Abstract
RAG1 and RAG2 play a central role in V(D)J recombination, a process for antigen receptor gene assembly. The truncated ‘core’ regions of RAGs are sufficient to catalyze the recombination reaction, although with lower joining efficiency than full-length proteins. To investigate the role of the non-core regions of RAGs in the end-joining phase of antigen receptor rearrangement, we analyzed recombination products isolated from core RAG1 and core RAG2 knock-in mice. Here, we report that the truncation of RAGs increases the frequency of aberrant recombination in vivo. Signal joints (SJs) associated with V-to-D recombination of core RAG1 knock-in mice were normal, whereas those of core RAG2 knock-in mice were highly imprecise, containing large deletions and additions, and in some cases coding sequences. In contrast, we found an elevated level of imprecise D-to-J associated SJs for both core RAG1- and RAG2-expressing mice. Likewise, sequences of coding joints (CJs) were also affected by the expression of core RAGs. Finally, sequences found at the junctions of rearranged T-cell receptor loci were highly influenced by differences in rearranging recombination signal sequence pairs. We provide the first evidence that the non-core regions of RAGs have critical functions in the proper assembly and resolution of recombination intermediates in endogenous antigen receptor loci.
INTRODUCTION
During lymphocyte development, immunoglobulin (Ig) and T-cell receptor (TCR) genes are assembled from germline V, D and J gene segments to generate antigen receptor diversity, by a process known as V(D)J recombination (1,2). V(D)J recombination is initiated by RAG1 and RAG2 proteins that introduce double-strand breaks (DSBs) between two coding segments and flanking recombination signal sequences (RSSs). Subsequent repair via the non-homologous end-joining (NHEJ) pathway generates precise signal joints (SJs) and imprecise coding joints (CJs). RSSs are composed of two consensus elements, a heptamer and nonamer, along with an intervening spacer sequence of 12 or 23 bp. Cleavage occurs in a coupled fashion that requires the pairing of a 12 bp RSS with that of a 23 bp RSS. Depending on the transcriptional orientation of the gene segments, recombination results in either deletion or inversion of intervening DNA.
RAG-mediated DNA cleavage produces two types of DNA ends: blunt signal ends (SEs) and covalently sealed hairpin coding ends (CEs). Following DNA cleavage, the ends are held together in a cleaved signal complex (CSC), wherein CJ formation is thought to occur, leaving behind a stable signal end complex (SEC) (3–5). The opening and processing of RAG-generated hairpinned CEs are required for CJ formation, and are often accompanied by nucleotide loss and addition. The activity of terminal deoxynucleotidyl transferase (TdT) further increases diversity via N nucleotide addition. In contrast, the blunt SEs within the SEC are usually re-ligated in a precise manner.
Genetic analyses of various mutant cells and mice have revealed important roles for several NHEJ factors in the repair of DSBs introduced by RAGs during V(D)J recombination (1,6). The Ku70/80 heterodimer, XRCC4 and DNA ligase IV are required for the formation of both SJ and CJ. The catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) and Artemis are more important for CJ formation. Recent data suggest that there can be additional component(s) required for V(D)J recombination (7).
Core RAG1 (384–1008 amino acids) and core RAG2 (1–383 amino acids) have been defined as the minimal regions sufficient for recombination of extrachromosomal substrates (8–12). Although core RAGs are capable of recognizing RSS and perform coupled DNA cleavage in vitro, several studies have shown that the missing regions have important functions for normal V(D)J recombinational activity, including suppression of aberrant rearrangements (13–17), regulation of protein degradation (18–20), control of nuclear localization (21,22) and regulation of proper lymphocyte development (23–26).
In addition to these specific effects on V(D)J recombination, studies have shown that the truncated RAGs have decreased overall recombination efficiency compared to the full-length RAGs when tested with extrachromosomal substrates (9,11,12). The higher levels of SE recombination intermediates generated using core RAGs suggest that the non-core regions may be required for effective joining, since there is a reduction in the formation of SJ and CJ (27). The identification of RAG mutations within the core regions of RAG1 and RAG2 that affect the second phase of the reaction supports an architectural role of the post-cleavage complex to facilitate the end-joining reaction (28–31). However, a role for RAGs in the end-joining phase of V(D)J recombination at endogenous antigen receptor loci has never been addressed.
To elucidate potential functions of non-core regions of RAGs in end processing and joining of endogenous loci, we have compared the DNA sequences of recombination products isolated from lymphocytes of core RAG1, core RAG2 and wild-type (WT)-RAG expressing mice. We find a reduction in the fidelity of SJ isolated from core RAG1 or core RAG2 mice compared to controls. Expression of core RAG1 alters the features of SJ associated with D-to-J joining but not of V-to-D joining. On the other hand, expression of core RAG2 severely affects the fidelity of V-to-D joining, and to a lesser extent D-J joining. In addition, characteristics of junctional sequences are highly dependent on particular RSS pairings. SJ containing D-coding sequences suggest aberrant cleavage, indicating a possible role of non-core regions of RAGs for controlling proper alignment of the synaptic complex. Our results suggest that the non-core regions of RAGs influence the structure of the synaptic complex and stability of the post-cleavage complex, thereby influencing end-joining products.
MATERIALS AND METHODS
Mice and preparation of genomic DNA
The generation of knock-in mice with the core RAGs have been described (23,24). All the mice used in this study were of the 129SvEv background, and housed under specific pathogen-free conditions. Single cell suspensions of homogenized thymus or spleen were prepared from 8 to 12 weeks old mice. CD4+ CD8+ (DP) T cells were isolated by staining thymocytes with FITC anti-CD4 and PE anti-CD8 antibodies and sorting on a MoFlo (DakoCytomation). Genomic DNA was prepared as described previously (32).
Analysis of SJs and CJs
An aliquot of 500 ng of genomic DNA for SJ and 150–180 ng for CJ were subjected to 35 cycles of PCR amplification using the following combinations of primers. To amplify SJ associated with V-to-D recombination, Vβ14 SIGNAL and Dβ1.1 SIGNAL for Vβ14-Dβ1 SJ; SR1 and SR2 for Vδ5-Dδ2 SJ; and Vβ8.3 SIGNAL and Dβ1.1 SIGNAL for Vβ8.3-Dβ1 SJ were used. To amplify SJ associated with D-to-J recombination, SR5 and SR6 for Dδ2-Jδ1 SJ; Jβ1-F and Dβ1R420 for Dβ1-Jβ1.1 SJ; and SR14 and SR15 for Dβ2-Jβ2.1 SJ were used. To amplify Vβ14- Jβ1.1 CJ, Vβ14-S and Jβ1.1 CODING primers were used. The primers SR7 and SR8 were used to recover Vβ10-Jβ2.1 CJ. PCR products containing SJ and CJ were cloned using the TA cloning kit (Invitrogen). For SJ, inserts were identified using PCR and products were digested with ApaLI to detect precise joints. Plasmid DNA was prepared from ApaLI resistant SJ clones and all the CJ clones. Inserts were sequenced using DYEnamic™ ET terminator kit (Amersham Pharmacia Biotech) and analyzed by ABI prism 3100 genetic analyzer (Applied Biosystems).
Oligonucleotides
The following oligonucleotides were used in this study: Vβ14 SIGNAL, 5′-CAGACTTCAACTTGACTATC-3′; Dβ1.1 SIGNAL, 5′-ACCTTCCTTATCTTCAACTC-3′; SR1, 5′-GC1;AAGTCTGGCCTGAACTAA-3′; SR2, 5′-CAACCTGGCA1;TGTGACTTTC-3′; Vβ8.3 SIGNAL, 5′-AGCTAGAAACCC1;ATCCTGCA-3′; SR5, 5′-CTTGTCCAGTCAACTTCCTG-3′; SR6, 5′-AAGTCATGATGAGCCAGCTG-3′; Jβ1-F, 5′-CT1;GTGATGCACACAAAGCGA-3′; Dβ1R 420, 5′-CATTCTGGATCTAAACAC-3′; SR14, 5′-AAGACCTTGTGAGTCCA1;CTC-3′; SR15, 5′-GCCTCATGCAAGGTCAAGAT-3′; Vβ14-S, 5′-AGAGTCGGTGGTGCAACTGAACCT-3′; Jβ1.1 CODING, 5′-TGCTTTGTCCGAAGAGAGACCTG-3′; SR7, 5′-GCTTCTCACCTCAGTCTTCA-3′; and SR8, 5′-TGCTAA GGTTTTCTGCTCCG-3′.
