Abstract
DNA helicases participate in virtually all aspects of cellular DNA metabolism by using ATP-fueled directional translocation along the DNA molecule to unwind DNA duplexes, dismantle nucleoprotein complexes, and remove non-canonical DNA structures. Post-translational modifications and helicase interacting partners are often viewed as determining factors in controlling the switch between bona fide helicase activity and other functions of the enzyme that do not involve duplex separation. The bottleneck in developing a mechanistic understanding of human helicases and their control by post-translational modifications is obtaining sufficient quantities of the modified helicase for traditional structure-functional analyses and biochemical reconstitutions. This limitation can be overcome by single-molecule analysis, where several hundred surface-tethered molecules are sufficient to obtain a complete kinetic and thermodynamic description of the helicase-mediated substrate binding and rearrangement. Synthetic oligonucleotides site-specifically labeled with Cy3 and Cy5 fluorophores can be used to create a variety of DNA substrates that can be used to characterize DNA binding, as well as helicase translocation and duplex unwinding activities. This chapter describes “single-molecule sorting”, a robust experimental approach to simultaneously quantify, and distinguish the activities of helicases carrying their native post-translational modifications. Using this technique, a DNA helicase of interest can be produced and biotinylated in human cells to enable surface-tethering for the single-molecule studies by total internal reflection fluorescence microscopy. The pool of helicases extracted from the cells is expected to contain a mixture of post-translationally modified and unmodified enzymes, and the contributions from either population can be monitored separately, but in the same experiment providing a direct route to evaluating the effect of a given modification.
Keywords: Single-molecule, Total Internal Reflection Fluorescence Microscopy, Förster resonance energy transfer (FRET), DNA helicase, FBH1, FANCJ, BRCA1, phosphorylation, ubiquitylation
1. Introduction
Single-molecule fluorescence imaging has revolutionized our ability to study nucleoprotein transactions in real time and in unprecedented detail. The ever-growing repertoire of single-molecule methodologies provides two distinct advantages over traditional ensemble experiments. First, single-molecule assays are designed to consume at least 103 to 106 times less material; this can be a limiting factor when the sample is difficult to produce in sufficient quantity for rigorous biochemical analysis. More importantly, examining the activities of individual molecules can reveal characteristics that are otherwise hidden in ensemble experiments due to averaging [1, 2]. Stochastic behaviors, for instance, would need to be synchronized and evenly distributed among a population of molecules to be detectable in bulk studies. For these reasons, single-molecule experiments using surface-immobilized proteins are especially useful to study how post-translational modifications (PTMs) affect their biochemical properties since the repeating cycles of activity originating from the same modified or unmodified protein molecule can be monitored.
1.1. Post-translational modifications
PTMs are covalent protein modifications that form either during or after protein synthesis. PTMs can include adding small functional groups such as phosphate, methyl, and acetate, or even protein moieties such as ubiquitin and SUMO, onto the amino acid side chains (or termini) of a target protein [3, 4]. Such modifications are widely associated with signal transduction and also provide a mechanism to regulate protein functions. This article will specifically address PTMs of the proteins involved in DNA metabolism, and in particular, a class of proteins known as helicases.
1.2. DNA helicases
DNA helicases are molecular motors that separate double-stranded (ds) DNA in order to form the single-stranded (ss) intermediates needed for DNA replication, recombination, and repair. Helicases can also translocate along ssDNA or dsDNA to remove or remodel protein complexes and unconventional structures [5, 6]. Helicase activity is therefore essential during all stages of DNA metabolism. Not surprisingly, mutations in human helicases often contribute to the onset of genetic diseases associated with premature aging and chromosome instability [7]. DNA helicases may act individually or as essential components of multiprotein DNA repair machineries. Many DNA helicases are subject to PTMs that drastically, albeit transiently, alter their biochemical properties and interactions. For example, the BLM helicase (a RecQ-family helicase, mutations in which are associated with Bloom syndrome) can be phosphorylated, ubiquitylated, and SUMOylated; these modifications have an impact on the BLM cellular localization, overall stability, and interactions with protein partners as described in this review [8]. When the FBH1 helicase (F-box containing helicase 1, which switches between pro- [9] and anti-recombinogenic [10–13] activities at the damaged replication forks) is poly-ubiquitylated, it is converted into a pro-recombinase that no longer binds to a RAD51 nucleoprotein filament [14]. Despite their expected importance, the functional consequences for many helicase PTMs are not known. The FANCJ helicase (Fanconi Anemia complementation group J), for example, interacts with the BRCA1 tumor suppressor protein when FANCJ is phosphorylated on serine residue 990 [15–18]. It is not clear whether the FANCJ-BRCA1 complex possesses unique properties distinct to FANCJ alone, or if it combines the DNA structure selective binding properties of the two proteins. Traditional biochemical characterization of this complex is not feasible since FANCJ and BRCA1 are difficult to produce and only a small fraction of FANCJ is phosphorylated on serine 990. As a result, the vast majority of purified FANCJ will not be active in BRCA1-binding. Single-molecule fluorescence imaging provides an elegant solution to both of these experimental constraints.
1.3. Single-molecule sorting overview
This article describes using “single-molecule sorting” to investigate how PTMs influence the activities of the FBH1 and FANCJ helicases. This approach can be adapted to the analyses of a wide variety of systems, with a caveat that the protein of interest must be immobilized on the slide surface for fluorescence imaging and subsequent sorting of the activities; hence, this method is readily applied only to the proteins that either form stable complexes or act as monomers. If the assembly state of the protein is not known, it can be determined empirically by fluorescently labeling the protein site-specifically [19] prior to surface-immobilization and visualizing the number of photobleaching steps in the fluorescence trajectories of the immobilized protein alone. Surface-tethering is commonly achieved by labeling the protein with biotin (described in Methods) and placing it on a neutravidin-coated slide surface; alternatively, it can also be achieved via a “single-molecule pull down” (SiMPull), an elegant method developed by the Taekjip Ha’s group [20, 21]. The positions of the individual protein molecules on the microscope slide can then be determined using a fluorescently labeled binding partner (e.g. DNA, antibody, peptide), and protein molecules possessing a particular PTM can be similarly identified with a fluorescently labeled interaction partner that is specific for that epitope. Single-molecule activity assays can be carried out with the entire pool of tethered proteins, either before or after probing for a specific PTM, and their activities can be correlated with the presence or absence of the modification.
