ABSTRACT
The yeast homolog of DJ-1, Hsp31, is a multifunctional protein that is involved in several cellular pathways including detoxification of the toxic metabolite methylglyoxal and as a protein deglycase. Prior studies ascribed Hsp31 as a molecular chaperone that can inhibit α-Syn aggregation in vitro and alleviate its toxicity in vivo. It was also shown that Hsp31 inhibits Sup35 aggregate formation in yeast, however, it is unknown if Hsp31 can modulate [PSI+] phenotype and Sup35 prionogenesis. Other small heat shock proteins, Hsp26 and Hsp42 are known to be a part of a synergistic proteostasis network that inhibits Sup35 prion formation and promotes its disaggregation. Here, we establish that Hsp31 inhibits Sup35 [PSI+] prion formation in collaboration with a well-known disaggregase, Hsp104. Hsp31 transiently prevents prion induction but does not suppress induction upon prolonged expression of Sup35 indicating that Hsp31 can be overcome by larger aggregates. In addition, elevated levels of Hsp31 do not cure [PSI+] strains indicating that Hsp31 cannot intervene in a pre-existing prion oligomerization cycle. However, Hsp31 can modulate prion status in cooperation with Hsp104 because it inhibits Sup35 aggregate formation and potentiates [PSI+] prion curing upon overexpression of Hsp104. The absence of Hsp31 reduces [PSI+] prion curing by Hsp104 without influencing its ability to rescue cellular thermotolerance. Hsp31 did not synergize with Hsp42 to modulate the [PSI+] phenotype suggesting that both proteins act on similar stages of the prion cycle. We also showed that Hsp31 physically interacts with Hsp104 and together they prevent Sup35 prion toxicity to greater extent than if they were expressed individually. These results elucidate a mechanism for Hsp31 on prion modulation that suggest it acts at a distinct step early in the Sup35 aggregation process that is different from Hsp104. This is the first demonstration of the modulation of [PSI+] status by the chaperone action of Hsp31. The delineation of Hsp31's role in the chaperone cycle has implications for understanding the role of the DJ-1 superfamily in controlling misfolded proteins in neurodegenerative disease and cancer.
KEYWORDS: Amyloids, Chaperone, DJ-1, Hsp31, Hsp104, Neurodegenerative diseases, Small Heat Shock Protein, Sup35, Yeast prion, [PSI+]
INTRODUCTION
Amyloids are highly ordered cross β-sheet protein polymers that are associated with a broad range of neurodegenerative diseases including Parkinson, Alzheimer, Huntington and Prion diseases.1,2 Growth of amyloids occurs by the nucleated polymerization of soluble proteins of a particular sequence.3,4 Many proteins can polymerize to form amyloid when provided an appropriate environment in vitro, indicating this as inherent characteristic of polypeptides. Indeed, many recent studies have shown the existence of beneficial amyloids that are important for survival of a host organism.5 Furthermore, the highly rigid structure of self-propagating amyloids provides a possible tool in designing a unique nanomaterial.6 Therefore, it is important to study the process of amyloid formation as well as its modulation.
