Abstract
Hepatitis C virus (HCV) uniquely affects desmosterol homeostasis by increasing its intracellular abundance and affecting its localization. These effects are important for productive viral replication since inhibition of desmosterol synthesis has an antiviral effect that can be rescued by the addition of exogenous desmosterol. Here, we use subgenomic replicons to show that desmosterol has a major effect on replication of HCV JFH1 RNA. Fluorescence recovery after photobleaching (FRAP) experiments performed with synthetic supported lipid bilayers demonstrate that substitution of desmosterol for cholesterol significantly increases lipid bilayer fluidity, especially in the presence of saturated phospholipids and ceramides. We demonstrate using LC-MS that desmosterol is abundant in the membranes upon which genome replication takes place and that supported lipid bilayers derived from these specialized membranes also exhibit significantly higher fluidity compared to negative control membranes isolated from cells lacking HCV. Together, these data suggest a model in which the fluidity-promoting effects of desmosterol on lipid bilayers play a crucial role in the extensive membrane remodeling that takes place in the endoplasmic reticulum during HCV infection. We anticipate that the supported lipid bilayer system described can provide a useful model system in which to interrogate the effects of lipid structure and composition on the biophysical properties of lipid membranes as well as their function in viral processes such as genome replication.
Keywords: hepatitis C virus, desmosterol, supported lipid bilayer, lipid structure-function, membrane fluidity, viral replication
Introduction
All viruses rely on the host cell to provide the lipids that serve essential structural, energetic, and regulatory functions in viral infection and the production of progeny virions, a process known as “viral replication.” Many viruses utilize specific subcellular membranes during entry, expression and replication of the viral genome, and the assembly and egress of progeny virions. These membranes are generally thought to serve as physical barriers that protect the virus from the environment or from host defense mechanisms and/or as platforms that reduce the entropic barrier for viral processes by bringing together the components required for biosynthetic or signaling reactions. Our understanding of these membrane-associated viral processes has relied heavily on the identification and functional characterization of the viral and host proteins associated with these membranes; comparatively less is known about how lipids affect intrinsic membrane properties and membrane function in the viral process. Despite this, numerous transcriptomic and proteomic studies have documented virus-induced changes in the steady-state expression of host lipid biosynthetic, metabolic, and trafficking enzymes; moreover, lipidomic profiling studies have directly demonstrated that viruses cause significant changes in the steady-state abundance of distinct classes of lipids1-5 as well as changes to specific lipid molecules within those classes6-9 Understanding how these changes relate to the composition and functional properties of subcellular membranes and their associated viral processes thus presents major challenges but also exciting opportunities for investigation.
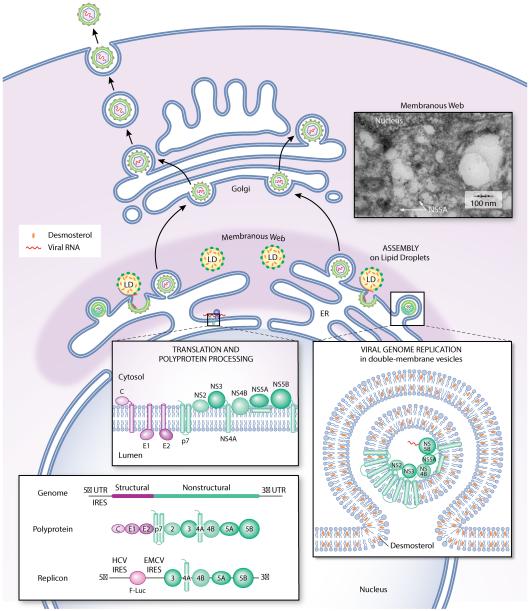
Hepatitis C virus (HCV) is the cause of both acute and chronic hepatitis, with the world’s 150 million chronically infected patients at high risk for cirrhosis and hepatocellular carcinoma leading to approximately 500,000 deaths annually10. Individuals with chronic HCV often exhibit hyperlipidemia, hepatic steatosis, and fatty liver disease, telltale signs that the virus affects steady-state lipid metabolism in the host. While these clinical manifestations of HCV’s impact on host lipid metabolism may be by-products with no direct effect on viral replication, the molecular mechanisms underlying these changes have likely evolved because they promote viral replication. An enveloped virus with a positive-strand RNA genome, HCV has been shown to utilize the host lipid metabolism, storage, and trafficking machinery throughout its replication cycle2, 11-15 and to interact with many factors comprising this machinery. In particular, replication of the HCV genome and viral assembly occur within a specialized, membranous environment dubbed the “membranous web” (Figure 1). While it is well-established that HCV induces formation of the membranous web by altering the endoplasmic reticulum (ER)16-19 less is known about the lipid composition of this specialized, membranous environment and how this composition affects the viral processes that occur therein.
Figure 1. The membranous web is a specialized, membranous environment for hepatitis C virus genome replication and assembly.
Translation of the HCV genome and processing of the HCV polyprotein occurs on the ER. Genome replication occurs in double-membrane vesicles. Virion assembly is initiated by the association of the viral genomic RNA with core protein on the surface of lipid droplets that are coated with the NS5A protein and that are usually proximal to sites of genome replication. The double-membrane vesicles and lipid droplets where genome replication and assembly, respectively, occur are derived from the ER and together appear as a “membranous web” (see electron micrograph inset). Subgenomic replicons derived from the HCV genomic RNA provide useful tools for studying translation and replication of the HCV RNA in the absence of viral entry, virion assembly, and egress. Desmosterol has been detected in replicase-containing membranes by LC-MS analysis (Figure 2) and in NS5A-associated lipid droplets by stimulated Raman scattering microscopy11.
We previously performed untargeted lipidomic profiling of HCV JFH1 infected Huh7.5 cells and discovered that the virus causes a greater than ten-fold increase in the steady-state abundance of desmosterol, the penultimate intermediate in the Bloch branch of cholesterol biosynthesis, without changing the steady-state abundance of cholesterol or other late-stage sterols6, 11 This perturbation of desmosterol homeostasis appears to be functionally important for HCV JFH1 replication since the virus is severely inhibited upon pharmacological inhibition or RNAi-mediated depletion of 7-dehydrocholesterol reductase (DHCR7), the enzyme that directly catalyzes synthesis of desmosterol; moreover, viral replication is rescued by the addition of exogenous desmosterol and only to a lesser extent rescued by cholesterol 6, 11. Since desmosterol and cholesterol differ only by the presence of a double bond between carbons 24 and 25 (Figure 2A), this provokes the question of how this subtle structural difference results in the significantly different effects of these sterols on HCV JFH1 replication in cell culture. Here, we show that desmosterol is present in the membranes where replication of the HCV RNA genome occurs and that the antiviral effect observed upon pharmacological inhibition of desmosterol biosynthesis has a significant inhibitory effect on HCV RNA replication. We further report the development of a supported lipid bilayer system and its use to demonstrate that desmosterol increases the fluidity of synthetic bilayers as well as bilayers derived from HCV replication membranes. Collectively, these data suggest a model in which desmosterol directly affects HCV RNA replication by increasing membrane fluidity and facilitating formation of the membranous web.
