Summary
Mutations in enhancer-associated chromatin-modifying components and genomic alterations in non-coding regions of the genome occur frequently in cancer and other diseases pointing to the importance of enhancer fidelity to ensure proper tissue homeostasis. In this review, I will use specific examples to discuss how mutations in chromatin-modifying factors might affect enhancer activity of disease-relevant genes. I will then consider direct evidence from single nucleotide polymorphisms, small insertions or deletions but also larger genomic rearrangements such as duplications, deletions, translocations and inversions of specific enhancers to demonstrate how they have the ability to impact enhancer activity of disease genes including oncogenes and tumor suppressor genes. Considering that the scientific community only fairly recently has begun to focus its attention on “enhancer malfunction” in disease, I propose that multiple new enhancer-regulated and disease-relevant processes will be uncovered in the near future that will constitute the mechanistic basis for novel therapeutic avenues.
Keywords: cancer, enhancer deregulation, enhancer mutations and genomic alterations, Kabuki syndrome, MLL3/KMT2C, MLL4/KMT2D, UTX/KDM6A
Introduction
History of enhancers
The term “enhancer” was initially coined in the early eighties based on studies of a viral DNA element from Simian virus 40 (SV40) when it was demonstrated that this particular DNA sequence had the ability to convey increased activity towards a T-antigen or β-globin reporter in mammalian cells [1]. Based on these seminal studies the hallmarks of an enhancer were defined as having the capacity to work with different promoters, independent of genomic location and distance from the transcriptional start site and regardless of orientation. This standard enhancer definition paved the way for functional studies in the metazoan system and, for the most part with certain restrictions, is still the prevalent working model within the enhancer field today. Shortly thereafter other virus enhancers with similar properties albeit at times with higher tissue or host specificity were also described [2].
Subsequent studies on the mouse immunoglobulin heavy chain locus uncovered the first eukaryotic enhancer –the Eµ enhancer- thus confirming the existence of operatively similar genomic elements in metazoans [3]. Additional insight into the mechanisms of enhancer function particularly as it pertains to their importance in organismal development was provided by genetic studies in the fruit fly Drosophila melanogaster. These studies demonstrated that important developmental genes can often be regulated by several enhancers in time and space, that enhancers can function in a combinatorial and modular manner and have the ability to act over very large distances (summarized in [4]).
Enhancers are bound by transcription factors and chromatin-modifying co-activators/co-repressors
Both viral and metazoan enhancers are bound by activating and/or repressing transcription factors on specific sites which are often characterized by factor-specific DNA binding motifs [5] (summarized in [6, 7]) (Fig. 1A). Additionally, transcription factors often form a platform for the recruitment of co-activators and co-repressors (Fig. 1A). While transcription factors often appear to be “master regulators” with profound effects on enhancer activation or repression, co-activators and co-repressors in many instances occupy a modulatory role in this process. Additionally, they often constitute proteins with chromatin-modifying capabilities which generally fall under the categories (1) DNA or histone-modifying enzymes, (2) chromatin-associated factors (“readers” of DNA or histone modifications) and (3) chromatin-remodeling proteins (summarized in [8]) (Box 1). Additionally, the enzymatic activities and chromatin-altering abilities of these co-activators and co-repressors also directly affect the DNA methylation, histone modification and DNA accessibility patterns within and around enhancers and thus sets them apart from other regulatory elements within the genome (Box 1). Thus in summary, the combinatorial binding of transcription factors along with these chromatin-modifying components to enhancers results in so called “enhancer signatures” which can serve as a readout to define enhancers in a tissue-specific manner and on a global scale.
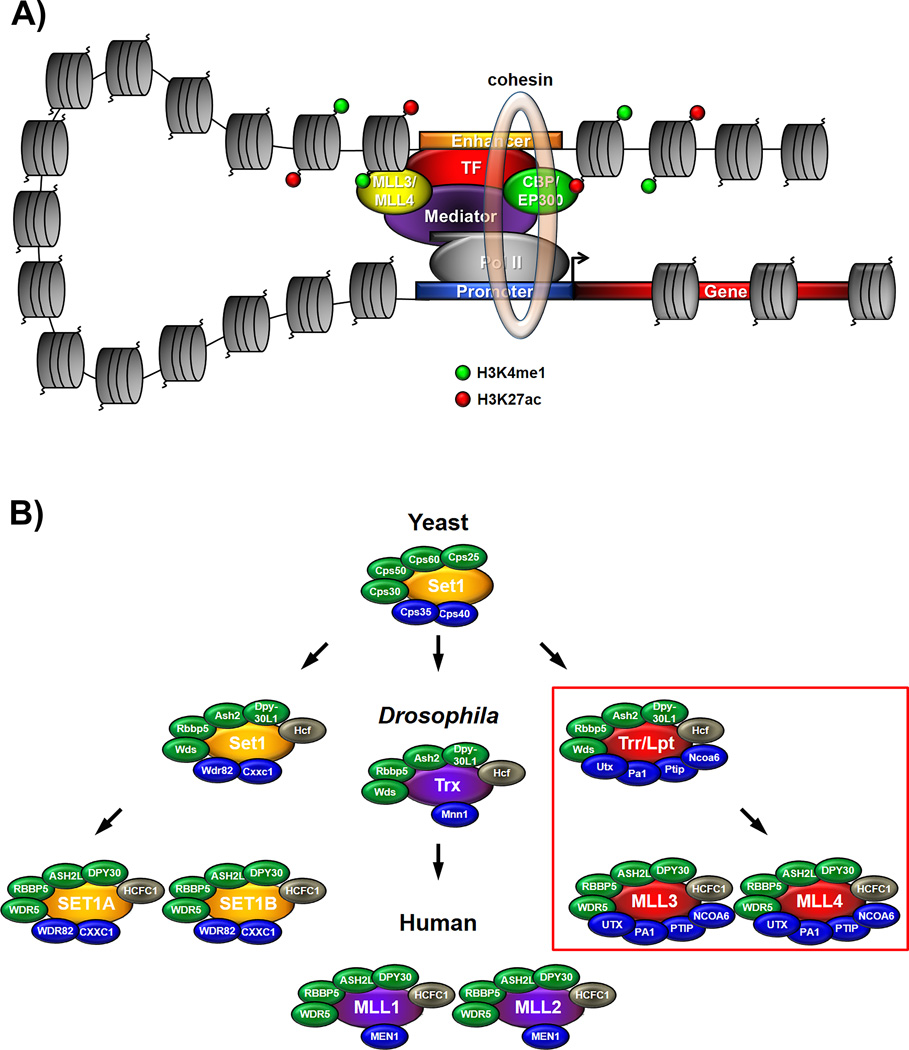
Figure 1.
A: Factors involved in enhancer-promoter interaction and communication. Tissue-specific transcription factors (TF, red) recruit co-activators to enhancers which often constitute chromatin-modifying proteins such as the histone H3 lysine 4 (H3K4) monometyltransferases MLL3/KMT2C or MLL4/KMT2D (yellow) and the histone H3 lysine 27 (H3K27) acetyltransferases CBP or EP300 (green). MLL3/MLL4 and CBP/EP300 implement H3K4 monomethylation (H3K4me1, green circles) and H3K27 acetylation (H3K27ac, red circles) on enhancers, respectively. Interactions of these enhancer-associated factors with the general transcription machinery at the promoter including RNA Polymerase II (gray) are mediated by the Mediator complex (purple) and further stabilized by the ring-shaped cohesin complex (light brown transparent ring). B: The family of histone H3K4 methyltransferases in yeast, Drosophila and mammals. Core subunits which are commonly shared among all complexes are highlighted in green. Complex-specific subunits are only contained within one out of three branches and are highlighted in blue. UTX/KDM6A is an H3K27 demethylase and constitutes a complex-specific subunit within the Trithorax-related (Trr) branch (red box) while MLL3/KMT2C and MLL4/KMT2D form the catalytic core as H3K4 methyltransferases within the mammalian complexes of the Trr branch.
