Abstract
Background
Oral vancomycin remains the mainstay of therapy for severe infections produced by Clostridium difficile, the most prevalent cause of healthcare-associated infectious diarrhoea in developed countries. However, its short- and long-term effects on the human intestinal microbiota remain largely unknown.
Methods
We utilized high-throughput sequencing to analyse the effects of vancomycin on the faecal human microbiota up to 22 weeks post-antibiotic cessation. The clinical relevance of the observed microbiota perturbations was studied in mice.
Results
During vancomycin therapy, most intestinal microbiota genera and operational taxonomic units (OTUs) were depleted in all analysed subjects, including all baseline OTUs from the phylum Bacteroidetes. This was accompanied by a vast expansion of genera associated with infections, including Klebsiella and Escherichia/Shigella. Following antibiotic cessation, marked differences in microbiota resilience were observed among subjects. While some individuals recovered a microbiota close to baseline composition, in others, up to 89% of abundant OTUs could no longer be detected. The clinical relevance of the observed microbiota changes was further demonstrated in mice, which developed analogous microbiota alterations. During vancomycin treatment, mice were highly susceptible to intestinal colonization by an antibiotic-resistant pathogen and, upon antibiotic cessation, a less-resilient microbiota allowed higher levels of pathogen colonization.
Conclusions
Oral vancomycin induces drastic and consistent changes in the human intestinal microbiota. Upon vancomycin cessation, the microbiota recovery rate varied considerably among subjects, which could influence, as validated in mice, the level of susceptibility to pathogen intestinal colonization. Our results demonstrate the negative long-term effects of vancomycin, which should be considered as a fundamental aspect of the cost–benefit equation for antibiotic prescription.
Introduction
The intestinal microbiota is essential for the proper development of the intestinal tract and maturation of the immune and nervous system. In addition, commensal bacteria confer resistance to infection by suppressing the growth of intestinal pathogens.1–4 Consequently, alterations of the microbiota composition induced by antibiotics can promote pathology, including obesity, asthma or infectious diseases.2,5–7 Thus, understanding the effects that specific antibiotics have on the human intestinal microbiota is crucial for clinicians in order to choose the most efficacious (and otherwise less deleterious) of the therapeutic options available for treating infections.
Vancomycin, a glycopeptide antibiotic mainly active against Gram-positive bacteria, is often used to treat infections produced by multidrug-resistant Staphylococcus aureus and Clostridium difficile. Indeed, among the different antibiotics currently available, oral vancomycin remains the treatment of choice for severe C. difficile infection (CDI). Despite its effectiveness against CDI (cure rate of ∼90%), a subset of cured patients (14%–21%) develop recurrent infections, which are thought to be enhanced by microbiota changes promoted by vancomycin.8,9 In addition, microbiota alterations induced by vancomycin may promote intestinal colonization by other pathogens, including VRE, Klebsiella pneumoniae or Escherichia coli.10 Moreover, oral vancomycin therapy may predispose to other microbiota-related disorders, including obesity, asthma or diabetes.7,11,12
Despite the potential negative effects caused by vancomycin administration, the impact that this antibiotic has on the human microbiota is not well characterized. Indeed, most prior studies analysing intestinal microbiota changes upon vancomycin administration have focused on CDI patients.13–16 Importantly, the majority of these patients have received antibiotics before vancomycin administration, preventing a true understanding of the vancomycin effects on intestinal commensal populations. Moreover, subjects recruited for these studies were only followed for up to one month after antibiotic discontinuation, rendering the long-lasting effects of vancomycin unknown. In addition, these reports have only partially characterized the changes in the microbiota upon vancomycin administration. Indeed, only changes at the family level,16 or changes in particular bacterial groups have been defined.13–15 High-throughput sequencing of the 16S rRNA gene currently allows for an in-depth taxonomic survey of bacterial communities beyond the genera level, enhancing our capability to understand how vancomycin alters the ecological communities inhabiting the intestinal tract.
To better characterize the effects of vancomycin on the intestinal microbiota, we have analysed, using 16S rRNA high-throughput sequencing, the faecal microbiota of patients with treatment-naive, new-onset rheumatoid arthritis (RA) who underwent a 2 week course of oral vancomycin. These patients had not been exposed to any other antibiotic at least 3 months prior to vancomycin administration, nor were they exposed to immunosuppressive therapies, allowing us to clearly define the microbiota changes solely produced by this antibiotic.