RESULTS
Increased frequency of aberrant SJ in V-to-D recombination in core RAG2 knock-in mice
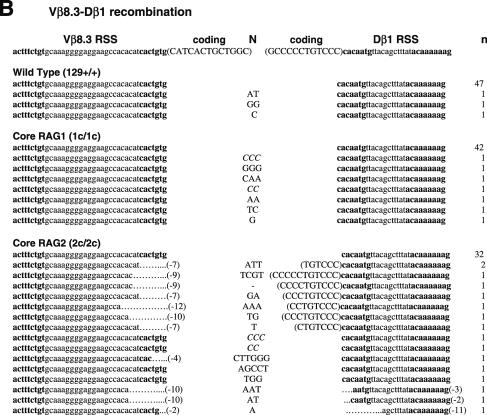
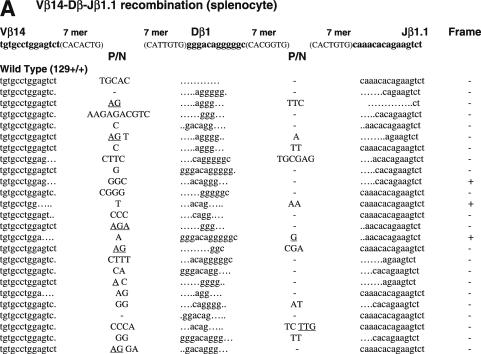
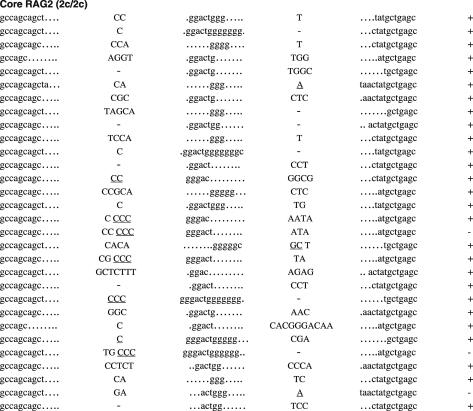
Unlike CJ, SJ is free from selective forces, leaving the evidence of the joining reaction behind. The majority of SJ isolated from WT lymphocytes do not contain nucleotide additions or deletions (33). Thus, an elevated frequency of imprecise SJ provides evidence of defects in the end-joining process. To compare the sequences of SJ produced in WT, core RAG1 and core RAG2 knock-in mice, we first analyzed SJ formed during Vβ14-Dβ1 joining (Figure 1 and Table 1). Subcloned PCR fragments containing Vβ14-Dβ1 SJ were tested for the presence of a new restriction site, ApaLI, which is generated upon precise head-to-head ligation of RSS heptamers. Genomic thymus DNA was prepared from at least two independent animals. Since no significant difference was observed in the data among these animals, the results were combined to calculate each frequency. As predicted, 90% of clones (43 out of 48 independent clones) isolated from WT thymus were sensitive to ApaLI digestion, demonstrating that the majority of recombination products form precise SJ (Table 1). Similarly, 82% (40 out of 49) of SJ recovered from core RAG1 mice were also precise. On the other hand, only 62% (29 out of 47) of SJ recovered from core RAG2 mice were precise. Chi-square analysis shows that the reduction in the frequency of precise joints in core RAG2 mice is statistically significant (P < 0.01). To further analyze the quality of SJ, ApaLI-insensitive clones were sequenced and compared. Imprecise joints produced in WT and core RAG1 mice contained additions of several nucleotides but no deletions were observed (Figure 1A). On the other hand, imprecise joints produced in core RAG2 mice contained additions and deletions. Two clones contained patches of deletions displaying microhomology at their break points that could have come from either joining partner. Furthermore, three clones contained additional sequences identical to coding sequences originally located adjacent to the RSS.
Figure 1.
Sequences of SJs formed by V-D recombination. (A) PCR fragments containing SJs formed by Vβ14-Dβ1 recombination were compared among WT (RAG1+/+ RAG2+/+), RAG1 core (RAG1c/c RAG2+/+) and RAG2 core (RAG1+/+ RAG2c/c) mice. The original sequences are indicated by lowercase letters and possible insertions are indicated in capital letters. Heptamer and nonamer sequences are in boldface letters. Nucleotide additions containing more than three continuous bases identical to either CEs were considered as coding sequence and indicated in parentheses. Nucleotide additions assigned as N additions with 2 or 3 nt homology with either CE are italicized. Numbers of deleted bases are shown in parentheses at the right end of each RSS. Microhomologies are underlined. ‘n’ indicates the number of junctions with the indicated sequence. (B) SJs formed by Vβ8.3-Dβ1 recombination. (C) SJ formed by Vδ5-Dδ2 recombination.
Table 1. Comparison of V-D SJs.
| WT | Core RAG1 | Core RAG2 | |
|---|---|---|---|
| Vβ14-Dβ1 | |||
| Precise joints | 43/48 (90%) | 40/49 (82%) | 29/47 (62%) |
| Imprecise joints | 5/48 (10%) | 9/49 (18%) | 18/47** (38%)a |
| N addition | 5/5 (100%) | 9/9 (100%) | 15/18 (83%) |
| Coding sequences | 0/5 (0%) | 0/9 (0%) | 3/18 (17%) |
| Deletion | 0/5 (0%) | 0/9 (0%) | 3/18 (17%) |
| Microhomology | 0/5 (0%) | 0/9 (0%) | 2/18 (11%) |
| Vβ8.3-Dβ1 | |||
| Precise joints | 47/50 (94%) | 42/49 (86%) | 32/48 (67%) |
| Imprecise joints | 3/50 (6%) | 7/49 (14%) | 16/48** (33%)a |
| N addition | 3/3 (100%) | 7/7 (100%) | 15/16 (94%) |
| Coding sequences | 0/3 (0%) | 0/7 (0%) | 8/16 (50%) |
| Deletion | 0/3 (0%) | 0/7 (0%) | 12/16 (75%) |
| Microhomology | 0/3 (0%) | 0/7 (0%) | 0/16 (0%) |
| Vδ5-Dδ2 | |||
| Precise joints | 55/66 (83%) | 54/67 (81%) | 15/56 (27%) |
| Imprecise joints | 11/66 (17%) | 13/67 (19%) | 41/56** (73%)a |
| N addition | 11/11 (100%) | 13/13 (100%) | 23/41 (56%) |
| Coding sequences | 0/11 (0%) | 1/13 (8%) | 1/41 (2%) |
| Deletion | 0/11 (0%) | 0/13 (0%) | 37/41 (90%) |
| Microhomology | 0/11 (0%) | 0/13 (0%) | 13/41 (32%) |
Number of each joint out of total number analyzed was shown. The frequency of each joint is shown in parentheses. Significant differences versus WT mice, when compared by chi-squared test, are indicated by ** (P < 0.01). Junctions were sequences from two independent animals, and the results were combined to calculate the number of each case.
aDifferences from WT mice are still statistically significant even when the data containing coding sequences are excluded, to exclude possible non-12/23 HJ from SJ (see Discussion).