Budding yeast Saccharomyces cerevisiae, provides a useful model to understand the formation, modulation and disaggregation of amyloids including prions.7 One of the most extensively studied yeast prions is formed from the translation termination factor Sup35, which has the normal function of releasing polypeptide chains from the ribosome upon encountering a stop codon. The prion form of Sup35 involves a self-perpetuating conformational change that results in stop codon suppression and translational read-through.8 This termination defect can be visualized easily in vivo by nonsense suppression of a designed premature stop codon in a gene that affects colony color, thus providing a convenient phenotypic assay to monitor [PSI+] prion.8
The propagation of [PSI+] prion in yeast is rigorously controlled by molecular chaperone machinery including, Hsp104, Hsp70, Hsp40 and their co-chaperones.9-12 Hsp104 is a member of AAA+ ATPase superfamily and its expression is induced under stress to facilitate refolding and dissociation of protein aggregates.13,14 A moderate level of Hsp104 is required for [PSI+] prion propagation as overproduction or deletion of Hsp104 cures the [PSI+] prion in yeast.15,16 Since, Hsp104 disaggregates the misfolded protein after heat shock, it is postulated that Hsp104 generates “propagons” by fragmenting the prion polymers that are available for further polymerization and therefore maintain the prion propagation.7 Another model proposes that elevated levels of Hsp104 cure [PSI+] prion by dissolution of the prion seeds, the evidence for this model came from the observation of the diffuse expression of Sup35 tagged with GFP when Hsp104 was overexpressed in [PSI+] cells,17 and a large fraction of soluble Sup35 was observed in the cell lysate of [PSI+] with excess of Hsp104 expression.18 Recent studies also suggests that dissolution of prion seed might be due to trimming activity of Hsp104 in which Sup35 dissociates from the end of the prion seed thus reduces its size without generating new seeds.19,20 Trimming activity of Hsp104 is still present even when severing activity is inhibited by treatment of guanidine.20 Hsp104 collaborates with Hsp70 and Hsp40 families in dissolution of heat-damaged proteins as well as prion propagation.21-26 Interestingly, different Hsp70 homologues have opposing effects on [PSI+] in which the Ssa proteins antagonize while the Ssb proteins potentiate [PSI+] curing by elevated levels of Hsp104.10,27,28 Deletion of co-chaperones of Hsp70/90 such as Sti1, Cpr7 and Sis1, also inhibit [PSI+] curing by Hsp104 overexpression.10,25,29
In addition to Hsp104 and its assistant chaperones, small heat shock proteins such as Hsp31, Hsp26 and Hsp42 also play a role in disaggregation of misfolded proteins in yeast.30-36 These proteins are highly expressed under moderate stress and during late growth phase for transition to stationary phase.35,37 Hsp42 and Hsp26 work synergistically to inhibit prion formation and potentiate dissolution of Sup35 prion aggregates by distinct mechanisms.30 Furthermore, Hsp26 or Hsp42 collaborate with Hsp70 and or Hsp104 to reduce the SDS-resistant polyglutamine aggregation.31,38 Hsp31 is an ortholog of DJ-1, which has been well established to have a role in neurodegenerative disease and more recently in cancer.39,40 Hsp31 inhibits the formation of α-Syn aggregates in vitro and toxicity in vivo.41,42 It was also demonstrated that Hsp31 inhibits Sup35 aggregation formation in yeast41 however, it is unknown whether Hsp31 interferes with [PSI+] prion induction and propagation and if like other small heat shock proteins it can coordinate with Hsp104.
In the present study, we aimed to explore the role of Hsp31 in prion propagation and induction using [PSI+] prion model. First, we investigated the ability of Hsp31 to inhibit Sup35 prion aggregation and induction by overexpression of the prion-forming domain (PrD) in [psi−] strain. In this study we have delineated the collaboration between Hsp31 and Hsp104 on [PSI+] prion curing and prion associated toxicity. These results are the first evidence that Hsp31 acts as a chaperone protein that coordinates with Hsp104 to rescue cells from prion toxicity.
RESULTS
Hsp31 Antagonizes Sup35 Aggregation Formation in [psi−PIN+] strain background
We previously reported that Hsp31 has chaperone activity against Sup35 aggregates when tested in the wild-type (WT) W303 yeast strain. In this study, we validated the ability of Hsp31 to inhibit Sup35 fibril formation in the [psi− PIN+] strain background. [PIN+], an epigenetic element, is required to induce [PSI+] formation spontaneously or by overexpression of Sup35 or its PrD.43 The presence of [PIN+] is necessary at early stages of prion formation but is not needed for maintenance and propagation of [PSI+].7 We overexpressed Hsp31-DsRed under the GPD promoter concomitantly with PrD-Sup35-EYFP under GAL expression in a [psi− PIN+] 74D-694 strain. We have previously shown the DsRed-tagged Hsp31 is nearly functionally equivalent to an untagged protein because it can complement a hsp31Δ strain.41 When examined by confocal microscopy, PrD-Sup35 fluorescent foci were greatly reduced in cells co-expressing pAG415-GPD-HSP31-DsRed and pAG424-GAL-PrD- Sup35-EYFP as compared to vector control (Fig. 1A-B). We further confirmed reduced foci formation by measuring fluorescence intensity using flow cytometry and obtained similar results as fluorescence microscopy (Fig. 1C). We have previously observed and established that Sup35 aggregates are associated with increased fluorescence and can be quantified using flow cytometry.41 Finally, we performed semi-denaturing detergent agarose electrophoresis (SDD-AGE) to determine the level of SDS-resistant aggregate forms. Consistent with the previous results, elevated Hsp31 greatly reduced the level of Sup35 aggregates as measured by SDD-AGE but did not greatly reduce the overall steady-state level of Sup35 in SDS-PAGE (Fig. 1D). These results show that Hsp31 can reduce de novo [PSI+] aggregate formation, as can be detected by SDD-AGE, in the presence of the [PIN+] genetic element. It should be noted that additional lower molecular weight prion forms, that might not be detected by SDD-AGE may persist and still be infectious.
