Abstract
The wMel Wolbachia strain was known for cytoplasmic incompatibility (CI)-induction and blocking the transmission of dengue. However, it is unknown whether it can establish and induce CI in a non-dipteran host insect. Here we artificially transferred wMel from Drosophila melanogaster into the whitefly Bemisia tabaci. Fluorescence in situ hybridisation demonstrated that wMel had successfully transfected the new host. Reciprocal crossing was conducted with wMel-transfected and wild-type isofemale lines, indicating that wMel could induce a strong CI without imposing significant cost on host fecundity. We then determined the maternal transmission efficiency of wMel in the offspring generations, showing a fluctuating trend over a period of 12 generations. We thus detected the titre of wMel during different developmental stages and in different generations by using real-time quantitative PCR, revealing a similar fluctuating mode, but it was not significantly correlated with the dynamics of transmission efficiency. These results suggest that wMel can be established in B.tabaci, a distantly related pest insect of agricultural importance; moreover, it can induce a strong CI phenotype in the recipient host insect, suggesting a potential for its use in biological control of B. tabaci.
Maternally inherited microorganisms are common and have diverse effects on invertebrate biology1. Wolbachia are the most widely studied of these endosymbionts, and 40% of terrestrial arthropod species and approximately 66% of all insect species are infected with Wolbachia2,3. They are master manipulators of arthropod reproduction and induce a number of reproductive abnormalities, including cytoplasmic incompatibility (CI), feminization, male killing and parthenogenesis induction, all of which enhance the spread of Wolbachia in host populations4,5,6. Among these, CI is the most commonly reported phenotype caused by Wolbachia, which has caught the attention of many researchers for its potential as a useful strategy for biological control of pest insects of medical and agricultural importance7,8,9.
CI occurs when conspecific insects with different Wolbachia infection status mate, which causes embryonic mortality in diploid species but results in sex distortion in haplodiploid species, such as in Bemisia tabaci, where male offspring is produced from unfertilised eggs10,11. CI can be either unidirectional or bidirectional. The former occurs when an uninfected female mates with an infected male, while the reciprocal mating is compatible; the latter is expressed between conspecifics infected with different Wolbachia strains. Wolbachia-induced CI was exploited as a method for pest population suppression in a very early time12. The prerequisite for a CI-based pest control strategy is the obtainment of incompatible male insects. In early attempts, incompatible insects were obtained through introgressive hybridisation and released to control disease vectors in a way analogous to the sterile insect technique13,14. However, this process is time-consuming and laborious. Since then, artificial interspecies transinfection of Wolbachia strains has been developed through embryonic microinjection15,16. To date, experimental transinfection of Wolbachia endosymbionts between different host species has been repeatedly achieved through either embryonic or nymphal injection17,18,19,20,21,22,23,24. Artificial transinfection has greatly facilitated the application of CI-based technologies in pest population suppression25,26,27. The most famous case was the successful establishment of the wMel Wolbachia strain from Drosophila melanogaster in Aedes populations to block dengue transmission due to its effective interference with RNA viruses28,29,30,31. The Wolbachia strain wMel originating from D. melanogaster has spread globally within the last century32. Despite the knowledge that wMel could induce CI and invade the dipteran Aedes vectors33,34,35, it is still unknown whether it can establish and induce CI in a distantly related host insect.
B. tabaci (Hemiptera:Aleyrodidae) is a species complex with a global distribution, which has caused considerable damages to ornamental, vegetable, grain legume and cotton production36, particularly the Middle East-Asia Minor 1 (MEAM1 or B biotype) and Mediterranean (MED or Q biotype)37. Due to the serious problem of insecticide resistance in this species, alternative control approaches are needed38, and Wolbachia-induced CI might be a useful strategy since the natural populations of B. tabaci (especially the B and Q biotypes) are widely infected with Wolbachia and antibiotic treatment could induce strong CI in B biotype39,40. Nevertheless, before any biological control programs can really be tested, an isofemale line stably infected with a heterologous CI-inducing Wolbachia strain should be established. Here we isolated the wMel strain from a local D. melanogaster population and then transferred it into B. tabaci through nymphal microinjection. After localisation of wMel in the new host, transfected isofemale lines were established and reciprocal crossing experiments were thus conducted. Subsequently, we measured the titre of wMel at different developmental stages of the new host over a total of 12 generations by using real-time quantitative PCR (qPCR), and determined the transmission efficiency of wMel in the offspring population. The aims of this study were to determine (i) whether wMel could establish in a distantly related host insect; (ii) the CI-inducing capability of wMel in a pest insect of agricultural importance; (iii) the dynamics of the titre of wMel, and (iv) the correlation between titre and transmission rate. Our experimental data suggested that wMel could establish in the new host and induce a strong CI; while the titre and transmission rate shared a similar fluctuating mode at different developmental stages and between generations, the transmission rate of wMel was not necessarily determined by its titre.