Box 1. Enhancer-associated chromatin modifying components, “enhancer signatures” and mediators of enhancer-promoter communication.
To date many chromatin-modifying proteins have been identified to associate with enhancers and contain the ability to alter the DNA methylation, histone modification and DNA accessibility patterns within and around enhancers (see Introduction). For example some silent enhancers are marked by DNA methylation which is implemented by the DNA cytosine-5-methyltransferases DNMT1, DNMT3A and DNMT3B [29–32, 102] while on the other hand the family of TET enzymes including TET1, TET2 and TET3 is recruited to many active enhancers and catalyzes the oxidation of 5-methylcytosine to 5-hydroxy-metylcytosine [19, 103–106]. Many histone-modifying enzymes including histone lysine methyltransferases and demethylases also play a prominent role on enhancers (summarized in [8]) and particularly the histone modifications H3 lysine 4 monomethylation (H3K4me1), H3 lysine 9 acetylation (H3K9ac) and H3 lysine 27 acetylation (H3K27ac) are prominently enriched on many enhancers. While H3K4me1, H3K9ac and H3K27ac are usually strongly enriched on active enhancers some H3K4me1 and the repressive mark histone H3 lysine 27 trimethylation (H3K27me3) can be found on certain incactive and/or “poised” enhancers while H3K27 dimethylation (H3K27me2) occupies a protective role on non-cell-type-specific enhancers [107–111]. Drosophila Trithorax-related (Trr) and its mammalian homologs MLL3 and MLL4 in a redundant fashion constitute major H3K4 monomethyltransferases on enhancers (Fig. 1B) while the H3K4 demethylase LSD1/KDM1A can be responsible for enhancer “decommissioning” by removing H3K4me1 from enhancers [39–41, 112]. Additionally, a complex consisting of the chromatin “reader” RACK7 and the H3K4 trimethyl-specific demethylase SMCX/JARID1C/KDM5C prevents enhancer overactivation by keeping already active enhancers in a balanced state [113, 114]. All H3K27 methylation (including mono-, di- and trimethylation) is carried out by EZH1 and EZH2 the catalytic subunits of polycomb repressive complex 2 (PRC2). Interestingly, PRC2’s suppressive function on enhancer activity is antagonized by UTX an H3K27 demethylase which – either with MLL3 or MLL4 and additional components – forms a large macromolecular complex (Fig. 1B) [38]. Thus, the MLL3/MLL4 complexes with their ability to demethylate H3K27 and monomethylate H3K4 unite two enzymatic activities that are important for the transition from inactive/”poised” enhancers to active enhancers [39–41]. The GCN5L2 and PCAF containing histone acetyltransferase (HAT) complex ATAC catalyzes the deposition of H3K9ac [115–117] while the CBP and EP300 HATs implement the majority of H3K27ac on enhancers [117–119]. All these unique “enhancer signatures” implemented by DNA and histone modifying enzymes hold the potential to specifically recruit chromatin-associated factors. Among these are H3K4me1-binders such as TIP60 (also a HAT) [120], BROMO domain-containing proteins which recognize acetylated histones including BRD4 [81], but also components of ATP-dependent chromatin remodeling complexes such as CHD7 and BRG1 which interact with H3K4me1 and acetylated histones, respectively and CHD8, SMARCB1/BAF47, BAF155 and BAF170 [90, 121–125]. Some chromatin remodelers such as the NuRD complex can also be required to keep enhancers in a repressed state and thus prevent inappropriate enhancer activation [126].
The cohesin complex is well known for its function in sister chromatid cohesion during meiosis and mitosis and postreplicative DNA damage repair but also functions in regulating enhancer activity. Members of the cohesin complex and their loading factors were first described as effectors of enhancer-promoter communication in Drosophila studies before this role was also confirmed in the mammalian system [127–130]. Another large complex, Mediator, assists in bringing together enhancer-associated transcription factors with the general transcription machinery including RNA polymerase II on promoters and the cohesin complex in this context is thought to further stabilize these long-range enhancer-promoter interactions (Fig. 1A) [129, 131] (summarized in [10, 132]). Cohesin also interacts with the insulator protein CTCF albeit not on enhancers. Instead, cohesin is involved in linking CTCF-bound loci called insulator elements with each other. These insulator elements are important in restricting enhancer activity to a given “neighborhood” often involving only a smaller subset of genes and thus preventing ectopic activation of other genes that otherwise might come under the influence of a given enhancer [10, 100, 131].
Mediators of enhancer-promoter communication
Early on, factors with the potential to mediate between enhancer- and promoter-bound factors were discovered and were shown to play a major role in enhancer-promoter communication (Fig. 1A, Box 1). In order to effectively explain enhancer-promoter communication particularly over large distances a looping mechanism was proposed by which enhancers are brought into close proximity to their cognate promoters by these mediating factors (summarized in [9, 10]). More recently, enhancer-promoter looping has been experimentally validated by chromosome confirmation capture (3C) technology-based approaches confirming this initial model [11–13]. Additionally, work on the β-globin locus has confirmed that looping between the β-globin enhancer region (the β-globin locus control region) and the β-globin promoter causally underlies induction of β-globin transcription [14, 15].
Mutations in chromatin-modifying enhancer-associated factors as well as germline and somatic variants in enhancers occur frequently in cancer
Over the past few years whole genome sequencing (WGS) and genome wide association (GWAS) studies across many different cancer types, including solid tumors and different forms of leukemia, have made it increasingly clear that many of the abovementioned chromatin-modifying and remodeling proteins play a central role in cancer pathogenesis and other diseases [16, 17] (summarized in [18–22]). At the same time similar WGS and GWAS studies have also revealed that the majority of germline and somatic variants in cancer occur in non-coding regions of the genome including enhancers and thus point to the importance of enhancer misregulation in tumorigenesis [23–27] (summarized in [28]).
In this review I focus on the most recent advances in the field that connect enhancer “malfunction” to various diseases including cancer. I will discuss several possible mechanisms that result in enhancer misregulation of disease-relevant genes. In particular, I will first consider mutations in enhancer-bound chromatin-modifying proteins and will then move on to describe how genomic alterations such as single nucleotide polymorphisms (SNPs), small insertions or deletions but also larger genomic rearrangements such as duplications, deletions, translocations and inversions can impact enhancer activity of disease genes including oncogenes and tumor suppressor genes.