Vβ14 rearranges via inversional joining, whereas other Vβs undergo deletional recombination, therefore we next asked if the quality of SJ could be affected by transcriptional orientation of RSSs. Vβ8.3-Dβ1 SJ, produced by deletional recombination, were readily recovered by PCR amplification of total thymus DNA, which contains excision circles formed by deletional recombination. We detected a significant decrease in the frequency of precise Vβ8.3-Dβ1 SJ derived from core RAG2 mice (67%) compared to WT or core RAG1 mice (94 and 86%, respectively; Table 1). Vβ8.3-Dβ1 SJ was similar in feature to those isolated from Vβ14-Dβ1, with nucleotide deletions and additions occurring, and some containing homologous sequences (Figure 1B). However, the frequency of deletions and presence of coding sequences were elevated when 5′Dβ1 RSS was paired with Vβ8.3 RSS (Table 1). Thus, the characteristics of SJ appeared sensitive to the differences in RSS partners.
To determine if the reduced ability of core RAGs to mediate precise SJ extended to other TCR loci, we analyzed Vδ5-Dδ2 SJ that occur by inversional joining (Figure 1C and Table 1). A total of 83 and 81% of clones recovered from thymus DNA of WT and core RAG1 knock-in mice, respectively, contained precise Vδ5-Dδ2 SJ. All imprecise joints were found to contain nucleotide additions, with no deletions observed. In contrast, only 27% of Vδ5-Dδ2 SJ isolated from core RAG2 thymus DNA were precise (Table 1). Strikingly, 90% (37 of 41) of imprecise Vδ5-Dδ2 SJ from core RAG2 thymus DNA contained deletions (average = −17.1 ± 2.8 bp). Furthermore, 13 imprecise joints exhibited microhomologies at the junction. These results indicate that the non-core region of RAG2, but not the non-core region of RAG1, is important for precise Vδ5-Dδ2 SJ formation.
These findings demonstrate that a recombinase complex containing truncated RAG2 more frequently produces imprecise SJ than does full-length RAG2. In contrast, core RAG1 does not affect the characteristics of SJ produced by VD joining. Furthermore, the quality of SJ produced in the presence of truncated RAG2 was not dependent on the genomic configuration of RSS pairs, as we detected reduced fidelity in SJ formed during both deletional and inversional recombination. However, all three different sets of VD joints tested from core RAG2 knock-in mice showed distinct preferences for modifications: Vβ14-Dβ1 SJ displayed fewer deletions (3 out of 18 imprecise SJ) though with a high frequency of N additions (15 out of 18). Vβ8.3-Dβ1 SJ often contained Dβ1 coding sequences (8 out of 16 imprecise SJ), nucleotide additions (15 out of 16) and deletions (12 out of 16). Most Vδ5-Dδ2 SJ had deletions (37 out of 41 imprecise SJ), and frequently contained short stretches of microhomology (13 out of 41) (Table 1). Thus, the characteristics of end processing mediated by core RAG2 when producing imprecise SJ seemed to be affected by the combination of different RSSs.
Increased frequency of aberrant SJ in D-to-J recombination in core RAG1 and core RAG2 knock-in mice
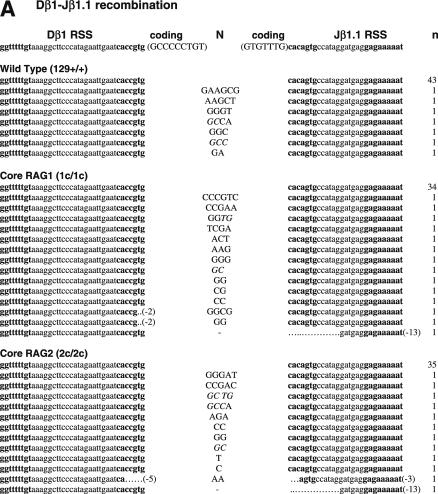
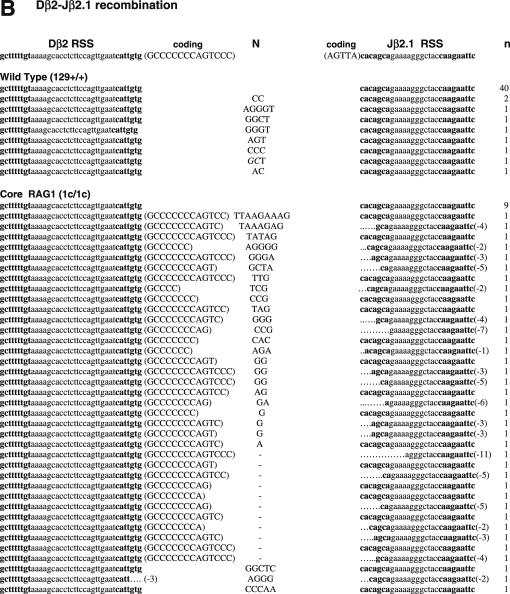
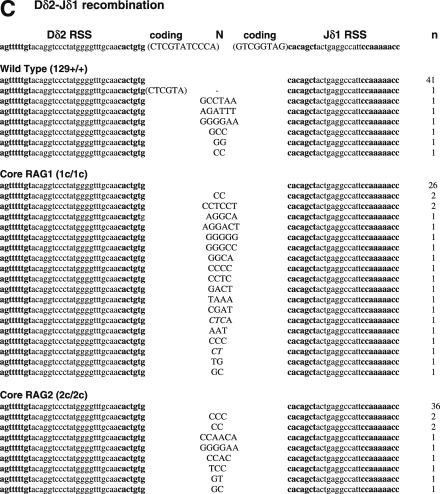
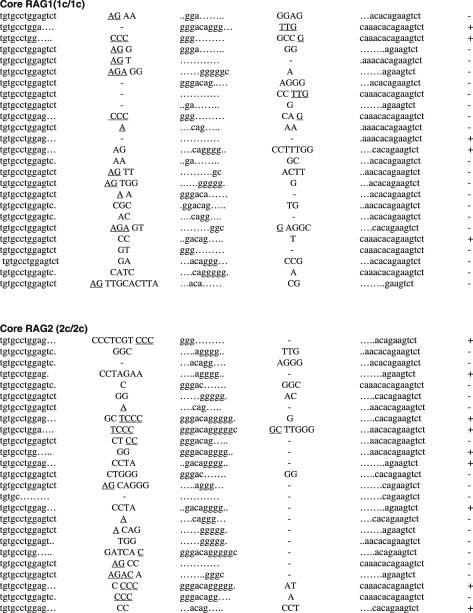
To determine if non-core regions of RAGs differentially influence V-to-D and D-to-J joining steps, we next analyzed SJ associated with Dβ1-Jβ1.1, Dβ2-Jβ2.1 and Dδ2-Jδ1 joining, all produced by deletional recombination (Figure 2 and Table 2). PCR amplification of total thymus DNA isolated from WT mice demonstrated 14, 18 and 15% of Dβ1-Jβ1.1, Dβ2-Jβ2.1 and Dδ2-Jδ1 SJ, respectively, were imprecise joints (Table 2). In contrast, we detected a 2–4-fold increased frequency (for Dβ1-Jβ1.1, Dβ2-Jβ2.1 and Dδ2-Jδ1 SJ: 29, 80 and 43% for core RAG1 and 26, 69 and 22% for core RAG2, respectively) of imprecise SJ from both core RAG1- and core RAG2-expressing mice. The higher frequency of imprecise joints associated with core RAG1 expression was statistically significant for Dβ2-Jβ2.1 and Dδ2-Jδ1 SJ (Table 2). Although the overall frequency of Dβ1-Jβ1.1 and Dδ2-Jδ1 imprecise joints were elevated in core RAG2 expressing mice, only the level of Dβ2-Jβ2.1 imprecise joints was statistically significant.
Figure 2.
Sequences of SJs formed by D-J recombination. (A) PCR fragments containing SJs formed by Dβ1-Jβ1.1 recombination were compared among WT (RAG1+/+ RAG2+/+), core RAG1 (RAG1c/c RAG2+/+) and core RAG2 (RAG1+/+ RAG2c/c) expressing mice. Details are as in Figure 1 legend. (B) SJ formed by Dβ2-Jβ2.1 recombination. (C) SJ formed by Dδ2-Jδ1 recombination.