Results
MLST typing, transinfection and establishment of isofemale line
The Wolbachia strain isolated from D. melanogaster was identified by gene sequencing and MLST typing41. A batch sequence query in the PubMLST database revealed that its allelic profile for gatB, coxA, hcpA, ftsZ and fbpA was 1, 1, 1, 1 and 1 (corresponding to ST-1), and its HVR profile for HVR1, HVR2, HVR3 and HVR4 was 1, 12, 21 and 24 (corresponding to ST-31) (Supplementary Table S1). A comparison of its sequence types with the STs in the database showed that the MLST and HVR profiles of this strain exactly matched those of the strain Dmel_A_wMel from D. melanogaster (PubMLST ID: 1), belonging to Supergroup A42. Dmel_A_wMel is a typical CI-inducing Wolbachia strain. We also examined the sub-strain status of the strain, showing that it is wMel but not wMelCS since only the primers32 targeting IS5-WD0516/7 could produce amplicon of expected size (~2500 bp).
The purified Wolbachia (wMel) was directly transferred into the 4th instar nymphs (pseudopupae) of B. tabaci by microinjection. Approximately 80–100 4th-instar whitefly nymphs (pseudopupae) were microinjected for each batch, and the survival rate was 50–60%. The wMel-transfected nymphs were maintained in a climate incubator until eclosion after approximately 72 h (Supplementary Fig. S1). When the whiteflies emerged (G0), they were confirmed for positive infection with wMel strain by using Wolbachia wsp-based PCR detection, and infected adults were moved to a nylon mesh cage for establishment of isofemale lines.
Localisation of wMel
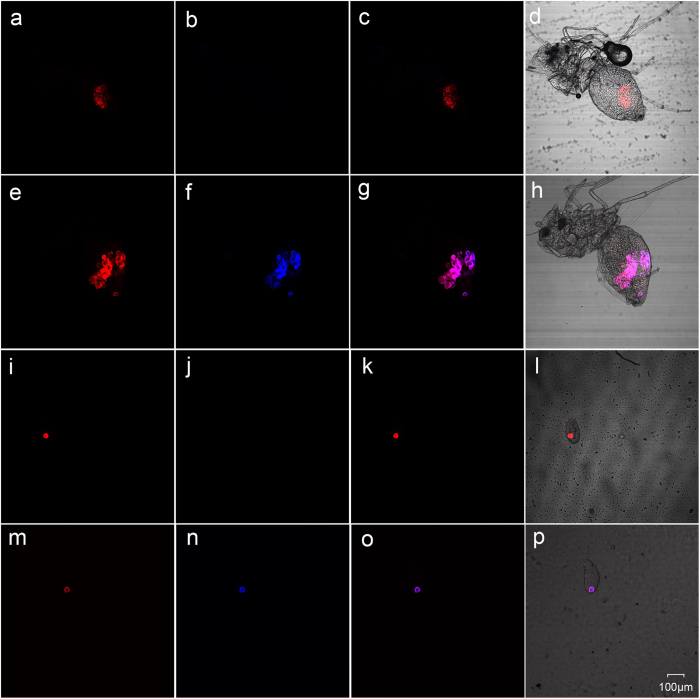
FISH was employed to localize the wMel strain in the new host. The results showed that both Portiera and Wolbachia were localized inside the bacteriocytes; wMel existed along with the primary endosymbiont Portiera throughout the life cycle of transfected B. tabaci (egg, nymph and adult), though the titres of wMel were different during different developmental stages (Fig. 1). Wolbachia gave a specific blue light, while Portiera released a strong red signal. The wild-type B. tabaci hosted only Portiera without wMel being observed. Our FISH data suggested that the wMel strain had been successfully transfected into the recipient host, and also demonstrated that it could be transmitted from mother to offspring through eggs (see Supplementary Fig. S2 for a full set of FISH images).
Figure 1. FISH analysis of wMel-transfected and wild-type B. tabaci with Portiera-specific probe (red) and Wolbachia-specific probe (blue).
(a–d) Wild-type female adult; (e–h) Transfected female adult; (i–l) Wild-type egg; (m–p) Transfected egg. (a,e,i,m) Portiera channel only; (b,f,j,n) Wolbachia channel only; (c,g,k,o) Merged images showing overlap of Wolbachia and Portiera channels in dark field; (d,h,l,p) Merged images showing overlap of Wolbachia and Portiera channels in bright field.
Crossing and CI analysis
Totally four different crossing experiments were conducted: (WT♀ × WT♂); (WT♀ × TI♂); (WT♂ × TI♀), and (TI♀ × TI♂). The whiteflies are taken from G6. The results showed that there was significant difference in the number of offspring per female between (WT♀ × TI♂) (16.1 ± 0.53) and (TI♀ × TI♂) (19.8 ± 0.97) (SNK, P = 0.006), but no significant difference was found between the other crossings. There was an extremely significant difference in the number of male offspring among the four crossings (SNK, P = 0.0001), though no significant difference was observed between (WT♀ × WT♂) (53.56 ± 2.49) and (TI♀ × TI♂) (58.35 ± 1.89) (SNK, P = 0.074), both of which gave approximately equal mean number of male and female progenies (Table 1). A simple criterion for judging CI level in B. tabaci (a haplodiploid species) is to calculate the male/female ratio in the progenies: if CI is induced, the percentage of male offspring will increase as the unfertilised eggs produce male offspring. In the present study, the highest mean percentage of male offspring (97.46%) was observed in the crossing (WT♀ × TI♂), but surprisingly the reciprocal crossing (WT♂ × TI♀) (n = 11) also produced more male (68.13%) than female progenies. We conducted a Chi-square test to analyse the male-biased reciprocal crossing result, suggesting that the difference between the observed and expected (1:1) numbers of male offspring is not significant (Pearson χ2 = 1.029, df = 1, P = 0.310). Our crossing data indicated that nearly complete CI was induced between TI♂ and WT♀ since the offspring from this crossing was almost all male, but the partial CI in the reciprocal direction (WT♂ × TI♀) is not statistically supported.
