Abstract
We have developed a method for genomic representation using Type IIB restriction endonucleases. Representation by concatenation of restriction digests, or RECORD, is an approach to sample the fragments generated by cleavage with these enzymes. Here, we show that the RECORD libraries may be used for digital karyotyping and for pathogen identification by computational subtraction.
INTRODUCTION
High-throughput genome analysis can be facilitated by the use of fractional representations of the genome. Several methods for representational analyses of concatenated sequence tags have been described, including serial analysis of gene expression (SAGE) and long SAGE (1,2), tandem arrayed ligation of expressed sequence tags (TALEST) (3), digital karyotyping (4,5) and genomic signature tags (6). These ‘SAGE-like’ methods rely on sequencing libraries of concatenated short DNA fragments generated using Type IIS restriction enzymes, which cleave outside their recognition sequence (7). Type IIS enzymes are used as ‘tagging enzymes’; tags can be generated by ligating linkers containing a Type IIS recognition sequence to DNA or cDNA that has been digested with a frequently cutting restriction enzyme, such as NlaIII. The DNA is then digested using the specific Type IIS enzyme, and the fragment flanking the linker site is released. SAGE-like protocols require several ligation and restriction enzyme digestion steps in addition to cloning, PCR and multiple purifications. PCR amplification of ditags has been shown to create bias in SAGE-like protocols, but this phenomenon is not believed to affect cloning-based amplification of tags (3). The maximum length of tags generated is limited by the availability of Type IIS restriction enzymes and the upper limit using commercially available enzymes is currently 21 bases (including 4 bases from the NlaIII site).
Genomic representations are useful for those applications that will benefit from increasing the throughput of sequencing. For example, we have recently described a new sequence-based method for pathogen discovery. ‘Computational subtraction’ eliminates sequences that match the human genome in silico from sequenced libraries of human genomic DNA or cDNA (8,9). Briefly, DNA- or mRNA-based libraries are made using specimens from diseases believed to be of infectious origin, but where the causative agent has not been identified (10). The sequences generated from these libraries are compared with databases of human nucleic acid sequences. After sequences with a significant match to the human genome are subtracted, the presence of ‘non-human’ sequences in such libraries can be linked to the presence of a pathogen. This approach requires accurate high-throughput sequencing and effective analytical tools for in silico sequence filtering. The increased throughput of concatenated libraries could increase the power of computational subtraction-based pathogen discovery.
Numerous methods have been developed for high resolution karyotyping. Digital karyotyping uses sequence data from concatenated libraries made using genomic DNA and Type IIS restriction enzymes. Unique sequence tags can be matched to their corresponding site in the genome, and statistical analyses of tag density can be used to assess the relative ploidy of different loci (4). Other methods rely on the hybridization of a fragmented and labeled representation of genomic DNA to arrays, and single nucleotide polymorphism (SNP) arrays have been used for this purpose (11–13).
Type IIB restriction endonucleases cleave both strands of template DNA upstream and downstream of a specific recognition site (7). Similar to most bacterial restriction endonucleases, Type IIB enzymes are part of the restriction/methylation systems that protect host bacteria from foreign DNA (14,15). The target base for methylation is likely to be the adenine nucleotide symmetrically flanking the six or seven base pair core of the recognition sequence [(16) and Table 1]. All described Type IIB enzymes leave a 3′ overhang after cutting, and released tags range in size from 29 to 40 bases including the cohesive ends and from 21 to 33 bases without the cohesive ends. Recognition sequences are generally interrupted and range from five to seven bases long. Some recognition sites are palindromically symmetrical whereas others are not. The enzymes listed in Table 1 have a predicted cutting frequency ranging from one site per 8192 bases to one site per 512 bases.
Table 1. List of Type IIB restriction enzymes.
| Enzyme and recognition sequence | Cutting frequencya | Blunt tag length |
|---|---|---|
| CspCI(11/13)b CAANNNNNGTGG(12/10) | 8192 | 33 |
| AloI(7/12) GAACNNNNNNTCC(12/7) | 8192 | 27 |
| PpiI(7/12) GAACNNNNNCTC (13/8) | 8192 | 28 |
| PsrI(7/12) GAACNNNNNNTAC(12/7) | 8192 | 27 |
| BplI(8/13) GAGNNNNNCTC (13/8) | 4096 | 27 |
| FalI(8/13) AAGNNNNNCTT (13/8) | 4096 | 27 |
| Bsp24I(8/13)c GACNNNNNNTGG(12/7) | 2048 | 27 |
| BsaXI(9/12) ACNNNNNCTCC(10/7) | 2048 | 27 |
| HaeIV(7/13)c GAYNNNNNRTC (14/9) | 1024 | 27 |
| CjeI(8/14)c CCANNNNNNGT (15/9) | 512 | 28 |
| CjePI(7/13)c CCANNNNNNNTC (14/8) | 512 | 27 |
| Hin4I(8/13) GAYNNNNNVTC (13/8) | 512 | 27 |
| BaeI(10/15) ACNNNNGTAYC(12/7) | 4096 | 28 |
| AlfI(10/12) GCANNNNNNTGC (12/10) | 4096 | 32 |
| BcgI(10/12) CGANNNNNNTGC (12/10) | 2048 | 32 |
| BslFI(6/10) GGGAC (10/14) | 512 | 21 |
Boldfaced adenines, in addition to the symmetric adenine base pairing with the boldfaced thymidines, indicate possible target nucleotides for methylation.
aAverage, theoretical distance between cut sites assuming random sequence with no base composition bias.
bNumber of bases away from recognition sequence where enzyme cuts in upper/lower strand.
cEnzyme currently not commercially available.
