
- 35.Palmer JT, de Crespigny AJ, Williams S, Busch E, van Bruggen N. High-resolution mapping of discrete representational areas in rat somatosensory cortex using blood volume-dependent functional MRI. Neuroimage 1999;9:383-92. [DOI] [PubMed] [Google Scholar]
- 36.van Bruggen N, Busch E, Palmer JT, Williams SP, de Crespigny AJ. High-resolution functional magnetic resonance imaging of the rat brain: mapping changes in cerebral blood volume using iron oxide contrast media. J Cereb Blood Flow Metab 1998;18:1178-83. [DOI] [PubMed] [Google Scholar]
- 37.Beckmann N, Mueggler T, Allegrini PR, Laurent D, Rudin M. From anatomy to the target: contributions of magnetic resonance imaging to preclinical pharmaceutical research. Anat Rec 2001;265:85-100. [DOI] [PubMed] [Google Scholar]
- 38.Marota JJ, Mandeville JB, Weisskoff RM, Moskowitz MA, Rosen BR, Kosofsky BE. Cocaine activation discriminates dopaminergic projections by temporal response: an fMRI study in Rat. Neuroimage 2000;11:13-23. [DOI] [PubMed] [Google Scholar]
- 39.Wang S, Melhem ER, Poptani H, Woo JH. Neuroimaging in amyotrophic lateral sclerosis. Neurotherapeutics 2011;8:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 2006;51:527-39. [DOI] [PubMed] [Google Scholar]
- 41.Caschera L, Lazzara A, Piergallini L, Ricci D, Tuscano B, Vanzulli A. Contrast agents in diagnostic imaging: Present and future. Pharmacol Res 2016;110:65-75. [DOI] [PubMed] [Google Scholar]
- 42.De Leon-Rodriguez LM, Martins AF, Pinho MC, Rofsky NM, Sherry AD. Basic MR relaxation mechanisms and contrast agent design. J Magn Reson Imaging. 2015;42(3):545-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boschi F, Marzola P, Sandri M, Nicolato E, Galie M, Fiorini S, et al. Tumor microvasculature observed using different contrast agents: a comparison between Gd-DTPAAlbumin and B-22956/1 in an experimental model of mammary carcinoma. Magn Reson Mater Phy 2008;21:169-76. [DOI] [PubMed] [Google Scholar]
- 44.Farace P, Merigo F, Fiorini S, Nicolato E, Tambalo S, Daducci A, et al. DCE-MRI using small-molecular and albumin-binding contrast agents in experimental carcinomas with different stromal content. Eur J Radiol 2011;78:52-9. [DOI] [PubMed] [Google Scholar]
- 45.Ritelli R, Ngalani Ngaleu R, Bontempi P, Dandrea M, Nicolato E, Boschi F, et al. Pancreatic cancer growth using magnetic resonance and bioluminescence imaging. Magn Reson Imaging 2015;33:592-9. [DOI] [PubMed] [Google Scholar]
- 46.Bull E, Madani SY, Sheth R, Seifalian A, Green M, Seifalian AM. Stem cell tracking using iron oxide nanoparticles. Int J Nanomedicine 2014;9:1641-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Xu C, Ow H. Commercial nanoparticles for stem cell labeling and tracking. Theranostics 2013;3:544-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L, Jiang W, Luo K, Song H, Lan F, Wu Y, et al. Superparamagnetic iron oxide nanoparticles as MRI contrast agents for non-invasive stem cell labeling and tracking. Theranostics 2013;3:595-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Busato A, Bonafede R, Bontempi P, Scambi I, Schiaffino L, Benati D, et al. Magnetic resonance imaging of ultrasmall superparamagnetic iron oxide-labeled exosomes from stem cells: a new method to obtain labeled exosomes. Int J Nanomedicine 2016;11:2481-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bigini P, Diana V, Barbera S, Fumagalli E, Micotti E, Sitia L, et al. Longitudinal tracking of human fetal cells labeled with super paramagnetic iron oxide nanoparticles in the brain of mice with motor neuron disease. PLoS One 2012;7:e32326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willenbrock S, Knippenberg S, Meier M, Hass R, Wefstaedt P, Nolte I, et al. In vivo MRI of intraspinally injected SPIO-labelled human CD34+ cells in a transgenic mouse model of ALS. In Vivo 2012;26:31-8. POST intra-muscle injection. [PubMed] [Google Scholar]


