Summary
During DNA double strand break (DSB) repair, the ring-shaped Ku70/80 complex becomes trapped on DNA and needs to be actively extracted, but it has remained unclear what provides the required energy.
By means of reconstitution of DSB repair on beads, we demonstrate here that DNA-locked Ku rings are released by the AAA-ATPase, p97. To achieve this, p97 requires ATP hydrolysis, cooperates with the Ufd1-Npl4 ubiquitin adapter complex and specifically targets Ku80 that is modified by K48-linked ubiquitin chains. In U2OS cells, chemical inhibition of p97, or siRNA-mediated depletion of p97 or its adapters impairs Ku80 removal after non-homologous end-joining of DSBs. Moreover, it attenuates early steps in homologous recombination consistent with p97-driven Ku release also affecting repair pathway choice.
Thus, our data solve a central question regarding regulation of Ku in DSB repair, and illustrate the ability of p97 to segregate even tightly bound protein complexes for release from DNA.
Introduction
DNA double strand breaks (DSBs) represent the most devastating type of DNA damage. Hence, cells have evolved sophisticated repair mechanisms that involve the coordinated assembly and disassembly of repair protein complexes (Ciccia & Elledge, 2010; Jackson & Bartek, 2009). The initial sensor protein for DSBs is the Ku70/80 (Ku) heterodimer with a central cavity exactly large enough to accommodate a B-form DNA helix (Walker et al., 2001). Its toroidal shape not only confers high affinity to open ends (Kd ~2 nM; Blier et al., 1993) by threading the DNA through the rigid channel but also ensures selective binding, as intact DNA is not able to enter the closed ring. Upon binding to DNA, Ku initiates non-homologous end-joining (NHEJ) by recruiting the additional NHEJ factors, including DNA-PKcs, XLF, Artemis, and DNA ligase IV (reviewed in Lieber et al., 2010). Concomitantly, Ku binding to DNA prevents extensive end resection and thereby inhibits the competing error-free homologous recombination repair (HRR) pathway (Sun et al., 2012). However, as each subunit of Ku fully encircles the DNA (Walker et al., 2001), DNA ligation results in the Ku70/80 rings being sterically interlocked with the DNA. Since the Ku ring does not possess a clasp, this excludes opening the ring to leave the DNA by dissociation of the discrete subunits, as seen for other DNA binding proteins (e. g. PCNA). Consequently, Ku needs to be actively extracted by structural remodelling (Postow et al., 2008). Extraction of Ku is triggered by modification with K48-linked ubiquitin chains and three different E3 ligases, SCFFbxl12, RNF8 and RNF138 have been identified to mediate Ku80 ubiquitination (Postow & Funabiki, 2014; Feng & Chen, 2012; Ismail et al., 2015; Schmidt et al., 2015). However, it is unclear what provides the required energy for Ku extraction.
Valosin-containing protein (VCP)/p97 is a hexameric AAA+-type ATPase that targets ubiquitinated substrate proteins through ubiquitin adapter proteins. It uses the energy of ATP hydrolysis to segregate the substrate proteins from binding partners or cellular structures often, but not always, for downstream degradation by the proteasome (Jentsch & Rumpf, 2007). While its best-studied role is in ER-associated degradation (Stolz et al., 2011), p97 was shown to extract different ubiquitinated proteins from chromatin in processes such as cell cycle regulation, transcriptional and replication stress responses, nucleotide excision repair or replication (reviewed in Meyer et al., 2012; Dantuma et al., 2014). Recently, p97 was found to be involved in DSB repair (Meerang et al., 2011; Acs et al., 2011) with one function at a late stage of HRR in controlling association of the Rad51-Rad52 complex (Bergink et al., 2013). In addition, p97 is involved in NHEJ and compromising p97 function leads to accumulation of K48-linked ubiquitin chains on DSB sites (Meerang et al., 2011). However, so far, it is unclear what the relevant ubiquitinated target of p97 is and, therefore, the major mechanism by which p97 contributes to the NHEJ pathway is unsolved.
In an in vitro mass spectrometry approach, we identify Ku as a major substrate of p97 during DSB repair. By reconstituting Ku extraction in vitro, we demonstrate that p97 along with its cofactor Ufd1 extracts Ku rings that are trapped during DNA repair and show that p97-mediated release of Ku is relevant in living cells. Moreover, we provide evidence consistent with p97-mediated Ku extraction facilitating HRR initiation and thus contributing to repair pathway choice.