2. Methods
2.1. Construction of protein expression vectors
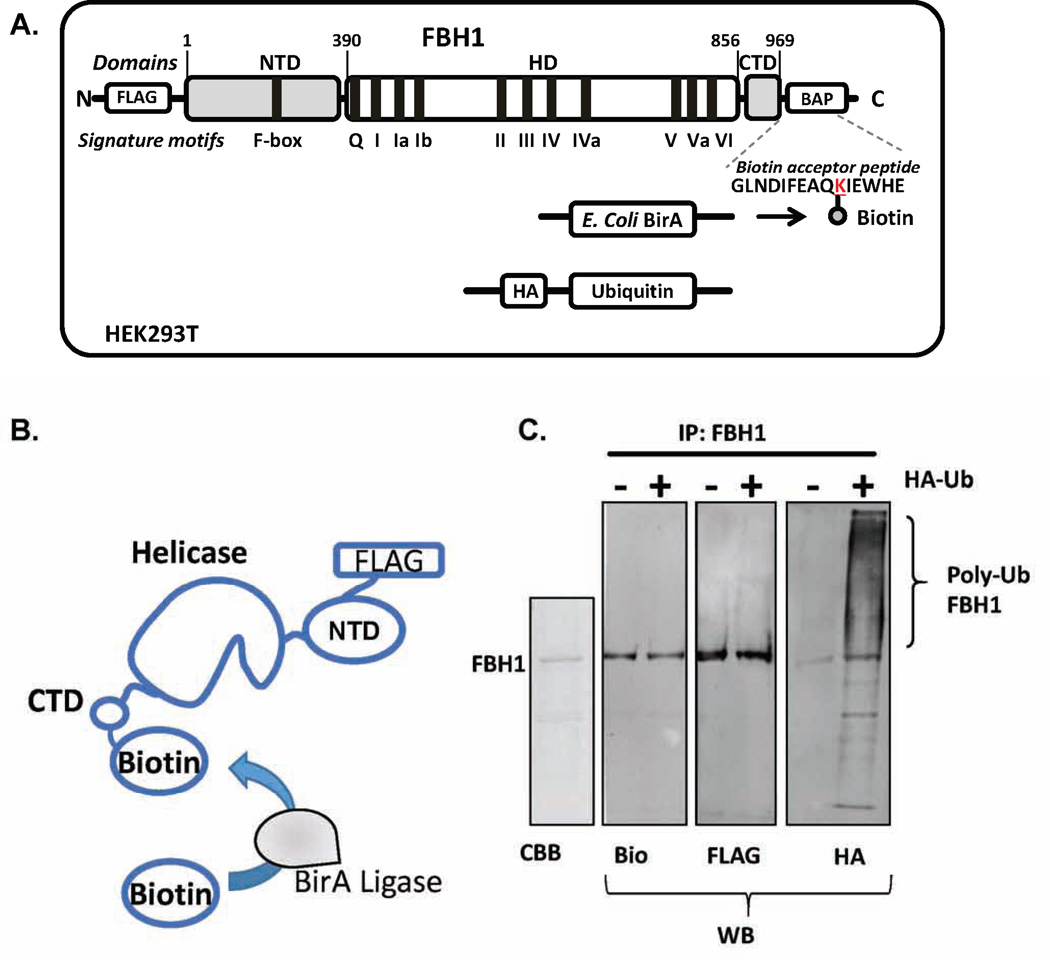
A biotin acceptor peptide (BAP) sequence (GLNDIFEAQKIEWHE) is engineered into the protein expression vector. E. coli BirA ligase recognizes and covalently labels the lysine residue within this sequence (Figure 1A). The BAP can be incorporated into a solvent exposed loop if the three dimensional structure of the protein of interest is known, or at the N- or C-terminus if structural data are not available. Activity assays can be carried out using protein with different anchoring positions as well as with the free protein to test whether surface-immobilization has altered its biochemical properties. Additionally, a FLAG tag (DYKDDDDK) is engineered into the protein expression vector. This tag not only facilitates protein purification via affinity based methods, but also provides an epitope for binding fluorescently labeled antibodies during single-molecule imaging. As an example of “single-molecule sorting” application, this article focuses on the human FBH1 and FANCJ helicases. The open reading frames of these proteins are cloned into a mammalian expression vector (e.g. pcDNA3) for production in human cells so that they can undergo their native PTMs.
Figure 1. Production of dually labeled FBH1 for single-molecule TIRFM experiments.
(A) FBH1 was expressed and purified from HEK293 cells following transient transfection. FBH1 was cloned into a pcDNA3 vector. A FLAG tag was also incorporated on the N-terminus of the protein and a biotin acceptor peptide was added to the C-terminus. HEK293 cells were transfected with the FBH1 construct along with pcDNA3-BirA, which encoded the E. coli BirA ligase. To produce ubiquitylated FBH1, HEK293 cells were additionally transfected with a pcDNA3 vector that expressed HA-tagged ubiquitin. (B) BirA ligase covalently biotinylates the lysine residue within the biotin acceptor peptide with high efficiency and specificity in vivo, thereby producing biotinylated FBH1. (C) Coomassie staining (CBB) and western blot (WB) analysis of purified FBH1. The blot of FBH1 using anti-HA antibody (HA) showed that FBH1 is poly-ubiquitylated in vivo. The blot of FBH1 using anti-biotin (bio) and anti-FLAG (FLAG) antibodies are also shown. Modified from Ref.14.
2.2. Producing Flag-FBH1-Biotin from HEK293 cells
The following procedure pertains to the expression and purification of the Flag-tagged biotinylated FBH1, but this method can be applied to study any protein of interest. For the initial, prove-of-principle studies we chosen to express human proteins in HEK293 cells (ATCC CRL-1573). HEK293 cells are adherent and display high transfection capacity, thereby allowing transfect the cell with multiple plasmids simultaneously. One, however, needs to exercise judgement with respect to selection of the most appropriate cell lines for each intended study.
2.2.1. HEK293 cell growth
HEK293 cells are seeded in a T75 culture flask using pre-warmed (37°C) Dulbecco’s modified eagle medium (DMEM) that is supplemented with 10% fetal bovine serum (FBS), 1% penicillin, 1% streptomycin, and 1 mM sodium pyruvate. The flask is kept at 37°C, 5% CO2 for 3 hours, at which time the starting media is exchanged for fresh DMEM to remove any non-adhering cells. Upon reaching 95% surface confluence (usually achieved after 72 hrs), the media is aspirated out and the cells are washed with Dulbecco’s phosphate buffered saline (DPBS). The cells are detached from the culture flask by treating with 1 mL of 0.05% Trypsin-EDTA for 5 minutes at 37°C. Trypsin is then inactivated with 9 mL of supplemented DMEM and the cells are further diluted and re-seeded at 1:10 (v/v cells to DMEM) into ten T150 culture flasks. After splitting, the flasks are kept at 37°C, 5% until the cells reach 90% confluence and are ready for transfection.
2.2.2. Transfection of HEK293 cells
HEK293 cells are transiently transfected using branched polyethylenimine (PEI) (MW: 25,000, Sigma Aldritch) [22, 23]. 10 mg/ml PEI is added in a 3:1 (v/w) ratio with plasmid DNA. Typically, 60 µg of plasmid is used for each T150 culture flask. The pcDNA3-Flag-FBH1-BAP (a plasmid containing the expression construct for human FBH1 with the N-terminal Flag tag and a C-terminal biotin acceptor peptide) and pcDNA3-BirA (a plasmid for expression of E. coli BirA ligase) are added in a 1:1 ratio (30 µg of each plasmid for a T150 flask). PEI and plasmid DNA are separately diluted in Opti-MEM I Reduced Serum Medium (Gibco) and incubated for 5 minutes at room temperature. DNA and PEI are then mixed and incubated for an additional 20 minutes at room temperature. The transfection media is supplemented with 100 µM final concentration of biotin (Sigma-Aldritch) to promote FBH1 biotinylation (Figure 1B). PEI-DNA mixture is added to HEK293 cells and the culture flask is incubated at 37°C, 5% CO2 for 24 hours. For FBH1-biotin (bio-FBH1) and HA-ubiquitin co-expression, cells are transfected with pcDNA3-Flag-FBH1-BAP, pcDNA3-BirA, and pcDNA3-HA-Ubiquitin in a 1:1:1 ratio, following the method above. Transfected cells are washed twice with DPBS and harvested with 0.05% Trypsin-EDTA treatment for 5 minutes at 37°C. The detached HEK293 cells are pooled from the ten T150 culture flasks and are collected into a 50 mL centrifuge tube. Cells are centrifuged at 1000 rpm for 3 minutes to remove any remaining DPBS and media, and the pellet is frozen and stored at −80°C until protein purification.
