Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A-rearrangements, and monosomy 7 are associated with poor outcome; RBM15/MKL1 and others fare better.
Screening for NUP98/KDM5A, RBM15/MKL1, CBFA2T3/GLIS2, and KMT2A rearrangements combined with conventional karyotyping is advisable.
Abstract
Genetic abnormalities and early treatment response are the main prognostic factors in acute myeloid leukemia (AML). Acute megakaryoblastic leukemia (AMKL) is a rare subtype of AML. Deep sequencing has identified CBFA2T3/GLIS2 and NUP98/KDM5A as recurrent aberrations, occurring in similar frequencies as RBM15/MKL1 and KMT2A-rearrangements. We studied whether these cytogenetic aberrations can be used for risk group stratification. To assess frequencies and outcome parameters of recurrent cytogenetic aberrations in AMKL, samples and clinical data of patients treated by the Associazione Italiana Ematologia Oncologia Pediatrica, Berlin-Frankfurt-Munster Study Group, Children’s Oncology Group, Dutch Childhood Oncology Group, and the Saint Louis Hôpital were collected, enabling us to screen 153 newly diagnosed pediatric AMKL cases for the aforementioned aberrations and to study their clinical characteristics and outcome. CBFA2T3/GLIS2 was identified in 16% of the cases; RBM15/MKL1, in 12%; NUP98/KDM5A and KMT2A rearrangements, in 9% each; and monosomy 7, in 6%. These aberrations were mutually exclusive. RBM15/MKL1-rearranged patients were significantly younger. No significant differences in sex and white blood cell count were found. NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A–rearranged lesions and monosomy 7 (NCK-7) independently predicted a poor outcome, compared with RBM15/MKL1-rearranged patients and those with AMKL not carrying these molecular lesions. NCK-7-patients (n = 61) showed a 4-year probability of overall survival of 35 ± 6% vs 70 ± 5% in the RBM15/MKL1-other groups (n = 92, P < .0001) and 4-year probability of event-free survival of 33 ± 6% vs 62 ± 5% (P = .0013), the 4-year cumulative incidence of relapse being 42 ± 7% and 19 ± 4% (P = .003), respectively. We conclude that these genetic aberrations may be used for risk group stratification of pediatric AMKL and for treatment tailoring.
Introduction
Pediatric acute myeloid leukemia (AML) is a heterogeneous disease characterized by recurrent genetic aberrations. The most important factors predicting clinical outcome are presence of (molecular) genetic aberrations and early response to treatment.1,2 Acute megakaryoblastic leukemia (AMKL) is a rare subtype of AML and is diagnosed mostly in myeloid leukemia of Down syndrome (ML-DS) patients, where it is associated with mutations in the transcription factor GATA1.3 In pediatric non-Down syndrome AML, AMKL accounts for ∼10% of the cases and has been reported to be associated with poor outcome.4-7 Most study groups consider pediatric AMKL as high-risk AML, in contrast to the outcome of ML-DS, and some consider it to have the indication for allogeneic hematopoietic stem cell transplantation (HSCT) in first complete remission (CR1).4,8-10 However, risk group stratification and treatment protocols are not yet optimized for this subtype of pediatric AML.11
Translocation t(1;22)(p13;q13), resulting in a chimeric fusion of RBM15 and MKL1, formerly known as OTT/MAL, until recently was the only recurrent aberration described in pediatric AMKL, occurring in ∼10% of the patients.12-14 Conflicting results on the prognostic relevance of RBM15/MKL1 have been reported, as some series suggested the outcome to be poor and others favorable.5,6,13,15 Recently, the cytogenetically cryptic translocation t(11;12)(p15;p13), resulting in the chimeric fusion gene NUP98/KDM5A, was identified as a recurrent aberration found in ∼10% of AMKL cases.5,7,16 Moreover, another cytogenetically cryptic event, inv(16)(p13q24), resulting in a fusion between CBFA2T3 and GLIS2 and accounting for ∼15% of the cases, was identified as a recurrent aberration in this specific subtype of AML.5,7,16,17 Both NUP98/KDM5A and CBFA2T3/GLIS2 fusion transcripts were reported to confer poor outcome, but numbers of patients analyzed to support this conclusion were small. Moreover, CBFA2T3/GLIS2 was also reported to behave as an aggressive leukemia in mouse models.7,16 Another recurrent abnormality consists of KMT2A rearrangements, which are seen in ∼10% of the pediatric AMKL cases.5 The clinical outcome of KMT2A-rearranged leukemia is dependent on the fusion-partner gene.18,19 NUP98/KDM5A, CBFA2T3/GLIS2, and RBM15/MKL1 are rarely or never seen in other pediatric AML subtypes, with CBFA2T3/GLIS2 being the least specific for AMKL.5,17,20 In addition, Inaba et al described AMKL risk groups based on cytogenetic data obtained with conventional karyotyping, proposing 3 groups: good risk including patients carrying 7p abnormalities; poor risk including cases with monosomy 7, or 9p abnormalities, including KMT2A/MLLT3, −13/13q-, and −15, and intermediate risk, which encompassed all other AKML patients.11
In this study, we present the clinical, cell-biological, and genetic characteristics of 153 pediatric AMKL patients from the Associazione Italiana Ematologia Oncologia Pediatrica (AIEOP; Italian Association of Pediatric Hematology and Oncology, Italy), the Berlin-Frankfurt-Munster-Study Group (BFM-SG; Essen, Germany), the Dutch Childhood Oncology Group (DCOG; The Hague, The Netherlands), the Saint Louis Hôspital (SLH; Paris, France), and the Children’s Oncology Group (COG; Philadelphia, PA), which were fully characterized for the aforementioned genetic aberrations, and karyotypes were analyzed, We also investigated the frequencies and prognostic relevance of these recurrent aberrations.
