Abstract
Objectives
The pharmacodynamics of polymyxin/carbapenem combinations against carbapenem-resistant Acinetobacter baumannii (CRAB) are largely unknown. Our objective was to determine whether intensified meropenem regimens in combination with polymyxin B enhance killing and resistance suppression of CRAB.
Methods
Time–kill experiments for meropenem and polymyxin B combinations were conducted against three polymyxin B-susceptible (MIC of polymyxin B = 0.5 mg/L) CRAB strains with varying meropenem MICs (ATCC 19606, N16870 and 03-149-1; MIC of meropenem = 4, 16 and 64 mg/L, respectively) at 108 cfu/mL. A hollow-fibre infection model was then used to simulate humanized regimens of polymyxin B and meropenem (2, 4, 6 and 8 g prolonged infusions every 8 h) versus N16870 at 108 cfu/mL over 14 days. New mathematical mechanism-based models were developed using S-ADAPT.
Results
Time–kill experiments were well described by the mathematical mechanism-based models, with the presence of polymyxin B drastically decreasing the meropenem concentration needed for half-maximal activity against meropenem-resistant populations from 438 to 82.1 (ATCC 19606), 158 to 93.6 (N16870) and 433 to 76.0 mg/L (03-149-1). The maximum killing effect of combination treatment was similar among all three strains despite divergent meropenem MIC values (Emax = 2.13, 2.08 and 2.15; MIC of meropenem = 4, 16 and 64 mg/L, respectively). Escalating the dose of meropenem in hollow-fibre combination regimens from 2 g every 8 h to 8 g every 8 h resulted in killing that progressed from a >2.5 log10 cfu/mL reduction with regrowth by 72 h (2 g every 8 h) to complete eradication by 336 h (8 g every 8 h).
Conclusion
Intensified meropenem dosing in combination with polymyxin B may offer a unique strategy to kill CRAB irrespective of the meropenem MIC.
Introduction
Acinetobacter baumannii is increasingly plaguing the global healthcare system as a nosocomial pathogen responsible for a myriad of infections including pneumonia, urinary tract infections, meningitis and bacteraemia.1,2 Particularly troubling are ventilator-associated pneumonias (VAPs) due to A. baumannii, which are characterized by high bacterial burdens and correspondingly high mortality rates. The frequency of A. baumannii isolation during VAP treatment may range from ∼20% to >50% of reported cases.3–6 Carbapenems were traditionally the drug of choice for countering ESBL-producing organisms such as A. baumannii. However, the acquisition of non-chromosomally mediated oxacillinase enzymes and other carbapenem resistance mechanisms has obscured the optimal treatment for A. baumannii VAPs.2
Although meropenem and imipenem are both approved for use in VAP,7 meropenem has a lower affinity for certain oxacillinase enzymes and a comparatively lower seizure threshold than imipenem,8–14 making meropenem a rational choice for VAPs due to A. baumannii. Unfortunately, rising MICs of meropenem substantially decrease the probability of achieving a %T>MIC of ≥40% [the target value of the relevant pharmacokinetic (PK)/pharmacodynamic (PD) index correlating with bactericidal activity] with routine dosing regimens of 1 g every 8 h (q8h).15–18 Prior investigations suggest that a 2 g q8h regimen with a prolonged infusion is more likely to achieve the desired %T>MIC target, but the spread of carbapenem-resistant A. baumannii (CRAB) frequently necessitates the use of alternative antimicrobials.
Polymyxins now represent the drug class that most consistently retains activity against CRAB.19–21 However, reports of polymyxin heteroresistance and treatment failures have surfaced in the wake of widespread colistin and polymyxin B utilization.22 In lieu of newer agents with activity against CRAB, synergy achieved by pairing a polymyxin with meropenem may encourage the use of combination therapy despite pre-existing β-lactam resistance mechanisms.23 Unlike colistin, polymyxin B is not converted from a prodrug into an active moiety, resulting in plasma concentrations of polymyxin B that more quickly reach target levels.24 Despite the favourable PK of polymyxin B, dose-related nephrotoxicity (up to 60%) presents a dose ceiling that limits the concentration of polymyxin B used in combination therapy.25–27 In contrast, meropenem has a wide therapeutic index that allows for comparatively safer dose modulation to optimize killing during combination therapy.28
In the current study, we investigated the relationship between the concentration of meropenem and the killing of CRAB during combination treatment with polymyxin B in time–kill experiments. A mechanism-based mathematical model was utilized to characterize the PD of combinations against three A. baumannii strains with various levels of meropenem resistance to inform regimen selection. A hollow-fibre infection model (HFIM) was then used to simulate plasma concentrations of meropenem and polymyxin B achieved in humans during combination therapy. In the HFIM, the dose of meropenem was progressively increased to define the exposure effect relationship of meropenem in combination with polymyxin B. Such an understanding may help identify the optimal carbapenem and polymyxin B combination regimen that maximizes killing and minimizes the emergence of resistance in CRAB.
Methods
Bacterial isolates and oxacillinases
One laboratory A. baumannii strain (ATCC 19606) and two clinical isolates (N16870 and 03-149-1) were utilized for the investigation. N16870 and 03-149-1 were obtained from critically ill patients infected with A. baumannii in a recently completed NIH study.29 MICs of polymyxin B and meropenem were determined for all three strains in quadruplicate per CLSI standards. In addition, a previously published methodology for detecting oxacillinase enzymes in A. baumannii using PCR was used to detect the presence of the chromosomal oxacillinase OXA-51, as well as the plasmid-mediated oxacillinases OXA-23, OXA-24 and OXA-58.30
Time–kill experiments
To assess how bacterial killing is influenced by the concentration of meropenem present in combination regimens, time–kill experiments were conducted over 48 h using all three A. baumannii isolates. Each strain was grown overnight in Mueller–Hinton broth adjusted with magnesium (12.5 mg/L) and calcium (25 mg/L), and the turbidity of the bacterial suspension was adjusted to achieve a 108 cfu/mL starting inoculum that approximated a difficult-to-treat high-burden infection.31 On the first day of each experiment, a 1.55 mg/L solution of polymyxin B and meropenem solutions consisting of 50, 100, 200, 300, 400 and 500 mg/L was prepared from analytical grade powder purchased from Sigma Aldrich (St Louis, MO, polymyxin B lot number WXB734470V). All three strains were exposed to the meropenem concentration array alone and in combination with polymyxin B during constant shaking in a water bath at 37°C. To compensate for the degradation of meropenem, reaction vessels were supplemented with meropenem at 8.5, 23.5 and 32.5 h to maintain static drug concentrations. At 0, 1, 2, 4, 6, 8, 24, 28, 32 and 48 h, samples were collected and plated onto Mueller–Hinton agar (MHA) after serial dilutions with saline.32 Following 24 h of incubation, the number of colonies present on the agar was enumerated to quantify total population counts (limit of detection = 100 cfu/mL).