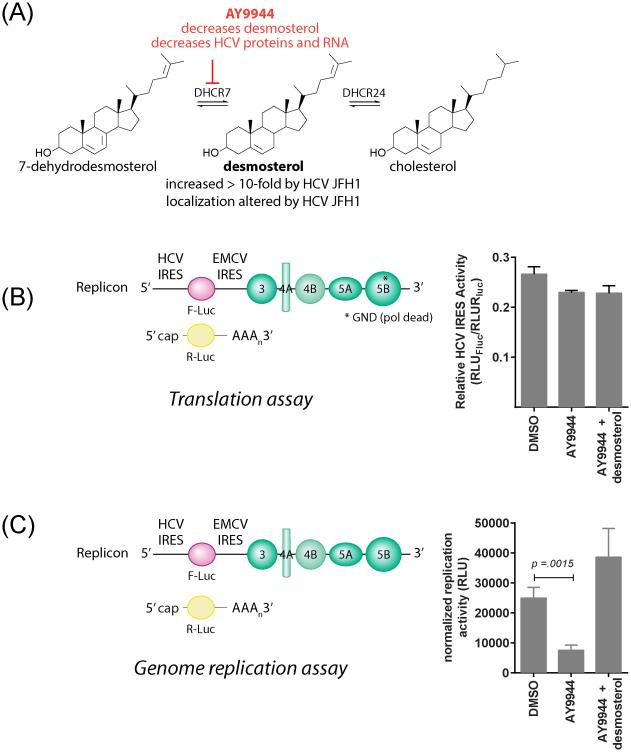
Figure 2. Desmosterol affects HCV RNA replication but has minimal effects on translation of the HCV genome.
(A) Desmosterol is the immediate precursor of cholesterol in the Bloch pathway. HCV JFH1 affects both desmosterol abundance6 and localization11. Pharmacological inhibition of DHCR7 leads to depletion of intracellular desmosterol and a profound reduction in both HCV proteins and genomic RNA that is rescued by the addition of exogenous desmosterol.6 (B and C) Effects of sterols on HCV translation and RNA replication were assessed by electroporation of a polymerase-dead mutant replicon RNA (“HCV-NS5B-GND”) (B) or wildtype HCV replicon RNA (C) along with a capped renilla luciferase mRNA into Huh7.5 cells pretreated with AY9944 to deplete intracellular desmosterol. (B) Relative translation of the HCV-NS5B-GND replicon RNA was determined by quantification of luciferase activities at 4 hours post-electroporation and normalization of the firefly luciferase signal (RLUFluc) using the renilla luciferase (RLURluc) signal to account for differences in electroporation efficiency. (C) Relative replication of the wildtype replicon RNA was assessed by normalizing the RLUFluc measured at 72 hours post-electroporation to RLUFluc/RLURLuc measured at 4 hours post-electroporation to account for differential electroporation efficiencies and decay of the input RNA. Translation of viral RNA is relatively unaffected by the absence of desmosterol, but RNA replication exhibits significant inhibition by AY9944 that is rescued by the addition of exogenous desmosterol. Representative data from an experiment independently performed n ≥ 2 times are shown.
RESULTS AND DISCUSSION
Desmosterol has a significant effect on steady-state replication of HCV RNA
In order to examine how desmosterol might function in the life cycle of HCV, we first sought to determine which of the component processes of this cycle – viral entry, translation of the viral RNA genome, replication of the viral RNA genome, or assembly and egress of progeny virions – is primarily affected by the presence or absence of desmosterol. Although our original discoveries were made using the live virus HCVcc20-22, which recapitulates the entire HCV life cycle, we have also shown that desmosterol affects the steady-state abundance of HCV proteins in a subgenomic replicon system that has been widely used as a model for studying HCV gene expression and genome replication in the absence of viral entry and virion assembly/egress23-25. We therefore initially focused our efforts on using well-established subgenomic replicon experiments to distinguish effects of desmosterol on translation of the viral genome from effects on steady-state replication of the viral genome.
The HCV subgenomic replicon is a bicistronic RNA that expresses a firefly luciferase reporter gene under control of the HCV internal ribosomal entry site (IRES) while the IRES of the encephalomyocarditis virus (EMCV) is used to drive expression of the HCV nonstructural proteins 3, 4A, 4B, 5A, and 5B (NS3, NS4A, NS4B, NS5A, NS5B)26 (Figure 1). Since this subgenomic replicon lacks the region of the genome encoding the HCV structural proteins -- core, E1, E2, and p7 – it cannot make new viral particles. Instead, the replicon RNA is synthesized by in vitro transcription and delivered to the cytosol by transfection or electroporation, thus by-passing the normal viral entry process. Firefly luciferase activity predominantly reflects translation of the input RNA at early times post-electroporation (< 8 hours) but at later time points reflects steady-state accumulation of newly synthesized replicon RNA. To ensure that we could resolve effects on translation versus RNA replication, we made use of a replicon bearing mutations in the NS5B gene (“HCV-NS5B-GND”) that render the viral RNA-dependent RNA polymerase inactive, thus providing a system in which firefly luciferase activity can unequivocally be taken as a readout of translation of the input replicon RNA.
Huh7.5 cells were pretreated with AY9944, a small molecule inhibitor of DHCR7, under conditions empirically established to result in the depletion of intracellular desmosterol 6 and then electroporated with the HCV-NS5B-GND mutant replicon RNA along with a capped renilla luciferase mRNA as an internal control for electroporation efficiency. Luciferase activities measured at four hours post-electroporation show that translation of the NS5B-GND replicon RNA is slightly affected by depletion of intracellular desmosterol; however, this slight reduction is not rescued by the addition of exogenous desmosterol (Figure 2B). These results demonstrate that translation of the replicon RNA is largely unaffected by the presence or absence of desmosterol and that an effect on translation is unlikely to explain to HCV’s sensitivity to desmosterol depletion. To confirm this, we performed analogous experiments with the wildtype replicon but quantifying luciferase activities at 72 hours post-electroporation as a measure of steady-state RNA replication. Depletion of intracellular desmosterol using AY9944 causes a significant decrease in RNA replication that is prevented in the presence of exogenously added desmosterol (Figure 2C). The correlation of these results using the replicon system with our previous results using live HCVcc provides strong evidence that desmosterol’s effect on viral RNA replication is a significant factor in its overall effect on HCV JFH1. Although these results do not preclude the possibility that desmosterol has additional effects on other steps of the HCV life cycle, we focused our efforts on investigating how desmosterol may affect HCV RNA replication.
