ABSTRACT
Analysis of gene expression and whole-genome features of 64 human epidermal growth factor 2 (HER2)-positive breast tumors supports the idea that their intrinsic heterogeneity actually reflects their cell of origin, suggesting that HER2 amplification is an embedded event in the natural history of these tumors. Possible mechanisms for this event involve breakage-fusion-bridge and chromothripsis.
KEYWORDS: Breast cancer, Breakage-Fusion-Bridge, cancer genomics, ERBB2 amplification, mammary epithelial hierarchy
Human epidermal growth factor 2-positive (HER2+) breast cancers (BC) represent a clinically well delineated group of tumors, characterized by amplification and overexpression of the erb-b2 receptor tyrosine kinase 2 gene (ERBB2, also known as HER2) on chromosome arm 17q. This feature makes them amenable to efficient anti-HER2 targeted therapies (e.g., trastuzumab), which are now routinely used in patients with HER2+ tumors and have completely changed the outcome of HER2+ BC over the last 2 decades.1 However, the situation is strikingly different on the biological side. This group of tumors is now well recognized as being highly heterogeneous from different, but related, standpoints, including expression subtypes,2 DNA alterations,3 immune microenvironment,4 and response to therapy.5 In this context, and in the framework of the International Cancer Genome Consortium (ICGC), we undertook a comprehensive study of 99 HER2+ primary invasive carcinomas, including whole-genome sequencing (WGS) of 64 tumor–normal paired samples.6 This provided further insight into their genomic architecture using high dynamic copy number analysis and detection of large-scale rearrangements.
First, we used array-based gene expression as an operational basis to classify HER2+ tumors into 4 groups (A to D in Fig. 1), which were further characterized in terms of interdependent biological and genomic variables. Groups A and B included most ER-positive (ER+) tumors, whereas groups C and D mostly contained ER-negative (ER-) tumors. In terms of PAM50 intrinsic subtypes, the tumors were predominantly luminal B (in groups A and B) and HER2-enriched (in groups C and D), with only a marginal number of luminal A and basal tumors. These four groups displayed specific genomic alterations in terms of mutations, amplifications, and rearrangements. All samples in group D and none in group A displayed mutations in the tumor protein p53 gene (TP53). Conversely, only one sample in group D harbored a mutation in phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α gene (PIK3CA) whereas the other groups displayed an equal number of such mutations. A similar gradient, from A to D, was also observed in terms of genomic (homologous recombination deficiency-associated genomic instability, fraction of genome altered) and cell of origin transcriptomic signatures. Group D displayed more genomic instability and a progenitor luminal signature whereas group A was more stable and displayed a typical mature luminal signature. All these observations are concordant with the cell of origin scheme,7,8 in which the intertumoral heterogeneity reflects the developmental stage of the epithelial mammary cells. Thus, groups D and C, with basal-like features, arise from luminal progenitors whereas groups A and B, with typical luminal features, are associated with mature, differentiated luminal cells. This suggests that ERBB2 amplification, although probably strongly selected, is an embedded event that is superimposed on the standard time course of breast carcinogenesis.
Figure 1.
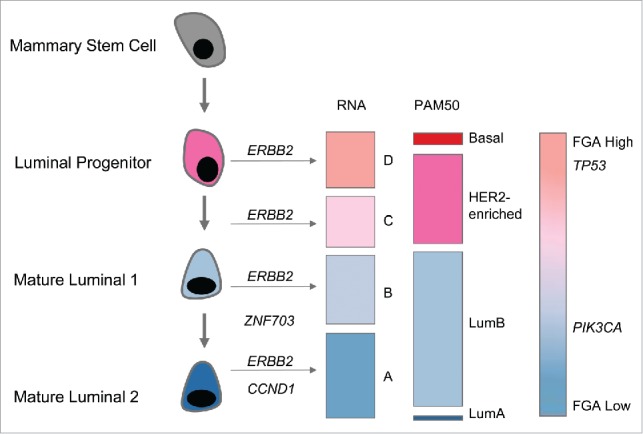
Mammary epithelial hierarchy and genomic features of epidermal growth factor 2-positive breast cancer. From left to right: developmental stage of normal epithelial mammary cells; amplification events: erb-b2 receptor tyrosine kinase 2 (ERBB2), cyclin D1 (CCND1), and zinc finger protein 703 (ZNF703); RNA expression groups (A, B, C, and D); PAM50 intrinsic subtypes (luminal A, luminal B, HER2-enriched, and basal); genomic features: fraction of genome altered (FGA); mutations in tumor protein p53 (TP53) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α (PIK3CA).