PK/PD analyses
An integrated PK/PD model was used to interpret the results of the time–kill studies.32 Following the enumeration of bacterial counts over 48 h, the log10 cfu/mL of each strain was plotted against time for each concentration of meropenem alone and in combination with polymyxin B. The area under the cfu/mL curve (AUCFU0–48) was then calculated for each concentration of meropenem alone and when combined with polymyxin B and normalized by the AUCFU0–48 of the growth control to obtain the log ratio area [LRA; Equation (1)]. In a separate analysis, the log ratio change (LRC) was determined as the log10 change in cfu between 0 and 48 h for each drug treatment [(Equation (2)].
| (1) |
| (2) |
After plotting the LRA and LRC as a function of the meropenem concentration, combination treatments were fitted with a Hill-type model to characterize the PK/PD of the combination (version 12, Systat Software Inc., San Jose, CA, USA).32 Using Equation (3), E represents the LRA or LRC, E0 is the LRA or LRC in the absence of drug, Emax is the maximum effect elicited by the escalating concentration (C) of meropenem in combination with polymyxin B, EC50 is the meropenem concentration displaying half the maximum effect and H is the sigmoidicity constant. Data from experiments involving meropenem alone were described by either a linear model or Hill-type function to visualize the trends in dose escalation. The coefficient of determination (R2) was used to gauge overall model fits.
| (3) |
HFIM
An HFIM was utilized to simulate the time-course of polymyxin B and meropenem concentrations expected in patients as previously described.33 Against strain N16870 (MIC of meropenem = 16 mg/L), antibiotic combinations were administered over 14 days. Briefly, fresh cation-adjusted Mueller–Hinton broth and meropenem were infused into a central reservoir, while the outflow from the reservoir replicated a 2.5 h meropenem half-life that mirrored meropenem PK in critically ill patients.34 Using cellulosic cartridges (C3008; FiberCell Systems Inc., Fredrick, MD, USA), a starting inoculum of 108 cfu/mL was introduced into the extracapillary space of each cartridge. During each experiment, samples were collected at 0, 24, 48, 72, 96, 144, 192, 240, 288 and 336 h. Total bacterial populations were quantified via plating on MHA, whereas population analysis profiles (PAPs) were determined by plating on MHA containing 16, 32, 64 and 128 mg/L meropenem or 0.5, 1, 2 and 4 mg/L polymyxin B to profile less-susceptible subpopulations. Amplification of meropenem or polymyxin B resistance during the PAP analysis was then confirmed with MIC testing on isolates collected after 336 h of antibiotic exposure in the HFIM. PK samples collected from the HFIM experiments were placed in microcentrifuge tubes and immediately stored at −80°C until analysis whereby polymyxin B concentrations were determined by a liquid chromatography single quadrupole MS (LC-MS) method adapted as detailed previously.35 Meropenem concentrations were quantified using a liquid chromatography tandem MS (LC-MS/MS) method (Agilent 1200 and Agilent 6430, Santa Clara, CA, USA). The meropenem calibration curve was linear with a R2 > 0.999 with good reproducibility (relative standard deviation ≤3.57%) and accuracy (99.7%–109.4%). The limit of detection was 0.05 mg/L. The observed versus targeted concentrations for both polymyxin B and meropenem were linear (R2 > 0.90).
The following meropenem regimens were simulated with a 3 h prolonged infusion (regimens i–iv) alone and in combination with polymyxin B (t1/2 = 8 h, regimen v) using an approach described by Blaser.36 To simulate clinically relevant polymyxin B concentrations in critically ill patients, the population PK study conducted by Sandri et al.24 was used to derive a regimen using a median unbound polymyxin B fraction of 0.42 in human plasma. A prior investigation utilizing a thigh infection model observed that an fAUC/MIC of 20 resulted in a 2 log10 reduction in A. baumannii counts.37 Monte Carlo simulations performed by Sandri et al.24 found that a dosing scheme of ∼1.5 mg/kg every 12 h (q12h) of polymyxin B results in an fAUC/MIC of 20 in ∼50% of patients if the causative organism's MIC is 2 mg/L and that a loading dose is appropriate to achieve target concentrations more quickly.
Meropenem 2 g q8h (fCmax = 49.0 mg/L, %fT>MIC = 88.9%, fAUC24 = 707 mg·h/L)
Meropenem 4 g q8h (fCmax = 98.0 mg/L, %fT>MIC = 100%, fAUC24 = 1410 mg·h/L)
Meropenem 6 g q8h (fCmax = 147 mg/L, %fT>MIC = 100%, fAUC24 = 2120 mg·h/L)
Meropenem 8 g q8h (fCmax = 196 mg/L, %fT>MIC = 100%, fAUC24 = 2830 mg·h/L)
Polymyxin B 2.22 mg/kg × 1 dose, then 1.43 mg/kg q12h (fCmax = 2.41 mg/L, fAUC24 = 35.9 mg·h/L)
Mechanism-based modelling of antibiotic combinations
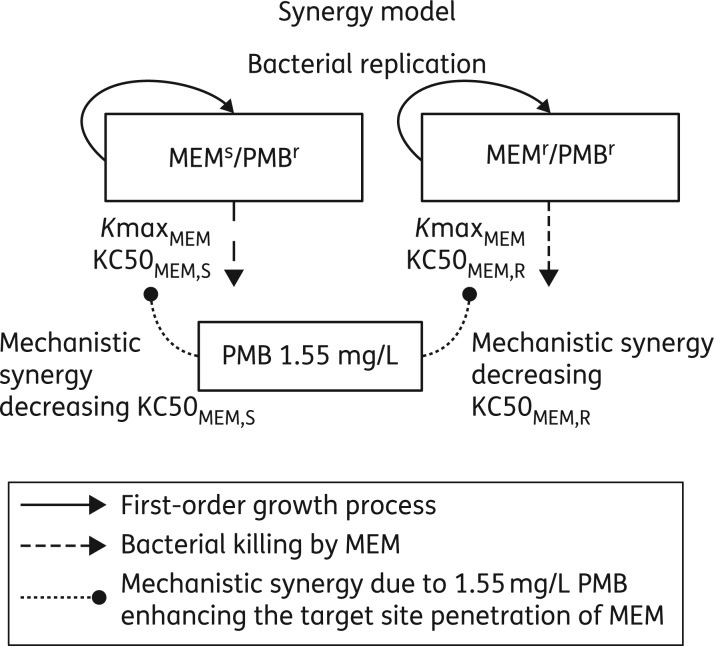
We characterized the extent, time-course and potential synergy mechanisms for the combination of meropenem and polymyxin B by mechanism-based models. Readers unfamiliar with the use of mathematical models to describe the PD of antimicrobials may benefit from reading a well-written review by Nielsen and Friberg.38 Bacterial growth and killing by meropenem and polymyxin B monotherapies and combinations of both agents were described by models with two pre-existing populations (Figure 1). As polymyxin B showed no bacterial killing in monotherapy at the studied polymyxin B concentration, both populations were assumed to be polymyxin B-resistant at the high initial inoculum. The first population was meropenem susceptible and the second population meropenem resistant (Figure 1). Bacterial killing by meropenem was assumed to follow a Hill-type function with different meropenem concentrations resulting in 50% of maximal killing for the meropenem-susceptible and meropenem-resistant populations. We assumed that polymyxin B decreased the meropenem concentrations required for 50% of maximal killing of the meropenem-susceptible population (monotherapy: KC50MEM,SM; combination: KC50MEM,SC) and of the meropenem-resistant population (monotherapy: KC50MEM,RM; combination: KC50MEM,RC). We estimated all PD model parameters simultaneously based on the total population viable counts of the respective strain. We used previously published procedures and methods for estimation, model development and evaluation.33,39–49 Details on the modelling methods are available as Supplementary data at JAC Online.
Figure 1.
Mechanism-based model for the synergy of meropenem and polymyxin B against three A. baumannii strains studied at high bacterial inocula of ∼108 cfu/mL. The model contained two populations: the first population was meropenem susceptible and polymyxin B resistant (MEMs/PMBr), whereas the second population was resistant to both antibiotics (MEMr/PMBr). Bacterial killing terms (i.e. arrows) were only included for the effect of meropenem as 1.55 mg/L polymyxin B displayed negligible killing in time–kill experiments. Synergy was implemented by assuming that polymyxin B permeabilized the outer membrane of A. baumannii and thereby enhanced the target site concentration of meropenem. The presence of 1.55 mg/L polymyxin B decreased the meropenem concentration required to achieve half of maximal killing against the meropenem-susceptible and meropenem-resistant populations as listed in Table 2. MEM, meropenem and PMB, polymyxin B.
Results
Antibacterial MICs and oxacillinases
The MICs of polymyxin B and meropenem for each A. baumannii strain and the type of oxacillinase enzymes detected are listed in Table 1. All the strains demonstrated a susceptible polymyxin B MIC of 0.5 mg/L, whereas the meropenem MICs were all non-susceptible and included 4 mg/L (ATCC 19606), 16 mg/L (N16870) and 64 mg/L (03-149-2). The chromosomally encoded oxacillinase enzyme OXA-51 was present in all three A. baumannii isolates, whereas the plasmid-mediated β-lactamase OXA-23 was detected in N16870 and 03-149-2.
