Abstract
Background
The clinical development of antimicrobial peptides (AMPs) is currently under evaluation to combat the rapid increase in MDR bacterial pathogens. However, many AMPs closely resemble components of the human innate immune system and the ramifications of prolonged bacterial exposure to AMPs are not fully understood.
Objectives
We show that in vitro serial passage of a clinical USA300 MRSA strain in a host-mimicking environment containing host-derived AMPs results in the selection of stable AMP resistance.
Methods
Serial passage experiments were conducted using steadily increasing concentrations of LL-37, PR-39 or wheat germ histones. WGS and proteomic analysis by MS were used to identify the molecular mechanism associated with increased tolerance of AMPs. AMP-resistant mutants were characterized by measuring in vitro fitness, AMP and antibiotic susceptibility, and virulence in a mouse model of sepsis.
Results
AMP-resistant Staphylococcus aureus mutants often displayed little to no fitness cost and caused invasive disease in mice. Further, this phenotype coincided with diminished susceptibility to both clinically prescribed antibiotics and human defence peptides.
Conclusions
These findings suggest that therapeutic use of AMPs could select for virulent mutants with cross-resistance to human innate immunity as well as antibiotic therapy. Thus, therapeutic use of AMPs and the implications of cross-resistance need to be carefully monitored and evaluated.
Introduction
Antibiotic resistance is one of the most urgent threats facing modern medicine. Many pathogens are acquiring resistance mechanisms at an alarming rate making options for treatment limited.1 Antimicrobial peptides (AMPs) are currently under evaluation as a therapeutic solution to treat MDR pathogens.2,3 AMPs are a diverse class of small, typically cationic peptides, which display broad-spectrum bactericidal activity. All kingdoms of life (bacteria, archaea and eukaryotes)4 produce AMPs as defence mechanisms to protect against viral, bacterial, protozoan and fungal infections. The antibacterial activity of AMPs can be direct by pore formation, which occurs following the interaction of positively charged, amphipathic AMPs with negatively charged bacterial cell wall components such as LPS in Gram-negative and teichoic acids (TA) in Gram-positive bacteria.5 AMPs have also displayed bactericidal effects following translocation across the cell wall followed by inhibition of essential cellular processes.6
As a result of this broad-spectrum bactericidal activity, at least 10 AMPs are in various stages of clinical development, the majority of which are indicated for topical use.7 Several of these AMPs are derivatives of components of the human innate immune system. OP-145 (also known as P60.4Ac8), derived from the human LL-37 defence peptide, is currently in phase 2 clinical trials to treat chronic bacterial middle ear infections.9 LL-37 is the only human cathelicidin AMP, is a key component of the human innate immune system and serves as a first line of defence against many bacterial pathogens.10,11 The human-derived lactoferrin 1-11 (hLF1–11) is the only AMP in clinical trials indicated for intravenous administration to treat life-threatening infections in stem cell transplantation patients.12 Although no human-derived AMPs have been FDA approved, routine administration of such AMPs would result in prolonged bacterial exposure ultimately selecting for resistant mutants.13–15
Resistance development to AMPs was initially thought to be unlikely due to the fact that these peptides kill bacteria rapidly and typically have multiple cellular targets.16 However, it has become increasingly evident that bacteria are able to evade AMP killing through a variety of mechanisms including cell surface modifications, external sequestering of AMPs, efflux, proteolytic degradation and even by down-regulation of host AMP production (reviewed in detail by Nizet17). These modifications are often reversible (not stable) and only expressed under specific environmental conditions to enable bacterial pathogens to evade AMP killing during brief AMP exposure in a host. However, the mechanisms of stable resistance to AMPs are much less understood. Several recent studies have shown that serial passage of bacterial pathogens in the presence of AMPs selects for stably acquired resistance, which typically occurred at a substantial fitness cost to the bacterium.13–15,18 These studies support the notion that AMP resistance can be acquired in bacteria to yield cross-resistant pathogens. However, AMP exposure was performed in rich medium conditions that do not accurately reflect a natural environment in which AMPs would be encountered. The influence that the host environment may have on the development of stable AMP resistance has not been characterized.
The purpose of this study was to evaluate the consequences of prolonged AMP exposure under conditions more similar to what would occur if AMPs were used therapeutically. The pathogen we selected for our studies is one of the largest burdens on modern medicine, MRSA.19,20 This opportunistic pathogen is often associated with multidrug resistance, which greatly increases morbidity, mortality and hospital-associated costs.21 With limited therapeutic options for XDR isolates, clinical use of AMPs to treat MRSA infections has been proposed.7 However, with little information available about the development of resistance to these agents, the ramifications of widespread clinical use are largely unknown. Here, we describe an evolution experiment conducted in mammalian ionic environment medium (MIEM) in which a human MRSA USA300 isolate was serially passaged in the presence of three different AMPs derived from humans, pigs and plants.
Materials and methods
Bacterial strains, growth conditions and peptides
Bacterial strains used in this study are derivatives of the highly characterized community-associated MRSA USA300 strain isolated from the Los Angeles County jail (LAC)22 cured of several small plasmids to yield strain JE2, which has been described previously23 (termed DA28823 in our collection). Insertional mutations were obtained from the Nebraska Transposon Mutant Library.24,25 The primary growth medium used in this study is a MIEM as described by Dorschner et al.26 made without NaCl and adjusted to pH 7.4. Other bacterial growth media conditions include tryptic soy broth/agar (TSB/A) for routine culturing, Lysogeny broth (LB) for in vitro fitness assays and Mueller–Hinton II (cation-adjusted) broth/agar for antibiotic susceptibility testing. All strains were grown at 37°C. Dilutions and bacterial cell washes were performed using sterile PBS (13 mM phosphate with 137 mM NaCl at pH 7.4). For haemolysis assessments, bacterial strains were streaked on TSA plates containing 5% v/v veal blood and incubated for 24 h. AMPs were obtained from Innovagen AB (Lund, Sweden) with the exception of a mixture of wheat germ histones (referred to as WGH), which were a kind gift from Lars-Olof Hedén (Lund University, Sweden) and isolated as described.27 CNY100HL, a modified AMP derived from the human C3 complement peptide CNY21,28 was also obtained from Innovagen. Amino acid sequences of all peptides used in this study are listed in Table S1 (available as Supplementary data at JAC Online).
Serial passage experiment with AMPs
The serial passage experiment was done with WT MRSA JE2 (DA28823). The experiment was initiated at a peptide concentration equal to a growth reduction of 30% for the DA28823 strain with the respective peptide, as measured in a Bioscreen C reader (Oy Growth Curves AB Ltd, Finland) with MIEM. The following concentrations of peptides were used as starting points for the serial passage experiment: 0.75 mg/L of PR-39, 50 LL-37, 25 mg/L WGH and a combination of 6.25 mg/L WGH with 12.5 mg/L LL-37. WT MRSA JE2 was serially passaged without peptide as a media-adaptation control. The experiment was initiated by inoculating eight linages of 100 μL of MIEM (with or without peptide) with 1 μL from eight independent overnight cultures (from eight single colonies) for each peptide treatment (or control without peptide) in a 96-well plate (Thermo Fisher Scientific Nunc A7S, Roskilde, Denmark). The cell cultures were passaged (1 μL in 100 μL fresh medium with or without peptide) every day for 3–4 days with the initial peptide concentration. On the fourth or fifth day the peptide concentration was increased 50% and the cell cultures where serial passaged for another 3–4 days. In the case of poor cell growth the cell volume transferred was either increased from 1 to 2–5 μL or the concentration of peptide was reduced 50% to continue passage in the previous concentration. If no growth was observed the culture was re-inoculated from the previous day (each daily plate was saved at −80°C). These steps were repeated until the initial concentration of peptide was increased 10 times for LL-37 (500 mg/L) and WGH (250 mg/L), 12 times for the combination treatment of WGH/LL-37 (75 and 150 mg/L) and 23 times for PR-39 (17 mg/L). At various timepoints throughout the cycling, single colonies were isolated on LB agar from two cell lineages per peptide, which were then whole-genome sequenced. Four control populations passaged in MIEM without peptide were collected at the termination of the experiment as media-adapted controls.