Results
Ku70/80 accumulates on DSBs in vitro when p97 is compromised
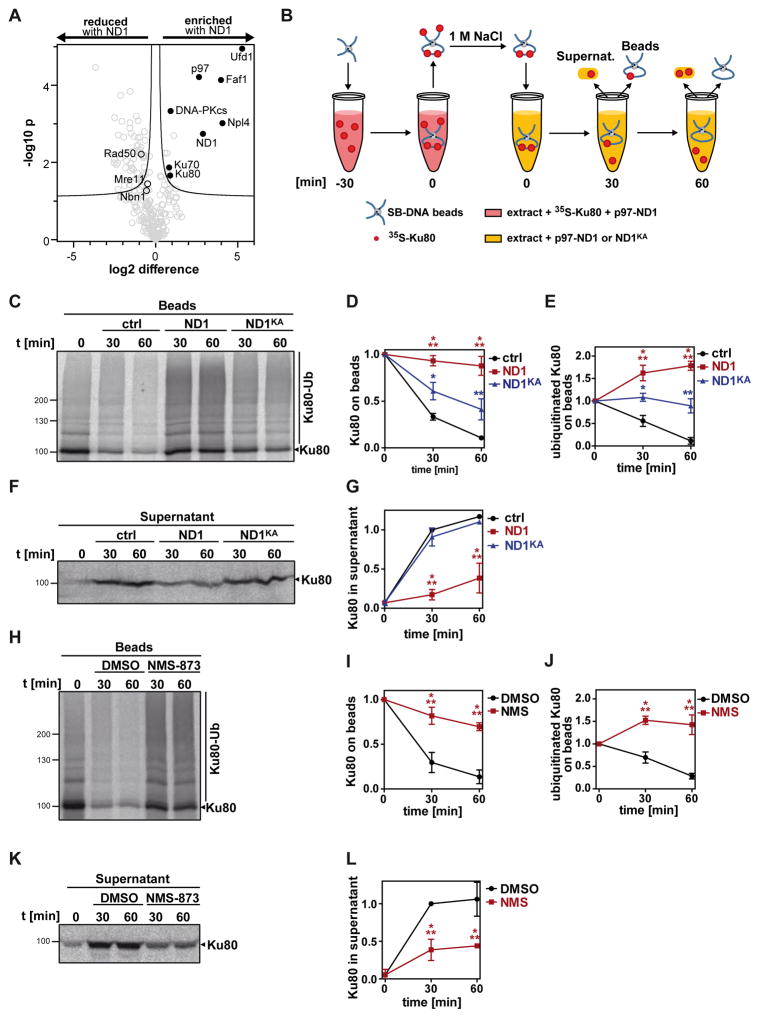
To recapitulate DNA DSB repair in vitro, we adopted a method using CSF (cytostatic factor) arrested Xenopus laevis egg extracts (Postow et al., 2008). These extracts are capable of repairing damaged DNA via NHEJ using very similar mechanisms to those found in mammalian cells (Labhart, 1999; Di Virgilio & Gautier, 2005). For the isolation of DNA repair proteins from egg extracts, we selectively biotinylated DNA fragments on one end (single-biotinylated, SB-DNA) and coupled them to streptavidin beads (Figure S1A). As expected, the free ends of the SB-DNA beads were recognized as DSBs and readily repaired by the extract, resulting in a doubling of DNA length (Figure S1B). In contrast, double-biotinylated DNA fragments (DB-DNA) with both ends attached to the beads were not ligated. Consistently, Ku70/80 was found with much higher abundance on SB-DNA beads (Figure S1C). In line with the efficient DNA repair, we identified the major NHEJ and HRR factors on SB-DNA beads by mass spectrometry, including Ku70, Ku80, DNA-PKcs, XLF, Mre11, Rad50, and the Xenopus orthologue of Nbs1, Nbn1 (Table S1).
For an unbiased approach to uncover the relevant target of p97 in DSB repair, we applied label-free quantitative mass spectrometry (LFQ-MS) to identify those proteins that specifically accumulated on SB-DNA beads when p97 activity was compromised. To inhibit p97, we used the p97-ND1 truncation that is dominant-negative because it lacks the second, most active ATPase domain, D2, and therefore traps ubiquitinated substrate proteins (Ye et al., 2003). Along with the endogenous Xenopus p97 on SB-DNA beads, we detected the NHEJ components Ku70/80 and the regulatory kinase DNA-PKcs among the enriched proteins in presence of p97-ND1 (Figure 1A and Table S2). Conversely, components of the competing HRR-initiating MRN complex, Rad50, Mre11 and Nbn1/Nbs1 were less abundant, albeit the latter two were below the applied threshold.
Figure 1. p97 is required for Ku80 release from DSBs in Xenopus egg extracts.
A Ku70/80 accumulates on DSBs when p97 is compromised. DSB repair was reconstituted in Xenopus egg CSF extract with linear DNA immobilised at one end on streptavidin beads (SB-DNA beads) such that free ends were ligated (see Figure S1A). Beads were incubated with or without the dominant-negative p97-ND1 mutant (8 μM hexamer) for 45 min and bound proteins were compared by label-free quantitative mass spectrometry. Proteins above the 5% false discovery rate threshold (black line) were regarded as significantly enriched (positive x values; closed circles) or reduced (negative x values; open circles = MRN complex) upon addition of p97-ND1.
B Experimental scheme of the Ku80 release assay. SB-DNA beads were first incubated in egg extract supplemented with 35S-labelled Ku80 and p97-ND1 (8 μM). After 30 min (t0), Ku80 from open ends was removed by salt-wash and the beads were transferred to fresh extract without 35S-Ku80 but supplemented with buffer alone, 8 μM p97-ND1 (ND1), or the inactive p97-ND1 harbouring the K251A suppressor mutation (ND1KA). At indicated times, samples of supernatant and beads were taken and analysed by SDS gels and autoradiography.
C Autoradiograph of Ku80 on SB-DNA beads from the assay in B.
D Quantification of C. Ku80 retained on beads was normalised to t0.
E Quantification of C. Total ubiquitinated Ku80 with was normalised to t0.
F Autoradiograph of the supernatants from the Ku80 release assay.
G Quantification of F. Released Ku80 was normalised to t30 of the control.
H Autoradiograph of SB-DNA beads from 35S-Ku80 release assay after treatment with the p97 inhibitor NMS-873 (20 μM) or DMSO alone.
I Quantification of H. Ku80 retained on beads was normalised to t0.
J Quantification of H. Total polyubiquitinated Ku80 was normalised to t0.
K Autoradiograph of the supernatants from the Ku80 release assay as in H.
L Quantification of K. Released Ku80 was normalised to t30 of the control.
Error bars in all quantifications denote sd (n=3). * p<0.05; ** p<0.01; *** p<0.001. See also Figure S1, Tables S1 and S2.
In addition, we observed an enrichment of the p97 ubiquitin adapter proteins Ufd1, Npl4 and FAF1, suggesting that they were trapped on p97-ND1-stabilised ubiquitin conjugates.
p97 is essential for Ku extraction from DSBs in Xenopus egg extracts
Given that Ku was previously shown to be released from chromatin in a ubiquitin-dependent manner (Postow et al., 2008; Ismail et al., 2015), we reasoned that p97 might extract Ku from DNA. To follow Ku extraction in a quantitative manner, we first loaded in vitro synthesized 35S-radiolabelled Ku80 onto SB-DNA beads in egg extract. We subsequently applied a high-salt wash that was previously shown to remove all Ku complexes that are not sterically trapped on DNA (Paillard & Strauss, 1991), and that also dissociated non-trapped proteins from DNA beads in this assay (Figure S1D). We then monitored the release of the remaining Ku80 in extract that lacked radiolabelled Ku80, as previously established (Figure 1B; Postow et al., 2008).