Table 1.
MICs of polymyxin B and meropenem for each A. baumannii strain, as well as the presence of the chromosomal oxacillinase OXA-51 and plasmid-mediated oxacillinase enzymes OXA-23, OXA-24 and OXA-58
| Strain | Polymyxin B MIC (mg/L) | Meropenem MIC (mg/L) | Oxacillinases present |
|---|---|---|---|
| ATCC 19606 | 0.5 | 4 | OXA-51 |
| N16870 | 0.5 | 16 | OXA-51, OXA-23 |
| 03-149-1 | 0.5 | 64 | OXA-51, OXA-23 |
Time–kill experiments
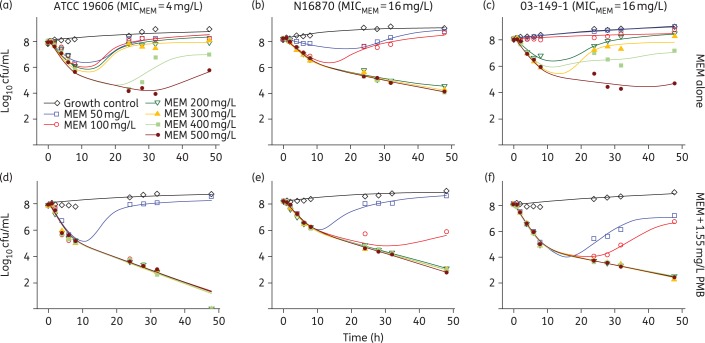
The PD of meropenem alone and in combination with polymyxin B against the three strains of A. baumannii are summarized in Figure 2. Treatment with meropenem alone and in combination with polymyxin B resulted in killing that was dependent on the concentration of meropenem. When in combination with polymyxin B, meropenem concentrations of 50, 100 and 200 mg/L resulted in the following respective net changes in bacterial counts at 48 h (log10 cfu/mL): +0.71, −7.90 and −7.82 (ATCC 19606), +0.41, −2.38 and −5.11 (N16870), and −0.95, −1.36 and −5.58 (03-149-1). Meropenem concentrations of 50, 100 and 200 mg/L in the absence of polymyxin B resulted in regrowth by 24 h in strains ATCC 19606 (MIC = 4 mg/L) and 03-149-1 (MIC = 64 mg/L), whereas 300 mg/L meropenem resulted in regrowth by 48 h in both strains, 400 mg/L meropenem achieved ∼1 log10 cfu/mL reductions by 48 h and 500 mg/L meropenem achieved 2.17 and 3.33 log10 cfu/mL reductions at 48 h for ATCC 19606 and 03-149-1, respectively. For N16870 (MIC = 16 mg/L), meropenem concentrations ≥200 mg/L resulted in a ≥3 log10 cfu/mL reduction by 48 h. Polymyxin B alone was unable to achieve >0.5 log10 cfu/mL reduction in any strain, with counts that largely paralleled the growth control (data not shown). Although 100 mg/L meropenem in the combination regimen achieved a ≥3 log10 cfu/mL reduction for all three strains, 200 mg/L meropenem was necessary for maximum activity against N16870 and 03-149-1.
Figure 2.
Observed (symbols) and model fitted (lines) viable counts for time–kill experiments involving meropenem alone (a–c) and in combination with 1.55 mg/L polymyxin B (d–f) against A. baumannii strains ATCC 19606 (a and d), N16870 (b and e) and 03-149-1 (c and f). MIC values of meropenem are listed for each strain. MEM, meropenem; PMB, polymyxin B. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The mechanism-based model provided excellent curve fits for the static time–kill experiments. The parameter estimates from the synergy model indicated that meropenem activity was comparable in all three strains when meropenem was used in combination with polymyxin B (Table 2, all relative standard errors ≤33%). In the absence of polymyxin B, the concentration of meropenem required for half of maximal killing (KC50MEM) ranged from 43.0 to 199 mg/L against meropenem-susceptible A. baumannii populations and from 158 to 438 mg/L against the meropenem-resistant populations of all three strains. In the presence of polymyxin B, the KC50MEM values were very consistent across strains and varied from 2.41 to 3.78 mg/L against meropenem-susceptible populations and from 76.0 to 93.6 mg/L against meropenem-resistant populations. Given the large disparity in the meropenem MICs for each strain (4, 16 and 64 mg/L), the activity of meropenem in combination with polymyxin B did not appreciably relate to the individual meropenem MIC values for each strain.
Table 2.
Parameter estimates for ATCC 19606, N16870 and 03-149-1 obtained from the mechanism-based model used to describe the static time–kill and the HFIM data
| Parameter | Symbol | Unit | Population mean (relative standard error) |
|||
|---|---|---|---|---|---|---|
| ATCC 19606 | N16870 | 03-149-1 | N16870 (HFIM) | |||
| Mean generation time (MGT) | MGT12 | min | 64.4 (4.0%)a | 91.5 (4.5%)a | 64.3 (4.7%)a | 61.1 (6.6%) |
| MGT for the resistant population in the presence of polymyxin B | MGT12 | min | 68.6 (6.3%) | |||
| Maximum population size | log10(cfumax) | 9.29 (1.9%) | 9.02 (1.5%) | 9.93 (2.6%) | 10.2 (0.5%) | |
| Initial inoculum | log10(cfuo) | 8.16 (1.0%) | 8.30 (0.6%) | 8.17 (0.5%) | 8.23 (2.4%) | |
| Mutation frequency for the meropenem-resistant population | log10(MFMEM) | −5.25 (3.5%) | −4.01 (10.6%) | −5.12 (3.5%) | −4.67 (6.5%) | |
| Maximum rate of bacterial killing by meropenem | KmaxMEM | 1/h | 1.01 (6.9%)a | 0.768 (4.3%)a | 0.986 (4.9%)a | 2.48 (8.1%) |
| KmaxMEM for the meropenem-resistant population | KmaxMEM,R | 1/h | 1.28 (6.9%) | |||
| Meropenem resulting in 50% of maximal killing of the | ||||||
| susceptible population in monotherapy | KC50MEM,SM | mg/L | 43.0 (9.2%) | 50.9 (13.0%) | 199 (9.7%) | 101 (21.6%) |
| susceptible population in the presence of 1.55 mg/L polymyxin B | KC50MEM,SC | mg/L | 3.64 (24.1%) | 3.78 (13.3%) | 2.41 (33.0%) | 2.38 (14.9%) |
| resistant population in monotherapy | KC50MEM,RM | mg/L | 438 (6.1%) | 158 (9.0%) | 433 (6.5%) | 111 (24.8%) |
| resistant population in the presence of 1.55 mg/L polymyxin B | KC50MEM,RC | mg/L | 82.1 (5.3%) | 93.6 (8.4%) | 76.0 (7.2%) | 7.97 (18.4%) |
| Hill coefficient for bacterial killing by meropenem | HillMEM | 5.90 (8.2%) | 4.97 (16.3%) | 4.30 (24.0%) | 1 (fixed) | |
| Maximum fractional reduction in the growth rate at high signal molecule concentrations | ImaxSig12 | 0.988 (8.8%) | 0.755 (47.0%) | 0.969 (20.1%) | b | |
| Signal molecule concentration associated with 50% of maximal inhibition of bacterial growth rate | log10(IC50,Sig) | 6.98 (1.8%) | 6.87 (2.7%) | 6.85 (2.4%) | b | |
| Mean degradation time of hypothetical signal molecules | MTTSig | h | 1.76 (28%) | 1.42 (22.7%) | 2.17 (16.8%) | b |
| Polymyxin B concentration yielding half-maximal decrease of the KC50MEM | C50PMB,SYN | c | c | c | 0.5 (fixed) | |
| Hill coefficient for synergistic effect of polymyxin B | HillSYN | c | c | c | 5 (fixed) | |
| Additive residual error on log10 scale | SDCF | 0.310 (8.4%) | 0.158 (7.3%) | 0.192 (7.5%) | 0.346 (6.3%) | |
aEstimate applies to all populations in monotherapy and combination therapy.
bThe effect of signal molecules was estimated to be minimal in the dynamic HFIM. Thus, these parameters were removed from the model for the hollow-fibre dataset.
cSynergy was handled via an IF condition for the static time–kill data.
