Abstract
Purpose
To determine whether some of the most often used uveal melanoma cell lines resemble their original tumor.
Methods
Analysis of the literature, patient charts, histopathology, mutations, chromosome status, HLA type, and expression of melanocyte markers on cell lines and their primary tumors. We examined five cell lines and the primary tumors from which they were derived.
Results
Four of the five examined primary tumors were unusual: one occupied the orbit, two were recurrences after prior irradiation, and one developed in an eye with a nevus of Ota. One cell line did not contain the GNA11 mutation, but it was present in the primary tumor. Three of the primary tumors had monosomy 3 (two of these lacked BAP1 expression); however, all five cell lines showed disomy 3 and BAP1 expression. All of the cell lines had gain of 8q. Two cell lines lacked expression of melanocyte markers, although these were present in the corresponding primary tumor.
Conclusions
All cell lines could be traced back to their original uveal melanoma. Four of the five primary tumors were unusual. Cell lines often differed from their primary tumor in chromosome status and melanocyte markers. However, their specific chromosome aberrations and capacity to continue proliferation characterize them as uveal melanoma cell lines.
INTRODUCTION
Cancer treatment is becoming increasingly individualized, and genetic changes in a tumor may influence the sensitivity to therapeutic drugs. Mutations in important regulator genes may make tumor cells sensitive to drugs: in cutaneous melanoma, tumors with a BRAFV600E mutation respond very well to a specific BRAF inhibitor.1 Metastases of cutaneous melanoma nowadays undergo specific mutation testing prior to deciding which drug may be the best treatment. In vitro experiments indicate that a similar situation may exist for uveal melanoma, as specific drug responses have been described in relation to mutations in two genes, GNAQ and GNA11, which are specifically associated with uveal melanoma.2–5 These mutations are considered essential early changes en route to malignancy of uveal melanocytes, but they are not related to the formation of metastases.6
Uveal melanoma are characterized not only by these mutations, but also by specific chromosome aberrations, such as gain at chromosome 8q, chromosome 3 monosomy, and additional material at chromosome 6p.7–11 Chromosome 3 monosomy in the tumor is related to many phenotypical differences, such as a large tumor size, location in the ciliary body, and the presence of epithelioid cells and an inflammatory infiltrate. Chromosome 3 monosomy is associated with a bad prognosis, and in some studies, but not in others, the addition of 8q material further worsens prognosis.7,11–16 Addition of material at 6p is associated with a better prognosis.12
Two studies suggest that gain of additional copies on 8q precedes chromosome 3 monosomy. De Lange and associates17 observed some tumors that had gain in 8q in most cells and monosomy 3 in some, providing evidence of sequential events. At the same time, Singh and colleagues18 studied the evolutionary tree (clades) of uveal melanoma and observed that tumors can be divided into those that gain 8q (and subsequently lose chromosome 3) and those that gain 6p. Still, it is unknown why uveal melanomas often lose this chromosome 3, but it is clear that this loss affects prognosis. Particularly the BAP1 gene, located on this chromosome, is associated with a bad prognosis: mutations in the BAP1 gene on the remaining chromosome 3 are associated with the development of metastases.19 Clear differences exist in the characteristics of tumors with and without BAP1 expression. These are the same associations as previously described for chromosome 3 monosomy, as chromosome 3 monosomy and loss of BAP1 expression are strongly correlated.20 While loss of BAP1 expression is probably due to loss of one chromosome 3 together with a mutation in the BAP1 gene,21 this correlation is not absolute: according to Kalirai and colleagues,22 chromosome 3 monosomy and loss of BAP1 expression carry, independently, a bad prognosis. Another mutation seen in uveal melanoma occurs in EIF1AX. Combining information on the chromosome 3 status with information on the mutation status of BAP1 and EIF1AX provides very good prognostic value.5
One can look for associations between the sensitivity to drugs and specific mutations in cell lines. However, only a few cell lines of uveal melanoma are in existence, and one may wonder why. My laboratory tried to grow uveal melanoma from primary tumors, but failed in 21 of the 22 attempts. The only cell line that grew out, 92-1, was derived from an unusual primary tumor, which had led to destruction of the eye and gave rise to some unusually located metastases years later.23 One wonders what factors determine this difficulty to grow uveal melanoma cell lines and whether the cell lines that are available are derived from tumors that have been exposed to any specific treatments, such as irradiation, which may have led to new mutations or chromosome aberrations.24
The few uveal melanoma cell lines that are available vary in genetic backgrounds and mutations.25,26 However, while mutations in GNAQ/GNA11 are considered important early changes in the development of a uveal melanoma, no GNAQ or GNA11 mutations have yet been identified in some of the available cell lines, such as Mel285 and Mel290. Additionally, chromosome 3 monosomy is uncommon in uveal melanoma cell lines,26,27 and it is hard to find cell lines that lack BAP1 expression.28
When cell lines are being used in research, one often questions how representative they are of the original tumor and whether mutations or chromosome aberrations of the cell lines correspond to the aberrations of the primary tumor. Specific characteristics may be lost or gained during culturing. We hypothesize that the unusual lack of GNAQ/GNA11 mutations and chromosome 3 monosomy in uveal melanoma cell lines is due to outgrowth of selected clones from the original tumor.
Another reason why cell lines may not represent their original tumor may be accidental exchanges: genetic studies have revealed that several cell lines that were originally supposed to be derived from different patients share the same origin.29 Furthermore, some cell lines that were considered to be derived from metastases of a uveal melanoma lacked GNAQ and GNA11 mutations and carried BRAF mutations, which are characteristic of cutaneous melanoma.26,29–31 This suggests that in these cases we are dealing with cutaneous melanoma–derived cell lines instead of uveal melanoma–derived cell lines. This could have happened when the cell line came from a tumor in a patient carrying both a uveal melanoma and a cutaneous melanoma, in which case the cutaneous melanoma may have been the origin of the metastases. Another possibility is an accidental laboratory exchange of cell lines.
Knowing the genetic polymorphisms, such as the HLA antigens or short tandem repeats (STRs), of the original tumor would enable the proper identification of real lineage–derived cultured cells and would help to identify which cell lines have been derived from admixed cells from another patient and are no longer original. As described by Folberg and colleagues,29 DNA technology showed that several cell lines that were considered to be from different backgrounds, in reality, shared the same background (cell line OCM1 = MUM2C, OCM3 = OCM8, M619 = C918 = MUM2B). Nowadays, authors of manuscripts submitted for publication are asked to provide, for instance, the STRs of the cell lines that they used in their study to ensure proper identification of the cell lines.32,33 This is even required when the cells have been obtained from reputable sources, as exchanges may have happened early on. Thus one should preferably have the information on the genetic markers of the original tissue from which the cell line was derived.
With regard to uveal melanoma cell lines in general, little is known about the clinical and genetic background of the original tumors. Recently, several new cell lines were created in Paris, either from primary tumors or from xenografts derived from primary uveal melanoma or liver or skin metastases. Detailed information was provided regarding mutation and chromosome status: the cell lines’ expression markers and chromosome status were compared to the original tumors.34 Two of the cell lines came from a primary uveal melanoma, and both differed in their chromosome status from this tumor. Only limited information was provided about the clinical background of the two primary tumors that gave rise to these cell lines. We also know little about the origin of cell lines Mel202, Mel270, Mel285, and Mel290, all of which were grown from material obtained from eyes that had been enucleated in Miami.35 While more is known about the characteristics and background of cell line 92-1 developed in Leiden, The Netherlands, only a limited comparison between the genetic data from the cell line and the original tumor has taken place. Two different STR patterns have been published for this cell line.
In order to see whether the cultured cells represent their original tumor and whether the original tumor harbored any special characteristics that would have favored outgrowth of tumor cells, we set out to obtain information regarding the original tumors that gave rise to the five uveal melanoma cell lines. Information was available on the original pathology numbers, and with permission of the Medical Ethical Committee of the University of Miami, the clinical history of the cases was traced back and primary tumor material obtained. Data on the clinical history of the patients, including tumor treatments, were collected and histologic and genetic data were (when possible) determined in the cell lines as well as in their corresponding primary tumor.
We hypothesize that choroidal melanoma that give rise to cell lines must already carry cells that are highly proliferative. In vitro selection of minority clones may lead to outgrowth of unusual cells that do not show mutations in GNAQ/GNA11, chromosome 3 monosomy, and loss of BAP1 expression, even though these are present in the primary tumors.
Determining the characteristics of the primary tumors that gave rise to uveal melanoma cell lines may provide an answer to our hypothesis; additionally, such data may help future researchers to select specific tumors for developing new cell lines. We studied the clinical history of these five cases and were surprised to see that only one (Mel285) of the five studied cases was a “normal” uveal melanoma; two (Mel202 and Mel270) were recurrences after prior irradiation (one with a 16-year history), another (Mel290) occurred in an eye with a nevus of Ota, and another tumor (92-1) had been so massive that it had destroyed the eye and orbit and led to metastases, although this tumor had disomy of chromosome 3 and expressed BAP1. The cell line showed an EIF1AX mutation, which has been associated with the development of metastases in disomy 3 uveal melanoma.5,23,28,36
Overall, the current study combines historical histopathology reports with the most modern genetic data to get a better insight into the nature of the original tumors that have produced some of the most frequently used cell lines in uveal melanoma research. It tries to provide an answer to our hypothesis that the lack of monosomy 3, lack of GNAQ or GNA11 mutations, and the loss of melanocyte markers in uveal melanoma cell lines are due to clonal selection.
METHODS AND MATERIALS
CLINICAL ANALYSIS AND HISTOLOGY
We studied five cell lines derived from primary tumors: 92-1 (from Leiden),23 and Mel202,37 Mel270,35,38 Mel285, and Mel290 (all from Miami).35 Cell lines OMM2.3 and OMM2.5 (originally named OMM1.3 and OMM1.5) were derived from metastases.
Analysis of material of the original tumor of 92-1 was allowed in accordance with the Dutch regulations, conforming to the FEDERA (Federation of Dutch Medical Scientific Societies) agreements. Following prospective permission from the Ethical Committee of the Bascom Palmer Eye Institute and the University of Miami and using the original pathology numbers as the linking code, the original patient charts and blocks were retrieved from the Florida Lions Ocular Pathology Laboratory of the Bascom Palmer Eye Institute. All studies followed the tenets of the Declaration of Helsinki.
MUTATION ANALYSIS
The presence of a mutation in either the GNAQ or GNA11 gene in the primary tumors was analyzed using hydrolysis probes in a multiplex Droplet Digital polymerase chain reaction (ddPCR), using a QX100 droplet generator and DG8 cartridges (Bio-Rad Laboratories, Inc) and a QX100 droplet reader (Bio-Rad Laboratories, Inc).11 Digital PCR (dPCR) software (QuantaSoft) reads the positive and negative droplets in each sample and plots the fluorescence droplet by droplet. The positive droplets represent the concentration of the target allele in the samples. Droplet PCR was performed in Leiden (Dr P. A. van der Velden, Dr M. Versluis).
For identification of the BRAF and GNAQ/GNA11 mutation status in the cell lines, Sanger sequencing was performed on the cell lines by PCR using a Sybr green premixture from Bio-Rad Laboratories, Inc.11 Mutations in EIF1AX, SF3B1, and BAP1 were determined by Sanger sequencing in Rotterdam, The Netherlands (Dr A. de Klein).