We now show that Type IIB restriction enzyme digests can be used to construct concatenated libraries. The RECORD method for library construction has several advantages over SAGE-like methods. It is PCR-independent and provides an alternative way to perform karyotyping that should be particularly useful when working with systems where array-based platforms are not available.
As a proof of principle, we have made two different libraries from human genomic DNA using BsaXI as a tag-generating enzyme. We show that the databases of genomic tags can be used to investigate genomic contents by mapping tags to their positions on the genome and inferring copy number from mapped tag density using a hidden Markov model (HMM) approach. Tags can also be used for the detection of pathogens, and a nested ‘vectorette PCR’ method is described that allows efficient amplification of sequences flanking genomic BsaXI tags.
METHODS
Overview of method
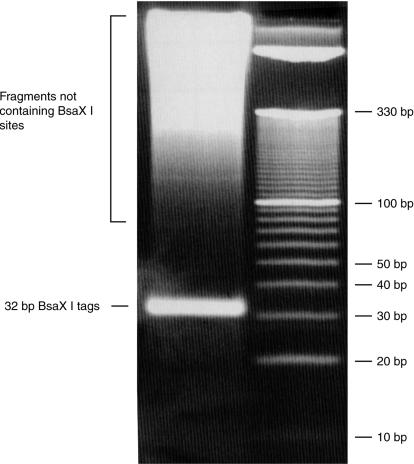
An example of digestion with a Type IIB restriction enzyme, BsaXI, is shown in Figure 1. Note that the 32 bp restriction digest tag is clearly separated from the smear of higher molecular weight fragments that do not contain the recognition site.
Figure 1.
BsaXI digested human genomic DNA with 10 bp ladder (Invitrogen). Acrylamide gel (8%), stained with GelStar (Cambrex).
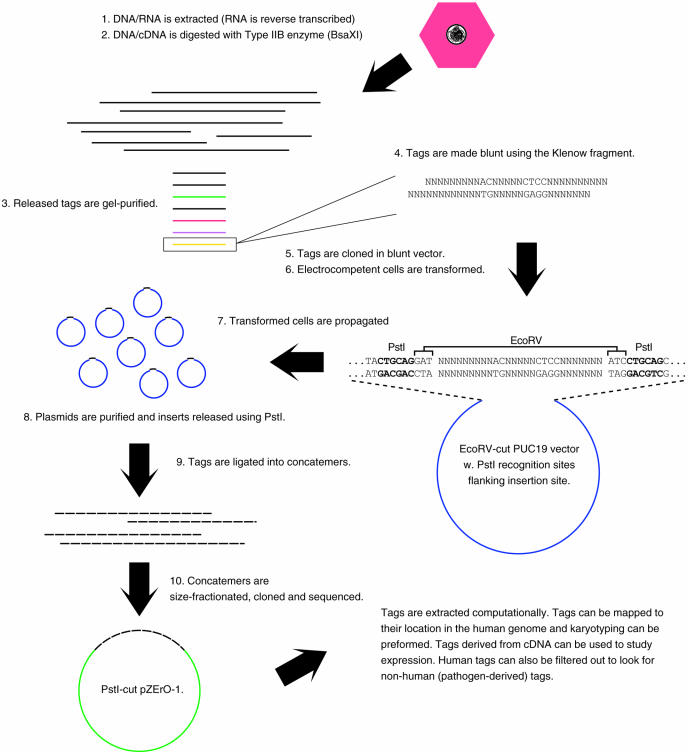
The first step in library construction is the digestion of DNA with a Type IIB restriction enzyme such as BsaXI (Figure 2, steps 1–3). After digestion, the reaction mixture is phenol/chloroform extracted and ethanol/ammonium acetate precipitated (17). The digest is then run on an 8% polyacrylamide gel using TBE buffer (200 V for 2.5 h). The gel is stained using GelStar (Cambrex, East Rutherford, NJ), and the band corresponding to the BsaXI tags excised. Tags are purified using the crush and soak method (17), and dissolved in 39.5 μl of dH2O and 5 μl of Eco Pol Buffer (New England Biolabs, Beverly, MA). Blunting of tags is done by adding 5 U of large fragment DNA polymerase I (‘Klenow’; New England Biolabs) in the presence of 33 μM of each dNTP for 15 min at 25°C in a total volume of 50 μl (Figure 2, step 4).
Figure 2.
Overview of method for the construction of Type IIB restriction enzyme tag libraries.