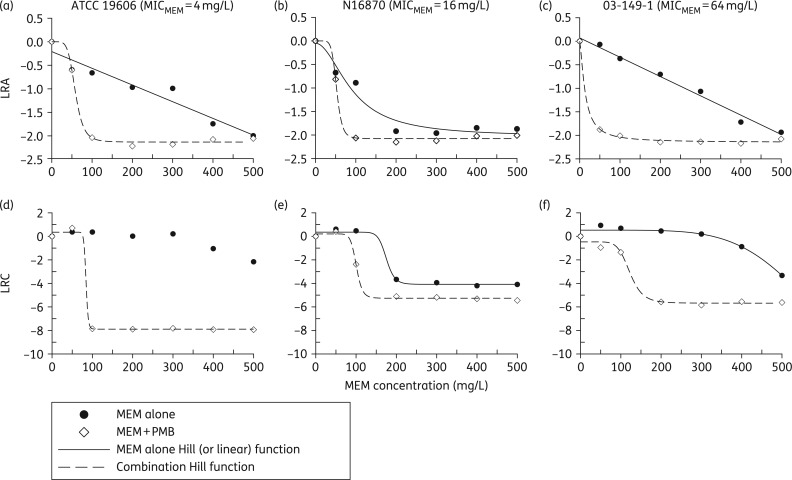
Figure 3 displays the results of the LRA and LRC analyses of the data from the time–kill studies for meropenem alone. In the LRA analysis (Figure 3a–c), strains ATCC 19606 and 03-149-1 each exhibited a linear decrease in the LRAs as the meropenem concentration was increased. In contrast, a Hill-type function best described both the LRA data of strain N16870 and the LRC data for all three strains (Figure 3d–f). In the LRC analysis of meropenem alone, strain N16870 produced the lowest EC50 of 175 mg/L, whereas the EC50 values for ATCC 19606 and 03-149-1 were 399 and 626 mg/L, respectively.
Figure 3.
PK/PD analyses from the time–kill experiments for each strain plotted as LRA (a–c) or LRC (d–f) as a function of meropenem concentration. Data for meropenem alone are represented by black circles, whereas data for meropenem in combination with 1.55 mg/L polymyxin B are represented by white diamonds. Each plot was fitted with a Hill-type function or linear equation (panels a and c for LRA). Parameter estimates for combination treatments are listed in Table 3. MEM, meropenem; PMB, polymyxin B.
For the meropenem and polymyxin B combinations, the Hill-type function provided an excellent fit to the data analysed using both the LRA and the LRC metrics (Figure 3), with R2 values >0.97 for each model (parameter estimates listed in Table 3). Despite a range of meropenem MICs (4, 16 and 64 mg/L), the maximal activity of combination treatment was similar regardless of the strain when using the LRA metric (Emax = 2.13, 2.08 and 2.15, respectively). Using the LRC metric the corresponding Emax values were 8.25, 5.47 and 5.21; however, the LRC for the ATCC 19606 strain was confounded by counts that were below the limit of detection. Whereas strains N16870 and 03-149-1 experienced similar maximal reductions from baseline (5.47 and 5.21 log10 cfu/mL, respectively), ATCC 19606 was undetectable at meropenem concentrations ≥100 mg/L in combination with polymyxin B. The EC50 values of meropenem and polymyxin B combinations did not appreciably relate to the MICs in the LRA analysis (EC50 = 58.7, 52.8 and 13.6 mg/L), but the EC50 values did trend towards higher meropenem concentrations when applying the LRC metric (EC50 = 84.3, 101 and 123 mg/L; MIC of meropenem = 4, 16 and 64 mg/L, respectively).
Table 3.
Parameter estimates for combinations evaluated in the time–kill studies (standard estimates are listed parenthetically); the data were analysed using LRA and LRC
| Strain |
||||||
|---|---|---|---|---|---|---|
| ATCC 19606 |
N16870 |
03-149-1 |
||||
| Meropenem MIC (mg/L) | 4 | 16 | 64 | |||
| PK/PD metric | LRA | LRC | LRA | LRC | LRA | LRC |
| Emax | 2.13 (4.19) | 8.25 (3.96) | 2.08 (3.84) | 5.47 (4.40) | 2.15 (3.40) | 5.21 (7.29) |
| EC50 (mg/L) | 58.7 (5.22) | 84.3 (>100.0) | 52.8 (6.45) | 101 (37.4) | 13.6 (82.6) | 123 (17.3) |
| R2 | 0.992 (8.04) | 0.995 (29.4) | 0.993 (7.16) | 0.993 (22.6) | 0.996 (5.01) | 0.976 (41.5) |
HFIM
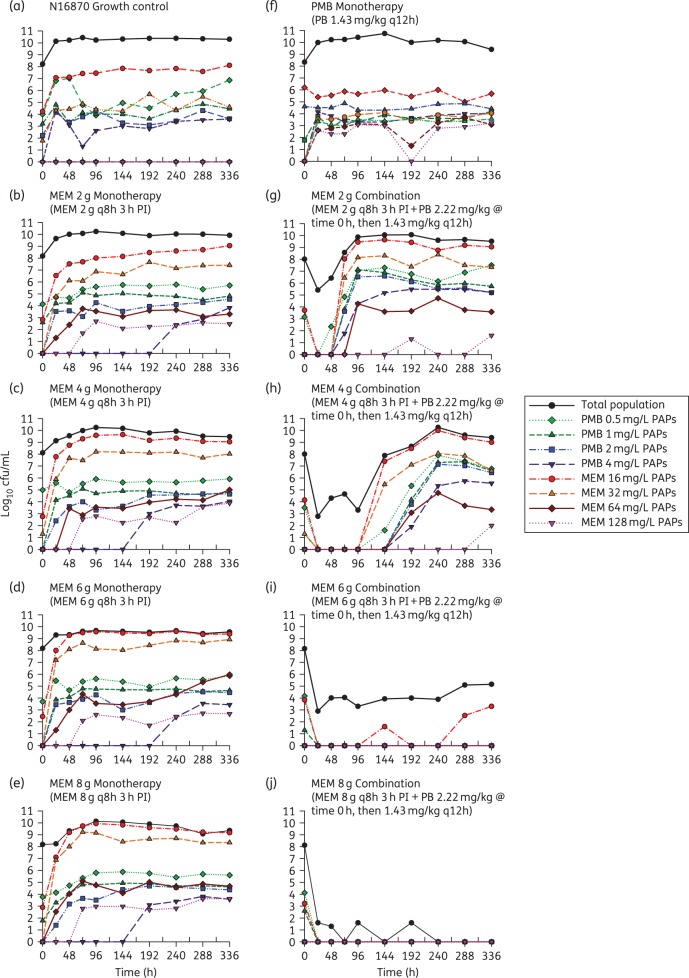
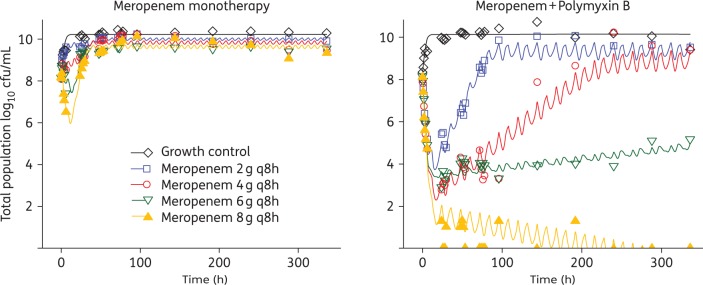
The results of the HFIM experiments conducted with strain N16870 are presented in Figure 4 and include both the total population and PAPs for each regimen. Although meropenem concentrations of 200 mg/L were able to achieve a ≥3 log10 reduction in the time–kill study, all of the HFIM regimens involving meropenem alone, including the 8 g q8h regimen with an fCmax of 196 mg/L, were unable to prevent bacterial growth by 48 h. The meropenem dose of 8 g q8h was the only monotherapy capable of maintaining the total population at a static 108 cfu/mL bacterial load for 24 h, then steady growth following the 24 h timepoint occurred that plateaued at ∼1 × 1010 cfu/mL at 96 h. Despite achieving an fAUC/MIC of 71.8 and utilizing a loading dose, the polymyxin B monotherapy was unable to substantially reduce N16870 counts, with growth to 1 × 1010 cfu/mL observed by 24 h. Surprisingly, the rise of polymyxin-resistant subpopulations was not observed during polymyxin B monotherapy in the PAP analysis and MIC testing conducted after 336 h of polymyxin B exposure confirmed that the polymyxin B MIC only shifted a single dilution from 0.5 to 1.0 mg/L. The amplification of polymyxin B resistance may have been relatively minor due to the inability of polymyxin B to kill enough of the A. baumannii to shift the population toward a resistant phenotype.
