ABSTRACT
The Suv39h lysine methyltransferases, known as key enzymes responsible for histone H3 lysine 9 methylation, are critical for heterochromatin protein 1 enrichment at constitutive heterochromatin. Our recent findings reveal a new role for the Suv39h1 paralog that links it to SUMO pathway function at constitutive heterochromatin.
KEYWORDS: Heterochromatin, HP1, nuclear organization, SUMO pathway, suv39h1
Heterochromatin has long been considered an inert part of the genome, merely gathering repetitive sequences. The last 10 y of research in chromatin and epigenetics have highlighted the importance of heterochromatin features for transcriptional regulation, nuclear organization, and genomic stability in a variety of species. In particular, histone marks characteristic of constitutive heterochromatin such as histone H3 lysine 9 methylation (H3K9me3) established by distinct “writers” (i.e., the Suv39h lysine methyltransferases [KMTs] in mammals) that protein “readers” (such as heterochromatin protein 1 [HP1]) can recognize provide a paradigm for a mechanism involved in and/or associated with a plethora of genomic functions affecting cell physiology and homeostasis. Notably, these functions include global genomic stability/heritability over cellular division, with a key role at the centromere and telomeres, developmental programs and cell fate, aging and senescence, and cancer.1-4 Whether these functions originate from the same or distinct features of constitutive heterochromatin, such as solely transcriptional repression or nuclear organization, remains to be documented. It therefore became of the utmost importance to decipher how the dynamics of heterochromatin organization are controlled and how they can be linked to changes in cellular functions during development and most critically in the context of human health. A logical approach has been to focus on the already well-identified players Suv39h, H3K9me3, and HP1 that contribute to various levels of heterochromatin organization. The Suv39h KMTs5 ensure H3K9me3 marks at nucleosomes in heterochromatin, which in turn provide binding/anchoring sites for the HP1 proteins; the capacity of Suv39h to bind HP1 further contributes to propagation of methylation of H3K9 on neighboring nucleosomes. Once an initiating site has been established, this latter “self-sustaining” loop provides a simple maintenance system for constitutive heterochromatin at specific genomic loci.6
Using the constitutive heterochromatin at pericentric domains (chromosomal domains flanking the microtubule attachment sites of the sister chromatids during mitosis) in mouse cells as a model, our previous studies showed that the SUMO pathway is involved in the initiation of this loop. We identified that the HP1α isoform was targeted at these domains when post-translationally modified by sumoylation, even in the absence of Suv39h-dependent H3K9me3. Importantly, we revealed a specific interaction between SUMO-HP1α and pericentric RNA transcripts7 (Fig. 1). Interestingly, in addition to sumoylation, desumoylation proved to be equally important as another means of regulation. In this respect, the SUMO-specific protease SENP7 localized at pericentric heterochromatin is a likely candidate, in particular given its ability to bind to HP1 proteins8 and its reported involvement in tumorigenesis.9
Figure 1.
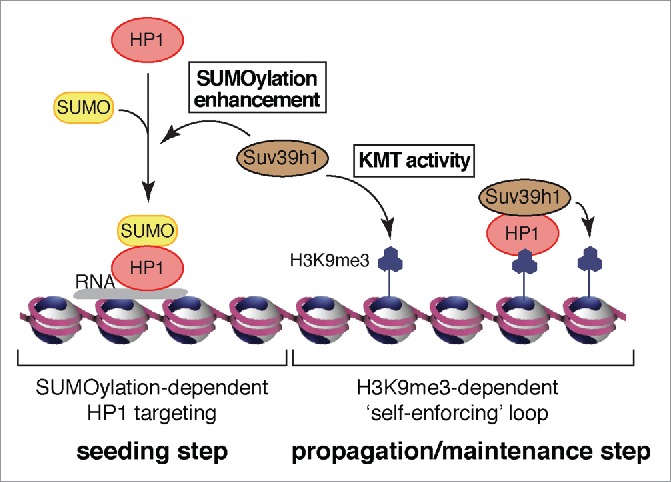
Dual function of Suv39h1 in constitutive heterochromatin dynamics. Suv39h1 enhances HP1α sumoylation and promotes its targeting to pericentric domains independently of H3K9me3 (seeding step). Suv39h1 KMT activity promotes H3K9me3, providing binding sites for HP1α and ensuring H3K9me3-dependent maintenance of constitutive heterochromatin (propagation/maintenance step).
Therefore, 2 steps emerge as critical in the organization of constitutive heterochromatin: a “seeding” step requiring HP1α sumoylation and a “propagation/maintenance” step involving H3K9me3. This sequential mechanism provides a molecular basis for both the establishment and maintenance of pericentric constitutive heterochromatin (Fig. 1). In this scheme, on one hand the SUMO pathway acting early would be most critical for de novo establishment or major heterochromatin rearrangements associated with a transition in status. On the other hand, the propagation/maintenance step would secure genomic loci already established in constitutive heterochromatin, or perhaps in some cases amplify a pre-existing marking. The concerted action of both steps thus represents a means to confer or restrict “plasticity” of constitutive heterochromatin to adapt to cell type, physiology, and environmental stimuli. Misregulation and/or loss of coordination in the functioning of these 2 distinct steps could easily lead to abnormal cellular phenotypes as encountered in pathologic situations and exemplified in cancer cells. Considering that methylation status can reflect the metabolic status of the cell whereas sumoylation is rather independent of this aspect, the challenge was to understand how coordination of these 2 branches enabled proper control of heterochromatin organization.