MS analysis
The Proteomics Core Facility at Sahlgrenska Academy (Gothenburg University) performed relative quantification of peptides. All proteomics analyses were performed with two biological replicates of each strain. Peptide levels for each strain are normalized on a protein median of the WT MRSA JE2 (DA28823) replicates. A detailed description of the MS analysis is available as Supplementary data at JAC Online. All proteomics fold change data and statistical analysis are also available as Supplementary data at JAC Online. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE29 partner repository with the dataset identifier PXD004036.
Antimicrobial susceptibility assays
MIC determination
The MIC was determined using the broth microdilution method30,31 for AMPs and by Etest for antibiotics. In both cases, the MIC was interpreted as the lowest concentration at which no bacterial growth was observed. Values given are the consensus MIC derived from at least two independent determinations.
Microdilution
MIC measurements for AMPs were performed using the broth microdilution method in MIEM using 96-well plates (round bottom; Nunc A/S). A total volume of 100 μL with a bacterial inoculum of 5 × 105 cfu/mL was incubated with various dilutions of peptides at 37°C for 18–20 h without shaking.
Etest
WT MRSA JE2 (DA28823) and mutants strains were grown overnight shaking at 37°C in Mueller–Hinton II broth and then diluted to ∼108 cfu/mL in Mueller–Hinton II broth and distributed on Mueller–Hinton II agar using a sterile cotton swab according to manufacturer's instructions. An Etest strip (bioMérieux, Marcy-l′Étoile, France) was applied to the plate and then incubated at 37°C for 18–20 h.
Bacterial cell survival assays
Bacterial strains were grown overnight shaking at 37°C in TSB.
LL-37
Stationary phase cells were washed once and re-suspended in 10 mM sodium phosphate buffer at pH 7.4 (termed NaPB). Cells were subsequently diluted in NaPB to ∼5 × 105 cfu/mL, combined with 3.3 mg/L LL-37 in a final volume of 100 μL and incubated in 96-well plates in a sealed and humidified container shaking at 37°C for 2 h. Bacteria were plated for cfu on TSA. Following overnight incubation at 37°C, viable cells were enumerated and percentage cell survival was calculated as cfuwith peptide/cfuno peptide × 100 at 2 h post-peptide exposure; values given are the mean ± SEM derived from at least three independent determinations.
Human defensins
Exponential phase cells were obtained by diluting overnight grown cells 1:100 in pre-warmed TSB and shaking at 37°C for 2 h. Bacterial cells were washed once and re-suspended in NaPB. Cells were subsequently diluted further in NaPB to 0.69−4.7 × 104 cfu/mL, combined with 1 μM (hBD3) or 10 μM (all others) of peptide in a final volume of 100 μL and incubated in 96-well plates in a sealed and humidified container shaking at 37°C for 2 h. Bacteria were plated and percentage cell survival was calculated as for LL-37.
Assessment of membrane potential using flow cytometry
Differences in membrane potential between the strains were estimated using a flow cytometry assay based on the BacLight Bacterial Membrane Potential Kit according to the manufacturer (Life Technologies®, cat. no. B34950). Briefly, bacterial strains were grown overnight shaking at 37°C in TSB. Fifty μL of cells from each overnight culture was inoculated into 10 mL of TSB in 100 mL Erlenmeyer flasks and grown with shaking at 37°C to an OD600 of 0.2. Then 15 μL of culture was transferred to 1 mL of PBS and 10 μL of fluorescent membrane potential indicator dye, DiOC2(3) was added. Cells were stained for 30 min at room temperature followed by analysis on a BD Biosciences Accuri C6 flow cytometer (Becton, Dickinson and Company), with emission filters suitable for detecting red and green fluorescence. Fifty-thousand events were recorded at a forward scatter threshold of 15 000 and medium flow rate. Gating of stained cell population and analysis of flow cytometry data were performed in CFlow® (Becton, Dickinson and Company). As an indicator of membrane potential, the ratio of red to green fluorescence intensity was calculated. The assay was verified using the protonophore CCCP and two strains derived from the Nebraska Transposon Mutant Library25 that display reduced membrane potential (ΔmenD, NE1345 and ΔhemB, NE184532).
Fitness measurements
At least four independent overnight cultures of each bacterial strain were grown overnight in each of three different medium conditions: MIEM, TSB or LB. Cells were diluted 1:1000 in the same medium as overnight growth and 300 μL was transferred into BioscreenC plates. Growth of the samples was monitored at 37°C with shaking for 16–20 h in a BioscreenC Analyzer (Oy Growth Curves AB Ltd) with OD600 measurements taken every 4 min. Calculations of maximum growth rate were based on OD600 values where growth was observed to be exponential. Relative growth rates were calculated as generation timeWT/generation timemutant. Values given are the mean ± SEM derived from at least four independent determinations performed in technical triplicates.
Carotenoid pigment extraction
Bacterial strains were grown overnight shaking at 37°C in TSB for 24 h. Methanol extraction of carotenoid was performed as previously described33 with the modification that 750 μL of the culture was harvested by centrifugation and cells were suspended in 500 μL of methanol. OD465 was measured with a spectrophotometer (GENESYS 10vis; Thermo Scientific). Values presented are the mean ± SEM derived from three independent determinations.
Ethics
The Uppsala Animal Experiments Ethics Review Board in Uppsala, Sweden approved all mouse protocols undertaken in this study under reference no. 154/14.
Mouse infections
Female BALB/c mice (6–8 weeks old) were used in all virulence studies (Charles River Laboratories, distributed by Scanbur). A single colony of each Staphylococcus aureus strain was grown overnight in TSB, then diluted 1:100 in pre-warmed TSB and incubated shaking at 37°C for 2 h to achieve exponential phase growth. Cells were pelleted, washed once in and re-suspended in PBS (13 mM phosphate with 137 mM NaCl at pH 7.4). Female BALB/c mice were administered 100 μL containing 0.5–2 × 107 cfu via the intraperitoneal route. Three to four mice were used per bacterial strain. Eight hours post-infection, mice were humanely sacrificed by cervical dislocation, bacterial cells were recovered from the spleen, liver and kidney of acutely infected animals and enumerated by direct colony count following plating on TSA with overnight incubation at 37°C. Values given are normalized per gram of tissue (cfu/g).
Statistical analyses
General
Data were analysed as mean ± SEM unless otherwise stated. Raw data (membrane potential, relative fitness and carotenoid pigment) or log-transformed data (bacterial survival) were analysed using one-way analysis of variance (ANOVA) with Fisher's least significant difference test used for post hoc analysis to determine statistical significance relative to WT. Bacterial viability counts (cfu/g in tissue) were analysed relative to WT (DA28823) using the Mann–Whitney U-test. For all statistical analyses of P < 0.05, values were considered significant and the degrees of statistical significance are presented as ***P < 0.001, **P < 0.01 or *P < 0.05.