Table 2. Comparison of D-J SJs.
| WT | Core RAG1 | Core RAG2 | |
|---|---|---|---|
| Dβ1-Jβ1.1 | |||
| Precise joints | 43/50 (86%) | 34/48 (71%) | 35/47 (74%) |
| Imprecise joints | 7/50 (14%) | 14/48 (29%) | 12/47 (26%) |
| N nucleotides | 7/7 (100%) | 13/14 (93%) | 11/12 (92%) |
| Deletion | 0/7 (0%) | 3/14 (21%) | 2/12 (17%) |
| Dβ2-Jβ2.1 | |||
| Precise joints | 40/49 (82%) | 9/46 (20%) | 14/45 (31%) |
| Imprecise joints | 9/49 (18%) | 37/46** (80%)b | 31/45** (69%)a |
| N nucleotides | 9/9 (100%) | 26/37 (70%) | 28/31 (90%) |
| Coding sequences | 0/9 (0%) | 34/37 (92%) | 13/31 (42%) |
| Deletion | 0/9 (0%) | 20/37 (54%) | 7/31 (23%) |
| Dδ2–Jδ1 | |||
| Precise joints | 41/48 (85%) | 26/46 (57%) | 36/46 (78%) |
| Imprecise joints | 7/48 (15%) | 20/46** (43%) | 10/46 (22%) |
| N nucleotides | 6/7 (86%) | 20/20 (100%) | 10/10 (100%) |
| Coding Sequences | 1/7 (14%) | 0/20 (0%) | 0/10 (0%) |
| Deletion | 0/7 (0%) | 0/20 (0%) | 0/10 (0%) |
Number of each joint out of total number of analyzed was shown. The frequency of each joint is shown in parentheses. Significant differences versus WT mice, when compared using chi-squared test, are indicated by ** (P < 0.01). Junctions were sequences from two independent animals, and the results were combined to calculate the number of each case.
aDifferences from WT mice are statistically significant even when the data containing coding sequences are excluded, to exclude possible non-12/23 HJ from SJ (see Discussion).
bDifference from WT mice is not significant when the data containing coding sequences are excluded.
Thus, we conclude that the expression of core RAG1 affected the accuracy of recombination associated with DJ joining in general. Expression of core RAG2 also influences DJ associated SJ, however, it seems to be more sensitive to different RSS pairs. Most imprecise Dβ1-Jβ1.1 and Dδ2-Jδ1 joints contained N nucleotide additions, although some deletions were detected in a few Dβ1-Jβ1.1 SJ isolated from both core RAG1 and core RAG2 knock-in mice. Dβ2-Jβ2.1 SJ was even more severely affected, containing deletions, N nucleotide additions and sequences homologous to coding gene segments at the junction (Figure 2B). The long Dβ2 coding sequences attached to modified Jβ2.1 SE strongly suggests that the involvement of aberrant cleavage during Dβ2-Jβ2.1 recombination.
Non-core regions of RAGs influence the quality of CJ in vivo
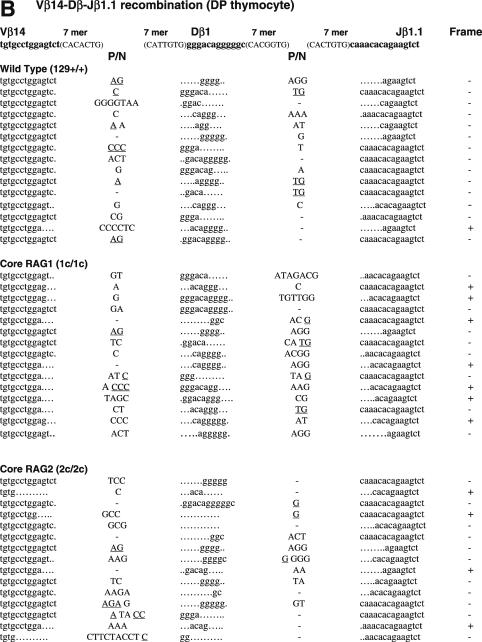
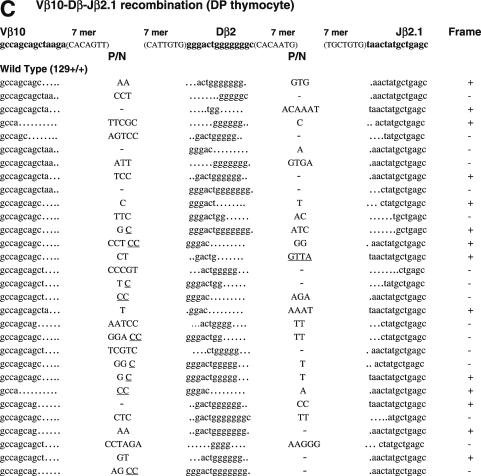
To ask if non-core regions of RAGs play a critical role in normal CJ formation, we next analyzed CJ associated with Vβ14-Dβ-Jβ1.1 and Vβ10-Dβ-Jβ2.1 (Figure 3). The Vβ14 RSS was selected as an example of an RSS that rearranges by inversion. Vβ10 RSS was chosen as an example of an RSS whose recombination with downstream gene segments results in the deletion of genomic DNA, since it showed the closest match with consensus 23-RSS of VH/Vβ origin (25). Vβ14-Dβ-Jβ1.1 CJ was analyzed from genomic DNA of splenocytes using PCR (Figure 3A). To exclude effects biased by the negative/positive selection of productive rearrangements (34), Vβ14-Dβ-Jβ1.1 CJ was also analyzed from genomic DNA of CD4+CD8+ (DP) thymocytes (Figure 3B). Vβ10-Dβ-Jβ2.1 CJ was analyzed from DP thymocytes (Figure 3C). Frequency and average length of nucleotide modifications are summarized in Table 3.
Figure 3.
Sequences of CJs formed by V-D-J recombination. (A) CJs formed by Vβ14-Dβ-Jβ1.1 recombination in splenocytes were compared among WT (RAG1+/+ RAG2+/+), core RAG1 (RAG1c/c RAG2+/+) and core RAG2 (RAG1+/+ RAG2c/c) expressing mice. In Vβ14-Dβ-Jβ1.1 recombination, Dβ1 within the Dβ1-Jβ1 gene cluster is expected to be utilized. Original coding sequences of Vβ14, Dβ1 and Jβ1.1 (boldface lowercase letters) and their flanking heptamer sequences (uppercase letters in parentheses) are indicated on the top. Nucleotide insertions are indicated by capital letters. Presumptive P nucleotides are underlined. All the clones contained different junctional sequences to each other. In-frame or out-of-frame rearrangement is denoted as ‘+’ or ‘−’ at the right end of each sequence. (B) CJ formed by Vβ14-Dβ-Jβ1.1 recombination in DP thymocytes. (C) CJ formed by Vβ10-Dβ-Jβ2.1 recombination in DP thymocytes. Original coding sequences of Vβ10, Dβ2 and Jβ2.1 (boldface lowercase letters) and their flanking heptamer sequences (uppercase letters in parentheses) are indicated on the top. Although Dβ1 can be used instead of Dβ2 in at least one-fourth of all Vβ10-Dβ-Jβ2.1 CJ (69), but they could not be assigned due to similarity. See (A) and (B) for Dβ1 coding sequence.
