Abstract
Pyridinols and pyridinamines are important intermediates with many applications in chemical industry. The pyridine derivatives are in great demand as synthons for pharmaceutical products. Moreover, pyridines are used either as biologically active substances or as building blocks for polymers with unique physical properties. Application of enzymes or whole cells is an attractive strategy for preparation of hydroxylated pyridines since the methods for chemical synthesis of pyridinols, particularly aminopyridinols, are usually limited or inefficient. Burkholderia sp. MAK1 (DSM102049), capable of using pyridin-2-ol as the sole carbon and energy source, was isolated from soil. Whole cells of Burkholderia sp. MAK1 were confirmed to possess a good ability to convert different pyridin-2-amines and pyridin-2-ones into their 5-hydroxy derivatives. Moreover, several methylpyridines as well as methylated pyrazines were converted to appropriate N-oxides. In conclusion, regioselective oxyfunctionalization of pyridine derivatives using whole cells of Burkholderia sp. MAK1 is a promising method for the preparation of various pyridin-5-ols and pyridin-N-oxides.
The pyridine ring is found in various man-made compounds, such as dyes, industrial solvents, herbicides, pesticides as well as in many natural metabolites. Among the N-heterocyclic rings, pyridin-2-ol has considerable chemical and pharmacological importance. Due to a peptidomimetic functionality of 1H-pyridin-2-one tautomer, it plays an essential role as a scaffold in drug design1,2. Pharmacophores containing 1H-pyridin-2-ones are found in various therapeutic agents, including reverse transcriptase inhibitors3, antibiotics and antifungals4,5,6, anti-allergic drugs7 and analgesics8. Moreover, 1H-pyridin-2-ones are promising compounds for the preparation of modified nucleotides and oligonucleotides9,10,11.
A novel class of phenolic antioxidants, 6-aminopyridin-3-ols, are more effective than many other phenolic-class compounds reported to date12. Pyridin-2-amines serve as a starting material for production of fused heterocycles, including imidazo-derivatives that possess significant biological activities similar to those of antiviral and immunosuppressive agents13,14,15. Moreover, 3-amino-imidazo[1,2-a]pyridines were identified as a novel class of Mycobacterium tuberculosis glutamine synthetase inhibitors16, and 6-aminopyridin-3-ol was applied for the synthesis of new antibiotics17.
Preparation of some 5-hydroxy-2-pyridones is achievable by both organo-chemical and biocatalytic approaches18, whereas no satisfactory synthetic methods leading to 6-aminopyridin-3-ols have been described thus far. In addition, only a few synthetic methods to aminopyridinol structures may be found in literature to date12,19,20. Recently, a chemical synthesis of pyridine-3,5-diol derivatives from renewable carbohydrates has been demonstrated21.
Oxyfunctionalization of chemical compounds by using enzymes or whole cells is an attractive strategy to obtain the desired products22,23,24. The degradation of N-heterocyclic compounds, especially with regard to the production of metabolic intermediates, has received considerable attention in biotechnology as the starting process for the synthesis of fine and commodity chemicals, e.g., pyridine-2,5-diol (an intermediate for production of 5-aminolevulinic acid), 6-hydroxynicotinic acid and other pyridine derivatives25,26,27,28. Recently, the hydroxylation of the pyridine ring has been achieved using tetramethylpyrazine-degrading bacteria27. It has been also shown that pyridine N-oxides can be prepared using microbial cells and enzymes; however, this has been accomplished using Methylococcus capsulatus monooxygenase29, Agrocybe aegerita peroxygenase30, and Verticillium sp. GF39 cells31 only.
Several bacteria belonging to the genera Arthrobacter, Achromobacter, Rhodococcus, and Nocardia are able to grow on pyridin-2-ol32,33,34,35. The first common steps in the microbial metabolism of pyridin-2-ol involve the hydroxylation of the ring yielding di- or trihydroxypyridine intermediates32,35,36 that are promising synthons for the preparation of substituted pyridines.
In this study, the oxyfunctionalization of the pyridine ring by whole bacterial cells was investigated. The pyridin-2-ol-degrading Burkholderia sp. MAK1 was found to be an efficient biocatalyst for the hydroxylation of various pyridin-2-ols and pyridin-2-amines. Moreover, Burkholderia sp. MAK1 was capable of oxidising several N-heterocyclic ring systems to corresponding N-oxides.
Results and Discussion
Isolation of pyridin-2-ol-degrading bacteria
The gram-negative bacterial isolate MAK1, capable of using pyridin-2-ol as a sole carbon and energy source, was isolated from soil. The 16S rRNA gene sequence of MAK1 showed similarity to that of bacteria belonging to Burkholderia sordidicola. Based on the results of 16S rRNA gene sequence analysis (Supplementary information Fig. S-I) and biochemical characterization (Supplementary information Table S-1) the strain MAK1 was identified as Burkholderia sp. MAK1.
In bacteria, pyridin-2-ol may be catabolized by two different pathways. The first pathway proceeds via formation of pyridine-2,3,6-triol, which spontaneously oxidises and dimerises to a blue pigment, 4,5,4′,5′-tetrahydroxy-3,3′-diazadiphenoquinone-(2,2′)32,35,37. The other known catabolic pathway proceeds via formation of pyridine-2,5-diol, maleamic acid, maleic acid, and fumaric acid33.
In the case of Burkholderia sp. MAK1 described here, pyridin-2-ol was catabolized without the formation of a blue pigment. Assuming that pyridine-2,5-diol is an intermediate in pyridin-2-ol catabolic pathway, the activity of pyridine-2,5-diol 5,6-dioxygenase detected in the pyridin-2-ol-induced cells of Burkholderia sp. MAK1 suggested that this strain possesses an inducible pyridin-2-ol 5-monooxygenase.
Selection of pyridine derivatives as substrates for hydroxylation with Burkholderia sp
As we found out that Burkholderia sp. MAK1 consumes pyridine-2-ol via pyridine-2,5-diol by supposedly pyridine-2-ol inducible pyridin-2-ol 5-monooxygenase we wanted to test whether Burkholderia sp. MAK1 is capable of hydroxylating other pyridine derivatives. In this study, more than 100 of pyridine, pyrimidine, and pyrazine derivatives were screened for the hydroxylation using Burkholderia sp. MAK1 as a whole-cell biocatalyst (Supplementary information Table S-2). The pyridin-2-ol-induced Burkholderia sp. MAK1 cells were incubated with a potential substrate as described in the Methods section. The progress of the reaction was followed by HPLC-MS. The efficiency of conversion of several compounds by whole cells of Burkholderia sp. MAK1 is presented as Supplementary information Table S-3.
It is worth mentioning that induction of Burkholderia sp. MAK1 hydroxylation activity was observed only in the presence of pyridin-2-ol. Several other tested compounds (pyridine, pyridine-2,5-diol, pyridin-2-amine) were not able to trigger the induction. Also no hydroxylation occurred when cells were cultivated with other sole carbon source (glucose or succinate) instead of pyridin-2-ol.
Optimization of cultivation and reaction conditions
Burkholderia sp. MAK1 grew poorly in rich nutrient medium, but the growth was observed in mineral medium (EFA or Koser) with pyridin-2-ol as a sole carbon source. The growth reached its peak after 40 h of incubation in EFA medium (OD600 = 0.4). The optimal temperature for cultivation of Burkholderia sp. MAK1 appeared to be 30 °C. At higher tested temperature (37 °C), Burkholderia sp. MAK1 cells were not able to grow. Although bacterial growth was observed at 25 °C it was rather slow compared to 30 °C. The effect of temperature on Burkholderia sp. MAK1-mediated synthesis of hydroxylated pyridine derivatives was also investigated (Fig. 1). For this experiment 4-chloropyridin-2-amine was selected due to its great conversion percentage and definite product (Table 1). During the first hour of the experiment, the bioconversion of 4-chloropyridin-2-amine was most rapid at 30 °C and 35 °C with 6-amino-4-chloro-pyridin-3-ol production rate of 7 mg (g biomass)-1 h-1 and 7.4 mg (g biomass)-1 h-1, respectively. Higher temperatures (40–45 °C) were found to be unfavorable for the synthesis, probably because of the inactivation of the biocatalyst. The conversion reached near completion (~97%) after six hours at 30 °C.