Indirect evidence of enhancer malfunction in disease
Epigenetic alteration of the “enhancer landscape” in cancer
Based on already known “enhancer signatures” (see Introduction) several studies have started to explore the hypothesis that changes in the “enhancer landscape” including DNA methylation and histone modification patterns might correlate with tumorigenesis. Evidence for DNA methylation changes on enhancers in cancer comes from several studies in breast, cervical, lung and prostate cancer [29–32]. Gains and losses in H3K4 monomethylation (H3K4me1) on many enhancers have been reported in colon cancer [33] and similar findings were reported for changes in the DNA accessibility pattern on cis-regulatory elements in various cancers [34]. Interestingly, two recent studies also suggest that changes in the “enhancer landscape” can underlie therapy resistance of cancer cells. Endocrine therapy-resistant breast cancer cells for example rely on the NOTCH signaling pathway and are characterized by gains and losses in H3 lysine 4 dimethylation (H3K4me2) on many enhancers with a concomitant change in chromatin accessibility [35]. Furthermore, in NOTCH1-dependent T cell acute lymphoblastic leukemia (T-ALL) with a resistance to γ-secretase inhibitors increased chromatin compaction and reduced H3 lysine 27 acetylation (H3K27ac), an active enhancer mark, can be observed on various enhancers [36].
Enhancer-associated factors are often mutated in cancer and other diseases
In addition to changes of the “enhancer landscape” in cancer which could be an indirect effect of various processes, enhancer-associated chromatin-modifying proteins (Box 1) and factors that mediate enhancer-promoter interaction (Box 1) are often mutated and/or misexpressed in many different cancer types (Table 1) but also play a role in other diseases (Table 2). Based on the enhancer-associated nature of these factors, it is therefore very likely that their misregulation will directly affect enhancer activity. Plausible scenarios that could explain how mutations in enhancer-associated factors drive tumorigenesis or certain diseases include inappropriate activation of oncogenic enhancers, deactivation of enhancers of tumor suppressor genes and ectopic activation or deactivation of enhancers of disease-relevant genes (other than cancer genes). Below I will discuss these possibilities in more detail based on a “case study” of the enhancer-associated histone H3 lysine 27 demethylase UTX/KDM6A and the histone H3 lysine 4 (H3K4) methyltransferases MLL3/KMT2C and MLL4/KMT2D and their roles in cancer and a genetic disease called Kabuki syndrome (Fig. 1B, 2).
Table 1.
Mutations in mediators of enhancer-promoter interaction or chromatin-modifying proteins in cancer (with a frequency of >3%).
| Gene | Molecular Function |
Cancer Type | References |
|---|---|---|---|
| CTCF | Chromatin insulator | Endometrial, head and neck | [16, 17] |
| RAD21/SCC1 | Cohesin complex | Acute myeloid leukemia | [16] |
| SMC1A | Cohesin complex | Acute myeloid leukemia, endometrial |
[16, 17] |
| SMC3 | Cohesin complex | Acute myeloid leukemia | [16] |
| STAG2/SCC3B | Cohesin complex | Acute myeloid leukemia, bladder, glioblastoma |
[16, 17, 133] |
| ARID1A/BAF250 | Chromatin remodeling (SWI/SNF complex) |
Bladder, colorectal, endometrial, esophageal, gastric, kidney, liver, lung, ovarian |
[16, 17, 133– 137] |
| ARID1B/BAF250B | Chromatin remodeling (SWI/SNF complex) |
Liver | [135] |
| ARID2/BAF200 | Chromatin remodeling (SWI/SNF complex) |
Liver, lung, melanoma | [16, 135] |
| PBRM1/BAF180 | Chromatin remodeling (SWI/SNF complex) |
Bladder, kidney | [17, 138] |
| SMARCA4 | Chromatin remodeling (SWI/SNF complex) | Esophageal, medulloblastoma, lung |
[16, 139, 140] |
| SMARCB1/BAF47/SNF5 | Chromatin remodeling (SWI/SNF complex) | Rhabdoid tumor | [16, 141] |
| CHD4 | Chromatin remodeling |
Endometrial | [16] |
| CHD6 | Chromatin remodeling |
Bladder | [134] |
| CHD8 | Chromatin remodeling |
Glioblastoma | [16] |
| BCOR | Corepressor complex | Endometrial | [16] |
| NCOR1 | Corepressor complex | Breast, head and neck, melanoma |
[16] |
| DNMT3A | DNA methyltransferase |
Acute myeloid leukemia | [16, 17] |
| TET2 | DNA demethylase | Acute myeloid leukemia | [16, 17] |
| MLL1/KMT1A | H3K4 methyltransferase |
Bladder, liver | [16, 134, 135] |
| MLL2/KMT1B | H3K4 methyltransferase |
Bladder, endometrial, head and neck |
[16, 17] |
| MLL3/KMT2C | H3K4 methyltransferase |
Bladder, breast, colorectal, endometrial, gastric, head and neck, lung, liver, medulloblastoma |
[16, 17, 134, 135, 137, 139, 142, 143] |
| MLL4/KMT2D | H3K4 methyltransferase | Non-Hodgkin lymphoma, bladder, breast, endometrial, head and neck, kidney, lung, medulloblastoma, squamous cell carcinoma |
[16, 17, 133, 139, 140, 144–146] |
|
SMCX/JARID1C/ KDM5C |
H3K4 demethylase | Kidney, lung | [16, 17, 145] |
| CBP | H3K27 acetyltransferase |
Acute lymphoblastic leukemia, B cell lymphoma, bladder, non-Hodgkin lymphoma |
[16, 134, 144, 146, 147] |
| EP300 | H3K27 acetyltransferase |
B cell lymphoma, bladder, endometrial, head and neck, lung |
[16, 17, 133, 134, 146] |
| EZH2 | H3K27 methyltransferase |
Non-Hodgkin lymphoma, B- cell lymphoma |
[144, 146] |
| UTX/KDM6A | H3K27 demethylase | Bladder, medulloblastoma, renal cell carcinoma |
[16, 17, 133, 134, 140, 145] |
Table 2.
Mutations in enhancer-associated chromatin-modifying proteins in other diseases.
| Gene | Molecular Function | Disease | References |
|---|---|---|---|
| MLL4/KMT2D | H3K4 methyltransferase | Kabuki Syndrome | [54, 55] |
| SMCX/JARID1C/KDM5C | H3K4 demethylase | X-linked mental retardation |
[148] |
| CBP | H3K27 acetyltransferase | Rubinstein-Taybi Syndrome |
[149, 150] |
| EP300 | H3K27 acetyltransferase | Rubinstein-Taybi Syndrome |
[149] |
| UTX/KDM6A | H3K27 demethylase | Kabuki Syndrome | [55, 56] |
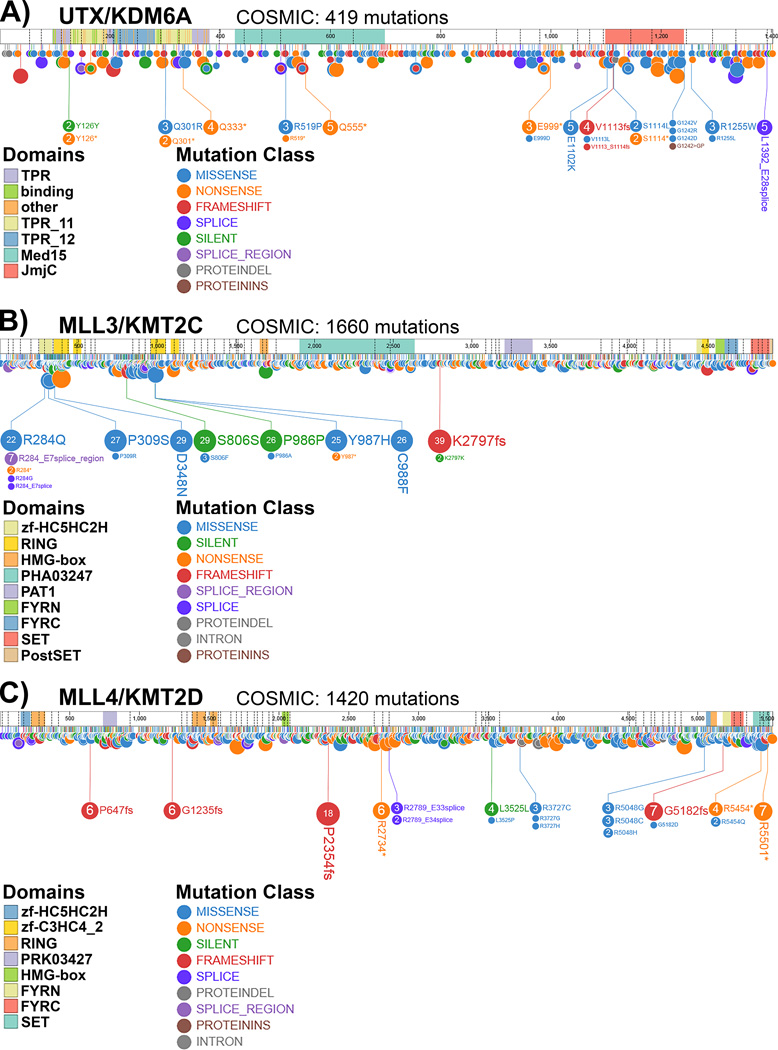
Figure 2.