Radiolabelled Ku80 was detected on beads as a distinct band in addition to higher molecular weight species that indicated polyubiquitination of Ku80 (Figure 1C). Over the course of 60 min, Ku80 was released from the beads (Figures 1C–1E) and unmodified Ku80 emerged in the supernatant as a distinct band at 90 kDa (Figures 1F and 1G), demonstrating the Ku80 was efficiently extracted from the beads and suggesting that it was subsequently deubiquitinated. We used two approaches to demonstrate p97 requirement for Ku80 extraction. First, addition of the dominant-negative p97-ND1 variant significantly delayed extraction of Ku80 and led to persistence of the ubiquitinated form (Figures 1C–1E). Consistently, release into the supernatant was inhibited (Figures 1F and 1G). This effect was specific, because the same concentration of a control variant of p97-ND1, p97-ND1-KA affected Ku80 extraction and release into the supernatant to a lower degree. p97-ND1-KA harbours the K251A suppressor mutation rendering it deficient in substrate binding and therefore less inhibitory (Ye et al., 2003).
As an independent approach to confirm p97 requirement, we applied the allosteric p97 ATPase inhibitor NMS-873 (Magnaghi et al., 2013). The inhibitor significantly delayed extraction of Ku80 from the DNA beads comparable to the effect of p97-ND1 addition (Figures 1H and 1I) and again led to accumulation of ubiquitinated Ku80 (Figures 1H and 1J). Consistently, it also inhibited release of Ku80 in the supernatant (Figures 1K and 1L). Ku80 extraction is a prerequisite for downstream degradation by the proteasome, which can be observed in extract when larger amounts of free linear DNA is added (Postow et al., 2008). We confirmed DNA-induced degradation of Ku80 upon addition of DNA, and found that Ku80 degradation was blocked when p97 was inhibited by NMS-873 (Figures S1E and S1F) as expected if p97 is required for Ku extraction. Thus, these data demonstrate that p97 and its ATPase activity are required for extraction of Ku80 from DNA at DSBs in vitro.
p97 targets Ku80 modified with K48-linked ubiquitin chains on DNA beads
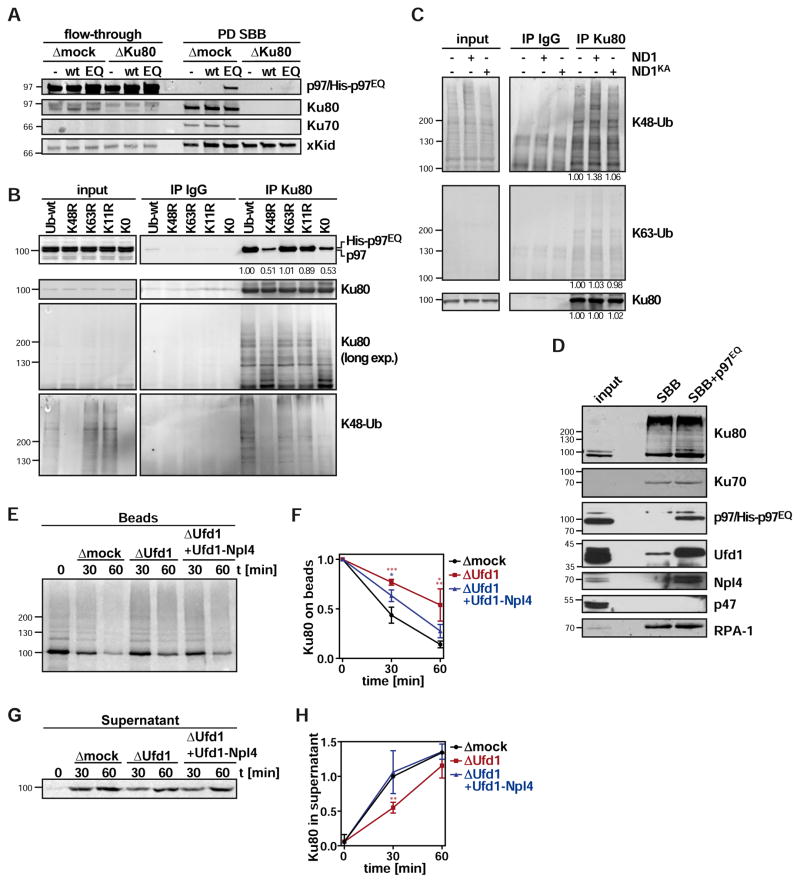
We next asked whether the role of p97 in Ku extraction was direct and therefore examined p97 recruitment to DNA beads in egg extract by Western blotting. Endogenous or added wild-type p97 was not detected on beads, consistent with a transient interaction of p97 with its substrates (Figure 2A). However, when we supplemented the extract with the ATPase-deficient p97 substrate-trapping mutant, in this case with full-length p97-EQ (E578Q mutation in the D2 ATPase domain; Ye et al., 2003), p97 was readily detectable on beads (Figure 2A). To explore if the recruitment of p97 to DNA beads was dependent specifically on Ku80, we immuno-depleted Ku80 from extract before addition of the DNA-beads. Of note, Ku80 depletion co-depleted Ku70 and also abolished p97-EQ recruitment (Figure 2A), suggesting that p97 localised to the sites of DSBs primarily through binding to Ku.
Figure 2. p97 targets Ku80 modified with K48-linked ubiquitin chains on DSB beads.
A Interaction of p97 with SB-DNA beads (SBB) is ATPase regulated and depends on Ku80. SB-DNA beads were incubated in extract that was mock-depleted or immuno-depleted of Ku80, and supplemented with buffer alone, p97-wild-type (wt; 0.33 μM) or the same concentration of the substrate-trapping mutant p97E578Q (EQ) as indicated for 30 min. Beads were recovered (PD SBB) and bound proteins analysed by Western blot with indicated antibodies. xKid was probed as recovery control.