2.2.3. Purification of Flag-FBH1-Biotin and Flag-ubFBH1-Biotin
The HEK293 cell pellet is thawed on ice and re-suspended in lysis buffer (50mM Hepes pH 7.4, 250mM NaCl, 1mM EDTA, 1% Nonidet P-40, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mini-Complete EDTA free protease inhibitor tablet per 10mL Buffer). The cells are lysed by freeze-thaw and are agitated at 4°C for 20 minutes. The lysate is then centrifuged at 13,000 rpm for 10 minutes at 4°C to remove any cellular debris. The supernatant is passed through a 0.2 µm filter and added to anti-FLAG magnetic beads (M8823, Sigma Aldritch). The lysate-bead mixture is incubated for 2 hours at 4°C with rotation to promote binding after which the resulting mixture is placed on a magnetic separator (Qiagen). The beads are attracted to the magnet for 1 minute, and the supernatant is removed by pipetting. The beads are gently washed twice with wash buffer (50mM Hepes pH 7.4, 250mM NaCl, 1mM EDTA, 1% Nonidet P-40, 10% glycerol), and Flag-FBH1 is eluted from the beads by incubation in elution buffer (50mM Hepes pH 7.4, 250mM NaCl, 1mM EDTA, 1% Nonidet P-40, 10% glycerol, and 150ug/uL 3×FLAG peptide) for 30 minutes at 4°C with rotation. The purified protein is then flash frozen in liquid nitrogen and stored at −80°C. If necessary, additional purification steps could be applied to increase protein purity and to remove the 3×FLAG peptide (Figure 1C). Size-exclusion or anion exchange chromatography may be used with most of the DNA helicases.
2.3. DNA substrates
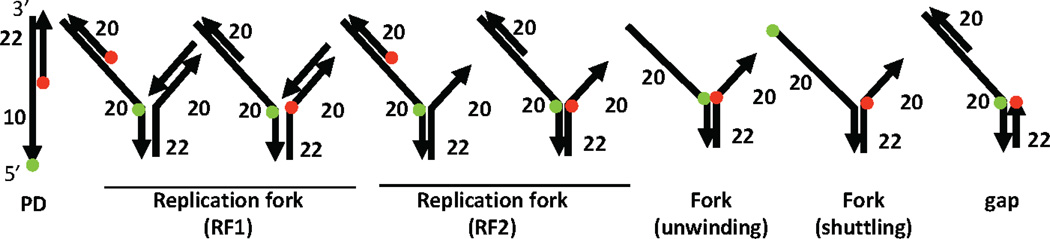
The individual DNA oligos used in this study are procured from Integrated DNA Technologies. Depending on the application, a number of DNA substrates can be selected (Figure 2). Helicases often display selectivity for distinct DNA structures resembling the intermediates of DNA metabolism. Upon binding to a cognate DNA substrate, helicases use ATP to fuel two important biochemical activities (i) strand separation (aka bona fide helicase), where dsDNA is unwound to produce transient single-stranded intermediates; and (ii) directional translocation, which can be coupled to remodeling of the nucleoprotein complexes or unorthodox DNA structures, such as a G-quadruplex or triplex DNA. Many DNA helicases require an ssDNA landing site in order to bind to the substrate and to stimulate its motor activity. This requirement is related to the helicase directionality: FBH1, for example, is a 3’→5’ DNA helicase, and therefore requires a 3’-overhang to unwind the duplex downstream. In most cases, the analysis of helicase-mediated duplex unwinding starts with the construction of a partial duplex DNA by annealing two complementary ssDNA oligonucleotides with an appropriate single-strand overhang. This is done by combining 3 µM (molecules) of complementary ssDNA oligonucleotides in annealing buffer (10mM Tris-HCl pH 7.0, 100mM NaCl). The DNA mixture is heated to 95°C for 5 minutes and then slowly cooled to room temperature over a 2 hour period. The annealed DNA is aliquoted in amber tubes to reduce exposure to light and stored at −20°C until use. Other substrates that prove useful include structures resembling a stalled replication fork, 3-way and 4-way junctions, D-loops (the central intermediate of homologous recombination), as well as G-quadruplex containing DNA substrates. These substrates are formed by annealing the ssDNA oligos as described for a simple partial duplex although DNA substrates comprised of 3 or more ssDNA oligos are gel-purified to remove any partially annealed products.
Figure 2. Repertoire of fluorescently labeled DNA substrates used to monitor helicase-related activities.
Depending on the application, a variety of DNA substrates can be assembled from the individual ssDNA components. Several configurations of DNA substrates that mimic different intermediates of a stalled replication fork are shown. Structural information of the helicase-DNA initiation complex is especially useful to determine the placement of the fluorophores. Direction of the arrow indicates 3’–5’ directionality of the DNA substrate.
2.4. Single-Molecule TIRF Microscope
Single-molecule experiments described below were performed on a prism-type TIRF microscope as previously described [24]. The protocols below may be combined with a broad range of fluorophores and corresponding excitation sources. The examples below are for the Cy3, TAMRA and Cy5 dyes. Cy3 or TAMRA fluorophores are excited by a DPSS laser (532nm, 100mW, Coherent), while Cy5 is excited directly with a diode laser (641nm, 100mW, Coherent). Alternatively, Cy5 can be excited as a result of FRET when it is in close proximity to Cy3. Fluorescent signals are originated by a 60× water-immersion objective (Olympus). Note that if using an objective-based TIRF microscope, an oil-immersion objective high numerical aperture (NA > 1.45 NA) is used to achieve TIR and the refractive index of the immersion oil has to closely match that of the coverslip. Cy3 and Cy5 signals are then separated in a dual-view (DV2, Photometrics) via 630nm dichroic mirror and 550nm long-pass filter, and collected via EMCCD (Andor). Time scale resolution is typically set to 30–100 ms per frame, but can be enhanced by reducing the imaging field.
2.5. Slide preparation for single-molecule experimentation
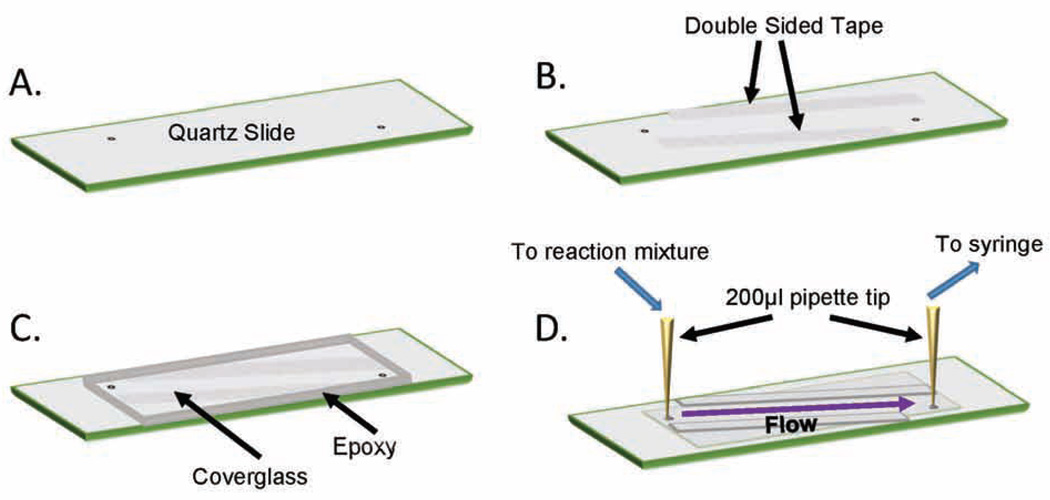
Quartz Slides (25mm×75mm×1mm) and cover glass (24mm×60mm-1.5, Fisherbrand) are prepared for experimentation following the previously described protocol [25]. Slides are drilled (0.75mm diamond tip bit, Lasco) (Figure 3A) and washed via sonication in 10% Alconox for 20 minutes. The slides are rinsed with double-distilled water until all of the Alconox has been removed, the slides are next sonicated in acetone for 30 minutes and rinsed three additional times. Both, the slides and cover glass will be used in the subsequent steps. Sonicate in 1M KOH for 40 minutes and rinse at least 5 times. Using a Bunsen burner, burn both sides of the slides for three minutes combined with focus on the drilled holes and center of the slides, and burn the both sides of the cover glass, for a total of 1 minute. Aminosilanize the slides and cover glass by incubating in aminosilane mixture (4.7% v/v glacial acetic acid and 1% v/v aminosilane (N-(2-aminoethyl)-3-aminopropyltrimethoxysilane; United Chemical Technologies) in methanol) at room temperature and in darkness for 10 minutes. Sonicate in the aminosilane mixture for 1 minute and incubate in darkness at room temperature for an additional 10 minutes. Rinse with methanol thrice. Dry slides with N2 gas and add PEG mixture (8.4mg/mL NaHCO3, 3.2×:2× ratio of biotinylated-PEG to m-PEG (MW 5000)) to the top of the slide. Assemble the cover glass onto the top of the slide and incubate in darkness at room temperature overnight. Rinse slides with double-distilled water and dry with N2 gas. Slides and cover glass are individually placed in 50 mL falcon tubes that are then vacuum sealed within a Food Saver bag, and stored at −20°C in the dark until use.