Desmosterol is present in the membranes where HCV RNA replication occurs
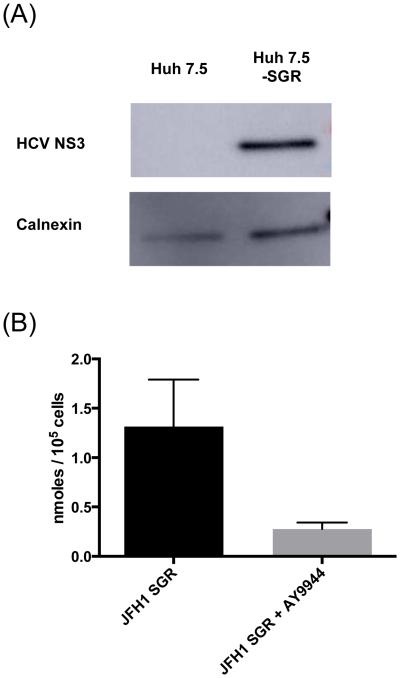
To investigate whether desmosterol might affect HCV RNA replication directly, we first examined whether it is present in the membranes where this viral process occurs. Synthesis of the viral RNA occurs in double-membrane vesicles (DMVs) that are often proximal to lipid droplets within the membranous web (Figure 1)19. The viral replicase includes all of the viral nonstructural proteins, including the NS3/4A protease, NS3 helicase, NS5B RNA-dependent RNA polymerase as well as the NS5A protein, which is required for and localizes at the sites of both HCV genome replication and virion assembly. We used previously established methods27 to isolate crude HCV replication membranes from Huh7.5 cells stably replicating the HCV JFH1 subgenomic replicon (Huh7.5-SGR) and performed Western blot analysis for the HCV NS3 protein to confirm successful isolation of replicase-containing membranes (Figure 3A). Liquid chromatography – mass spectrometry (LC-MS) analysis of lipidomic samples extracted from these replicase-containing membranes revealed that they have abundant desmosterol whereas desmosterol is significantly reduced in samples isolated from Huh7.5-SGR cells treated with AY9944 (Figure 3B) and undetectable in samples from uninfected cells (data not shown). Desmosterol’s presence in HCV JFH1-containing membranes demonstrates that it is a component of the membranous web and suggests that it may directly affect HCV RNA replication as well as other viral processes that occur therein rather than influencing HCV via signal transduction or other indirect mechanisms.
Figure 3. Desmosterol is abundant in HCV replication membranes.
(A) Western blot analysis confirms successful isolation process of replicase-containing membranes from Huh7.5-SGR cells. Both replicase-containing membranes and negative control membranes isolated from naïve Huh7.5 cells are positive for the ER marker calnexin, consistent with derivation of the replication membrane from ER. Only the membrane isolated from Huh7.5-SGR cells is positive for the HCV NS3 protein. (B) LC-MS was used to quantify desmosterol in replicase-containing membranes. Extracted ion chromatograms at m/z = 367.3367 were integrated and quantified according to the internal standard cholesterol D7 present in each sample at known quantities. Treatment of cells with AY9944 results in a decrease in desmosterol in replicase-containing membranes. The data are shown for n ≥ 3 independent experiments.
Desmosterol increases the fluidity of synthetic supported lipid bilayers (SLBs)
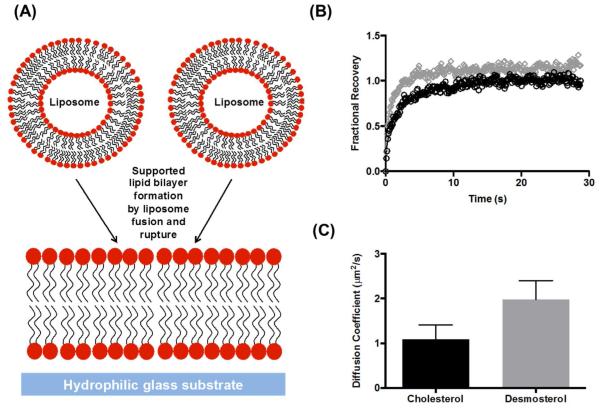
Since our experiments indicate that desmosterol is present in replicase-containing membranes (Figure 3) and has a significant effect on steady-state replication of HCV RNA (Figure 2C), we next considered how desmosterol might affect the biophysical properties of these membranes. Previous work has demonstrated that cholesterol is significantly better than desmosterol at promoting the formation of lipid-ordered domains in model membranes and inducing or stabilizing detergent-resistant membranes in mammalian cells28. While we are unaware of studies that quantitatively compare the effects of desmosterol and cholesterol on membrane fluidity and diffusivity, molecular dynamics simulations predict that desmosterol increases the surface area and fluidity of bilayers and, conversely, decreases order relative to those containing cholesterol and 7-dehydrocholesterol29, 30 To test this prediction experimentally, we turned to supported lipid bilayers (SLBs)31, 32. Since SLBs can be generated from synthetic liposomes, their composition is chemically “tunable” and can be modulated to interrogate how lipid composition affects the biophysical and biochemical properties of membranes33, 34. In particular, we sought to compare the effects of desmosterol and cholesterol on membrane fluidity using fluorescence recovery after photobleaching (FRAP)35 to determine diffusion coefficients as a metric of bilayer fluidity for SLBs with precisely controlled lipid content. Performing analogous experiments in cellulo is challenging due to the complexity and lack of planarity of cellular membranes. In addition, the reagents used to modulate lipid abundance in cellulo typically affect multiple species within a given lipid class making it difficult to precisely alter abundance of an individual species. For simplicity, we initially examined SLBs containing only unsaturated phospholipids and sterols and then progressively varied the sterol (desmosterol versus cholesterol), the diversity of lipid headgroups (phosphatidylcholines, phosphatidylethanolamines, phosphatidylserines, ceramides), and the saturation of hydrocarbon tails. Since the exact lipid composition of the membranous web and the ER membrane from which it is derived are not well-defined, we chose phospholipid mixtures that mimic the estimated content of the ER36.