Somatic mutations reported for UTX/KDM6A (A), MLL3/KMT2C (B) and MLL4/KMT2D (C) according to the Catalogue of Somatic Mutations in Cancer (COSMIC) based on the Pediatric Cancer (PeCan) Data Portal from St. Jude Children’s Research Hospital.
UTX and MLL3/MLL4 function as histone H3 lysine 27 demethylases and lysine 4 monomethyltransferases on enhancers
MLL3 and MLL4 are both mammalian homologs of Set1 which constitutes the sole H3K4 methyltransferase in yeast, implements all three H3K4 methylation states -H3 lysine 4 mono-, di- and trimethylation (H3K4me1, -me2 and –me3)- and exists in a large protein complex termed COMPASS (Complex of Proteins Associated with Set1) (summarized in [37, 38]). In the fruit fly Drosophila three Set1-related proteins exist: Set1, Trithorax (Trx) and Trithorax-related (Trr). The mammalian genome contains six yeast Set1-related proteins: SET1A/SET1B (homologous to Drosophila Set1), MLL1/MLL2 (homologous to Drosohila Trx) and MLL3/MLL4 (homologous to Drosophila Trr) (Fig. 1B). All metazoan complexes share identical core subunits (Fig. 1B, green) but also contain complex-specific subunits (Fig. 1B, blue) that are conserved only within the Set1, Trx and Trr branches. For example Utx (Drosophila)/UTX (mammals) exists as a complex-specific subunit only in the Drosophila Trr and mammalian MLL3 or MLL4 complexes. Trr in Drosophila and MLL3/MLL4 in a redundant fashion in mammals constitute major H3K4 monomethyltransferases on enhancers while Utx/UTX acts as an H3K27 demethylase removing H3K27 trimethylation (H3K27me3), an inhibitory histone modification, from enhancers [39–41]. This suggests a model in which prior removal of H3K27me3 via Utx/UTX is required on inactive/”poised” enhancers before they can transition to an activated state via addition of H3K4me1 through Trr/MLL3/MLL4 (see also Box 1).
UTX, MLL3 and MLL4 are frequently mutated across a broad spectrum of cancers
WGS studies across many different cancers have revealed that UTX, MLL3 and MLL4 are frequently mutated within many different forms of solid tumors but also certain leukemias such as non-Hodgkin lymphoma (Fig. 2, Table 1). Interestingly, this is not equally the case for other subunits of the MLL3/MLL4 complexes. The functional requirement for core subunits within H3K4 methyltransferase complexes other than MLL3/MLL4 might render the effects of their mutation too deleterious in cancer cells to administer a competitive advantage. However, this would not explain why other complex-specific subunits are not mutated at comparable frequencies suggesting that UTX, MLL3 and MLL4 might actually have tumor suppressive/oncogenic roles outside the canonical MLL3/MLL4 complexes. Matters are additionally complicated by the fact that depending on the context UTX, MLL3 and MLL4 can act as tumor suppressors or oncogenes. For example, Drosophila studies have shown that Utx and trr display tumor suppressive properties in the eye [42, 43]. In contrast, trr acts as an oncogene in collaboration with the Hippo signaling pathway in another context [44, 45]. Similar findings have been reported in the mammalian system. In most cases UTX, MLL3 and MLL4 appear to have tumor suppressive properties [46–48] but have also been implicated in oncogenesis [49–52]. Furthermore, it has not been fully elucidated to date whether haploinsufficiency of UTX, MLL3 and MLL4 is sufficient to drive tumorigenesis or whether homozygous mutations are required to achieve the same effect. For example, in homozygous TP53 mutant acute myeloid leukemia (AML) MLL3 was validated as a haploinsufficient tumor suppressor [48] whereas most studies to date have mainly investigated the effects of homozygous UTX, MLL3 and MLL4 deletions or siRNA/shRNA-mediated knock-down in various tumor contexts and thus not specifically addressed heterozygous versus homozygous mutations. An interesting connection between transcription-associated DNA damage and enhancer malfunction might be made by a recent study which suggests that MLL4 functions as a guardian of genome stability by regulating RNA polymerase II fidelity over the bodies of actively transcribed genes [53]. While this study only focused on the effects of MLL4 deletion over actively transcribed genes, it is very likely that due to the enhancer-associated nature of MLL4 similar effects could also occur on transcribed enhancers resulting in accumulation of enhancer mutations over time. Thus, MLL4 (and possibly UTX and MLL3) might potentially be involved in enhancer regulation by directly affecting enhancer activity and indirectly through co-transcriptional DNA damage-inducing mechanisms which might alter transcription factor recruitment on specific enhancers of tumor suppressor and/or oncogenes. This hypothesis however needs to be further tested in the future.
UTX and MLL4 in Kabuki syndrome
MLL4 is also very frequently mutated (>56%) in the genetic disease Kabuki syndrome which is characterized by craniofacial anomalies but also other clinical features (Table 2) [54, 55]. Mutations in UTX have also been described albeit with lower frequency (Table 2) [55, 56]. All identified mutations are heterozygous dominant thus suggesting haploinsufficiency for MLL4 and UTX in Kabuki syndrome. To date it has not been investigated whether Kabuki syndrome mutations result in changes of enhancer activity but based on the role of UTX and MLL4 in regulating enhancer activity this is a very plausible scenario. However, the lack of a clear association between Kabuki syndrome and an increased cancer risk seems to imply that MLL4 haploinsufficiency is an unlikely event in most cancers with MLL4 mutations.
In summary, this “case study” on UTX, MLL3 and MLL4 in cancer and Kabuki syndrome provides us with valuable insight how mutations in enhancer-associated chromatin-modifying factors might result in disease. Firstly, it implies that based on the tissue- and context-dependent expression of transcription factors mutations in enhancer-associated chromatin-modifiers might either have tumor suppressive or oncogenic potential and secondly that their role of haploinsufficiency needs to be more rigorously tested in the future.