After blunting, tags are phenol/chloroform extracted and ethanol/ammonium acetate precipitated before resuspension in 7 μl of dH2O with 1 μl of 10× T4 DNA ligase buffer (New England Biolabs). A primary ligation is done by adding 200 ng of vector, 1 μl of high concentration T4 DNA ligase (New England Biolabs) and incubating overnight at 16°C. The vector used is an EcoRV cleaved, dephosphorylated pUC19 plasmid (Invitrogen, Carlsbad, CA) that has been modified to contain two PstI sites immediately flanking an EcoRV site (Figure 2, step 5). Ligations are phenol/chloroform extracted and ethanol/ammonium acetate precipitated, and electrocompetent E.cloni™ 10G Elite cells (Lucigen, Middleton, WI) transformed in accordance with the manufacturer's recommendations (Figure 2, step 6). After electroporation and 1 h incubation, the transformations are transferred to 250 ml TB medium containing 75 μg/ml ampicillin. Cells are grown until OD600 reaches ∼1.6 (∼13 h) and plasmids purified using a QIAfilter Plasmid Maxi kit (Qiagen, Valencia, CA) (Figure 2, step 7). An aliquot of 200 μg of plasmids is digested using 1000 U of PstI (New England Biolabs) (Figure 2, step 8). Digests are phenol/chloroform extracted and ethanol/ammonium acetate precipitated and run on an 8% polyacrylamide gel (200 V for 20 min—just sufficient to separate released inserts from opened vector). Released tags are crush and soak gel purified, and dissolved in 8 μl of dH2O and 1 μl of 10× T4 DNA ligase buffer. An aliquot of 1 μl of high concentration T4 DNA ligase is added and the concatenation reaction incubated for 1 h at 16°C (Figure 2, step 9).
Concatenates are loaded directly on a 13 cm, agarose gel (1.5%) containing GelStar and electrophoresed for 1.5 h (125 V). Concatenation products between 1200 and ∼3000 bp can be gel-purified using the MinElute gel extraction kit (Qiagen). Concatenates are cloned in a PstI-cleaved p-ZeRO-1 vector, and secondary transformations done using E.cloni™ 10G Elite cells as described above (Figure 2, step 10). Clones are sequenced, and the tags can be extracted computationally and stored in a database for analyses.
Construction of karyotyping libraries
Two experimental libraries were made using DNA purchased from ATCC (American Type Culture Collection, Manassas, VA). One library was made for karyotyping using DNA from a breast cancer cell line (primary ductal carcinoma, HCC38) and a matched normal library was made from the corresponding Epstein–Barr virus (EBV) transformed blood cell line (HCC38 BL). An aliquot of 15 μg of DNA was digested using 60 U of BsaXI (New England Biolabs), and 2208 clones from each concatenated library were sequenced unidirectionally by SeqWright (Houston, TX).
Digital karyotyping
To determine whether Type IIB restriction enzyme tags could be used for digital karyotyping (4), we generated a virtual BsaXI tag map of the human genome. Computational analysis of build 34 of the human genome (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens) identified 1 303 799 virtual BsaXI cut sites. When comparing the 27 bases that comprise the double-stranded part of the tags, 1 067 617 had unique sequences and could thus be unambiguously placed in the human genome. The average distance between unique tags was 2449 bp (median: 1507 bp). Using this in silico digest, tags from the experimental libraries were mapped to their positions on the genome by matching their sequences to those obtained from the virtual cut sites.
Applying the principle that the mapped density of experimentally derived tags should be higher in regions of genomic amplification than in normal regions (and similarly, should be lower in regions of deletion), we developed a HMM-based approach (18) to perform digital karyotyping of the RECORD data.
The data resulting from mapping the experimentally derived tags to their positions on the genome may be thought of as a string of 1 067 617 non-negative integers—one integer for each in silico tag—each indicating the number of times that the corresponding tag was found in the library. If we let n denote the number of library tags sequenced, then the integers in the data string sum to n. The scale of sequencing in our experiments yielded integer strings overwhelmingly comprised of zeroes. The data string was transformed in two steps in order to make the data as compact as possible for efficient and accurate HMM estimation procedures. First, any integers larger than 1 (quite rare for our data) was truncated to 1, and the difference was ‘spread’ to the nearest zeroes so that some of these zeroes became ones. Second, this new string was again transformed to indicate the lengths of uninterrupted runs of zeroes. For example, the two-step procedure applied to an initial data string
0 1 0 0 0 1 1 0 2 0 0 0 0 1 0 1…
would yield
0 1 0 0 0 1 1 0 1 1 0 0 0 1 0 1…
for the first step, followed by
1 3 0 1 0 3 1…
The zeroes in this final string indicate consecutive positive integers in the second string.
It is relatively straightforward to show that integers in a data string, after the two-step transformation, follow an approximately geometric distribution with parameter (kn)/(2 135 234), where k represents the copy number (normal = 2) of the region containing the in silico tags corresponding to the integer. For simplification, we assumed that each integer corresponded to one of four states: normal, amplification, heterozygous deletion and homozygous deletion. Before applying the HMM, the expectation-maximization (EM) algorithm (19) was employed, assuming a mixture of four geometric distributions (one for each state), to find initial estimates for the HMM parameters. The HMM was then run, using the implementation in the R software (http://www.R-project.org) package developed by J. K. Lindsey (http://popgen0146uns50.unimaas.nl/~jlindsey/rcode.html). The resulting estimates were used to infer copy number k at each in silico tag site.
Computational subtraction
For computational subtraction, tags from the HCC38 BL library were filtered with regard to sequence quality and tag length. Tags that appeared shorter than 27 bases or had bases with phred scores lower than 20 were removed. To test the accuracy of the subtraction, a control dataset was made by computationally extracting 9989 tags from 1292 complete viral genomes (ftp://ftp.ncbi.nih.gov/release/viral) and 9953 tags from 164 complete bacterial genomes (ftp://ftp.ncbi.nih.gov/genomes/bacteria).