Figure 4.
HFIM counts for the total bacterial population of N16870, as well as polymyxin B- and meropenem-resistant subpopulations, during 14 days of antibiotic exposure in monotherapy and in combination. A polymyxin B regimen simulating 1.43 mg/kg q12h alone was investigated. Meropenem dosing schemes of 2, 4, 6 and 8 g q8h administered as a 3 h prolonged infusion (3 h PI) were investigated as monotherapies and in the presence of the polymyxin B regimen. MEM, meropenem; PMB, polymyxin B; PAPs—counts on agar imbued with meropenem or polymyxin B. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
In contrast to the monotherapies investigated in the HFIM, the high-dose combination regimens were able to achieve extensive killing that was largely maintained for the duration of the experiment. The combination regimen of 2 g q8h meropenem with polymyxin B reduced bacterial counts by >2.5 log10 cfu/mL at 24 h, followed by complete regrowth by 72 h. Increasing the meropenem dose in the combination regimen to 4 g resulted in a >5 log10 cfu/mL reduction by 24 h and full regrowth did not occur until 144 h. In both the 2 g and 4 g q8h combination regimens, the proportion of the population capable of growing on 4 mg/L polymyxin B was >40 times higher relative to the corresponding meropenem monotherapies by 336 h. The amplification of polymyxin B resistance was also confirmed by polymyxin B MICs that shifted from 0.5 mg/L at baseline to 4 mg/L by 336 h in the 2 g and 4 g q8h combination regimens. The 6 g q8h meropenem combination regimen was able to maintain the bacterial load at ∼104 cfu/mL for the majority of the experiment, until the total count re-stabilized at ∼105 cfu/mL at 288 h and consistent growth on plates containing meropenem 16 mg/L was observed. Finally, the combination regimen with 8 g q8h meropenem maintained the bacterial load at the limit of detection until no colonies were observed past 192 h. The 8 g q8h regimen also completely suppressed the emergence of resistance, with no growth occurring on MHA imbued with meropenem or polymyxin B beginning at 24 h.
Similar to the time-killing experiments, the mechanism-based model provided excellent curve fits for the HFIM dataset (Figure 5). The maximal rate of N16870 killing in the HFIM was nearly twice as fast comparing the meropenem-susceptible versus meropenem-resistant populations (KmaxMEM 2.48 and 1.28 1/h). The KC50MEM for meropenem in the presence of polymyxin B was also >13.9 times lower for the combination versus monotherapy for both the meropenem-susceptible population (KC50MEM 2.38 versus 101 mg/L) and meropenem-resistant population (KC50MEM 7.97 versus 111 mg/L).
Figure 5.
Observed (symbols) and model fitted (lines) total population counts for strain N16870 investigated in the HFIM. Meropenem was studied as either a 2, 4, 6 or 8 g q8h regimen in the absence (left) or presence (right) of a polymyxin B regimen consisting of 2.22 mg/kg at time 0 h, followed by 1.43 mg/kg q12h thereafter. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
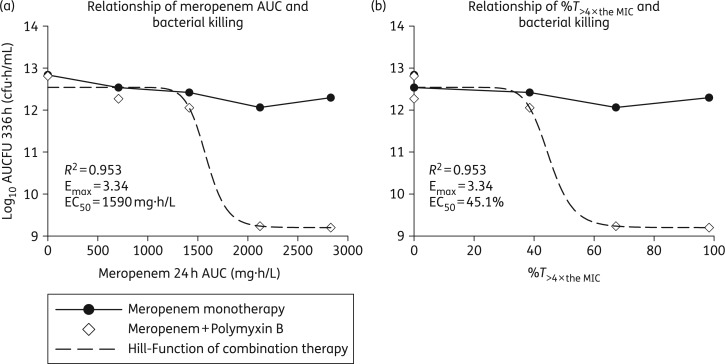
In place of the traditional %T>MIC index used to describe and predict bacterial killing of β-lactam monotherapy, the performance of each combination regimen paralleled the AUC of meropenem over 24 h (R2 = 0.953, Figure 6a). Without concomitant polymyxin B, total bacterial populations were largely constant in the face of escalating meropenem concentrations, whereas meropenem AUCs of 2120 and 2830 mg·h/L conferred log10 AUCFU reductions of 3.58 and 3.61 cfu·h/mL in combination therapy. The meropenem AUC that achieved half of the maximal effect when used in combination was 1590 mg·h/L in the HFIM (Figure 6), which was similar to the meropenem AUC of 1270 mg·h/L (51.8 mg/L × 24 h) that produced half of the maximal effect for the combination in the LRA analysis of the time–kill data for strain N16870. In addition to the meropenem AUC, killing of A. baumannii in combination regimens was also well described by the percentage of time that meropenem concentrations were >4× the MIC (Figure 6b). In the time-dependent killing analysis, achieving a %T>4× the MIC of 45.1% resulted in half-maximal killing.
Figure 6.
The relationship between meropenem AUC over 24 h and the AUCFUs of the total bacterial populations throughout the 14 day HFIM experiments is shown for meropenem alone (black circles) and meropenem in combination with polymyxin B (white diamonds) in (a), whereas (b) displays the relationship between bacterial killing and the percentage of time the meropenem concentration was >4× the MIC (MIC = 16 mg/L, 4× the MIC = 64 mg/L). Combination regimens were fitted with a Hill-type function, whereas meropenem monotherapies are shown as line plots. PAPs—counts on agar imbued with meropenem.
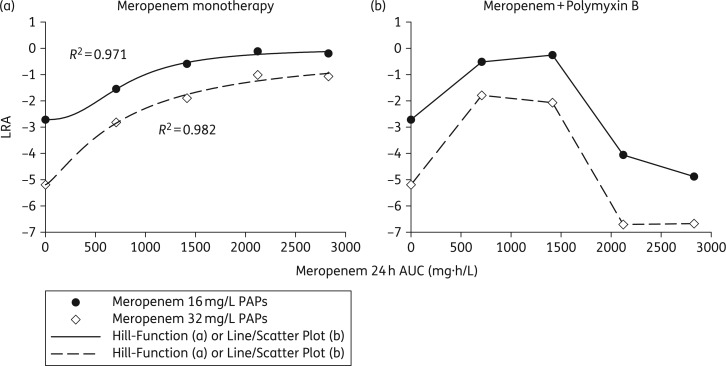
Unlike regimens with meropenem alone and combination regimens utilizing meropenem doses of 2 g or 4 g q8h, meropenem doses of 6 g and 8 g q8h suppressed carbapenem-resistant subpopulations when in combination with polymyxin B. Increasing the dose of meropenem alone amplified carbapenem-resistant subpopulations by 48 h, with >90% of the population growing on 16 mg/L meropenem in the 6 g treatment arm, whereas <0.4% of the population exposed to the 2 g q8h regimen grew on 16 mg/L meropenem by 48 h. MIC testing conducted on A. baumannii exposed to meropenem monotherapy for 336 h verified that meropenem MICs shifted from 16 mg/L at baseline to 128 mg/L at 336 h regardless of the meropenem dose. The ability of A. baumannii to augment carbapenem resistance upon exposure to meropenem was confirmed in an analysis that tracked the LRAs of meropenem-resistant subpopulations (Figure 7). As exposure to meropenem in the HFIM increased from 0 to 2830 mg·h/L in monotherapy, the LRA of subpopulations growing on 16 and 32 mg/L meropenem increased by 2.52 and 4.13, respectively. Similar to the meropenem monotherapies, a meropenem dose of 2 g q8h in combination with polymyxin B amplified the growth of meropenem-resistant subpopulations by 2.20 and 3.40 (meropenem PAPs, 16 and 32 mg/L), which exceeded the LRA increases of 1.17 and 2.37 observed during the corresponding monotherapies. However, intensified meropenem doses of 6 g and 8 g q8h in combination with polymyxin B suppressed carbapenem resistance, with LRA reductions of 1.34 and 1.48 conferred by the 6 g q8h combination regimen (meropenem PAPs, 16 and 32 mg/L, respectively).