Methods
Ethics
This study was approved by the Institutional Review Board of New York University School of Medicine, protocol number 09-0658. Further details are published on the www.ClinicalTrials.gov web site, identifier NCT01198509. All included patients gave their consent to participate in the study. Mice procedures were approved by University of Valencia Animal Care Committee, protocol number 2015/VSC/PEA/00082.
Vancomycin administration in RA patients: study design
All RA patients were recruited from a previously described study.17 Patient characteristics and inclusion and exclusion criteria are described in detail in the supplementary methods (available as Supplementary data at JAC Online). After consent was obtained, we randomly divided new-onset RA patients into two groups. The first group (the vancomycin-treated group) received vancomycin orally (250 mg four times a day) for 2 weeks, followed by treatment with methotrexate starting 6 weeks after discontinuation of antibiotic therapy. The second group (control group) received methotrexate from the beginning of the study and did not receive vancomycin. This control group was included to identify changes in the microbiota due to methotrexate administration. Importantly, as described below, no alterations in the gut microbiota composition were observed at any studied timepoint in the control group.
Microbiota analysis
Bacterial DNA extraction, amplification of 16S rRNA gene and high-throughput sequencing analysis, including confirmation of the obtained results using different subset of sequences, are indicated in the supplementary methods and Figures S7–S10 (available as Supplementary data at JAC Online).
Statistics
The two-tailed Student's t-test was applied to identify significant differences in the number of operational taxonomic units (OTUs), genera or Shannon index.18
The two-tailed Wilcoxon non-parametric test was applied to identify significant microbiota taxonomic changes that occur in patients after vancomycin therapy. The false discovery rate (FDR) approach was applied to adjust for multiple hypothesis testing.19 Very low abundance taxa and OTUs (<10 counts in the two groups of samples under comparison) were not included in the statistical analysis. Changes with a P < 0.05 and FDR < 0.2 were considered significant.
Correlation between the microbiota recovery rate and susceptibility to intestinal colonization by VRE were analysed using the Spearman test. Correlations between pairs of variables were considered to be significant when P values were <0.05.
Mouse model of pathogen intestinal colonization
Details on the mouse experimental procedures are described in the supplementary methods.
Results
Vancomycin treatment alters the human intestinal microbiota structure
Twenty-one subjects were included in the study. Nine of the patients received vancomycin orally for 2 weeks, followed by methotrexate (beginning 6 weeks after vancomycin cessation), while 12 subjects were only treated with oral methotrexate from day 1 (control group; no vancomycin). No significant differences in clinical or demographic baseline characteristics were observed between both groups (Table S1, available as Supplementary data at JAC Online). Faecal samples were obtained immediately before the initiation of vancomycin treatment, the day the treatment was ended (week 2), and 2, 6, 14 and 22 weeks after antibiotic discontinuation. Samples from control patients were obtained at the same timepoints. With only a few exceptions, samples for most timepoints were included in the study for all vancomycin-treated (4.9 ± 0.9 out of 6 timepoints; mean ± SD) and control patients (4.2 ± 1.1) (Figure S1).
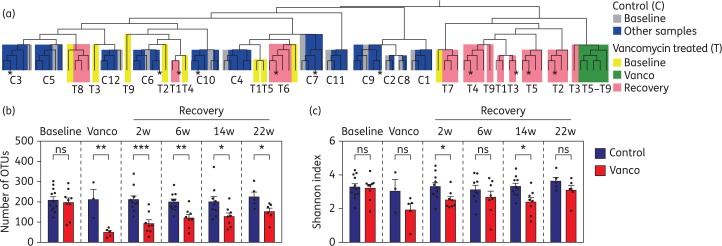
First, to obtain a global view of the changes induced by vancomycin, we applied hierarchical clustering (see the supplementary methods) to group samples by microbiota similarity (samples with a more similar microbiota will be those grouped together in the same branch of the tree). As shown in Figure 1(a), oral methotrexate alone did not produce major changes in the faecal microbiota, as all prospective samples from control patients cluster with their respective baseline samples. In contrast, oral administration of vancomycin for 2 weeks consistently altered the faecal microbiota structure, with all faecal samples obtained immediately after vancomycin treatment clustering together and away from their respective baseline samples. After antibiotic cessation, a few vancomycin-treated patients recovered their overall baseline microbiota structure (i.e. samples after antibiotic cessation from patients T6 and T8 cluster together with their respective baseline samples). However, the faecal microbiota structure for most vancomycin-treated patients remained altered, even 22 weeks after antibiotic cessation (i.e. samples after antibiotic cessation from patients T2, T3, T4 and T5 do not cluster with their respective baseline samples).