CHROMOSOME ANALYSIS
Data on the chromosome constitution of the cell lines was obtained from the literature (Table 1). Fluorescence in situ hybridization (FISH) analysis was performed in Rotterdam on sections obtained from paraffin blocks.13 A probe was used for 3p13 (RP11-522N9) and for the centromere (P×3.5) Chromosome analysis was also performed by using the multiplex Droplet PCR (Dr P. A. van der Velden, Dr M. Versluis, Leiden) and by single-nucleotide polymorphism (SNP) array.
TABLE 1.
CHARACTERISTICS OF PRIMARY TUMORS AND CELL LINES*
| MATERIAL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| CHARACTERISTIC | PRIMARY TUMOR 92-1 (IIA) | CELL LINE 92-1 | PRIMARY TUMOR E-143-90 (MEL202) | CELL LINE MEL202 | PRIMARY TUMORE-1268-94 (MEL270) | CELL LINE MEL270 | PRIMARY TUMOR E-994-95 (MEL285) | CELL LINE MEL285 | PRIMARY TUMOR E-131-96 (MEL290) | CELL LINE MEL290 |
| Exenteration | I-125 | I-125 | nevus of Ota | |||||||
| choroidal, very pigmented | choroidal, large, into optic nerve; nevoid area | ciliochoroidal, mushroom | choroidal, dome, many vessels | |||||||
| GNA11ex4, ex5 | WTL,R | WT26,28,R | WT28 | WTL,R | WT28,R | Q209 LR,L | WT26,R | WTL | WT26,R | |
| GNAQ ex4, ex5 | Q209L 626 A>T | Q209L 626 A>T26,R | Q209L 626 A>T R210K26 629 G>A28 | Q209PL,R | Q209P 626 A>C28,R | WTL,R | WT26,R | WTL | WT26,R | |
| BAP1 mutation | WTR | WTR | WTR | WTR | ||||||
| BAP1 protein | yes28 | yes28 | Yes (OMM2.5 yes)28 | yes, low† | yes, low† | |||||
| BAP1 IHC | positive | positiveR | negative | positive R | negative | positiveR | positive | negative R | ||
| Chromosome 3 | disomy 31 | disomy 31,42 | disomy 327,46,L
51 chrom |
monosomy 3 (50%)R,L | disomy 3L loss 3p24, loss 3q21.2–3q2445 | monosomy 3L | disomy 327 loss 3p26-pter42 | monosomy 3 (80%)R,L | disomy 3R,L loss 3p26-pter42,44 | |
| Chromosome 6 | tetrasomy 6pL | gain 6p42,L | gain 6p loss 6q |
gain 6pL | tetrasomy 6p42 | disomy 6pL | 6q loss42 | disomy 3pL | disomy 642 | |
| Chromosome 8 | tetrasomy 8R,L | gain 8q42,L | gain 8q (6 copies)L | disomy 8R,L | disomy 8qL extra 842 | disomy 8qL | disomy 8pR tetrasomy 8qR,L | disomy 8R,L | disomy 8R,L /8q24.1-24.2 gain42 | |
| EIF1AX | c.17G/A28,R | WT28 | WTR | WTR | WTR | |||||
| SF3B1 exon 12–16 | WT28,R | C1793c>T28 | WTR | WTR | WTR | |||||
| Cell type | 90% epithelioid | mixed, many vessels | mixed, 20% epithelioid | 95% epithelioid, many vessels | ||||||
| HMB45 (gp100) | positive | positive40,43 | positive40,43 | 99% 3+ nevus 50% | positive41 | 99% 3+ | absent43 | 99% 3+ | absent43 | |
| Melan-A | positive26,43 | 80% 1+ nevus 0% | positive26,28 | 80% 2–3+ | absent43 | 95% 3+ | absent43 | |||
| CD68 | 3+ | 3+ | 3+ | |||||||
| CD3 | 1–2+ | 1+ | 1+ | 2+ | ||||||
| Pigment macrophages | many | many | sporadic | many | ||||||
Data regarding the primary choroidal melanoma and the choroidal melanoma cell lines were collected from published papers. A cell is empty when information is not available. The presence of mutations in exons 4 and 5 of GNAQ and GNA11 was studied in the primary choroidal melanomas and in the choroidal melanoma cell lines (R = Rotterdam, L = Leiden). Mutations in E-1268-94 and E-994-95 were also determined in Liverpool with the same results. Mutations in exons 1 and 2 of EIF1AX and exons 12–16 of SF3B1 were determined in 92-1, Mel270, Mel285, and Mel290 (Rotterdam). The chromosome composition of the primary tumors and the choroidal melanoma cell lines was obtained from the literature or obtained by FISH analysis or Droplet PCR (primary tumors) or SNP analysis (cell lines). Immunohistochemical analyses were performed for expression of BAP1, HMB45, Melan-A, and the presence of macrophages (CD8) and T cells (CD3) on sections of the primary uveal melanoma.
As determined by Dr. A. G. Jochemsen, LUMC, Leiden, The Netherlands.
SHORT TANDEM REPEAT PROFILES
Short tandem analysis was carried out by BaseClear BV (Leiden) using the AmpFLSTR Identifiler PCR Amplification Kit (Life Technologies) based on the procedure recommended by the International Cell Line Authentication Committee (ICLAC) (Table 2).
TABLE 2.
SHORT TANDEM REPEAT PROFILES OF THE DIFFERENT CHOROIDAL MELANOMA CELL LINES*
| LOCUS | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CELL LINE | D8S1179 | D21S11 | D7S820 | CSF1PO | D3S1358 | TH01 | D13S317 | D16S539 | D2S1338 | D19S433 | VWA | TPOX | D18S51 | AM | D5S818 | FGA |
| 92-1 Leiden | 15 | 30 | 10;11 | 10;11 | 14;15† | 9;9.3 | 11;12 | 12 | 19;23 | 13;16 | 16 | 8;9† | 12;13 | X | 9;11 | 21;23 |
| 92-1 EBML | 11 | 10;11 | 14;15 | 9;9.3 | 10;11 | 16 | 8;9 | X | 12;13 | 21;23 | ||||||
| 92-1 Griewank | 10;11 | 10;11 | 14;15 | 9;9.3 | 11;12 | 11;12 | 16 | 8;9 | X | 9;11 | 21;23 | |||||
| Mel202 Leiden | 13;16† | 30.2;31.2 | 11;12 | 11†‡ | 16 | 6;7 | 11;13 | 11;12 | 18;25 | 14 | 18;19 | 8 | 12;15 | X | 11;12 | 23;24 |
| Mel202 EBML | 11;12 | 10;11 | 16 | 6;7 | 11:13 | 18;19 | 8 | X | 11;12 | 23;24 | ||||||
| Mel270 Leiden | 8;13 | 28;29 | 8;9 | 11 | 16 | 6;9 | 12 | 12 | 25 | 13 | 17;18 | 10 | 19 | X;Y† | 12 | 24;26 |
| Mel270 Griewank | 8;9 | 11 | 16 | 6;9 | 12 | 12 | 17;18 | 10 | X;Y | 12 | 24;26 | |||||
| OMM2.3 Leiden | 8;13 | 28;29 | 8;9 | 11 | 16 | 6;9 | 12 | 12 | 25 | 13 | 17;18 | 10 | 19 | X;Y | 12 | 26 |
| OMM2.5 Leiden | 8;13 | 28;29 | 8;9 | 11 | 16 | 6;9 | 12 | 12 | 25 | 13 | 17;18 | 10 | 19 | X;Y | 12 | 26 |
| Mel285 Leiden | 11;15 | 29 | 8;13 | 10;13 | 15;17 | 8;9.3 | 11 | 11;14 | 18;24 | 14 | 19 | 11 | 16;17 | X | 13 | 20;22 |
| Mel285 Griewank | 8;13 | 10;13 | 15;17 | 8;9.3 | 11 | 11;14 | 19 | 11 | X | 13 | 20;22 | |||||
| Mel290 Leiden | 11;13 | 28;32.2 | 11;12 | 10;11 | 15;18 | 6;9 | 8;11 | 11;13 | 20;24 | 12;14 | 14;16 | 8;11 | 14;15 | X | 10;12 | 18;20 |
For Mel202, both EBML and the Curie Institute, Paris, France, observed 10;11 for CSF1PO in Mel202. Otherwise, the LUMC in Leiden, The Netherlands, Griewank at M.D. Anderson Cancer Center, and the Curie Institute (not shown) reported the same STRs.
STR profiles were determined in Leiden, published by M.D. Anderson Cancer Center, Houston Texas,26 and reported by the EBML-EBI Web site for ESTDAB cell line profiles (www.ebi.ac.uk/ipd/estdab/directory.html). Differences between EBML and the other centers were observed for D7S820 and D5S818 for 92-1. Empty cells indicate an STR was not tested.
Red indicates likely assignment
IMMUNOHISTOCHEMISTRY
Expression of Melan-A/MART-1, HMB-45, and S100B was determined in Miami on sections obtained from paraffin blocks. Immunohistochemical staining for BAP1 was performed in Rotterdam (Dr R. Verdijk) using the Ventana BenchMark ULTRA fully automated staining system (Ventana Medical Systems Inc) with an alkaline phosphatase red detection kit. Sections were deparaffinized and consecutively heated using heat-induced epitope retrieval for 64 minutes at 97°C. A titration experiment was performed to determine the optimal antibody dilution. The sections were then incubated for 32 minutes at 37°C with the primary BAP1 antibody (sc-28383, concentration 1:50, Santa Cruz Biotechnology, Inc). Target amplification was performed and followed by incubation with Hematoxylin II counterstain for 8 minutes. An additional counterstain was performed with blueing reagent (Ventana Medical Systems Inc). Tumors stained either unequivocally positive or negative.6,20
RESULTS
CLINICAL HISTORIES
The clinical history and histopathologic description of the cases that gave rise to cell lines 92-1,23 Mel202,37 Mel270,35,38 Mel285,38 and Mel29038 were obtained and described. Clinical photographs were retrieved for case Mel202. The blocks of all cases except of Mel202 were retrieved and used for specific laboratory tests as indicated in the following sections.
Case 92-1
The history of this patient has been described previously.23,39 She was a 76-year-old woman who was referred to the Department of Ophthalmology at Leiden University Medical Center because of a large tumor in the right orbit, for which she incurred delayed consultation. The right eyeball had been displaced superotemporally by a large tumor. Computed tomography showed a small and deformed eyeball with tumor outgrowth into the orbit. The tumor mass included the optic nerve, the inferior rectus muscle, the lateral rectus muscle, and the superior rectus muscle. Exenteration of the orbit was performed. The patient died 2½ years later and an autopsy was performed. Multiple melanoma metastases were present at various sites, including the heart, which may have led to cardiac arrest.
We were able to recover the paraffin blocks containing material from the primary tumor that had given rise to cell line 92-1 and paraffin blocks of several metastases. Blocks H92-1223 IIA and IIB contained material from the nasal orbit; H92-1223 IIIA and IIIB were derived from material of the temporal orbit.