Table 1. Crossing between wMel-transinfected (TI) and wild-type (WT) whiteflies.
| Cross type (♀ × ♂) | No. of crosses (N) | No. of offspring per female | Percentage of male offspring (%) |
|---|---|---|---|
| WT × WT | 11 | 17.1 ± 0.87 ABab | 53.56 ± 2.49 Aa |
| WT × TI | 16 | 16.1 ± 0.53 Bb | 97.46 ± 1.33B |
| TI × WT | 11 | 17.7 ± 0.84 ABab | 68.13 ± 1.24 C |
| TI × TI | 14 | 19.8 ± 0.97 Aa | 58.35 ± 1.89 Aa |
The whiteflies are taken from G6. The data are mean ± SE. Different letters in the same column indicate significant difference based on SNK test of One-way ANOVA (lowercase letter, P < 0.05; uppercase letter, P < 0.01).
Maternal transmission rate
Transmission rate is an important parameter for estimating the transmission efficiency of a Wolbachia strain, which determines its potential as a biocontrol agent. Our data showed that the transmission rates of wMel fluctuated between generations: reaching a lowest point at G2 (10.72%), then gradually climbing up to a peak point at G6 (87.5%) and heading down again, followed by a slight rebound (Fig. 2). It seems that the transmission rate of wMel may fluctuate around the equilibrium level (~50%) in the next generations if more generations are investigated. In a period of 12 generations, the artificially transfected wMel was shown to be transmitted in the offspring populations with a fluctuating mode.
Figure 2. Infection dynamics of the wMel strain in the recipient host B. tabaci at different developmental stages and in different generations.
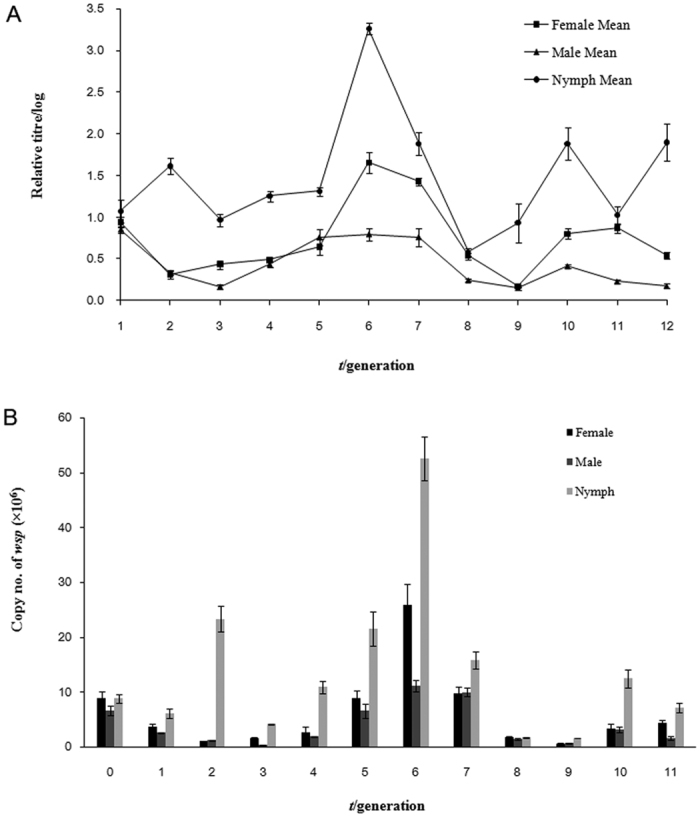
(A) Log relative titre of wMel in B. tabaci. (B) The copy number of wMel in B. tabaci.
Dynamics of the titre of wMel
The fluctuating transmission rate of wMel observed above may be the consequence of a series of complicated interactions, and one of the possible measurable factors is the titre of wMel. In the present study, we measured the relative titre and copy number of wsp gene of wMel in the new host during different developmental stages and different generations after transinfection by using qPCR. The results showed that wMel could be transmitted from generation to generation, but the relative titre of wMel changed drastically between different developmental stages and different generations. Specifically, the titre of wMel reached a low level during different developmental stages in the 2nd generation (G2), and then climbed to the peak in the 6th generation (G6), from where the titre of wMel dropped again to a very low level in the 7th and 8th generations, followed by a weak rebound (Fig. 3A). The titre of wMel was persistently low in male adults, but at a higher level and with a more fluctuating mode in female adults; the titre of wMel was higher during nymphal stage than adult stage. Correspondingly, the copy number of wMel in nymphs was also higher than that in adults, and male adults had lower copy number of wMel than female adults (Fig. 3B). The copy number of wMel in female adults climbed up from the low point at G2, reaching to a peak at G6, and then declined. Similar trend occurred in male adults and nymphs. The highest copy number of wMel in nymphs (52.6 × 106 copies/μl at G6) was nearly five times that in male adults (11.1 × 106 copies/μl at G6). After G6, the copy numbers of wMel in the host gradually decreased and maintained at a comparatively low level.
Figure 3. Maternal transmission rate of the wMel strain in offspring generations.