For the initial computational subtraction of tags, MegaBLAST (20,21) was used with word size 12, score for match 1 and penalty for mismatch −2. Tag sequences were compared against phase0, 1, 2 and 3 of the human genome (ftp://ftp.ncbi.nih.gov/genbank/genomes/H_sapiens/), build 34 of the human genome (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/) and the mitochondrial genome (accession number NC_001807.4) sequentially. High-scoring tags were removed, and a second round of subtraction was carried out using BLAST (22). BLAST was run using word size 7, E-value 1000, cost to open gap 5, cost to extend gap 2 and −3 as penalty for nucleotide mismatch. The BLAST database comprises phase0, 1, 2 and 3, build 34 and the mitochondrial genome. Tags were ranked according to bit scores and high-scoring tags were sequentially subtracted.
Vectorette PCR
Several methods exist for PCR amplification of cDNA regions flanking SAGE tags (23,24), but no such method has been described for tags derived from genomic DNA. We designed a modified ‘vectorette PCR’ protocol based on a previously published method for genome walking in Drosophila (25). Vectorette templates were made by digesting 1 μg of HCC38 and HCC38 BL DNA with 10 U of NlaIII (New England Biolabs), and ligating vectorettes with NlaIII compatible cohesive ends to the digested DNA as described by Ko et al. (25).
Vectorette PCR was performed using a nested approach. Primers for first round PCR were designed from BsaXI tags by converting tag sequences into the form ‘NNNNNNNNNACNNNNNCTCCNNNNNNN’. Primers corresponding to the first 22 bases were made, and nested primers were designed by shifting the 22 bases three bases toward the 3′ end of the tag sequences.
First round PCR was done using a ‘touchdown’ method with 1.5 mM [Mg2+] and AmpliTaq Gold hot start polymerase (Applied Biosystems, Foster City, CA). Cycles were as follows: 95°C for 5 min (denaturing and activation of polymerase), 5 cycles of 95°C for 30 s, 72°C for 30 s, 72°C for 30 s; 5 cycles of 95°C for 30 s, 68°C for 30 s, 72°C for 30 s; 15 cycles of 95°C for 30 s, 64°C for 30 s, 72°C for 30 s; and 15 cycles of 95°C for 30 s, 64°C for 30 s, 72°C for 30 s. Products were then diluted 1000-fold and used as a template for the second round PCR. Secondary PCR was done using AmpliTaq Gold polymerase in combination with uracil N-glycosylase (AmpErase) (Applied Biosystems) to avoid contamination. Amplifications were done using uracil instead of thymidine, 2 mM [Mg2+] and the following cycle: 37°C for 10 min (for the AmpErase); 95°C for 5 min (to denature template, activate hot start polymerase and inactivate AmpErase); 25 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 45 s; and a final elongation step of 5 min (72°C). PCR products were electrophoresed on a 1.8% agarose gel stained with GelStar.
RESULTS
Digital karyotyping
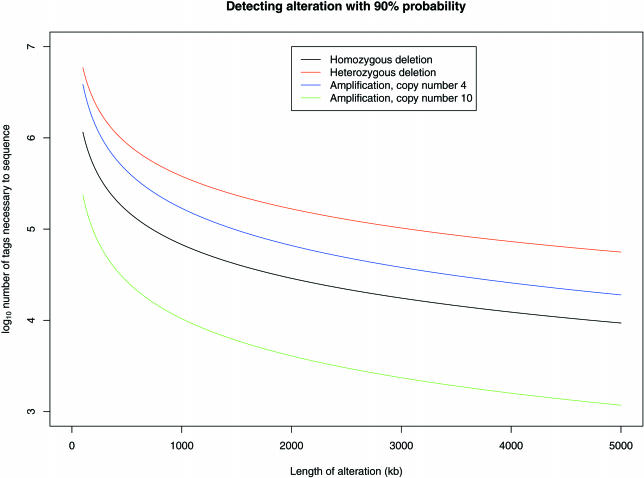
To check the theoretical resolving power of mapped BsaXI tags, a simulation study was performed to determine the number of tags one would have to sequence in order to detect, with 90% probability using the HMM approach (see Methods above), various types of alterations with a range of lengths. As expected, the detection of smaller alterations requires a dramatically larger number of sequenced tags, and heterozygous deletions are the most challenging alterations to detect with high degree of certainty (Figure 3). A related simulation of a normal genome to determine false positive rates demonstrated strong specificity for the approach, even for modest numbers of sequenced tags (data not shown).
Figure 3.
The results of a simulation study analyzing number of sequenced tags necessary to detect various types of alterations, as a function of alteration length. For each of the four types of alterations shown, digital karyotyping experiments were simulated for a range of alteration lengths. These simulations were conducted by randomly sampling from a set of integers representing the in silico tags, with the randomly placed altered region simulated by increasing (for amplification) or decreasing (for deletion) the integer value representing the number of copies of the corresponding tags. Simulations were repeated 100 times for each combination of alteration type and length, and isomeric curves were fit to 90% detection rates for each alteration type.
From the 2208 clones sequenced, 21 122 full-length tags with phred scores >20 for all bases could be extracted from the HCC38 concatenates, and 22 397 tags could be extracted from the HCC38 BL sequence reads. These tags were compared with databases of human genomic sequences (phase0, 1, 2 and 3, build 34 and the mitochondrial genome), and 16 997 (80.5%) of the HCC38 tags matched perfectly. Tags from the HCC38 BL library were compared to the human genomic sequences and the EBV genome (accession number NC_001345), and 19 894 tags (88.8%) were perfect matches.