The histopathologic description of H92-1223 IIA and IIB was as follows: “Pieces of tissue consisting almost entirely of tumor. There are monomorphic cells, round/oval nuclei and enlarged nucleoli, with most cells showing eosinophil cytoplasm. Numerous cells contain melanin pigmentation. Connective tissue is located focally. There are numerous mitoses.” Different areas of the primary tumor that had given rise to cell line 92-1 were studied (IIA, IIB, IIIB), and all showed 2–3+ infiltrating macrophages and 1–2+ infiltrating CD3 cells (Table 1). Analysis by ddPCR of the primary tumor that gave rise to cell line 92-1 showed the presence of a GNAQ Q209L mutation. This is the same mutation that has been reported to occur in cell line 92-1.26
We previously reported that the primary tumor that gave rise to cell line 92-1 showed the presence of disomy 3 as well as aneuploidy of other chromosomes, with tetrasomy of 6p and tetrasomy of 8 (FISH testing23). The disomy 3 status of the primary tumor was for this thesis confirmed by FISH on material from the orbit (parts IIIA and IIIB, Rotterdam), with 84% to 86% of the cells carrying two signals for probe 3p13 and 86% for probe cen3. There were 3 to 4 copies of 8p and of 8q. BAP1 expression was analyzed on different parts of the primary tumor of 92-1 (1223 IIIA and IIIB) and on metastases in the liver, heart, and adrenal gland and was found to be positive in all of these tissues. Immunohistochemical staining showed expression of HMB45 on the primary tumor.
In 1995, the cell line showed a complex karyotype: 47, −X, der (X) t (X;6) (q28;p11), +8, +8, der (17) t(6;17)(p11;25). In another study, the cell line showed disomy 3, with loss of 6q and gain at 6p21.2–p21.3 and of 8q24.1–24.3.27,42 Amirouchene-Angelozzi and colleagues28 reported an EIF1AX mutation in cell line 92-1; this is now confirmed in Rotterdam, except we have not been able to analyze this in the primary tumor. The cell line does not carry a mutation in BAP1 and expresses BAP1.
The HLA type was originally determined using the leukocytes of the patient (Table 3); this can always be used to determine whether a cultured cell line is derived from this patient.
TABLE 3.
HLA TYPE OF PATIENTS AND CELL LINES*
| HLA POLYMORPHISMS | ||||||
|---|---|---|---|---|---|---|
| CELL LINE | HLA-A | HLA-B | HLA-C | HLA-DR | HLA-DRw | HLA-DQ |
| 92-1 (PBLs) | 2, 3 | 44 (12), 51 (5), 7B, Bw4 | 501, 14 | 4 | 53 | 3,7, 8 |
| 92-1 cell line (DNA) | 0201, 0301 | 4402, 5101 | 0501, 1402 | 0401 | 4 | 03 |
| Expressed | 2, 3 | B5 (not B44) | ||||
| Mel202 (PBLs) | 1, 3 | 16, 38, 52, Bw4 | -,- | 13, 15 | 51, 53 | 5, 6 |
| Mel202 cell line (DNA) | 0101, 0301 | 3801, 5201 | 1202, 1203 | 1301, 1501 | 3, 5 | 0601, 0603 |
| Expressed | 1, 3 | B38 (not B5) | ||||
| Mel270 (primary tumor E-1268-94) | 11, 29 | 7, 52, Bw4, Bw6 | 13, 15 | 51, 52 | 6 | |
| Mel270 cell line (DNA) | 1101, 2901 | 0705, 5201 | 1202, 1505 | 1301, 1501 | 3, 5 | 0601, 0603 |
| Expressed | 11, 29 | 5, 27 | ||||
| Mel285 (primary tumor E-994-95) | 24 | 7, (53), Bw4, Bw6 | Cw1 | 4 | 51, 53 | 3, 6 |
| Mel285 cell line (DNA) | 2402 | 1302 | 0602 | 0701 | 4 | 0201 |
| Expressed | 24 | 5, 27 (not 7) | ||||
| Mel290 (primary tumor E-131-96) | 1, 2 | 57, Bw4 | Cw6, Cw12 | 13 | 52 | (6), (7) |
| Mel290 cell line (DNA) | 0101, 0201 | 3801, 5701 | 0602, 1203 | 1301, 1305 | 3 | 0301, 0603 |
| Expressed | 1, 2 | 5 (not 7) | ||||
HLA types as determined in Leiden on peripheral blood leukocytes (92-1), in Boston on peripheral blood leukocytes (Mel202, HLA Class II of Mel 270), or on material acquired from the primary choroidal melanoma (according to the nomenclature of 1997, Verbik35), and on the cell lines (in Leiden). Empty cells indicate that a certain HLA type has not been tested.
-,- indicates an unsuccessful test.
Genotyping of the cell lines in Leiden47 showed for 92-1: A*0206, A*03, B*05, B*44, B*04; for Mel202 A*01, A*03, B*05, B*38, B*04; for Mel270 A*11, A*29, B*7, B*52 (5), B*04, B*06; for Mel285 A824, B*7, B*53, B*04, B*06; for Mel290 A81, A*2, B*57 (17), B*04.
Case Mel202, Pathology Case E-143-90
The patient was an 81-year-old Caucasian woman who noticed a decrease in the vision of her right eye in March 1988. She had been seen by her own ophthalmologist, who had seen her for follow-up examinations yearly since 1983; at the last examination, in July 1987, there was no evidence of a choroidal melanoma. Her past surgical history included removal of two benign colon polyps in 1981, a basal cell carcinoma on her nose in 1988, and hypertension. According to her family history, her father died at age 55 of a cerebral hemorrhage and her mother at age 89 of old age. A brother had diabetes, a sister had undergone a mastectomy, and a second brother had had stomach cancer. The patient was first seen at the Bascom Palmer Eye Institute in July 1988, after a 5-week history of blurred central vision in the right eye. The patient’s visual acuity was 20/25 OU. Slit-lamp examination showed no anterior segment abnormalities. Intraocular tension was 16 mm Hg OD and 17 mm Hg OS. Funduscopic examination of the right eye revealed an inferonasal elevated pigmented tumor with a collar-button configuration and an inferotemporal choroidal nevus (Figure 1). The base of the tumor was 10.5 mm. Echography showed a solid dome-shaped, regularly structured, low-reflective lesion inferiorly at the 5:30 position, with a maximal elevation of 5.4 mm. General workup revealed no evidence for metastases. The tumor was treated with a radioactive iodine plaque in August 1988. The tumor received a dose of 10,000 rad.
FIGURE 1.
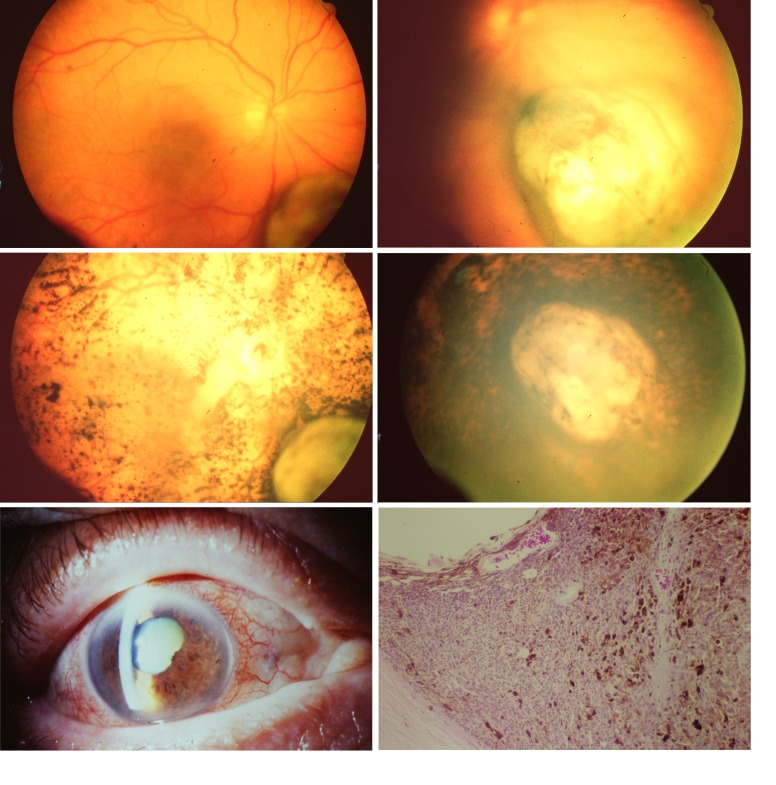
Clinical photographs of the eye and the choroidal melanoma that had been treated with an iodine-125 plaque in August 1988 and after enucleation gave rise to cell line Mel202. Top row, fundus and choroidal melanoma at the time of diagnosis in July 1988. Middle row, fundus and choroidal melanoma in January 1989 (left) and in March 1989 (right). Bottom row, eye just prior to enucleation in 1990 (left) and histologic section of the scleral part of the tumor (right), illustrating the high density of blood vessels and pigment macrophages (hematoxylin-eosin, ×10).
In January 1990, the patient was seen for follow-up examination, and visual acuity was found to be light perception. Slit-lamp examination showed rubeosis and angle neovascularization in the right eye and intraocular tension of 36 mm Hg. There was no view of the fundus. Ultrasound indicated the tumor had increased in height to 5.6 mm, up from 4.9 mm after radiation therapy. The basal diameter was now 9.0 mm (previously 6.5 mm) laterally and 11.0 mm (previously 8.5 mm) radially. Enucleation of the right eye was performed that month.
The pathology report for case E-143-90 (Dr V. T. Curtin) was as follows: “The gross specimen shows a diffuse variable transillumination defect to the sclera, but a more concentrated one inferonasally with an irregular outline of about 10×14 mm. Externally, the eye is normal. After sectioning, a pigmented tumor is seen inferonasally with a base of about 8 mm and an elevation of 5 mm. Microscopic examination reveals a globe with cornea with nonkeratinized, stratified, squamous epithelium which is focally artifactitiously absent. Basement membrane and Bowman’s layer are intact. The stroma is free of infiltrate. Descemet’s membrane is intact and endothelial cells are normal in number. The anterior chamber is free of cells. The angle is open and displays few extravasated red blood cells. The iris leaflets exhibit neovascularization. The ciliary body is mildly atrophic and there is mild hyalinization of ciliary processes. The lens is present and exhibits minimal nuclear sclerotic changes. There are occasional extravasated red blood cells, debris and pigment clumps in the vitreous. The retina and choroid are focally factitiously detached. There is a large basophilic mass arising from the posterior pole choroid consisting of numerous tightly packed basophilic spindle shaped cells with interspersed vessels and extensive areas of pigment deposition and necrosis. There are areas of more epithelioid-appearing tumor cells. The tumor has broken through Bruch’s membrane. There is extensive thinner epiretinal extension of the tumor temporally over much of the remaining retina, which displays varying degrees of disruption and tumor invasion. Four mitotic figures were seen in ten 40 high power fields. No tumor extension into the sclera is seen, however, there are a few scleral chronic inflammatory cells adjacent to the tumor mass. Some giant cells and foreign material is seen here. Cross section of the optic nerve exhibits hypercellularity and widening of pial septa, but no tumor extension is seen. Tumor extends into the disc but not through the lamina cribrosa. Conclusions: (1) choroidal malignant melanoma, mixed cell type with necrosis, 60% epithelioid cells, (2) retinal surface extension and invasion, (3) iris neovascularization, (4) status post radioactive plaque with episcleral foreign body reaction, (5) vitreous pigment phagocytosis, (6) optic atrophy.”
The last contact with the patient was in January 1991, when she indicated that she wanted to be seen by an ophthalmologist closer to home.
As the original block could not be retrieved, no genetic analysis could be performed on the primary tumor. When analyzing the cell line that was derived from this tumor, Mel202, a Q209L 626 A>T mutation in GNAQ was found. The cell line also showed a SF3B1 C1793 C>T mutation, with a normal EIF1AX and BAP1 gene.