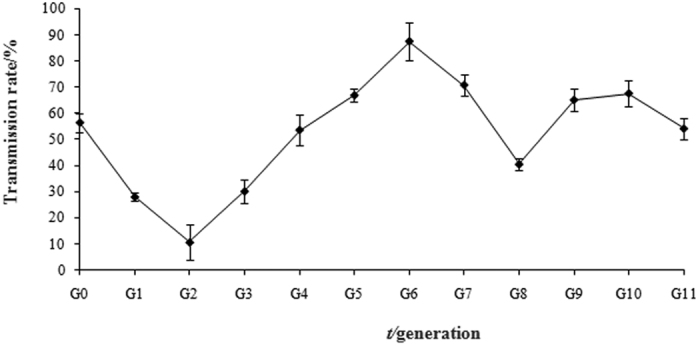
Data are mean ± S.E., derived from three independent isofemale lines.
Correlation between the titre and transmission rate of wMel
Linear regression analyses showed that wMel titre was not significantly correlated with its transmission rate over a period of 12 generations (R2 = 0.10–0.35) (Table 2). We also analysed the correlation between the copy number of wsp gene and the titre of wMel, showing that the copy number of wsp gene was positively correlated with the titre (R2 = 0.60–0.80) in adults and nymphs (Supplementary Fig. S3). These data indicate that the relative titre can largely represent the copy number of wMel, and the transmission rate of wMel was most probably not determined by its titre.
Table 2. Correlation between copy no. of wsp, relative titre and transmission rate of the wMel strain in Bemisia tabaci.
| Copy no. of wsp |
Relative titre |
||||||
|---|---|---|---|---|---|---|---|
| Female | Male | Nymph | Female | Male | Nymph | ||
| Transmission rate | 0.464 | 0.475 | 0.189 | 0.347 | 0.123 | 0.310 | |
| Copy no. of wsp | Female | 0.705 | |||||
| Male | 0.618 | ||||||
| Nymph | 0.788 | ||||||
Transmission rates (%) are transformed by arcsine square root before analysis. All data are R2 values based on linear regression analysis.
Discussion
The wMel Wolbachia strain was known for its CI induction and blocking of dengue transmission after transferred from its original host D. melanogaster into the dipteran Aedes host28. In this study, we showed that the wMel strain could establish and induce a strong CI phenotype in a distantly related insect host of agricultural importance after transferred into B. tabaci through microinjection. FISH analysis demonstrated that wMel was localised inside the bacteriocytes along with the primary endosymbiont Portiera and could be transmitted into the offspring through the egg; qPCR and transmission efficiency analyses indicated that wMel could comparatively rapidly be adapted to a phylogenetically distantly related host and establish in the new host after only several generations. Moreover, our study revealed that the relative titre and transmission rate of wMel shared a similar fluctuating mode during different developmental stages over more than 10 generations, but the transmission rate is probably not determined by the titre.
FISH is a useful technique for identification and localisation of bacterial endosymbionts in insect species as it can not only indicate whether the symbiont is present but also where it is. Our FISH results were significant because they provided hard evidence for the presence of wMel strain in the new host at various developmental stages (Supplementary Fig. S2) and also for the vertical maternal transmission of wMel strain through eggs between generations. Our finding that wMel was located with the primary symbiont Portiera inside the bacteriocyte is consistent with what has been reported previously43. The wMel strain and other two Wolbachia strains artificially transfected in our previous studies were all found to share the bacteriocyte with Portiera in B. tabaci11,20, which may be an efficient mechanism for vertical transmission of these maternally inherited endosymbionts. Nevertheless, cohabitation of different symbionts (exotic and native; secondary and primary) also enhances their interactions that can significantly influence the dynamics of their co-existence.
The wMel strain has previously been shown to induce a strong CI phenotype in the dipteran Aedes mosquito after transinfection33, but it is still unclear whether wMel can establish and cause CI in a more distantly related host insect. Here we showed that the wMel strain could induce nearly complete CI between wMel-transfected male and wild-type female whiteflies. The same Wolbachia strain can induce totally different reproductive phenotypes in different hosts. For instance, the wCauA strain from the almond moth, Cadracautella, induced CI in C. cautella but male killing in the Mediterranean flour moth, Ephestia kuehniella44. Our data combined with previous reports indicated that wMel could induce a strong CI phenotype not only in its native host and other dipteran hosts but also the non-dipteran host insect without a significant host effect. Interestingly, a low-level sex-biased ratio (68.13% male offspring) was also observed in the reciprocal crossing between wMel-transfected female and wild-type male whiteflies (WT♂ × TI♀). Nevertheless, data analysis revealed that the difference between the observed and expected (1:1) numbers of male offspring is not significant, and therefore the partial CI in the reciprocal direction (WT♂ × TI♀) is not statistically supported in the present study. Theoretically, sex bias in the reciprocal direction should not have taken place if the wild-type male is not infected with a native Wolbachia strain (as shown in FISH analysis; Fig. 1a–d); however, bidirectional CI was observed in two of our previous studies using traninsfected B. tabaci11,20. One possible explanation for this may be that the wild-type B. tabaci is likely infected with a low-titre Wolbachia strain. The presence of Wolbachia infection in natural B. tabaci populations was supported by FISH analysis showing a very weak Wolbachia-specific signal in wild-type B. tabaci20 and CI induction between wild-type B. tabaci males and antibiotic-treated females (Wolbachia−)40. In this study, no native Wolbachia strain was detected in the wild-type B. tabaci in both FISH and qPCR analyses, and thus no statistically significant bidirectional CI has been induced.