Figure 3. Assembly of a flow-cell for single-molecule TIRFM experiments.
(A) A quartz slide was drilled and passivated as described in section 2.6. (B) Double-sided tape was applied to the slide as shown to form the borders of the sample chamber. (C) A passivated coverglass was applied onto the slide covering the pre-drills holes, and the edges of the coverglass were sealed with epoxy. (D) Tubing and pipette tips were secured onto each of the holes of the slide on the reverse side using epoxy. One end of the tubing was connected to a syringe while the other tubing was placed in a centrifuge tube containing sample. Drawing from the syringe initiated the flow of sample onto the microscope slide. The solution can be exchanged by pausing flow and placing the tubing into another sample tube before re-establishing flow.
2.6. Flow Chamber assembly for single-molecule experiments
When ready to use a slide and cover glass for experimentation, first remove them from −20°C storage and allow to equilibrate to room temperature in darkness. Once equilibrated, use double-sided tape (Figure 3B) to adhere the cover glass to the top of the slide, creating the chamber for experimentation. From both ends of the chamber add 10 µL double-distilled water. This will prevent epoxy in the next step from flowing into the chamber via capillary action and plugging the pre-drilled holes necessary for sample injection. Use 5-minute epoxy (Devcon®Home) glue around the four edges of the cover glass (Figure 3C) and allow to dry in darkness for 20 minutes, until epoxy is no longer tacky. Insert a 200 µL pipette tip into each of the pre-drilled holes and epoxy in place. Once dried, insert 1.0 mm tubing into one of the pipette tips until it can no longer be pushed in. Verify by eye that the tubing is facing down toward the slide chamber, then insert a small amount of epoxy into the pipette tip in order to glue the tubing in place, repeat for the other pipette tip (Figure 3D). Using a 1 mL syringe with a 0.5 inch, 26-gauge needle (PrecisionGlide™, BD) draw TE buffer into chamber to wash, repeat three times. Incubate chamber in 0.2 mg/mL neutravidin for 3 minutes. This provides the matrix to which biotinylated protein, or a DNA substrate, can tether. Rinse chamber 3 additional times with TE buffer to remove excess neutravidin. Clean slide exterior for use, with a small amount of ethanol on a kimwipe.
2.7. Assembly of the flow cell for the single-molecule sorting experiment
The “sorting” experiment is carried out in two steps. First, the biotinylated DNA helicase molecules are immobilized on the surface of the TIRFM cell and are incubated with the Cy3/Cy5-labeled substrate (DNA construct or a nucleoprotein complex). While invisible by itself, each immobilized helicase molecule yields an activity trajectory (time-based appearance and disappearance of the Cy3/Cy5 signals in a particular location on the slide, which corresponds to the location of the surface-tethered molecule). Each trajectory may contain tens of individual substrate binding/processing events. The second step is sorting of the posttranslationally modified molecules from unmodified enzymes. This step requires exchanging the fluorescently-labeled substrate for the fluorescently-labeled probe specific to the modification of interest (a specific antibody or a protein that interacts only with the modified form of the helicase) without altering the view of the flow cell. This is important, because the activity signal needs to be matched to the modification. Therefore, the experiment needs to be carried out in the flow cell. Data are collected in a flow experiment using the open-ended fluid chamber [14, 26] described previously. Using this method allows for collection of data while sample solutions are being exchanged.
2.8. Single-molecule oxygen scavenging system
In order to increase the life span of fluorophores in single-molecule experiments it is necessary to implement an oxygen scavenging system with the purpose of reducing reactive oxygen species (ROS). ROS interact with fluorophores in a manner that causes rapid photobleaching, thus it is desirable to decrease ROS presence in order to increase the time that data can be collected. To reduce the effects of ROS 12 mM Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid) and Gloxy (catalase and glucose oxidase solution) are used. Trolox is prepared in 10 ml batches and stored in 1 mL aliquots. To prepare Trolox, add 60 mg of Trolox powder to a 15 mL sterile tube. Add 10 mL of double-distilled water and before Trolox is completely dissolved add 60 µL of 2M NaOH. NaOH neutralizes the pH change induced by the addition of Trolox, increase solubility of the compound. Invert the tube to mix, then incubate the mixture on a rotating shaker at room temperature, under constant fluorescent light, for 3 days. After three days the solution will be yellow in color and cloudy. Filter the Trolox solution with a 0.22µm filter and store at 4°C in darkness. Trolox can be used for 1 month, or until noticeable color change can be detected by eye. Gloxy is prepared as a mixture of 4mg/mL catalase and 100mg/mL glucose oxidase. All steps of preparation must be carried out at 4°C. Add 40mg of catalase to 1mL of T50 buffer (10mM Tris-HCl pH 7.5, 50mM NaCl) and mix until catalase is dissolved. Add 10mg of glucose oxidase to 90uL T50 buffer and 10uL catalase solution. Gently mix and centrifuge for 1 minute at 1000 xg. Collect the supernatant and store at 4°C, storing for no longer than one month.