Nareyeck and colleagues27 described a complex karyotype for cell lines Mel202, but also mentioned that they observed disomy for chromosome 3. Director-Myska and colleagues42 analyzed the karyotype (Table 1) and observed 52 chromosomes and disomy of chromosome 3. The complete karyotype was 52, XX, der(1) t(1;9)(q11;11), der (6) t(6;170(q16;q22), +der(6) t(6;17)(q16;q22), +7, +8, +8, +8, der(11) t(11;12)(q22;q21), inv(18)(q12;q23), der(20) t(8;20)(q22;q13.2). BAP1 is expressed on the cell line. The cell line displays HMB45.
Case Mel 270, Pathology Case E-1268-94
In 1977, the patient, aged 62, had eye movement problems following dysthyroid ophthalmopathy. Strabismus surgery was performed, which led to an infectious necrotizing pyogenic scleritis in the left eye with a serous retinal detachment. A nevus was observed in the right eye and stereo-photographs were taken. The description was “pigmented nevus with drusen, no serous fluid, 1.5 disc diameters (DD) in size, superior of the disc,” and visual acuity was 20/20. His history indicated benign prostate hypertrophy, shingles, and the fact that he had smoked five packs a day for 20 to 30 years but had quit 20 years ago. Alcohol intake was occasional. The patient’s father had abdominal cancer. In 1984, when the patient was 69 years old, Dr D. Gass at the Bascom Palmer Eye Institute ordered fluorescein photographs for the lesion in the right eye, which was now 3.5×4.5 DD with an elevation of 3 mm, with orange pigment, drusen, and trace subretinal fluid. Following a discussion between Dr Gass and Dr R. M. Ellsworth in New York, it was advised that treatment should be delayed until a clear change was documented, or until vision began to decline.
In March 1987, 10 years after a nevus had first been observed in the right eye, examination by Dr P. Finger in New York showed a lesion that by indirect ophthalmoscopy was considered to be 11×8×6 mm, located superior to the optic nerve, and surrounding the optic nerve along the superior 180°. The tumor edge extended to within 1.5 mm of the fovea. Ultrasonography showed a tumor height of 6.0 mm. An iodine-125 plaque was used as treatment for this choroidal melanoma.
The patient was seen again at the Bascom Palmer Eye Institute in November 1992, when an echographic examination was performed because of a 3-day history of metamorphopsia secondary to a submacular hemorrhage in the right eye. Echography showed mild vitreous opacities. The lesion located in the superior pole had changed shape and had developed a more irregular, peaked appearance, suggesting the possibility of a collar-button shape. The retina was thickened over the apex of the tumor. The internal structure was slightly irregular with medium reflectivity and moderate vascularity. The maximum elevation was 4.5 mm. The basal diameter was unchanged. On January 11, 1993, echography showed a lesion that now measured 3.5 mm in height, with a base measurement of 11×9 mm (unchanged). The internal structure continued to be somewhat irregular with medium to high reflectivity. A mild internal vascularity was noticed. No extrascleral extension was detected, although the sclera was less reflective than prior to the treatment. There was only a slight elevation at the macula, probably due to resorption of the submacular hemorrhage. In September 1993, a shallow retinal detachment was present and the tumor displayed a collar-button shape with an increased size of 5.2×14.5×12.5 mm. It remained low to medium in reflectivity, mildly vascular, and had a slightly irregular internal structure.
Everything went relatively well until further radiation-associated retinal alterations occurred with formation of a subretinal neovascular membrane, leading to a massive subretinal and intravitreal hemorrhage, reducing vision in the right eye to light perception. In January 1994, very dense vitreous opacities were seen, and the lesion measured 8.2×18×15 mm, with a collar-button shape, mildly vascular on A-scan, with only a few vessels on color Doppler. In April 1994, the lesion was irregularly structured and lobulated. Height equaled 8.0 mm at the 11-o’clock position, and the lesion was highly reflective and irregular in structure. A low reflective, dome-shaped, vascularized region was located at the 9-o’clock position. The height here equaled 4.3 mm, with a radial base of approximately 10 mm. There was also a low reflective elevation, from approximately the 3-o’clock position clockwise to the lesion at the 9-o’clock position. It was uncertain whether or not this area was solid. No vascularity was seen in this region with A-scan or Doppler. The height measurement was approximately 2.5 mm, and it extended anterior to the equator. The conclusion was that there was further growth of the lesion, with extension into the macular area and possibly extensive involvement of the inferior fundus. This inferior region could possibly be subretinal blood. Echography performed in August 1994 showed a dome-shaped, medium-low reflective, regularly structured, highly vascular lesion, involving the entire temporal fundus. The lesion’s height was 12.7 mm, still without extraocular extension (Figure 2).
FIGURE 2.
Histologic overviews of three choroidal melanoma (hematoxylin-eosin, original magnification ×12.5), expression of Melan-A/MART-1, HMB45, and S100 (original magnification ×400) for tumors E-1268-94 (Mel270) (top), E-994-95 (Mel285) (middle), and 131-96 (Mel290) (bottom) (original magnification ×400). The morphology and immunophenotypic profile of the three tumors are consistent with choroidal melanoma. The aspect of the iris of 131-96 (lower left) is consistent with a nevus of Ota.
An enucleation was performed that month. The pathology report by Dr V. T. Curtin was as follows: “A large, variably pigmented but primarily brown mass arises from the choroid, occupying about half of the vitreous cavity (24×12.7 mm height). On sectioning, one noticed a large, vascularized pigmented mass composed of basophilic cells with a necrotic center arising from the temporal choroid and extending from the ciliary body to and surrounding the optic nerve head. Part of the tumor facing the vitreous had the characteristics of a nevus. The tumor contained highly anaplastic cells of varying morphology including spindle-shaped cells with syncytial cytoplasm and elongated nuclei with prominent nucleoli, and epithelioid cells with eosinophilic cytoplasm and dark nuclei with multiple nucleoli. There were 11 mitotic figures per ten 40× high powered fields. The tumor contained many PAS channels and vessels. There was an extension of tumor through the emissary channel to the episclera. The retina was completely detached in a funnel pattern and was necrotic and atrophic. There were areas of retinal hemorrhage overlying the tumor. There was prelaminar invasion of the tumor and the optic nerve was atrophied. The sclera was otherwise unremarkable. Conclusions: (1) malignant melanoma of the choroid and ciliary body, mixed cell type (70% epithelioid), with extrascleral extension, (2) marked tumor necrosis, (3) secondary retinal detachment, (4) optic atrophy, (5) status post I-125 radiation plaque.”
In June 1995, the then 80-year-old patient developed pain across his abdomen. Ultrasound and a CT scan showed multiple liver lesions, and a biopsy proved the presence of liver metastases of the uveal melanoma. Other metastases included those in a rib and the iliac wing. Several biopsies were obtained. Following one course of DTIC, BCNU, and cisplatin, he became terminally ill in July 1995.
Sanger sequencing (Liverpool, Rotterdam) and ddPCR analysis (Leiden) of the primary tumor showed a GNAQ Q209P mutation. Chromosome analysis by FISH (Rotterdam) showed heterogeneity with only one signal of chromosome 3 in 50% of the cells, with two copies of 8q and 8p. Immunohistochemical analysis showed no BAP1 staining. We do not have an analysis of BAP1 mutations in the primary tumor. Staining for HMB45 showed 99% of the cells to be positive (3+), whereas Melan-A was expressed on 80% (1+) and S100 on 10% (1% to 2%). In the nevus-like part along the vitreous, HMB45 was positive on 50%, Melan-A on 1%, and S100 on 1%. The tumor contained many CD68+ macrophages and moderate numbers of CD3+ T cells.
Cell lines Mel270, OMM2.3, and OMM2.5, derived from the primary tumor of this patient and two metastases of the same case, respectively, are known to harbor a 626 A > C mutation in GNAQ Q209P.26 This was confirmed by Rotterdam, and no mutations were found in BAP1, EIF1AX, or SF3B1. Chromosome analysis of the cell line showed disomy of chromosome 3, with tetrasomy of 6p and extra chromosomes 8.42 The cell line was positive for HMB45 and Melan-A. The HLA type of the cell line as determined in Leiden corresponded to the HLA type of primary tumor material as analyzed in Boston (Table 3).
Case Mel285, Pathology Case E-994-95
A 49-year-old Hispanic woman was referred to the Bascom Palmer Eye Institute in July 1995. One year earlier, the patient complained of dizziness, but her doctor ruled out any ocular or ENT-region involvement. In June 1995, she noticed blurred vision, floaters, and flashes in her left eye and consulted her local ophthalmologist, who referred her to the Bascom Palmer Eye Institute because of a large ciliary body mass. She was known to have had hypercholesterolemia for 2 years and had undergone a hysterectomy. Visual acuity was 20/20 OD and 20/40 OS (correction S-0.75 C+ 0.75×155°), without improvement when using a pinhole. On the Amsler grid, the left eye noticed central metamorphopsia. Pupil size was 5.0 mm in the right eye and 4.5 mm in the left eye, without an afferent pupillary defect. The eye pressure was 16 mm Hg in both eyes. The extraocular motility was intact. Slit-lamp examination with a dilated pupil showed an extensive pigmented mass inferonasally, extending for 2.5 to 3 clock hours, with pigmented cells in the vitreous. The peripheral lesion was dome-shaped and located at the ciliary body. The diagnosis was a large anterior uveal melanoma for which enucleation was indicated. Echography (July 1995) for an elevated ciliary body lesion showed a dome-shaped lesion between the 5:30 and 8:30 positions in the ciliary body. It was low to medium reflective, regularly structured, and markedly vascular on both A-scan and B-scan and with color flow Doppler. The dimensions were 12.0×10.5×13.0 mm (radial dimension includes the overhang). No extraocular extension was detected, but the ciliary body location made it difficult to be certain. The conclusion was that the echograms were typical for melanoma. At this time her liver functions were normal: aspartate aminotransferase (AST) 37 U/L (normal range, 5–40), alanine aminotransferase (ALT) 52 U/L (normal range, 7–56), and lactate dehydrogenase (LDH) 485 U/L (normal range, 297–618). A chest x-ray did not reveal abnormalities. She underwent an enucleation of the left eye with a hydroxyapatite implant in July 1995 (Figure 2).
The pathology report (Dr R. L McDonough and Dr V. T. Curtin) was as follows: “The specimen consists of a left eye, already sectioned intraoperatively, measuring 26×25.5×25 mm with 7 mm of attached optic nerve. The cornea is clear, the anterior chamber formed. The lens is cataractous. There is a large brown pigmented mass arising from choroid and ciliary body abutting the lens, measuring 11×9 mm (base). The vitreous is clear and the retina, choroid and sclera are otherwise unremarkable. The optic nerve is not cupped. Microscopic evaluation reveals a nonkeratinized stratified squamous epithelium with patchy areas of denuded epithelium. Bowman’s membrane is intact and the corneal stroma, Descemet’s membrane and endothelium are unremarkable. The trabecular meshwork and angle are within normal limits. On the side of the tumor the trabecular meshwork contains a small amount of blood. On the side of the tumor there is perivascular infiltrate mixed with chronic inflammatory cells. There is a small amount of cataractous changes, chronic iritis, small amount of degenerative material of the posterior capsule. There is a large hyperpigmented, hypercellular, vascularized circular mass arising from the choroid infiltrating the ciliary body. This tumor mass contains poorly differentiated cells with prominent nucleoli and indistinct cell borders with areas of focal necrosis. The vitreous appears unremarkable. The retina has areas of secondary retinal detachment which may be artifactitious. The sclera is unremarkable and without tumor infiltration. The optic nerve appears normal. Conclusion: (1) ciliochoroidal melanoma, mushroom shaped, with sporadic melanophages, of mixed cell type with about 25% epithelioid cells. Slight invasion in the sclera.”