The CI level or strength of reproductive incompatibility, fitness costs associated with Wolbachia infection, and transmission rate from mother to offspring are among the major determinants for the rate and extent of spread of a Wolbachia strain in a target population45. Our studies showed that wMel could induce a strong CI in the recipient host without imposing a significant cost on host fecundity. Furthermore, the transmission rate of wMel returned to a relatively stable level (>50%) after a drastic fluctuation within less than 10 generations. These experimental results suggest that B. tabaci is a highly permissive host for exotic Wolbachia strains and CI expression as this insect pest was also shown to easily accept other two artificially transfected Wolbachia strains from the distantly related hosts Scleroderma guani (wSguBJ) and Corcyra cephalonica (wCcep)11,20. All arthropods are not permissive for exotic Wolbachia strains. For example, experimental interspecific transfer of Wolbachia in terrestrial isopods indicated that isopod Wolbachia were highly adapted to their hosts and could hardly establish in another host species46. A highly permissive host provides us more opportunities to regulate its population by CI-based strategy. Here, wMel-transinfected male whiteflies can induce a nearly complete CI in B. tabaci populations, producing almost all male offspring, which seriously distorts the sex ratio of the host population and leads to suppression of the target population.
The titre or infection density of an endosymbiont is among the major parameters to understand the dynamics of its establishment process in the recipient host after transinfection. The titre of a Wolbachia strain represents the consequence from the interaction between Wolbachia and its host during the infection process. Our qPCR results indicated that, during the initial period of infection (G0-G3), wMel seemed to be poorly adapted to the internal environment of the host, leading to a relatively low titre of wMel. As infection progressed, wMel became better adapted to the new host, which promoted the titre of wMel to a peak level at G6. However, the titre of wMel dropped dramatically after G6, which might be caused by an induced “counter-attack” from the host. Different from other report47, our study showed that the nymphs persistently hosted higher titre of wMel than adults, indicating a complex mechanism regulating the infection progress of an exotic Wolbachia invader in the recipient host over different developmental stages. From G10, the titre of wMel began to recover, but it remained at a low level in adults, particularly in male adults, indicating that a weak equilibrium has been reached between the host and wMel for some unknown mechanism. Possible reasons include regulation by the host immune system and symbiont-symbiont interactions48,49. Identification of the specific factors (molecules) that determine the titre of Wolbachia in the host during different developmental stages would help us understand the precise molecular mechanisms underlying the endosymbiont-host interaction. Monitoring the infection dynamics of a Wolbachia strain in the target host is relevant to the Wolbachia-based strategy for biological control of pest insects. For one thing, the infection density can remarkably affect CI level50; for another, a potential Wolbachia strain as a biocontrol agent should be highly invasive with a persistent infection and transmission capability. Our correlation analysis suggested that the transmission rate of wMel was not determined by its titre, though they shared a similar dynamic trend over a period of 12 generations, indicating that the transmission mechanism of transfected Wolbachia might be much more complicated than expected. For further studies, we will determine the correlation between titre and CI level by conducting more crossing experiments by using transinfected whiteflies from different generations with different Wolbachia titres.
In conclusion, the wMel strain isolated from D. melanogaster could be transmitted and established in the whitefly B. tabaci, a distantly related insect host, in a relatively short period of time. This Wolbachia strain could induce a strong CI without imposing a significant cost on host fecundity, suggesting a potential for its use for biological control of B. tabaci, a worldwide agricultural insect pest.
Methods
Insect rearing
The wMel Wolbachia strain for transinfection was isolated from the fruit fly D. melanogaster in China Agricultural University, Beijing, China in 2014. The fruit flies were maintained on Maize-Agarose-Yeast culture medium at 25 °C, 65% relative humidity (RH) in a climate incubator under a 12-h light-dark cycle. The recipient host insect B. tabaci was maintained on the cotton plants (L14:D10 at 28 °C and 60–80% RH).
Isolation and MLST typing of wMel
Genomic DNA was extracted from individual insects by using the potassium acetate method as previously described40. The purified DNA was used as the template for PCR amplification of the six Wolbachia genes (gatB, coxA, hcpA, ftsZ, fbpA, and wsp) with the specific primers reported in Baldo et al.41 (Table 3); the PCR products were purified and subcloned for sequencing, and the sequences were then used for MLST typing according to the protocol described in the MLST database (http://pubmlst.org/Wolbachia/). We also used the previously reported primers32 targeting IS5-WD0516/7 (F: 5′-CCAT CAAGGTCTCTTTCA; R: 5′-TGCAAGGAAAACTAAACCAG; expected size: 2488 bp) and IS5-WD1310 (F: 5′-AGGAGAACTGGTCTACGC; R: 5′-TGTTGCTGAGCTTTG CT; expected size: 745 bp) to determine the sub-strain status of the strain isolated in our study.