Direct evidence of enhancer malfunction in disease
Mutations and genomic alterations of enhancers associated with diseases other than cancer
Specific enhancer mutations or genomic alterations of enhancers have been associated with or directly implicated in campomelic dysplasia, celiac disease, cleft palate, coronary heart disease, Crohn’s disease, Hirschsprung’s disease, multiple sclerosis, preaxial polydactyly, rheumatoid arthritis, systemic lupus, ulcerative colitis, Van Buchem disease and X-linked deafness which supports the idea that enhancer misregulation in these cases might confer disease susceptibility [23, 57–61] (summarized in [62]). For example, aniridia which is characterized by an absence of the iris and generally caused by heterozygous null mutations in the PAX6 coding sequence can also result from genomic rearrangements downstream of the PAX6 locus. It was shown that these rearrangements are responsible for the inactivation of enhancer elements that have the ability to drive eye-specific expression of PAX6 [63, 64]. Furthermore, X-linked deafness can be caused by deletion of an enhancer element located nearly one megabase (Mb) upstream of POU3F4 [58, 65]. Similarly, in Van Buchem disease, a bone sclerosing dysplasia, a non-coding region ∼35 kilobases (kb) downstream of SOST is homozygously deleted [66]. Additionally, in a skeletal malformation syndrome called campomelic dysplasia genomic translocations interrupt putative cis-regulatory elements upstream of SOX9 [67]. Even single nucleotide changes in enhancers have been implicated in disease. Point mutations in a long-range limb bud-specific enhancer located ∼1 Mb upstream of the sonic hedgehog gene (SHH) in the intron of a neighboring gene results in overactivation of SHH and preaxial polydactyly [61, 68]. Furthermore, a SNP within an IRF6 enhancer results in disruption of an AP-2α transcription factor binding site and is associated with cleft lip [69]. In summary, these studies provide direct evidence that enhancer mutations or genomic alterations of enhancers are directly causative of disease.
Select examples of enhancer mutations and genomic alterations of enhancers in cancer
Similar mechanisms including different types of genomic enhancer alterations as described for some genetic disorders above are also operative in cancer. Below I will discuss select examples of individual misregulated enhancers in different cancer types in more detail. This certainly does not constitute an exhaustive list but will provide a representative cross section of the most recently reported mechanisms that can result in altered enhancer activity of oncogenes and tumor suppressor genes. I will attempt to cover a broad spectrum of genomic alterations including SNPs, small insertions or deletions and larger genomic rearrangements including duplications, deletions, translocations and inversions.
Point mutations in enhancers resulting in increased/decreased affinity of transcription factor binding
Multiple genome-wide association studies have revealed several SNPs located in a gene desert upstream of MYC and have associated individual SNPs with an increased risk to develop certain cancer types (summarized in [70]). The investigation of individual cancer risk loci in the context of specific cancers such as colorectal cancer, prostate cancer and breast cancer showed an increased enrichment of known enhancer marks such as histone H3K4me1. Some of these SNP-bearing regions were also demonstrated to drive reporter gene expression, and showed differential binding of transcription factors such as TCF7L2 and an ability to form 3D interactions with the MYC promoter (Fig. 3) [71–75]. Remarkably, studies in a mouse model containing a deletion in a non-coding region 500 kb upstream of MYC that usually harbors a SNP associated with many cancers show that these mice are resistant to the development of intestinal tumors [76]. This suggests that the gene desert upstream of MYC contains enhancer elements with the ability to tissue-specifically regulate MYC expression (Fig. 3).
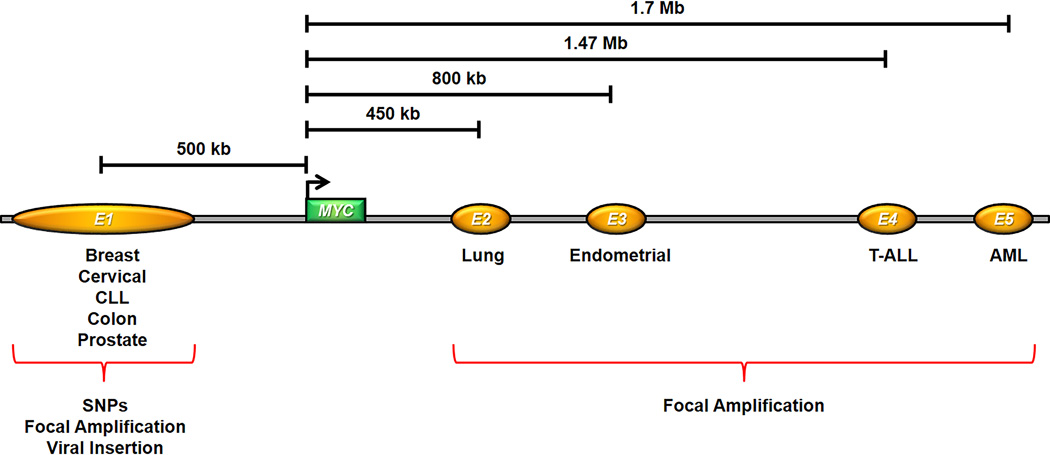
Figure 3.
Regions containing tissue-specific Myc enhancers. At least five regions (E1-5) that often contain several tissue-specific enhancer clusters have been reported for the MYC locus. The approximate distance from the center of each region to the MYC promoter is indicated by a black bar. E1 shows a high enrichment for SNPs in various cancers but is also affected by focal amplification or viral insertion. Regions E2-5 have been shown to be focally amplified in lung cancer (E2), endometrial cancer (E3), T-ALL (E4) and AML (E5).
In prostate cancer a risk-associated SNP on chromosome 6 is located in a non-coding region in the vicinity of the RFX6 gene and contains a binding site for the homeodomain-containing transcription factor HOXB13. The region of the risk-associated SNP displays increased binding of HOXB13 compared to control tissue resulting in allele-specific upregulation of RFX6. Prostate cancers bearing this SNP essentially require RFX6 for proper proliferation, migration and invasion. Additionally, increased RFX6 levels appear to be a more general hallmark of prostate cancers as they are indicative of poor clinical outcome [77].
The identification of a highly risk-associated SNP located within the first intron of the LMO1 gene in neuroblastoma affirms that SNPs can also result in reduced transcription factor binding to enhancers. This particular SNP located within a super-enhancer highly enriched for H3K27ac alters a consensus GATA binding motif to a TATA motif resulting in loss of GATA3 binding on the TATA allele and decreased LMO1 expression [78].
Deletions within enhancers
Deletions in non-coding regions can also promote tumorigenesis, if affecting enhancers of tumor suppressor genes. Some indirect evidence for this was provided by studies on a deletion within a non-coding region on chromosome 15. This region is characterized by enrichment for H3K4me1, increased DNAse hypersensitivity and recruitment of the transcription factor c-JUN as evidenced by the presence of multiple activating protein 1 (AP-1) binding motifs and long range interactions with distant genes [79]. However, the exact mechanism by which this deletion might potentially affect tumor progression remains unresolved.
Small insertions resulting in de novo creation of transcription factor binding sites
Super-enhancers which are also known as locus control regions or stretch enhancers are defined as regions that contain clusters of individual enhancers and are prominently bound by many different transcription factors, chromatin-modifying proteins or enhancer-promoter mediating factors (see Introduction) [80–83]. They regulate genes that play important roles in cell identity and within disease-relevant cell types often show increased enrichment for mutations within regions that have been associated with certain diseases such as Alzheimer’s disease, type 1 diabetes and systemic lupus erythematosus [81]. Super-enhancers are also often enriched at/around important oncogenes including MYC in various cancers such as diffuse large B cell lymphoma, multiple myeloma, glioblastoma multiforme and small-cell lung cancer compared to normal tissue [81, 84, 85].