Table 3. Frequencies and average length in nucleotides modification in CJ.
| WT | Core RAG1 | Core RAG2 | |
|---|---|---|---|
| Vβ14-Dβ-Jβ1.1 | |||
| Deletion within Vβ | 24/40 (60%, −1.79) | 21/40 (53%, −2.86**) | 26/40 (65%, −3.34**) |
| P/N addition at VD junction | 36/40 (90%, +2.72) | 33/40 (83%, +2.72) | 35/40 (88%, +3.87*) |
| Dβ ≤4 nt | 17/40 (43%, −6.61) | 18/40 (45%, −7.20) | 17/40 (43%, −7.18) |
| P/N addition at DJ junction | 20/40 (50%, +2.25) | 34/40** (85%, +3.03) | 18/40 (45%, +2.55) |
| Deletion within Jβ | 23/40 (58%, −4.82) | 27/40 (68%, −3.74) | 30/40 (75%, −4.23) |
| Vβ10-Dβ-Jβ2.1 | |||
| Deletion within Vβ | 30/30 (100%, −4.77) | 30/30 (100%, −4.83) | 30/30 (100%, −4.73) |
| P/N addition at VD junction | 26/30 (87%, +3.19) | 27/30 (90%, +3.74) | 25/30 (83%, +3.20) |
| Dβ ≤4 nt | 3/30 (10%, −6.03) | 15/30** (50%, −8.83**) | 6/30 (20%, −7.56) |
| P/N addition at DJ junction | 20/30 (67%, +2.50) | 25/30 (83%, +2.60) | 24/30 (80%, +2.96) |
| Deletion within Jβ | 25/30 (83%, −2.60) | 26/30 (87%, −3.81) | 28/30 (93%, −3.93*) |
Sequence data obtained from two independent genomic DNA samples were combined. Data obtained from spleen and DP thymocytes were combined in Vβ14-Dβ-Jβ1.1. Number of each event out of total number of analyzed were shown. The frequency and the average length of addition (+) and deletion (−) of each joining are shown in parentheses. Significant differences versus WT mice, when compared by chi-squared test (frequencies) or t-test (average length of addition and deletion), are indicated by * (P < 0.05) and ** (P < 0.01).
Although the sequences of CJ were heavily modified in all clones analyzed, CJ isolated from core RAG1 and core RAG2 knock-in mice frequently contained a higher degree of modification than those of WT mice (Table 3). The average length of Vβ14 segment deletions were larger in core RAG1 and core RAG2 than in WT (−2.86, −3.34 and −1.79 bp, respectively), although we did not observe a difference in the frequency of such deletions. Similarly, the average length of nucleotide additions at the VD junction of Vβ14-Dβ-Jβ1.1 CJ was also larger in core RAG2 samples than WT (+3.87 and +2.72 bp, respectively). On the other hand, no significant differences were observed in the frequency or degree of modifications of Vβ or VD junctions for Vβ10-Dβ-Jβ2.1 CJ between core RAG1, core RAG2 and WT samples. Instead, the length of Dβ coding sequence was frequently shorter in Vβ10-Dβ-Jβ2.1 CJ when core RAG1 was expressed, with 50% of clones (15 out of 30) containing 4 or less nucleotides of potential Dβ coding sequence origin. Only 10% (3 out of 30) of CJ isolated from WT samples contained Dβ segments consisting of four or fewer nucleotides, and only 20% (6 out of 30) in core RAG2 samples. In addition, core RAG1 Dβ segments in Vβ10-Dβ-Jβ2.1 CJ had an average length deletion of −8.83 bp, compared to an average of −6.03 bp in WT Dβ segments. Thus, core RAG1 expression resulted in more frequent and extensive deletions within the Dβ segment of Vβ10-Dβ-Jβ2.1 CJ. In contrast, we observed no significant differences in the frequency or length of Dβ within Vβ14-Dβ-Jβ1.1 CJ of core RAG1, core RAG2 and WT samples. The frequency of nucleotide additions at DJ junctions of Vβ14-Dβ-Jβ1.1 CJ was significantly elevated when core RAG1 was expressed (50 and 85% for WT and core RAG1, respectively), although we did not observe a difference in the size of such additions. The increased frequency of nucleotide additions at DJ junctions was less apparent in Vβ10-Dβ-Jβ2.1 CJ. However, core RAG2 Vβ10-Dβ-Jβ2.1 CJ exhibited more extensive deletion of Jβ coding sequences compared to WT (−3.93 and −2.6 bp, respectively). Vβ10-Dβ-Jβ2.1 CJ from core RAG1 T cells also included deletions of Jβ (−3.81 bp), although these were not statistically different from WT. Vβ14-Dβ-Jβ1.1 CJ showed no significant difference among samples in the frequency or extent of deletions of Jβ sequences. Therefore, though expression of core RAGs influenced the overall quality of CJ, the characteristics of these modifications tended to be associated with specific V-D and D-J junctions.
DISCUSSION
It has been proposed that the post-cleavage complex facilitates DNA repair by serving as a scaffold for the four broken ends generated by RAG-mediated cleavage (28–31). RAG proteins have also been shown to affect the formation of precise SJ in a yeast system (35). Although studies using artificial substrates are useful for understanding the molecular mechanism of V(D)J recombination, they may not completely recapitulate all aspects of antigen receptor rearrangement in vivo. Here, we provide the first evidence that the non-core regions of RAGs have critical functions in the proper assembly and resolution of recombination intermediates in endogenous loci. RAG2 truncation affects SJ associated with VD joining more severely than that of DJ joining, whereas core RAG1 affects DJ joining, as shown by previous studies focused on the efficiency of VD and DJ rearrangement associated with lymphocyte development (23,24). Moreover, we demonstrate a variety of CJ features that are influenced by the truncation of RAGs.
Since DSBs are the most lethal form of cellular DNA damage, there are at least three different pathways to repair a DSB, including classical NHEJ, microhomology-directed end-joining (MHEJ, or alternative NHEJ) and homologous recombination repair (HRR) (36–39). Although DSBs associated with V(D)J recombination are normally repaired by NHEJ, alternative pathways must take part in completing the repair when conventional pathways are not available or not effective (40). When either RAG1 or RAG2 were truncated, we observed SJ that exhibit features consistent with repair via a pathway other than NHEJ.
Imprecise joints isolated from mice producing truncated RAG proteins frequently contained nucleotide deletions. A total of 90% of Vδ5-Dδ2 imprecise joints isolated from core RAG2 mice contained deletions, with an average loss of 17 nt, and the largest being a 74 bp deletion (Figure 1C). Thus, truncation of RAGs might directly influence the stability of the SEC in certain RSS pairs, allowing exonuclease attack of unprotected SE. The occurrence of deletions also suggests an end-joining defect, as end-joining deficiencies are known to lead to large deletions. In fact, these joints are somewhat similar to SJ from artificial substrates introduced into XRCC4-deficient cells, as they tend to have large deletions with short stretches of homology at the junction (41). Either joining partner could have contributed these nucleotides, the presence of which is consistent with joining mediated by a homologous stretch of nucleotides between the two joining partners. Such MHEJ is thought to occur as a back-up to conventional NHEJ repair, since defects in NHEJ result in elevated MHEJ (38,39). In addition, CJ isolated from core RAG knock-in mice also appeared to have an increased frequency of nucleotide deletions, although the effect was less obvious than in SJ due to the imprecise nature of CJ.
Sequences homologous to the germline coding sequence were frequently observed in SJ isolated from core RAG-expressing mice. The presence of these coding sequences can be explained by a number of mechanisms. Oligonucleotide capture is a mechanism by which short DNA segments cleaved at both 5′ and 3′ ends can be incorporated into the junction (42,43). CJ with short oligonucleotides that could have been derived in this manner have been reported previously (44–46). The Dδ2 coding sequence trapped within a Vδ5-Dδ2 SJ of a core RAG2 sample (Figure 1C) could have resulted from this.
The frequent detection of long Dβ coding flank sequence attached directly to RSS cannot be explained by oligonucleotide capture, but is more likely the result of aberrant cleavage. For instance, RSS-like sequences found within coding gene segments may be mistakenly used as RAG substrates. In this regard, heptamer-like sequences containing CAC trinucleotide or nonamer-like sequences can serve as substrates for RAG proteins in vitro (47,48), and evidences indicating aberrant recombination using such sites have been reported not only at the breakpoint of chromosomal translocations associated with lymphoid neoplasia (33,49) but also in the TCRδ locus of normal mice (46).