To maximize the resolving power of our dataset, all tags that matched build 34 of the human genome were used to perform the digital karyotyping analysis. Tags were extracted without any quality filtering and compared to build 34. A total of 16 606 tags from the HCC38 library and 18 868 tags from the HCC38 BL library could be extracted and matched perfectly to unique sites in the human genome (3251 and 3802 from the respective libraries matched non-unique loci; note that the totals of the unselected tags exceed those with phred scores > 20 for every base). The HMM analysis of the tag data from the normal HCC38 BL library falsely identified 3.8% of the 1 067 617 virtual cut sites as being in altered regions, a false positive rate that matches closely with that predicted from the simulation study. This concordance speaks well for the fidelity of our procedures. Tags derived from the HCC38 BL library were also used as a normal control to make sure that there were no systematic biases in our library construction protocol. Initial analysis of ploidy indicated that HCC38 BL had no bias in numbers of tags obtained from each chromosome when compared to the expected numbers of tags (given the number of virtual cut sites), but chromosome 20 appeared overrepresented for HCC38. The cell line has been reported to have aberrant ploidy [modal chromosome number: 75 (with a range of 65–79); polyploidy rate: 22%; and the number of cells examined: 59, information from www.atcc.org], and subsequent spectral karyotyping analysis (26) of HCC38 cell line DNA showed a complex karyotype with all observed chromosomes comprising fragments of different genomic origins (Supplementary Figure 1).
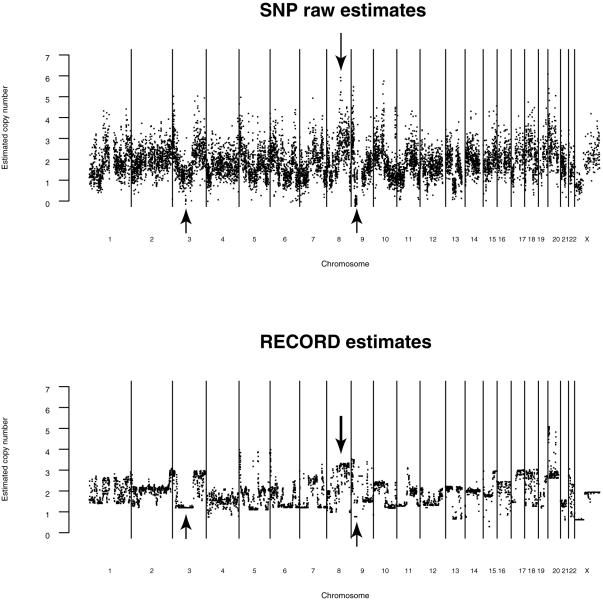
The HMM analysis of the tags from the HCC38 RECORD library uncovered many suspected specific regions of chromosomal alteration (Figure 4, lower panel). We compared these results to a previous karyotyping study performed on the same HCC38 cell line, in which a similar HMM approach was applied to SNP array data in order to estimate copy number (12) (Figure 4, upper panel). Three of the predicted alterations had been confirmed by PCR, comprising deletions within chromosomes 3 and 9 and an amplification within chromosome 8, all of which were identified by HMM analysis of our RECORD library (Figure 4, arrows). Comparisons of SNP copy number estimates versus the RECORD library copy number estimates indicated a significant positive correlation (Pearson correlation ≈ 0.57, P < 10−15 and for scatter plot, see Supplementary Figure 2). This correlation is somewhat lower than that seen between array methods (0.62–0.76), most probably due to the resolution limits of the analysis at this sequencing depth, and to the discrete nature of the HMM data.
Figure 4.
Predicted copy numbers using SNP arrays (top panel) and RECORD libraries (bottom panel). Arrows indicated PCR-confirmed deletions of the ROBO1 gene (base position ∼79.1 Mb in chromosome 3) and the CDKN2A tumor suppressor gene (base position ∼22.0 Mb in chromosome 9) and a PCR-confirmed amplification of the region containing the MYC oncogene (base position ∼128.7 Mb in chromosome 8).
Pathogen detection using RECORD library analysis of genomic DNA
To test the feasibility of using RECORD libraries for pathogen detection and discovery, we used the 22 397 27 bp tags (19 934 different tag sequences) of high sequence quality from our HCC38 BL dataset (2208 clones sequenced in one direction). Of these tags, 44 different tags (77 tags total) were perfect matches to the EBV genome.
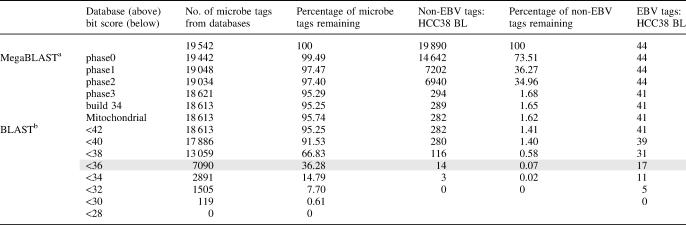
For MegaBLAST and BLAST-based subtraction, three different datasets were analyzed: 19 542 tags extracted from known, sequenced viral and bacterial genomes (‘microbe tags’); the 19 890 different HCC38 BL tag sequences; and the 44 EBV tags. In an initial analysis, subtraction was performed using MegaBLAST against the various phases of human genome sequence and build 34 of the human genome as well as human mitochondrial sequences. After removing tags that matched the databases with bit scores above 40 (roughly equivalent to 20 consecutive identical nucleotides), we found that 4.26% of the in silico-derived microbe tags were removed (95.74% remaining), 98.38% of the HCC38 BL non-EBV tags were removed after subtraction (1.62% remaining) and 3 of the HCC38BL EBV-derived tags were removed (41 remaining) (Table 2, top section).