One mechanism of this “de novo” creation of super-enhancers has recently been described by Look and colleagues in a subset of T-ALL. In these cases heterozygous insertions upstream of the TAL1 oncogene result in a new binding site for the transcription factor MYB thus creating a super-enhancer which drives allele-specific TAL1 expression [86].
Focal amplification of enhancers
An increase in oncogenic enhancer activity can also be achieved by focal amplification, and copy number gains of non-coding regions have also been described for super-enhancers across various tumor types [87]. In particular, the MYC locus appears to constitute a hotspot for focal amplifications which are often confined to non-coding regions excluding the MYC gene itself, thus pointing to a prominent role of tissue-specific MYC enhancers in tumorigenesis (Fig. 3).
For example, a focal amplification located 500 kb upstream of MYC was described in approximately 5% of chronic lymphocytic leukemia (CLL) cases [88] and also occurs in the cervical HeLa cancer cell line where it was caused by integration of the human papillomavirus and is thought to have been the tumor-initiating event (Fig. 3) [89]. More direct evidence that focal amplifications containing enhancers are directly involved in enhanced/aberrant oncogene activation comes from studies on enhancers located downstream of MYC (Fig. 3) [87, 90, 91].
Two non-coding amplifications were detected 450 kb and 800 kb downstream of MYC in approximately 2% of lung adenocarcinoma and 4% of uterine corpus endometrial carcinoma, respectively [87]. Both amplified regions in lung adenocarcinoma and endometrial carcinoma encompass a cluster of enhancers with high enrichment for H3K27ac and form stable chromatin interactions with the MYC promoter (Fig. 3). Enrichment for H3K27ac on the amplified region in lung adenocarcinoma and enhancer-promoter interaction are tissue-specific as they cannot be observed in an endometrial carcinoma cell line and vice versa [87]. A small sequence of 150 base pairs (bp) within the amplified lung adenocarcinoma region is responsible for the majority of enhancer activity, contains binding motifs for several important transcription factors and depends on the recruitment of NFE2L2 and CEBPβ. Furthermore, repression or deletion of this enhancer element via CRISPR/Cas9 in two lung adenocarcinoma cell lines resulted in a reduction of MYC expression and a reduced ability of anchorage-independent and clonogenic growth [87].
A long-range MYC enhancer situated 1.47 Mb downstream of the MYC promoter is recurrently duplicated in 5% of T-ALL and was demonstrated to depend on the NOTCH signaling pathway and to be essentially required for proper thymocyte development and leukemogenesis (Fig. 3) [91].
Another enhancer region located 1.7 Mb downstream of MYC contains a cluster of five enhancers and is amplified in 3–5% of AML cases (Fig. 3). For example, in mouse RN2 AML cells the analogous enhancer region is occupied by the SWI/SNF chromatin remodeling complex, the enhancer mark H3K27ac and the histone lysine acetyl reader Brd4. Work in mouse and human leukemia cell lines confirmed that enhancer activity and recruitment of Brg1/BRG1, the catalytic subunit of SWI/SNF, are restricted to the hematopoietic lineage. Brg1/BRG1 is also required for chromatin looping of these enhancers to the Myc/MYC promoter and for appropriate recruitment of hematopoietic transcription factors [90].
Structural genomic rearrangements/enhancer hijacking
In three recent studies “enhancer hijacking” was described as one means of ectopic oncogene activation [92–94]. “Enhancer hijacking” describes a process by which an enhancer, through genomic rearrangements, is removed from its natural genomic context and brought into proximity of another gene to activate it ectopically.
The first description of this phenomenon dates back more than thirty years describing a translocation between chromosome 8 and 14 in Burkitt’s lymphoma presumably as a faulty result of class switch recombination thus bringing an enhancer of the immunoglobulin H (IgH) gene into close proximity of the MYC gene resulting in MYC overexpression [95].
A similar phenomenon was described in follicular lymphoma for the BCL2 locus. Here, a translocation between chromosome 14 and 18 places the IgH enhancer downstream of the antiapoptotic BCL2 gene. This leads to overexpression of BCL2 and suppression of apoptosis [96, 97].
For example, in glioblastoma a translocation from chromosome 10 to chromosome 5 brings a putative super-enhancer into close proximity to the telomerase reverse transcriptase (TERT) promoter. The functional consequence of this event has not been investigated. However, TERT as the catalytic subunit of telomerase is known to be overexpressed in cancer. Furthermore, recurrent mutations in the TERT promoter which create new binding motifs for transcription factors and thus result in TERT overexpression have been reported in several tumor types (summarized in [98]).
In AML, cases with a particular inversion or translocation on chromosome 3 are characterized by the repositioning of a distal GATA2 enhancer close to the EVI1 (also known as PRDM3 or MECOM) locus thus resulting in inactivation of the rearranged GATA2 allele while simultaneously activating ectopic expression of the oncogene EVI1 [93].
Furthermore, within two particular subgroups of pediatric medulloblastoma somatic structural variants including tandem duplications, deletions or inversions can place potent enhancers located within the DDX31 locus proximal to GFI1B. These enhancers which generally function to presumably activate DDX31 or one of its neighboring genes now in a different context start to drive expression of the oncogene GFI1B. Similarly, interchromosomal translocations and tandem duplications were also found to drive ectopic expression of the GFI1B paralog GFI1 [92].
Further support that “enhancer hijacking” might be a more generally employed mechanism by which oncogenes are activated in cancers was provided by the finding that binding sites of the insulator protein CTCF and the enhancer-promoter mediating cohesin complex are frequently mutated in colorectal cancer (Box 1) [99]. CTCF and cohesin are also involved in the creation of loops that isolate subgroups of genes within so-called insulated neighborhoods, thus creating boundaries to prevent ectopic activation via enhancers from other genes that lie outside these loops [100]. Indeed, in T-ALL some deletions involve CTCF/cohesin boundaries that flank insulated neighborhoods which contain proto-oncogenes. Specifically, deletion of a CTCF/cohesin boundary element via CRISPR/Cas9 in human embryonic kidney (HEK-293T) cells and primary human T cells demonstrated ectopic activation of the oncogene TAL1. Additional data supports a model in which an active enhancer element that is located outside the insulated TAL1-containing neighborhood and is usually prevented from interaction with the TAL1 locus is now able to drive TAL1 expression. A similar phenomenon in HEK-293T cells for the proto-oncogene LMO2 was reported with a larger deletion containing several CTCF sites, and data mining of esophageal and liver carcinoma samples confirms an increased enrichment for mutations in CTCF boundary sites [101].