Figure 1.
Vancomycin induces persistent changes in the structure and richness of the human microbiota. Patients received vancomycin treatment for 2 weeks (T, vancomycin treated) or did not receive vancomycin (C, control group). Faecal samples were obtained immediately before treatment (Baseline), the day of antibiotic cessation (Vanco), and 2, 6, 14 and 22 weeks (w) after antibiotic cessation (Recovery). As control, faecal samples from patients that did not receive vancomycin were obtained at similar timepoints. (a) Hierarchical clustering based on microbiota similarity (see the supplementary methods) among the faecal samples analysed from vancomycin-treated and control patients. Samples with a more similar microbiota are clustered within the same branch of the tree. Colours indicate the time frame where the faecal sample was obtained. Numbers indicate the patient ID. Those samples obtained at the last timepoint (22 weeks post-antibiotic withdrawal) are labelled with asterisks. (b) Number of OTUs (a close estimate of bacterial species) and (c) Shannon index of microbial diversity calculated from the microbiota of faecal samples collected at baseline and at different timepoints after vancomycin treatment. For comparison, the same indices were calculated from the faecal samples collected at similar timepoints from patients who did not receive vancomycin (control). Bar graphs represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed t-test. ns, not significant. n = 5–12 per group and timepoint except for the second timepoint from control patients where n = 3. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Vancomycin treatment diminishes the richness and diversity of the human microbiota
Subsequently, we analysed the impact of vancomycin treatment on microbiota richness, measured as the number of identified OTUs, a sequencing approach that closely defines the number of bacterial species within a sample. The number of OTUs did not differ over time in the control patients not receiving vancomycin (Figure 1b and Figure S2). By contrast, microbiota richness was greatly reduced upon vancomycin administration. Following antibiotic cessation, microbiota richness gradually increased, although it never recovered to baseline levels. A similar result was obtained when the Shannon diversity index was calculated (Figure 1c and Figure S3), which takes into account the number of OTUs and their relative proportion. In this case, however, baseline levels were recovered 22 weeks after antibiotic withdrawal.
Abundance of most intestinal bacterial taxa is altered during vancomycin treatment
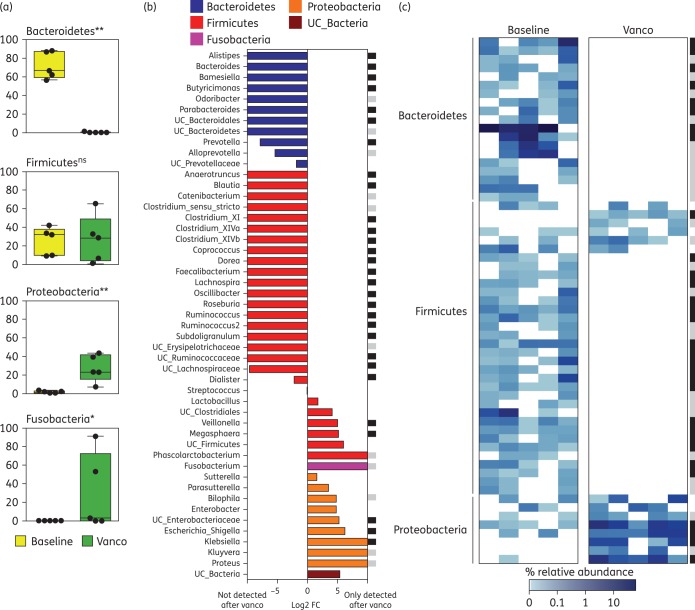
Considering the large effects of vancomycin on the overall microbiota structure and richness, we next decided to examine its effects on specific taxa. We focused on those taxa that were present in at least 50% of the patients (either at baseline or after treatment). Vancomycin treatment greatly reduced the levels of the Bacteroidetes phylum (Figure 2a). In contrast, the Proteobacteria and Fusobacteria phyla underwent a drastic expansion after vancomycin therapy. The abundance of most analysed genera and OTUs was also significantly altered by vancomycin treatment (Figure 2b and c). Indeed, most abundant genera and OTUs from the Bacteroidetes or Firmicutes phyla could not be detected in any of the patients after vancomycin treatment. Nonetheless, some Firmicutes increased after vancomycin treatment (i.e. Megasphaera, Veillonella), likely explaining why, overall, the Firmicutes phylum did not diminish upon vancomycin treatment (Figure 2a). Importantly, several genera belonging to the Proteobacteria phylum (i.e. Escherichia/Shigella and Klebsiella), which have been associated with infectious processes, increased after antibiotic administration. In contrast, no statistically significant changes in any analysed taxa or OTU (P < 0.05, FDR < 0.2) were detected at any timepoint in patients who did not receive vancomycin.