Table 2. Computational subtraction of BsaXI tags.
Primers for vectorette PCR were designed from HCC38 BL tags that had bit scores below 36 after BLAST analysis (highlighted in grey).
aMegaBLAST analyses (word size 12, score for match 1 and penalty for mismatch −2) were done using phase0, phase1, phase2 and phase3 of the human genome, build 34 of the human genome and the mitochondrial genome. Tags were sequentially compared to the databases, and tags with bit scores above 40 removed.
bBLAST analyses (word size 7, E-value 1000, cost to open gap 5, cost to extend gap 2 and −3 as penalty for nucleotide mismatch) were performed using a composite database comprising phase1, 2 and 3 of the human genome, build 34 and the mitochondrial genome. Tags with bit scores above specific values were subtracted.
We then performed BLAST analyses of the 18 613 remaining database-derived microbial tags, the 282 remaining non-EBV tags from HCC38 BL and the 41 EBV tags under more stringent conditions (Table 2, bottom section). Different bit score thresholds led to different selection of microbial versus human tags. None of the human tags had bit scores below 32, whereas the lowest score for the microbial tags was approximately 28 (119 tags). Based on these results, we selected a maximum bit score of 36 (corresponding to 18 identical consecutive nucleotides or the equivalent) as our cut-off for vectorette PCR confirmation of HCC38 BL tags (see below). We recognize that this threshold will eliminate a significant fraction of microbial tags and intend that the threshold be raised as we optimize experimental methods and as human genome sequencing proceeds toward completion.
Vectorette PCR
For experimental follow-up of tags with weak BLAST matches to the human genome, we designed vectorette PCR primers for a set of EBV and non-EBV tags with maximum BLAST bit scores of less than 36. These included 6 of the 17 unsubtracted EBV tags and 6 of the 14 unsubtracted non-EBV tags (for primer and vectorette linker sequences, see Supplementary Material).
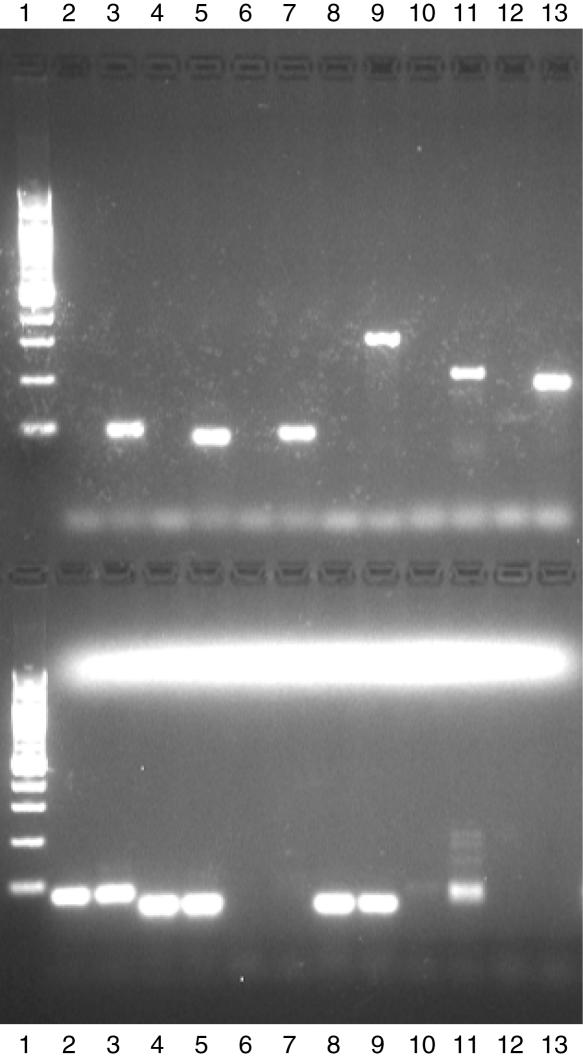
All of the vectorette PCRs using EBV primers gave visible products after second round of amplification only when vectorette library of HCC38 BL DNA was used as template (Figure 5, upper panel, lanes 3, 5, 7, 9, 11 and 13). PCR using primers designed from the non-matching tags gave visible amplification in both HCC38 and HCC38 BL for three primer sets, two primer sets did not amplify either, and the remaining nested primer pair only gave strong amplification when HCC38 BL was used as a template (Figure 5, lower panel, lane 11).
Figure 5.
Vectorette PCR. A 2% agarose gel stained with GelStar. First lane upper and lower panel: 100 bp ladder (Invitrogen). Upper panel: lanes 2–13, six sets of EBV tag-specific primers (HCC38 and HCC38 BL). Lower panel: lanes 2–13, six sets of primers designed from non-matching tags (HCC38 and HCC38 BL).
PCR products from amplifications using the EBV-specific primers were purified using a MinElute PCR purification kit (Qiagen) and subsequent sequencing showed that all primers had annealed specifically and amplified the correct loci.