In March 1997, the patient noticed problems with the fitting of the prosthesis, which led to repetitive ocular irritation and infections. She was refitted with a new prosthesis. In August 1999, no abnormalities were noticed regarding the left socket, and there were no signs of metastasis. Vision was 20/20 OD, with a normal pressure and no abnormalities in the posterior segment. The patient’s AST was 37 U/L (normal, up to 29 U/L) and alkaline phosphatase was 117 U/L (normal, up to 111 U/L). In February 1999, all thyroid values were increased: triiodothyronine (T3) was 39.0 (reference 24.3%), thyroxine (T4) was 12.0 (reference, 4.5 μg/dL), thyrotropin was 6.0 (reference, 0.5 μIU/ml), and the free thyroxine index (FT4) was 3.7 (reference, 1.7%). No subsequent information is available on this patient.
The original tumor has a GNA11 Q209 mutation (Liverpool, Rotterdam). The primary tumor was found by ddPCR to have a chromosome 3 monosomy and was negative for BAP1 expression. HMB45 stained almost all cells of the primary tumor. The iris and ciliary body were negative. Melan-A was more positive in the basic part of the tumor that was primarily made up of spindle cells (80%, 2–3+). Melanophages were Melan-A–positive, with vessels staining negative. S100 stained the optic nerve, 70% of the spindle part (2+), and 70% of the epithelioid part (1+). One local lymphocytic infiltrate was observed. MITF was highly expressed in melanophages.
While the original tumor has a GNA11 Q209 mutation, this is not present in the cell line26 (confirmed in Rotterdam and Leiden). The cell line had chromosome 3 disomy, with a reported loss at chromosome 3p26-pter42 and four copies of chromosome 8q (ddPCR Leiden). Cell line Mel285 is known not to express HMB45 or Melan-A.43
Case Mel290, Pathology Case E-131-96
In January 1996, a 67-year-old Caucasian woman visited an ophthalmologist because of decreased vision in the right eye for approximately 1 month. She had noticed a spot right in the middle of her sight, without flashes. As she described it when she visited the Bascom Palmer Eye Institute, she had been born with a brown right eye and a blue left eye. Her history showed the presence of emphysema. She smoked a package of tobacco a day. Her family history was unremarkable.
At examination, visual acuity was 20/40- OD with metamorphopsia and 20/20 OS; pressure was 10 mm Hg OD and 14 mm Hg OS. The right visual field of the right eye was restricted. The right eye showed a nevus of Ota with discoloration of the sclera and conjunctiva temporally and superiorly. The anterior segment showed no other abnormalities, except the presence of a right afferent pupillary defect. Fundus examination revealed a juxtapapillary ocular melanoma in the right eye, with a collar-button configuration, located just superior to the optic nerve. There was an exudative retinal detachment surrounding the lesion. An intrinsic pigmentation was present with orange flecks within the lesion itself.
Echographic examination showed a large, solid, and slightly peaked mass superiorly at the optic disc, extending to the equator, with a maximum size of 10.5×10.5 mm, 6.5 mm in height, with an overlying neurosensory detachment and a baseline hemorrhage in the area of the collar-button rupture. The lesion was regularly structured, low reflective, and moderately vascular, without evidence of extrascleral extension. The retrobulbar optic nerve was within normal limits.
Liver function tests and chest x-ray were normal. An enucleation was performed in January 1996, and a hydroxyapatite implant wrapped in donor sclera was implanted (Figure 2).
The pathology report stated the following: “Gross histopathological examination showed a tumor with a diameter of 12×10 mm, 5 mm in height, with a brown, tan colour. The sclera contained numerous foci of pigmented cells. The iris was heavily pigmented. The ciliary body was atrophic with hyalinization. There was an anterior subcapsular cataract. In the posterior pole was a tumor, arising from the choroid and in close proximity to the optic nerve, with prominent nucleoli with numerous pigmented cells. There were no mitoses. The choroid contained numerous pigmented cells (melanophages and pigment-containing tumor cells) with a focal area of chronic inflammatory cells. The tumor was highly vascularized. Conclusions: (1) a malignant melanoma of the choroid, of the mixed cell type (30% epithelioid cells), (2) melanosis oculi with a history of a nevus of Ota, (3) a detached retina, secondary to melanoma, (4) cataract.”
In February 2000, the patient informed her doctor at the Bascom Palmer Eye Institute that she was fine but no longer wished to travel to visit him.
E-131-96 developed in an eye with a nevus of Ota. ddPCR showed the presence of monosomy 3 in primary tumor E-131-96. Chromosome analysis of the primary tumor by FISH showed only one chromosome 3 in over 80% of the cells and two copies of 8q and 8p (Rotterdam). BAP1 staining of primary tumor 131-96 was positive. Hematoxylin-eosin sections of the primary tumor showed a lot of pigment-containing cells, including melanophages. HMB45 was expressed on 99% of the cells (3–4+). Melan-A was expressed on 95% of the cells (3+). S100 was expressed in the optic nerve and on up to 10% of the cells in the tumor (1+).
Cell line Mel290 has been reported 26 not to have a GNAQ or a GNA11 mutation. Neither was a mutation found in BRAF, SF3B1 (exon 12, 13, 14, 15), EIF1AX, or BAP1 (A. de Klein, Rotterdam). This cell line does not show LOH at chromosome 3, and carries 46 chromosomes.44 SNP analysis showed only one chromosome 1, with two copies of chromosome 3 and chromosome 8 (Rotterdam). Director-Myska and colleagues42 observed that the cell line lacks 3p26-pter, and has gained 8q24.1-q24.2, where the oncogene C-MYC is located. BAP1 immunohistochemical testing showed a loss of BAP1 expression in this cell line, although the protein was found by Western blotting. Cell line Mel290 is known not to express HMB45 and Melan-A/MART-1.43 The presence of several chromosome abnormalities in the cell line suggests that it may still be derived from uveal melanoma cells, in spite of the lack of a GNAQ or GNA11 mutation in the cell line itself.
GNAQ, GNA11, BRAF MUTATIONS
As BRAF mutations may identify tumors derived from cutaneous melanoma, cell lines were analyzed in Leiden for this mutation. Not one of the cell lines tested contained a BRAF mutation, as was previously also reported by Griewank and colleagues26 (Table 1).
SHORT TANDEM REPEAT PROFILES
We were able to determine the STR profiles in our cell lines that have either been grown in our laboratory (92-1) or been sent directly to us from Boston (Mel202, Mel270, Mel285, Mel290) and subsequently have been maintained in our laboratory (Table 2). We did not have enough material of good-quality DNA to determine STR profiles of the primary tumors. Griewank and colleagues26 mention that one cell line, 92-1, in their hands, had a different STR profile than the one that had been reported for this cell line by the EMBL-EBI file. Our analysis of the STR profiles agrees with Griewank’s.
HLA POLYMORPHISMS
HLA polymorphisms have been determined in Leiden on the leukocytes of the patient whose tumor gave rise to cell line 92-1 in Boston on primary tumor material or leukocytes (four cases), and in Leiden on the cell lines by DNA typing45 (Table 3). As the combination of the different alleles is quite unique, one can identify the five cell lines described in this thesis purely on the basis of their HLA genotype. Cell lines Mel270 and the two cell lines that were derived from metastases from the same patient (OMM2.3 and OMM2.5), of course, have the same HLA polymorphism.
DISCUSSION
The clinical histories of the cases showed five different backgrounds: Cell line 92-1 is derived from a large, slowly growing melanoma, and even after exenteration, the patient lived 2½ years before succumbing to metastatic diseases. This specific cell line carries an EIF1AX mutation, which in some studies has been related to the development of late metastases. Mel202 and Mel270 were derived from tumors that had previously been irradiated and subsequently recurred. Of the five original tumors, Mel285 was the only “normal” straightforward ciliochoroidal melanoma, with loss of BAP1 expression. The fifth tumor, E-131-96, which gave rise to cell line Mel290, developed in a heavily pigmented eye with a nevus of Ota.
Two of the five patients had hyperthyroidism, and several were heavy smokers. A recent report analyzed the impact of thyroid hormones in a B16F10 murine model of ocular melanoma48: mice given thyroxine developed metastases faster than mice with induced hypothyroidism, although the numbers were quite small, and thyroxine-treated mice did not differ significantly from control mice. The impact of smoking on the development of uveal melanoma and of its metastases warrants more study. We previously observed that high-risk uveal melanomas contain an inflammatory phenotype, with many tumor-infiltrating lymphocytes and macrophages.49 One of the effects of smoking is the induction of ocular inflammation, in which especially bone marrow–derived cells such as macrophages are involved.50 We do not know how hyperthyroidism influences human uveal melanoma, but it might be worth investigating further.
Two of the five cell lines were derived from previously irradiated tumors. We have not found information regarding irradiation treatment in the cases described by Nareyeck and colleagues27; in their cases, tumors displayed complex karyotypes, which may indicate prior irradiation. We recently observed that although it was more difficult to perform karyotyping and FISH on previously irradiated tumors, tumors that had been irradiated often showed complex karyotypes.24 Némati and associates34 described cell lines that have been derived from xenografts that grew after placement of human tumor material in SCID mice; it would be interesting to know how many of these xenografts were derived from previously irradiated tumors.
Our data are consistent with the idea that when trying to grow out uveal melanoma cell lines, one should not select the most straightforward tumors. Heegaard and colleagues51 tried to grow uveal melanoma xenografts in mice but were not successful. The work in Paris showed that material obtained from metastases has a higher chance of becoming xenografts.34
We tried to compare characteristics of five often-used cell lines with the characteristics of their primary tumors. All five tumors were of mixed-cell type, with a large epithelioid component. However, the laboratory data showed unexpected results, such as a normal chromosome 3 status as well as a normal BAP1 expression in the tumor that gave rise to the highly aggressive cell line 92-1 and the presence of a GNA11 mutation in the original tumor of cell line Mel285, although no GNA11 or GNAQ mutation is present in the Mel285 cell line. Two cell lines that lack GNA11 and GNAQ mutations, ie, Mel285 and Mel290, also lack pigment markers, although both of the primary tumors express Melan-A/MART-1 and HMB-45 at very high levels. This suggests that the pathways that are stimulated by GNA11/GNAQ mutations may influence the expression of pigmentation markers in uveal melanoma. This would be consistent with the findings of Van Raamsdonk and colleagues,52 who were studying the role of GNAQ/GNA11 in murine skin pigmentation, which ultimately led to the identification of the role of these two genes in uveal melanoma.2,3 The influence of epigenetic markers next to mutations might also be relevant.