Figure 7.
Resistance plots that graphically summarize either the beneficial suppression of carbapenem resistance or the deleterious amplification of resistance conferred with meropenem dose escalation. The AUCFU of A. baumannii subpopulations capable of growing on 16 mg/L meropenem (black circles) and 32 mg/L meropenem (white diamonds) throughout the 14 day HFIM experiments were normalized by the AUCFU of the total population for each meropenem regimen. Using Equation (1), the LRAs were then calculated for meropenem monotherapies (a) and polymyxin B combinations (b) and plotted as a function of meropenem 24 h AUC. Increasing meropenem exposure without polymyxin B induced the expansion of meropenem-resistant subpopulations. In contrast, escalating meropenem concentrations in combination with polymyxin B increased meropenem resistance at lower exposures (2 and 4 g q8h regimens) and reduced meropenem resistance at higher exposures (6 and 8 g q8h regimens).
Discussion
A paucity of agents active against CRAB are available. Here, we investigated approaches to combat carbapenem resistance by increasing the dose intensity of meropenem alone and in combination with polymyxin B. Although achieving a high %T>MIC during carbapenem monotherapy is widely accepted for obtaining bactericidal activity,50 neither the time-killing studies nor the HFIM data support the use of %T>MIC for the investigated A. baumannii strains. Indeed, in time–kill experiments a meropenem concentration of 100 mg/L (%T>MIC = 100% for all three strains) was unable to cause a reduction in bacterial counts by 48 h for the two meropenem-resistant strains (MIC = 16 and 64 mg/L) or the meropenem-intermediate strain (MIC = 4 mg/L).51,52 In the HFIM analysis, meropenem monotherapies of 4, 6 and 8 g q8h achieved a %T>MIC of 100% but produced negligible killing by 24 h, demonstrating that simply increasing the dose of a carbapenem relative to the MIC for the strain may not be enough to overcome A. baumannii resistance mechanisms. Given the inability of the current study to distinguish between AUC and %T>4×MIC as predictive indices for bacterial killing achieved by meropenem in combination with polymyxin B, dose fractionation studies are needed to help clarify whether AUC, time or a hybrid index best describes the combinatorial PD.
Despite the limited bacterial killing of meropenem monotherapies in the HFIM, the addition of polymyxin B resulted in bacterial killing in a meropenem dose-dependent manner. A. baumannii infections are commonly treated with empirical meropenem doses of 0.5–1 g q8h,7,53 with 2 g q8h being the most aggressive regimen utilized during life-threatening infections.54 There have also been reports of clinicians escalating meropenem regimens beyond the traditional 2 g q8h limit to combat organisms with elevated meropenem MICs,55,56 including a 3 g every 6 h regimen that utilizes the same daily dose (12 g/day) as the 4 g q8h regimen.57 In the present study, the lowest meropenem dose used in the HFIM was 2 g q8h, which corresponds to the highest meropenem dose investigated in a clinical trial,54 and resulted in full regrowth of A. baumannii by 72 h when combined with polymyxin B. Although the 6 and 8 g meropenem combination regimens produced the most killing, a 4 g combination regimen may be sufficiently bactericidal to an A. baumannii isolate to allow the host immune system to clear the infection. The meropenem concentration dependency supports the notion that regardless of the magnitude of a dose increase, amplifying the amount of meropenem used in combination with polymyxin B against CRAB infections may increase the likelihood of bacterial eradication.
Not only were the combination regimens with elevated meropenem doses capable of substantial killing of CRAB, but the 6 and 8 g q8h regimens also suppressed the emergence of carbapenem resistance. In monotherapy, increasing the dose of meropenem was counterproductive, with more extensive meropenem exposure leading to the increased amplification of meropenem-resistant subpopulations. When the meropenem dose was escalated in combination with polymyxin B, an ‘inverted-U’ relationship was observed in which suboptimal meropenem exposure amplified carbapenem resistance and more extreme meropenem exposures killed the entire CRAB population.58 Unexpectedly, the combination regimen utilizing the lowest meropenem dose of 2 g q8h amplified more carbapenem resistance than its corresponding monotherapy. Combination regimens that use inadequate carbapenem doses may therefore be at the highest risk for augmenting carbapenem resistance, emphasizing the potential value of intensified carbapenem dosing schemes.
Taken together, these data support the use of a meropenem and polymyxin B combination against CRAB in which the meropenem dose is optimized to increase the bacterial killing. While the dose-dependent nephrotoxicity of polymyxin B may prevent substantial exposure increases over a long duration,25,59 the concentration of polymyxin B was sufficient to confer synergistic killing with meropenem. Due to the favourable safety profile of meropenem, modulation of the meropenem dose may be a practical solution to increase the activity of the combination.54 It is also noteworthy that the meropenem combination performed similarly against three strains of A. baumannii with varying levels of nominal carbapenem resistance as indicated by the MIC. Considering the absence of plasmid-mediated oxacillinases in the ATCC 19606 strain, the similar killing profiles observed in time–kill experiments cannot be ascribed to the presence of specific β-lactamase enzymes. Due to the inability of MIC measurements to accurately forecast the susceptibility of drug-resistant subpopulations that emerge during antibiotic treatment, empirical doses of intensified meropenem may be warranted whenever carbapenem resistance mechanisms are suspected. However, the safety and tolerability of intensified regimens must be validated prior to clinical utilization. The use of only three A. baumannii strains may also prevent an accurate prediction of how intensified combination regimens will perform against other clinical strains. In the mechanism-based modelling, we conservatively assumed near-maximal (>99%) synergy was achieved by ∼1.5 mg/L polymyxin B. Further studies are needed to determine whether higher polymyxin B concentrations achieve more extensive synergy.
While the activity of elevated meropenem doses in combination with polymyxin B may offer a new approach to combating multidrug-resistant A. baumannii infections, the hypothetical risk of carbapenem-induced seizures may jeopardize any mortality benefit afforded by an intensified meropenem regimen.28 However, a meta-analysis of seizure risk in carbapenems found the odds ratio of a seizure was 1.04 (95% CI 0.61, 1.77) for meropenem compared with ceftazidime or cefotaxime, suggesting that high meropenem doses may be tolerable.28 Also, because convulsions induced by β-lactams are associated with peak drug concentrations,60 administering meropenem as a 3 h prolonged infusion reduces peak meropenem concentrations and minimizes the likeliness of a seizure. According to the manufacturer of meropenem, healthy volunteers given 1 g of meropenem as a 5 min infusion experienced a mean peak plasma concentration of 112 mg/L, with a maximum value of 140 mg/L.61 In the present study, fCmax concentrations for the 4 g q8h and 6 g q8h regimens were 98.0 and 147 mg/L, respectively, which are comparable to plasma concentrations observed during meropenem boluses of traditional doses. In an animal study, an extreme dose of 100 mg/kg meropenem was quickly infused intravenously into rats and intracerebroventricularly into mice and did not result in EEG changes or convulsions, whereas intracerebroventricular injections in dogs resulted in a localized hippocampal discharge with no corresponding behavioural changes or convulsions.60 If a patient is at a high risk for a seizure, phenytoin and phenobarbital are options for seizure prophylaxis as neither agent shares a PK interaction with carbapenems and both have demonstrated anticonvulsant activity in rat models of imipenem-induced seizures.62
In addition to the hypothetical seizure risk of intensified meropenem regimens, doses of meropenem that exceed 2 g q8h may carry other unforeseen risks for patients. As the objective of the current investigation was to fully define the PD of meropenem in combination with polymyxin B, it was necessary to escalate the dose of meropenem beyond traditional limits to explore whether additional bacterial killing was conferred by the intensified dosing schemes. The meropenem regimens of 4, 6 and 8 g q8h therefore represent theoretical regimens that have unknown applicability in the clinical setting. The safety and tolerability of such intensified regimens was not assessed in the current in vitro investigation and meropenem doses above 2 g q8h must first be validated with preclinical studies to fully characterize potential side effects and risks for patients before intensified meropenem regimens can be advocated in the clinic.