Although RSS-like sequences can occur anywhere in the genome, we failed to find such a sequence in the coding sequences near the cleavage site, except for the bonafide RSS present on the opposite end of Dβ gene segments. The majority of Dβ2-Jβ2.1 SJ contained unmodified Dβ2 coding flank directly attached to the 3′Dβ2 RSS, while there were frequent modifications observed between the end of Dβ2 coding sequence and the Jβ2.1 RSS (Figure 2B). The structure of these aberrant joints suggests that the cleavage has occurred at the 5′Dβ2 RSS instead of the 3′Dβ2 RSS. The frequent deletion of Dβ coding sequence within Vβ10-Dβ-Jβ2.1 CJ isolated from core RAG1 samples (Figure 3C and Table 3) also supports the involvement of aberrant cleavage, since corresponding Dβ2 coding sequences appeared in reciprocal SJ. Although the formation of a 12/12 deletional hybrid joints (HJs) between the CE of one element and the SE of another may sound unlikely, we speculate that short Dβ coding sequence flanked by 3′ and 5′ RSSs in the presence of core RAGs may result in such HJ. Indeed, HJ in apparent violation of the 12/23 rule has similarly been described in the TCRδ locus of WT mice, where Dδ elements were involved (46). During V(D)J recombination, a RAG complex is first thought to form on a single RSS, followed by subsequent capture of a second RSS (50,51). Though the correct 12/23 pair may be favored by full-length RAGs, a complex formed with core RAGs may allow improper alignment with an RSS only 1–1.5 turns of the helix away. Or, the synaptic complex may initially form correctly with a 12/23 pair, but core RAGs could form an unstable complex that might lead to improper cleavage of a nearby heptamer. These possibilities may also be related to the finding that the RAG complex cleaves a non-B-DNA structure at the Bcl-2 major breakpoint region (52). If RAG binding to the 3′ end of a D gene segment can induce the formation of non-B-form DNA that extends to the 5′ end of the D gene segment, this might provide a target for RAG nicking. Indeed, recent results showed that at least 12 bp of coding region, proximal to the heptamer, are protected in the synaptic complex formed by core RAG proteins (53). Thus the RSS flanking the other side of a D segment may be directly adjacent or partially contained within the synaptic complex, likely affecting the structure of the corresponding DNA.
Unlike recombination involving the Dβ2-Jβ2.1 cluster, imprecise SJ associated with the Dβ1-Jβ1.1 cluster contained no nucleotides with coding sequence homology (Figure 2A). We speculate that subtle differences in RSS pairs and/or length of Dβ segments (12 and 14 bp for Dβ1 and Dβ2, respectively) might significantly affect the DNA structure and/or stability of the synaptic complex, resulting in dramatic differences in how the synaptic complex can be formed and/or resolved by core RAGs. Similarly, Dβ1 coding sequences detected within Vβ8.3 to Dβ1 rearrangements in core RAG2 samples could have arisen through deletional rearrangements between Vβ and 3′Dβ1 RSS. Previous studies have shown that D-J rearrangement was significantly reduced when core RAG1 or core RAG2 was expressed, with nearly half of mature B cells containing germ line alleles (23,24). Since Dβ-to-Jβ rearrangement is not required for Vβ-Dβ joining (54), Vβ-to-Dβ rearrangement can occur in core RAG-expressing mice even before the completion of Dβ-Jβ rearrangement. Thus in the presence of core RAG2, a 3′Dβ1 RSS might be mistakenly cleaved within a VD synaptic complex. Increased accumulation of HJ has been reported in cells expressing core RAG1 and/or RAG2 (15). The increased rate of HJ formation in core RAGs in combination with the structure of short D segments might account for HJ violating the12/23 rule. To confirm that the SJ results were not all due to the presence of HJ, we excluded data potentially arising from HJ by removing all data containing coding sequences from the calculation of frequencies (see legends for Tables 1 and 2). Consistent with most of the original analyses, we still found significant increases in imprecise SJ, except for data involving Dβ2-Jβ2.1 SJ of core RAG1 samples. This is likely due to an increase in the rate of Dβ2-Jβ2.1 associated HJ in the presence of core RAG1. Thus, we conclude that the expression of core RAG increases the frequency of imprecise SJ, in a manner highly influenced by specific RSS partners.
The coding sequences observed in Vβ14-Dβ1 SJ of core RAG2 samples could still involve a different mechanism, since they contained not only Dβ coding flank, but also Vβ coding flank sequences (Figure 1A). Similar aberrant joints have been reported previously in Vβ14-Dβ1 SJ isolated from transgenic mice expressing a T490A mutant of RAG2. T490 phosphorylation is known to promote cell-cycle-dependent RAG2 degradation (19). Since core RAG2 lacks this phosphorylation site due to C-terminal truncation, the protein is expected to behave like the T490A mutant, remaining active into S phase where HRR is the prevalent DSB repair pathway. Although homologous sequences observed within these SJ are shorter than those usually used in HRR, we cannot completely rule out the possibility that factors involved in HRR might participate in the repair of these. Importantly, T490A mutant expressed on a RAG2−/− background does not affect the frequency of precise Vβ14-Dβ1 SJ, thus, it is unlikely this point mutation affects complex stability or increases aberrant cleavage (19). On the other hand, core RAG2 expression reduced the frequency of precise joints associated with Vβ14-Dβ1 recombination from 90% in WT to 62% in core RAG2 (Table 1). Increased N additions and possible microhomology-mediated joining are likely responsible for the low fidelity of these joints. Microhomology was never observed in imprecise SJ isolated from T490A transgenic mice (19). This suggests that deletion of the C-terminal RAG2 region is responsible for lowering the frequency of precise joints, and not its aberrant expression in S phase, presumably due to an inability to provide a proper architectural role during the end-joining phase.
Importantly, the reduced accuracy of SJ formed via core RAG is sensitive to nucleotide sequences and/or the structure of RSS. The RSS analyzed in this study are listed in Table 4. As RAGs undergo coupled RSS recognition and cleavage, both individual RSS sequences as well as their combinations are likely to contribute to the structure of the synaptic complex. SJ from different RSS combinations had quite distinct characteristics for both sides of SE. Although it has been reported that the quality of RSS affects accessibility to RAGs (54,55) and recombination frequencies (56,57), it may also affect the quality of recombination, especially when core RAGs are involved. The post-cleavage synaptic complex is known to be stable (3–5), however, certain RSS combinations such as Vδ5-Dδ2, may be inherently sensitive to structural change and become unstable when formed with truncated RAGs. Indeed, expression of mutant RAG1 proteins that form unstable post-cleavage complexes allow DNA ends to participate in both HRR and alternative NHEJ pathway (58). Thus, non-core regions of RAG might also have important roles for stabilizing the post-cleavage synaptic complex by minimizing the effects from RSS variations, since truncated RAGs result in higher frequency of imprecise SJ and modest increase in modification of CJ in vivo.
Table 4. Comparison of RSS analyzed in this study.
| Position | 7mer | 12/23 spacer | 9mer |
|---|---|---|---|
| 3′Vβ14 | CACACTG | AGTAGGGTGGGGCAGACATCTGT | GCAAAAACC |
| 3′Vβ8.3 | CACAGTG | ATGTGTGGCTTCCTCCCCTTTGC | ACAGAAAGT |
| 3′Vβ10 | CACAGTT | GTGCAGAGTCACTGTTTCCCTGT | GCACAAACC |
| 3′Vδ5 | CACTGTG | GTGCAGGTGCCCAGGGAGCCTGT | ACCCAAACC |
| 5′Dβ1 | CACAATG | TTACAGCTTTAT | ACAAAAAAG |
| 3′Dβ1 | CACGGTG | ATTCAATTCTATGGGAAGCCTTT | ACAAAAACC |
| 5′Dβ2 | CACAATG | TTACATCGTGAT | ACAAAAAAG |
| 3′Dβ2 | CACAATG | ATTCAACTGGAAGAGGTGCTTTT | ACAAAAAGC |
| 5′Dδ2 | CACGGTG | CTACAGAGCTTT | GCAAAAACC |
| 3′Dδ2 | CACAGTG | TTGCAAACCCCATAGGGACCTGT | ACAAAAACT |
| 5′Jβ1.1 | CACAGTG | CCATAGGATGAG | GAGAAAAAT |
| 5′Jβ2.1 | CACAGCA | GAAAAGGGCTAC | CAAGAATTC |
| 5′Jδ1 | CACAGCT | ACTGAGGCCATT | CCAAAAACC |
Nucleotides different from the consensus sequences (heptamer, CACAGTG and nonamer, ACAAAAACC) are underlined.