Table 3. The primers used for sequencing, PCR-based detection and real-time qPCR analysis.
| Gene name | Primer sequence (5′–>3′) | Fragment size (bp) |
|---|---|---|
| gatB | F:GAKTTAAAYCGYGCAGGBGTTR:TGGYAAYTCRGGYAAAGATGA | 471 |
| coxA | F:TTGGRGCRATYAACTTTATAG R:CTAAAGACTTTKACRCCAGT | 487 |
| hcpA | F:GAAATARCAGTTGCTGCAAA R:GAAAGTYRAGCAAGYTCTG | 515 |
| ftsZ | F:ATYATGGARCATATAAARGATAG R:TCRAGYAATGGATTRGATAT | 524 |
| fbpA | F:GCTGCTCCRCTTGGYWTGAT R:CCRCCAGARAAAAYYACTATTC | 509 |
| wsp | 81 F:TGGTCCAATAAGTGATGAAGAAAC 691 R:AAAAATTAAACGCTACTCCA | 632 |
| β-actin | F:CTTCCAGCCATCCTTCTTG R:CGGTGATTTCCTTCTGCATT | 130 |
| wspQ384 wspQ513 | F:TGGAACCCGCTGTGAATGAT R:GCACCATAAGAACCGAAATAACG | 130 |
Transinfection by microinjection
Wolbachia was purified from 10 fruit flies by using the Percoll density-gradient centrifugation method11. Microinjection was performed on the 4th instar nymphs (pseudopupae). The amount of Wolbachia for each injection (Nanoliter 2000, World Precision Instruments, USA) was 46 nL in SPG buffer (220 mM sucrose, 4 mM KH2PO4, 9 mM Na2HPO4, 5 mM L-glutamate, pH7.4) with a glass needle (Φ0.3 mm, CFT-8201, Jiangsu Rich Life Science Instruments Co., Ltd., China). Approximately 80–100 4th-instar whitefly nymphs were injected for each batch. After injection, the nymph in the dish was placed in a climate incubator until adult emergence (L14:D10 at 28 °C and 60–70% RH). A pair of newly emerged ♀/♂ adults (G0) was separately maintained on potted cotton plants for establishment of isofemale lines. Five independent single-pairs were constructed, from which three transinfected isofemale lines (G1) were selected for further maintenance based on molecular detection of the parental whiteflies (G0) (only the parents detected positive for wMel infection were used). The offspring of transfected (TI) whiteflies was detected for the presence of wMel strain by using the primers wsp81F/691R targeting the wsp gene of Wolbachia (Table 3).
Localisation by FISH
The egg, nymph and adult (male and female) of transfected whiteflies were prepared for FISH analysis and the wild-type (WT) whitefly was used as the control. The methods used were essentially the same as described20. Briefly, whitefly samples were fixed in the Carnoy’s fixative overnight. The fixed samples were immersed in 6% H2O2 for 6 h for decoloration, and then hybridised overnight with the fluorescent probes, BTP1-Cy3 (5′-Cy3-TGTCAGTGTCAGCCCAGAAG-3′), targeting the 16 S rRNA of the primary symbiont Portiera, and W2-Cy5 (5′-Cy5-CTTCTGTGAGTACCGTCATTATC-3′) specific to Wolbachia 16 S rRNA43. Stained samples were viewed under an Olympus FluoViewFV 1000 confocal microscope (Olympus, Japan).
Reciprocal crossing and CI analysis
Male and female TI and WT whiteflies were selected for crossing experiments. TI whiteflies were taken from the sixth generation (G6) when the infection rate was high. Four different crossing experiments were designed: WT♀ × WT♂; WT♀ × TI♂; WT♂ × TI♀, and TI♀ × TI♂. The mating pair was confined in a leaf-clip cage on a cotton plant for 5 days, and then both male and female were removed for molecular detection as described20. The positive male or female was designated as TI♂ or TI♀. The eggs laid on cotton plants were placed into climate incubator for further development till adult emergence (L14:D10 and 65% RH at 27 °C). The progenies were collected, and the number of offspring per female and the percentage of males were calculated. CI level was assessed as the proportion of male progenies.
Maternal transmission rate
Maternal transmission rate was measured as the proportion of infected adult progenies from infected mothers over a total of 12 generations (G0 to G11). B. tabaci is a haplodiploid species, and CI will not increase the proportion of infected offspring because unfertilised eggs will not die but produce male adults. Wolbachia infection was detected by using wsp-specific primers with total genomic DNA as template11. The PCR program was set as follows: 95 °C for 4 min, followed by 2 cycles of touchdown amplification (94 °C for 35 s, 62 °C → 53 °C for 30 s, 72 °C for 30 s), and 20 cycles of 94 °C for 45 s, 42 °C for 45 s, 72 °C for 45 s, and a final extension at 72 °C for 10 min. Ten individuals were detected (n = 10) for each generation, with three biological replicates (N = 3).
Titre analysis by qPCR
The relative titre and copy number of wMel in B. tabaci at different developmental stages (egg, nymph and adult) were measured by using real-time quantitative PCR over 12 generations after transinfection. The qPCR assay was conducted based on the single-copy gene wsp encoding the surface protein of Wolbachia. Each DNA sample was extracted from 20 individuals, and the target gene (wsp) was amplified, purified, sequenced and then ligated into pGEM-T vector as previously described11. The relative titre was measured with β-actin as the internal control. The qPCR primers wspQ384/wspQ513 were designed to specifically detect wMel strain (Table 3). The qPCR reactions were performed by using 10 μl of the Platinum SYBR Green qPCR Supermix-UDG (Invitrogen), 0.4 μl of each primer (10 μM), 1.0 μl gDNA and nuclease-free water in a final volume of 20 μl. For absolute qPCR, a standard curve was drawn through five consecutive dilutions with the fusion plasmids (plasmid + insert). The number of wsp gene copies (N) per microliter was determined by using the protocol as described51. To ensure the validity of the data, each measurement was performed in three independent biological replicates and the wild-type whitefly was used as the control. The cycling conditions were: 2 min activation at 95 °C, 40 cycles of 15 s at 95 °C, 30 s at 60 °C. A melting curve was generated under the thermal conditions from 60 °C to 99 °C with a 1-°C rise at each step and a waiting period of 5 s between steps (ABI 7500).