B Extracts containing p97EQ (0.33 μM) were supplemented with linear DNA or buffer alone, and incubated in the presence of 0.75 mg/ml ubiquitin (wt) or ubiquitin with K48R, K63R, K11R, or K0 (all lysines mutated to arginines) mutations as indicated for 30 min prior to immunoprecipitation (IP) with Ku80 or control antibodies as indicated. Signal intensities were quantified and normalized to the wild-type control.
C Extracts were supplemented with linear DNA, and p97-ND1 or p97-ND1KA (8 μM) as indicated, and incubated for 1 h prior to immunoprecipitation (IP). Western blot as indicated. Signal intensities were quantified and normalised to control.
D SB-DNA beads were incubated in egg extract supplemented with buffer (SBB) or 0.33 μM p97E578Q (SBB + p97EQ) for 30 min and bound proteins analysed by immunoblot.
E Ku80 release assay as in Figure 1B in extracts that were mock-depleted (Δmock), Ufd1-depleted (ΔUfd1), or Ufd1-depleted and supplemented with 2 μM recombinant Ufd1-Npl4 complex (ΔUfd1 + Ufd1-Npl4).
F Quantification of E. Retained Ku80 on SB-beads was quantified and normalised to t0.
G Autoradiograph of supernatant from bead release assay as in E.
H Quantification of G. Released Ku80 was normalised to t30 of the control.
All error bars denote sd of three independent experiments. * p<0.05; ** p<0.01; *** p<0.001. See also Figure S2.
It was previously shown that release of Ku from DNA is triggered by modification of Ku80 specifically with K48-linked ubiquitin chains (Postow et al., 2008; Feng & Chen, 2012), and we confirmed increased levels of K48-linked ubiquitination on SB-DNA beads compared to DB-DNA beads (Figure S1C). To test whether this modification was critical for p97 targeting to Ku, we added a ubiquitin variant with the K48R mutation, which blocks extension of K48 chains, and immunoprecipitated Ku80. Despite the presence of endogenous wild-type ubiquitin, addition of recombinant ubiquitin-K48R shifted Ku80 modification to a profile with shorter chains compared to the control with recombinant wild-type ubiquitin (Figure 2B), confirming previous results (Postow et al., 2008). Notably, ubiquitin-K48R, but not wild-type ubiquitin reduced p97 recruitment to the Ku complex (Figure 2B) showing that p97 targets K48-modified Ku80 on DNA. The effect of K48R on ubiquitination and p97 binding was equally strong as for the K0 variant of ubiquitin that lacks all lysines, but was not observed for the K63R and K11R ubiquitin mutants confirming that K48-linked ubiquitin chains are the relevant chain species for this process. Conversely and consistently, inactivation of p97 by p97-ND1 (Figure 2C) or NMS-873 (Figure S2A) led to specific accumulation of K48, but not K63 chains. As expected, this accumulation was not observed in the presence of the p97-ND1-KA control (Figure 2C). These data provide direct biochemical evidence that p97 targets and extracts Ku80 that is modified with K48-linked ubiquitin chains.
p97 cooperates with the Ufd1-Npl4 ubiquitin adapter for Ku extraction
p97-mediated extraction typically requires ubiquitin adapters to facilitate substrate binding and processing (Stolz et al., 2011). We reasoned that this involved the major adapter Ufd1, because it is required for a number of p97-mediated functions on chromatin and accumulated on SB-DNA-beads in our mass spectrometry approach along with its binding partner Npl4 (Figure 1A). We first confirmed by Western blotting the enrichment of Ufd1 and Npl4 on SB-DNA beads in egg extract supplemented with p97-EQ compared to buffer control (Figure 2D). In contrast, an alternative p97 cofactor, p47 was not recruited in either of the conditions. We depleted Ufd1 from egg extracts with a Ufd1-specific antibody. As expected, this led to partial codepletion of Npl4 but not p47 or p97 (Figure S2B). Of note, Ufd1-Npl4 depletion reduced binding of p97 to Ku80 in co-immunoprecipitation experiments (Figure S2C) suggesting that Ufd1-Npl4 functions as a substrate adapter for ubiquitinated Ku80. Importantly, depletion of Ufd1-Npl4 decreased the rate of Ku80 extraction from SB-DNA beads (Figures 2E–2H), and this was restored by re-addition of purified Ufd1-Npl4 complex (Figures 2E–2H). These data show that p97 needs to cooperate with ubiquitin adapters including the major cofactor Ufd1 in the extraction of Ku80 from DNA DSBs.