Proteomics
We used the relative fold change in protein abundance (determined as described in the MS analysis section) to calculate the mean of the biological duplicates for each strain. These average fold change values were subject to a log transformation and, to adjust for sample variation, we standardized the values over each strain. We then performed t-tests between the WTs and the mutants to identify differentially expressed accessions. The corresponding P values were corrected for multiple testing using the Benjamini–Hochberg method to control for false discovery rate. Since no accessions had a false discovery rate <0.05 we selected the accessions with an unadjusted P < 0.05 to explore pathways and clusters. We used DAVID Bioinformatics Resources34,35 to find possible pathways for our dataset and the GENE-E package in R36 to create heatmaps.
Results and discussion
Prolonged AMP exposure results in a stable AMP-resistant phenotype
To assess the ability of S. aureus to become stably resistant to AMPs, the community-associated MRSA USA300 LAC JE2 strain was serially passaged in steadily increasing concentrations of three different host-derived AMPs. The peptides used for selection were the human cathelicidin peptide LL-37,2 a mixture of histones and histone peptides derived from wheat germ (WGH) shown previously to have antimicrobial properties,37 and a well-characterized cathelicidin defence peptide PR-39 derived from pigs.38 These three peptides were chosen to represent various AMP structures and host species. Experimental evolution was conducted using MIEM to simulate a mammalian epithelial environment.26 Seven AMP-resistant single colony isolates that were derived from independently passaged lineages were selected for further analysis.
Stable resistance evolved rapidly within as few as 168 generations. The mutants isolated displayed increased resistance of up to 16-fold using broth microdilution to determine the MIC of peptide in MIEM (Table 1). Cross-resistance was also observed for all of the mutants isolated when tested against peptides other than that were used in serial passage. We also tested susceptibility to a modified human C3 complement CNY21 derivative termed CNY100HL28 and found that all mutants also displayed a slight reduction in susceptibility to this human complement peptide, another component of the innate immune system. The control strains passaged in MIEM without peptide (media-adapted) retained WT susceptibility to all peptides tested.
Table 1.
AMP-resistant S. aureus display cross-resistance to various AMPs
| Strain no. | Passaged with | Approximate no. of generationsa | MIC (mg/L)b |
|||
|---|---|---|---|---|---|---|
| LL-37 | WGH | PR-39 | CNY100HLc | |||
| DA28823 | WT, no passage | none | 192 | 16 | 4 | 24 |
| DA30560 | no peptide | 322 | 192 | 16 | 4 | 24 |
| DA29677 | LL-37 | 189 | >512 | 64 | 4 | 48 |
| DA29680 | LL-37 | 189 | >512 | 128 | 8 | 32 |
| DA30557 | WGH | 322 | >512 | 192 | 12 | 48 |
| DA30559 | WGH | 322 | >512 | 256 | 12 | 32 |
| DA36424 | LL-37 and WGH | 210 | >512 | 192 | 12 | 32 |
| DA35920 | PR-39 | 168 | >512 | 128 | 8 | 32 |
| DA37296 | PR-39 | 168 | >512 | 96 | 8 | 32 |
aCalculated based on number of generations achieved in MIEM without peptide.
bMIC was determined in MIEM using the broth microdilution method.
cCNY100HL is a modified AMP derived from the human C3 complement peptide CNY21.28
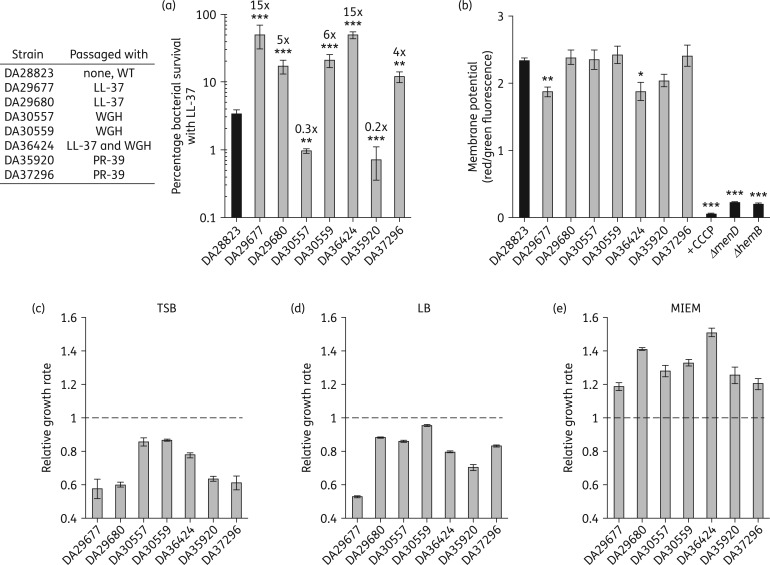
The solubility limit of LL-37 in MIEM (∼550 mg/L) hindered determination of resistance levels by MIC; therefore, the full extent of LL-37 resistance was assessed using time–kill assays (Figure 1a). Two mutants passaged with LL-37 displayed a 15-fold increase in LL-37 resistance, which was associated with a reduced membrane potential compared with WT (Figure 1b). This parallels the observation that small-colony variant (SCV) S. aureus, which are known to have severely defective membrane potential, display reduced susceptibility to human AMPs.39 To determine whether high-level resistance coincided with a growth defect, as is seen with SCVs, we determined the fitness cost of AMP resistance in MIEM, TSB and LB conditions relative to the WT strain (DA28823). A growth defect was observed for all AMP-resistant mutants in both rich medium conditions, TSB and LB (Figure 1c and d). However, in MIEM conditions all AMP-resistant mutants grew significantly faster than the non-passaged WT strain (Figure 1e). These results indicate that high-level AMP resistance can coincide with adaptation to growth in mammalian environmental medium conditions and thereby occur in the absence of a fitness cost.
Figure 1.
AMP resistance in S. aureus can evolve without a fitness cost. (a) Extent of LL-37 resistance was determined by calculating percentage bacterial survival after LL-37 exposure. Fold changes over WT (DA28823) are indicated above each bar. (b) Differences in membrane potential are presented as the ratio of red/green fluorescence intensity following incubation with the fluorescent membrane potential indicator dye DiOC2(3). Assay was verified using the protonophore CCCP and two strains derived from the Nebraska Transposon Mutant Library that display reduced membrane potential (ΔmenD, NE1345 and ΔhemB, NE184532). Fitness was determined in (c) TSB, (d) LB or (e) MIEM growth conditions. Relative fitness is presented as the maximum exponential generation time relative to WT with the broken line representing the relative WT growth rate. All values given are the mean ± SEM derived from at least three independent determinations. Relative fitness assays were also performed in technical triplicate. Statistical significance relative to WT (DA28823) was determined by one-way ANOVA with Fisher's least significant difference test used for post hoc analysis (***P < 0.001, **P < 0.01 or *P < 0.05). All relative fitness values were statistically significant with P < 0.001.