FIGURE 1.
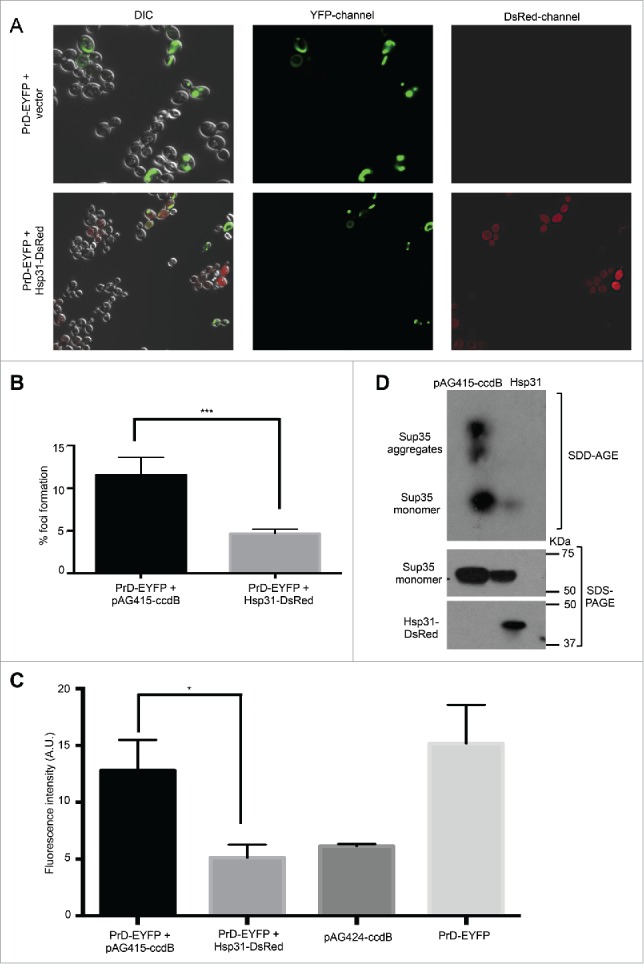
Hsp31 overexpression decreases the level of PrD-Sup35 aggregates. (A) GAL-driven PrD-Sup35-EYFP was overexpressed for 24 h at 30°C in [psi− PIN+] cells with or without overexpression of DsRed tagged Hsp31. PrD-Sup35-EYFP aggregates appeared as ribbon-like vacuolar peripheral rings. Hsp31 remains cytoplasmically diffuse in these cells. Elevated levels of Hsp31 decreased the presence of Sup35 aggregates in individual cells. (B) Quantitation of the number of cells with one or more Sup35-EYFP foci. The average of at least 3 independent experiments was plotted; error bars represent mean ±SEM. (*** unpaired Student's t-test; p ≤ 0.001). (C) Quantitation of the level of Sup35-EYFP fluorescence aggregates using flow cytometry. Aggregates are associated with higher fluorescence. Elevated levels of Hsp31 lowered the median fluorescence intensity (FI – arbitrary units) of Sup35-EYFP compared to vector control. Values represent mean ±SEM of 3 independent biological replicates (* unpaired Student's t-test; p ≤ 0.01). (D) Cellular lysate of cells describe in A and B was analyzed by semi-denaturing agarose electrophoresis and SDS-PAGE. Overexpression of Hsp31 suppresses the level of SDS-resistant Sup35 aggregates as detected with anti-GFP antibody. Lower panel shows the expression of Hsp31 detected by anti-DsRed antibody.