2.9. Single-molecule characterization of FBH1
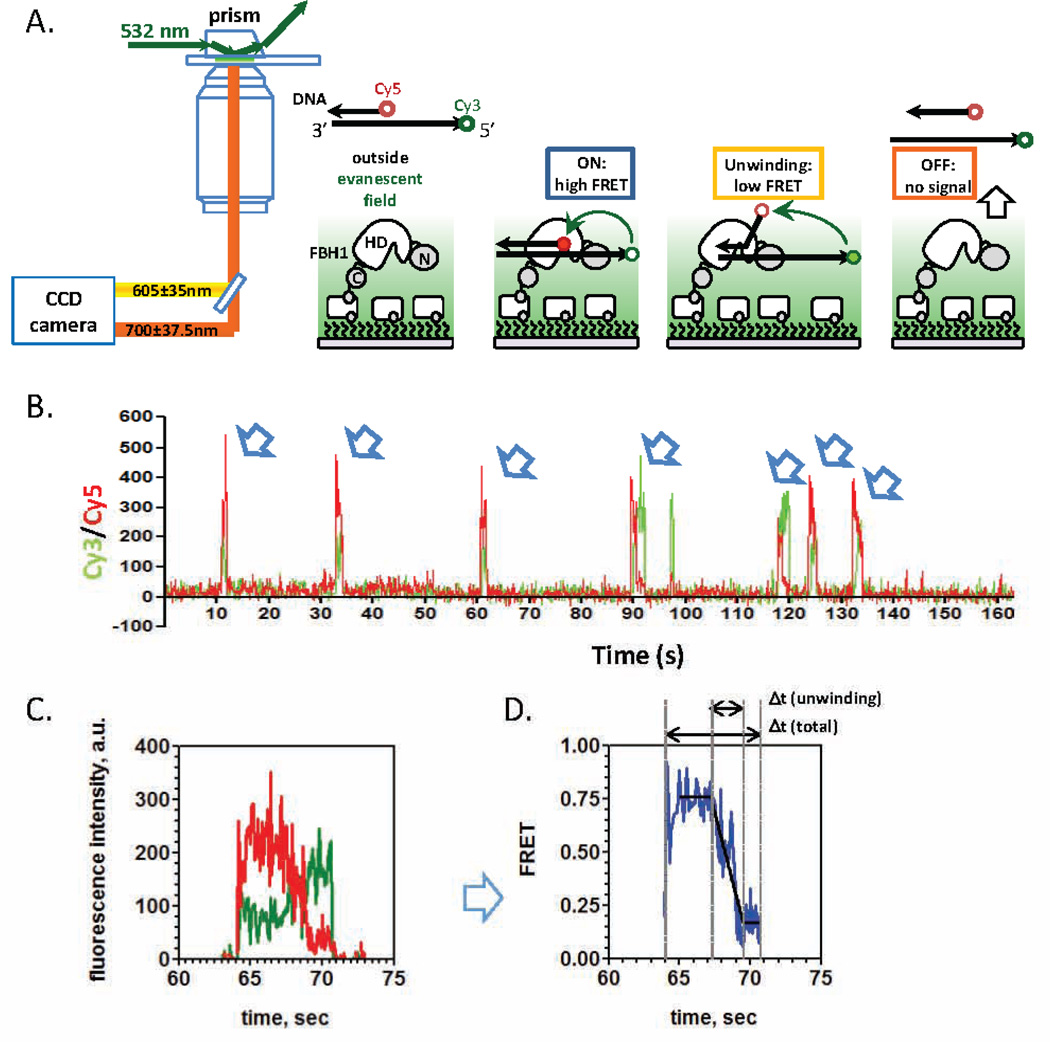
Biotinylated FBH1 is tethered to the passivated slide chamber and then treated with 50 pM Cy3 and Cy5 labeled partial duplex DNA in SM buffer supplemented with 1mM ATP (Figure 4A). Data collection begins immediately upon injection of the DNA substrate. No observable DNA binding should be detected in a control experiment in the absence of biotinylated FBH1. This important control insures that each activity trajectory in actual experiments originates from the surface-tethered helicase molecule and not from a “sticky” spot on the slide.
Figure 4. TIRFM to monitor FBH1 catalyzed DNA unwinding.
(A) Biotinylated FBH1 is surfaced-immobilized on a microscope slide via interaction with neutravidin. A partial duplex DNA substrate, containing Cy3/Cy5 fluorophores which can undergo FRET, is introduced into the chamber along with ATP. FBH1 can bind to the dually labeled substrate and then use the ATP to unwind the dsDNA in the 3’ to 5’ direction. (B) Representative DNA binding and unwinding trajectory from a single FBH1 molecule. The binding of the labeled partial duplex to the surface-tethered FBH1 results in the appearances of both Cy3 (green trace) and Cy5 fluorescence signals (red trace). Multiple DNA binding events are observed for the same FBH1 molecule as indicated by the arrows. (C) A close up view of one of the binding events show that after binding to the partial duplex, FBH1 can use ATP to unwind the DNA which results in anti-correlated changes in Cy3/Cy5 signals. (D) The changes in Cy3/Cy5 fluorescence intensities during FBH1 catalyzed DNA unwinding is converted into changes in FRET. DNA unwinding results in a monotonic decrease in FRET as the two strands become separate. The FRET trajectory is fit to a three segmented line to determine the total unwinding time for each DNA unwinding event. Modified from Ref.14. Direction of the arrow indicates 3’–5’ directionality of the DNA substrate.
2.10. Single-molecule data acquisition and analysis
IDL version 6.2 (Exelis Visual Information Systems, Boulder, Colorado) is used to extract trajectories from the raw video file. After the trajectories are extracted from the movie (Figure 4B), visually inspect all trajectories and then proceed to analysis. Scripts for extraction and trace analysis were developed and generously shared by the HA lab (download information provided in Appendix 1). For the simple binding events, a single-color measurement may be sufficient since the fluorescently labeled DNA binding to the surface-tethered protein would result in the appearance and disappearance of fluorescence signal. The individual ssDNA binding trajectories are fit to a two-state binding model using QuB [27, 28] software (University of Buffalo) from which the “on-states” and “off-states” for the individual binding events are determined. These values are binned (see below) and plotted as histograms using GraphPad Prism 6.0 software. In the case of a simple two-state model, these histograms are well described by a single-exponential decay function. Time constant obtained from fitting the distribution of the “on-times,” ton, indicate the average time the protein remains associated with the labeled DNA and is also inversely related to the dissociation rate constant, koff. The time constant determined analysis of the “off-times” distribution provides the average time between successive DNA binding events and is related to the on rate von. Since von is expected to be dependent on the free concentration of ssDNA, TIRFM experiments are performed at multiple DNA concentrations in order to extract the association rate constant, kon (M−1s−1) from the concentration dependence. The equilibrium dissociation constant can then be calculated as KD = koff/kon (M). An important consideration in such dwell-time analysis is the selection of a bin size. If the bin size is too small, the histograms appear noisy with too few events in each narrow bin, and their fit with an exponential function produces large Χ2. More importantly, the low boundary of the bin size selection should take into account the uncertainty in determining the dwell times set by the camera resolution. A large bin size, on the other hand, may yield the fits of seemingly good quality, but with the time constant for the exponential decay residing within the first bin, therefore resulting in erroneous rate constants. Several approaches can be applied to determine the optimal bin size for the ordinary dwell time distributions. Szoszkiewicz and colleagues, for example, proposed a procedure that iteratively increases the bin size and fits the histogram ordinate to determine the Χ2 [29]. With each iteration, the quality of the fit increases until Χ2 reaches a global minimum, the bin size corresponding to this global minimum is optimum. This approach works well for the large data sets of dwell times. Practically, and especially if the number of collected dwell times is limited, it is prudent to check that not only does the selected bin size results in an optimal Χ2, but also the rate constants determined from exponential fits do not change between the selected bin size and smaller bin sizes.
The data for Cy3 and Cy5 intensities for each “active” location on the slide are recorded in different channels. When the substrate is dually labeled with Cy3 and Cy5 and substrate rearrangement is taking place, the FRET efficiencies are calculated as the ratio of acceptor intensity (Cy5) to total intensity (sum of donor (Cy3) and acceptor (Cy5) intensities). The following equation describes the method by which FRET is calculated:
Where I*Cy3 and I*Cy5 are the background corrected donor and acceptor intensities, respectively. β is the measured leakage intensity from the donor to the acceptor channel. Determination of donor linkage requires a single-molecule trace in which the acceptor fluorophore photobleaches prior to the donor, any acceptor signal that remains after photobleaching is from donor excitation. In order to correct for this leaking signal, donor leakage is then subtracted from acceptor intensity and added back into donor intensity. Variable γ is the calculated ratio of the change in acceptor intensity (ΔICy5) to the change in donor intensity (ΔICy3) upon acceptor photobleaching (γ= ΔICy5/ ΔICy3), in the case of the Cy3/Cy5 FRET pair γ = 1 [26, 30]. Each dsDNA unwinding event is analyzed separately (Figure 4C). The FRET trajectories trimmed to the beginning and the end of the DNA binding/rearrangement event (Figure 4D), plotted in GraphPad Prism and fitted to three segment line, where the first and the third segment are parallel to the x-axis and represent the times before and after the active unwinding of the substrate. The second segment represents the actual duplex unwinding phase. More complex FRET trajectories may be observed when the helicase shuttles on the DNA substrate [14, 31, 32].