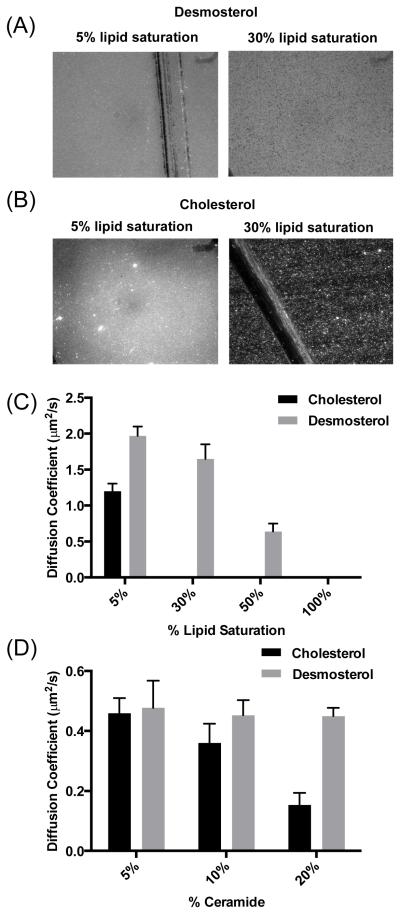
For simple unsaturated phospholipid SLBs (46 mol% POPC, 25 mol% POPE, 4 mol% POPS 13 mol% POPI, 0.01 mol% Texas red-DHPE, 5 mol% sterol), the presence of desmosterol was associated with slightly faster recovery of photobleached lipids (Figure 4B) and correspondingly larger diffusion coefficients (Figure 4C) when compared to SLBs containing cholesterol (1.1 ± 0.3 μ2/s to 1.98 ± 0.42 μ2/s, respectively). This functional difference in bilayer fluidity was even more pronounced when we examined more complex SLBs that included phosphatidylethanolamine and that varied the degree of saturation to better recapitulate the composition of the ER and HCV replicase-containing membranes. In examining a series of SLBs (Table 1) in which unsaturated phospholipids were systematically replaced with a saturated analog, we found that saturated phospholipid tail groups amplify the effects of sterol structure on bilayer formation and mobility. For example, at 5-mol% saturation (DPPS), both cholesterol- and desmosterol-containing SLBs exhibit a uniform fluorescent field after liposome incubation and subsequent washes (Figure 5A and 5B), indicating formation of a contiguous lipid bilayer. In contrast, when phospholipid saturation is increased to approximately 30% by the replacement of POPE with DPPE, only liposomes that contain desmosterol rupture and form a contiguous lipid bilayer (Figure 5A and 5B). Lipid diffusion measurements performed on these bilayers (Figure 5C) quantitatively demonstrate the effects of phospholipid saturation and sterol structure on membrane fluidity: whereas both desmosterol- or cholesterol-containing SLBs are mobile at 5% lipid saturation, only SLBs containing desmosterol exhibit lipid mobility when saturation of the phospholipid tail groups is increased to 30 and 50%.
Figure 4. Effect of desmosterol on SLB mobility.
(A) SLBs are formed upon incubation liposomes on hydrophilic glass substrates. (B). Fractional recovery of fluorescence in the photobleached spot plotted as a function of time for SLBs containing desmosterol (grey diamonds) or cholesterol (black circles) shows that membranes containing desmosterol recover at a faster rate after photobleaching. (C) Diffusion coefficients extracted from fitting data in (B) for membranes containing cholesterol (black) or desmosterol (gray) demonstrate slightly increased diffusivity for the desmosterol-containing bilayer. Representative data are shown or n ≥3 independent experiments. Error bars represent the standard deviation of the mean calculated for 3 replicates, p-value = 0.043.
Table 1.
Liposomes compositions for SLB experiments in Figure 5C. The degree of acyl chain saturation (16:0 or 16:0-18:1) is shown for each phospholipid headgroup class present in liposomes.
| PC | PE | PS | Sterol |
%
Saturation |
|
|---|---|---|---|---|---|
| 1 | 16:0 (DPPC) |
16:0 (DPPE) |
16:0 (DPPS) |
Des/Chol | 100% |
| 2 | 16:0 (DPPC) |
16:0-18:1 (POPE) |
16:0-18:1 (POPS) |
Des/Chol | 55% |
| 3 | 16:0-18:1 (POPC) |
16:0 (DPPE) |
16:0-18:1 (POPS) |
Des/Chol | 30% |
| 4 | 16:0-18:1 (POPC) |
16:0-18:1 (POPE) |
16:0 (DPPS) |
Des/Chol | 5% |
Figure 5. Formation and fluidity of SLBs containing saturated lipids is promoted by desmosterol.
(A and B) Epifluorescence image of SLB containing 5% and 30% saturated lipids with desmosterol (A) or cholesterol (B). Scratches in images are used to ensure focus is in the correct plane. Liposomes containing cholesterol and 30% saturated lipids do not rupture and appear as punctate fluorescent dots on the glass slide. (C) Diffusion coefficients extracted from FRAP data of SLBs containing increasing lipid tail saturation and desmosterol (grey) or cholesterol (black) show that increasing saturation reduces diffusivity and that desmosterol counteracts this effect by maintaining bilayer fluidity even at high levels of saturation whereas cholesterol does not. (D) Diffusion coefficients derived from FRAP data of SLBs containing increasing concentration of C18 ceramide similarly show that increasing C18 ceramide abundance reduces bilayer diffusivity. Desmosterol-containing SLBs retain bilayer diffusivity whereas cholesterol- containing SLB do not. Representative data are shown or n ≥3 independent experiments. Error bars represent the standard deviation of the man calculated for 3 replicates.
The examination of ceramide-containing SLBs was prompted by several reasons. First, the membranous web and double-membrane vesicles wherein HCV RNA replication occurs are derived from the ER, the site of ceramide biosynthesis. Second, the abundance of ceramide species in Huh7 cells increases during HCV infection1, and we detected a doubling of ceramide-PI (16:0) levels in our prior lipidomic profiling of HCV JFH1-infected Huh7.5 cells6 Third, ceramides are known to exert profound effects on lipid packing and order, segregating into regions of highly ordered lipids with decreased membrane fluidity (“lipid rafts")37. We therefore questioned whether desmosterol restores membrane fluidity of bilayers with increasing concentrations of saturated ceramide. Analysis of a series of SLBs containing POPC (55%) POPE (35%), 5% sterol, and increasing concentrations (5-20%) of saturated C-18 ceramide showed that desmosterol- and cholesterol-containing SLBs exhibit similar diffusion coefficients when ceramide concentration is low (5%), but differ significantly as the concentration of ceramide increases (Figure 5D). Whereas bilayer diffusivity decreases significantly with increasing ceramide content for cholesterol-containing SLB, the desmosterol-containing SLBs maintain diffusivity despite increasing ceramide content. Together, these results demonstrate that desmosterol increases the diffusivity and hence fluidity of lipid bilayers; moreover, this effect appears to be somewhat generalizable since we observe it in bilayers containing unsaturated phospholipids, saturated phospholipids, and ceramides.