DISCUSSION OF INDIVIDUAL CELL LINES
Cell Line 92-1
The 92-1 cell line has been used in many experiments concerning uveal melanoma worldwide and has been shown to have a high proliferation rate and to give rise to many metastases in the original patient and in animal experiments.43,53,54 Cell line 92-1 had been derived from a very large tumor that had destroyed the eye and filled most of the orbit. Two-and-a-half years after exenteration, the patient died and autopsy revealed multiple highly pigmented metastases. The primary tumor, the metastases, and the cell line all show two signals for chromosome 3, expression of BAP1, and gain of 8q, which are not the typical characteristics associated with high-risk uveal melanoma. Kalirai and colleagues22 analyzed the chromosome status and BAP1 expression of four groups of uveal melanoma that were either typical (early death associated with monosomy 3, or survival after 6 years in disomy 3 cases) or nontypical (survival after 6 years in monosomy 3 cases, or death within 6 years due to metastases in disomy 3 cases). In the latter group, as concerning our case, both nuclear BAP1-positive and BAP1-negative cases were identified. Our case had a high number of mitoses (35 per 15 high-power fields)53; similarly, in Kalirai’s group with disomy 3 and early death, high numbers of mitoses were associated with death.22 While unusual, our case is thus not unique, and an association between a high HLA expression and death due to metastases has been seen in several studies.53 Amirouchene-Angelozzi and colleagues28 reported the presence of an EIF1AX mutation in cell line 92-1. EIF1AX mutations have been associated with the development of metastases in disomy 3 uveal melanoma.5 Further research into the function of EIF1AX as an inducer of cell growth and the late development of metastases is warranted. Cell line 92-1 does not carry a mutation in exon 14 of SF3B1 or in BAP1 (Rotterdam).28
Field and colleagues55 reported that a subgroup of the generally benign Class I tumors did result in metastases and these tumors were characterized by expression of PRAME; such tumors did not show chromosome 3 monosomy, but most did show extra copies of 8q, as is the case in the primary tumor that gave rise to cell line 92-1.
Cell Line Mel202
The origin of this cell line has been described by Ksander.37 The tumor was diagnosed in 1988 in an 81-year-old Caucasian woman and irradiated with an iodine-125 plaque. Seventeen months later, the tumor had grown and the eye showed neovascular glaucoma. A striking phenomenon was the heavy pigmentation of the fundus after irradiation (Figure 1). We have only 1 year of follow-up on this patient, at which time the patient had not developed metastases. We do not have any further material of this tumor. While analysis of cell line Mel202 showed disomy 3, this was probably a case of isodisomy, as homozygosity for all chromosome 3 loci has been observed.27 However, this cell line does express the BAP1 protein.28 Additionally, the cell line contains five copies of chromosome 8. An increased number of chromosome 8 in the primary tumor is associated with a worse prognosis than when only one extra chromosome 8 is present.11,15, This cell line, together with cell lines Mel270, Mel285, and Mel290, has been used in many experiments to determine what factors play a role in the development of metastases, especially which immunologic influences.56–58 This cell line is being applied in drug testing, as it carries a GNAQ Q209L mutation and therewith represents many uveal melanomas.3,28
Cell Line Mel270
Another extensively used cell line is Mel270; as an addition to this primary tumor–derived cell line, several metastases from the same patient have been cultured, providing cell lines as well. These metastasis-derived cell lines are now known as OMM2.3 and OMM2.5, but they were originally called OMM1.3 and OMM1.5, respectively.38 Their STR profiles have been described by Griewank and colleagues,26 whose findings correspond with the STR profiles we obtained from these cell lines. We also have the HLA types, which had also originally been obtained in Boston from the original ocular tumor (Table 3).
This case itself is remarkable for its link with so many famous ophthalmologists. The clinical history was followed from the early identification of a nevus/small melanoma in 1977 until the death of the patient in 1995. The original lesion was first observed by Dr D. Gass and subsequently followed by Dr R. M. Ellsworth in New York. When a clear change was noticed in 1987, Dr P. Finger irradiated the lesion with a cobalt-60 plaque. In 1994, the tumor had regrown and vision had been reduced to light perception, at which time the eye was enucleated. In the summer of 1995, the patient developed metastases. Pathologic examination showed a mixed-cell type tumor that occupied half of the vitreous. Part of the tumor had the characteristics of a nevus. All three cell lines were already known to carry a GNAQ Q209P mutation, and recent studies in Liverpool and in Leiden identified the same mutation in the primary tumor material. Prior chromosome testing of cell lines Mel270 and OMM2.3 showed the presence of two copies of chromosomes 3, with contradicting results on chromosome 8q, and four copies of 6p with loss of 6q.42 The primary tumor had lost one copy of chromosome 3, did not express BAP1, and carried two copies of chromosome 8 (Leiden, Rotterdam). Interestingly, Mel270 and the metastasis-derived cell line OMM2.3 display BAP1 expression, with both Amirouchene-Angelozzi and colleagues28 and our own laboratory reporting this.
Cell Line Mel285
The original tumor was a large ciliochoroidal melanoma that had not been irradiated at any time. While ddPCR indicated that the primary tumor had a monosomy of chromosome 3 and did not express BAP1, divergent reports exist with regard to the chromosome 3 status of the cell line: Nareyeck and colleagues27 did not find loss of heterozygosity for chromosome 3, whereas Director-Myska and colleagues,42 using comparative genomic hybridization, observed a loss of chromosome 3p26-pter, and Rotterdam, using a SNP array, noticed a partial deletion at 3p. BAP1 was expressed on the cell line, and no mutation was found. Currently, the techniques are not yet good enough to determine the mutation status of BAP1 in the preserved paraffin-embedded tissue.
Director-Myska and colleagues42 already reported a gain at 8q24.1–q24.2. A. de Klein similarly noticed two copies of 8p and 4 copies of 8q. This suggests that a gene located on 8q may play a role in the propagation of this cell line. The lack of pigment in this cell line is related to methylation of the NruI regulator site, which regulates a series of pigment genes (see discussion under “Melanocyte Lineage Markers”).
Cell Line Mel290: Nevus of Ota
Similar to the previous two cell lines, Mel290 is one of the workhorses of ocular oncology research, being used in immunologic pathway and drug experiments. The tumor originated in an eye with a nevus of Ota: when the patient (a 67-year-old Caucasian woman) visited the Bascom Palmer Eye Institute in 1996, she mentioned that she had been born with a blue eye and a brown eye. On examination, her right eye showed the typical nevus of Ota discoloration of the sclera and conjunctiva temporally and superiorly. A juxtapapillary melanoma with a collar-button configuration was present, with an exudative retinal detachment. On pathologic examination, the iris was found to be heavily pigmented. The tumor was a choroidal melanoma of the mixed-cell type. Four years later, the patient was in good condition.
It is well known that the presence of a nevus of Ota carries a significant risk of uveal melanoma.59 In one of the original reports, Van Raamsdonk and colleagues3 studied several cases of a nevus of Ota and reported the presence of both a GNA11 and a GNAQ mutation in this disease. Analysis of the primary tumor by ddPCR did not reveal a mutation in exons 4 or 5 of GNA11 or GNAQ. When techniques become more sophisticated, we hope to find whether the development of a nevus of Ota is determined by a mutation or by epigenetic regulation. It is fascinating that in spite of the lack of monosomy 3 in the cell lines and the absence of known mutations, the cell line proliferates well. ddPCR (Leiden) and FISH analysis (Rotterdam) showed loss of one chromosome 3 in the primary tumor. Ambrosini and colleagues60 reported overexpression of MYC. When studying the effect of drugs, this cell line can (like Mel280) be used as the control GNAQ/GNA11 wild type, with, for example, 92-1 and Mel202 as mutated cell lines (both with GNAQQ209L mutations).61
STR PROFILES
Griewank and colleagues26 mention that the STR profile that they found for cell line 92-1 differed from the profile reported by the European Molecular Biology Laboratory–European Bioinformatics Institute (http://www.ebi.ac.uk/ipd/estdab/directory.html). Otherwise, profiles were similar.
HLA POLYMORPHISMS
A variety of HLA polymorphisms were observed in the uveal melanoma patients, and the only remarkable characteristic was that all five cell lines carried the Bw4 antigen. A large study from our laboratory on the distribution of polymorphic HLA antigens among 235 uveal melanoma patients did not identify any genetic associations between HLA genes and the development of uveal melanoma.62 Bw4 was not studied at that time. A prior study on expression of HLA antigens on cell lines demonstrated that some antigens were lost during progression to cell lines: 92-1 expresses HLA-A2, A3, and B5 but not B44, and Mel202 expresses HLA-A1, A3, and B38 but not B5, which does not respond to interferon alfa or gamma.47 The presence of HLA-A2 in two cell lines, ie, 92-1 and Mel290, allows many immunologic investigations, as tumor antigen–specific cytotoxic T cell responses are often directed against HLA-A2–associated antigens. Determining whether the antigens are really being expressed and not only genetically determined is essential for proper evaluation of cytotoxic responses.
MELANOCYTE LINEAGE MARKERS
Several studies investigated the expression of melanocyte lineage markers on uveal melanoma cell lines. Van Dinten and colleagues43 studied the expression of a wide range of melanocyte-related antigens on cell lines 92-1, Mel270, Mel285, Mel290, OMM2.3, and OMM 2.5. Two groups of cell lines were identified: 92-1, Mel270, OMM2.3, as well as OMM2.5 expressed melanocyte markers, whereas Mel285 and Mel290 did not express Melan-A/MART-1, tyrosinase, or tyrosinase-related protein. A corresponding difference in methylation of the NruI regulator site was noticed, suggesting that different methylation states of the DNA encompassing the Melan-A/MART-1 regulatory region played a role in the expression patterns.
We looked at the corresponding primary tumors of Mel285 and Mel290 and observed expression of Melan-A/MART-1 and HMB45 on the majority of cells of these two tumors, with S100 expressed on 70% of the cells of the primary tumor of Mel285 but not of Mel290. That cell lines can derive from subsets of cells in a primary tumor has been known for a long time63; our laboratory was one of the first to also show heterogeneity for the presence of monosomy 3.64 The staining results of the different primary tumors show that the melanocyte antigens that we studied are often expressed on a majority of the cells, but not on all.
When studying the presence of GNA11 and GNAQ mutations, we noticed that the original GNA11 mutation that was present in primary tumor Mel285 was not present in the cell line. Griewank and colleagues26 previously stated the following: “It is unclear whether the absence of these melanoma-associated antigens is a product of the expansion of these cells in culture or represents the antigenic nature of the tumors from which they were derived; however, it is of interest that this phenotype was observed in uveal melanoma cell lines lacking mutations in GNAQ or GNA11.” Clearly, the absence of the melanoma-associated genes is not a representation of the original nature of the tumors from which they were derived. Cell line Mel285 must be derived from a clone of tumor Mel285, not representing the majority of the original tumor cells. That the cell line was not switched somewhere and comes from the original tumor can be ascertained by its HLA typing, which is highly characteristic for this individual. Van Raamsdonk and colleagues52 suggested, following the first studies on skin color in mice, that mutations in GNAQ and GNA11 might be relevant to dermal skin color, and we propose that the absence of mutations in GNAQ and GNA11 in cell lines Mel285 and Mel290 and the lack of expression of melanocyte lineage markers are related. How the methylation of the NruI regulator site is regulated is unknown, but an early abnormality in epigenetic regulation may also be involved in providing a proliferation signal different from that of the GNAQ/GNA11 mutations. Griewank and colleagues26 also mentioned in their report that it may be difficult to culture tumor cells from tumors that consist mainly of GNAQ/GNA11 mutated cells, and that may have happened here; it may therefore not come as a surprise, when more sophisticated techniques might one day demonstrate the presence of a GNAQ or GNA11 mutation in the original tumor of Mel290 (associated with the nevus of Ota).
ARE WE DEALING WITH UVEAL MELANOMA CELL LINES?
Since GNA11 and GNAQ mutations are regarded as the typical mutations of uveal melanoma, just as BRAF mutations are typical for cutaneous melanoma, one may wonder whether cell lines Mel285 and Mel290 are malignant, or are derived from, for example, tumor fibroblasts. The absence of melanocyte markers may support such a background. The chromosomal abnormalities in cell line Mel285 and Mel290 support a malignant background, but are these cell lines derived from their primary choroidal melanoma?