Reduced AMP uptake results in stable AMP resistance in S. aureus
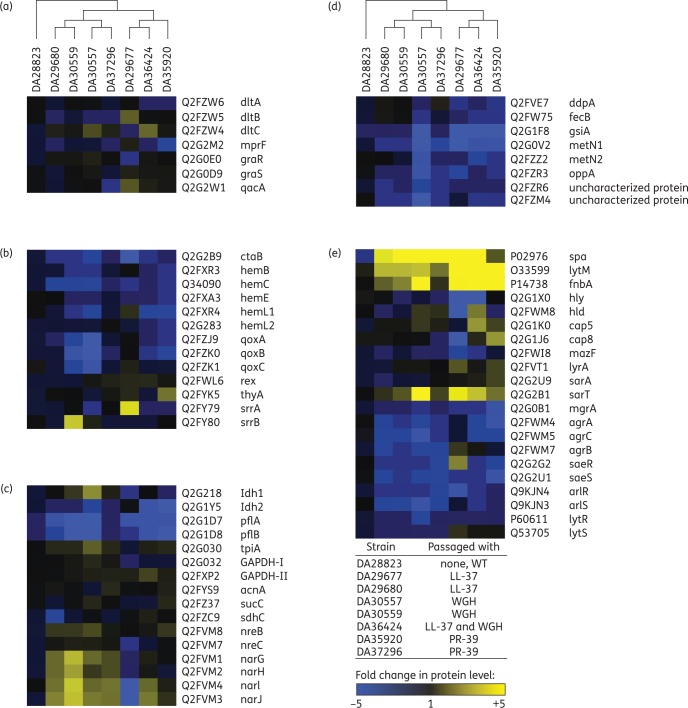
Bacteria are able to resist the killing of AMPs through membrane modifications (reducing the net negative charge), active efflux, proteolytic degradation and reduced AMP uptake. To identify the mechanism by which all seven S. aureus mutants convey a stable AMP-resistant phenotype in the absence of selective pressure, each mutant was subject to both whole-proteome analysis by MS (∼2000 proteins) and WGS and compared with the WT, AMP-susceptible parental strain (DA28823).
No sequence alterations were found in common S. aureus AMP resistance determinants (dltABC, mprF, graRS, qacA; Table S2).40 Three (of seven) mutants displayed increased DltB or DltC expression (Figure 2a); however, the other factors remained unchanged or displayed reduced expression in all seven mutants. Instead, protein analysis revealed two major patterns that all seven AMP-resistant mutants displayed but were not observed in the AMP-susceptible WT (DA28823) or media-adapted control strain (cycled without peptide, DA30560), i.e. (i) reduced haem/menaquinone biosynthesis, and (ii) reduced expression of various transporters.
Figure 2.
Stable AMP resistance is conferred by reduced AMP uptake. Heatmaps of selected protein expression profiles grouped into categories based on their biological roles: (a) AMP resistance determinants; (b) electron transport chain; (c) metabolism; (d) transporters; and (e) virulence factors. S. aureus strains (AMP-resistant mutants or WT) are indicated above each column and protein expression is shown in rows. Proteomics analysis was performed using MS in bacterial cells derived from exponential growth in TSB medium in the absence of AMP selection as described in the Materials and methods section. Each column represents the average fold change calculated relative to WT (DA28823) for the indicated protein. All expression experiments were performed with two biological replicates. Changes in protein expression levels are represented in colour: yellow indicates increased protein levels and blue indicates decreased protein levels relative to WT. Data are available via ProteomeXchange with identifier PXD004036.
Defects in electron transport have been previously associated with reduced uptake of AMPs and consequently AMP resistance in S. aureus mutants displaying a SCV phenotype.41 All seven AMP-resistant mutants displayed slight reductions in the expression of components in the menaquinone or haem biosynthesis pathways required for synthesizing electron transport chain components (Figure 2b). An electrochemical gradient is necessary for the import of positively charged molecules such as AMPs and aminoglycosides. Therefore, reduced electron transport is a mechanism by which stable AMP resistance can be achieved in S. aureus.
None of the seven AMP-resistant mutants display genetic mutations (i.e. haemin/menadione biosynthesis, cytochrome c assembly, thyA) or growth defects commonly associated with an SCV phenotype. With a complete interruption of the electron transport chain, neither oxygen nor nitrate would be utilized as electron acceptors and there would be an up-regulation of anaerobic respiration. Contrarily, these AMP-resistant mutants display a reduced expression of fermentation enzymes (ldh1 and ldh2) and anaerobic metabolism (pflA, pflB) while expression of glycolysis and citric acid cycle components remained unaltered (tpiA, gapdh, acnA, sucC and sdhC). Nitrate respiration (narGHIJ) was also increased in six of seven AMP-resistant mutants (Figure 2c). Therefore, there does not appear to be a complete disruption in electron transport chain in these AMP-resistant mutants.
ATP-binding cassette (ABC) transporters actively transport various substrates across cell membranes. In bacteria, ABC transporters are involved in many processes including nutrient acquisition, environmental sensing, virulence (adhesion and protein secretion) and antimicrobial resistance (efflux and influx).42 All seven AMP-resistant S. aureus mutants displayed reduced expression of ABC transporters involved in transferring peptides (ddpA, gsiA, oppA, hypothetical protein Q2FZR6), iron (fecB, hypothetical protein QRFZM4) and amino acids (metN1, metN2) across the bacterial membrane (Figure 2d). Reducing cellular uptake of peptides is a direct mechanism by which AMP resistance can be achieved in S. aureus to convey a stable cross-protective phenotype against a variety of AMPs. Reduced iron uptake can indirectly confer AMP resistance by resulting in a disruption of many cellular processes including assembly of the electron transport chain.43
The genetic basis of reduced AMP uptake in AMP-resistant S. aureus mutants appears to be multifactorial. Each mutant was found to contain between one and three mutations relative to the sequence of our WT JE2 strain, the vast majority of which were single amino acid substitutions (Table S2). Mutations were identified in a variety of cellular pathways including global regulators (srrA, srrB, walK, rpoB, phoR), nucleotide synthesis/signalling (gmk, guaA), nutrient uptake (ptsI, trkA) and virulence factors (cspA, ebh, mazF). Protein expression profiles indicated a repression of staphylococcal respiratory response (srrAB, also known as srhSR) regulated genes in all seven AMP-resistant mutants of S. aureus (Figure 2b). SrrAB is a two-component regulatory system involved in sensing and maintaining the electron transport chain by activating cytochrome and quinol-oxidase assembly (ctaB, cydAB, qoxABCD) and haem biosynthesis (hemACDX) in response to impaired electron flow.44
Reconstruction of these mutations singly in a clean background failed due to the inherent difficulty in S. aureus allelic replacement and lack of an easily selectable phenotype for transduction (i.e. AMP activity is highly dependent on medium conditions and cost-prohibitive). Insertional knockouts of genes that were available from the Nebraska Transposon Mutant Library24,25 did not display AMP resistance (data not shown). Therefore, the individual contribution of each mutation towards the AMP resistance phenotype and/or adaptation to growth in MIEM is unknown.
Media adaptation in control versus AMP-resistant populations
Mutations were identified in three of seven AMP-resistant mutants in the uncharacterized gene SAUSA300_1112 (predicted to encode a phosphatase 2C domain-containing protein). Two of these mutations result in inactivation via frameshift or introduction of an early stop codon. However, an insertional knockout mutant of SAUSA300_1112 from the Nebraska Transposon Mutant Library does not display AMP resistance (data not shown) indicating a likely role of this mutation in adaptation to growth in MIEM.