2.11. Single-molecule sorting of post-translationally modified FBH1
Bio-FBH1 is tethered to the neutravidin-coated chamber. All experiments are carried out in SM buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 10% glycerol, 1mg/mL Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid), 1mg/mL glucose oxidase, 0.4% D-Glucose, 1% v/v 2-mercaptoethanol, and 0.04 mg/mL catalase (Calbiochem). All chemicals sourced from Sigma-Aldrich unless otherwise stated. 100 pM bioFBH1 is added to the slide, and 100 pm – 1 nM Cy3 and Cy5 labeled DNA substrate is used for all experiments in the presence/absence of 1mM MgCl2 and 1mM ATP. FBH1 activity is assayed by flowing fluorescently labeled DNA substrate (Figure 5A) or RAD51-ssDNA nucleoprotein filament (Figure 5B) into the chamber. To detect post-translationally ubiquitylated FBH1, TMR-TUBE2 is used to bind the ubiquitylated species of FBH1. Commercially available (LifeSensors) TUBE2 is labeled with a single-TAMRA fluorophore (λEM, MAX = 540nm, λEX, MAX = 578nm) attached to the 6× His tag of the TUBE2 tandem ubiquitin binding domains. The advantage of this reagent is that it is commercially available in its labeled form, and it binds poly-ubiquitin chains with high specificity and affinity. Other reagents that can be used in place of TUBE2 are antibodies specific for a particular type of ubiquitin chain. Exchange 1:5000 TMR-TUBE2 with the DNA solution and incubate for 5 minutes. This dilution of TMR-TUBE2 was determined by testing a range of dilutions and corresponds to the point at which lower concentration yields underestimation of ubFBH1 and increased concentration yields increased background fluorescence, but does not increase the fraction of the TMR-TUBE positive trajectories. Free TUBE2 is removed via wash with SM buffer. Trajectories of ub-FBH1 are distinguished from unmodified FBH1 by the presence of fluorescence of the TAMRA fluorophore. TMR-TUBE2 selectivity is tested by flowing 1:5000 TMR-TUBE2 into a chamber in the absence of FBH1 or in the presence of non-ubiquitylated FBH1. There should be no non-specific surface tethering of TUBE2 seen in the chamber, as indicated by no TMR fluorescent signal. Similarly, DNA substrate specificity is examined by flowing 100pM-1nM of DNA substrate into a single-molecule chamber that does not have FBH1 present. There should be no fluorescent signal produced by DNA in the absence of FBH1.
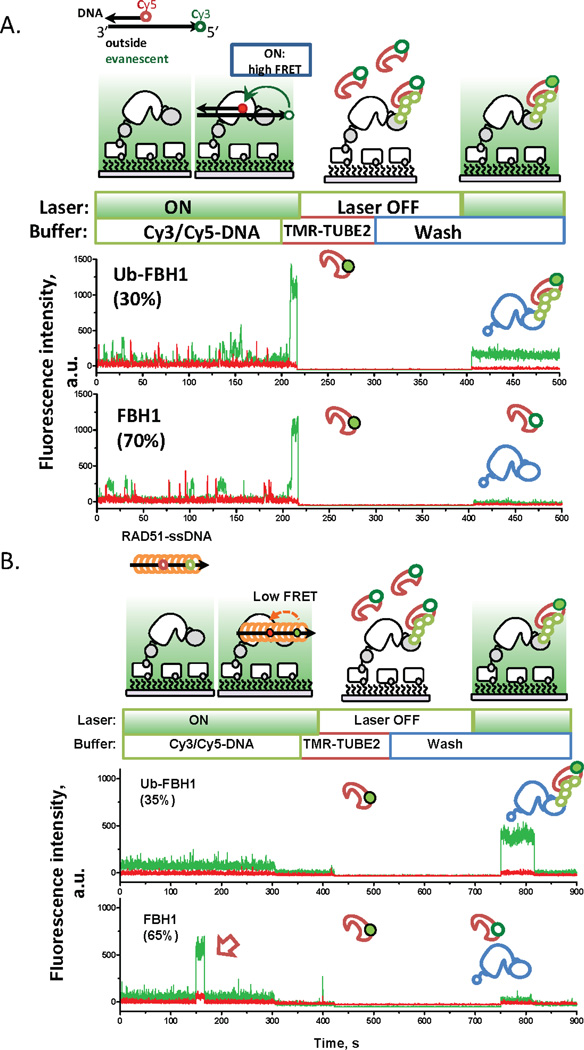
Figure 5. Single-molecule sorting experiment using FBH1.
(A) DNA unwinding assays are performed with surface immobilized FBH1. After the experiment, the laser is turned off, and the slide is washed and TMR-TUBE2 is introduced into the chamber. Excess TMR-TUBE2 is washed away and the laser is turned back on. When TMR-TUBE2 binds to poly-ubiquitylated FBH1 on the surface, fluorescence signal from the green laser excitation is recovered. If the FBH1 molecule is not modified, then no fluorescence signal was recovered after washing. (B) A pre-formed RAD51 filament using a Cy3/Cy5 labeled ssDNA oligo is added to surface immobilized FBH1. When FBH1 binds to the nucleoprotein filament, the simultaneous appearance of Cy3 and Cy5 signals were observed. Similar to panel A, the laser was switched off after the experiment and TMR-TUBE2 was introduced into the slide chamber. The chamber was then gently washed to remove excess TMR-TUBE2 and the laser was turned back on. The recovery of fluorescence signal indicates the presence of a poly-ubiquitylated FBH1 molecule bound to TMR-TUBE2. Interestingly, ubiquitylated FBH1 failed to bind a RAD51 filament while non-modified FBH1 molecules bound to the preassembled nucleoprotein filament. Modified from Ref.14. Direction of the arrow indicates 3’–5’ directionality of the DNA substrate.
Due to the tethered protein not bearing a fluorescent label, photobleaching is not a primary concern in this experiment, particularly in experiments where binding events are on the order of a few seconds. Previous studies that used conditions described in this section, have shown that Cy3 and Cy5 fluorophores photobleach on the order of several minutes [33], indicating that dwell time estimations are expected to be free from photobleaching effects. In order to verify that photobleaching effects are not contributing to experimental data, the experiments are performed at several different laser intensities. Binding constants independent of laser intensity indicate that photobleaching does not contribute to experimental signal.
In the single-molecule sorting experiments, the activity of FBH1 is assayed first, following by the TMR-TUBE visualization of the ubiquitylated molecules. The reverse order of the experiment may result in TMR-TUBE remaining associated with the helicase and interfering with the fluorescent signal and/or activity. Harsher washing conditions that insure the TMR-TUBE dissociation may inactive the helicase.