Figure 1. The dependence of the rate of 6-amino-4-chloro-pyridin-3-ol biosynthesis on temperature.
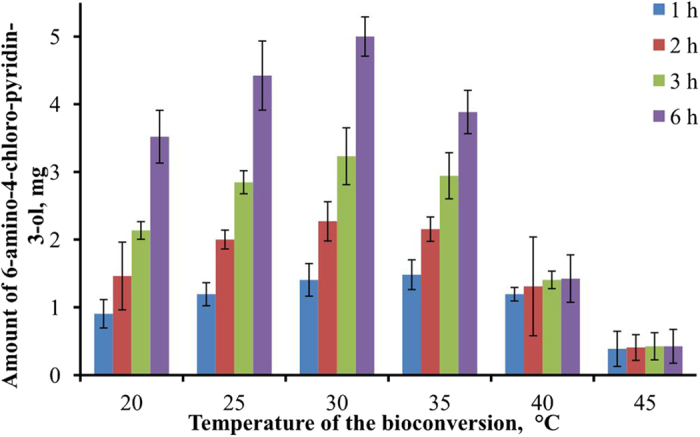
2-hydroxypyridine-induced Burkholderia sp. MAK1 cells were used as biocatalyst. The values represent the average of three independent experiments ± standard deviation.
Table 1. The bioconversion features of substituted pyridine-2-amines for which reactions products were detectable.
| Substrate |
Product |
Conversion % | |||
|---|---|---|---|---|---|
| Name | [M + H]+ | [M + H]+ | NMR | Name/Possible outcome | |
| Pyridin-2-amine | 95 | 111 | − | Hydroxylated at the 5-position | 99 |
| 3-Fluoropyridin-2-amine | 113 | 129 | − | Hydroxylated at the 5-position | 43 |
| 3-Bromopyridin-2-amine | 173 and 175 | 189 and 191 | − | Hydroxylated at the 5-position | 48 |
| 3-Chloropyridin-2-amine | 129 | 145 | + | 6-amino-5-chloro-pyridin-3-ol | 88 |
| Pyridin-2,6-diamine | 110 | 214 | − | Hydroxylation followed by dimerization | 88 |
| 4-Bromopyridin-2-amine | 173 and 175 | 189 and 191 | − | Hydroxylated at the 5-position | 48 |
| 4-Fluoropyridin-2-amine | 113 | 129 | + | 6-amino-4-fluoro-pyridin-3-ol | 70 |
| 4-Chloropyridin-2-amine | 129 | 145 | + | 6-amino-4-chloro-pyridin-3-ol | 96 |
| 4-methyl-pyridin-2-amine | 109 | 125 | + | 6-Amino-4-methyl-pyridin-3-ol | 74 |
| 6-Bromopyridin-2-amine | 173 and 175 | 189 and 191 | − | Hydroxylated at the 5-position | 53 |
| 6-Chloropyridin-2-amine | 129 | 145 | + | 6-amino-2-chloropyridin-3-ol | 89 |
For the NMR analysis approximately 10 mg of purified bioconversion product was used.
Biotransformation of various pyridin-2-ols by Burkholderia sp. MAK1 cells
The study of N-alkylpyridine transformation revealed that 1-methyl-, 1-ethyl- and 1-propylpyridin-2-ol were transformed to the final dihydroxy products by Burkholderia sp. MAK1 cells. In the chromatogram of 1-ethylpyridin-2-ol bioconversion, two dominant peaks A and B were detected (Supplementary information Fig. S-II) corresponding to the newly formed compound and the residual substrate, respectively. The absorption maximum of the product, compared with that of the substrate, shifted to longer wavelengths (~30 nm), which is characteristic of compounds with additional hydroxy group. Also, the mass of the molecular ion of the product was 16 Da higher than that of the parent compound, supporting the hydroxylation of 1-ethylpyridin-2-ol. Similar results were obtained with 1-methyl- and 1-propylpyridin-2-ol. In all cases, the formation of a single product was observed indicating the position-specific hydroxylation. Moreover, the apparent equivalence with pyridin-2-ol transformation suggested that 1-alkylpyridin-2-ols were hydroxylated at the 5-position. Of all the compounds tested, only 1-butylpyridin-2-ol remained unchanged, which is most likely due to its bulkiness. In summary, pyridin-2-ols containing small 1-alkyl substituent are hydroxylated regioselectively, but further pyridine ring opening reaction does not occur. Thus, Burkholderia sp. MAK1 is capable of producing 1-alkylpyridine-2,5-diols.
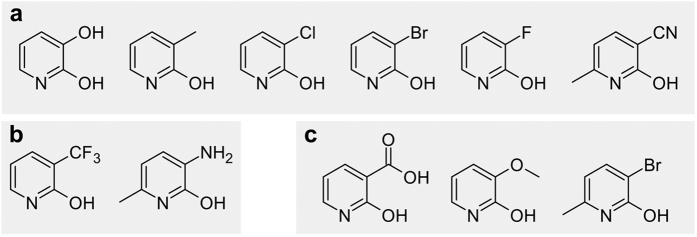
Another group of potential Burkholderia sp. MAK1 substrates comprised pyridin-2-ols substituted at position 3 (Fig. 2). HPLC-MS analysis revealed that compounds containing hydroxyl, methyl, bromo, chloro, or fluoro functional groups were completely catabolized by Burkholderia sp. MAK1 cells since no significant peaks corresponding to any hydroxylated products were detected. The latter suggests that the hydroxylated metabolites were likely further metabolized to aliphatic products. However, 3-(trifluoromethyl)pyridin-2-ol was slowly converted into a detectable new compound whose molecular mass was 16 Da higher than that of the substrate. Burkholderia sp. MAK1 cells were not able to hydroxylate pyridin-2-ols containing carboxyl or methoxy groups at position 3.
Figure 2. The 2-hydroxypyridines substituted at the third position, which were used as potential Burkholderia sp. MAK1 substrates.
(a) Substrate consumption occurs, no products detected, (b) substrate consumed, product detected, (c) no reaction observed. The 2-hydroxypyridine induced cells were suspended in 10 mM potassium phosphate buffer, pH 7.2, supplemented with 15 mM glucose and 0.25 mM of corresponding substrate. Reactions were carried out at 30 °C. The progress of each reaction was observed by HPLC-MS.
Pyridin-2-ols carrying substituents at positions 3 and 6 were also examined. The pyridin-2-ol-induced cells were able to metabolize 2-hydroxy-6-methyl-pyridine-3-carbonitrile: substrate concentration decreased over time, and no new products were detectable by HPLC-MS. After incubation of Burkholderia sp. MAK1 with 3-amino-6-methyl-pyridin-2-ol, a new compound with a molecular mass of 278 Da accumulated in the reaction mixture. Since the molecular mass of the expected 3-amino-6-methyl-pyridin-2-ol hydroxylation product is 140 Da, it is likely that the oxidation of the substrate is followed by the spontaneous dimerization. When Burkholderia sp. MAK1 cells were incubated with 3-bromo-6-methyl-pyridin-2-ol, neither hydroxylation, nor any other transformation occurred suggesting that 3-bromo functional group disrupted the proper orientation of the substrate.
Pyridin-2-ols substituted at positions 4 and/or 6 were also used as substrates in this study (Fig. 3). Pyridine-2,4-diol was completely oxidized by Burkholderia sp. MAK1 cells after 20 hours of incubation. However, the intermediate product accumulating in the reaction mixture was detected by HPLC-MS and its absorption spectra as well as molecular mass ([M + H]+ = 128.05, [M + H2O + H]+ = 146.10, [2M+ H]+ = 255.05) were consistent with those of hydroxylated pyridine-2,4-diol (Supplementary information Fig. S-III). Using 4-cyano, 4-chloro, 4-bromo, or 4-trifluomethyl substituted pyridin-2-ols, hydroxylation of the pyridine ring did not occur suggesting that the nature of a substituent at position 4 is important for the hydroxylation process.