Increased frequency in aberrant joints associated with the expression of core RAGs have never been reported in studies using extrachromosomal substrates (8–12,28,29,31,59–61). There are several reasons why these effects have never been found in transfection studies. First, RSS tested for joining in plasmid substrates tended to be limited to so-called ‘consensus’ RSS. Our results demonstrate that not all combinations of RSS result in extensive changes to CJ or SJ. Although endogenous RSS have been tested for ex vivo recombination and in vitro binding/cleavage assays (56,57,62,63), specific features of these joints were not addressed in these studies. Thus, it will be important to compare core and full-length RAGs for quality of end-joining between different RSS pairs formed ex vivo. In addition, LM–PCR should be performed with and without T4 DNA polymerase treatment, since we cannot completely rule out the possibility that truncated RAGs might produce aberrant SE with certain pairs of RSS. Second, artificial substrates are composed of limited coding flank sequences. Since an importance of coding flank sequence to recombination frequency has been shown in the earlier study (64), it has shown to play a critical role in binding (53,65,66), cleavage (47,48,67) and accessibility control by RAGs (68). Thus, coding flank sequences might also affect the joining reaction. Third, the structure of short D gene segments with RSS at both 3′ and 5′ ends might account for the aberrant cleavage or misalignment of core RAGs. From this point of view, D segments attached to 3′ and 5′RSS should be tested in artificial substrates to compare the end-processing of full-length RAGs versus core RAGs. Fourth, chromatin structure in endogenous loci might make core RAGs more sensitive to subtle difference in RSS. To further address these issues, the structure of cleaved ends, junctions and stability of the SEC will need to be compared in full-length versus core RAGs, and with different combinations of RSS, including natural D coding segments, in naked or chromatinized substrates. Our results indicate that the non-core regions of RAGs are critical for tolerating variations between RSS and to suppress aberrant joints in endogenous loci. In combination with our findings, future analyses focusing on the above aspects in vitro should reveal the complete role of RAG proteins in V(D)J joining.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Craig H. Bassing and Yan Dai for critical reading of the manuscript, Hiroshi Nakase for stimulating discussions and Kaori Gotoh for help with our initial work. This study was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan (Y.T. and Y.A.) and NIH grants (F.W.A.). F.W.A. is the investigator of the HHMI.
REFERENCES
- 1.Bassing C.H., Swat,W. and Alt,F.W. (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell, 109 (Suppl.), S45–S55. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M. (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem., 71, 101–132. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal A. and Schatz,D.G. (1997) RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell, 89, 43–53. [DOI] [PubMed] [Google Scholar]
- 4.Hiom K. and Gellert,M. (1998) Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell, 1, 1011–1019. [DOI] [PubMed] [Google Scholar]
- 5.Jones J.M. and Gellert,M. (2001) Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl Acad. Sci. USA, 98, 12926–12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt V.L. and Roth,D.B. (2002) A recombinase diversified: new functions of the RAG proteins. Curr. Opin. Immunol., 14, 224–229. [DOI] [PubMed] [Google Scholar]
- 7.Dai Y., Kysela,B., Hanakahi,L.A., Manolis,K., Riballo,E., Stumm,M., Harville,T.O., West,S.C., Oettinger,M.A. and Jeggo,P.A. (2003) Nonhomologous end joining and V(D)J recombination require an additional factor. Proc. Natl Acad. Sci. USA, 100, 2462–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuomo C.A. and Oettinger,M.A. (1994) Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res., 22, 1810–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirch S.A., Sudarsanam,P. and Oettinger,M.A. (1996) Regions of RAG1 protein critical for V(D)J recombination. Eur. J. Immunol., 26, 886–891. [DOI] [PubMed] [Google Scholar]
- 10.Sadofsky M.J., Hesse,J.E., McBlane,J.F. and Gellert,M. (1993) Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res., 21, 5644–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadofsky M.J., Hesse,J.E. and Gellert,M. (1994) Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res., 22, 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silver D.P., Spanopoulou,E., Mulligan,R.C. and Baltimore,D. (1993) Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl Acad. Sci. USA, 90, 6100–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkin S.K., Matthews,A.G. and Oettinger,M.A. (2003) The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J., 22, 1931–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melek M. and Gellert,M. (2000) RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell, 101, 625–633. [DOI] [PubMed] [Google Scholar]
- 15.Sekiguchi J.A., Whitlow,S. and Alt,F.W. (2001) Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell, 8, 1383–1390. [DOI] [PubMed] [Google Scholar]
- 16.Swanson P.C., Volkmer,D. and Wang,L. (2004) Full-length RAG-2, and not full-length RAG1, specifically suppresses RAG-mediated transposition, but not hybrid joint formation or disintegration. J. Biol. Chem., 279, 4034–4044. [DOI] [PubMed] [Google Scholar]
- 17.Tsai C.L. and Schatz,D.G. (2003) Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J., 22, 1922–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones J.M. and Gellert,M. (2003) Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc. Natl Acad. Sci. USA, 100, 15446–15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J. and Desiderio,S. (1999) Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity, 11, 771–781. [DOI] [PubMed] [Google Scholar]
- 20.Yurchenko V., Xue,Z. and Sadofsky,M. (2003) The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev., 17, 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corneo B., Benmerah,A. and Villartay,J.P. (2002) A short peptide at the C-terminus is responsible for the nuclear localization of RAG2. Eur. J. Immunol., 32, 2068–2073. [DOI] [PubMed] [Google Scholar]
- 22.Spanopoulou E., Cortes,P., Shih,C., Huang,C.M., Silver,D.P., Svec,P. and Baltimore,D. (1995) Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity, 3, 715–726. [DOI] [PubMed] [Google Scholar]
- 23.Akamatsu Y., Monroe,R., Dudley,D.D., Elkin,S.K., Gartner,F., Talukder,S.R., Takahama,Y., Alt,F.W., Bassing,C.H. and Oettinger,M.A. (2003) Deletion of the RAG2 C-terminus leads to impaired lymphoid development in mice. Proc. Natl Acad. Sci. USA, 100, 1209–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudley D.D., Sekiguchi,J., Zhu,C., Sadofsky,M.J., Whitlow,S., DeVido,J., Monroe,R.J., Bassing,C.H. and Alt,F.W. (2003) Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J. Exp. Med., 198, 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang H.E., Hsu,L.Y., Cado,D., Cowell,L.G., Kelsoe,G. and Schlissel,M.S. (2002) The ‘dispensable’ portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity, 17, 639–651. [DOI] [PubMed] [Google Scholar]
- 26.Villa A., Sobacchi,C., Notarangelo,L.D., Bozzi,F., Abinun,M., Abrahamsen,T.G., Arkwright,P.D., Baniyash,M., Brooks,E.G., Conley,M.E. et al. (2001) V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood, 97, 81–88. [DOI] [PubMed] [Google Scholar]
- 27.Steen S.B., Han,J.O., Mundy,C., Oettinger,M.A. and Roth,D.B. (1999) Roles of the ‘dispensable’ portions of RAG-1 and RAG-2 in V(D)J recombination. Mol. Cell Biol., 19, 3010–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu J.X., Kale,S.B., Yarnell Schultz,H. and Roth,D.B. (2001) Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell, 7, 77–87. [DOI] [PubMed] [Google Scholar]