Interestingly, none of the four media-adapted controls passaged without peptide contained mutations in SAUSA300_1112 (Table S3). The four independent control populations passaged in MIEM without peptide displayed a very different mutational profile. In fact, only one AMP-resistant mutant (DA30557) contained a mutation in a gene that was identified in the control populations and could be attributed to MIEM adaptation (SAUSA300_2326, a transcription regulatory protein). Instead, all four control populations contained mutations in either the SaeRS and/or ArlRS two-component virulence regulators, the majority of which included either a frame shift or early stop codon indicating an inactivation of these global virulence regulators. None of the AMP-resistant mutants displayed mutations in either of these virulence determinants. These findings indicate that the selective pressure imposed by the presence of AMPs results in alternate pathways for adaptation in MIEM conditions.
AMP resistance reduces susceptibility to clinically prescribed antibiotics
The impact of AMP resistance on the susceptibility of S. aureus to antibiotics was tested against 15 drugs representing five classes indicated for the treatment of MRSA infections (Table 2). MICs were determined using Mueller–Hinton medium conditions, which are the clinically accepted standards for determination of antimicrobial susceptibility. All seven AMP-resistant mutants displayed diminished susceptibility to teicoplanin, a glycopeptide antibiotic structurally related to vancomycin, yet all seven mutants displayed little to no change in susceptibility to vancomycin. All seven AMP-resistant mutants were also less susceptible to daptomycin, a lipopeptide antibiotic. Cross-resistance between daptomycin and human defence peptides in MRSA has been previously observed.45 DA29677 (passaged with LL-37) and DA35920 (passaged with PR-39) displayed an 8- and 10-fold increase in daptomycin MIC, respectively, which is approaching clinical resistance (>1 mg/L) according to breakpoints defined by EUCAST. Four of seven mutants also displayed reduced susceptibility to gentamicin, a protein synthesis inhibitor, with an 8-fold increase in MIC for DA30557 (passaged with WGH), which is also at the clinical resistance level (>1 mg/L, EUCAST). Additionally, AMP-resistant S. aureus also displayed diminished susceptibility to tetracycline (six of seven mutants), polymyxin B (three of seven), erythromycin (two of seven) and tigecycline (two of seven). These results indicate that AMP-resistant S. aureus are less susceptible to clinically prescribed antibiotics from three of the five drug classes tested, which will further limit effective therapeutic options to treat these infections.
Table 2.
AMP-resistant S. aureus display increased resistance to clinically prescribed antibiotics
| Strain no. | Cycled with | Peptidoglycan synthesis inhibitors/clinical breakpointa (mg/L) |
Detergent-like peptide antibiotics/clinical breakpointa (mg/L) |
Protein synthesis inhibitors/clinical breakpointa (mg/L) |
Folate synthesis inhibitors/clinical breakpointa (mg/L) | RNA synthesis inhibitor/clinical breakpointa (mg/L) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VAN |
TEC |
CFT |
DAP |
PMB |
GEN |
CHL |
CLI |
ERY |
LZD |
TGC |
TET |
DOX |
SXT |
RIF |
||
| R >2 | R > 2 | R > 1 | R > 1 | ND | R > 1 | R > 8 | R > 0.5 | R > 2 | R > 4 | R > 0.5 | R > 2 | R > 2 | R > 4 | R > 0.5 | ||
| MIC (mg/L)b | ||||||||||||||||
| DA28823 | WT | 1.5 | 0.5 | 0.25 | 0.094 | 96 | 0.125 | 4 | 0.094 | 0.19 | 2 | 0.094 | 0.094 | 0.19 | 0.094 | 0.012 |
| DA30560 | no peptide | 1.5 | 0.5 | 0.25 | 0.094 | 64 | 0.125 | 4 | 0.094 | 0.19 | 2 | 0.094 | 0.094 | 0.19 | 0.094 | 0.012 |
| DA29677 | LL-37 | 2 | 1.5 | 0.19 | 0.75 | 96 | 0.094 | 6 | 0.047 | 0.19 | 1.5 | 0.094 | 0.19 | 0.125 | 0.047 | 0.008 |
| DA29680 | LL-37 | 2 | 2 | 0.25 | 0.19 | 96 | 0.094 | 4 | 0.094 | 0.19 | 1.5 | 0.19 | 0.25 | 0.094 | 0.064 | 0.012 |
| DA30557 | WGH | 2 | 1 | 0.38 | 0.19 | 192 | 1 | 4 | 0.064 | 0.125 | 2 | 0.094 | 0.25 | 0.094 | 0.064 | 0.012 |
| DA30559 | WGH | 1.5 | 1.5 | 0.38 | 0.19 | 192 | 0.38 | 4 | 0.064 | 0.125 | 1 | 0.094 | 0.19 | 0.125 | 0.094 | 0.016 |
| DA36424 | LL-37 and WGH | 2 | 1.5 | 0.38 | 0.38 | 96 | 0.19 | 4 | 0.125 | 0.25 | 2 | 0.19 | 0.25 | 0.25 | 0.094 | 0.012 |
| DA35920 | PR-39 | 2 | 2 | 0.19 | 1 | 128 | 0.25 | 3 | 0.094 | 0.38 | 1 | 0.125 | 0.125 | 0.094 | 0.094 | 0.016 |
| DA37296 | PR-39 | 2 | 1.5 | 0.25 | 0.38 | 384 | 0.5 | 6 | 0.125 | 0.38 | 2 | 0.125 | 0.19 | 0.125 | 0.064 | 0.008 |
VAN, vancomycin; TEC, teicoplanin; CFT, ceftaroline; DAP, daptomycin; PMB, polymyxin B; GEN, gentamicin; CHL, chloramphenicol; CLI, clindamycin; ERY, erythromycin; LZD, linezolid; TET, tetracycline; TGC, tigecycline; DOX, doxycycline; SXT, trimethoprim/sulfamethoxazole; RIF, rifampicin.
Light grey, 2–5-fold MIC increase; dark grey, >5-fold MIC increase.
aDerived from EUCAST Clinical Breakpoint Table v. 5.0, valid from 1 January 2015 for Staphylococcus spp. (R, resistant; ND, no value determined).
AMP-resistant S. aureus maintain virulence
Antimicrobial resistance in bacterial pathogens has often been associated with a fitness cost and/or alterations in virulence capacity.43,46,47 Virulence factors are required for bacterial survival in a host. In S. aureus, the expression of cell wall adhesins occurs during initial colonization while the production of toxins occurs later during dissemination in tissue.48 Expression of virulence factors in S. aureus is depending on environment and growth phase under the control of many often overlapping global regulatory systems (i.e. agrAC, sarA, srrAB, saeRS, arlRS, lytRS). Protein expression profiles of AMP-resistant S. aureus mutants reveal an increase in colonization-related regulators and virulence factors including immunoglobulin G-binding protein (spa), autolysin (lytM) and fibronectin-binding protein A (fnbA) (Figure 2e).