p97 is essential for Ku80 extraction during DSB repair in U2OS cells
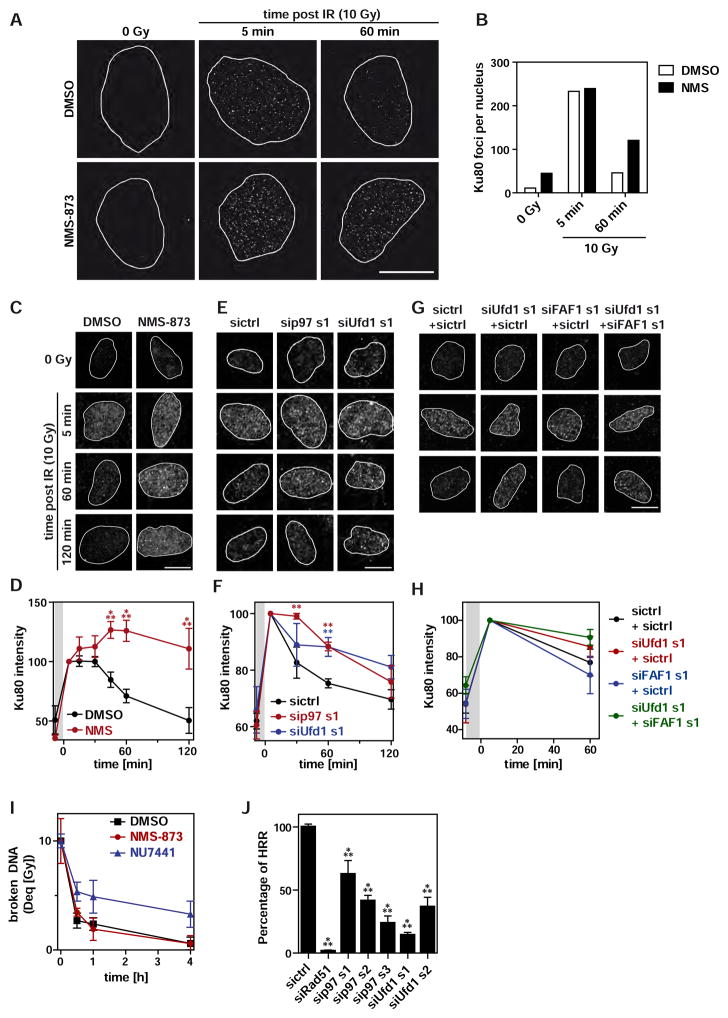
To further confirm that p97-mediated extraction of Ku is relevant in living cells, we monitored Ku80 foci dynamics at DSBs by super-resolution microscopy using a pre-extraction technique to remove non-chromatin bound Ku80 before cell fixation (Britton et al., 2013). As expected, ionizing radiation (IR; 10 Gy) caused formation of Ku80 foci that were largely resolved within 60 min (Figures 3A and 3B). In contrast, in cells treated with the p97 inhibitor NMS-873, Ku80 foci persisted after 60 min. For quantification of larger sample numbers, we analysed Ku release by determining total Ku80 signal intensities using conventional confocal microscopy. Consistent with our super-resolution approach, Ku80 persisted on chromatin upon p97 inhibition even 2 h after irradiation (Figures 3C and 3D).
Figure 3. p97 mediates Ku release from chromatin in U2OS cells.
A Ku80 foci assay. U2OS cells were irradiated with 10 Gy or mock treated (0 Gy), left to recover in the presence of NMS-873 (10 μM) or DMSO, and fixed at indicated times. Cells were pre-extracted with RNase containing buffer (CSK+R), fixed and Ku80 was visualised by immunofluorescence and 3D-SIM super-resolution microscopy.
B Automated image quantification of A (mean of 5 cells per condition).
C U2OS cells were treated as in A with or without NMS-873 and Ku80 was visualised by conventional confocal microscopy.
D Mean Ku80 signal intensity in the nucleus was quantified by automated image analysis and normalised to t5. Grey area indicates the time of irradiation.
E U2OS cells were treated with indicated siRNA targeting p97, Ufd1, or control (sictrl) for 48 h. See Figure S4 for depletion efficiencies and additional siRNAs. Cells were irradiated with 10 Gy or mock treated (0 Gy), fixed at indicated times, and chromatin-bound Ku80 was visualised as in C.
F Automated image quantification of E.
G U2OS cells were transfected with siRNA targeting Ufd1, FAF1 or control (sictrl) in indicated combinations and treated as in E.
H Automated image quantification of G as in F.
I Quantification of pulsed-field gel electrophoresis. U2OS cells were irradiated (10 Gy) and treated with NMS-873 (10 μM), NU7441 (DNA-PKcs inhibitor; 5 μM) or DMSO alone. The amount of broken DNA in each sample was compared to a reference set of samples irradiated with different doses (Dose equivalent, Deq). NU7441 served as positive control for inhibiting NHEJ.
J Readout of the homologous recombination repair (HRR) reporter assay DR-GFP with indicated siRNAs. siRad51 served as positive control. GFP fluorescence was measured by flow cytometry.
Mean ± sd; n= 3 with ≥ 50 cells per condition; ** p<0.01; *** p<0.001. All scale bars: 10 μm. See also Figure S3.
To confirm this result, we performed siRNA-mediated depletion of p97 or its adapters (for depletion efficiencies see Figures S4B–S4F). Depletion of p97 or Ufd1 with different siRNAs significantly reduced the removal of Ku80 at later time points compared to control-depleted cells (Figures 3E, 3F, S3A and S3B). This was confirmed by a depletion/restoration setup for Ufd1 (Figures S3C and S3D).
Depletion of the Ufd1 partner Npl4 also affected Ku80 removal although to a lower degree (Figures S3E and S3F) suggesting that Npl4 is less critical than Ufd1 for the process, which has been also observed for other p97 substrates before (Riemer et al., 2014; Raman et al., 2011). Depletion of the p97-adapter FAF1, which also accumulated on the SB-DNA-beads in vitro (Figure 1A), had only a mild effect when depleted alone (Figures S3E and S3F). Intriguingly, however, codepletion of Ufd1 and FAF1 more strongly reduced Ku80 extraction than Ufd1 depletion alone (Figures 3G and 3H) suggesting that Ufd1 and FAF1 can function partially redundantly as ubiquitin adapters in the process. Depletion of the p97 adapter UBXD7, which was not detected on SB-DNA-beads, had no effect (Figures S3E and S3F).
The experiments in cells so far did not formally demonstrate that the persisting Ku observed upon p97 inhibition was trapped on repaired DSBs rather than bound to unrepaired open DNA ends. In fact, we observed persistent H2AX phosphorylation upon NMS-873 treatment (Figure S3G) indicating on-going DNA damage signalling. We therefore applied pulsed-field gel electrophoresis (PFGE) to monitor the extent of DNA repair in the absence or presence of NMS-873. This revealed that DNA was efficiently repaired even in the presence of the p97 inhibitor 2 h after irradiation (Figure 3I), when Ku80 still largely persisted (*********Figures 3C and 3D). This demonstrates that, in the absence of p97 activity, the majority of Ku persists on re-joined DNA, and that Ku persistence upon p97 inhibition is not a consequence of failure to repair DNA. While the repair by NHEJ was not affected by p97 inhibition, HRR as monitored in a reporter assay was reduced by depletion of p97 or Ufd1 (Figure 3J), consistent with previous results (Meerang et al., 2011; Bergink et al., 2013). Together, these data support the notion that p97 extracts the majority of Ku in cells after DNA repair by NHEJ and, and that it is also involved in HRR.