WGS data allowed us to gain more information about the amplification process itself and provided some indications about how (and maybe when) it arose. To this purpose, whole-genome paired end sequencing provides 2 important experimental clues: (1) dynamic and high-resolution analysis of copy numbers; and (2) the ability to pinpoint large-scale structural rearrangements using clipping and abnormal mapping of read pairs. We could show that, in several cases, the observed sequence of copy numbers as well as the orientation of clipped reads was consistent with a breakage-fusion-bridge (BFB) folding mechanism.9 However, the observation of long distance and inter-chromosomal rearrangements further showed that the amplification is a complex event (or sequence of events), likely involving multiple amplicons on the same or different chromosomes and several intertwined mechanisms. Indeed, one of the features of HER2+ tumors is the ubiquitous presence of firestorms, corresponding to multiple closely spaced amplicons on highly rearranged chromosomal arms.10 It is therefore tempting to explain the complex amplification patterns observed by combining 2 mechanisms: chromothripsis, which will generate a mosaic of fragments (but no amplification per se), followed by a BFB amplification of chromosomal arm(s). When limited within the 17q arm, this gives rise to the observed co-amplification of the 17q12 region (ERBB2) with 17q21 and 17q23. These 17q co-amplifications are more frequent in group A. It is important to note that here the term “co-amplification” does not only refer to the co-occurrence of amplicons but also denotes that rearrangement breakpoints (detected by abnormal mapped reads) are observed between them, suggesting they arose from a common molecular process. When involving other chromosomes, this gives rise to important co-amplifications with cyclin D1 (CCND1) on 11q13, zinc finger protein 703 (ZNF703) on 8p12, and v-myc avian myelocytomatosis viral oncogene homolog (MYC) on 8q24. Both CCND1 and ZNF703 co-amplifications are more frequently observed in ER+ tumors (groups A and B), whereas MYC co-amplifications seem to be ubiquitous.
One can speculate that combining phenotypic (such as gene expression groups) and mechanistic (such as co-amplifications) features may improve our current classification of HER2+ BCs and lead to rational therapeutic strategies by targeting additional pathways and/or genes co-amplified with ERBB2. This is of course important for patients who show an initial poor response or exhibit resistance to HER2-targeted drugs. From this latter point of view, a critical aspect that was not addressed in this first paper is intra-tumoral heterogeneity. This is clearly a question that should be tackled in the near future.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work could not have been done without the impulse and scientific insight of Gilles Thomas, who left us in February 2014. All of the authors remember him.
References
- 1.Slamon D, Eiermann W, Robert N, Pienkowski T, Martín M, Press M, Mackey J, Glaspy J, Chan A, Pawlicki M, et al.. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med 2011; 365:1273-83; PMID:21991949; http://dx.doi.org/ 10.1056/NEJMoa0910383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prat A, Carey LA, Adamo B, Vidal M, Tabernero J, Cortés J, Parker JS, Perou CM, Baselga J. Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast cancer. J Natl Cancer Inst 2014; 106:dju152; PMID:25139534; http://dx.doi.org/ 10.1093/jnci/dju152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staaf J, Jönsson G, Ringnér M, Vallon-Christersson J, Grabau D, Arason A, Gunnarsson H, Agnarsson BA, Malmström P-O, Johannsson OT, et al.. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Res 2010; 12:R25; PMID:20459607; http://dx.doi.org/ 10.1186/bcr2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchini G, Pusztai L, Pienkowski T, Im Y-H, Bianchi GV, Tseng L-M, Liu M-C, Lluch A, Galeota E, Magazzù D, et al.. Immune modulation of pathologic complete response after neoadjuvant HER2-directed therapies in the NeoSphere trial. Ann Oncol 2015; 26:2429-36; PMID:26387142; http://dx.doi.org/ 10.1093/annonc/mdv395 [DOI] [PubMed] [Google Scholar]
- 5.Carey LA, Berry DA, Cirrincione CT, Barry WT, Pitcher BN, Harris LN, Ollila DW, Krop IE, Henry NL, Weckstein DJ, et al.. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol 2015; 34:542-9; PMID:26527775; http://dx.doi.org/ 10.1200/JCO.2015.62.1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari A, Vincent-Salomon A, Pivot X, Sertier A-S, Thomas E, Tonon L, Boyault S, Mulugeta E, Treilleux I, MacGrogan G, et al.. A whole-genome sequence and transcriptome perspective on HER2-positive breast cancers. Nat Commun 2016; 7:12222; PMID:27406316; http://dx.doi.org/ 10.1038/ncomms12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev 2014; 28:1143-58; PMID:24888586; http://dx.doi.org/ 10.1101/gad.242511.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skibinski A, Kuperwasser C. The origin of breast tumor heterogeneity. Oncogene 2015; 34:5309-16; PMID:25703331; http://dx.doi.org/ 10.1038/onc.2014.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenman CD, Pleasance ED, Newman S, Yang F, Fu B, Nik-Zainal S, Jones D, Lau KW, Carter N, Edwards PAW, et al.. Estimation of rearrangement phylogeny for cancer genomes. Genome Res 2012; 22:346-61; PMID:21994251; http://dx.doi.org/ 10.1101/gr.118414.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al.. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 2006; 16:1465-79; PMID:17142309; http://dx.doi.org/ 10.1101/gr.5460106 [DOI] [PMC free article] [PubMed] [Google Scholar]