Conclusions and prospects
The amassment of genome-wide sequencing data via GWAS and WGS studies across many different cancer types and various genetic diseases over the past few years has revealed the importance of maintaining the integrity of non-coding regulatory elements, a fact that was previously largely underappreciated. It has become increasingly clear that tumorigenesis or disease state is often associated with epigenetic changes of the “enhancer landscape” and that mutations in or misregulation of enhancer-associated chromatin modifiers can have profound effects on enhancer activity holding the potential to cause inappropriate activation of oncogenic enhancers, inactivation of tumor-suppressive enhancers or inappropriate activation/inactivation of disease-relevant genes. As discussed here, this notion is strongly supported by multiple concrete studies on disease-relevant enhancers or enhancers of oncogenes and tumor suppressor genes. I predict that the next few years will see a further surge in the discovery of new disease-relevant enhancers. Despite a huge amount of disease-relevant data from GWAS and WGS studies our mechanistic understanding of individual enhancer-mediated processes in cancer and other diseases is strongly lacking behind and should lie at the forefront of future research investigations. Efforts are currently being made to target misregulated enhancers in cancer. For example, BROMO domain inhibitors are now tested in clinical cancer trials. They target the histone acetyl-binding BROMO domain of the enhancer-associated and -activating factor BRD4, thus preventing its recruitment (Box 1). BRD4 has a strong preference to bind to enhancers of oncogenic and lineage-specific genes also known as super enhancers (see above) making it a promising target [84, 85]. Time will tell whether this will prove a successful strategy to target individual tumors as BROMO-domain inhibitors apparently indiscriminately target most super enhancers. Thus, combination therapeutic approaches including BROMO domain inhibitors and the development of more enhancer-specific therapies should be a major focus of future clinical treatment regimens.
Acknowledgments
We thank all the members of the Herz laboratory for helpful discussions. We apologize to all whose work could not be discussed or referenced due to space limitations. This work was supported by a grant from the National Institutes of Health 4R00CA181506-02 to Hans-Martin Herz.
Abbreviations
- AML
acute myeloid leukemia
- GWAS
genome-wide association studies
- SNP
single nucleotide polymorphism
- T-ALL
T cell acute lymphoblastic leukemia
- WGS
whole genome sequencing studies
Footnotes
The author has declared no conflict of interest.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.de Villiers J, Olson L, Tyndall C, Schaffner W. Transcriptional ‘enhancers’ from SV40 and polyoma virus show a cell type preference. Nucleic Acids Res. 1982;10:7965–7976. doi: 10.1093/nar/10.24.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 4.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Zhuang J, Iyer S, Lin X, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 7.Yanez-Cuna JO, Kvon EZ, Stark A. Deciphering the transcriptional cis-regulatory code. Trends Genet. 2013;29:11–22. doi: 10.1016/j.tig.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol. 2013;25:327–333. doi: 10.1016/j.ceb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 11.de Laat W, Dekker J. 3C–based technologies to study the shape of the genome. Methods. 2012;58:189–191. doi: 10.1016/j.ymeth.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes JR, Roberts N, McGowan S, Hay D, et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 13.Li G, Ruan X, Auerbach RK, Sandhu KS, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng W, Lee J, Wang H, Miller J, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W, Rupon JW, Krivega I, Breda L, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandoth C, McLellan MD, Vandin F, Ye K, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell. 2014;26:309–317. doi: 10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20:662–671. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Herz HM, Hu D, Shilatifard A. Enhancer malfunction in cancer. Mol Cell. 2014;53:859–866. doi: 10.1016/j.molcel.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurano MT, Humbert R, Rynes E, Thurman RE, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khurana E, Fu Y, Colonna V, Mu XJ, et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinhold N, Jacobsen A, Schultz N, Sander C, et al. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet. 2014;46:1258–1263. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- 27.Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet. 2015;47:710–716. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khurana E, Fu Y, Chakravarty D, Demichelis F, et al. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17:93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 29.Yegnasubramanian S, Wu Z, Haffner MC, Esopi D, et al. Chromosome-wide mapping of DNA methylation patterns in normal and malignant prostate cells reveals pervasive methylation of gene-associated and conserved intergenic sequences. BMC Genomics. 2011;12:313. doi: 10.1186/1471-2164-12-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14:R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aran D, Hellman A. DNA methylation of transcriptional enhancers and cancer predisposition. Cell. 2013;154:11–13. doi: 10.1016/j.cell.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 32.Taberlay PC, Statham AL, Kelly TK, Clark SJ, et al. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 2014;24:1421–1432. doi: 10.1101/gr.163485.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–739. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stergachis AB, Neph S, Reynolds A, Humbert R, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnani L, Stoeck A, Zhang X, Lanczky A, et al. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc Natl Acad Sci USA. 2013;110:E1490–E1499. doi: 10.1073/pnas.1219992110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knoechel B, Roderick JE, Williamson KE, Zhu J, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herz HM, Garruss A, Shilatifard A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem Sci. 2013;38:621–639. doi: 10.1016/j.tibs.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shilatifard A. The COMPASS Family of Histone H3K4 Methylases: Mechanisms of Regulation in Development and Disease Pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herz HM, Mohan M, Garruss AS, Liang K, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu D, Gao X, Morgan MA, Herz HM, et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33:4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JE, Wang C, Xu S, Cho YW, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herz HM, Madden LD, Chen Z, Bolduc C, et al. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol. 2010;30:2485–2497. doi: 10.1128/MCB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanda H, Nguyen A, Chen L, Okano H, et al. The Drosophila ortholog of MLL3 and MLL4, trithorax related, functions as a negative regulator of tissue growth. Mol Cell Biol. 2013;33:1702–1710. doi: 10.1128/MCB.01585-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh H, Slattery M, Ma L, White KP, et al. Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Rep. 2014;8:449–459. doi: 10.1016/j.celrep.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qing Y, Yin F, Wang W, Zheng Y, et al. The Hippo effector Yorkie activates transcription by interacting with a histone methyltransferase complex through Ncoa6. eLife. 2014;3:e02564. doi: 10.7554/eLife.02564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ortega-Molina A, Boss IW, Canela A, Pan H, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21:1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Dominguez-Sola D, Hussein S, Lee JE, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21:1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, Liu Y, Rappaport AR, Kitzing T, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25:652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santos MA, Faryabi RB, Ergen AV, Day AM, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514:107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim JH, Sharma A, Dhar SS, Lee SH, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo C, Chen LH, Huang Y, Chang CC, et al. KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget. 2013;4:2144–2153. doi: 10.18632/oncotarget.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kantidakis T, Saponaro M, Mitter R, Horswell S, et al. Mutation of cancer driver MLL2 results in transcription stress and genome instability. Genes Dev. 2016;30:408–420. doi: 10.1101/gad.275453.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bogershausen N, Wollnik B. Unmasking Kabuki syndrome. Clin Genet. 2013;83:201–211. doi: 10.1111/cge.12051. [DOI] [PubMed] [Google Scholar]
- 56.Lederer D, Grisart B, Digilio MC, Benoit V, et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Kok YJ, Vossenaar ER, Cremers CW, Dahl N, et al. Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene POU3F4. Hum Mol Genet. 1996;5:1229–1235. doi: 10.1093/hmg/5.9.1229. [DOI] [PubMed] [Google Scholar]
- 59.Emison ES, McCallion AS, Kashuk CS, Bush RT, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 60.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lettice LA, Horikoshi T, Heaney SJ, van Baren MJ, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci USA. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noonan JP, McCallion AS. Genomics of long-range regulatory elements. Annu Rev Genom Hum Genet. 2010;11:1–23. doi: 10.1146/annurev-genom-082509-141651. [DOI] [PubMed] [Google Scholar]