Figure 2.
The abundance of the majority of taxa and OTUs is altered during vancomycin treatment. (a) Relative abundance of different phyla in patients treated with vancomycin at baseline or 2 weeks after treatment initiation (Vanco). Only phyla that are present in at least 50% of the patients at baseline or after vancomycin treatment are shown. **P < 0.01, *P < 0.05, FDR < 0.2, two-tailed Wilcoxon test. ns, not significant. (b) Log2 average fold change (FC) between the genera abundance from samples obtained immediately after vancomycin treatment compared with their respective baseline samples. Only genera that are present in at least 50% of the patients at baseline or after vancomycin treatment are shown. Genera are sorted by phyla, FC difference and then alphabetically. UC, unclassified; vanco, vancomycin. (c) Heatmap representing the relative abundance (%) of OTUs present in at least 50% of the patients at baseline or after vancomycin treatment, showing a depletion of all prevalent Bacteroidetes OTUs and most Firmicutes OTUs. Additional analysis including all detected OTUs confirmed the depletion of all baseline OTUs from the Bacteroidetes phylum (not shown). For both (b) and (c): black squares indicate significant changes (P < 0.05, FDR < 0.1); grey squares indicate close to significance changes (P < 0.073, FDR < 0.1); two-tailed Wilcoxon test. n = 5. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Incomplete and individualized microbiota recovery after vancomycin withdrawal
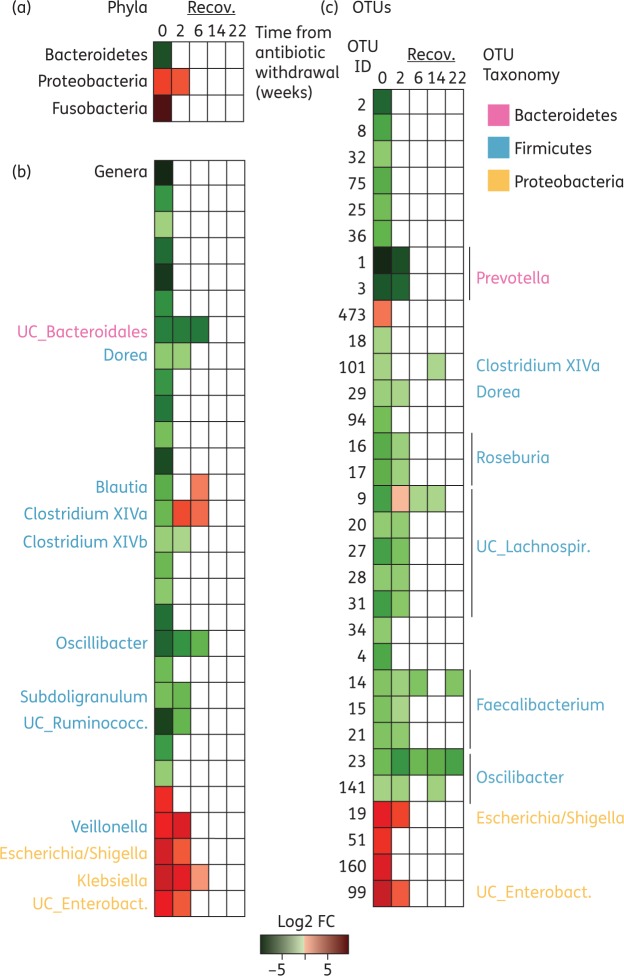
We next examined the capacity of the different commensal bacteria to recover after antibiotic cessation. We focused on those commensals whose relative abundance significantly changed during vancomycin therapy. As shown in Figure 3, the speed of recovery was different depending on the phyla, genera or OTU analysed. Nevertheless, 22 weeks post-antibiotic cessation, all genera and phyla had recovered their baseline levels (with the exception of only two OTUs which still showed significant alterations).
Figure 3.