Desmosterol increases the fluidity of HCV replicase-containing membranes
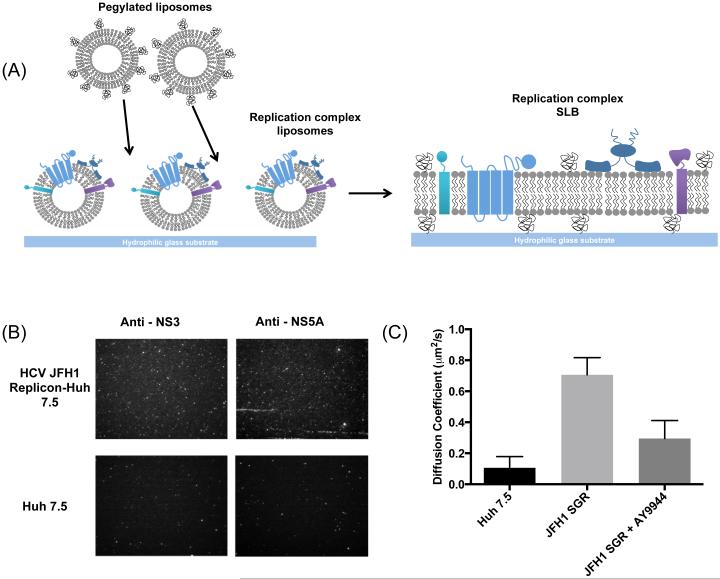
To validate the effects of desmosterol that we observed on purely synthetic SLBs in a more biologically relevant system, we compared authentic replicase-containing membranes from Huh7.5-SGR cells with the fluidity of membranes isolated by the same protocol from naïve Huh7.5 cells lacking the HCV replicon. In particular, we sought to correlate the presence of desmosterol in HCV JFH1 replicase-containing membranes with increased bilayer fluidity and diffusivity. Since the complete inventory and stoichiometry of protein components in the HCV replicase are not known, we generated SLBs using HCV replicase-containing membranes isolated from Huh7.5-SGR cells to ensure the presence of the authentic replicase. For this, we built on our prior success using plasma membrane-derived proteoliposomes to generate SLBs containing membrane proteins amenable to characterization in biophysical and enyzmatic assays 38, 39. Briefly, HCV replicase-containing membranes were isolated from Huh7.5-SGR cells and negative control membranes were isolated from Huh7.5 cells using the same protocol, as described for Figure 3. These isolated membranes were used to generate proteoliposomes that were labeled with octadecyl rhodamine (R18), a lipophilic dye, and then deposited onto a hydrophilic glass substrate. Proteoliposome rupture was triggered by the addition of synthetic liposomes, leading to the formation of a proteinaceous, planar SLB. The presence of viral proteins in these SLB was confirmed by immunofluorescence detection of the HCV NS3 and NS5A proteins (Figure 6A). Although some staining is apparent in the negative control SLB derived from Huh7.5 cells, this signal is less abundant than for the SLB derived from Huh7.5-SGR cells and is likely due to background staining.
Figure 6. Characterization of SLBs derived from replicase-containing membranes.
(A) Schematic of bilayer fo1mation containing viral replicase (multicoloured proteins). Replicase-containing membranes are deposited on a hydrophilic glass substrate. Liposomes containing POPC and PEG are added to induce mpture of the protein-rich liposomes and fo1mation of an SLB. (B) Immunofluorescence detection of the HCV NS3 and NS5A proteins confom the presence of viral replicase proteins in the membranes. ER vesicles isolated from naive Huh 7.5 cells using the same procedure were used as a negative control. (C) Diffusion coefficients dete1mined from FRAP experiments demonstrate that replicase-containing SLBs derived from Huh7.5-SGR cells have significantly increased lipid mobility relative to negative control SLBs (p < 0.0014) . SLBs derived from desmosterol-depleted replicase membranes exhibit significantly decreased mobility compared to wild type membranes (p < 0.0114). Representative data are shown or n ≥3 independent experiments. Error bars represent the standard deviation of the man calculated for 3 replicates.
FRAP microscopy experiments performed to measure diffusion coefficients on these SLBs demonstrated a striking difference between those derived from the desmosterol-containing HCV replication membranes versus the negative control SLB lacking desmosterol (Figure 6B). Specifically, the diffusion coefficient measured for the Huh7.5-SGR SLB was 0.7 ± 0.04 μm2/s, seven times greater than that of the Huh7.5 SLB (0.11 ± 0.06 μm2/s). To further determine if the increase in fluidity in Huh7.5-SGR-derived SLBs is due to increased desmosterol, we prepared SLBs from crude replication membranes isolated from cells treated with AY9944 to deplete desmosterol. The diffusion coefficient of desmosterol-depleted replication membranes were calculated to be 0.29 ± 0.11 μm2/s, two-fold slower than wild type replication membranes. These data provide strong evidence that increased desmosterol in HCV JFH1 replication membranes contributes to significantly increased bilayer fluidity. Since the exact composition of the biological membranes used to produce these SLBs cannot be known in the absence of untargeted lipidomic analysis, we cannot currently exclude the possibility that other lipid species present in these membranes also modulate bilayer fluidity and thus the fluidity of the SLBs. Despite this caveat, our analysis of replicase-containing and negative control membranes in Figures 3 and 6 demonstrates a striking difference in desmosterol content that is correlated here with a major effect on membrane fluidity (Figure 6).
Desmosterol’s effect on membrane fluidity influences HCV replication by promoting formation of the membranous web
Although there is growing appreciation for the importance of membranes in viral replication, understanding how the lipid composition of membranes affects their biophysical properties and function in viral processes remains a major challenge. Investigations in this area require, first, knowledge of the specific lipid molecules (versus classes of lipids) present in the membranes where viral processes occur, and second, experimental systems in which biophysical and biochemical measurements can be made on membranes whose lipid composition is chemically well-defined and precisely controlled. HCV’s well-known perturbation of the host cell to induce formation of membranous web as a site for viral genome replication and assembly provides an apt virological system in which to pursue these questions.
We previously demonstrated that HCV JFH1 induces a ten-fold or greater increase in intracellular desmosterol6 and also causes desmosterol to accumulate in lipid droplets associated with the viral NS5A protein11. These HCV-induced perturbations of desmosterol are functionally important for viral replication since experimental conditions that reduce intracellular desmosterol cause a profound reduction in HCV proteins and RNA that is rescued upon the addition of exogenous desmosterol6, 11 Importantly, HCV JFH1’s interaction with desmosterol appears to be sterol-specific since the intracellular abundance of cholesterol and other late-stage sterols is unaffected by the virus6 and stimulated Raman scattering (SRS) microscopy experiments show that HCV JFH1-induced lipid droplets containing desmosterol are visually distinct from those containing cholesterol11. Together, these findings have suggested a model in which HCV JFH1 perturbs desmosterol homeostasis to promote viral replication, likely by affecting a viral process or processes directly. The work presented here builds upon this model by mapping desmosterol’s effects to the step of viral RNA replication, and by beginning to investigate the biochemical and biophysical mechanism underlying desmosterol’s effect on this viral process.