A shared characteristic of all of these five cell lines is disomy of chromosome 3 and extra 8q copies in four of the five cell lines. In the steps toward tumor metastases, the GNAQ/GNA11 mutation is considered an early event, and a mutation in BAP1 in combination with loss of chromosome 3 is considered an essential step in the pathway toward the prognostically bad Class 2 tumor type vs the prognostically-good Class 1 phenotype. Two recent studies on the sequence of events in uveal melanoma show that it is likely that addition of 8q precedes loss of one chromosome 8.17,18 The association between gain of 8q and the development of metastases, in tumors with or without loss of chromosome 3, shows that this chromosome aberration by itself plays a role in setting the stage for malignancy. We suggest that the gain of 8q was important in the process of making the tumor cells proliferate and give rise to cell lines. A study on cell lines derived from primary tumors or uveal melanoma xenografts similarly showed gain of 8q in all seven cell lines.28
One of the primary tumors that we studied did not have a mutation in GNAQ/GNA11 and neither did the cell line. Whether the cells in the other GNAQ/GNA11-negative cell line lost their GNAQ/GNA11 mutations during growth in vitro or whether there was selection of primary tumor cells that never carried this mutation is unknown. In a French study on two cell lines derived from primary uveal melanoma, both retained the same mutation as the primary tumor.28,34,65
CONCLUSION
Four of the five tumors that had given rise to metastases are unusual, having been irradiated or developing from a nevus of Ota. This finding may provide clues to which tumors to select when one wants to obtain cell lines from uveal melanoma, such as using previously irradiated tumors. We were able to compare several cell lines with their primary tumor. Clearly, all of the cell lines share only some characteristics with their primary choroidal melanoma. In some instances, we were surprised by the results, for example, that the primary tumor of cell line 92-1 had a normal disomy 3 status and BAP1 expression, as this cell line is highly metastatic in animal models. This cell line contains an EIF1AX mutation, which has been shown to be associated with metastases development in disomy 3 tumors.
Several of the cell lines show no mutations in GNAQ or GNA11, disomy for chromosome 3, and a lack of melanocyte antigens. We hypothesize that while part of the primary tumor contained cells with a GNAQ/GNA11 mutation, not all proliferating cells may have carried this characteristic. The original tumors of Mel270 and Mel285 show monosomy of chromosome 3 and lack BAP1 expression, but the cell lines have disomy of chromosome 3, with small local losses; two cell lines lack melanocyte markers, whereas their original tumors display a variety of antigens and in at least one of these cell lines, a mutation in GNA11 has been lost. It is very hard to grow cell lines of uveal melanoma: chromosome 3 monosomy and lack of BAP1 expression seem to inhibit the growth of cells in culture, although especially these characteristics are clinically highly associated with the development of metastases. In Paris, cultures were set up of 73 primary uveal melanomas, resulting in outgrowth of two cell lines only. Interestingly, both of these cell lines carry a BAP1 mutation, and both are known to grow very slowly.28
Current HLA typing of our cell lines showed that we were still working (after 20 years) with the correct cell lines, although we do not know whether any unknown parameters have changed. Authors who publish articles about cell lines should always state the original source where their cell line was first described and mention through which laboratories they acquired their cell lines. Overall, our data support our hypothesis that unusual findings in uveal melanoma cell lines may be caused by (1) selection (through, for example, irradiation) of cells from specific choroidal melanoma, and (2) outgrowth of specific clones of choroidal melanoma cells that do not resemble the primary tumor in, for example, chromosome composition, either through in vivo or in vitro selection. Dividing cells may or may not have retained the GNAQ/GNA11 mutation that was present in a subpopulation of the primary tumor. Extra material at 8q was observed in all cell lines, and one may assume that this is a major factor that determines proliferation. A study on two cell lines created from primary tumors and five cell lines derived from uveal melanoma xenografts in Paris similarly showed gain at 8q in all cases.28,34,65 Most likely, cells carrying extra copies of 8q had an advantage and thus helped to continue the proliferation of these five uveal melanoma cell lines. As suggested recently,17,18 following a mutation in GNAQ/GNA11, multiplication of material at 8q may be the second step toward proliferation of a melanocyte, and it may well be that this modification similarly sustains the development of uveal melanoma cell lines.
As cell lines are being used to test drugs, one wonders whether these cell lines represent uveal melanoma. The best model would be to have cell lines with all the characteristics of a highly malignant primary tumor, thus having monosomy of chromosome 3 and loss of BAP1. Only one of the two French cell lines has these characteristics, and it is known to grow very slowly. We do not know whether this tumor had been irradiated prior to enucleation. In our experience, only cells from unusual tumors continued to grow. Overall, they corresponded with their primary tumor in many, but not all, aspects and can be used for drug effects on specific pathways or immunologic studies.
ACKNOWLEDGMENTS
Funding/Support: Supported in part by a grant from the Stichting Nederlands Oogheelkundig Onderzoek (Netherlands Foundation for Eye Research).
Financial Disclosures: Dr Jager, Dr de Klein, and Dr Verdijk have received a grant from the Stichting Nederlands Oogheelkundig Onderzoek for BAP1 analysis and genetic testing. Dr Jager was among the recipients of a Horizon 2020 grant for the UM Cure project.
Author Contributions: Conceiving and designing the study (M.J.J., J.A.B.M., B.R.K., S.R.D.); Writing the manuscript or providing critical revisions (M.J.J., J.A.B.M., B.R.K., S.R.D.); Final approval of the manuscript (M.J.J., J.A.B.M., B.R.K., S.R.D.); Data collection and provision of materials (M.J.J., J.A.B.M., B.R.K., S.R.D.).
Other Acknowledgments: We thank Dr T. Murray for providing tumor material to Dr Ksander; Drs M. Wessels and R. Smesseim for retrieving material; Dr S. van Duinen, Leiden, for the histopathologic description of tumor and metastases of 92-1; Prof S. Coupland, Liverpool, for determining the GNAQ/GNA11 mutations in the original tumors of MEL270 and Mel290; Dr R. Verdijk for performing BAP1 staining; Dr A. de Klein for FISH testing; Dr J. Cao for testing BRAF and GNAQ/GNA11 mutations in the cell lines and in tumors; Dr M. Versluis and Dr P. A. van der Velden for performing the ddPCR; Prof Dr F. H. J. Claas for providing the HLA typing of the cell lines; and the personnel of the Laboratory of Pathology of the Bascom Palmer Eye Institute for all their help.
REFERENCES
- 1.Flaherty KT, Puzanow I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue nevi. Nature. 2009;457(7229):599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Eng J Med. 2010;363(23):2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalili JS, Yu X, Wang J, et al. Combination small molecule MEK and Pl3K inhibition enhances uveal melanoma cell death in a mutant GNAQ- and GNA11-dependent manner. Clin Cancer Res. 2012;18(16):4345–4355. doi: 10.1158/1078-0432.CCR-11-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ewens KG, Kanetsky PA, Richards-Yutz J, et al. Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastases in uveal melanoma. Invest Ophthalmol Vis Sci. 2014;55(6):5160–5167. doi: 10.1167/iovs.14-14550. [DOI] [PubMed] [Google Scholar]
- 6.Koopmans AE, Vaarwater J, Paridaens D, Naus NC, Kilic E, de Klein A. Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br J Cancer. 2013;109(2):493–496. doi: 10.1038/bjc.2013.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White VA, Chambers JD, Courtright PD, Chang WY, Horsman DE. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer. 1998;83(2):354–359. [PubMed] [Google Scholar]
- 8.Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jöckel KH, Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347(9010):1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 9.Sisley K, Rennie IG, Parsons MA, et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer. 1997;19(1):22–28. doi: 10.1002/(sici)1098-2264(199705)19:1<22::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Kilic E, van Gils W, Lodder E, et al. Clinical ad cytogenetic analyses in uveal melanoma. Invest Ophthalmol Vis Sci. 2006;47(9):3703–3707. doi: 10.1167/iovs.06-0101. [DOI] [PubMed] [Google Scholar]
- 11.Versluis M, de Lange MJ, Van Pelt SI, Ruivenkamp CAL, Kroes WGM, Cao J, et al. Digital PCR validates 8q dosage as a prognostic tool in uveal melanoma. PLoS One. 2015;10(3):e0116371. doi: 10.1371/journal.pone.0116371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aalto Y, Eriksson L, Seregard S, Larsson O, Knuutila S. Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6 and 8 in metastasizing primary uveal melanoma. Invest Ophthalmol Vis Sci. 2001;42(2):313–317. [PubMed] [Google Scholar]
- 13.Kilic E, Naus NC, van Gils W, et al. Concurrent loss of chromosome arm p and chromosome 3 predicts a decreased diseasefree survival in uveal melanoma patients. Invest Ophthalmol Vis Sci. 2005;46(7):2253–2257. doi: 10.1167/iovs.04-1460. [DOI] [PubMed] [Google Scholar]
- 14.Trolet J, Hupe P, Huon I, et al. Genomic profiling and identification of high-risk uveal melanoma by CGH analysis of primary tumors and liver metastases. Invest Ophthalmol Vis Sci. 2009;50(6):2572–2580. doi: 10.1167/iovs.08-2296. [DOI] [PubMed] [Google Scholar]
- 15.Cassoux N, Rodrigues MJ, Plancher C, et al. Genome-wide profiling is a clinically-relevant and affordable prognostic test in posterior uveal melanoma. Br J Ophthalmol. 2014;98(6):769–774. doi: 10.1136/bjophthalmol-2013-303867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caines R, Eleuteri A, Kalirai H, et al. Cluster analysis of multiplex ligation-dependent probe amplification data in choroidal melanoma. Mol Vis. 2015;21:1–11. [PMC free article] [PubMed] [Google Scholar]
- 17.De Lange MJ, Van Pelt SI, Versluis M, Jordanova ES, Kroes WGM, Ruivenkamp C, et al. Heterogeneity revealed by integrated genomic analysis uncovers a molecular switch in malignant uveal melanoma. Oncotarget. 2015;6(35):37824–37835. doi: 10.18632/oncotarget.5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh N, Singh AD, Hie W. Inferring an evolutionary tree of uveal melanoma from genomic copy number aberrations. Invest Ophthalmol Vis Sci. 2015;56(11):6801–6809. doi: 10.1167/iovs.15-16822. [DOI] [PubMed] [Google Scholar]