Changes in the human microbiota following vancomycin withdrawal. Heatmap representing the average fold change of (a) phyla, (b) genera and (c) OTUs that were significantly increased (red) or decreased (green) (P < 0.05, FDR < 0.2, two-tailed Wilcoxon test) at a given timepoint as compared with the baseline. Time 0 represents the sample obtained the day of vancomycin withdrawal. n = 6–9 per timepoint. UC, unclassified; Enterobact., Enterobacteriaceae. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
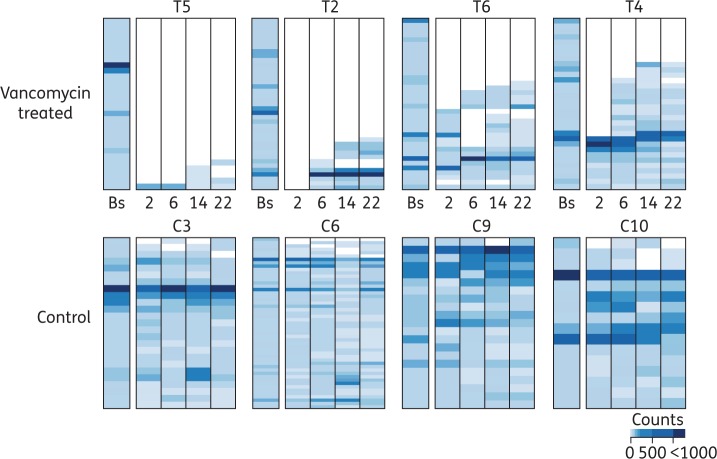
The performed analysis allowed us to identify perturbation patterns common to all patients after antibiotic cessation. For example, OTU23 abundance was significantly reduced 22 weeks post-antibiotic cessation, since this OTU was not recovered in any of the patients analysed (Figure 3c and Figure S4). However, this type of analysis does not allow for the identification of inter-individual differences in the microbiota recovery. For example, OTU15 mean abundance was diminished 22 weeks post-antibiotic cessation, but this change was not significant due to the variable recovery of this OTU among different patients (Figure S4). Considering that a previous analysis (Figure 1a) had suggested that major shifts in the microbiota of some patients still occurred 22 weeks post-antibiotic cessation, we decided to characterize the degree of taxa recovery at the individual patient level. To this end, we identified and plotted the most abundant OTUs present at baseline for each patient (≥10 counts), and analysed the recovery of those OTUs after antibiotic withdrawal (Figure 4, Figures S5 and S6). As expected, there was a high inter-individual variation in the recovery pattern. While some patients recovered most of their abundant OTUs (e.g. T6 and T4), in other patients as few as 10.7% of their most abundant OTUs could be detected 22 weeks post-antibiotic withdrawal (i.e. T5). Control patients (methotrexate-treated only) did develop a few changes over time, and some OTUs were absent at a given timepoint. However, in most cases, the missing OTUs were detected at subsequent sampling. Overall, in control patients, 90.6 ± 8.1% of the most abundant OTUs at baseline could be detected at any timepoint (Figure S6a). By contrast, in vancomycin-treated patients, we could only detect 39 ± 21.9% of the baseline most abundant OTUs at the last timepoint analysed. A similar result was obtained at the genus level, although the recovery rate was greater than the one observed for OTUs (Figure S6b).
Figure 4.
Incomplete and individualized recovery of the human microbiota after vancomycin withdrawal. Relative abundance at baseline (Bs) and at different weeks post-antibiotic cessation of OTUs whose abundance was ≥10 counts in the baseline sample of the analysed patient. Note that each patient baseline OTUs are distinct. The ID of each patient is indicated. As control, samples at similar timepoints were analysed from patients who did not receive vancomycin. Four representative patients from each group are shown (the four control patients from whom we were able to collect and analyse faecal samples at every timepoint and four out of the five vancomycin-treated patients for whom we were able to collect and analyse faecal samples at every timepoint). The rest of the patients are shown in Figure S5. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
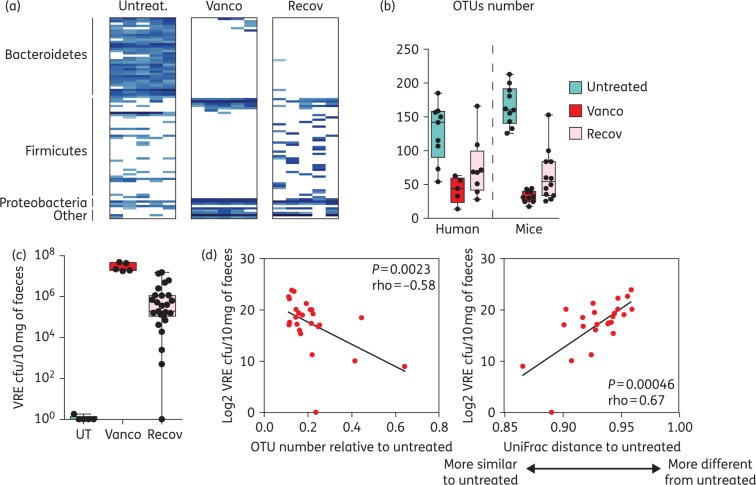
Microbiota recovery rate after vancomycin cessation influences the level of susceptibility to pathogen intestinal colonization