Hsp31 Transiently Inhibits Sup35 Prion Induction in vivo
The ability of Hsp31 to inhibit Sup35 aggregation in the [psi− PIN+] background led us to investigate its ability to inhibit [PSI+] induction. Spontaneous de novo [PSI+] induction frequency is extremely low unless Sup35 or its PrD is overexpressed.8,44 This system is widely used to investigate the process of prion induction in yeast. To determine whether Hsp31 can inhibit prion induction we used a [psi− PIN+] strain that forms red color colonies on ¼ YPD plates with limited adenine. This strain forms white colonies in the presence of [PSI+] because soluble Sup35 is depleted by aggregate formation resulting in suppression of a premature stop codon and restoration of adenine prototrophy. We observed that overexpression of Hsp31 antagonized [PSI+] prion induction triggered by overexpression of Sup35 PrD for 6 h (Fig. 2A-B). However, when Sup35 PrD was expressed for 12 or 24 h, the [PSI+] prion induction level between vector control and Hsp31 was statistically similar (Fig. 2C). In addition, we have previously shown that Hsp31 overexpression does not significantly alter Hsp104 expression levels.41 These findings suggested that Hsp31 antagonizes prion induction transiently but can be overcome by excess production of PrD or cannot intervene when cells have established a full and more mature prion cycle. In addition, the transient and non-permanent effect on [PSI+] by overexpression of Hsp31 suggests that the [PIN+] prion is not cured. These results are consistent with our previous proposal that Hsp31 intervenes early in the protein misfolding processes.41,45
FIGURE 2.
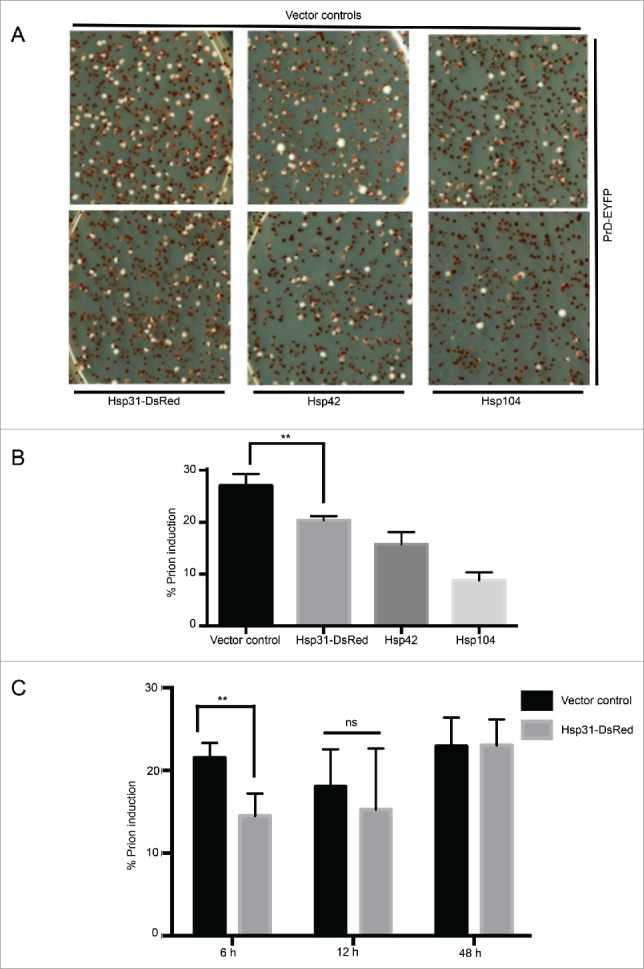
Hsp31 transiently inhibits Sup35 prion induction. (A) To induce prion formation, a GAL-driven vector expressing PrD-Sup35-EYFP was expressed in the [psi− PIN+] strain containing the ade1-14 nonsense mutation. Constitutive expression of Hsp31 was driven by the GPD promoter and [PSI+] formation was scored by quantifying white color colonies on ¼ YPD plates. Top panel represents the vector controls for the expression plasmids in the bottom panel where Hsp31-DsRed, Hsp42 and Hsp104 were overexpressed. (B) PrD-Sup35-EYFP was expressed for 6 h to transiently induce prion formation. Hsp31 overexpression decreased the prion induction level. Only one vector control is shown because other vectors had similar levels of prion induction. Error bars represent ±SEM (** unpaired Student's t-test; p ≤ 0.001, n = 3). (C) Time course of prion formation by pAG424-GAL-PrD-Sup35-EYFP with varied expression times in the presence of GPD-Hsp31. Error bars represent mean ±SEM (** unpaired Student's t-test; p ≤ 0.001, n = 3) ns = not significant. All experiments in this figure were biological replicates.