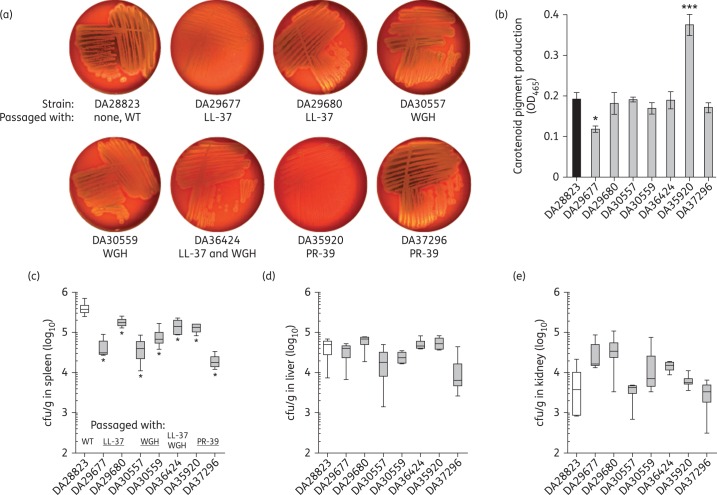
With an increase in colonization factors, we determined whether AMP-resistant S. aureus mutants were capable of expressing virulence factors required for later stages of infection. Haemolysis is considered to be one of the major factors contributing to the transition of S. aureus from a commensal organism to an invasive pathogen.49 To evaluate the haemolytic activity of AMP-resistant S. aureus, we isolated the mutant and WT strains on veal blood agar plates (Figure 3a). The WT displayed both α- and β-haemolysis on veal blood, which are indicated by green and yellow discoloration, respectively. AMP-resistant mutant DA29677 (passaged with LL-37) displayed greatly reduced haemolytic activity, which was confirmed by proteomic analysis (Figure 2e). Four of seven mutants displayed moderate to high-level β-haemolytic activity similar to WT; however, a reduction in α-haemolysis was observed. The PR-39 passaged mutant DA37296 displayed both α- and β-haemolytic activity similar to the virulent WT strain (DA28823) indicating that AMP resistance can be achieved in the absence of a defect in haemolysis.
Figure 3.
AMP-resistant S. aureus are virulent. (a) Haemolytic activity was assessed following isolation on veal blood agar. (b) Carotenoid pigment production was assayed using methanol extraction. Values presented are the mean ± SEM derived from three independent determinations and statistical significance relative to WT was analysed using one-way ANOVA with Fisher's least significant difference test for post hoc analysis (***P < 0.001 or *P < 0.05). To assay in vivo virulence capacity, BALB/c mice were infected intraperitoneally with 0.5–2 × 107 cfu of S. aureus and bacterial cells were enumerated from the (c) spleen, (d) liver and (e) kidney. Values presented are bacterial cfu counts normalized per gram of tissue (cfu/g) from three to four mice per bacterial strain. Boxes show the median and 25th and 75th percentiles, while error bars show the minimum and maximum values. Significant differences in bacterial colonization of AMP-resistant S. aureus strains were determined relative to WT using the Mann–Whitney U-test (*P < 0.05).
Another virulence determinant of S. aureus is its golden pigmentation. The production of yellow carotenoid pigment has been shown to promote virulence by protecting the bacterium from phagocyte killing.50 Levels of carotenoid pigment production were assessed using methanol extraction (Figure 3b) and notably the PR-39 passaged mutant DA35920 displayed a marked elevation in carotenoid pigment production. An increase in carotenoid pigment production in S. aureus has previously been associated with a thickening of the bacterial cell wall resulting in resistance to antibiotics targeting the cell wall,33 which corresponds with the daptomycin and teicoplanin resistance found in this AMP-resistant mutant (Table 2). Together, these results indicate that AMP resistance in S. aureus can result in enhanced pathogenic properties.
Previous studies have shown that stable AMP resistance mutations can result in attenuation of S. aureus in an invasive disease model.51,52 To determine whether AMP-resistant S. aureus was capable of causing invasive disease we used a mouse model of sepsis. Mice were infected intraperitoneally with 0.5–2 × 107 cfu and the ability of S. aureus AMP-resistant mutants to disseminate invasively was assessed at 8 h post-infection. Bacterial colonization of the spleen (Figure 3c), liver (Figure 3d) and kidney (Figure 3e) were used as indicators of virulence capacity compared with the fully virulent WT strain. Fewer AMP-resistant S. aureus mutants were recovered from the spleen of infected mice compared with WT for all mutants (Figure 3c). However, all AMP-resistant mutant strains achieved similar levels of colonization to WT in both the liver and kidney (Figure 3d and e), indicating that AMP-resistant S. aureus are fully capable of disseminating and causing invasive disease in mice. Thus, it is also important to note that fitness evaluations in rich medium conditions may not accurately reflect the capacity of AMP-resistant mutants to persist in vivo.
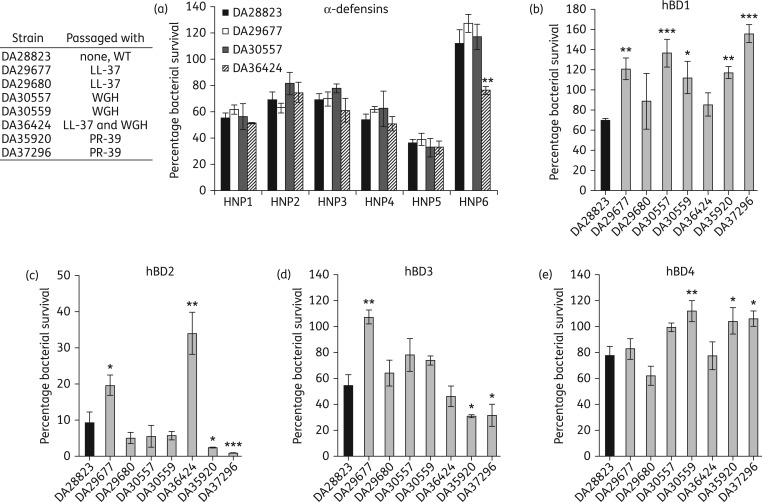
AMP-resistant S. aureus resist killing by human defensins
In humans, the skin provides a substantial barrier for S. aureus to cause invasive disease. Persistent S. aureus colonization of the nasal passages occurs in ∼20% of healthy adults53 increasing the risk of invasive infection in these individuals.54 The production of AMPs in the skin, particularly in the nasal passages, plays a major role in preventing S. aureus persistent colonization and consequently people displaying defects in β-defensin 3 (hBD3) production show enhanced nasal colonization of S. aureus.55 To evaluate the capacity of AMP-resistant S. aureus to evade the human defensins required to control colonization, we determined the susceptibility of these mutants to all 10 of the human defensins. Highly AMP-resistant mutants displayed WT susceptibility to the six α-defensins (Figure 4a) with one high-level LL-37 resistant mutant (DA36424) displaying significantly reduced survival when exposed to HNP6. In contrast, six of seven mutants showed enhanced resistance to at least one of the human β-defensins (Figure 4b–e). The other high-level LL-37 resistant mutant DA29677 showed increased resistance to hBD1, hBD2 and hBD3. These results indicate that human innate immune peptides, including hBD3, are less capable of killing AMP-resistant S. aureus.
Figure 4.
AMP-resistant mutants are less susceptible to human defensins. (a) Susceptibility to the human α-defensins was determined by calculating percentage bacterial survival following 2 h of exposure to 10 μM peptide. Susceptibility to the human β-defensins (b) hBD1, (c) hBD2, (d) hBD3 and (e) hBD4 was determined as for α-defensins with 10 μM (hBD1, hBD2, hBD4) or 1 μM (hBD3) peptide concentrations. Limit of detection of the assay was 1% survival. Values given are the mean ± SEM derived from at least three independent determinations. Statistical significance relative to WT was determined by performing one-way ANOVA with Fisher's least significant difference test used for post hoc analysis on log-transformed data (***P < 0.001, **P < 0.01 or *P < 0.05).