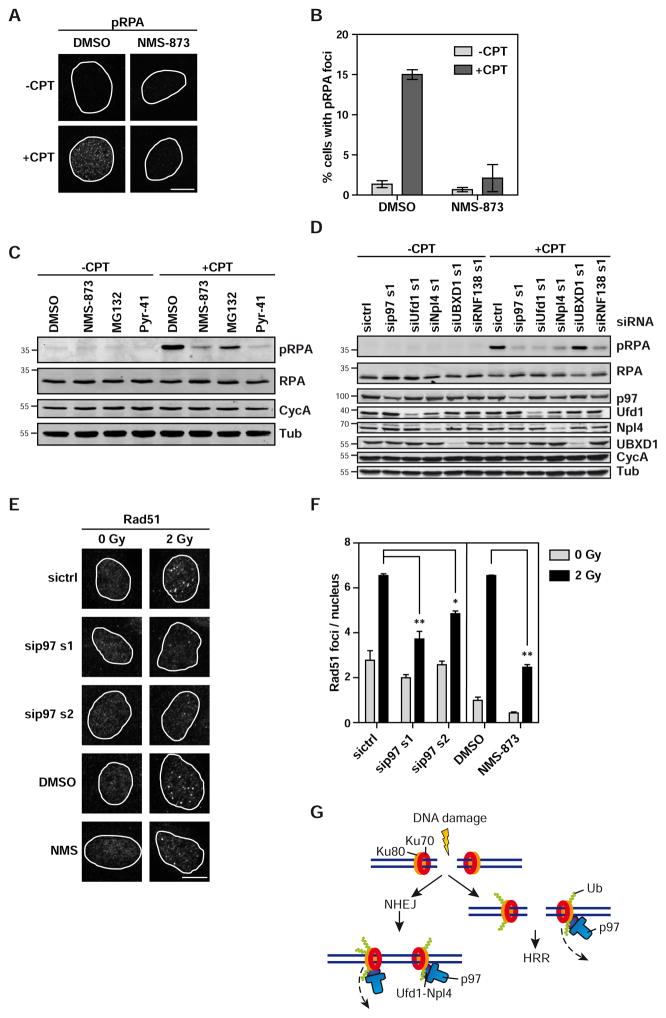
p97 inhibition attenuates HRR-associated phosphorylation of RPA
It was recently shown that a fraction of Ku can also be extracted from DNA ends before DSB repair, and that this is triggered by the ubiquitin ligase RNF138 as an element of repair pathway choice to favour the error-free HRR over NHEJ (Ismail et al., 2015; Schmidt et al., 2015). We induced S-phase specific DSBs using the topoisomerase I inhibitor camptothecin in U2OS cells and monitored the DSB-dependent phosphorylation of the ssDNA binding protein RPA32 at Ser4/8 as an early marker for HRR activity. Confocal microscopy revealed that p97 inhibition by NMS-873 largely reduced phospho-RPA foci formation indicating attenuation of HRR (Figures 4A and 4B). This was reflected by a reduction of total levels of damage-induced RPA phosphorylation upon NMS-873 treatment as detected by Western blot (Figure 4C). As expected (Ismail et al., 2015), the proteasome inhibitor MG132 only had a mild effect in this setting, whereas Pyr-41, an inhibitor of the E1 ubiquitin-activating enzyme, abolished RPA phosphorylation (Figure 4C). To confirm this effect, we used RNAi. As expected, depletion of RNF138 decreased RPA phosphorylation (Figure 4D). Notably, also depletion of p97, Ufd1 or Npl4, but not of the alternative cytoplasmic UBXD1 adapter as control, reduced RPA phosphorylation (Figure 4D and S4A). Consistently, p97 depletion by two siRNAs, or chemical inhibition of p97 by NMS-873 also reduced downstream formation of Rad51 foci after irradiation (Figures 4E–4F), in line with previous reports (Meerang et al., 2011; Bergink et al., 2013). This finding suggests that p97 promotes commitment to HRR and is consistent with a role in extracting Ku also from open DNA ends during DNA repair pathway choice.
Figure 4. p97 inhibition attenuates an early step in homologous recombination repair.
A Inhibition of p97 reduces phospho-RPA foci formation. U2OS cells were treated with DMSO or NMS-873 (5 μM) briefly before and during treatment with 1 μM camptothecin (+CPT) for 60 min to induce DSBs or with DMSO (−CPT) as control. Phospho-RPA (Ser4/8) was visualised by immunostaining and confocal microscopy.
B Automated quantification of images in A. Mean (n=2), error bars denote range of values (n=2 with at least 100 cells per condition).
C Immunoblot analysis of RPA phosphorylation (Ser4/8) 60 min after induction of DSBs as in A on cell lysates from U2OS cells treated with indicated inhibitors (MG132: proteasome inhibitor, 20 μM; Pyr-41: E1 ubiquitin ligase inhibitor, 50 μM). Detection of Cyclin A (CycA) to confirm cell cycle progression into S/G2.
D Immunoblot analysis of cells treated as in C after siRNA-mediated depletion of indicated proteins.
E U2OS cells were transfected with siRNA targeting p97 or control (sictrl) or treated with DMSO or 5 μM NMS-873 and irradiated with 2 Gy or mock treated (0 Gy). Cells were left to recover in presence of the siRNA or inhibitor and fixed 8 h after irradiation. Rad51 was visualised by confocal microscopy.
F Mean number of Rad51 foci per nucleus was quantified by automated image analysis. Mean ± sd; n = 3 with at least 50 cells per condition and experiment.