Our experiments demonstrate that the presence of desmosterol has a major effect on replication of HCV JFH1 RNA (Figure 2) and that this is correlated with an increase in membrane fluidity (Figures 4-6). Although additional work is needed to understand how desmosterol’s enhancement of membrane fluidity promotes viral RNA replication, several non-mutually exclusive possibilities are worth mention. First, desmosterol’s presence in both replicase-containing membranes (Figure 3) and HCV-induced lipid droplets11, which together comprise the membranous web16-18 suggests that the increased fluidity conferred by desmosterol is important for the massive structural rearrangement and reorganization of cellular lipids required to form this specialized, membranous environment for viral genome replication and assembly. Increased membrane fluidity may facilitate membrane curvature and bending. Alternatively, increased lateral diffusivity of the double-membrane vesicles where HCV genome replication occurs may facilitate trafficking of replicase components to this site. A third possibility is that increased membrane fluidity may promote RNA replication by enhancing the structural dynamics needed for efficient catalysis.
Interestingly, the importance of membrane fluidity for replication of the HCV genome has also been highlighted by recent studies demonstrating that HCV is exquisitely sensitive to lipid peroxidation and suggesting that increasing lipid peroxidation during HCV infection has an inhibitory effect on formation (or maintenance) of the membranous web40. This sensitivity to lipid peroxidation coincides with previous work showing that oxidized phospholipids decrease membrane fluidity41. HCV JFH1 is unique among HCV isolates in its relative insensitivity to lipid peroxidation40, 42. While the biochemical mechanism(s) that enable HCV JFH1 to overcome the detrimental effects of lipid peroxidation have yet to be defined, one possibility suggested by our data is that the HCV JFH1-induced accumulation of desmosterol in replication membranes reduces the membrane ‘stiffening’ effect resulting from oxidative stress. Analogous studies performed on cells, replicase-containing membranes, and SLBs isolated from cells bearing subgenomic replicons derived from other HCV strains may help to determine whether desmosterol is critical for HCV’s resistance to lipid peroxidation and to test the hypothesis that its effects on membrane fluidity are responsible.
Our experiments also illustrate the potential of synthetic and cell-derived supported lipid bilayers as experimental systems for interrogating how lipid composition affects membrane properties and function. While our efforts here focused on investigation of the effects of desmosterol on bilayer fluidity, we anticipate that this and other SLB systems will provide a useful platform for additional studies aimed at interrogation of the effects of lipid composition on other functional properties of membranes (e.g., membrane depth, elasticity) and the function of these membranes in viral replication. Experimental models that recapitulate individual viral processes outside of the cell are needed for these efforts since properties like membrane fluidity may have differential effects on viral entry, gene expression, genome replication, and viral assembly/egress. For example, whereas our results correlate desmosterol’s enhancement of membrane fluidity with successful RNA replication, Ye and colleagues have shown that increased membrane fluidity has a deleterious effect on HCV fusion during viral entry 42. Generation of SLBs from replication membranes isolated from HCV SGR and HCV-infected cells allows study of the authentic viral replicase on membranes amenable to a variety of biochemical, biophysical, and imaging methods. We anticipate that systems such as the one we describe, along with minimalist, biochemical reconstitution systems,44 will enable elucidation of how lipid structure and membrane composition affect HCV RNA replication.
Materials and Methods
Replicon experiments
Huh7.5 cells s were generous gifts from Dr. Charles Rice (Rockefeller University) via Apath, LLC. Replicon plasmids pSGR-JFH1and pSGR-JFH1-NS5B-GND were gifts from Dr. Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan). Huh 7.5 cells, that had been treated 24 hours previously with 4 μM AY9944 were trypsinized and washed twice with Accugene PBS. Cells were resuspended in Accugene PBS at 5 × 106 cells/mL. In vitro transcription (IVT) was performed on pSGR-JFH1and pSGR-JFH1-NS5B-GND using an Ampliscribe kit following the manufacturer’s protocol. 500 μL of Huh 7.5 cells were electroporated (4 mm gap cuvette; 5 pulses of 820 V at 100 milliseconds for each pulse with a BTX ECM830 electroporator) in the presence of 5-10 μg of HCV-NS5B-GND or HCV-SGR mRNA transcript and capped renilla luciferase as a control. Cells were allowed to recover for 10 minutes and then treated with 10 μM of cholesterol or desmosterol. Cells electroporated with HCV-NS5B-GND for analysis of translation were harvested after 4 hours. Cells electroporated with HCV-SGR wildtype for analysis of RNA replication were treated with sterol every 24 hours and harvested 72 hours after electroporation. Luciferase signal for renilla and firefly luciferase was detected using Stop and Glow luciferase detection kit (Promega) on a Biotek Syngery plate reader. Translation of the input replicon RNA was measured by dividing firefly luciferase activity (RLUFLuc) by renilla luciferase activity (RLURLuc). Relative RNA replication was assessed by calculating RLUFluc/RLURluc at 72 hours and dividing this value by RLUFluc/RLURluc measured at 4 hours post-electroporation to normalize for differential electroporation efficiencies and decay of the input RNA.
Isolation of crude replication complexes
Naïve Huh 7.5 cells lacking HCV or Huh7.5-HCV-SGR stably replicating the HCV JFH1 subgenomic replicon cells were harvested at 70% confluency by trypsinization followed by centrifugation (1000×g for 10 minutes at 4°C). Cells treated with AY9944 to deplete desmosterol were harvested 3 days after treatment with 4 μM AY9944. The cell pellets were washed in PBS and then resuspended in hypotonic buffer. Cells were lysed with 75 strokes of a Dounce homogenizer. The lysate was then centrifuged at 1200×g for 10 minutes at 4°C. The supernatant was removed and centrifuged at 10,000×g for 10 mins at 4°C. The supernatant was collected and then centrifuged at 67000×g for 75 minutes at 4°C. The pellet was resuspended in PBS and stored in aliquots at −80°C. The presence of HCV NS3 and/or the ER marker calnexin in replication complexes was confirmed by western blot using primary antibodies against Calnex (Abcam) and HCV NS3 (Abcam) and goat anti-mouse HRP secondary antibody (Biorad).