Figure 3. 2-Hydroxypyridine derivatives substituted at the fourth, the sixth or both the fourth and the sixth positions, which were used as potential substrates for regioselective oxidation by Burkholderia sp. MAK1.
(a) Substrate consumption occurs; (b) no reaction observed. The 2-hydroxypyridine induced cells were suspended in 10 mM potassium phosphate buffer, pH 7.2, supplemented with 15 mM glucose and 0.25 mM of corresponding substrate. Reactions were carried out at 30 °C. The progress of each reaction was observed by HPLC-MS.
Pyridine-2,6-diol was transformed by Burkholderia sp. MAK1 to a blue pigment. Previously, Holmes with colleagues described dimerization of pyridine-2,3,6-triol, which led to the formation of a blue pigment38. Following this observation, the hydroxylation of the symmetric pyridine-2,6-diol by Burkholderia sp. MAK1 cells likely occurred at position 3 of the pyridine ring and the resulting pyridine-2,3,6-triol spontaneously dimerized to a blue compound. Moreover, if the sixth position of pyridin-2-ol was occupied by a small and uncharged functional group, the pyridine ring cleavage probably followed the hydroxylation event.
Summarizing experiments with substituted pyridin-2-ols we can make the statement that most of the substrates were consumed without detectable products. Although we were unable to provide any data about structures of the detectible product there were strong evidences suggesting regioselective hydroxylation at 5-position (Table 2).
Table 2. The bioconversion features of substituted pyridine-2-ols for which reactions products were detectable.
| Substrate |
Product |
Conversion % | ||
|---|---|---|---|---|
| Name | [M+H]+ | [M+H]+ | Possible outcome | |
| 1-Methylpyridin-2-one | 110 | 126 | Hydroxylated at the 5-position | 20 |
| 1-Ethylpyridin-2(1H)-one | 124 | 140 | Hydroxylated at the 5-position | 17 |
| 1-Propylpyridin-2(1H)-one | 138 | 154 | Hydroxylated at the 5-position | Traces of product |
| 3-(Trifluoromethyl) pyridin-2-ol | 164 | 178 [M+H]− | Hydroxylated at the 5-position | 46 |
| 3-amino-6-methyl-pyridin-2-ol | 125 | 279 | Hydroxylation followed by dimerization | 88 |
| Pyridine-2,6-diol | 112 | — | Hydroxylation followed by dimerization (blue pigment) | — |
Screening of pyridin-2-amines as potential substrates for regioselective hydroxylation by Burkholderia sp. MAK1 cells
The ability of Burkholderia sp. MAK1 to transform various pyridin-2-ols encouraged us to study pyridin-2-amines as another group of potential substrates. During the initial experiments, the cells were incubated with pyridin-2-amine for 20 hours. HPLC-MS analysis revealed that pyridin-2-amine was completely consumed, and the new peak in the chromatogram belonged to the expected product. The molecular mass of the product, which was 16 Da higher than that of pyridin-2-amine, confirmed the notion that hydroxylation of the substrate occurred. The UV-Vis spectrum of the product was compared with spectra of commercially available reference standards (pyridin-2-amine hydroxylated at position 3, 4, or 6), yet none of these spectra matched that of the product (Supplementary information Fig. S-IV). From this we presume that in the case of Burkholderia sp. MAK1, pyridin-2-amine undergoes hydroxylation at position 5.
Next, pyrazin-2-amine, a homolog of pyridin-2-amine containing two nitrogen atoms in the aromatic ring, was chosen as a substrate for the bioconversion. HPLC-MS analysis showed that the molecular mass of the biotransformation product was 16 Da higher than that of pyrazin-2-amine, suggesting that Burkholderia sp. MAK1 cells are also capable of pyrazin-2-amine hydroxylation.
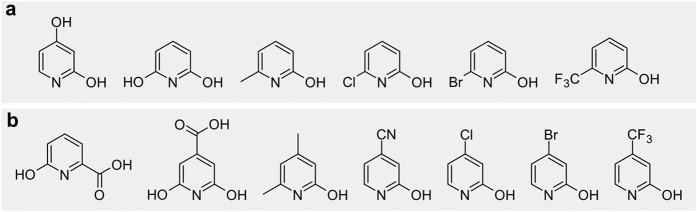
Pyridin-2-amines with methyl, nitro, chloro, bromo, or fluoro substituent at position 3 (Fig. 4a) were all transformed by Burkholderia sp. MAK1. Moreover, the pyridin-2-ol-induced cells were also capable of hydroxylating ethyl-2-aminopyridine-3-carboxylate, a compound with a bulky functional group at the 3-position. The conversion product of 3-chloropyridin-2-amine was purified as described in the Materials and Methods section, and its structure was analysed by 1H NMR, 13C NMR, and HPLC-MS analyses. The molecular mass of the product (145 Da) corresponded to that of 6-amino-5-chloro-pyridin-3-ol. The compound showed four peaks in the 1H NMR spectrum (DMSO-d6, ppm): δ = 5.51 (s, 2 H, NH2), 7.11 (d, J = 2.6 Hz, 1H, CH), 7.56 (d, J = 2.6 Hz, 1H, CH), 9.24 (brs, 1H, OH), and five peaks in the 13C NMR spectrum (DMSO-d6, ppm): δ = 113.58, 125.21, 133.73, 146.21, 149.46), identifying the product as 6-amino-5-chloro-pyridin-3-ol. The production yield of 6-amino-5-chloro-pyridin-3-ol was 34%.
Figure 4. The 2-aminopyridines, which were tested as potential substrates for conversion by 2-hydroxypyridine-induced whole cells of Burkholderia sp. MAK1.
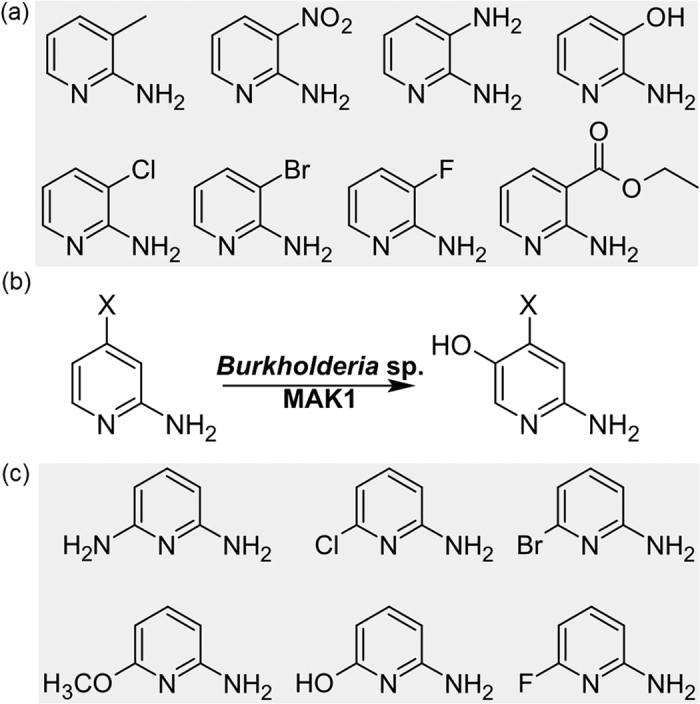
(a) The 2-aminopyridine derivatives substituted at the third position; (b) the regioselective hydroxylation of 2-aminopyridines substituted at the fourth position (X = CH3, Cl, F); (c) the 2-aminopyridines substituted at the sixth position.