Material and methods
Patient inclusion
We collected data on clinical characteristics and outcome of newly diagnosed pediatric (younger than 18 years at disease onset) patients with non-Down syndrome AMKL. These patients were diagnosed between 1990 and 2014 and had to have available RNA or complementary DNA for molecular studies and a blast percentage >20%. They were from 3 consortia: (1) AIEOP; (2) BFM-SG, DCOG, and SLH; and (3) COG. Approval for this study had been obtained from the institutional review board of the participating centers. Morphological classification and karyotyping, including the presence of KMT2A rearrangements, monosomy 7, or a hyperdiploid karyotype, were centrally reviewed by each study group.
Screening for recurrent translocations
Screening for molecular aberrations was performed in the Erasmus Medical Center–Sophia Children’s Hospital for samples provided by the AML BFM-SG, COG, DCOG, and the SLH, whereas samples provided by the AIEOP group were screened at the University of Padova and the University of Bologna. All patients were screened for NUP98/KDM5A, CBFA2T3/GLIS2, and RBM15/MKL1 with reverse transcription polymerase chain reaction (RT-PCR), using specific primers for each fusion gene, as previously described (Primers; supplemental Table 1, available on the Blood Web site).5,17 In case of a positive sample, purified PCR product was directly sequenced on a PRISM 3100 genetic analyzer (Applied Biosystems) and analyzed using CLCWorkbench (Version 3.5.1; CLC Bio) to confirm the translocation. A higher incidence of AMKL is seen in Down syndrome patients, and these children are usually characterized by a GATA1 mutation.21-23 All 153 samples, including the positive KMT2A-rearranged cases (diagnosis being based on either KMT2A split signal fluorescence in situ hybridization or cytogenetic analysis as performed by each study group), were screened for KMT2A rearrangements for selected partner genes using specific primers for KMT2A/MLLT3, KMT2A/MLLT10, KMT2A/MLLT4(MLL/AF6), KMT2A/MLLT1(MLL/ENL), KMT2A/ELL, and KMT2A/MLLT6(MLL/AF17), and the purified PCR product was directly sequenced (Primers; supplemental Table 1).18,19
Statistical analyses
Statistical analyses were performed with SPSS Statistics 20 (IBM, Armonk, NY). To assess the outcome, the following parameters were used: (1) probability of achieving CR1, (2) probability of event-free survival (pEFS), (3) probability of overall survival (pOS), and (4) cumulative incidence of relapse (pCIR). CR was defined as <5% blasts in the bone marrow, with regeneration of trilineage hematopoiesis, and absence of leukemic cells elsewhere. pEFS was defined as the time between diagnosis and first event, including relapse, death because of any cause, and second malignancy, whichever occurred first. For calculation of EFS, lack of achieving CR was considered an event occurring on day 0. pOS was defined as the time between diagnosis and death. Both pEFS and pOS were estimated by the Kaplan-Meier method, and groups were compared with the log-rank test. pCIR was defined as time between diagnosis and relapse, with nonresponders being attributed an event on day 0, and was analyzed by the Kalbfleisch and Prentice method taking into account death and second malignancy as competing event; groups were compared with the Gray’s test. A Cox regression analysis was performed for EFS, OS, and for the probability of relapse-free survival (RFS) considering the following covariables: cytogenetic subgroup, age, sex, white blood cell (WBC) count at diagnosis, and HSCT (as time-dependent variable), as well as the previously described poor prognostic factors for pediatric AMKL “normal karyotype” and monosomy 7.11,24,25 Furthermore, we included a hyperdiploid karyotype and 7p-abnormalities in the Cox regression analysis, based on the association with pediatric AMKL.11,26,27 Statistical analysis was restricted to groups with >5 cases. Statistical significance was considered if P values were <.05.
Results
Clinical characteristics
The AIEOP group included 47 patients; the BFM-SG, DCOG, and SLH consortium included 45 patients; and COG provided 61 patients. All 153 patients were eligible for screening of the 4 molecular aberrations, and 119 patients for cytogenetics. Median age was 1.6 years (range 0.1-17.1), median WBC count at diagnosis was 13.7 × 109/L (range 1.1 × 109/L to 378.5 × 109/L), and 46% of the included patients were males (Table 1). Patients were treated with different treatment protocols according to their respective study group. All protocols consisted of intensive chemotherapy using an anthracycline and cytarabine backbone for both induction and consolidation; in addition, 41% of patients received HSCT in CR1. HSCT was performed in 66% of the patients reported by the AIEOP group, in 44% of the COG cases, and in 11% of the cases provided by DCOG, BFM, and SLH. Patient characteristics are detailed in Table 1.
Table 1.