DISCUSSION
Tags generated by Type IIB enzymes are generally longer than the distance from recognition sequence to cut site for Type IIS restriction enzymes. The longest SAGE-like tags that have been described can only be obtained by using a Type III restriction enzyme (23). No Type III restriction enzymes are currently commercially available, and these enzymes also require that the template DNA contains two inversely oriented copies of their recognition sequence to cut efficiently. Longer tags generally mean that more tags are unique and can thus be mapped unambiguously to their chromosomal position (27).
Increased tag length makes it easier to design primers for the amplification of regions flanking a sequence tag, and the length of Type IIB tags is sufficient for making nested primers. Nested PCR is a lot more sensitive and specific than PCR using only one primer pair, and our data using EBV primers based on BsaXI tags show that a modified vectorette PCR gives very efficient and specific amplification. Using two nested 22 bp primers that are three bases shifted over to amplify the region flanking a BsaXI site, the amplicon should contain the twenty-four 3′ bases of the original tag, as well as the region flanking the tag and part of the vectorette linker. So, in addition to the 22 bases corresponding to tag primer from the second round of PCR, there are two bases flanking the primer that should correspond to the original BsaXI tag. These bases can be used to check that the primer annealed correctly, and that the intended locus was amplified.
It has also been proposed that SAGE-like approaches can be used to study CpG methylation by deploying CpG methylation-sensitive enzymes for fragmentation of DNA (4). Certain Type IIB enzymes are also sensitive to CpG methylation (i.e. BcgI, information from http://www.neb.com/). A human genomic DNA library made using BcgI should only yield tags from unmethylated CpG sites, and a matched library with tags from all possible sites can be made from DNA where methylation has been erased, for instance by using whole genome amplification.
RECORD libraries can also be used to look for pathogens. We have shown that computational subtraction on Type IIB tag libraries can detect pathogens. This approach provides higher throughput for the same number of sequencing reactions, compared to the use of full-length libraries, but efficient subtraction of 27 base tags might be confounded by genetic differences between available databases of human nucleic acid sequences and clinical specimens. Three of the seven nested primer sets from tags that failed to match our available databases of human nucleic acid sequences were able to amplify both the HCC38 and HCC38 BL vectorette templates, suggesting limitations in the completeness of the human genome databases. Three other primer pairs failed to amplify vectorette templates from either library, suggesting the possibility of sequencing errors (phred score above 20 indicates that there is a 1% chance of any given base being incorrectly called), or mutations that might be introduced by our cloning protocols. Further completion of human genome sequencing efforts and refinements in our library construction techniques should alleviate these issues. In addition, it should be noted that the RECORD libraries can also be generated from reverse transcribed RNA simply by using cDNA instead of DNA as substrate for the Type IIB enzyme digest (data not shown), for both pathogen discovery and also transcriptional profiling experiments.
The feasibility of doing high resolution karyotyping based on sequencing of concatenated libraries of DNA tags is currently limited by sequencing costs. The complexity of the human genome makes it more expensive to perform a detailed investigation of genomic content using conventional sequencing technologies relative to array technologies, but alternative higher-throughput and more cost-effective sequencing methods such as polony sequencing (28) have the potential to alter the relative cost and effort of these methods. In the meantime, we believe that genomes of organisms for which arrays are not readily available could be effectively analyzed through representational analyses of concatenated sequence tags.
Furthermore, the Type IIB representations will in turn be well suited for analysis on arrays. Representational methods are widely used for copy number analysis. Type IIB tags would be well-suited for such an approach in that they can easily be obtained in pure form for labeling, and they are long enough for specific and effective hybridization.
The short tags generated by Type IIB enzyme digestion should be ideally suited for the analysis of DNA extracted from paraffin-embedded fixed tissue, the most common form of human clinical tissue preservation. Genomic representations from fixed tissue have been challenging and new methods based on Type IIB restriction enzyme digestion should find wide application. Array CGH arrays that represent Type IIB tags could be particularly useful for cancer genome analysis from fixed tissue biopsy specimens.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online. Databases with all virtual BsaXI tags and mapped HCC38 and HCC38 BL tags are available from the RECORD web site: http://research.dfci.harvard.edu/meyersonlab/RECORD/temp.htm.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Xiaojun Zhao, Cheng Li, Yuki Yuza and Jiajun Gu for helpful discussions, and Alexei Protopopov for assistance with spectral karyotyping. This work was supported by NIH NCI grant 1R01CA098185-01A1, Emerald Foundation, Joan's Legacy, Pardee Foundation, Mallinckrodt Foundation and National Multiple Sclerosis Society to M.M., as well as NIH grant T32-AI-07358 to T.L.