We have demonstrated that upon vancomycin cessation, the human microbiota recovers to varying extents in a subject-specific manner. It is conceivable, therefore, that antibiotic-driven dysbiosis and the microbiota recovery rate observed after vancomycin cessation could impact susceptibility to infection, with ‘slow recoverers’ being at higher risk. Because vancomycin also induces long-lasting intestinal microbiota perturbations in mice, allowing for intestinal colonization by pathogens such as VRE,10 we used this mouse model to investigate if a different microbiota recovery pattern upon vancomycin cessation could influence susceptibility to intestinal colonization by this pathogen. As shown in Figure 5, vancomycin induced changes in the murine intestinal microbiota strikingly similar to those observed in patients, including: decrease in microbiota richness, depletion of all Bacteroidetes and most Firmicutes OTUs and increase in Proteobacteria OTUs. Moreover, 2 weeks post-vancomycin cessation, mice partially recovered the baseline OTU numbers, and like in humans, the level of recovery was variable depending on the mouse. We next evaluated, using this mouse model, the extent to which a different microbiota recovery rate after vancomycin administration could impact VRE intestinal colonization. As shown in Figure 5(c) and previously reported,10 untreated mice were resistant to VRE colonization, while mice that received oral vancomycin were highly susceptible to VRE colonization. Two weeks after antibiotic cessation, mice were still susceptible to VRE colonization, but with a high degree of inter-individual variability. Importantly, correlation analysis between the level of microbiota recovery and the level of VRE intestinal colonization (Figure 5d), indicated that mice that recovered a higher number of OTUs (or a microbiota more similar to that of untreated mice) were colonized with lower levels of VRE.
Figure 5.
Microbiota recovery rate after vancomycin cessation influences the level of susceptibility to VRE intestinal colonization in mice. (a) Heatmap showing the most prevalent OTUs (≥10 counts per group of mice) found in the faeces of mice treated with vancomycin for 7 days (Vanco), 2 weeks after antibiotic cessation (Recov) or in mice that remained untreated (Untreat.). n = 5 per group. (b) Number of OTUs identified in faecal samples from untreated mice or humans, or mice/humans that were treated with oral vancomycin, the day the treatment was stopped (Vanco) or 2 weeks post-antibiotic cessation (Recov). Boxes extend from the 25th percentile to the 75th percentile. The line in the middle of the box represents the median. Whiskers extend from the minimum value to the maximum value. n = 5–12 per group. (c) VRE cfu/10 mg of faeces 2 days post-VRE inoculation in untreated mice (UT), mice that received vancomycin for 1 week and were inoculated with VRE immediately after vancomycin therapy (Vanco) or were inoculated 2 weeks post-antibiotic cessation (Recov). n = 5 for the UT and Vanco groups and n = 25 for the Recov group. (d) Correlation analysis between the y-axis variable (log2 VRE cfu/10 mg of faeces detected the second day after VRE inoculation in mice that recover for 2 weeks after vancomycin administration) and the x-axis variable, which is either (i) the number of OTUs identified in mice, the day of VRE inoculation, divided by the average number of OTUs identified in untreated mice, or (ii) the microbiota similarity between VRE-colonized mice, the day of VRE inoculation and untreated mice (based on UniFrac distance, see the supplementary methods). n = 25. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discussion
Using a high-throughput sequencing approach, we have investigated for the first time both the short- and long-term impact of vancomycin on the human intestinal microbiota. Our results have revealed that vancomycin depletes most bacterial OTUs found in the intestinal tract, including all detected baseline OTUs from the phylum Bacteroidetes. Furthermore, several bacterial genera from the Proteobacteria phylum that have been associated with human infections (i.e. Klebsiella and Escherichia/Shigella) increased after treatment. Our results showing Bacteroidetes depletion and a concomitant expansion of Proteobacteria are in agreement with previous studies utilizing lower-resolution techniques to study the effects of vancomycin in CDI patients.14,15 Importantly, our high-throughput sequencing approach allowed us to define that essentially all highly prevalent OTUs belonging to the Bacteroidetes phylum were affected by vancomycin. Consistent with our results but using a microarray lower-throughput approach, Vrieze et al.20 also observed a decrease in the levels of Faecalibacterium and Ruminococcus.