Presenting clinical characteristics of pediatric AMKL patients (n=153)
| Characteristics | CBFA2T3/GLIS2 (n = 24) | NUP98/KDM5A (n = 14) | KMT2A-r (n = 14) | RBM15/MKL1 (n = 18) | Monosomy 7 (n = 9) | Other (n = 74) | All (n = 153) |
|---|---|---|---|---|---|---|---|
| Median age, y (range) | 1.5 (0.5-4.0) | 1.9 (0.8-8.5) | 1.9 (0.7-12.0) | 0.7* (0.1-2.7) | 1.5 (0.5-17.1) | 1.6 (0.1-15.1) | 1.6 (0.1-17.1) |
| Median WBC count (range) | 17.3 × 109/L (7.5-300.1) | 14.0 × 109/L (5.8-188.0) | 7.4 × 109/L (1.1-31.0) | 13.8 × 109/L (5.6-32.7) | 14.5 × 109/L (8.7-90.2) | 14.4 × 109/L (1.1-378.5) | 13.7 × 109/L (1.1-378.5) |
| Male, % | 25 | 43 | 71 | 33 | 22 | 51 | 46 |
| Origin | |||||||
| AIEOP, % | 42 | 21 | 36 | 17 | 22 | 32 | 31 |
| DCOG/BFM/SLH, % | 17 | 50 | 36 | 28 | 33 | 29 | 29 |
| COG, % | 42 | 29 | 29 | 56 | 45 | 39 | 40 |
| HSCT | 11 (46%) | 4 (29%) | 9 (64%) | 9 (50%) | 5 (56%) | 30 (36%) | 63 (41%) |
| 4-y pEFS, % | 33 ± 10 | 36 ± 13 | 36 ± 13 | 59 ± 12 | 33 ± 16 | 62 ± 6 | 51 ± 4 |
| 4-y pOS, % | 38 ± 10 | 36 ± 13 | 36 ± 13 | 70 ± 11 | 33 ± 16 | 70 ± 5 | 56 ± 4 |
| 4-y pCIR, % | 42 ± 10 | 36 ± 14 | 50 ± 14 | 6 ± 6 | 33 ± 18 | 22 ± 5 | 29 ± 4 |
KMT2A-r, KMT2A rearrangement.
Significantly lower age at diagnosis (P = .038).
Twenty-four (16%) samples were positive for CBFA2T3/GLIS2, 18 (12%) samples harbored the RBM15/MKL1 fusion gene, NUP98/KDM5A was present in 14 (9%) of cases, and 14 samples were positive for a KMT2A rearrangement, all diagnosed with fluorescence in situ hybridization. Three KMT2A-positive cases had no karyotype available, and in the other 11, the KMT2A rearrangement was also detected through karyotype analysis. Different fusion partners were identified; 9/14 were positive for KMT2A/MLLT3, 3 were identified with fusion KMT2A/MLLT10, and 1 case each harbored either KMT2A/MLLT1 or KMT2A/MLLT6 (supplemental Table 2).
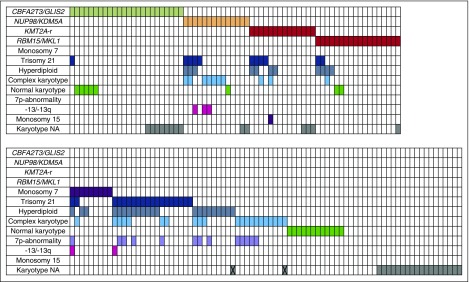
Detailed data on karyotype were lacking in 34/153 cases. Twenty cases were cytogenetically normal. Cytogenetics showed 30 cases with an acquired trisomy 21, another 9 cases were identified to carry monosomy 7, 33 cases were hyperdiploid with 49 to 84 chromosomes, and 31 showed a complex karyotype (defined as 3 or more aberrations excluding trisomy’s). A 7p-abnormality was identified in 13 cases. Some of these cases belonged to >1 of the cytogenetic or molecular subgroups (see also Figure 1 and supplemental Table 3 for details).
Figure 1.
Overview of cytogenetic aberrations and aberrations found in the karyotypes of pediatric AMKL (n = 153). Each column represents a single case. A colored cells means positive for the aberration corresponding with that particular row. X indicates that the karyotype is not available, the only information given is hyperdiploid, or complex karyotype. -13/13q, monosomy 13 or del(13q); NA, not available.
Age at diagnosis was significantly lower in the RBM15/MKL1-positive group, compared with the other pediatric AMKL cases (0.7 years, range 0.1-2.7, P = .035). Seventy-four patients were characterized as “other,” as they were negative for any of the above-mentioned translocations and monosomy 7. Other karyotypes identified were t(1:22)(p13;q13), but this case was negative for RBM15/MKL1 using RT-PCR, and t(16;21)(q24;q22.1), which was positive for RUNX1/CBFA2T3 (supplemental Table 3). The “other” subgroup had a median age of 1.6 years (range 0.1-15.1), and a median WBC count of 14.4 × 109/L (range 1.1 × 109/L to 378.5 × 109/L; Table 1).
Survival analyses
The median follow-up of surviving patients was 67 months (range 2.7-259.0). The 4-year pOS of the entire pediatric AMKL cohort was 56 ± 4%, the 4-year pEFS was 51 ± 4%, and the 4-year pCIR was 29 ± 4%. To test whether our cohort (n = 146, excluding the 7 cases present in both this study and the study published by Inaba et al11) had a selection bias, our patients were compared for outcome with all AMKL cases in the BFM cohort recently published by Inaba et al (n = 97), and outcome was comparable for pOS (56 ± 4% vs 60 ± 5%, P = .51), pEFS (51 ± 4% vs 47 ± 5%, P = .62) and pCIR (29 ± 4% vs 32 ± 5%, P = .62).