Both pyridine-2,3-diamine and 2-aminopyridin-3-ol were transformed into colored compounds, with a molecular mass of 213 Da (yellow-brown) and 214 Da (yellow-green), respectively. The retention time, UV-Vis spectra, and ionisation profile of the oxidation product of 2-aminopyridin-3-ol matched those of the analytical standard (2-amino-3H-dipyrido[3,2-b:2′,3′-e][1,4]oxazine-3-one) suggesting that Burkholderia sp. MAK1 catalyzes the oxidative dimerization of 2-aminopyridin-3-ol. Also, although another analytical standard, pyridine-2,3-diamine derivative, is commercially unavailable, our results indicate, that MAK1 catalyzes dimerization of pyridine-2,3-diamine as well. These dimers are potential anticancer and antimicrobial drugs39.
Next, the ability of Burkholderia sp. MAK1 cells to transform pyridin-2-amines substituted at position 4 was investigated. Compounds with methyl, chloro, bromo, or fluoro substituents were hydroxylated. In all cases, the molecular mass of reaction products, as estimated by HPLC-MS, was 16 Da higher than that of parent compounds indicating that oxidation of substrates had occurred.
In the case of 4-methyl-pyridin-2-amine, 4-chloro-pyridin-2-amine, and 4-fluoro-pyridin-2-amine, the biotransformation catalyzed by the pyridin-2-ol-induced Burkholderia sp. MAK1 cells resulted in the formation of a single product. The products of all three reactions were purified by a reverse phase chromatography (C18 cartridges, water/methanol mixture, 10:0 → 10:5), and their structures were analysed by 1H NMR and 13C NMR. 6-Amino-4-methyl-pyridin-3-ol (1H NMR (DMSO-d6, ppm): δ = 2.18 (s, 3H, CH3), 6.41 (dd, J = 6.6, 2.3 Hz, 1H, CH), 6.61 (d, J = 2.3 Hz, 1H, CH), 6.70 (s, 2H, NH2), 7.87 (d, J = 6.6 Hz, 1H, CH), 13C NMR (DMSO-d6, ppm): δ = 20.52, 109.39, 113.72, 136.73, 138.00, 150.61), 6-amino-4-chloro-pyridin-3-ol (1H NMR (DMSO-d6, ppm): δ = 6.66 (dd, J = 7.0, 2.9 Hz, 1H, CH), 6.84–6.83 (m, 1H, CH), 7.0 (s, 2H, NH2), 8.04 (d, J = 7.0 Hz, 1H, CH), 13C NMR (DMSO-d6, ppm): δ = 108.17, 112.44, 131.29, 138.34, 151.70) and 6-amino-4-fluoro-pyridin-3-ol (1H NMR (DMSO-d6, ppm): δ = 6.54 (td, J = 7.3, 3.4 Hz, 1H, CH), 6.62 (dd, J = 9.1, 3.2 Hz, 1H, CH), 7.11 (s, 2H, NH2), 8.07 (dd, J = 7.2, 6.0 Hz, 1H, CH), 13C NMR (DMSO-d6, ppm): δ = 95.45, 100.74, 139.03, 158.91, 161.39) were formed by whole cells of Burkholderia sp. MAK1 with the yield of 34%, 50% and 68%, respectively (Fig. 4b). In addition, 4-chloropyrimidin-2-amine, pyrimidine-2,4-diamine, and 2-aminopyrimidin-4-ol were also hydroxylated by the pyridin-2-ol-induced Burkholderia sp.MAK1 cells. According to the 1H NMR and 13C NMR analyses, the purified product of 2-aminopyrimidin-4-ol conversion was 2-aminopyrimidine-4,5-diol, and the conversion yield was 18%. To our knowledge, biocatalytical production of 6-amino-4-methyl-pyridin-3-ol has never been described previously. Moreover, there is no available information concerning the synthesis of 6-amino-4-chloro-pyridin-3-ol or 6-amino-4-fluoro-pyridin-3-ol. By analogy to aminophenols, the new compounds described in this study have great potential as materials for the production of dyes, drugs, pesticides, and etc.40.
The compounds substituted at position 6 (Fig. 4c) were also transformed by Burkholderia sp. MAK1. HPLC-MS analysis showed that pyridine-2,6-diamine was consumed; however, no new compounds were detected. Nevertheless, in the case of pyridine-2,6-diamine, the reaction mixture turned brown suggesting that after oxidation, further transformations (e. g. polymerisation) occurred. The compounds with 6-chloro or 6-bromo substituents were converted to the corresponding hydroxylated products. The product of oxidation of 6-chloropyridin-2-amine, 6-amino-2-chloropyridin-3-ol, was purified and identified by 1H NMR (DMSO-d6, ppm): δ = 5.90 (s, 2H, NH2), 6.38 (d, J = 7.8 Hz, 1H, CH), 6.84 (d, J = 7.8 Hz, 1H, CH), 9.79 (brs 1H, OH). While 6-fluoropyridin-2-amine conversion was very slow, the transformation of 6-methoxypyridin-2-amine did not occur at all. The conversion of 6-aminopyridin-2-ol led to several compounds suggesting that the substrate is hydroxylated at position 3 and/or 5, so that a mixture of several products in varying proportions results.
Unlike the aforementioned pyridin-2-ols, the products of hydroxylation of 6-substituted pyridin-2-amines were not metabolised further suggesting that Burkholderia sp. MAK1 may be applied for the regioselective synthesis of 6-substituted 2-aminopyridinols (Table 1).
Oxyfunctionalization of pyridine, pyrazine and their derivatives using whole-cell biocatalyst
The study on pyridin-2-amine and pyridin-2-ol bioconversion by Burkholderia sp. MAK1 cells showed that the pyridin-2-ol-inducible pyridin-2-ol 5-monooxygenase has broad substrate specificity and strict regiospecificity since it catalyzes hydroxylation at position 5 on the aromatic ring. With very few exceptions, microbial hydroxylation of pyridine-2-amines has been scarcely studied. One such exception is the study on the biotransformation of 4-methyl-3-nitro-pyridin-2-amine using whole-cells of fungus Cunninghamella elegans ATCC 26269. During this biotransformation, a mixture of three products, 6-amino-4-methyl-5-nitropyridin-3-ol, 2-amino-4-hydroxymethyl-3-nitropyridine, and 2-amino-4-methyl-3-nitropyridine-1-oxide was obtained suggesting that both aromatic and aliphatic positions as well as the heterocyclic nitrogen atom undergo oxidation41. In the case of Burkholderia sp. MAK1 cells, oxidation of the heterocyclic nitrogen atom was not observed when pyridin-2-ols were used as substrates. To determine if these bacteria were capable of producing N-oxides, various pyridine and pyrazine compounds without amino or hydroxy group at position 2 were tested as substrates for pyridin-2-ol-induced Burkholderia sp. MAK1 cells. HPLC-MS analysis showed that pyridine was transformed into a single product whose molecular mass was 16 Da higher than that of the parent compound. The UV spectrum of the product was very similar to that of pyridine yet did not match with the spectra of 2-, 3-, or 4-hydroxy-substituted pyridines at position suggesting that the product of pyridine biotransformation is pyridine-1-oxide (pyridine-N-oxide). The retention time, UV spectrum and ionisation profile of the bioconversion product matched those of analytical standard, pyridine-N-oxide, suggesting that Burkholderia sp. MAK1 catalyzes pyridine oxidation at position 1. Induction of cells with pyridin-2-ol was necessary for the oxidation of pyridine as well as for pyridin-2-ol and pyridin-2-amine transformation indicating that the same enzyme of Burkholderia sp. MAK1 is responsible for all these biotransformations.
A group of pyridines and pyrazines containing a methyl group attached to the aromatic ring at different positions (Fig. 5) was studied as potential substrates for Burkholderia sp. MAK1. The test revealed that the whole cells of Burkholderia sp. MAK1 catalyzed the transformation of 2-methyl-, 3-methyl-, and 4-methylpyridine into corresponding N-oxides whose structures were confirmed by HPLC-MS using analytical standards (Table 3). Burkholderia sp. MAK1 was also capable of transforming di- and trimethyl pyridines, except those in which both positions adjacent to nitrogen were occupied.