- 29.Yarnell Schultz H., Landree,M.A., Qiu,J.X., Kale,S.B. and Roth,D.B. (2001) Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol. Cell, 7, 65–75. [DOI] [PubMed] [Google Scholar]
- 30.Tsai C.L., Drejer,A.H. and Schatz,D.G. (2002) Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev., 16, 1934–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huye L.E., Purugganan,M.M., Jiang,M.M. and Roth,D.B. (2002) Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell Biol., 22, 3460–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller S.A., Dykes,D.D. and Polesky,H.F. (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res., 16, 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis S.M. (1994) The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv. Immunol., 56, 27–150. [DOI] [PubMed] [Google Scholar]
- 34.Starr T.K., Jameson,S.C. and Hogquist,K.A. (2003) Positive and negative selection of T cells. Annu. Rev. Immunol., 21, 139–176. [DOI] [PubMed] [Google Scholar]
- 35.Clatworthy A.E., Valencia,M.A., Haber,J.E. and Oettinger,M.A. (2003) V(D)J recombination and RAG-mediated transposition in yeast. Mol. Cell, 12, 489–499. [DOI] [PubMed] [Google Scholar]
- 36.Kanaar R., Hoeijmakers,J.H.J. and van Gent,D.C. (1998) Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol., 8, 483–489. [DOI] [PubMed] [Google Scholar]
- 37.Baumann P. and West,S.C. (1998) DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA, 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabotyanski E.B., Gomelsky,L., Han,J.O., Stamato,T.D. and Roth,D.B. (1998) Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res., 26, 5333–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verkaik N.S., Esveldt-van Lange,R.E., van Heemst,D., Bruggenwirth,H.T., Hoeijmakers,J.H., Zdzienicka,M.Z. and van Gent,D.C. (2002) Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol., 32, 701–709. [DOI] [PubMed] [Google Scholar]
- 40.Roth D.B. (2003) Restraining the V(D)J recombinase. Nature Rev. Immunol., 3, 656–666. [DOI] [PubMed] [Google Scholar]
- 41.Li Z., Otevrel,T., Gao,Y., Cheng,H.L., Seed,B., Stamato,T.D., Taccioli,G.E. and Alt,F.W. (1995) The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell, 83, 1079–1089. [DOI] [PubMed] [Google Scholar]
- 42.Roth D.B., Proctor,G.N., Stewart,L.K. and Wilson,J.H. (1991) Oligonucleotide capture during end joining in mammalian cells. Nucleic Acids Res., 19, 7201–7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lieber M.R. (1992) The mechanism of V(D)J recombination: a balance of diversity, specificity, and stability. Cell, 70, 873–876. [DOI] [PubMed] [Google Scholar]
- 44.Lieber M.R., Hesse,J.E., Lewis,S., Bosma,G.C., Rosenberg,N., Mizuuchi,K., Bosma,M.J. and Gellert,M. (1988) The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell, 55, 7–16. [DOI] [PubMed] [Google Scholar]
- 45.Schuler W., Ruetsch,N.R., Amsler,M. and Bosma,M.J. (1991) Coding joint formation of endogenous T cell receptor genes in lymphoid cells from scid mice: unusual P-nucleotide additions in VJ-coding joints. Eur. J. Immunol., 21, 589–596. [DOI] [PubMed] [Google Scholar]
- 46.Carroll A.M., Slack,J.K. and Mu,X. (1993) V(D)J recombination generates a high frequency of nonstandard TCR D delta-associated rearrangements in thymocytes. J. Immunol., 150, 2222–2230. [PubMed] [Google Scholar]
- 47.Ramsden D.A., McBlane,J.F., van Gent,D.C. and Gellert,M. (1996) Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J., 15, 3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 48.Cuomo C.A., Mundy,C.L. and Oettinger,M.A. (1996) DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell Biol., 16, 5683–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tycko B. and Sklar,J. (1990) Chromosomal translocations in lymphoid neoplasia: a reappraisal of the recombinase model. Cancer Cells, 2, 1–8. [PubMed] [Google Scholar]
- 50.Jones J.M. and Gellert,M. (2002) Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J., 21, 4162–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mundy C.L., Patenge,N., Matthews,A.G. and Oettinger,M.A. (2002) Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell Biol., 22, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raghavan S.C., Swanson,P.C., Wu,X., Hsieh,C.L. and Lieber,M.R. (2004) A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature, 428, 88–93. [DOI] [PubMed] [Google Scholar]
- 53.Nagawa F., Hirose,S., Nishizumi,H., Nishihara,T. and Sakano,H. (2004) Joining mutants of RAG1 and RAG2 demonstrate impaired interactions with the coding-end DNA. J. Biol. Chem. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 54.Sleckman B.P., Bassing,C.H., Hughes,M.M., Okada,A., D'Auteuil,M., Wehrly,T.D., Woodman,B.B., Davidson,L., Chen,J. and Alt,F.W. (2000) Mechanisms that direct ordered assembly of T cell receptor beta locus V, D, and J gene segments. Proc. Natl Acad. Sci. USA, 97, 7975–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bassing C.H., Alt,F.W., Hughes,M.M., D'Auteuil,M., Wehrly,T.D., Woodman,B.B., Gartner,F., White,J.M., Davidson,L. and Sleckman,B.P. (2000) Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature, 405, 583–586. [DOI] [PubMed] [Google Scholar]
- 56.Montalbano A., Ogwaro,K.M., Tang,A., Matthews,A.G., Larijani,M., Oettinger,M.A. and Feeney,A.J. (2003) V(D)J recombination frequencies can be profoundly affected by changes in the spacer sequence. J. Immunol., 171, 5296–5304. [DOI] [PubMed] [Google Scholar]
- 57.Lee A.I., Fugmann,S.D., Cowell,L.G., Ptaszek,L.M., Kelsoe,G. and Schatz,D.G. (2003) A functional analysis of the spacer of V(D)J recombination signal sequences. PLoS Biol., 1, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee G.S., Neiditch,M.B., Salus,S.S. and Roth,D.B. (2004) RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell, 117, 171–184. [DOI] [PubMed] [Google Scholar]
- 59.Fugmann S.D., Villey,I.J., Ptaszek,L.M. and Schatz,D.G. (2000) Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell, 5, 97–107. [DOI] [PubMed] [Google Scholar]
- 60.Fugmann S.D. and Schatz,D.G. (2001) Identification of basic residues in RAG2 critical for DNA binding by the RAG1–RAG2 complex. Mol. Cell, 8, 899–910. [DOI] [PubMed] [Google Scholar]
- 61.Purugganan M.M., Shah,S., Kearney,J.F. and Roth,D.B. (2001) Ku80 is required for addition of N nucleotides to V(D)J recombination junctions by terminal deoxynucleotidyl transferase. Nucleic Acids Res., 29, 1638–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu K., Taghva,A. and Lieber,M.R. (2002) The cleavage efficiency of the human immunoglobulin heavy chain VH elements by the RAG complex. J. Biol. Chem., 277, 5040–5046. [DOI] [PubMed] [Google Scholar]
- 63.Olaru A., Patterson,D.N., Villey,I. and Livak,F. (2003) DNA–Rag protein interaction in the control of selective D gene utilization in the TCR beta locus. J. Immunol., 171, 3605–3611. [DOI] [PubMed] [Google Scholar]
- 64.Gerstein R.M. and Lieber,M.R. (1993) Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev., 7, 1459–1469. [DOI] [PubMed] [Google Scholar]
- 65.Eastman Q.M., Villey,I.J. and Schatz,D.G. (1999) Detection of RAG protein–V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell Biol., 19, 3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mo X., Bailin,T. and Sadofsky,M.J. (2001) A C-terminal region of RAG1 contacts the coding DNA during V(D)J recombination. Mol. Cell Biol., 21, 2038–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu K. and Lieber,M.R. (1999) Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol. Cell Biol., 19, 8094–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jung D., Bassing,C.H., Fugmann,S.D., Cheng,H.L., Schatz,D.G. and Alt,F.W. (2003) Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted VDJ recombination in nonlymphoid cells. Immunity, 18, 65–74. [DOI] [PubMed] [Google Scholar]
- 69.Livak F., Burtrum,D.B., Rowen,L., Schatz,D.G. and Petrie,H.T. (2000) Genetic modulation of T cell receptor gene segment usage during somatic recombination. J. Exp. Med., 192, 1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]