Data analysis
Statistical differences between groups were analysed by using Student Newman Keuls (SNK) multiple range test of One-way ANOVA at α = 0.01 and 0.05 levels. Error bars in all graphs represent standard error. Chi-square test was conducted to analyse the difference in the number of male offspring between the expected 1:1 sex ratio and the crossing results from transinfected and wildtype whiteflies. Linear regression analysis was run on SPSS 20.0 to quantify the relationships between the gene copy number, titre and transmission rate. Transmission rates (%) are transformed by arcsine square root before analysis.
Additional Information
How to cite this article: Zhou, X.-F. and Li, Z.-X. Establishment of the cytoplasmic incompatibility-inducing Wolbachia strain wMel in an important agricultural pest insect. Sci. Rep. 6, 39200; doi: 10.1038/srep39200 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work is supported by the National Science Foundation of China (Grant nos. 31071748, 31371940 and 31171845) and the Specialized Research Fund for the Doctoral Program of Higher Education of China (Grant no. 20130008110011).
Footnotes
Author Contributions X.-F.Z. conducted the experiments and wrote a draft report; X.-F.Z. and Z.-X.L. analysed the data; Z.-X.L. conceived the study, and wrote the manuscript.
References
- O’Neill S. L., Hoffmann A. A. & Werren J. H. Influential passengers: Inherited microorganisms and invertebrate reproduction. Oxford: Oxford University Press (1997). [Google Scholar]
- Hilgenboecker K., Hammerstein P., Schlattmann P., Telschow A. & Werren J. H. How many species are infected with Wolbachia?-a statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug R. & Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 7, e38544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer R., Breeuwer J. J. & Hurst G. D. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Ann. Rev. Microbiol. 53, 71–102 (1999). [DOI] [PubMed] [Google Scholar]
- Werren J. H., Baldo L. & Clark M. E. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751 (2008). [DOI] [PubMed] [Google Scholar]
- Werren J. H., Zhang W. & Guo L. R. Evolution and phylogeny of Wolbachia: Reproductive parasites of arthropods. Proc. R. Soc. Lond. B-Biol. Sci. 261, 55–63 (1995). [DOI] [PubMed] [Google Scholar]
- Bourtzis K. Wolbachia-based technologies for insect pest population control. Adv Exp. Med. Biol. 627, 104–113 (2008). [DOI] [PubMed] [Google Scholar]
- Christodoulou M. Biological vector control of mosquito-borne diseases. Lancet Infect. Dis. 11, 84–85 (2011). [DOI] [PubMed] [Google Scholar]
- Hancock P. A., Sinkins S. P. & Godfray H. C. Strategies for introducing Wolbachia to reduce transmission of mosquito-borne diseases. PLoS Negl. Trop. Dis. 5, e1024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhaar H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 36, 533–543 (2011). [Google Scholar]
- Zhong Y. & Li Z. X. Bidirectional cytoplasmic incompatibility induced by cross-order transinfection of Wolbachia: implications for control of the host population. Microb. Ecol. 68, 463–471 (2014). [DOI] [PubMed] [Google Scholar]
- Laven H. Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature 216, 383–384 (1967). [DOI] [PubMed] [Google Scholar]
- Heath B. D. et al. Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr. Biol. 9, 313–316 (1999). [DOI] [PubMed] [Google Scholar]
- Braig H. R. et al. Replacement of the natural Wolbachia symbiont of Drosophila simulans with a mosquito counterpart. Nature 367, 453–455 (1994). [DOI] [PubMed] [Google Scholar]
- Sasaki T. & Ishikawa H. Transinfection of Wolbachia in the mediterranean flour moth, Ephestia kuehniella, by embryonic microinjection. Heredity (Edinb) 85, 130–135 (2000). [DOI] [PubMed] [Google Scholar]
- Xi Z. et al. Generation of a novel Wolbachia infection in Aedes albopictus (Asian tiger mosquito) via embryonic microinjection. Insect Biochem. Mol. Biol. 35, 903–910 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier S. et al. Successful horizontal transfer of Wolbachia symbionts between Trichogramma wasps. Proc. R. Soc. B 265, 1441–1445 (1998). [Google Scholar]
- Hartmann N. et al. Trans-species transfer of Wolbachia: microinjection of Wolbachia from Litomosoides sigmodontis into Acanthocheilonema viteae. Parasitology 126, 503–511 (2003). [PubMed] [Google Scholar]
- Boyle L. et al. Inter- and intra-specific horizontal transfer of Wolbachia in Drosophila. Science 260, 1796–1799 (1993). [DOI] [PubMed] [Google Scholar]
- Hu H. Y. & Li Z. X. A novel Wolbachia strain from the rice moth Corcyra cephalonica induces reproductive incompatibility in the whitefly Bemisia tabaci: sequence typing combined with phenotypic evidence. Environ. Microbiol. Rep. l7, 508–515 (2015). [DOI] [PubMed] [Google Scholar]
- Hughes G. L. & Rasgon J. L. Transinfection: a method to investigate Wolbachia-host interactions and control arthropod-borne disease. Insect Mol. Biol. 23, 141–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S. et al. Transinfection of Wolbachia in planthoppers: nymphal injection of cultured Wolbachia and infection dynamics. Environ. Entomol. 38, 1626–1633 (2009). [DOI] [PubMed] [Google Scholar]