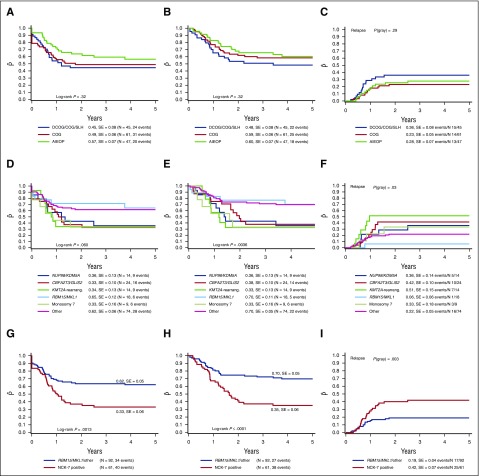
Outcome did not differ significantly between the different collaborative groups. The 4-year pEFS of all non-Down syndrome AMKL cases was 57 ± 7%, 45 ± 8%, and 49 ± 6% for the AIEOP, BFM/DCOG/SLH, and COG cohorts, respectively (P = .32; Figure 2A). The 4-year pOS was 60 ± 7%, 48 ± 8%, and 59 ± 6% (P = .32; Figure 2B), and 4-year pCIR was 28 ± 7%, 36 ± 8%, and 23 ± 5% (P = .29; Figure 2C).
Figure 2.
Survival curves of pediatric AMKL patients. The 4-year pEFS (A), 4-year pOS (B), and 4-year pCIR (C) comparing the outcome of patients of the different enrolled study groups. There is no significant difference in outcome among the study groups. The 4-year pEFS (D), 4-year pOS (E), and 4-year pCIR (F) comparing the described cytogenetic subgroups as identified in pediatric non-Down syndrome AMKL. RBM15/MKL1-positive cases and other pediatric AMKL have a favorable outcome compared with NUP98/KDM5A, CBFA2T3/GLIS, KMT2A rearrangements and monosomy 7 cases. The 4-year pEFS (G), 4-year pOS (H), and 4-year pCIR (I) of the NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A rearrangements and monosomy 7 cases compared with the RBM15/MKL1 and other pediatric AMKL cases. Harboring NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A rearrangements or monosomy 7 confers a poor outcome.
When we analyzed the outcome of patients according to the specific recurrent molecular lesions, we found that RBM15/MKL1 and pediatric AMKL without the aforementioned translocations showed a 4-year pOS of 70 ± 11% and 66 ± 5%, respectively, vs a poorer outcome in NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A–rearranged patients (4-year pOS 36 ± 13%, 38 ± 10%, and 33 ± 13%, respectively; P = .0036) with a similar trend for pEFS (P = .060; Figure 2D-E). The difference in outcome was mainly because of relapsed and refractory disease, as 22 and 9 of the 52 NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A–rearranged patients either relapsed or did not achieve CR, respectively (supplemental Table 2). The pCIR of RBM15/MKL1 translocated cases was 6 ± 6%, compared with 36 ± 14%, 42 ± 10%, and 51 ± 15% for the cases positive for NUP98/KDM5A, CBFA2T3/GLIS2, or KMT2A rearrangements, respectively (P = .03; Figure 2F). Next, we analyzed the poor prognostic cytogenetic aberrations as proposed by Inaba et al.11 The monosomy 7 cases also showed a poor outcome, confirming the previously found results. The 4-year pOS, pEFS, and pCIR were 33±%, 33 ± 16%, and 33 ± 18%, respectively (Figure 2D-F).
Altogether we concluded that NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A rearrangements and monosomy 7 (collectively referred to as NCK-7 positive) should be considered as poor risk, and RBM15/MKL1 and others as standard risk. Comparing the NCK-7-positive cases vs the RBM15/MKL1 and other cases, the pCIR differed significantly (4-year pCIR of NCK-7 cases being 42 ± 7% vs 19 ± 4% for RBM15/MKL1 and other cases, P = .003). This lower pCIR translated into a worse EFS (33 ± 6% for NCK-7 cases vs 62 ± 5% for RBM15/MKL1 and other cases, P = .0013) and a lower probability of OS (35 ± 6% vs 70 ± 5%, P < .0001; Figure 2G-I).
Apart from relapsed and refractory cases, other events leading to mortality among the patients with one of the identified fusions consisted of infections (n = 6; 3 CBFA2T3/GLIS2 positive cases and 3 other cases), toxicity (n = 3; a NUP98/KDM5A-positive, a monosomy 7, and an RBM15/MKL1-positive case), graft-versus-host disease after HSCT (n = 3; 1 KMT2A rearranged, an RBM15/MKL1-positive case, and 1 classified as other), hemorrhage (n = 3; an RBM15/MKL1 translocated case, a monosomy 7 case, and 1 “other”), and acute respiratory distress syndrome (n = 1) in a KMT2A-rearranged case, supplemental Table 2).
Multivariate analysis
Harboring NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A rearrangement or monosomy 7 was an independent risk factor for EFS, RFS, and OS (hazard ratio [HR] 1.69; 95% confidence interval [CI], 1.04-2.76, P = .035; HR 2.26, 95% CI, 1.15-4.48, P = .019; and HR 2.33, 95% CI, 1.36-3.99, P = .002, respectively; Table 2), when other risk factors such as sex, age at diagnosis, WBC count, HSCT, normal karyotype, 7p-aberrations, and hyperdiploid karyotype were included in the model.
Table 2.