REFERENCES
- 1.Saha S., Sparks,A.B., Rago,C., Akmaev,V., Wang,C.J., Vogelstein,B., Kinzler,K.W. and Velculescu,V.E. (2002) Using the transcriptome to annotate the genome. Nat. Biotechnol., 20, 508–512. [DOI] [PubMed] [Google Scholar]
- 2.Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 3.Spinella D.G., Bernardino,A.K., Redding,A.C., Koutz,P., Wei,Y., Pratt,E.K., Myers,K.K., Chappell,G., Gerken,S. and McConnell,S.J. (1999) Tandem arrayed ligation of expressed sequence tags (TALEST): a new method for generating global gene expression profiles. Nucleic Acids Res., 27, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T.L., Maierhofer,C., Speicher,M.R., Lengauer,C., Vogelstein,B., Kinzler,K.W. and Velculescu,V.E. (2002) Digital karyotyping. Proc. Natl Acad. Sci. USA, 99, 16156–16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T.L., Diaz,L.A.,Jr, Romans,K., Bardelli,A., Saha,S., Galizia,G., Choti,M., Donehower,R., Parmigiani,G., Shih Ie,M. et al. (2004) Digital karyotyping identifies thymidylate synthase amplification as a mechanism of resistance to 5-fluorouracil in metastatic colorectal cancer patients. Proc. Natl Acad. Sci. USA, 101, 3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn J.J., McCorkle,S.R., Praissman,L.A., Hind,G., Van Der Lelie,D., Bahou,W.F., Gnatenko,D.V. and Krause,M.K. (2002) Genomic signature tags (GSTs): a system for profiling genomic DNA. Genome Res., 12, 1756–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts R.J., Belfort,M., Bestor,T., Bhagwat,A.S., Bickle,T.A., Bitinaite,J., Blumenthal,R.M., Degtyarev,S., Dryden,D.T., Dybvig,K. et al. (2003) A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res., 31, 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y., Stange-Thomann,N., Weber,G., Bo,R., Dodge,S., David,R.G., Foley,K., Beheshti,J., Harris,N.L., Birren,B. et al. (2003) Pathogen discovery from human tissue by sequence-based computational subtraction. Genomics, 81, 329–335. [DOI] [PubMed] [Google Scholar]
- 9.Weber G., Shendure,J., Tanenbaum,D.M., Church,G.M. and Meyerson,M. (2002) Identification of foreign gene sequences by transcript filtering against the human genome. Nature Genet., 30, 141–142. [DOI] [PubMed] [Google Scholar]
- 10.Relman D.A. (1999) The search for unrecognized pathogens. Science, 284, 1308–1310. [DOI] [PubMed] [Google Scholar]
- 11.Lucito R., Healy,J., Alexander,J., Reiner,A., Esposito,D., Chi,M., Rodgers,L., Brady,A., Sebat,J., Troge,J. et al. (2003) Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res., 13, 2291–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X., Li,C., Paez,J.G., Chin,K., Janne,P.A., Chen,T.H., Girard,L., Minna,J., Christiani,D., Leo,C. et al. (2004) An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res., 64, 3060–3071. [DOI] [PubMed] [Google Scholar]
- 13.Bignell G.R., Huang,J., Greshock,J., Watt,S., Butler,A., West,S., Grigorova,M., Jones,K.W., Wei,W., Stratton,M.R. et al. (2004) High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res., 14, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piekarowicz A., Golaszewska,M., Sunday,A.O., Siwinska,M. and Stein,D.C. (1999) The HaeIV restriction modification system of Haemophilus aegyptius is encoded by a single polypeptide. J. Mol. Biol., 293, 1055–1065. [DOI] [PubMed] [Google Scholar]
- 15.Petrusyte M., Bitinaite,J., Menkevicius,S., Klimasauskas,S., Butkus,V. and Janulaitis,A. (1988) Restriction endonucleases of a new type. Gene, 74, 89–91. [DOI] [PubMed] [Google Scholar]
- 16.Vitkute J., Maneliene,Z., Petrusyte,M. and Janulaitis,A. (1997) BplI, a new BcgI-like restriction endonuclease, which recognizes a symmetric sequence. Nucleic Acids Res., 25, 4444–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J. and Russell,D. (2001) Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbour Press, Woodbury, NY. [Google Scholar]
- 18.Rabiner L.R. (1989) A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE, 77, 257–286. [Google Scholar]
- 19.Dempster A.P., Laird,N.M. and Rubin,D.B. (1977) Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. B, 39, 1–38. [Google Scholar]
- 20.Zhang Z., Schwartz,S., Wagner,L. and Miller,W. (2000) A greedy algorithm for aligning DNA sequences. J. Comput. Biol., 7, 203–214. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumura H., Reich,S., Ito,A., Saitoh,H., Kamoun,S., Winter,P., Kahl,G., Reuter,M., Kruger,D.H. and Terauchi,R. (2003) Gene expression analysis of plant host–pathogen interactions by SuperSAGE. Proc. Natl Acad. Sci. USA, 100, 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J.J., Rowley,J.D. and Wang,S.M. (2000) Generation of longer cDNA fragments from serial analysis of gene expression tags for gene identification. Proc. Natl Acad. Sci. USA, 97, 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko W.Y., David,R.M. and Akashi,H. (2003) Molecular phylogeny of the Drosophila melanogaster species subgroup. J. Mol. Evol., 57, 562–573. [DOI] [PubMed] [Google Scholar]
- 26.Schrock E., du Manoir,S., Veldman,T., Schoell,B., Wienberg,J., Ferguson-Smith,M.A., Ning,Y., Ledbetter,D.H., Bar-Am,I., Soenksen,D. et al. (1996) Multicolor spectral karyotyping of human chromosomes. Science, 273, 494–497. [DOI] [PubMed] [Google Scholar]
- 27.Unneberg P., Wennborg,A. and Larsson,M. (2003) Transcript identification by analysis of short sequence tags–influence of tag length, restriction site and transcript database. Nucleic Acids Res., 31, 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitra R.D., Shendure,J., Olejnik,J., Edyta Krzymanska,O. and Church,G.M. (2003) Fluorescent in situ sequencing on polymerase colonies. Anal. Biochem., 320, 55–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.