[PSI+] Prion State is not Affected by Hsp31 Overexpression or Deletion
Transient overexpression of small heat shock proteins such as Hsp26 and Hsp42 cure the [PSI+] prion, converting white or pink colonies that reflect the strength of the [PSI+] phenotype, into red [psi−] colonies on ¼ YPD plates with limited adenine.30 First, we tested if overexpression of Hsp31 can cure the [PSI+] prion. We used the 74D-694 [PSI+] strain which has a weak to moderate phenotype based on its pink color. Despite the fact that Hsp31 can prevent de novo prion aggregate formation in vivo as detected by SDD-AGE (Fig. 1), overexpression of Hsp31 is not sufficient to cure the moderate [PSI+] phenotype of the 74D-694 strain (Fig. 3A). In addition, Hsp42 was used as a control and was able to cure [PSI+] as previously reported.28 However, we found that Hsp26 overexpression using the identical plasmid vector from Duennwald et al.28 had 0 % curing of [PSI+] in this strain and under these experimental conditions in more than 3 biological replicates. Our results are more consistent with Wickner and colleagues46 who reported lack of prion curing by Hsp26 for both [Ure3-1] and [PSI+] phenotypes, which could be explained by differences in strain genotypes and experimental conditions.46 We tested different plasmid systems to express Hsp31 including the GPD and GAL promoters and none of them were able to modulate the [PSI+] prion phenotype (data not shown). Next, we determined if deletion of HSP31 influences the [PSI+] prion status. We constructed a [PSI+ PIN+] hsp31Δ strain carrying the reporter nonsense allele ade1-14, so that the Sup35 read-through caused by [PSI+] presence could be detected by development of white colonies. The phenotype of the [PSI+] prion was similar in the hsp31Δ strain compared to WT with no change in colony color (Fig. 3B), suggesting that Hsp31 cannot intervene in an established prion cycle.
FIGURE 3.
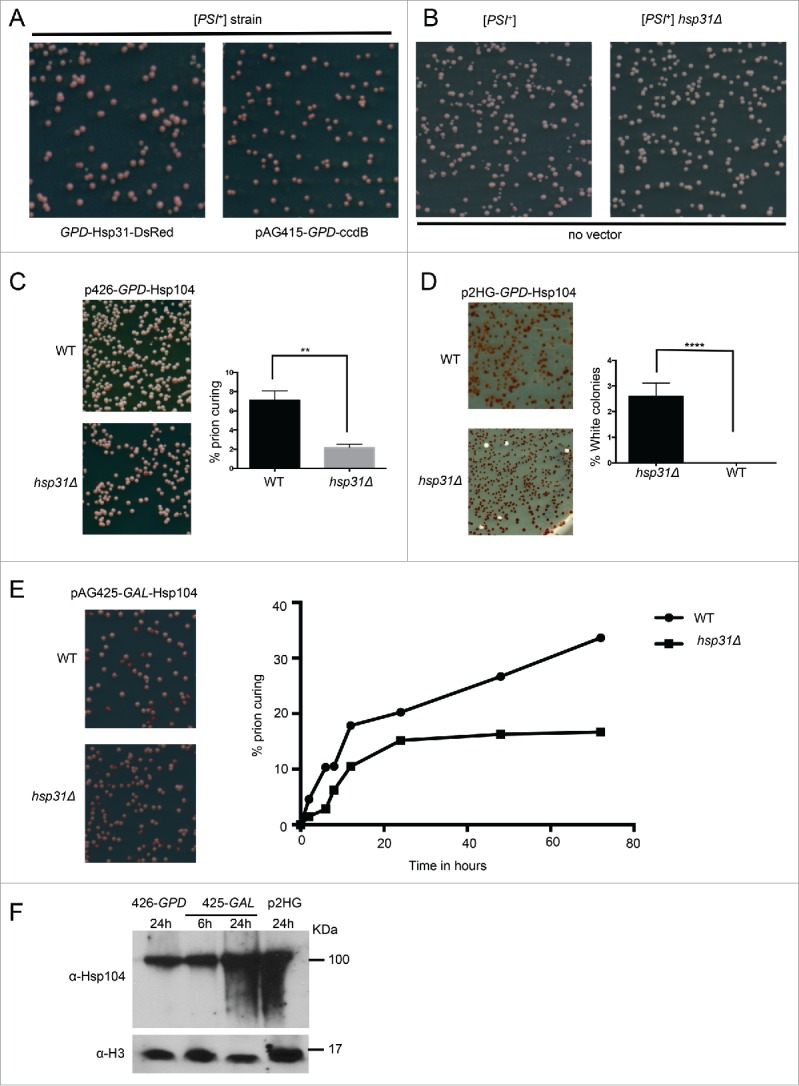
Hsp31 is required for optimal Hsp104-induced curing of the [PSI+] phenotype. (A) To determine the effect of Hsp31 on [PSI+] prion curing, the WT and hsp31Δ [PSI+] strains harboring the GPD-Hsp31 expression vector (pAG415-GPD-Hsp31-DsRed) or the vector (pAG415-GPD-ccdB-DsRed) were grown for 12 h at 30°C before plating on ¼ YPD plates. (B) The WT and hsp31Δ [PSI+] strains with no vector were also grown and treated in the same way. Plates were grown for 2–3 d at 30°C and transferred at 4°C for increased color development. No difference in colony color was observed in these strains. (C) Low-level overexpression