HRs of cytogenetic and clinical markers for event-free, relapse-free, and overall survival in multivariate analysis
| HR | 95% CI | P (χ2) | |
|---|---|---|---|
| NCK-7 positive (n = 61) | |||
| EFS | 1.69 | 1.04-2.76 | .035 |
| RFS | 2.26 | 1.15-4.48 | .019 |
| OS | 2.33 | 1.36-3.99 | .002 |
| Hyperdiploidy (n = 33) | |||
| EFS | 0.43 | 0.21-0.90 | .026 |
| RFS | 0.43 | 0.16-1.14 | .090 |
| OS | 0.44 | 0.20-1.01 | .053 |
| 7p-abnormality (n = 13) | |||
| EFS | 1.06 | 0.41-2.73 | .909 |
| RFS | 1.35 | 0.39-4.70 | .639 |
| OS | 1.10 | 0.38-3.17 | .860 |
| Normal karyotype (n = 20) | |||
| EFS | 0.85 | 0.43-1.70 | .651 |
| RFS | 0.98 | 0.40-2.43 | .966 |
| OS | 0.97 | 0.47-2.01 | .928 |
| Sex (male, n = 70) | |||
| EFS | 1.32 | 0.86-2.23 | .186 |
| RFS | 1.14 | 0.60-2.16 | .688 |
| OS | 1.41 | 0.84-2.38 | .193 |
| Age <2 y (n = 97) | |||
| EFS | 0.71 | 0.43-1.15 | .159 |
| RFS | 0.57 | 0.30-1.10 | .093 |
| OS | 0.82 | 0.49-1.38 | .453 |
| WBC count >20.0 × 109/L (n = 38) | |||
| EFS | 1.32 | 0.80-2.20 | .276 |
| RFS | 1.67 | 0.86-3.25 | .129 |
| OS | 1.62 | 0.95-2.76 | .079 |
| HSCT (n = 63) | |||
| EFS | 1.14 | 0.61-2.11 | .680 |
| RFS | 0.95 | 0.47-1.92 | .882 |
| OS | 1.18 | 0.67-2.08 | .575 |
Harboring a hyperdiploid karyotype or normal karyotype did have an overlap among the different cytogenetic subgroups (Figure 1). Presence of a hyperdiploid karyotype, including the cases harboring another cytogenetic or molecular aberration, was an independent good prognostic factor for EFS (HR 0.43, 95% CI, 0.21-0.90, P = .026), but not for RFS or OS (P = .09 and P = .053, respectively; Table 2). We could not confirm favorable outcome for cases harboring 7p-abnormalities (pOS 69 ± 13% vs 55 ± 4%, respectively, P = .45), neither did harboring a normal karyotype influence outcome (pOS 53 ± 11% vs 57 ± 4% in all other cases, P = .75; Table 2).
In the NCK-7 groups, 47% of the patients received HSCT, compared with 37% in the RBM15/MKL1 and other cases. HSCT did not influence the RFS (HR 0.95, 95% CI, 0.47-1.92, P = .0.882; Table 2).
Discussion
We report the results of an international collaborative study on the prognostic value of the recently identified, recurrent (cyto-) genetic aberrations found in pediatric AMKL. Pediatric AMKL is a rare entity, which comprises 5% to 10% of the pediatric AML cases and is considered to be characterized by poor outcome, in contrast to the outcome found in ML-DS cases. The high incidence of myelofibrosis may complicate cytogenetic diagnosis based on conventional karyotyping, and robust prognostic markers are still to be identified.1,4-6,8,11 In our cohort, the translocations CBFA2T3/GLIS2, NUP98/KDM5A, RBM15/MKL1 and rearrangements of KMT2A accounted for 46% of the pediatric AMKL cases, and harboring translocation CBFA2T3/GLIS2, NUP98/KDM5A, KMT2A rearrangement or monosomy 7 appeared to be an independent prognostic factor for poor outcome. This poor outcome was mainly because of a high incidence of nonresponse and relapse in these patients, as documented in 42% of these cases in our cohort. Our data confirm the poor prognosis for overall survival of monosomy 7 cases, which was previously described in pediatric AML and pediatric AMKL.11,24,25,28
Previous studies showed the association between a hyperdiploid karyotype and AMKL.26,27 In adult and pediatric AML hyperdiploidy was reported to result in intermediate or poor outcome, and other studies showed that hyperdiploid cases should be assessed for the presence of specific abnormalities to predict outcome.26,29,30 Based on our results a hyperdiploid karyotype should not be considered as risk factor in pediatric AMKL.
The outcome of RBM15/MKL1 positive cases is not uniform in different studies because of the nature of the events. The studies of Inaba et al and Schweitzer et al describe an intermediate overall survival and a poor EFS. 6,11 However, Inaba et al reported that most events were early deaths, and Schweitzer et al reported a high nonremitter rate.6,11 In our study, only 1/18 patients harboring RBM15/MKL1 suffered from early death, 2 cases died because of treatment-related mortality, and 2 cases died because of leukemia progression. This better outcome may be the result of differences in supportive care, as hypothesized by Inaba et al.11
Pediatric AMKL arises in very young patients, the median age at diagnosis being below 2 years. Our study shows that neither age, sex, nor WBC count at diagnosis is an independent prognostic factor in pediatric AMKL. In addition, there were no differences in presenting characteristics among the various genetic groups, with the exception of RBM15/MKL1-positive cases, which had a significant younger age at diagnosis.