In closing, combination therapy of polymyxin B with meropenem doses that exceed 2 g q8h may offer an intensified dosing strategy for combating life-threatening infections caused by CRAB. Mechanism-based modelling suggested that synergy was achieved due to polymyxin B greatly increasing the target site concentrations of meropenem, which was consistently observed across three strains. In lieu of a well-designed dose-fractionation study that identifies an index adequately predictive of the performance of the combination, clinicians should use caution when dosing meropenem based on the reported MIC for a clinical CRAB strain. It is important to note that intensified meropenem regimens have not yet been evaluated in humans and the clinical use of meropenem doses exceeding 2 g q8h cannot be advocated without fully understanding the safety of meropenem at elevated doses. The A. baumannii killing observed in the HFIM was also based on a single strain and requires further confirmation. Future investigations in animal models evaluating the toxicology of high-dose meropenem in combination with polymyxin B are needed before the risk-to-benefit of such a regimen can be fully appreciated.
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI111990. C. B. L. is an Australian National Health and Medical Research Council (NHMRC) Career Development Fellow. J. L. is an Australian NHMRC Senior Research Fellow. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Transparency declarations
None to declare.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary data
References
- 1.Kim UJ, Kim HK, An JH et al. Update on the epidemiology, treatment, and outcomes of carbapenem-resistant Acinetobacter infections. Chonnam Med J 2014; 50: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 2008; 21: 538–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed NH, Hussain T, Biswal I. Antimicrobial resistance of bacterial isolates from respiratory secretions of ventilated patients in a multi-specialty hospital. Avicenna J Med 2015; 5: 74–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Saed A, Balkhy HH, Al-Dorzi HM et al. Acinetobacter is the most common pathogen associated with late-onset and recurrent ventilator-associated pneumonia in an adult intensive care unit in Saudi Arabia. Int J Infect Dis 2013; 17: e696–701. [DOI] [PubMed] [Google Scholar]
- 5.Walkey AJ, Reardon CC, Sulis CA et al. Epidemiology of ventilator-associated pneumonia in a long-term acute care hospital. Infect Control Hosp Epidemiol 2009; 30: 319–24. [DOI] [PubMed] [Google Scholar]
- 6.Inchai J, Pothirat C, Liwsrisakun C et al. Ventilator-associated pneumonia: epidemiology and prognostic indicators of 30-day mortality. Jpn J Infect Dis 2015; 68: 181–6. [DOI] [PubMed] [Google Scholar]
- 7.Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 2005; 171: 388–416. [DOI] [PubMed] [Google Scholar]
- 8.Brown S, Young HK, Amyes SG. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect 2005; 11: 15–23. [DOI] [PubMed] [Google Scholar]
- 9.Norrby SR, Newell PA, Faulkner KL et al. Safety profile of meropenem: international clinical experience based on the first 3125 patients treated with meropenem. J Antimicrob Chemother 1995; 36 Suppl A: 207–23. [DOI] [PubMed] [Google Scholar]
- 10.Schmutzhard E, Williams KJ, Vukmirovits G et al. A randomised comparison of meropenem with cefotaxime or ceftriaxone for the treatment of bacterial meningitis in adults. Meropenem Meningitis Study Group. J Antimicrob Chemother 1995; 36 Suppl A: 85–97. [DOI] [PubMed] [Google Scholar]
- 11.Colardyn F, Faulkner KL. Intravenous meropenem versus imipenem/cilastatin in the treatment of serious bacterial infections in hospitalized patients. Meropenem Serious Infection Study Group. J Antimicrob Chemother 1996; 38: 523–37. [DOI] [PubMed] [Google Scholar]
- 12.Norrby SR. Carbapenems in serious infections: a risk-benefit assessment. Drug Saf 2000; 22: 191–4. [DOI] [PubMed] [Google Scholar]
- 13.Mouton JW, Touzw DJ, Horrevorts AM et al. Comparative pharmacokinetics of the carbapenems: clinical implications. Clin Pharmacokinet 2000; 39: 185–201. [DOI] [PubMed] [Google Scholar]
- 14.Smith CA, Antunes NT, Stewart NK et al. Structural basis for carbapenemase activity of the OXA-23 β-lactamase from Acinetobacter baumannii. Chem Biol 2013; 20: 1107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaruratanasirikul S, Sriwiriyajan S, Punyo J. Comparison of the pharmacodynamics of meropenem in patients with ventilator-associated pneumonia following administration by 3-hour infusion or bolus injection. Antimicrob Agents Chemother 2005; 49: 1337–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaruratanasirikul S, Kositpantawong N, Jullangkoon M et al. Pharmacodynamics of meropenem in critically ill patients with ventilator-associated pneumonia. J Med Assoc Thai 2013; 96: 1283–9. [PubMed] [Google Scholar]
- 17.Li C, Kuti JL, Nightingale CH et al. Population pharmacokinetic analysis and dosing regimen optimization of meropenem in adult patients. J Clin Pharmacol 2006; 46: 1171–8. [DOI] [PubMed] [Google Scholar]
- 18.Lodise TP Jr, Lomaestro B, Drusano GL. Piperacillin-tazobactam for Pseudomonas aeruginosa infection: clinical implications of an extended-infusion dosing strategy. Clin Infect Dis 2007; 44: 357–63. [DOI] [PubMed] [Google Scholar]
- 19.Lee HJ, Bergen PJ, Bulitta JB et al. Synergistic activity of colistin and rifampin combination against multidrug-resistant Acinetobacter baumannii in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 2013; 57: 3738–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagihara M, Housman ST, Nicolau DP et al. In vitro pharmacodynamics of polymyxin B and tigecycline alone and in combination against carbapenem-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 2014; 58: 874–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Li W, Feng Y et al. Efficacy and safety of polymyxins for the treatment of Acinectobacter baumannii infection: a systematic review and meta-analysis. PLoS ONE 2014; 9: e98091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai Y, Chai D, Wang R et al. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 2012; 67: 1607–15. [DOI] [PubMed] [Google Scholar]
- 23.Pankey GA, Ashcraft DS. The detection of synergy between meropenem and polymyxin B against meropenem-resistant Acinetobacter baumannii using Etest and time–kill assay. Diagn Microbiol Infect Dis 2009; 63: 228–32. [DOI] [PubMed] [Google Scholar]
- 24.Sandri AM, Landersdorfer CB, Jacob J et al. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 2013; 57: 524–31. [DOI] [PubMed] [Google Scholar]
- 25.Dubrovskaya Y, Prasad N, Lee Y et al. Risk factors for nephrotoxicity onset associated with polymyxin B therapy. J Antimicrob Chemother 2015; 70: 1903–7. [DOI] [PubMed] [Google Scholar]
- 26.Nation RL, Velkov T, Li J. Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin Infect Dis 2014; 59: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogue JM, Lee J, Marchaim D et al. Incidence of and risk factors for colistin-associated nephrotoxicity in a large academic health system. Clin Infect Dis 2011; 53: 879–84. [DOI] [PubMed] [Google Scholar]
- 28.Cannon JP, Lee TA, Clark NM et al. The risk of seizures among the carbapenems: a meta-analysis. J Antimicrob Chemother 2014; 69: 2043–55. [DOI] [PubMed] [Google Scholar]
- 29.Garonzik SM, Li J, Thamlikitkul V et al. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother 2011; 55: 3284–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodford N, Ellington MJ, Coelho JM et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 2006; 27: 351–3. [DOI] [PubMed] [Google Scholar]