G Model of p97 function in Ku80 extraction from DNA after DSB. Upon binding to damaged DNA, Ku initiates NHEJ (left side) and concomitantly prevents end resection to inhibit HRR. After completion of NHEJ, Ku is topologically trapped on the ligated DNA. It is ubiquitinated, and subsequently extracted by p97. As an element of repair pathway switch, a fraction of Ku can also be ubiquitinated, and targeted by p97 on open DNA ends to allow end resection and HRR (right side).
* p<0.05; ** p<0.01. All scale bars: 10 μm. See also Figure S4.
Discussion
By applying a combination of in vitro and in vivo approaches in this study, we gathered unequivocal evidence that the ubiquitin-directed AAA+-ATPase p97 provides the required driving force for the extraction of the key regulator of DSB repair, Ku, from DNA. In the past, a number of proteins have been identified that are extracted from chromatin by p97 in various processes (reviewed in Meyer et al., 2012, Dantuma et al., 2014). However, the Ku70/80 complex stands out among these factors because it forms a tightly bound ring that is sterically trapped on DNA after ligation of DSBs (Walker et al., 2001). Even on open DNA ends, Ku is tightly locked on DNA due to its oriented and thus one-directional binding mode (Krishna & Aravind, 2010). As a result, it requires energy-driven conformational changes to open the ring and release Ku from the DNA (see model Figure 4G).
Three ubiquitin ligases, SCFFbxl12, RNF8, and RNF138 have been shown to ubiquitinate Ku80 in different conditions that likely represent alternative pathways to trigger extraction (Postow & Funabiki, 2014; Feng & Chen, 2012; Ismail et al., 2015; Schmidt et al., 2015). This is in line with the observation that p97 is able to cooperate with diverse ligases (Alexandru et al., 2008) and is reminiscent of ER-associated degradation, where alternative ubiquitination pathways converge on p97 to mediate substrate extraction from the membrane (Stolz et al., 2011). Our in vitro data also support the notion that extraction of Ku80 is triggered specifically by ubiquitin chains that are linked via K48. This is consistent with earlier findings showing the role of K48-chains conjugated to Ku for at least two ligases, SCFFbxl12 in Xenopus egg extracts and RNF8 in cultured cells (Postow et al., 2008; Postow & Funabiki, 2014; Feng & Chen, 2012), and further highlights the role of K48-linked ubiquitin chains in DNA damage responses (Bekker-Jensen & Mailand, 2011; Jackson & Durocher, 2013). Consistently, we show that p97 cooperates with ubiquitin adapters during Ku extraction. We find that p97 requires the major Ufd1 adapter that also functions in other p97-mediated processes (Stolz et al., 2011). Moreover, it involves, in a partially redundant manner, the FAF1 ubiquitin adapter that, with respect to chromatin-related functions of p97, so far has only been linked to replication (Franz et al., 2016).
While we show that p97 extracts the salt-resistant Ku80 fraction that is sterically trapped after DNA repair, we present additional evidence consistent with a role of p97 in removing part of Ku before ligation from unrepaired DNA ends and thereby affecting repair pathway choice to favour error-free HRR over error-prone NHEJ (see model Figure 4G). However, the actual repair by the NHEJ pathway is not compromised by p97 inhibition. This mirrors the effect of depleting the ubiquitin ligase RNF138 that was previously implicated in repair pathway choice by ubiquitinating Ku and triggering its extraction before repair (Ismail et al., 2015; Schmidt et al., 2015). This suggests that p97 not only releases Ku after DSB repair but, along with RNF138, also contributes to a key decision process at initiation of repair and thereby affects the fidelity of repair.
Experimental Procedures
Antibodies, Proteins, and Inhibitors
Antibodies against Xenopus Ku70 and Xenopus Ku80 were used as described (Postow et al., 2008). Purified proteins were generated in bacteria as described (Ye et al., 2003). NMS-873 was obtained from Xcessbio, Pyr-41 and MG132 were from Merck-Millipore. See Supplemental Experimental Procedures for a full list.
Frog Egg Extract
CSF arrested Xenopus laevis egg extract was prepared as described previously (Postow et al., 2008). Egg extracts were supplemented with 10 μg/ml nocodazole (Sigma Aldrich).
Protein Isolation on DNA Beads
DNA beads were prepared as previously described (Postow et al., 2008). Briefly, pBluescript SK+ vector was cut and filled with Klenow (New England Biolabs), biotin-dATP and biotin-dUTP (Chemcyte) and bound to M280 dynabeads (Invitrogen). 25–50 μl extract was used for every 1 μg of DNA on beads. 35S-labelled Ku80 was generated in reticulocyte lysate (Promega) using X. laevis Ku80 cDNA (Postow et al., 2008) and diluted 1:10 in egg extract. Beads were removed from extract with a magnetic particle separator and eluted by denaturation. For Ku80 release assays, Ku80 was quantified using a phosphoimager (Fujifilm).
Immunoprecipitations
Egg extracts supplemented with linear DNA (600 μg/ml pBluescript SK+, digested with PvuII and SspI) and p97 mutants were incubated for indicated times at 22 °C. Extracts were diluted 1:5 in XB buffer (10 mM KCl, 10 mM Hepes, pH 8, 50 mM sucrose, and 1 mM MgCl2) supplemented with 0.5% Triton X-100 and 1 mg/ml N-ethylmaleimide and centrifuged at 17000 g (5 min; 4 °C). Protein G dynabeads coupled with Ku80 or normal IgG were incubated in supernatants for 1 h at 4 °C, eluted in SDS sample buffer. Samples were analysed by Western blot.
Immunodepletions in Egg Extract
Ufd1 was immunodepleted with 5E2 antibody (Abcam); Ku80 was immunodepleted as previously described (Postow & Funabiki, 2014). See also Supplemental Experimental Procedures.
pRPA Assay
U2OS cells were pre-treated with siRNAs for 48 h or inhibitors for 15 min before DNA damage was induced with 1 μM camptothecin. After 1 h treatment, whole cell lysates were analysed by Western blot. See also Supplemental Experimental Procedures.