2.12. Single-molecule sorting of post-translationally phosphorylated FANCJ
Dually labeled FANCJ (FLAG tagged and also biotinylated) is produced as described for FBH1. Fifty pM bioFANCJ in SM buffer is immobilized on a passivated microscope slide (Figure 6A). The positions of the individual bioFANCJ molecules on the slide surface are determined by flowing in 5 µM monoclonal anti-FLAG M2-Cy3 antibody (Sigma Aldrich). Similarly, Cy5-labeled BRCA1 BRCT domain is generated and used to identify phosphorylated FANCJ molecules (FANCJ-P) on the surface as follows. BRCT is readily expressed and purified from E. coli. Pure BRCT is labeled on the N-terminus using a Cy5 NHS ester dye pack (GE Healthcare) which labels primary amines. To selectively label the N-terminus and not a lysine residue, the reaction was carried out in 50 mM K2HPO4/KH2PO4 at pH 7.0, 1 M NaCl, 0.1 M DTT, 10% glycerol [34].
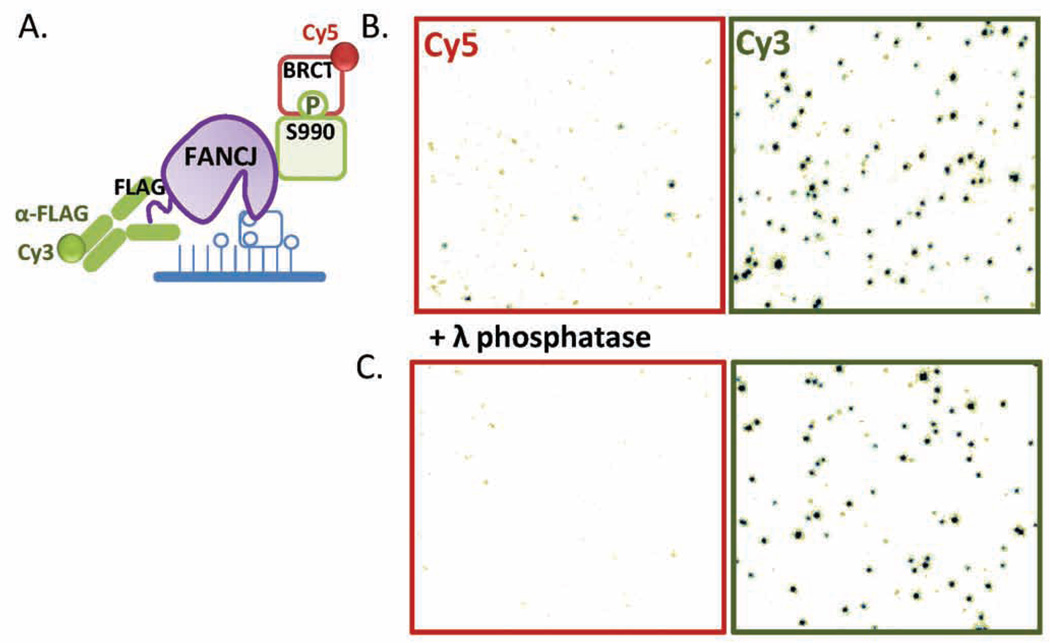
Figure 6. Single-molecule sorting experiment using FANCJ.
(A) Purified biotinylated FANCJ is immobilized on a microscope slide for single-molecule imaging. The tethered pool of FANCJ molecules is expected to contain both non-modified FANCJ as well as FANCJ that is phosphorylated on serine 990. The positions of all FANCJ molecules on the slide surface is determined using Cy3-anti-FLAG antibody while the locations of phosphorylated FANCJ (FANCJ-P) on the surface are identified with Cy5-BRCT, which binds to FANCJ-P with high affinity. (B) A camera field of view separated into a channel monitoring Cy3 fluorescence and a channel monitoring Cy5 fluorescence. The spots on the Cy3 channel correspond to individual surface-immobilized FANCJ molecules as a result of Cy3-antiFLAG binding, while the spots from the Cy5 channel indicate Cy5-BRCT binding to FANCJ-P. The co-localization of the two signals are used as a diagnostic for determining the locations of FANCJ-P on the slide. (C) FANCJ was pre-treated with λ phosphatase before it was immobilized on the slide surface. The same sorting experiment was carried out and the appearances of Cy5-BRCT loci (ie. FANCJ-P) was dramatically reduced.
A sample camera field of view of a single-molecule sorting TIRFM experiment is shown in (Figure 6B) where the same image is separated into two corresponding channels monitoring either Cy3 or Cy5 fluorescence. Each spot observed in the Cy3 channel indicate the binding of Cy3-antiFLAG on a surface-immobilized FANCJ monomer, while the Cy5 foci reflect Cy5-BRCT binding to phosphorylated FANCJ-P. The co-localization of the two signals is used as a diagnostic tool to identify FANCJ-P. The slide was then washed extensively with SM buffer containing 0.5 M NaCl followed by equilibration in SM buffer to remove the fluorescent probes. DNA binding or other activity assays using Cy3-labled substrates are then carried out with the entire pool of FANCJ molecules and the measured activities are correlated with either the presence or absence of FANCJ phosphorylation. When FANCJ is pre-treated with λ phosphatase before surface-immobilization, any phosphate group conjugated to FANCJ-P is expected to be removed. Indeed, when single-molecule sorting experiment is performed with this FANCJ sample, the number of Cy5 foci (ie: FANCJ-P molecules) is dramatically reduced while the number of Cy3 foci remains the same on average (Figure 6C).
3. Conclusion
Mammalian DNA repair helicases act within a robust adaptive network, in which a stochastic combination of overlapping and competing enzymatic activities and interactions ensures repair or bypass of the genotoxic lesions. Post-translational modifications may rapidly activate or inactivate these helicases and alter their interactions. Multiple PTMs may compete, appear co-dependent, or may be applied in a combinatorial manner. Subpopulations of a DNA helicase may be channeled into different pathways guided by PTMs and molecular associations, and multiple DNA repair pathways may compete for the same motor components. Deconvoluting the resulting networks requires novel methodologies capable of separating, quantifying, and evaluating subpopulations of a DNA helicase within heterogeneous cellular pools. The single-molecule sorting described in this chapter is an example of an emerging experimental strategy created for this task. Following a transient transfection, the helicase (or another protein of interest) is expressed and biotinylated in human cells and therefore contains native posttranslational modifications whose presence and ratios correlate to the conditions under which the protein is expressed (e.g. in the presence or absence of the DNA damaging agents). Incorporation of the biotin moiety is concomitant with the protein expression. The helicase molecules pulled from the cell can be therefore immediately surface-tethered for the single-molecule TIRFM investigation. Examining the activity trajectories originating from modified and unmodified enzymes allows us (i) to quantify the fraction of modified protein in the cell, and (ii) assess the effect of each modification or a combination of modifications on the target activity. The presence of unmodified protein allows for the internal control for reproducibility of the experimental outcomes. While we focused this chapter on the experimental system for the analysis of polyubiquitylated FBH1 and phosphorylated FANCJ helicases, any posttranslational modification can be analyzed in a similar fashion. The single-molecule sorting can be further extended to quantify and analyze multiprotein complexes containing the helicase and its stably bound protein partners.
Highlights.