- 31.Gadsby NJ, McHugh MP, Russell CD et al. Development of two real-time multiplex PCR assays for the detection and quantification of eight key bacterial pathogens in lower respiratory tract infections. Clin Microbiol Infect 2015; 21: 788.e1-e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenhard JR, von Eiff C, Hong IS et al. Evolution of Staphylococcus aureus under vancomycin selective pressure: the role of the small-colony variant phenotype. Antimicrob Agents Chemother 2015; 59: 1347–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ly NS, Bulitta JB, Rao GG et al. Colistin and doripenem combinations against Pseudomonas aeruginosa: profiling the time course of synergistic killing and prevention of resistance. J Antimicrob Chemother 2015; 70: 1434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munz MR, Bulitta JB, Höhl R et al. Individualizing meropenem prolonged infusions in intensive care unit patients via population modeling of renal function and infection biomarkers over time. In: Paracelsus Science Get Together, Nürnberg, Germany, 2015. [Google Scholar]
- 35.Cheah SE, Bulitta JB, Li J et al. Development and validation of a liquid chromatography-mass spectrometry assay for polymyxin B in bacterial growth media. J Pharm Biomed Anal 2014; 92: 177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaser J. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. J Antimicrob Chemother 1985; 15 Suppl A: 125–30. [DOI] [PubMed] [Google Scholar]
- 37.Dudhani RV, Turnidge JD, Nation RL et al. fAUC/MIC is the most predictive pharmacokinetic/pharmacodynamic index of colistin against Acinetobacter baumannii in murine thigh and lung infection models. J Antimicrob Chemother 2010; 65: 1984–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen EI, Friberg LE. Pharmacokinetic-pharmacodynamic modeling of antibacterial drugs. Pharmacol Rev 2013; 65: 1053–90. [DOI] [PubMed] [Google Scholar]
- 39.Bauer RJ, Guzy S, Ng C. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J 2007; 9: E60–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulitta JB, Yang JC, Yohonn L et al. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob Agents Chemother 2010; 54: 2051–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulitta JB, Ly NS, Yang JC et al. Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009; 53: 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bulitta JB, Landersdorfer CB, Forrest A et al. Relevance of pharmacokinetic and pharmacodynamic modeling to clinical care of critically ill patients. Curr Pharm Biotechnol 2011; 12: 2044–61. [DOI] [PubMed] [Google Scholar]
- 43.Bulitta JB, Bingolbali A, Shin BS et al. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J 2011; 13: 201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulitta JB, Landersdorfer CB. Performance and robustness of the Monte Carlo importance sampling algorithm using parallelized S-ADAPT for basic and complex mechanistic models. AAPS J 2011; 13: 212–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuji BT, Okusanya OO, Bulitta JB et al. Application of pharmacokinetic-pharmacodynamic modeling and the justification of a novel fusidic acid dosing regimen: raising Lazarus from the dead. Clin Infect Dis 2011; 52 Suppl 7: S513–9. [DOI] [PubMed] [Google Scholar]
- 46.Bulitta JB, Okusanya OO, Forrest A et al. Population pharmacokinetics of fusidic acid: rationale for front-loaded dosing regimens due to autoinhibition of clearance. Antimicrob Agents Chemother 2013; 57: 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landersdorfer CB, Ly NS, Xu H et al. Quantifying subpopulation synergy for antibiotic combinations via mechanism-based modeling and a sequential dosing design. Antimicrob Agents Chemother 2013; 57: 2343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulitta JB, Ly NS, Landersdorfer CB et al. Two mechanisms of killing of Pseudomonas aeruginosa by tobramycin assessed at multiple inocula via mechanism-based modeling. Antimicrob Agents Chemother 2015; 59: 2315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav R, Landersdorfer CB, Nation RL et al. Novel approach to optimize synergistic carbapenem-aminoglycoside combinations against carbapenem-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 2015; 59: 2286–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Craig WA. Interrelationship between pharmacokinetics and pharmacodynamics in determining dosage regimens for broad-spectrum cephalosporins. Diagn Microbiol Infect Dis 1995; 22: 89–96. [DOI] [PubMed] [Google Scholar]
- 51.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fourth Informational Supplement M100-S24. CLSI, Wayne, PA, USA, 2014. [Google Scholar]
- 52.EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 5.0, April 2015. http://www.eucast.org/clinical_breakpoints/.
- 53.Fishbain J, Peleg AY. Treatment of Acinetobacter infections. Clin Infect Dis 2010; 51: 79–84. [DOI] [PubMed] [Google Scholar]
- 54.Kaye KS, Pogue JM, Tran TB et al. Agents of last resort: polymyxin resistance. Infect Dis Clin North Am 2016; 30: 391–414. [DOI] [PubMed] [Google Scholar]
- 55.Cotta MO, Gowen B, Truloff N et al. Even high-dose extended infusions may not yield desired concentrations of β-lactams: the value of therapeutic drug monitoring. Infect Dis (Lond) 2015; 47: 739–42. [DOI] [PubMed] [Google Scholar]
- 56.Bulik CC, Quintiliani R Jr, Samuel Pope J et al. Pharmacodynamics and tolerability of high-dose, prolonged infusion carbapenems in adults with cystic fibrosis—a review of 3 cases. Respiratory Medicine CME 2010; 3: 146–9. [Google Scholar]
- 57.Taccone FS, Cotton F, Roisin S et al. Optimal meropenem concentrations to treat multidrug-resistant Pseudomonas aeruginosa septic shock. Antimicrob Agents Chemother 2012; 56: 2129–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tam VH, Louie A, Deziel MR et al. The relationship between quinolone exposures and resistance amplification is characterized by an inverted U: a new paradigm for optimizing pharmacodynamics to counterselect resistance. Antimicrob Agents Chemother 2007; 51: 744–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pike M, Saltiel E. Colistin- and polymyxin-induced nephrotoxicity: focus on literature utilizing the RIFLE classification scheme of acute kidney injury. J Pharm Pract 2014; 27: 554–61. [DOI] [PubMed] [Google Scholar]
- 60.Horiuchi M, Kimura M, Tokumura M et al. Absence of convulsive liability of doripenem, a new carbapenem antibiotic, in comparison with β-lactam antibiotics. Toxicology 2006; 222: 114–24. [DOI] [PubMed] [Google Scholar]
- 61.Merrem Package Insert. Wilmington, DE, USA: AstraZeneca Pharmaceuticals, Revised December 2013. [Google Scholar]
- 62.Zivanovic D, Stanojlovic O, Susic V et al. The effects of phenytoin and phenobarbital on seizures induced by imipenem/cilastatin in rats. Acta Neurol Belg 2004; 104: 20–6. [PubMed] [Google Scholar]
- 63.Maidhof H, Johannsen L, Labischinski H et al. Onset of penicillin-induced bacteriolysis in staphylococci is cell cycle dependent. J Bacteriol 1989; 171: 2252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bulitta JB, Li J, Poudyal A et al. Quantifying synergy of colistin combinations against MDR Gram-negatives by mechanism-based models. In: Abstracts of the Forty-ninth Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, USA, 2009. Abstract A1-573 American Society for Microbiology, Washington, DC, USA. [Google Scholar]
- 65.Kadurugamuwa JL, Clarke AJ, Beveridge TJ. Surface action of gentamicin on Pseudomonas aeruginosa. J Bacteriol 1993; 175: 5798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kadurugamuwa JL, Lam JS, Beveridge TJ. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effect. Antimicrob Agents Chemother 1993; 37: 715–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bulitta JB, Duffull SB, Kinzig-Schippers M et al. Systematic comparison of the population pharmacokinetics and pharmacodynamics of piperacillin in cystic fibrosis patients and healthy volunteers. Antimicrob Agents Chemother 2007; 51: 2497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.