Immunofluorescence Staining
U2OS cells were grown on glass coverslips and transfected with the indicated siRNA using RNAiMax (Life Technologies) for 48 h or treated with inhibitor for 15 minutes before exposure to 10 Gy ionizing radiation. Plasmid transfection for the Ufd1 restoration experiment was performed 24 h before IR. Ku80 staining was performed as reported previously (Britton et al., 2013). Images were taken at Nikon Eclipse Ti-E with confocal spinning disc unit, Leica TCS SP5 confocal laser scanning microscope or Zeiss Elyra PS.1 super-resolution microscope structured illumination (SIM) and automatic image analysis was performed with CellProfiler. See also Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
This work was supported by an EMBO Short-Term Fellowship to J. B. (596-2014), and DFG grants to H. M., M. K., and J. B. (CRC1093), to M. W. (GRK1739), to L. W. (GRK1431), and to M. K. (INST 20876/127-1 FUGG), and by an ERC starting grant to M. K. (258413). The authors acknowledge support by the Imaging Center Essen (IMCES).
Footnotes
Author Contributions:
J. B. performed experiments with Xenopus egg extracts with help of C. Z.; cell experiments were performed by M. W., L. W, and N. S.; A. R. established IR protocols; F. L. and G. I performed PFGE experiments; MS measurements were performed by F. K. and M. K. The study was designed and conceived by H. M. and H. F.; J. B. and H. M. wrote the manuscript.
References
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature Structural & Molecular Biology. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Letters. 2011;585:2914–2919. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- Bergink S, Ammon T, Kern M, Schermelleh L, Leonhardt H, Jentsch S. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51–Rad52 interaction. Nature Cell Biology. 2013;15:526–532. doi: 10.1038/ncb2729. [DOI] [PubMed] [Google Scholar]
- Blier PR, Griffith AJ, Craft J, Hardin JA. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. The Journal of biological chemistry. 1993;268:7594–7601. [PubMed] [Google Scholar]
- Britton S, Coates J, Jackson SP. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. The Journal of Cell Biology. 2013;202:579–595. doi: 10.1083/jcb.201303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Acs K, Luijsterburg MS. Should I stay or should I go: VCP/p97-mediated chromatin extraction in the DNA damage response. Experimental cell research. 2014;329:9–17. doi: 10.1016/j.yexcr.2014.08.025. [DOI] [PubMed] [Google Scholar]
- Di Virgilio M, Gautier J. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. The Journal of Cell Biology. 2005;171:765–771. doi: 10.1083/jcb.200506029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nature Structural & Molecular Biology. 2012;19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A, Pirson PA, Pilger D, Halder S, Achuthankutty D, Kashkar H, Ramadan K, Hoppe T. Chromatin-associated degradation is defined by UBXN-3/FAF1 to safeguard DNA replication fork progression. Nature communications. 2016;7:10612. doi: 10.1038/ncomms10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Gagné JP, Genois MM, Strickfaden H, McDonald D, Xu Z, Poirier GG, Masson JY, Hendzel MJ. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nature Cell Biology. 2015;17:1446–1457. doi: 10.1038/ncb3259. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. Regulation of DNA Damage Responses by Ubiquitin and SUMO. Molecular Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends in biochemical sciences. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Krishna SS, Aravind L. The bridge-region of the Ku superfamily is an atypical zinc ribbon domain. Journal of Structural Biology. 2010;172:294–299. doi: 10.1016/j.jsb.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P. Nonhomologous DNA end joining in cell-free systems. European journal of biochemistry/FEBS. 1999;265:849–861. doi: 10.1046/j.1432-1327.1999.00805.x. [DOI] [PubMed] [Google Scholar]
- Lieber, Gu J, Lu H, Shimazaki N, Tsai AG. Nonhomologous DNA end joining (NHEJ) and chromosomal translocations in humans. Sub-cellular biochemistry. 2010;50:279–296. doi: 10.1007/978-90-481-3471-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi P, D’Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nature Chemical Biology. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, Janscak P, Hübscher U, Meyer H, Ramadan K. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nature Cell Biology. 2011;13:1376–1382. doi: 10.1038/ncb2367. [DOI] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nature cell biology. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Paillard S, Strauss F. Analysis of the mechanism of interaction of simian Ku protein with DNA. Nucleic acids research. 1991;19:5619–5624. doi: 10.1093/nar/19.20.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Funabiki H. An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle. 2014;12:587–595. doi: 10.4161/cc.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break–induced ubiquitylation. The Journal of Cell Biology. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Havens CG, Walter JC, Harper JW. A Genome-wide Screen Identifies p97 as an Essential Regulator of DNA Damage-Dependent CDT1 Destruction. Molecular Cell. 2011;44:72–84. doi: 10.1016/j.molcel.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer A, Dobrynin G, Dressler A, Bremer S, Soni A, Iliakis G, Meyer H. The p97-Ufd1-Npl4 ATPase complex ensures robustness of the G2/M checkpoint by facilitating CDC25A degradation. Cell Cycle. 2014;13:919–927. doi: 10.4161/cc.27779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CK, Galanty Y, Sczaniecka-Clift M, Coates J, Jhujh S, Demir M, Cornwell M, Beli P, Jackson SP. Systematic E2 screening reveals a UBE2D–RNF138–CtIP axis promoting DNA repair. Nature Cell Biology. 2015;17:1458–1470. doi: 10.1038/ncb3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Hilt W, Buchberger A, Wolf DH. Cdc48: a power machine in protein degradation. Trends in biochemical sciences. 2011;36:515–523. doi: 10.1016/j.tibs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Sun J, Lee KJ, Davis AJ, Chen DJ. Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. The Journal of biological chemistry. 2012;287:4936–4945. doi: 10.1074/jbc.M111.306167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. Function of the p97–Ufd1–Npl4 complex in retrotranslocation from the ER to the cytosol. The Journal of Cell Biology. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.