We acknowledge several limitations of our study. First, we analysed the effects of vancomycin on RA patients, a population that demonstrates microbiota features usually absent in normal healthy subjects (i.e. higher levels of Prevotella copri).17 Therefore, it is possible that different changes could be observed in a healthy human population. Nevertheless, the fact that similar changes were observed in otherwise healthy mice and comparable alterations at the phylogeny level had been identified using lower-resolution techniques suggest that vancomycin effects are expected to be similar in healthy human subjects. Importantly, our study was performed in a human population that had not recently received other antibiotic treatments. This is in contrast to other studies performed in CDI patients, where most patients had received other antibiotics before vancomycin administration. This has allowed us to define the long-term effects of vancomycin administration in the absence of confounding factors such as exposure to other antibiotics. The rationale behind the use of vancomycin on RA patients was based on our pre-clinical observations in animal models.21 Our studies revealed that this antibiotic led to a dramatic reduction of segmented filamentous bacteria (SFB), a decrease in Th17 cell activation and proliferation, and the abrogation of inflammatory arthritis. Of note, this study was initiated several months prior to the realization that SFB was not a normal inhabitant of the human intestinal microbiota. Besides vancomycin treatment, RA patients also received methotrexate. For this reason, as a control group, we enrolled patients with RA who did not receive vancomycin but were treated with methotrexate from the very beginning of the study. We did not observe any significant intestinal microbiota changes in any of the analysed timepoints in this control group, suggesting that microbiota perturbations observed in the vancomycin-treated group were solely induced by vancomycin. Similarly to our results, a recent study showed that methotrexate has a minor effect on the faecal microbiome composition.22
A second limitation of our study may be the low number of participants included. Nevertheless, due to the drastic changes induced by vancomycin in the microbiota composition, even with this small number of participants, we were able detect large, statistically significant changes upon vancomycin administration. Thus the inclusion of further participants would not have substantially changed the conclusions of the study.
Beyond analysing the short-term effects of vancomycin, we also examined its long-lasting consequences on the intestinal microbiota. Despite the drastic microbiota disruption observed in the treated patients as a group, 22 weeks after cessation we could only identify changes in two OTUs, probably due to the variable microbiota recovery among patients. Similarly, a recent study identified significant changes in just two OTUs, 4 months after stopping either ciprofloxacin or clindamycin treatment.23 In that study, only changes in the microbiota common to all subjects were analysed. Here, in contrast, we further analysed the extent to which each subject was able to recover the baseline microbiota. Interestingly, we found a wide inter-individual variability in microbiota recovery after vancomycin cessation. This result is highly relevant because a subset of CDI patients develop secondary infections after vancomycin therapy, including those promoted by VRE.24–26 Although the reasons for developing these secondary infections are not completely understood, it has been proposed that alterations of the microbiota after vancomycin treatment may enhance intestinal colonization by bacterial pathogens and subsequent infections. This could have direct clinical implications, as a different microbiota recovery rate upon oral vancomycin therapy in hospital settings (i.e. CDI treatment) could ultimately influence the susceptibility to intestinal colonization by pathogens such as VRE. In fact, the results obtained in mice showing a significant negative correlation between the microbiota recovery rate and the intestinal VRE levels strongly support this hypothesis.
Altogether, our results demonstrate the negative long-term consequences of oral vancomycin administration, which should be taken into account in the decision-making prior to prescribing this antibiotic. In addition, our results highlight the potential value of monitoring microbiota dynamics for each patient before and after antibiotic administration. Microbiota tracking could lead to the identification of patients at higher risk of suffering the collateral negative effects of vancomycin (i.e. infections) and eventually inform the potential need for microbiota restoration through faecal transplantation or probiotic administration.
Funding
This work was supported by: the FP7 Marie Curie Actions (PCIG09-GA-2011-293894) and the Spanish Ministerio de Economía y Competitividad (SAF2014-60234-R) to C. U.; Tow Foundation and The National Institute of Health (RO1-AI042135 and P30-CA008748) to E. G. P.; The National Institute of Health (RC2-AR058986 and K23AR064318), American Recovery and Reinvestment Act, Judith and Stewart Colton Center of Autoimmunity and Riley Family Foundation to J. U. S. and S. B. A.; and a F.P.I predoctoral fellowship from the Spanish Ministerio de Economía y Competitividad to S. I.
Transparency declarations
None to declare.
Supplementary data
Acknowledgements
We thank E. A. Gomez and J. R. Penadés for infrastructure support.