Figure 5. Pyridine, pyrazine and their methylated derivatives as substrates for the 2-hydroxypyridine-induced Burkholderia sp. MAK1 cells.
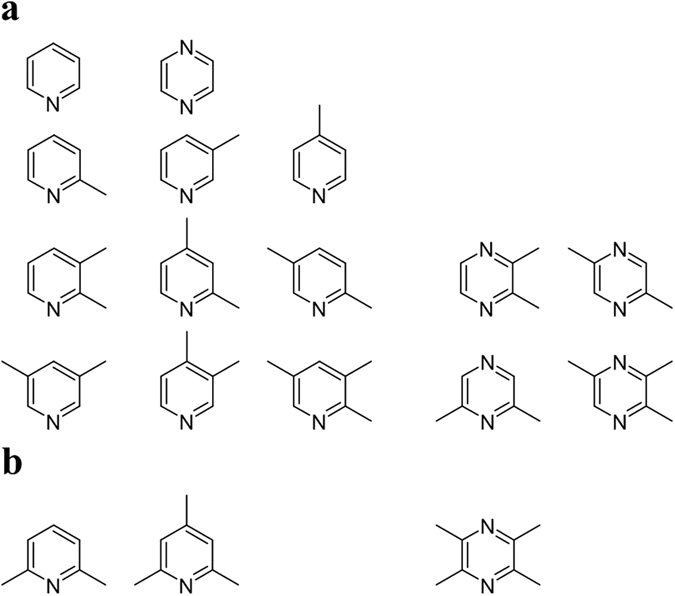
(a) Substrate consumption occurs; (b) no reaction observed.
Table 3. The bioconversion features of pyridine and pyrazine derivatives for which reactions products were detectable.
| Substrate |
Product |
|||
|---|---|---|---|---|
| Name | [M+H]+ | [M+H]+ | NMR | Name/Possible outcome |
| Pyridine | 80 | 96 | − | Hydroxylated at the 1-position |
| 2-Methylpyridine | 94 | 110 | − | Hydroxylated at the 1-position |
| 3-Methylpyridine | 94 | 110 | − | Hydroxylated at the 1-position |
| 4-Methylpyridine | 94 | 110 | − | Hydroxylated at the 1-position |
| Pyrazine | 81 | 97 and 113 | + | pyrazine-1-oxide and pyrazine-1,4-dioxide |
For the NMR analysis approximately 10 mg of purified bioconversion product was used.
Based on HPLC-MS analysis, the biotransformation of pyrazine resulted in the formation of two products with molecular masses that were 16 Da and 32 Da higher than that of the parent compound. 1H and 13C NMR analysis allowed identification of these products as pyrazine-1-oxide (1H NMR (DMSO-d6, ppm): δ = 8.34–8.36 (m, 2H, CH), 8.54–8.57 (m, 2H, CH); 13C NMR (DMSO-d6, ppm): δ = 134.85, 148.94) and pyrazine-1,4-dioxide (1H NMR (DMSO-d6, ppm): δ = 8.28 (s, 4H, CH); 13C NMR (DMSO-d6, ppm): δ = 137.21).
Our research revealed that Burkholderia sp. MAK1 has also the ability to oxidize various methylpyrazines. For the oxidation of methylated pyrazines the single free position adjacent to either one of nitrogen atoms was a sufficient condition, e. g. the cells could oxidize 2,3,5-trimethylpyrazine, but not 2,3,5,6-tetramethylpyrazine.
To date, only a few reports regarding the microbial N-hydroxylation of pyridines have been published. The formation of pyridine N-oxides has been observed in fungi Cunninghamella elegans ATCC 2626941, Verticillium sp. GF3931, and other fungi42 as well as in bacteria Methylococcus capsulatus29 and Diaphorobacter sp. J5-5143. Also, the purified aromatic peroxygenase from fungus Agrocybe aegerita has been found to be active towards pyridine and its derivatives30. In this context, the results of this study not only broaden our understanding of microbial transformation but also provide a versatile tool that can be used in a regioselective oxyfunctionalization of various pyridine derivatives.
Conclusions
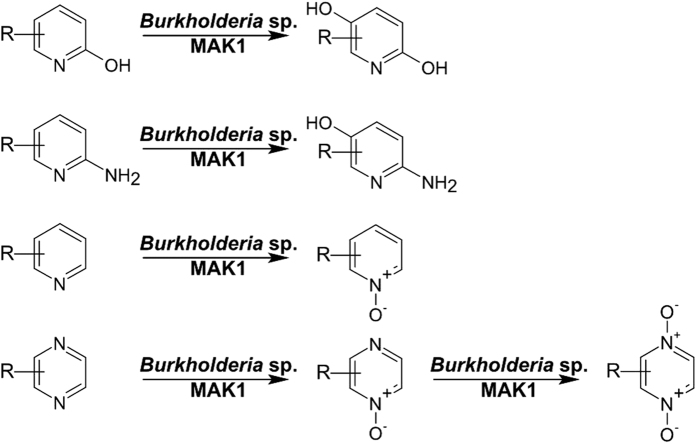
In summary, whole cells of Burkholderia sp. MAK1 have high activity towards pyridin-2-amines and pyridin-2-ols, and are applicable for the synthesis of pyridin-5-ols from the corresponding substrates. Moreover, unsubstituted pyridine and pyrazine as well as their methylated derivatives can be converted into the corresponding N-oxides using pyridin-2-ol-induced Burkholderia sp. MAK1 (Fig. 6). The approach presented here offers a promising alternative to chemical synthesis of hydroxylated pyridines.
Figure 6. The general scheme of oxyfunctionalization of pyridine and pyrazine derivatives using whole cells of Burkholderia sp. MAK1.