Posttranslational modifications alter activities and interactions of DNA helicases
Helicases with native modifications are produced and biotinylated in human cells
Single-molecule analyses is performed on surface-tethered helicases
Single-molecule sorting separates activities of modified and unmodified helicases
Relative abundance of the modification is quantified
Acknowledgments
This work was supported by the National Institutes of Health NIH R01 GM101167 to M.S. and American Cancer Society Postdoctoral Fellowship PF-11-243-01-DMC to C.G.W
Abbreviations
- ATP
adenosine triphosphate
- BAP
biotin acceptor peptide
- bio
biotinylated
- BLM
Bloom’s syndrome DNA helicase
- BRC1
Breast cancer type 1 protein
- BRCT
BRCA1 C-terminus
- DMEM
Dulbecco’s modified eagle medium
- DNA
deoxyribonucleic acid
- ds
double-stranded
- DPBS
Dubelcco’s phosphate buffered saline
- DPSS
Diode pumped solid state
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EMCCD
electon multiplying charge coupled device
- FANCJ
Fanconi anemia complementation group J protein
- FBH1
F-box containing helicase 1
- FBS
fetal bovine serum
- FRET
Förster resonance energy transfer
- HEK
human embryonic kidney
- Hepes
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- His
histidine
- NHS
N-hydroxysuccinimide esters
- -P
phosphorylated
- PEG
polyethylene glycol
- PEI
polyethylenimine
- PMSF
phenylmethylsulfonyl fluoride
- PTM
post-translational modification
- RAD51
radiation sensitive protein 51
- ROS
reactive oxygen species
- rpm
revolutions per minute
- SiMPull
single-molecule pull down
- sm
single-molecule
- ss
single-stranded
- SUMO
small ubiquitin-like modifier
- TE
Tris and EDTA
- TIRF
total internal reflection fluorescence
- TIRFM
total internal reflection fluorescence microscopy
- Trolox
6-hydroxy-2-5-7-8-tetramethylchroman-2-carboxylic acid
- TUBE
tandem ubiquitin binding entities
- ub
ubiquitylated
Appendix 1
General Supplies
| Fluorescently Labeled Oligonucleotides: |
| Integrated DNA Technologies, Inc. |
| Cy3/Cy5 Labeled Oligonucleotides |
| Eurofins: |
| Cy3/ 5’ Cy5 |
| Single-molecule Slide Chambers: |
| Chemglass Life Sciences |
| 25.0mm × 75.0mm × 1.0mm quartz slides |
| Fisher Scientific |
| 24 × 60 × 1.5 borosilicate cover glass |
| 3M |
| Double-sided tape |
| ITW Devcon |
| 5-minute epoxy |
| Single-molecule TIRF Microscope: |
| Chroma |
| Newport |
| Nikon |
| Olympus |
| Semrock |
| Thorlabs |
| Zeiss |
| EM-CCD Camera: |
| Andor |
| Software: |
| Microsoft |
| Visual C++ |
| ITT Visual Information Solutions |
| IDL |
| The MathWorks, Inc. |
| MatLab |
| Graphpad Prism Software, Inc. |
| GraphPad Prism 6.0 |
| Single-Molecule FRET data acquisition and analysis suite |
| https://cplc.illinois.edu/software |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 2.Xue Q, Yeung ES. Differences in the chemical reactivity of individual molecules of an enzyme. Nature. 1995;373(6516):681–683. doi: 10.1038/373681a0. [DOI] [PubMed] [Google Scholar]
- 3.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nature reviews. Molecular cell biology. 2006;7(7):473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 4.Jensen ON. Interpreting the protein language using proteomics. Nature reviews. Molecular cell biology. 2006;7(6):391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 5.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nature reviews. Molecular cell biology. 2008 doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 6.Wu CG, Spies M. Overview: What Are Helicases? Adv Exp Med Biol. 2013;973:1–16. doi: 10.1007/978-1-4614-5037-5_1. [DOI] [PubMed] [Google Scholar]
- 7.Brosh RM., Jr DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer. 2013;13(8):542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohm S, Bernstein KA. The role of post-translational modifications in fine-tuning BLM helicase function during DNA repair. DNA Repair (Amst) 2014;22:123–132. doi: 10.1016/j.dnarep.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, Bartek J, Lukas J, Mailand N. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. The Journal of cell biology. 2009;186(5):655–663. doi: 10.1083/jcb.200812138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osman F, Dixon J, Barr AR, Whitby MC. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Molecular and cellular biology. 2005;25(18):8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, Iwasaki H, Shinagawa H. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Molecular and cellular biology. 2005;25(18):8074–8083. doi: 10.1128/MCB.25.18.8074-8083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiolo I, Saponaro M, Baryshnikova A, Kim JH, Seo YS, Liberi G. The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles. Molecular and cellular biology. 2007;27(21):7439–7450. doi: 10.1128/MCB.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenz A, Osman F, Folkyte V, Sofueva S, Whitby MC. Fbh1 limits Rad51-dependent recombination at blocked replication forks. Molecular and cellular biology. 2009;29(17):4742–4756. doi: 10.1128/MCB.00471-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda-Ozawa T, Hoang T, Seo YS, Chen LF, Spies M. Single-molecule sorting reveals how ubiquitylation affects substrate recognition and activities of FBH1 helicase. Nucleic acids research. 2013;41(6):3576–3587. doi: 10.1093/nar/gkt056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105(1):149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 16.Botuyan MV, Nomine Y, Yu X, Juranic N, Macura S, Chen J, Mer G. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure. 2004;12(7):1137–1146. doi: 10.1016/j.str.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11(6):512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 18.Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell. 2004;14(3):405–412. doi: 10.1016/s1097-2765(04)00238-2. [DOI] [PubMed] [Google Scholar]
- 19.Rabuka D, Rush JS, deHart GW, Wu P, Bertozzi CR. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat Protoc. 2012;7(6):1052–1067. doi: 10.1038/nprot.2012.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aggarwal V, Ha T. Single-molecule pull-down (SiMPull) for new-age biochemistry: methodology and biochemical applications of single-molecule pull-down (SiMPull) for probing biomolecular interactions in crude cell extracts, BioEssays : news and reviews in molecular. cellular and developmental biology. 2014;36(11):1109–1119. doi: 10.1002/bies.201400090. [DOI] [PubMed] [Google Scholar]
- 21.Jain A, Liu R, Xiang YK, Ha T. Single-molecule pull-down for studying protein interactions. Nat Protoc. 2012;7(3):445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30(2):E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kichler A. Gene transfer with modified polyethylenimines. J Gene Med. 2004;6(Suppl 1):S3–S10. doi: 10.1002/jgm.507. [DOI] [PubMed] [Google Scholar]
- 24.Honda M, Park J, Pugh RA, Ha T, Spies M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol Cell. 2009;35(5):694–703. doi: 10.1016/j.molcel.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasnik I, McKinney SA, Ha T. Surfaces and orientations: much to FRET about? Acc Chem Res. 2005;38(7):542–548. doi: 10.1021/ar040138c. [DOI] [PubMed] [Google Scholar]
- 26.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milescu LS, Nicolai C, Bannen J. QuB Software. 2000–2013 [Google Scholar]
- 28.Nicolai C, Sachs F. SOLVING ION CHANNEL KINETICS WITH THE QuB SOFTWARE. Biophysical Reviews and Letters 08(03n04) 2013:191–211. [Google Scholar]
- 29.Szoszkiewicz R, Ainavarapu SR, Wiita AP, Perez-Jimenez R, Sanchez-Ruiz JM, Fernandez JM. Dwell time analysis of a single-molecule mechanochemical reaction. Langmuir : the ACS journal of surfaces and colloids. 2008;24(4):1356–1364. doi: 10.1021/la702368b. [DOI] [PubMed] [Google Scholar]
- 30.Joo C, Ha T. Single-molecule FRET with total internal reflection microscopy. Cold Spring Harb Protoc. 2012;2012(12) doi: 10.1101/pdb.top072058. [DOI] [PubMed] [Google Scholar]
- 31.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437(7063):1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 32.Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghoneim M, Spies M. Direct Correlation of DNA Binding and Single Protein Domain Motion via Dual Illumination Fluorescence Microscopy. Nano Letters. 2014;14(10):5920–5931. doi: 10.1021/nl502890g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443(7113):875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]