- 63.Lauderdale JD, Wilensky JS, Oliver ER, Walton DS, et al. 3’ deletions cause aniridia by preventing PAX6 gene expression. Proc Natl Acad Sci USA. 2000;97:13755–13759. doi: 10.1073/pnas.240398797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kleinjan DA, Seawright A, Schedl A, Quinlan RA, et al. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum Mol Genet. 2001;10:2049–2059. doi: 10.1093/hmg/10.19.2049. [DOI] [PubMed] [Google Scholar]
- 65.de Kok YJ, Merkx GF, van der Maarel SM, Huber I, et al. A duplication/paracentric inversion associated with familial X-linked deafness (DFN3) suggests the presence of a regulatory element more than 400 kb upstream of the POU3F4 gene. Hum Mol Genet. 1995;4:2145–2150. doi: 10.1093/hmg/4.11.2145. [DOI] [PubMed] [Google Scholar]
- 66.Loots GG, Kneissel M, Keller H, Baptist M, et al. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15:928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfeifer D, Kist R, Dewar K, Devon K, et al. Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am J Hum Genet. 1999;65:111–124. doi: 10.1086/302455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lettice LA, Heaney SJ, Purdie LA, Li L, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 69.Rahimov F, Marazita ML, Visel A, Cooper ME, et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grisanzio C, Freedman ML. Chromosome 8q24-associated cancers and MYC. Genes Cancer. 2010;1:555–559. doi: 10.1177/1947601910381380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia L, Landan G, Pomerantz M, Jaschek R, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci USA. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sotelo J, Esposito D, Duhagon MA, Banfield K, et al. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci USA. 2010;107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sur IK, Hallikas O, Vaharautio A, Yan J, et al. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 77.Huang Q, Whitington T, Gao P, Lindberg JF, et al. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat Genet. 2014;46:126–135. doi: 10.1038/ng.2862. [DOI] [PubMed] [Google Scholar]
- 78.Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418–421. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Demichelis F, Setlur SR, Banerjee S, Chakravarty D, et al. Identification of functionally active, low frequency copy number variants at 15q21.3 and 12q21.31 associated with prostate cancer risk. Proc Natl Acad Sci USA. 2012;109:6686–6691. doi: 10.1073/pnas.1117405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hnisz D, Abraham BJ, Lee TI, Lau A, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parker SC, Stitzel ML, Taylor DL, Orozco JM, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci USA. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Siersbaek R, Rabiee A, Nielsen R, Sidoli S, et al. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 84.Loven J, Hoke HA, Lin CY, Lau A, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chapuy B, McKeown MR, Lin CY, Monti S, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Choi PS, Francis JM, Imielinski M, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edelmann J, Holzmann K, Miller F, Winkler D, et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood. 2012;120:4783–4794. doi: 10.1182/blood-2012-04-423517. [DOI] [PubMed] [Google Scholar]
- 89.Adey A, Burton JN, Kitzman JO, Hiatt JB, et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med. 2014;20:1130–1137. doi: 10.1038/nm.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Northcott PA, Lee C, Zichner T, Stutz AM, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Groschel S, Sanders MA, Hoogenboezem R, de Wit E, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 94.Francis JM, Zhang CZ, Maire CL, Jung J, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4:956–971. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taub R, Kirsch I, Morton C, Lenoir G, et al. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bakhshi A, Wright JJ, Graninger W, Seto M, et al. Mechanism of the t(14;18) chromosomal translocation: structural analysis of both derivative 14 and 18 reciprocal partners. Proc Natl Acad Sci USA. 1987;84:2396–2400. doi: 10.1073/pnas.84.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cleary ML, Sklar J. Nucleotide sequence of a t(14;18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Proc Natl Acad Sci USA. 1985;82:7439–7443. doi: 10.1073/pnas.82.21.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. TERT promoter mutations in cancer development. Curr Opin Genet Dev. 2014;24:30–37. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 99.Katainen R, Dave K, Pitkanen E, Palin K, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 100.Dowen JM, Fan ZP, Hnisz D, Ren G, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hnisz D, Weintraub AS, Day DS, Valton AL, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ronnerblad M, Andersson R, Olofsson T, Douagi I, et al. Analysis of the DNA methylome and transcriptome in granulopoiesis reveals timed changes and dynamic enhancer methylation. Blood. 2014;123:e79–e89. doi: 10.1182/blood-2013-02-482893. [DOI] [PubMed] [Google Scholar]
- 103.Hon GC, Song CX, Du T, Jin F, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell. 2014;56:286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu F, Liu Y, Jiang L, Yamaguchi S, et al. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stroud H, Feng S, Morey Kinney S, Pradhan S, et al. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu M, Hon GC, Szulwach KE, Song CX, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heintzman ND, Stuart RK, Hon G, Fu Y, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 109.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ferrari KJ, Scelfo A, Jammula S, Cuomo A, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell. 2014;53:49–62. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 112.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shen H, Xu W, Guo R, Rong B, et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 2016;165:331–342. doi: 10.1016/j.cell.2016.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Outchkourov NS, Muino JM, Kaufmann K, van Ijcken WF, et al. Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell Rep. 2013;3:1071–1079. doi: 10.1016/j.celrep.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 115.Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, et al. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell. 2011;44:410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, et al. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jin Q, Yu LR, Wang L, Zhang Z, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Visel A, Blow MJ, Li Z, Zhang T, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jeong KW, Kim K, Situ AJ, Ulmer TS, et al. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat Struct Mol Biol. 2011;18:1358–1365. doi: 10.1038/nsmb.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.De S, Wurster AL, Precht P, Wood WH, 3rd, et al. Dynamic BRG1 recruitment during T helper differentiation and activation reveals distal regulatory elements. Mol Cell Biol. 2011;31:1512–1527. doi: 10.1128/MCB.00920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bossen C, Murre CS, Chang AN, Mansson R, et al. The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat Immunol. 2015;16:775–784. doi: 10.1038/ni.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ceballos-Chavez M, Subtil-Rodriguez A, Giannopoulou EG, Soronellas D, et al. The chromatin Remodeler CHD8 is required for activation of progesterone receptor-dependent enhancers. PLoS Genet. 2015;11:e1005174. doi: 10.1371/journal.pgen.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, et al. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011;7:e1002008. doi: 10.1371/journal.pgen.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Luo Z, Gao X, Lin C, Smith ER, et al. Zic2 is an enhancer-binding factor required for embryonic stem cell specification. Mol Cell. 2015;57:685–694. doi: 10.1016/j.molcel.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dorsett D, Eissenberg JC, Misulovin Z, Martens A, et al. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Faure AJ, Schmidt D, Watt S, Schwalie PC, et al. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res. 2012;22:2163–2175. doi: 10.1101/gr.136507.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gui Y, Guo G, Huang Y, Hu X, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fujimoto A, Totoki Y, Abe T, Boroevich KA, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 136.Jones S, Wang TL, Shih Ie M, Mao TL, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 138.Varela I, Tarpey P, Raine K, Huang D, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Parsons DW, Li M, Zhang X, Jones S, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jones DT, Jager N, Kool M, Zichner T, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 142.Watanabe Y, Castoro RJ, Kim HS, North B, et al. Frequent alteration of MLL3 frameshift mutations in microsatellite deficient colorectal cancer. PloS One. 2011;6:e23320. doi: 10.1371/journal.pone.0023320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ellis MJ, Ding L, Shen D, Luo J, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dalgliesh GL, Furge K, Greenman C, Chen L, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mullighan CG, Zhang J, Kasper LH, Lerach S, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jensen LR, Amende M, Gurok U, Moser B, et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Petrij F, Giles RH, Dauwerse HG, Saris JJ, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]