It is becoming increasingly evident that the interactions between host and pathogen can greatly impact the efficacy of clinical therapeutics and that the design of novel containment strategies should account for these dynamics.56,57 The host–pathogen interactions involved in the transition of S. aureus from a commensal organism to an invasive pathogen are complex and not yet fully characterized. It is known that host AMPs provide a significant barrier to S. aureus colonization in humans.55 Our results indicate that prolonged exposure to either human-, pig- or plant-derived AMPs results in the selection of stably resistant S. aureus mutants capable of evading host defensins. With a step-wise selection occurring at the typical mutation rates58 in the population sizes found in S. aureus during colonization,59 it is conceivable that resistant mutants could be selected. These mutants conveyed a resistance phenotype associated with reduced AMP uptake, which corresponded with increased expression of colonization factors. Consequently, widespread clinical usage of such peptides as therapeutics could inadvertently enrich for bacteria that are intrinsically resistant to a major component of the innate immune system and more capable of colonizing a host; and as a result, increase the fraction of the human population that is persistently colonized with S. aureus.60
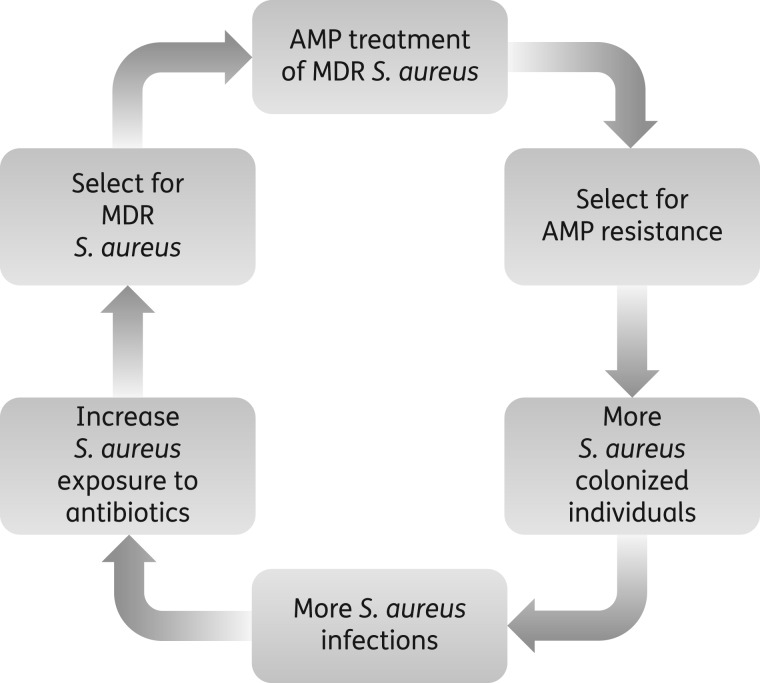
It is also known that individuals who are persistently colonized with S. aureus are at a greater risk of contracting symptomatic and invasive infections typically resulting from the same strain with which they were colonized.54 We have shown that AMP-resistant S. aureus is fully capable of causing invasive disease, which would require antibiotic treatment. However, these mutants also displayed reduced susceptibility to antibiotics, which would therefore limit therapeutic options to treat these infections. As a result, administration of precious last resort drugs such as vancomycin would provide further selection for antibiotic resistance. Clinical resistance to antibiotics such as daptomycin are associated with the development of a thicker cell wall, which in turn results in enhanced resistance to host AMPs,45 potentially creating a vicious cycle ultimately rendering S. aureus infections particularly difficult to treat (Figure 5). These findings highlight the importance of evaluating the development of bacterial resistance in the context of the host environment and have important implications for public health in the event that AMPs enter routine clinical use.
Figure 5.
Possible consequences of clinical AMP usage. Widespread clinical use of host-derived AMPs to treat MDR S. aureus may cause a vicious cycle further limiting treatment options. Continuous exposure to host-derived AMPs results in a stable AMP-resistant phenotype with reduced susceptibility to human defence peptides allowing for enhanced colonization of humans. Persistently colonized individuals are more susceptible to invasive S. aureus infections, which require antibiotic treatment. However, AMP-resistant S. aureus is associated with decreased susceptibility to antibiotics thereby increasing the use of last resort drugs. Enhanced exposure to these antibiotics will increase pressure for the evolution of bacterial resistance yielding highly drug-resistant pathogens.
Funding
This work was supported by grants from the Swedish Research Council and European Commission (project EvoTAR) to D. I. A. and by the Danish Council for Independent Research 12-127417 to H. I.
Transparency declarations
None to declare.
Author contributions
J. Z. K.-S., H. L., M. V. and D. I. A. designed experiments. J. Z. K.-S., H. L., M. V. and K. H. performed experiments. All authors analysed data. J. Z. K.-S. wrote the manuscript. All authors assisted in editing the manuscript.
Supplementary data
Acknowledgements
We thank Omar Warsi for assistance with statistical analysis. Proteomics analysis by MS was performed at the Proteomics Core Facility at Sahlgrenska Academy, Gothenburg University. Proteomics data analysis was performed at the Bioinformatics Core Facility at the Sahlgrenska Academy, Gothenburg University. Animal experiments were performed at the Swedish National Veterinary Institute (SVA) in Uppsala, Sweden.
References
- 1.WHO. Antimicrobial Resistance: Global Report on Surveillance. http://www.who.int/drugresistance/documents/surveillancereport/en/.