An important difference between the treatment protocols of the various collaborative groups in this study was the inclusion of HSCT in CR1 in case of an available donor. However, in multivariate analysis, HSCT did not influence the RFS. The added value of HSCT for pediatric AML in general is under discussion. There is clear evidence that allogeneic HSCT has a greater antileukemic potential than chemotherapy as postremissional treatment, but this favorable effect may be blunted by a higher risk of treatment-related mortality.9,10,31 Nevertheless, HSCT in CR1 is recommended in many AML treatment protocols for high-risk cases as defined by presence of genetic aberrations or early response to treatment (ie, high levels of minimal residual disease after 1 or 2 courses of induction chemotherapy). Based on the design of the current study, a benefit of HSCT could not be demonstrated in this retrospective analysis of pediatric AMKL.
AMKL is seen in >70% of ML-DS patients.6,11,32 ML-DS AMKL cases are characterized by mutations in the hematopoietic transcription factor GATA1, and these patients have a good prognosis when treated with reduced-intensity chemotherapy because of the unique sensitivity of leukemia cells to chemotherapeutic agents.21,22,32-36 Our group previously showed that RBM15/MKL1, CBFA2T3/GLIS2, NUP98/KDM5A, and KMT2A rearrangements were not present in ML-DS.5 GATA1 mutations have been described in non-Down syndrome AMKL, often seen with an acquired trisomy 21, and it was suggested that these cases fare well in terms of prognosis, although numbers were small.6,23,37-39 Cases of pediatric AMKL that we could not classify based on the 4 studied translocations may harbor a GATA1 mutation, because we were lacking DNA material of a large number of patients in this cohort. Next-generation sequencing techniques may be instrumental to unravel other recurrent aberrations in the near future in this relatively large group of AMKL cases, negative for any of the aforementioned aberrations.
Recently, Inaba et al described AMKL risk groups based on cytogenetic data obtained with conventional karyotyping, proposing 3 groups: good risk including patients carrying 7p abnormalities; poor risk including cases with monosomy 7, or 9p abnormalities, including KMT2A/MLLT3, −13/13q-, and −15; and intermediate risk, which encompasses all other AKML patients.11 This risk classification may be difficult to use, because AMKL is frequently associated with myelofibrosis, which contributes to difficulties in obtaining sufficient material for cytogenetic analysis. In our study, 34/153 (22%) patients lacked data on karyotype and could therefore not be included in any of the risk groups as proposed by Inaba et al. The percentage of missing karyotypes in our cohort is comparable to the findings of Inaba et al, although this is higher than that reported in pediatric AML including all morphology subtypes (3% to 13%).6,25,38 Moreover, conventional karyotyping will not reveal cryptic translocations, such as CBFA2T3/GLIS2 or NUP98/KDM5A. Through screening by RT-PCR, we were able to classify 38 cases with these cryptic events, of which 10 did not have an available karyotype, and the remaining cases did not show the occurrence of these events. Therefore, screening for the most frequent recurrent aberrations will provide additional information with respect to a risk stratification based on conventional karyotyping, revealing additional groups with poor outcome.
With international collaborations novel prognostic subgroups may be identified, which may lead to improved risk group stratification. Although the overall survival is poor for pediatric AMKL in general, with a 4-year pOS of 56% in this study, NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A rearrangements and monosomy 7 cases fare worse. We suggest that NUP98/KDM5A, CBFA2T3/GLIS2, KMT2A rearrangements and monosomy 7 cases in pediatric AMKL identify a high-risk patient subgroup, whereas children belonging to either RBM15/MKL1 or other pediatric AMKL should be considered as a standard risk subgroup.
Altogether, our results indicate that pediatric AMKL is a heterogeneous disease, and that presence of NUP98/KDM5A, CBFA2T3/GLIS2, or KMT2A rearrangements confers a poor clinical outcome, as well as cases with monosomy 7. Screening for the aforementioned translocations combined with conventional karyotyping in pediatric AMKL is advisable in future studies for improving risk group stratification and tailoring treatment intensity according to the biological characteristics of the leukemic clone.
Acknowledgments
This work was partly supported by grants from Stichting Kinderoncologisch Centrum Rotterdam (J.D.E.d.R.), Associazione Italiana Ricerca sul Cancro (special grant “5xmille”-9962) (F.L.), Ministero della Salute (RF-2010-2316606, Ricerca Corrente) (F.L.), and Ministero dell’Istruzione, Università e Ricerca (Grant Progetto di Rilevante Interesse Nazionale, PRIN 2010) (F.L.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.D.E.d.R., R.M., M.M.v.d.H.-E., R.P., C.M.Z., and F.L. designed the study; M.M.v.d.H.-E., J.-M.C., J.T., M.R., D.R., E.S., T.A.A., M.P., S.M., C.M.Z., and F.L. provided samples and clinical data; J.D.E.d.R. and R.M. performed the research; M.Z. and T.A.A. performed statistical analyses; J.D.E.d.R., R.M., M.M.v.d.H.-E., C.M.Z., and F.L. analyzed the data; J.D.E.d.R., C.M.Z., and F.L. wrote the manuscript; and all authors critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: C. Michel Zwaan, Department of Pediatric Oncology/Hematology, Erasmus MC–Sophia Children’s Hospital, P.O. Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: c.m.zwaan@erasmusmc.nl.