Methods
Chemicals
Pyridin-2-amine, 2-chloropyridine, pyridine-2-carboxylic acid, pyridine-N-oxide, pyridin-3-ol, 2-aminopyridin-3-ol, 3-nitropyridin-2-amine, 2-hydroxy-6-oxo-1H-pyridine-4-carboxylic acid, pyrazine, pyridine-4-carboxylic acid and pyridine-2,3-dicarboxylic acid were purchased from Merck (Darmstadt, Germany). Pyridin-2-ol, pyrimidin-2-ol, 2-methylpyridine, 3-methylpyridine, 4-methylpyridine, 2-pyridylmethanol, pyridine-2-thiol, 1-methylpyridin-2-one, 3-bromopyridin-2-ol, 3-methylpyridin-2-ol, 3-methoxypyridin-2-ol, pyridine-2,3-diol, pyridine-2,4-diol, pyridine-2,6-diol, 3-methylpyridin-2-amine, 3-methylpyridine-2-carbonitrile, 2,3-dimethylpyridine, 2,4-dimethylpyridine, 2,5-dimethylpyridine, 2,6-dimethylpyridine, 3,4-dimethylpyridine, 3,5-dimethylpyridine, 2,3,5-trimethylpyridine, pyridine-2,3-diamine, pyridine-2,6-diamine, 4-methyl-3-nitro-pyridin-2-ol, 4-methylpyridin-2-amine, 4,6-dimethylpyridin-2-amine, 3-methylpyridin-6-amine, pyridine, pyran-2-one, 2-hydroxy-6-methyl-pyridine-3-carbonitrile, 1-pyrazin-2-ylethanone, 2,3-dimethylpyrazine, 2,5-dimethylpyrazine, 2,6-dimethylpyrazine, 2,3,5-trimethylpyrazine, pyrazin-2-amine, tetramethylpyrazine, 2-(2-pyridyl)ethanol, 2-aminopyridine-3-carbonitrile, 3-methylpyridazine, 2-methylpyridin-3-ol, 4-methylpyridine-2-carbonitrile, pyridine-2,6-dicarbonitrile, 6,7-dihydro-5H-cyclopenta[b]pyridine, 5-methyl-6,7-dihydro-5H-cyclopenta[b]pyrazine, pyrimidine-2,4-diamine, 2-methylpyridin-5-ol, 2-aminopyridin-6-ol, 4-methyl-3-nitro-pyridin-2-amine were obtained from Sigma-Aldrich (St. Louis, MO). Pyridin-2-ol N-oxide, 2-hydroxypyridine-3-carboxylic acid, 6-hydroxypyridine-2-carboxylic acid, 3-hydroxypyridine-2-carboxylic acid, 6-methylpyridin-2-ol, 2-methylpyridine N-oxide, 2-ethylpyridine, 4-ethylpyridine, pyridine-3-carboxamide, 4-[(4-nitrophenyl)methyl]pyridine, pyridine-3-carbonitrile, 2,4,6-trimethylpyridine and pyridin-4-ol were purchased from Fluka (Buchs, Switzerland). 5-hydroxypyridine-2-carboxylic acid, 3-fluoropyridin-2-ol, 3-chloropyridin-2-ol, 6-chloropyridin-2-ol, 6-bromopyridin-2-ol, 6-methylpyridin-2-ol, 4-chloropyridin-2-ol, 2-oxo-1H-pyridine-4-carbonitrile, 4,6-dimethylpyridin-2-ol, 3-amino-6-methyl-pyridin-2-ol, 3-(trifluoromethyl)pyridin-2-ol, 4-(trifluoromethyl)pyridin-2-ol, 6-(trifluoromethyl)pyridin-2-ol, 6-(trifluoromethyl)pyridin-2-amine, 3-chloropyridin-2-amine, 4-chloropyridin-2-amine, 6-chloropyridin-2-amine, 3-bromopyridin-2-amine, 4-bromopyridin-2-amine, 6-bromopyridin-2-amine, 3,6-dibromopyridin-2-amine, 3-fluoropyridin-2-amine, 4-fluoropyridin-2-amine, 6-fluoropyridin-2-amine, 2-aminopyridin-4-ol, 3-methoxypyridin-2-amine, 6-methoxypyridin-2-amine, ethyl 2-aminopyridine-3-carboxylate, 2-aminopyrimidin-4-ol, 4-chloropyrimidin-2-amine, 4-bromopyridin-2-ol and 3-bromo-6-methylpyridin-2-ol were the products of Combi Blocks Inc (San Diego, USA). All reagents used in this study were of analytical grade. Nutrient agar (NA) medium and brain heart infusion (BHI) medium were obtained from Oxoid (Hampshire, UK). The 2-amino-3H-dipyrido[3,2-b:2’,3’-e][1,4]oxazine-3-one prepared by oxidation-dimerization of 2-amino-3-hydroxypyridine as described for 2-aminophenol44 was a gift from Dr. J. Šarlauskas. The alkylated pyridones were synthesized according to the published procedure45.
1-Ethylpyridin-2(1H)-one. Yield 170 mg (69%), colourless oil. MS (ESI+): m/z 124.15 [M + H]+. 1H NMR (CDCl3, ppm): δ = 1.37 (t, J = 7.2 Hz, 3H, CH3), 4.00 (q, J = 7.2 Hz, 2H, CH2), 6.21 (td, J = 6.7, 1.4 Hz, 1H, CH), 6.61 (ddd, J = 9.1, 1.4, 0.8 Hz, 1H, CH), 7.29–7.37 (m, 2H, CH); 13C NMR (CDCl3, ppm): δ = 14.69, 44.96, 106.47, 120.96, 137.01, 139.46, 162.51.
1-Propylpyridin-2(1H)-one. Yield 200 mg (73%), colourless oil. MS (ESI+): m/z 138.15 [M + H]+. 1H NMR (DMSO-d6, ppm): δ = 0.95 (t, J = 7.4 Hz, 3H, CH3), 1.56–1.69 (m, 2H, CH2), 3.75 (t, J = 7.4 Hz, 2H, CH2), 6.11 (td, J = 6.7, 1.4 Hz, 1H, CH), 6.42 (ddd, J = 9.1, 1.3, 0.6 Hz, 1H, CH), 7.35–7.46 (m, 1H, CH), 7.72 (ddd, J = 6.7, 2.1, 0.6 Hz, 1H, CH); 13C NMR (DMSO-d6, ppm): δ = 14.00, 29.23, 46.78, 105.55, 119.82, 138.93, 141.43, 162.15.
1-Butylpyridin-2(1H)-one. Yield 180 mg (60%), yellowish oil. MS (ESI+): m/z 152.20 [M + H]+. 1H NMR (DMSO-d6, ppm): δ = 0.90 (t, J = 7.4 Hz, 3H, CH3), 1.18–1.34 (m, 2H, CH2), 1.52–1.65 (m, 2H, CH2), 3.86 (t, J = 7.4 Hz, 2H, CH2), 6.19 (td, J = 6.7, 1.4 Hz, 1H, CH), 6.36 (ddd, J = 9.1, 1.4, 0.6 Hz, 1H, CH), 7.33–7.42 (m, 1H, CH), 7.66 (ddd, J = 6.7, 2.1, 0.6 Hz, 1H, CH); 13C NMR (DMSO-d6, ppm): δ = 14.05, 19.70, 31.25, 48.61, 105.50, 120.02, 139.53, 140.13, 161.82.
Microbial cultures and cultivation conditions
EFA (g/l): K2HPO4 10.0, KH2PO4 4.0, yeast extract 0.5, (NH4)2SO4 1.0, 2-hydroxypyridine 2.0, MgSO4 × 7H2O 0.2, salt solution 10 ml/l., pH 7.2; Salt solution (g/l): CaCl2 × 2H2O 2.0, MnSO4 × 4H2O 1.0, FeSO4 × 7H2O 0.5, all components were dissolved in 0.1N HCl and added in to EFA medium before cultivation; Koser mineral medium (g/l): NaCl 5.0, NH4H2PO4 1.0, K2HPO4 1.0, MgSO4 × 7H2O 0.4. The final pH was adjusted to 7.046. Koser agar medium was prepared adding agar to Koser mineral medium (15 g/l). Nutrient agar medium (g/l): 28.0; BHI (g/l): 37. All media and solutions were autoclaved at 1 atm for 30 min.
Bacteria were cultivated in liquid media with aeration at 30 °C.
For substrate specificity and bioconversion experiments Burkholderia sp. MAK1 was grown at 30 °C for 20 hours in 1 l flasks containing 200 ml EFA medium. The cells were harvested by centrifugation and washed twice with 10 mM potassium phosphate buffer, pH 7.2.
Isolation of 2-hydroxypyridine utilizing microorganisms
0.5 g of soil was suspended in 20 ml of Koser mineral medium. The aliquots (10–100 μl) were spread on Koser agar plates supplemented with 0.1% 2-hydroxypyridine and clotrimazole (20 μg/ml). Clotrimazole is known as cytochrome P450 inhibitor and was used to suppress growth of actinobacteria (e. g., Rhodococcus, Streptomyces, Mycobacterium) or fungi. After 3–5 days of aerobical cultivation at 30 °C the best growing colonies were selected for further work.
DNA analysis
DNA was extracted according to Woo et al.47. 16S rRNA encoding genes were amplified using universal primers w001 and w00248. The PCR product was cloned into the pTZ57R/T plasmid (Thermo Fisher Scientific, Lithuania) and sequenced using M13/pUC (-46) forward 22-mer and reverse 24-mer primers. The 16S rRNA sequence of MAK1 was analyzed using BLAST tool and The Ribosomal Database Project in the NCBI database. A phylogenetic tree was constructed and displayed using the neighbor-joining method with MEGA649. The Burkholderia sp. MAK1 16S rRNA gene sequence was deposited in GenBank under accession no. KU195413. Burkholderia sp. MAK1 was deposited to DSMZ German Culture Collection with accession no. DSM102049.
Bioconversion of pyridines, pyrimidines and pyrazines using the cells of Burkholderia sp. MAK1
0.05 g of wet biomass of Burkholderia sp. MAK1 cells was resuspended in 1 ml of 10 mM potassium phosphate buffer, pH 7.2. The suspension was supplemented with 15 mM glucose and 0.25 mM of corresponding substrate and incubated at 30 °C. The process of the conversion was followed by HPLC-MS.