- 2.van der Does AM, Bergman P, Agerberth B et al. . Induction of the human cathelicidin LL-37 as a novel treatment against bacterial infections. J Leukoc Biol 2012; 92: 735–42. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 2006; 24: 1551–7. [DOI] [PubMed] [Google Scholar]
- 4.Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 2016; 44: D1087–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epand RM, Walker C, Epand RF et al. . Molecular mechanisms of membrane targeting antibiotics. Biochim Biophys Acta 2016; 1858: 980–7. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh A, Kar RK, Jana J et al. . Indolicidin targets duplex DNA: structural and mechanistic insight through a combination of spectroscopy and microscopy. ChemMedChem 2014; 9: 2052–8. [DOI] [PubMed] [Google Scholar]
- 7.Fox JL. Antimicrobial peptides stage a comeback. Nat Biotechnol 2013; 31: 379–82. [DOI] [PubMed] [Google Scholar]
- 8.Nell MJ, Tjabringa GS, Wafelman AR et al. . Development of novel LL-37 derived antimicrobial peptides with LPS and LTA neutralizing and antimicrobial activities for therapeutic application. Peptides 2006; 27: 649–60. [DOI] [PubMed] [Google Scholar]
- 9.Malanovic N, Leber R, Schmuck M et al. . Phospholipid-driven differences determine the action of the synthetic antimicrobial peptide OP-145 on Gram-positive bacterial and mammalian membrane model systems. Biochim Biophys Acta 2015; 1848: 2437–47. [DOI] [PubMed] [Google Scholar]
- 10.Nizet V, Ohtake T, Lauth X et al. . Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 2001; 414: 454–7. [DOI] [PubMed] [Google Scholar]
- 11.Midorikawa K, Ouhara K, Komatsuzawa H et al. . Staphylococcus aureus susceptibility to innate antimicrobial peptides, β-defensins and CAP18, expressed by human keratinocytes. Infect Immun 2003; 71: 3730–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velden WJ, van Iersel TM, Blijlevens NM et al. . Safety and tolerability of the antimicrobial peptide human lactoferrin 1-11 (hLF1-11). BMC Med 2009; 7: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell G, Gouyon PH. Arming the enemy: the evolution of resistance to self-proteins. Microbiology 2003; 149: 1367–75. [DOI] [PubMed] [Google Scholar]
- 14.Lofton H, Pranting M, Thulin E et al. . Mechanisms and fitness costs of resistance to antimicrobial peptides LL-37, CNY100HL and wheat germ histones. PLoS One 2013; 8: e68875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perron GG, Zasloff M, Bell G. Experimental evolution of resistance to an antimicrobial peptide. Proc Biol Sci 2006; 273: 251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hale JD, Hancock RE. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther 2007; 5: 951–9. [DOI] [PubMed] [Google Scholar]
- 17.Nizet V. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr Issues Mol Biol 2006; 8: 11–26. [PubMed] [Google Scholar]
- 18.Habets MG, Brockhurst MA. Therapeutic antimicrobial peptides may compromise natural immunity. Biol Lett 2012; 8: 416–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg Infect Dis 2007; 13: 1840–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 2009; 7: 629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cosgrove SE, Qi Y, Kaye KS et al. . The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect Control Hosp Epidemiol 2005; 26: 166–74. [DOI] [PubMed] [Google Scholar]
- 22.Diep BA, Gill SR, Chang RF et al. . Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 2006; 367: 731–9. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy AD, Otto M, Braughton KR et al. . Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci USA 2008; 105: 1327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baba T, Bae T, Schneewind O et al. . Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 2008; 190: 300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fey PD, Endres JL, Yajjala VK et al. . A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 2013; 4: e00537–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorschner RA, Lopez-Garcia B, Peschel A et al. . The mammalian ionic environment dictates microbial susceptibility to antimicrobial defense peptides. FASEB J 2006; 20: 35–42. [DOI] [PubMed] [Google Scholar]
- 27.Simon JH, Becker WM. A polyethylene glycol/dextran procedure for the isolation of chromatin proteins (histones and nonhistones) from wheat germ. Biochim Biophys Acta 1976; 454: 154–71. [DOI] [PubMed] [Google Scholar]
- 28.Pasupuleti M, Walse B, Svensson B et al. . Rational design of antimicrobial C3a analogues with enhanced effects against staphylococci using an integrated structure and function-based approach. Biochemistry 2008; 47: 9057–70. [DOI] [PubMed] [Google Scholar]
- 29.Vizcaino JA, Csordas A, Del-Toro N et al. . 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 2016; 44: D447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 2008; 3: 163–75. [DOI] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically—Ninth Edition: Approved Standard M07-A9. CLSI, Wayne, PA, USA, 2012. [Google Scholar]
- 32.Baumert N, von Eiff C, Schaaff F et al. . Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb Drug Resist 2002; 8: 253–60. [DOI] [PubMed] [Google Scholar]
- 33.Morikawa K, Maruyama A, Inose Y et al. . Overexpression of sigma factor, sigmaB, urges Staphylococcus aureus to thicken the cell wall and to resist β-lactams. Biochem Biophys Res Commun 2001; 288: 385–9. [DOI] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gould J. GENE.E: Interact with GENE-E from R. http://www.broadinstitute.org/cancer/software/GENE-E.
- 37.Kawasaki H, Iwamuro S. Potential roles of histones in host defense as antimicrobial agents. Infect Disord Drug Targets 2008; 8: 195–205. [DOI] [PubMed] [Google Scholar]
- 38.Agerberth B, Lee JY, Bergman T et al. . Amino acid sequence of PR-39. Isolation from pig intestine of a new member of the family of proline-arginine-rich antibacterial peptides. Eur J Biochem 1991; 202: 849–54. [DOI] [PubMed] [Google Scholar]
- 39.Glaser R, Becker K, von Eiff C et al. . Decreased susceptibility of Staphylococcus aureus small-colony variants toward human antimicrobial peptides. J Invest Dermatol 2014; 134: 2347–50. [DOI] [PubMed] [Google Scholar]
- 40.Andersson DI, Hughes D, Kubicek-Sutherland JZ. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist Updat 2016; 26: 43–57. [DOI] [PubMed] [Google Scholar]
- 41.Samuelsen O, Haukland HH, Kahl BC et al. . Staphylococcus aureus small colony variants are resistant to the antimicrobial peptide lactoferricin B. J Antimicrob Chemother 2005; 56: 1126–9. [DOI] [PubMed] [Google Scholar]
- 42.Sutcliffe IC, Russell RR. Lipoproteins of Gram-positive bacteria. J Bacteriol 1995; 177: 1123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev 2009; 73: 233–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinkel TL, Roux CM, Dunman PM et al. . The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. MBio 2013; 4: e00696–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra NN, McKinnell J, Yeaman MR et al. . In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 2011; 55: 4012–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev 2013; 26: 185–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 2010; 8: 260–71. [DOI] [PubMed] [Google Scholar]
- 48.Cheung AL, Bayer AS, Zhang G et al. . Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol 2004; 40: 1–9. [DOI] [PubMed] [Google Scholar]
- 49.Graves SF, Kobayashi SD, DeLeo FR. Community-associated methicillin-resistant Staphylococcus aureus immune evasion and virulence. J Mol Med (Berl) 2010; 88: 109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu GY, Essex A, Buchanan JT et al. . Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med 2005; 202: 209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lofton H, Anwar N, Rhen M et al. . Fitness of Salmonella mutants resistant to antimicrobial peptides. J Antimicrob Chemother 2015; 70: 432–40. [DOI] [PubMed] [Google Scholar]
- 52.Strandberg KL, Richards SM, Tamayo R et al. . An altered immune response, but not individual cationic antimicrobial peptides, is associated with the oral attenuation of Ara4N-deficient Salmonella enterica serovar Typhimurium in mice. PLoS One 2012; 7: e49588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 1997; 10: 505–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Eiff C, Becker K, Machka K et al. . Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med 2001; 344: 11–6. [DOI] [PubMed] [Google Scholar]
- 55.Zanger P, Nurjadi D, Vath B et al. . Persistent nasal carriage of Staphylococcus aureus is associated with deficient induction of human β-defensin 3 after sterile wounding of healthy skin in vivo. Infect Immun 2011; 79: 2658–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubicek-Sutherland JZ, Heithoff DM, Ersoy SC et al. . Host-dependent induction of transient antibiotic resistance: A prelude to treatment failure. EBioMedicine 2015; 2: 1169–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thulin E, Sundqvist M, Andersson DI. Amdinocillin (mecillinam) resistance mutations in clinical isolates and laboratory-selected mutants of Escherichia coli. Antimicrob Agents Chemother 2015; 59: 1718–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young BC, Golubchik T, Batty EM et al. . Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci USA 2012; 109: 4550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu CM, Price LB, Hungate BA et al. . Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv 2015; 1: e1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown AF, Leech JM, Rogers TR et al. . Staphylococcus aureus colonization: modulation of host immune response and impact on human vaccine design. Front Immunol 2014; 4: 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.