- Riegler M., Charlat S., Stauffer C. & Merçot H. Wolbachia transfer from Rhagoletis cerasi to Drosophila simulans: investigating the outcomes of host-symbiont coevolution. Appl. Environ. Microbiol. 70, 273–279 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z. et al. Interspecific transfer of Wolbachia into the mosquito disease vector Aedes albopictus. Proc. R. Soc. B 273, 1317–1322 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson S. L., Fox C. W. & Jiggins F. M. The effect of Wolbachia-induced cytoplasmic incompatibility on host population size in natural and manipulated systems. Proc. Biol. Sci. 269, 437–445 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor L. et al. Open release of male mosquitoes infected with a Wolbachia biopesticide: field performance and infection containment. PLoS Negl. Trop. Dis. 6, e1797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalou S. et al. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc. Natl. Acad. Sci. USA 101, 15042–15045 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagrove M. S. et al. Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus. Proc. Natl. Acad. Sci. USA 109, 255–60 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagrove M. S. et al. A Wolbachia wMel transfection in Aedes albopictus is not detrimental to host fitness and inhibits Chikungunya virus. PLoS Negl. Trop. Dis. 7, e2152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A. et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476, 454–457 (2011). [DOI] [PubMed] [Google Scholar]
- Walker T. et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476, 450–453 (2011). [DOI] [PubMed] [Google Scholar]
- Riegler M., Sidhu M., Miller W. J. & O’Neill S. L. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr. Biol. 15, 1428–1433 (2005). [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A. et al. Stability of the wMel Wolbachia infection following invasion into Aedes aegypti populations. PLoS Negl. Trop. Dis. 8, e3115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert D. A. et al. Establishment of a Wolbachia superinfection in Aedes aegypti mosquitoes as a potential approach for future resistance management. PLoS Pathog. 12, e1005434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., Khoo C. C. & Dobson S. L. Wolbachia establishment and invasion in an Aedesaegypti laboratory population. Science 310, 326–328 (2005). [DOI] [PubMed] [Google Scholar]
- Perring T. M. The Bemisia tabaci species complex. Crop Prot. 20, 725–737 (2001). [Google Scholar]
- Sun D. B. et al. Competitive displacement between two invasive whiteflies: insecticide application and host plant effects. Bull. Entomol. Res. 103, 344–53 (2013). [DOI] [PubMed] [Google Scholar]
- Xie W. et al. Sensitivity of Bemisia tabaci (Hemiptera: Aleyrodidae) to several new insecticides in China: effects of insecticide type and whitefly species, strain, and stage. J. Insect Sci. 14, 1–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. X., Lin H. Z. & Guo X. P. Prevalence of Wolbachia infection in Bemisia tabaci. Curr. Microbiol. 54, 467–471 (2007). [DOI] [PubMed] [Google Scholar]
- Zhong Y. & Li Z. X. Influences of tetracycline on the reproduction of the B biotype of Bemisia tabaci. Appl. Entomol. Zool. 48, 241–246 (2013). [Google Scholar]
- Baldo L. et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72, 7098–7110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2, E69 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb Y. et al. Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J. 22, 2591–2599 (2008). [DOI] [PubMed] [Google Scholar]
- Sasaki T., Massaki N. & Kubo T. Wolbachia variant that induces two distinct reproductive phenotypes in different hosts. Heredity 95, 389–393 (2005). [DOI] [PubMed] [Google Scholar]
- Clancy D. J. & Hoffmann A. A. Behaviour of Wolbachia endosymbionts from Drosophila simulans in Drosophila serrata, a novel host. Am. Nat. 149, 975–988 (1997). [DOI] [PubMed] [Google Scholar]
- Rigaud T., Pennings P. S. & Juchault P. Wolbachia bacteria effects after experimental interspecific transfers in terrestrial isopods. J. Invertebr. Pathol. 77, 251–257 (2001). [DOI] [PubMed] [Google Scholar]
- McMeniman C. J. et al. Host adaptation of a Wolbachia strain after long-term serial passage in mosquito cell lines. Appl. Environ. Microbiol. 74, 6963–6969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N., Shimada M. & Fukatsu T. Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol. Lett. 1, 488–491 (2005). [DOI] [PMC free article] [PubMed]
- McGraw E. A. et al. Wolbachia density and virulence attenuation after transfer into a novel host. Proc. Natl. Acad. Sci. USA 99, 2918–2923 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda H. et al. Infection density of Wolbachia and incompatibility level in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem. Mol. Biol. 31, 727–737 (2001). [DOI] [PubMed] [Google Scholar]
- Whelan J. A., Russell N. B. & Whelan M. A. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods 27, 261–269 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.