Isolation and characterization of bioconversion products
~2 g of wet biomass of Burkholderia sp. MAK1 cells was resuspended in 100 ml of 10 mM potassium phosphate buffer, pH 7.2 supplemented with 15 mM of glucose and 0.25 mM of corresponding substrate and incubated at 30 °C. After bioconversion the cells of Burkholderia sp. MAK1 were separated by centrifugation. The supernatant liquid was vaporized to dryness under reduced pressure. The residue was dissolved in 5 ml of deionized water and purification of the product was carried out using reverse phase chromatography (12 g C-18 cartridge). Prior the purification the column was equilibrated with water. A mobile phase that consisted of water and methanol delivered in the gradient 10:0 → 10:5 elution mode. The collected fractions were analyzed by HPLC-MS. The fractions containing pure product were joined, and the solvent was removed under reduced pressure. 1H NMR spectra were recorded in DMSO-d6 or CDCl3 on Bruker Ascend 400, 400 MHz, and 13C NMR were recorded on Bruker Ascend 400, 100 MHz. Chemical shifts are reported in parts per million relative to the solvent resonance signal as an internal standard.
HPLC-MS analysis
Before the analysis the cells were separated from the reaction mixture by centrifugation. The resultant supernatant was mixed with an equal part of acetonitrile, centrifuged and analyzed using a high performance liquid chromatography system (CBM-20A controller, two LC-2020AD pumps, SIL-30AC auto sampler and CTO-20AC column oven; Shimadzu, Japan) equipped with a photo diode array (PDA) detector (SPD-M20A Prominence diode array detector; Shimadzu, Japan) and a mass spectrometer (LCMS-2020, Shimadzu, Japan) equipped with an ESI source. The chromatographic separation was conducted using a YMC Pack Pro column, 3 × 150 mm (YMC, Japan) at 40 °C and a mobile phase that consisted of 0.1% formic acid water solution (solvent A), and acetonitrile (solvent B) delivered in the 0 → 60% gradient elution mode. Mass scans were measured from m/z 10 up to m/z 700, at 350 °C interface temperature, 250 °C DL temperature, ±4,500 V interface voltage, neutral DL/Qarray, using N2 as nebulizing and drying gas. Mass spectrometry data was acquired in both the positive and negative ionization mode. The data was analyzed using the LabSolutions LCMS software.
Activity assay of pyridine-2,5-diol 5,6-dioxygenase from Burkholderia sp. MAK1
Burkholderia sp. MAK1 was grown at 30 °C for 20 hours in two 150 ml flasks, one containing 25 ml EFA medium (pyridin-2-ol induced cells), other containing 25 ml EFA medium where pyridin-2-ol is substituted for succinate (uninduced cells, negative control). The cells were harvested by centrifugation, washed twice with 10 mM potassium phosphate buffer (pH 7.2), suspended in 5 ml of the same buffer and sonicated. In 1.5 ml tubes three separate reaction mixtures were combined: internal control (990 μl 10 mM potassium phosphate buffer, pH 7.2 and 10 μl 2 mg/ml pyridine-2,5-diol solution), negative control (890 μl 10 mM potassium phosphate buffer, pH 7.2, 10 μl 2 mg/ml pyridine-2,5-diol solution and 100 μl cell-free extract of uninduced cells) and sample (890 μl 10 mM potassium phosphate buffer, pH 7.2, 10 μl 2 mg/ml pyridine-2,5-diol solution and 100 μl cell-free extract of induced cells). 100 μl of each reaction mixture was transferred to a 96 well plate and change in absorbance (λmax 320 nm) per 30 minutes was measured. Overall change in absorbance was evaluated by subtracting noise data (internal and negative controls) from sample data. We were able to achieve 200–250 mU per 1 l medium, where 1 enzyme unit (U) is an amount of enzyme that catalyzes depletion of 1 μmol pyridine-2,5-diol per minute. The measured molar extinction coefficient of pyridine-2,5-diol in ethanol was 9800 M−1∙cm−1.
Additional Information
How to cite this article: Stankevičiūtė, J. et al. Oxyfunctionalization of pyridine derivatives using whole cells of Burkholderia sp. MAK1. Sci. Rep. 6, 39129; doi: 10.1038/srep39129 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The work was supported by European Social Fund (ESF) under the Human Resources Development Action Programme, the Global Grant measure, project No. VP1-3.1-ŠMM-07-K-03-015. We are grateful to Dr. J. Šarlauskas for the valuable cooperation. We also thank Dr. L. Kalinienė for assistance with the preparation of the manuscript.
Footnotes
Author Contributions R.M. conceived the study. R.G. isolated bacterial strain. J.S., J.V. and V.P. carried out bioconversion experiments. D.T. synthesized the substrates. J.S. and D.T. conducted the analysis of compounds. J.S., J.V., V.P., R.G., D.T. and R.M. analyzed the data and prepared the manuscript. All authors read and approved the final manuscript.
References
- Loughlin W. A., Tyndall J. D. A., Glenn M. P. & Fairlie D. P. Beta-strand mimetics. Chem. Rev. 104, 6085–6117 (2004). [DOI] [PubMed] [Google Scholar]
- Costantino L. & Barlocco D. Privileged structures as leads in medicinal chemistry. Curr. Med. Chem. 13, 65–85 (2006). [PubMed] [Google Scholar]
- Goldman M. E. et al. Pyridinone derivatives - specific human-immunodeficiency-virus type-1 reverse-transcriptase inhibitors with antiviral activity. Proc. Natl. Acad. Sci. USA 88, 6863–6867 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Mitscher L. A. & Shen L. L. The 2-pyridone antibacterial agents: bacterial topoisomerase inhibitors. Med. Res. Rev. 20, 231–293 (2000). [DOI] [PubMed] [Google Scholar]
- Jessen H. J. & Gademann K. 4-Hydroxy-2-pyridone alkaloids: structures and synthetic approaches. Nat. Prod. Rep. 27, 1168–1185 (2010). [DOI] [PubMed] [Google Scholar]
- Desai N. C., Rajpara K. M. & Joshi V. V. Synthesis of pyrazole encompassing 2-pyridone derivatives as antibacterial agents. Bioorg. Med. Chem. Lett. 23, 2714–2717 (2013). [DOI] [PubMed] [Google Scholar]
- Charrier J. D. et al. Discovery and structure-activity relationship of 3-aminopyrid-2-ones as potent and selective interleukin-2 inducible T-cell kinase (Itk) inhibitors. J. Med. Chem. 54, 2341–2350 (2011). [DOI] [PubMed] [Google Scholar]
- Kusakabe K. et al. Design, synthesis, and binding mode prediction of 2-pyridone-based selective CB2 receptor agonists. Bioorgan. Med. Chem. 21, 2045–2055 (2013). [DOI] [PubMed] [Google Scholar]
- Urban M., Pohl R., Klepetarova B. & Hocek M. New modular and efficient approach to 6-substituted pyridin-2-yl C-nucleosides. J. Org. Chem. 71, 7322–7328 (2006). [DOI] [PubMed] [Google Scholar]
- Tauraite D., Razanas R., Mikalkenas A., Serva S. & Meskys R. Synthesis of pyridone-based nucleoside analogues as substrates or inhibitors of DNA polymerases. Nucleos. Nucleot. Nucl. 35, 163–177 (2016). [DOI] [PubMed] [Google Scholar]
- Rudra A. et al. Bromopyridone nucleotide analogues, anoxic selective radiosensitizing agents that are incorporated in DNA by polymerases. J. Org. Chem. 80, 10675–10685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijtmans M. et al. 6-Amino-3-pyridinols: towards diffusion-controlled chain-breaking antioxidants. Angew. Chem. Int. Edit. 42, 4370–4373 (2003). [DOI] [PubMed] [Google Scholar]