References
- 1.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F et al. . Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004; 118: 229–41. [DOI] [PubMed] [Google Scholar]
- 2.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 2013; 13: 790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanov II, Atarashi K, Manel N et al. . Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139: 485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heijtz RD, Wang S, Anuar F et al. . Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 2011; 108: 3047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ubeda C, Pamer EG. Antibiotics, microbiota, and immune defense. Trends Immunol 2012; 33: 459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ubeda C, Taur Y, Jenq RR et al. . Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120: 4332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Million M, Thuny F, Angelakis E et al. . Lactobacillus reuteri and Escherichia coli in the human gut microbiota may predict weight gain associated with vancomycin treatment. Nutr Diabetes 2013; 3: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zar FA, Bakkanagari SR, Moorthi KMLST et al. . Comparison of vancomycin and metronidazole for the treatment of Clostridium difficile-associated diarrhea, stratified by disease severity. Clin Infect Dis 2007; 45: 302–7. [DOI] [PubMed] [Google Scholar]
- 9.Johnson S, Gerding DN, Louie TJ et al. . Sustained clinical response as an endpoint in treatment trials of Clostridium difficile-associated diarrhea. Antimicrob Agents Chemother 2012; 56: 4043–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis B, Buffie CG, Carter RA et al. . Loss of microbiota-mediated colonization resistance to Clostridium difficile infection is greater following oral vancomycin as compared with metronidazole. J Infect Dis 2015; 212: 1656–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell SL, Gold MJ, Hartmann M et al. . Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep 2012; 13: 440–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candon S, Perez-Arroyo A, Marquet C et al. . Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS ONE 2015; 10: e0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tannock GW, Munro K, Taylor C et al. . A new macrocyclic antibiotic, fidaxomicin (OPT-80), causes less alteration to the bowel microbiota of Clostridium difficile-infected patients than does vancomycin. Microbiology 2010; 156: 3354–9. [DOI] [PubMed] [Google Scholar]
- 14.Louie TJ, Cannon K, Byrne B et al. . Fidaxomicin preserves the intestinal microbiome during and after treatment of Clostridium difficile infection (CDI) and reduces both toxin reexpression and recurrence of CDI. Clin Infect Dis 2012; 55 Suppl 2: S132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louie TJ, Byrne B, Emery J et al. . Differences of the fecal microflora with Clostridium difficile therapies. Clin Infect Dis 2015; 60 Suppl 2: S91–7. [DOI] [PubMed] [Google Scholar]
- 16.Abujamel T, Cadnum JL, Jury LA et al. . Defining the vulnerable period for re-establishment of Clostridium difficile colonization after treatment of C. difficile infection with oral vancomycin or metronidazole. PLoS ONE 2013; 8: e76269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scher JU, Sczesnak A, Longman RS et al. . Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013; 2: e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magurran AE. Measuring Biological Diversity. African J Aquatic Sci 2004; 29: 285–6. [Google Scholar]
- 19.Benjamini Y, Hockberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995; 57: 289–300. [Google Scholar]
- 20.Vrieze A, Out C, Fuentes S et al. . Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol 2014; 60: 824–31. [DOI] [PubMed] [Google Scholar]
- 21.Wu H-J, Ivanov II, Darce J et al. . Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010; 32: 815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D, Jia H, Feng Q et al. . The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 2015; 21: 895–905. [DOI] [PubMed] [Google Scholar]
- 23.Rashid MU, Zaura E, Buijs MJ et al. . Determining the long-term effect of antibiotic administration on the human normal intestinal microbiota using culture and pyrosequencing methods. Clin Infect Dis 2015; 60 Suppl 2: S77–84. [DOI] [PubMed] [Google Scholar]
- 24.Louie TJ, Peppe J, Watt CK et al. . Tolevamer, a novel nonantibiotic polymer, compared with vancomycin in the treatment of mild to moderately severe Clostridium difficile-associated diarrhea. Clin Infect Dis 2006; 43: 411–20. [DOI] [PubMed] [Google Scholar]
- 25.Pepin J, Valiquette L, Gagnon S et al. . Outcomes of Clostridium difficile-associated disease treated with metronidazole or vancomycin before and after the emergence of NAP1/027. Am J Gastroenterol 2007; 102: 2781–8. [DOI] [PubMed] [Google Scholar]
- 26.Falcone M, Russo A, Iraci F et al. . Risk factors and outcomes for bloodstream infections secondary to Clostridium difficile infection. Antimicrob Agents Chemother 2015; 60: 252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.