References
- 1.Creutzig U, van den Heuvel-Eibrink MM, Gibson B, et al. AML Committee of the International BFM Study Group. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. 2012;120(16):3187–3205. doi: 10.1182/blood-2012-03-362608. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29(5):551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xavier AC, Ge Y, Taub JW. Down syndrome and malignancies: a unique clinical relationship: a paper from the 2008 William Beaumont Hospital symposium on molecular pathology. J Mol Diagn. 2009;11(5):371–380. doi: 10.2353/jmoldx.2009.080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athale UH, Razzouk BI, Raimondi SC, et al. Biology and outcome of childhood acute megakaryoblastic leukemia: a single institution’s experience. Blood. 2001;97(12):3727–3732. doi: 10.1182/blood.v97.12.3727. [DOI] [PubMed] [Google Scholar]
- 5.de Rooij JD, Hollink IH, Arentsen-Peters ST, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia. 2013;27(12):2280–2288. doi: 10.1038/leu.2013.87. [DOI] [PubMed] [Google Scholar]
- 6.Schweitzer J, Zimmermann M, Rasche M, et al. Improved outcome of pediatric patients with acute megakaryoblastic leukemia in the AML-BFM 04 trial. Ann Hematol. 2015;94(8):1327–1336. doi: 10.1007/s00277-015-2383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiollier C, Lopez CK, Gerby B, et al. Characterization of novel genomic alterations and therapeutic approaches using acute megakaryoblastic leukemia xenograft models. J Exp Med. 2012;209(11):2017–2031. doi: 10.1084/jem.20121343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hama A, Yagasaki H, Takahashi Y, et al. Acute megakaryoblastic leukaemia (AMKL) in children: a comparison of AMKL with and without Down syndrome. Br J Haematol. 2008;140(5):552–561. doi: 10.1111/j.1365-2141.2007.06971.x. [DOI] [PubMed] [Google Scholar]
- 9.Hasle H. A critical review of which children with acute myeloid leukaemia need stem cell procedures. Br J Haematol. 2014;166(1):23–33. doi: 10.1111/bjh.12900. [DOI] [PubMed] [Google Scholar]
- 10.Passweg JR, Baldomero H, Peters C, et al. European Society for Blood and Marrow Transplantation EBMT. Hematopoietic SCT in Europe: data and trends in 2012 with special consideration of pediatric transplantation. Bone Marrow Transplant. 2014;49(6):744–750. doi: 10.1038/bmt.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inaba H, Zhou Y, Abla O, et al. Heterogeneous cytogenetic subgroups and outcomes in childhood acute megakaryoblastic leukemia: a retrospective international study. Blood. 2015;126(13):1575–1584. doi: 10.1182/blood-2015-02-629204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baruchel A, Daniel MT, Schaison G, Berger R. Nonrandom t(1;22)(p12-p13;q13) in acute megakaryocytic malignant proliferation. Cancer Genet Cytogenet. 1991;54(2):239–243. doi: 10.1016/0165-4608(91)90213-e. [DOI] [PubMed] [Google Scholar]
- 13.Carroll A, Civin C, Schneider N, et al. The t(1;22) (p13;q13) is nonrandom and restricted to infants with acute megakaryoblastic leukemia: a Pediatric Oncology Group Study. Blood. 1991;78(3):748–752. [PubMed] [Google Scholar]
- 14.Mercher T, Busson-Le Coniat M, Nguyen Khac F, et al. Recurrence of OTT-MAL fusion in t(1;22) of infant AML-M7. Genes Chromosomes Cancer. 2002;33(1):22–28. doi: 10.1002/gcc.1208. [DOI] [PubMed] [Google Scholar]
- 15.O’Brien MM, Cao X, Pounds S, et al. Prognostic features in acute megakaryoblastic leukemia in children without Down syndrome: a report from the AML02 multicenter trial and the Children’s Oncology Group Study POG 9421. Leukemia. 2013;27(3):731–734. doi: 10.1038/leu.2012.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber TA, Larson Gedman A, Zhang J, et al. An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell. 2012;22(5):683–697. doi: 10.1016/j.ccr.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masetti R, Pigazzi M, Togni M, et al. CBFA2T3-GLIS2 fusion transcript is a novel common feature in pediatric, cytogenetically normal AML, not restricted to FAB M7 subtype. Blood. 2013;121(17):3469–3472. doi: 10.1182/blood-2012-11-469825. [DOI] [PubMed] [Google Scholar]
- 18.Balgobind BV, Zwaan CM, Pieters R, Van den Heuvel-Eibrink MM. The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia. 2011;25(8):1239–1248. doi: 10.1038/leu.2011.90. [DOI] [PubMed] [Google Scholar]
- 19.Pigazzi M, Masetti R, Bresolin S, et al. MLL partner genes drive distinct gene expression profiles and genomic alterations in pediatric acute myeloid leukemia: an AIEOP study. Leukemia. 2011;25(3):560–563. doi: 10.1038/leu.2010.316. [DOI] [PubMed] [Google Scholar]
- 20.Mercher T, Courtois G, Berger R, Bernard OA. Molecular basis of the t(1;22)(p13;q13) specific for human acute megakaryoblastic leukemia [in French]. Pathol Biol (Paris) 2003;51(1):27–32. doi: 10.1016/s0369-8114(02)00354-1. [DOI] [PubMed] [Google Scholar]
- 21.Hitzler JK, Cheung J, Li Y, Scherer SW, Zipursky A. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood. 2003;101(11):4301–4304. doi: 10.1182/blood-2003-01-0013. [DOI] [PubMed] [Google Scholar]
- 22.Magalhães IQ, Splendore A, Emerenciano M, Figueiredo A, Ferrari I, Pombo-de-Oliveira MS. GATA1 mutations in acute leukemia in children with Down syndrome. Cancer Genet Cytogenet. 2006;166(2):112–116. doi: 10.1016/j.cancergencyto.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Reinhardt K, Zwaan C, Dworzak M, et al. High frequency of GATA1 mutations in childhood non-Down syndrome acute megakaryoblastic leukemia [abstract]. Blood. 2012;120(21). Abstract 888.