- 19.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Essen TH, van Pelt SI, Versluis M, et al. Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br J Ophthalmol. 2014;98(12):1738–1743. doi: 10.1136/bjophthalmol-2014-305047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koopmans AE, Verdijk RM, Brouwer RW, et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod Pathol. 2014;27(10):1321–1330. doi: 10.1038/modpathol.2014.43. [DOI] [PubMed] [Google Scholar]
- 22.Kalirai H, Dodson A, Faqir S, Damato BE, Coupland SE. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer. 2014;111(7):1–8. doi: 10.1038/bjc.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Waard-Siebinga I, Blom DJ, Griffioen M, et al. Establishment and characterization of a uveal melanoma cell line. Int J Cancer. 1995;62(2):155–161. doi: 10.1002/ijc.2910620208. [DOI] [PubMed] [Google Scholar]
- 24.Dogrusöz M, Kroes WGM, van Duinen SG, Creutzberg CL, Versluis M, Bleeker JC, et al. Radiation treatment affects chromosome testing in uveal melanoma. Invest Ophthalmol Vis Sci. 2015;56(10):5956–5964. doi: 10.1167/iovs.15-17092. [DOI] [PubMed] [Google Scholar]
- 25.Mitsiades N, Chew SA, He B, et al. Genotype-dependent sensitivity of uveal melanoma cell lines to inhibition of b-Raf, MEK, and Akt kinases: rationale for personalized therapy. Invest Ophthalmol Vis Sci. 2011;52(10):7248–7255. doi: 10.1167/iovs.11-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griewank KG, Yu X, Khalili J, et al. Genetic and molecular characterization of uveal melanoma cell lines. Pigment Cell Melanoma Res. 2012;25(2):182–187. doi: 10.1111/j.1755-148X.2012.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nareyeck G, Zeschnigk M, Bornfeld N, Anastassiou G. Novel cell lines derived by long-term culture of primary uveal melanomas. Ophthalmologica. 2009;223(3):196–201. doi: 10.1159/000201566. [DOI] [PubMed] [Google Scholar]
- 28.Amirouchene-Angelozzi N, Nemati F, Gentien D, et al. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol Oncol. 2014;8(8):1508–1520. doi: 10.1016/j.molonc.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folberg R, Kadkol SS, Frenkel S, et al. Authenticating cell lines in ophthalmic research laboratories. Invest Ophthalmol Vis Sci. 2008;49(11):4697–4701. doi: 10.1167/iovs.08-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kan-Mitchell J, Mitchell MS, Roa N, Ligett PE. Characterization of uveal melanoma cell lines that grow as xenografts in rabbit eyes. Invest Ophthalmol Vis Sci. 1989;30(5):829–834. [PubMed] [Google Scholar]
- 31.Calipel A, Lefevre G, Pouponnot C, Mouriaux F, Eychene A, Mascarelli F. Mutation of B-Raf in human choroidal melanoma cells mediates cell proliferation and transformation through the MEK/ERK pathway. J Biol Chem. 2003;278(43):42409–42418. doi: 10.1074/jbc.M308709200. [DOI] [PubMed] [Google Scholar]
- 32.The Editors of Molecular Vision. On authentication of cell lines. Mol Vis. 2013;19:1848–1851. [PMC free article] [PubMed] [Google Scholar]
- 33.American National Standards Institute. Authentication of human cell lines: Standardization of STR profiling. 2012. ANSI/ATCC:ASN-0002-2011. [Google Scholar]
- 34.Némati F, Sastre-Garau X, Laurent C, et al. Establishment and characterization of a panel of human uveal melanoma xenografts derived from primary and/or metastatic tumors. Clin Cancer Res. 2010;16(8):2352–2362. doi: 10.1158/1078-0432.CCR-09-3066. [DOI] [PubMed] [Google Scholar]
- 35.Verbik DJ, Murray TG, Tran JM, Ksander BR. Melanomas that develop within the eye inhibit lymphocyte proliferation. Int J Cancer. 1997;73(4):470–478. doi: 10.1002/(sici)1097-0215(19971114)73:4<470::aid-ijc3>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 36.Martin M, Masshofer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45(8):933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ksander BR, Rubsamen PE, Olsen KR, Cousins SW, Streilein JW. Studies of tumor-infiltrating lymphocytes from a human choroidal melanoma. Invest Ophthalmol Vis Sci. 1991;32(13):3198–3208. [PubMed] [Google Scholar]
- 38.Chen PW, Murray TG, Uno T, Salgallar ML, Reddy R, Ksander BR. Expression of MAGE genes in ocular melanoma during progression from primary to metastatic disease. Clin Exp Metastasis. 1997;15(5):509–518. doi: 10.1023/a:1018479011340. [DOI] [PubMed] [Google Scholar]
- 39.Blom DJR, Schurmans LRHM, De Waard-Siebinga I, De Wolff-Rouendaal, Keunen JEE, Jager MJ. HLA expression in a primary uveal melanoma, its cell line and four of its metastases. Br J Ophthalmol. 1997;81(11):989–993. doi: 10.1136/bjo.81.11.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luyten GPM, van der Spek CW, Brand I, et al. Expression of MAGE, gp100 and tyrosinase genes in uveal melanoma cell lines. Melanoma Res. 1998;8(1):11–16. doi: 10.1097/00008390-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 41.Kalirai H, Damato BE, Coupland SE. Uveal melanoma cell lines contain stem-like cells that self-renew, produce differential progeny, and survive chemotherapy. Invest Ophthalmol Vis Sci. 2011;52(11):8458–8466. doi: 10.1167/iovs.11-7379. [DOI] [PubMed] [Google Scholar]
- 42.Director-Myska AE, White JS, McLean IW. Molecular cytogenetic characterization of ten uveal melanoma cell lines. In: Jager MJ, Niederkorn JY, Ksander BR, editors. Uveal melanoma: A model for exploring fundamental cancer biology. London and New York: Taylor and Francis; 2004. pp. 89–114. [Google Scholar]
- 43.Van Dinten LC, Pul N, van Nieuwpoort AF, Out CJ, Jager MJ, van den Elsen PJ. Uveal and cutaneous melanoma: shared expression characteristics of melanoma-associated antigens. Invest Ophthalmol Vis Sci. 2005;46(1):24–30. doi: 10.1167/iovs.04-0961. [DOI] [PubMed] [Google Scholar]
- 44.Walker TM, van Ginkel PR, Gee RL, Ahmadi H, Subramanian L, Ksander BR, et al. Expression of angiogenic factors Cyr61 and tissue factor in uveal melanoma. Arch Ophthalmol. 2002;120(12):1719–1725. doi: 10.1001/archopht.120.12.1719. [DOI] [PubMed] [Google Scholar]
- 45.Van Gils W. Molecular prognostic markers in uveal melanoma: expression profiling and genomic studies [thesis] Erasmus University; Rotterdam: 2008. [Google Scholar]
- 46.Naus NC, van Drunen E, de Klein A, et al. Characterization of complex chromosomal abnormalities in uveal melanoma by fluorescence in situ hybridization, spectral karyotyping, and comparative genomic hybridization. Genes Chromosones Cancer. 2001;30(3):267–273. [PubMed] [Google Scholar]
- 47.Hurks HMH, Metzelaar-Blok JAW, Mulder A, Claas FHJ, Jager MJ. High frequency of allele-specific down-regulation of HLA Class I expression in uveal melanoma cell lines. Int J Cancer. 2000;85(5):697–702. doi: 10.1002/(sici)1097-0215(20000301)85:5<697::aid-ijc16>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 48.Fabian ID, Rosner M, Fabian I, et al. Low thyroid hormone levels improve survival in murine model for ocular melanoma. Oncotarget. 2015;6(13):11038–11046. doi: 10.18632/oncotarget.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maat W, Ly LV, Jordanova ES, de Wolff-Rouendaal D, Schalij-Delfos NE, Jager MJ. Monosomy of chromosome 3 and an inflammatory phenotype occur together in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49(92):505–510. doi: 10.1167/iovs.07-0786. [DOI] [PubMed] [Google Scholar]
- 50.Hou HY, Wang YS, Xu JF, Wang BR. Nicotine promotes contribution of bone marrow-derived cells to experimental choroidal neovascularization in mice. Exp Eye Res. 2008;86(6):983–990. doi: 10.1016/j.exer.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 51.Heegaard S, Spang-Thomsen M, Prause JU. Establishment and characterization of human uveal malignant melanoma xenografts in nude mice. Melanoma Res. 2003;13(3):247–251. doi: 10.1097/00008390-200306000-00004. [DOI] [PubMed] [Google Scholar]
- 52.Van Raamsdonk CD, Fitch KR, Fuchs H, de Angelis MH, Barsh GS. Effects of G-protein mutations on skin color. Nat Genet. 2004;36(9):961–968. doi: 10.1038/ng1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blom DJB, Luyten GPM, Mooy CM, Kerkvliet S, Zwinderman AH, Jager MJ. Human leucocyte antigen class I expression: marker of poor prognosis in uveal melanoma. Invest Ophthalmol Vis Sci. 1997;38(9):1865–1872. [PubMed] [Google Scholar]
- 54.Blanco PL, Marshall JC, Antecka E, et al. Characterization of ocular and metastatic uveal melanoma in an animal model. Invest Ophthalmol Vis Sci. 2005;46(12):4376–4382. doi: 10.1167/iovs.04-1103. [DOI] [PubMed] [Google Scholar]
- 55.Field MG, Decatur CL, Kurtenbach S, et al. PRAME as an independent biomarker for metastasis in low-risk uveal melanoma. Clin Cancer Res. 2016;22(5):1234–1242. doi: 10.1158/1078-0432.CCR-15-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He Y-G, Mayhew E, Mellon J, Niederkorn JY. Expression and possible function of IL-2 and IL-15 receptors on human uveal melanoma cells. Invest Ophthalmol Vis Sci. 2004;45:4240–4246. doi: 10.1167/iovs.04-0599. [DOI] [PubMed] [Google Scholar]
- 57.Ma D, Niederkorn JY. Transforming growth factor-beta downregulates major histocompatibility complex class I antigen expression and increases the susceptibility of uveal melanoma cells to natural killer cell-mediated cytolysis. Immunology. 1995;86(2):263–269. [PMC free article] [PubMed] [Google Scholar]
- 58.Ma D, Luyten GP, Luider TM, Niederkorn JY. Relationship between natural killer cell susceptibility and metastasis of human uveal melanoma cells in a murine model. Invest Ophthalmol Vis Sci. 1995;36(2):435–441. [PubMed] [Google Scholar]
- 59.Singh AD, De Potter P, Fijal B, Shields CL, Shields JA, Elston RC. Lifetime prevalence of uveal melanoma in Caucasian patients with oculo(dermal)melanocytosis. Ophthalmology. 1998;105(1):195–198. doi: 10.1016/s0161-6420(98)92205-9. [DOI] [PubMed] [Google Scholar]
- 60.Ambrosini G, Sawle AD, Musi E, Schwartz GK. BRD4-targeted therapy induces Myc-independent cytotoxicity in GNAQ/11-mutated uveal melanoma cells. Oncotarget. 2015;6(32):33397–33409. doi: 10.18632/oncotarget.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerne JZ, Hartig SM, Hamilton MP, et al. Protein kinase C inhibitors sensitize GNAQ mutant uveal melanoma cells to ionizing radiation. Invest Ophthalmol Vis Sci. 2014;55(4):2130–2139. doi: 10.1167/iovs.13-13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maat W, Haasnoot GW, Claas FH, Schalij-Delfos NE, Schreuder GM, Jager MJ. HLA Class I and II genotype in uveal melanoma: relation to occurrence and prognosis. Invest Ophthalmol Vis Sci. 2006;47(1):3–6. doi: 10.1167/iovs.05-1122. [DOI] [PubMed] [Google Scholar]
- 63.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418(6900):823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 64.Maat W, Jordanova ES, van Zelderen-Bhola SL, et al. The heterogeneous distribution of monosomy 3 in uveal melanoma: implications for prognostication based on fine-needle aspiration biopsies. Arch Pathol Lab Med. 2007;131(1):91–96. doi: 10.5858/2007-131-91-THDOMI. [DOI] [PubMed] [Google Scholar]
- 65.Laurent C, Gentian D, Piperno-Neumann S, Némati F, Nicolas A, Tesson B, et al. Patient-derived xenografts recapitulate molecular features of human uveal melanomas. Mol Oncol. 2013;7(3):625–636. doi: 10.1016/j.molonc.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]