LC-MS analysis of desmosterol content
Crude replication complexes were resuspended in 1 mL of PBS. Lipids were extracted using the Bligh-Dyer method. During extraction, all of the organic phase is collected. Cholesterol D7 (Avanti) was added to each sample as an internal standard before samples were dried down under nitrogen. Samples were dissolved in methanol for analysis. LC-MS was performed on an Agilent 6530 QTOF Mass Spectrometer with 1290 Infinity Binary LC in positive ion mode. For quantification of desmosterol in CRCs lipids were chromatographically separated on a UHPLC Phenomenex Kinetex 2.6μM XB-C18, 100Å column. Gradients using mobile phase A (85% Methanol, 15% water, 5mM ammonium acetate , 0.1% formic acid) and mobile phase B (100% methanol, 5mM ammonium acetate, 0.1% formic acid) were run as follows : 0-2 minutes 100% A, 2-15 mins 100% B, 15-25.5 100% B, 25.5-30 mins 100% A.
FRAP experiments
Lipid Vesicle Preparation
Lipids were purchased from Avanti Polar Lipids. The following lipids were used in these experiments: 1, 2 dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1-palmitoyl-2-oleoylsn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol (POPI), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) 1,2-dipalmitoyl-snglycero-3-phosphocholine (DPPC), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), 1,2-dipalmitoyl-sn-glycero-3-phosphoserine (DPPS), C18 ceramide (brain, procine), desmosterol, cholesterol, texas red DHPE (Molecular Probes). Lipids were thoroughly mixed in a scintillation vial at desired ratios and then the solvent was removed under a stream of high purity nitrogen gas. To ensure all solvent was removed, the vial was placed in a desiccator under vacuum for an additional 2.5 to 3 hours. 4 mL of PBS at pH 7.0 was then added to the dried lipid film and resuspended gently in a benchtop sonicator for twenty minutes on the lowest setting. The final lipid concentration was approximately 2mg/mL. Liposomes underwent two freeze thaw cycles and were then extruded twice through a polycarbonate filter with pore size 100 nm, and thirty times through a filter with a pore size of 50 nm.
Supported lipid bilayer formation
Glass microscope coverslips were cleaned by immersion in 150 mL of piranha solution (70% sulfuric acid, 30% hydrogen peroxide) for ten minutes. The slides were subsequently rinsed for 30 minutes with copious amounts of deionized water. Clean slides were stored under deionized water, then dried with a stream of ultra pure nitrogen gas prior to use. Integrity of bilayers and diffusion of the lipids within it were examined by fluorescence recovery after photobleaching (FRAP). A 0.2 mg/mL solution of liposomes in PBS was incubated in a PDMS well attached to a piranha cleaned slide. Liposomes were allowed to incubate for 20 minutes before being rinsed with PBS at pH 7.0 for one minute. The bilayer was scratched with a dissection tool to remove a thin line of bilayer to aid in focusing on the plane of the bilayer on the microscope. Following the scratching step, the bilayer was rinsed again for one minute with PBS to wash out any lipids removed by scratching. Bilayers were images on a Zeiss Axio Observer.Z1 with a 100x oil objective with a numerical aperture of 1.46. A circular region in the supported lipid bilayer was bleached with a 561 nm solid state laser for 100 ms. The recovery of the intensity of the photobleached spot was recorded for 3 minutes. The fluorescence intensity of the bleached spot was determined after background subtraction and normalization for each image. The recovery data was fit using a Bessel function following the method of Soumpasis45. The diffusion coefficient was then calculated using the following equation: , where w is the full width at half-maximum of the Gaussian profile of the focused beam.
Supported lipid bilayers derived from replicase-containing membranes
Crude replication complexes (CRCs) (0.1 μg/mL protein) diluted in PBS were fluorescently labeled with 0.18 mM lipophilic dye octadecyl Rhodamine (R18) for 15 minutes in a room temperature water bath with gentle sonication. 100 μL of fluorescently labeled CRC solution was incubated in a PDMS well on a piranha cleaned glass slide for 15 minutes. Unadsorbed CRCs were rinsed from the device with PBS. 100 μL of liposomes (0.2 mg/mL) containing POPC and 0.5 mol% DOPE PEG 2000 were incubated with the adsorbed CRCs to induce rupture for 20 minutes. Excess liposomes were rinsed using PBS. Bilayers were incubated with 1:3000 dilution of monoclonal anitbodies HCV NS3 (Abcam) or HCV NS5A (antibody 9E10; provided by Dr. Charles Rice, Rockefeller University) for 30 minutes. Unbound antibody was washed away with PBS and bilayers were incubated for 30 minutes with 1:3000 dilution of secondary goat-anti mouse conjugated to Oregon green (Molecular Probes).
ACKNOWLEDGEMENTS
The authors are thankful for support from NIH R01AI076442 and Diversity Supplement R01AI076442-S1 (PLY, VAV), NIH F32AI106178 (V.A.V), NIH R56AI112960 and R01AI112960 (J. Chou and P.L.Y.), and a John and Virginia Kaneb Fellowship (PLY). We thank Dr. Charles Rice (Rockefeller University via Apath LLC) for Huh7.5 cells and monoclonal antibody 9E10, and Dr. Takaji Wakita (National Institute of Infectious Diseases, Japan) for JFH1 plasmid. We gratefully acknowledge the Harvard ICCB-Longwood Screening Facility for use of plate readers, Jon Clardy and the Harvard ICCB-Longwood Analytical Chemistry Core for use of LC-MS, and Tomas Kirchhausen and the Microscopy Core of the Harvard Digestive Diseases Center (NIH DK034855, W. Lencer) for access to fluorescence microscopes.