In our recent article published in Nature Communications,10 we demonstrated that the Suv39h1 paralog (also known as KMT1A) enhances HP1α sumoylation both in vivo in a cellular system and in vitro. We identified a region in Suv39h1 responsible for promoting HP1α sumoylation that is distinct from the H3K9 methyltransferase domain. We further precisely delineated the binding domain for the SUMO E2 conjugating enzyme Ubc9 (also known as UBE2I). Of note, this region in Suv39h1 is distinct from the canonical RING (really interesting new gene)-type or HECT (homologous with E6-associated protein C-terminus)-type E3 ligases and harbors no obvious homology to any of the currently characterized SUMO E3 ligases. Future work should address how sumoylation is promoted via Suv39h1 and whether Suv39h1 can be considered to represent a novel SUMO E3 ligase, opening up the possibility of a whole new family. Importantly, we also find that intact binding to Ubc9 is critical for a mini-Suv39h1 to promote efficient targeting of HP1α at pericentric domains. In conclusion, our results not only identify a new control mechanism operating at the level of HP1α sumoylation but, most remarkably, also show that this involves a major histone-modifying enzyme Suv39h1. The integration within a single enzyme, Suv39h1, of both control of sumoylation of HP1 and the capacity to ensure H3K9me3, addresses the requirements to coordinate both seeding and maintenance steps in organizing heterochromatin (Fig. 1). Future work should address how Suv39h1-dependent HP1α sumoylation is itself regulated and, most importantly, whether this novel activity of Suv39h1 and its canonical KMT activity can also explain the series of observations in conditions where suv39h1 has been knocked down or knocked out.
In conclusion, by linking Suv39h1 activity as a histone methyltransferase with the SUMO pathway, we provide a new conceptual framework for the dynamics and the stability of constitutive heterochromatin, paving the way for exciting work in the field of nuclear organization and its role during normal development and pathologic situations.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by la Ligue Nationale contre le Cancer (Equipe labellisée Ligue), the European Commission Network of Excellence EpiGeneSys (HEALTH-F4-2010-257082), ERC Advanced Grant 2009-AdG_20090506 “Eccentric,” ANR-11-LABX-0044_DEEP, ANR “Epicure” ANR-14-CE16-0009 and ANR-10-IDEX-0001-02 PSL.
References
- 1.Fadloun A, Eid A, Torres-Padilla ME. Mechanisms and dynamics of heterochromatin formation during mammalian development: closed paths and open questions. Curr Top Dev Biol 2013; 104:1-45; PMID:23587237; http://dx.doi.org/ 10.1016/B978-0-12-416027-9.00001-2 [DOI] [PubMed] [Google Scholar]
- 2.Becker JS, Nicetto D, Zaret KS. H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends Genet 2016; 32:29-41; PMID:26675384; http://dx.doi.org/ 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandra T, Kirschner K. Chromosome organisation during ageing and senescence. Curr Opin Cell Biol 2016; 40:161-7; PMID:27101466; http://dx.doi.org/ 10.1016/j.ceb.2016.03.020 [DOI] [PubMed] [Google Scholar]
- 4.Morgan MA, Shilatifard A. Chromatin signatures of cancer. Genes Dev 2015; 29:238-49; PMID:25644600; http://dx.doi.org/ 10.1101/gad.255182.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al.. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406:593-9; PMID:10949293; http://dx.doi.org/ 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- 6.Maison C, Quivy JP, Probst AV, Almouzni G. Heterochromatin at mouse pericentromeres: a model for de novo heterochromatin formation and duplication during replication. Cold Spring Harb Symp Quant Biol 2010; 75:155-65; PMID:21209390; http://dx.doi.org/ 10.1101/sqb.2010.75.013 [DOI] [PubMed] [Google Scholar]
- 7.Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al.. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat Genet 2011; 43:220-7; PMID:21317888; http://dx.doi.org/ 10.1038/ng.765 [DOI] [PubMed] [Google Scholar]
- 8.Romeo K, Louault Y, Cantaloube S, Loiodice I, Almouzni G, Quivy JP. The SENP7 SUMO-Protease presents a module of two HP1 interaction motifs that locks HP1 protein at pericentric heterochromatin. Cell Rep 2015; 10:771-82.; PMID:25660026; http://dx.doi.org/23045645 10.1016/j.celrep.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 9.Bawa-Khalfe T, Lu LS, Zuo Y, Huang C, Dere R, Lin FM, Yeh ET. Differential expression of SUMO-specific protease 7 variants regulates epithelial-mesenchymal transition. Proc Natl Acad Sci U S A 2012; 109:17466-71; PMID:23045645; http://dx.doi.org/ 10.1073/pnas.1209378109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maison C, Bailly D, Quivy JP, Almouzni G. The methyltransferase Suv39h1 links the SUMO pathway to HP1alpha marking at pericentric heterochromatin. Nat Commun 2016; 7:12224; PMID:27426629; http://dx.doi.org/ 10.1038/ncomms12224 [DOI] [PMC free article] [PubMed] [Google Scholar]