- Mase T., Arima H., Tomioka K., Yamada T. & Murase K. Imidazo[2,1-b]benzothiazoles. 2. New immunosuppressive agents. J. Med. Chem. 29, 386–394 (1986). [DOI] [PubMed] [Google Scholar]
- Lhassani M. et al. Synthesis and antiviral activity of imidazo[1,2-a]pyridines. Eur. J. Med. Chem. 34, 271–274 (1999). [Google Scholar]
- Pires M. J. D., Poeira D. L. & Marques M. M. B. Metal-catalyzed cross-coupling reactions of aminopyridines. Eur. J. Org. Chem. 33, 7197–7234 (2015). [Google Scholar]
- Odell L. R. et al. Functionalized 3-amino-imidazo[1,2-a]pyridines: a novel class of drug-like Mycobacterium tuberculosis glutamine synthetase inhibitors. Bioorg. Med. Chem. Lett. 19, 4790–4793 (2009). [DOI] [PubMed] [Google Scholar]
- Radulovic N. et al. New 3,4-annelated coumarin derivatives: synthesis, antimicrobial activity, antioxidant capacity, and molecular modeling. Monatsh. Chem. 137, 1477–1486 (2006). [Google Scholar]
- Behrman E. J. Improved syntheses of 5-hydroxy-2-pyridones (2,5-dihydroxypyridines). Synthetic Commun. 38, 1168–1175 (2008). [Google Scholar]
- Moore J. A. & Marascia F. J. Heterocyclic studies. VII. The preparation and reactions of 2-amino-5-hydroxypyridines; the formation of an azaquinone. J. Am. Chem. Soc. 81, 6049–6056 (1959). [Google Scholar]
- Cai L. S. et al. Synthesis and structure - affinity relationships of new 4-(6-iodo-H-imidazo[1,2-a]pyridin-2-yl)-N-dimethylbenzeneamine derivatives as ligands for human beta-amyloid plaques. J. Med. Chem. 50, 4746–4758 (2007). [DOI] [PubMed] [Google Scholar]
- Gorantla J. N., Kovval D. & Lankalapalli R. S. An unusual synthesis of 2-pyridone and 3,5-dihydroxypyridine from a carbohydrate. Tetrahedron Lett. 54, 3230–3232 (2013). [Google Scholar]
- Schewe H., Mirata M. A., Holtmann D. & Schrader J. Biooxidation of monoterpenes with bacterial monooxygenases. Process Biochem. 46, 1885–1899 (2011). [Google Scholar]
- Schrewe M., Julsing M. K., Buhler B. & Schmid A. Whole-cell biocatalysis for selective and productive C-O functional group introduction and modification. Chem. Soc. Rev. 42, 6346–6377 (2013). [DOI] [PubMed] [Google Scholar]
- Urlacher V. B. & Girhard M. Cytochrome P450 monooxygenases: an update on perspectives for synthetic application. Trends Biotechnol. 30, 26–36 (2012). [DOI] [PubMed] [Google Scholar]
- Nakano H. et al. Purification, characterization and gene cloning of 6-hydroxynicotinate 3-monooxygenase from Pseudomonas fluorescens TN5. Eur. J. Biochem. 260, 120–126 (1999). [DOI] [PubMed] [Google Scholar]
- Brandsch R. Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69, 493–498 (2006). [DOI] [PubMed] [Google Scholar]
- Kutanovas S. et al. Bioconversion of methylpyrazines and pyridines using novel pyrazines-degrading microorganisms. Chemija 24, 67–73 (2013). [Google Scholar]
- Kutanovas S. et al. Isolation and characterization of novel pyridine dicarboxylic acid-degrading microorganisms. Chemija 27, 74–83 (2016). [Google Scholar]
- Colby J., Stirling D. I. & Dalton H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem. J. 165, 395–402 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich R., Dolge C., Kluge M. & Hofrichter M. Pyridine as novel substrate for regioselective oxygenation with aromatic peroxygenase from Agrocybe aegerita. FEBS Lett. 582, 4100–4106 (2008). [DOI] [PubMed] [Google Scholar]
- Mitsukura K., Hayashi M., Yoshida T. & Nagasawa T. Oxidation of aromatic N-heterocyclic compounds to N-oxides by Verticillium sp. GF39 cells. J. Biosci. Bioeng. 115, 651–653 (2013). [DOI] [PubMed] [Google Scholar]
- Cain R. B., Houghton C. & Wright K. A. Microbial metabolism of the pyridine ring. Metabolism of 2- and 3-hydroxypyridines by the maleamate pathway in Achromobacter sp. Biochem. J. 140, 293–300 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J. P., Feng Y. & Bollag J. M. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol. Rev. 60, 483–498 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin E. J., Sims G. K. & Traina S. J. Biodegradation of 2-methyl, 2-ethyl, and 2-hydroxypyridine by an Arthrobacter sp. isolated from subsurface sediment. Biodegradation 10, 93–104 (1999). [DOI] [PubMed] [Google Scholar]
- Stanislauskiene R. et al. Construction of Escherichia coli-Arthrobacter-Rhodococcus shuttle vectors based on a cryptic plasmid from Arthrobacter rhombi and investigation of their application for functional screening. FEMS Microbiol. Lett. 327, 78–86 (2012). [DOI] [PubMed] [Google Scholar]
- Fetzner S. Bacterial degradation of pyridine, indole, quinoline, and their derivatives under different redox conditions. Appl. Microbiol. Biotechnol. 49, 237–250 (1998). [Google Scholar]
- Vaitekunas J., Gasparaviciute R., Rutkiene R., Tauraite D. & Meskys R. A 2-hydroxypyridine catabolism pathway in Rhodococcus rhodochrous strain PY11. Appl. Environ. Microbiol. 82, 1264–1273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes P. E., Rittenberg S. C. & Knackmuss H. J. The bacterial oxidation of nicotine. 8. Synthesis of 2,3,6-trihydroxypyridine and accumulation and partial characterization of the product of 2,6-dihydroxypyridine oxidation. J. Biol. Chem. 247, 7628–7633 (1972). [PubMed] [Google Scholar]
- Tse H. et al. Production of 2-aminophenoxazin-3-one by Staphylococcus aureus causes false-positive results in beta-galactosidase assays. J. Clin. Microbiol. 50, 3780–3782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P. K. & Bae H. Bacterial degradation of chlorophenols and their derivatives. Microb. Cell. Fact. 13, 31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T. et al. Microbial transformation of 2-amino-4-methyl-3-nitropyridine. J. Ind. Microbiol. Biotechnol. 39, 1789–1799 (2012). [DOI] [PubMed] [Google Scholar]
- Parshikov I. A., Netrusov A. I. & Sutherland J. B. Microbial transformation of azaarenes and potential uses in pharmaceutical synthesis. Appl. Microbiol. Biotechnol. 95, 871–889 (2012). [DOI] [PubMed] [Google Scholar]
- Sun J. Q. et al. Bacterial pyridine hydroxylation is ubiquitous in environment. Appl. Microbiol. Biotechnol. 98, 455–464 (2014). [DOI] [PubMed] [Google Scholar]
- Nagasawa H. T. & Gutmann H. R. The oxidation of o-aminophenols by cytochrome c and cytochrome oxidase. I. Enzymatic oxidations and binding of oxidation products to bovine serum albumin. J. Biol. Chem. 234, 1593–1599 (1959). [PubMed] [Google Scholar]
- Kozikowski A. P., Tueckmantel W., Chellappan S., Kellar K. J. & Xiao Y. 10-Substituted cytisine derivatives and methods of use thereof. vol. 2010/0048606 A1. US2010.
- Koser S. A. & Baird G. R. Bacterial destruction of nicotinic acid. J Infect Dis. 75, 250–261 (1944). [Google Scholar]
- Woo T. H., Cheng A. F. & Ling J. M. An application of a simple method for the preparation of bacterial DNA. Biotechniques 13, 696–698 (1992). [PubMed] [Google Scholar]
- Godon J. J., Zumstein E., Dabert P., Habouzit F. & Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63, 2802–2813 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.