References
- 1.Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, Kohara M. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS pathogens. 2012;8:e1002860. doi: 10.1371/journal.ppat.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merz A, Long G, Hiet M-S, Brügger B, Chlanda P, Andre P, Bartenschlager R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. Journal of Biological Chemistry. 2011;286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu K, Nagy PD. RNA virus replication depends on enrichment of phosphatidylethanolamine at replication sites in subcellular membranes. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1418971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanner LB, Chng C, Guan XL, Lei Z, Rozen SG, Wenk MR. Lipidomics identifies a requirement for peroxisomal function during influenza virus replication. Journal of lipid research. 2014;55:1357–1365. doi: 10.1194/jlr.M049148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, Katze MG. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodgers MA, Villareal VA, Schaefer EA, Peng LF, Corey KE, Chung RT, Yang PL. Lipid metabolite profiling identifies desmosterol metabolism as a new antiviral target for hepatitis C virus. Journal of the American Chemical Society. 2012;134:6896–6899. doi: 10.1021/ja207391q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Bartenschlager R. Recruitment and Activation of a Lipid Kinase by Hepatitis C Virus NS5A Is Essential for Integrity of the Membranous Replication Compartment. Cell Host & Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyuncu E, Purdy JG, Rabinowitz JD, Shenk T. Saturated very long chain fatty acids are required for the production of infectious human cytomegalovirus progeny. PLoS Pathog. 2013;9:e1003333. doi: 10.1371/journal.ppat.1003333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Kuhn RJ. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012;8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet (London, England) 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villareal VA, Fu D, Costello DA, Xie XS, Yang PL. Hepatitis C Virus Selectively Alters the Intracellular Localization of Desmosterol. ACS chemical biology. 2016 doi: 10.1021/acschembio.6b00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. Journal of virology. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapadia SB, Barth H, Baumert T, McKeating JA, Chisari FV. Initiation of Hepatitis C Virus Infection Is Dependent on Cholesterol and Cooperativity between CD81 and Scavenger Receptor B Type I. Journal of virology. 2006;81:374–383. doi: 10.1128/JVI.01134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvisi G, Madan V, Bartenschlager R. Hepatitis C virus and host cell lipids: an intimate connection. RNA biology. 2011;8:258–269. doi: 10.4161/rna.8.2.15011. [DOI] [PubMed] [Google Scholar]
- 16.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of Hepatitis C Virus Proteins Induces Distinct Membrane Alterations Including a Candidate Viral Replication Complex. Journal of virology. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. Journal of virology. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nature Cell Biology. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 19.Romero-Brey I, Merz A, Chiramel A, Lee J-Y, Chlanda P, Haselman U, Bartenschlager R. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS pathogens. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 21.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature medicine. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 24.Targett-Adams P, McLauchlan J. Development and characterization of a transient-replication assay for the genotype 2a hepatitis C virus subgenomic replicon. The Journal of general virology. 2005;86:3075–3080. doi: 10.1099/vir.0.81334-0. [DOI] [PubMed] [Google Scholar]
- 25.Date T, Kato T, Miyamoto M, Zhao Z, Yasui K, Mizokami M, Wakita T. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. The Journal of biological chemistry. 2004;279:22371–22376. doi: 10.1074/jbc.M311120200. [DOI] [PubMed] [Google Scholar]
- 26.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 27.Quinkert D, Bartenschlager R, Lohmann V. Quantitative Analysis of the Hepatitis C Virus Replication Complex. Journal of virology. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vainio S, Jansen M, Koivusalo M, Rog T, Karttunen M, Vattulainen I, Ikonen E. Significance of Sterol Structural Specificity: DESMOSTEROL CANNOT REPLACE CHOLESTEROL IN LIPID RAFTS. Journal of Biological Chemistry. 2005;281:348–355. doi: 10.1074/jbc.M509530200. [DOI] [PubMed] [Google Scholar]
- 29.Róg T, Pasenkiewicz-Gierula M, Vattulainen I, Karttunen M. Ordering effects of cholesterol and its analogues. Biochimica et biophysica acta. 2009;1788:97–121. doi: 10.1016/j.bbamem.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Róg T, Vattulainen I, Jansen M, Ikonen E, Karttunen M. Comparison of cholesterol and its direct precursors along the biosynthetic pathway: effects of cholesterol, desmosterol and 7-dehydrocholesterol on saturated and unsaturated lipid bilayers. The Journal of Chemical Physics. 2008;129:154508. doi: 10.1063/1.2996296. [DOI] [PubMed] [Google Scholar]
- 31.Brian AA, McConnell HM. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc Natl Acad Sci U S A. 1984;81:6159–6163. doi: 10.1073/pnas.81.19.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castellana ET, Cremer PS. Solid supported lipid bilayers: From biophysical studies to sensor design. Surface Science Reports. 2006;61:429–444. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao L, Daniel S. Measuring the partitioning kinetics of membrane biomolecules using patterned two-phase coexistant lipid bilayers. Journal of the American Chemical Society. 2011;133:15635–15643. doi: 10.1021/ja205274g. [DOI] [PubMed] [Google Scholar]
- 34.Pace H, Simonsson Nystrom L, Gunnarsson A, Eck E, Monson C, Geschwindner S, Hook F. Preserved transmembrane protein mobility in polymer-supported lipid bilayers derived from cell membranes. Anal Chem. 2015;87:9194–9203. doi: 10.1021/acs.analchem.5b01449. [DOI] [PubMed] [Google Scholar]
- 35.Yguerabide J, Schmidt JA, Yguerabide EE. Lateral mobility in membranes as detected by fluorescence recovery after photobleaching. Biophysical journal. 1982;40:69–75. doi: 10.1016/S0006-3495(82)84459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holopainen JM, Subramanian M, Kinnunen PK. Sphingomyelinase induces lipid microdomain formation in a fluid phosphatidylcholine/sphingomyelin membrane. Biochemistry. 1998;37:17562–17570. doi: 10.1021/bi980915e. [DOI] [PubMed] [Google Scholar]
- 38.Costello DA, Hsia C-Y, Millet JK, Porri T, Daniel S. Membrane Fusion-Competent Virus-Like Proteoliposomes and Proteinaceous Supported Bilayers Made Directly from Cell Plasma Membranes. Langmuir. 2013;29:6409–6419. doi: 10.1021/la400861u. [DOI] [PubMed] [Google Scholar]
- 39.Costello DA, Millet JK, Hsia C-Y, Whittaker GR, Daniel S. Single particle assay of coronavirus membrane fusion with proteinaceous receptor-embedded supported bilayers. Biomaterials. 2013;34:7895–7904. doi: 10.1016/j.biomaterials.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamane D, McGivern DR, Wauthier E, Yi M, Madden VJ, Welsch C, Lemon SM. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nature Medicine. 2014;20:927–935. doi: 10.1038/nm.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borst JW, Visser NV, Kouptsova O, Visser AJ. Oxidation of unsaturated phospholipids in membrane bilayer mixtures is accompanied by membrane fluidity changes. Biochimica et biophysica acta. 2000;1487:61–73. doi: 10.1016/s1388-1981(00)00084-6. [DOI] [PubMed] [Google Scholar]
- 42.Huang H, Chen Y, Ye J. Inhibition of hepatitis C virus replication by peroxidation of arachidonate and restoration by vitamin E. Proc Natl Acad Sci U S A. 2007;104:18666–18670. doi: 10.1073/pnas.0708423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chamoun-Emanuelli AM, Pecheur EI, Simeon RL, Huang D, Cremer PS, Chen Z. Phenothiazines inhibit hepatitis C virus entry, likely by increasing the fluidity of cholesterol-rich membranes. Antimicrob Agents Chemother. 2013;57:2571–2581. doi: 10.1128/AAC.02593-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho NJ, Pham EA, Hagey RJ, Lévêque VJ, Ma H, Klumpp K, Glenn JS. Reconstitution and functional analysis of a full-length hepatitis C virus NS5B polymerase on a supported lipid bilayer. ACS Central Science. 2016;2:456–466. doi: 10.1021/acscentsci.6b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soumpasis DM. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys J. 1983;41:95–97. doi: 10.1016/S0006-3495(83)84410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]