- 24.Hasle H, Alonzo TA, Auvrignon A, et al. Monosomy 7 and deletion 7q in children and adolescents with acute myeloid leukemia: an international retrospective study. Blood. 2007;109(11):4641–4647. doi: 10.1182/blood-2006-10-051342. [DOI] [PubMed] [Google Scholar]
- 25.Harrison CJ, Hills RK, Moorman AV, et al. Cytogenetics of childhood acute myeloid leukemia: United Kingdom Medical Research Council Treatment trials AML 10 and 12. J Clin Oncol. 2010;28(16):2674–2681. doi: 10.1200/JCO.2009.24.8997. [DOI] [PubMed] [Google Scholar]
- 26.Chilton L, Hills RK, Harrison CJ, Burnett AK, Grimwade D, Moorman AV. Hyperdiploidy with 49-65 chromosomes represents a heterogeneous cytogenetic subgroup of acute myeloid leukemia with differential outcome. Leukemia. 2014;28(2):321–328. doi: 10.1038/leu.2013.198. [DOI] [PubMed] [Google Scholar]
- 27.Sandahl JD, Kjeldsen E, Abrahamsson J, et al. Ploidy and clinical characteristics of childhood acute myeloid leukemia: a NOPHO-AML study. Genes Chromosomes Cancer. 2014;53(8):667–675. doi: 10.1002/gcc.22177. [DOI] [PubMed] [Google Scholar]
- 28.von Neuhoff C, Reinhardt D, Sander A, et al. Prognostic impact of specific chromosomal aberrations in a large group of pediatric patients with acute myeloid leukemia treated uniformly according to trial AML-BFM 98. J Clin Oncol. 2010;28(16):2682–2689. doi: 10.1200/JCO.2009.25.6321. [DOI] [PubMed] [Google Scholar]
- 29.Luquet I, Laï JL, Barin C, et al. Francophone de Cytogenetique Hematologique. Hyperdiploid karyotypes in acute myeloid leukemia define a novel entity: a study of 38 patients from the Groupe Francophone de Cytogenetique Hematologique (GFCH). Leukemia. 2008;22(1):132–137. doi: 10.1038/sj.leu.2404974. [DOI] [PubMed] [Google Scholar]
- 30.Stölzel F, Mohr B, Kramer M, et al. Karyotype complexity and prognosis in acute myeloid leukemia. Blood Cancer J. 2016;6(1):e386. doi: 10.1038/bcj.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastos-Oreiro M, Perez-Corral A, Martínez-Laperche C, et al. Prognostic impact of minimal residual disease analysis by flow cytometry in patients with acute myeloid leukemia before and after allogeneic hemopoietic stem cell transplantation. Eur J Haematol. 2014;93(3):239–246. doi: 10.1111/ejh.12336. [DOI] [PubMed] [Google Scholar]
- 32.Taga T, Saito AM, Kudo K, et al. Clinical characteristics and outcome of refractory/relapsed myeloid leukemia in children with Down syndrome. Blood. 2012;120(9):1810–1815. doi: 10.1182/blood-2012-03-414755. [DOI] [PubMed] [Google Scholar]
- 33.Crispino JD. GATA1 mutations in Down syndrome: implications for biology and diagnosis of children with transient myeloproliferative disorder and acute megakaryoblastic leukemia. Pediatr Blood Cancer. 2005;44(1):40–44. doi: 10.1002/pbc.20066. [DOI] [PubMed] [Google Scholar]
- 34.Wechsler J, Greene M, McDevitt MA, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32(1):148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 35.Zwaan CM, Kaspers GJ, Pieters R, et al. Different drug sensitivity profiles of acute myeloid and lymphoblastic leukemia and normal peripheral blood mononuclear cells in children with and without Down syndrome. Blood. 2002;99(1):245–251. doi: 10.1182/blood.v99.1.245. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida K, Toki T, Okuno Y, et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013;45(11):1293–1299. doi: 10.1038/ng.2759. [DOI] [PubMed] [Google Scholar]
- 37.Gruber TA, Downing JR. The biology of pediatric acute megakaryoblastic leukemia. Blood. 2015;126(8):943–949. doi: 10.1182/blood-2015-05-567859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono R, Hasegawa D, Hirabayashi S, et al. Acute megakaryoblastic leukemia with acquired trisomy 21 and GATA1 mutations in phenotypically normal children. Eur J Pediatr. 2015;174(4):525–531. doi: 10.1007/s00431-014-2430-3. [DOI] [PubMed] [Google Scholar]
- 39.Reinhardt D, Reinhardt K, Neuhoff C, et al. GATA1-mutation associated leukemia in children with trisomy 21 mosaic [in German]. Klin Padiatr. 2012;224(3):153–155. doi: 10.1055/s-0032-1308988. [DOI] [PubMed] [Google Scholar]