of Hsp104 was used to induce prion curing in [PSI+] hsp31Δ and WT strains. Cells were grown in liquid media for 12 h at 30°C before plating on ¼ YPD plates. Significantly less prion curing was observed in the [PSI+] hsp31Δ strain (**unpaired Student's t-test; p ≤ 0.001, n = 3). (D) High-level overexpression of Hsp104 was used to induce prion curing in [PSI+] hsp31Δ and WT strains. A 100% curing level was observed in WT strain. In the [PSI+] hsp31Δ strain, 100% curing was never achieved. White color colonies were plotted for the WT and [PSI+] hsp31Δ strain (****unpaired Student's t-test; p ≤ 0.0001, n = 3 biological replicates). (E) Hsp104 expression under the GAL promoter for 2 to 72 h in WT and [PSI+] hsp31Δ strain. At each indicated time point, cells were plated on ¼ YPD plates. Percentage of prion curing was calculated at each point for both WT and [PSI+] hsp31Δ strain. The plotted graph is one representation of 3 independent biological repeats. (unpaired Student's t-test; p ≤ 0.001 at 24, 48 and 72 h; n = 3). (F) Western blot demonstrating the relative expression levels of Hsp104. Time points for the pGAL plasmid are represent time after switching the strain to inducing galactose media. Equal amount of cells lysates were loaded in each lane and anti-histone H3 antibody was used as a loading control.
Hsp31 Deletion Impairs [PSI+] Prion Curing by Hsp104 Overexpression
A moderate level of Hsp104 is required for maintenance of [PSI+]; either deletion or overexpression of Hsp104 cures the [PSI+] prion. Numerous chaperones such as Hsp70, Hsp40 and Hsp90 along with its co-chaperones Sti1 and Cpr7 are known to modulate prion curing by Hsp104.10,23 To determine if Hsp31 altered Hsp104-mediated [PSI+] prion curing, we transformed WT [PSI+] or hsp31Δ [PSI+] cells with p426-GPD-Hsp104 (low level of overexpression), pAG425-GAL-Hsp104-DsRed (medium overexpression) or p2HG-Hsp104 (high overexpression) plasmids (Fig. 3C-E). The varied overexpression level of Hsp104 under these different plasmid systems was confirmed by western blotting (Fig. 3F). The level of [PSI+] prion curing was correlated to the level of Hsp104 expression in WT [PSI+] cells. The level of [PSI+] prion curing was significantly reduced in hsp31Δ [PSI+] under the lowest overexpression condition of GPD-Hsp104 (2% compared to 7% in WT) (Fig. 3C). Under high overexpression of Hsp104, a 100% curing level was observed in the presence of HSP31 but the hsp31Δ strain never achieved 100% curing and clearly white colonies were observed at 3% frequency (Fig. 3D). Intriguingly, the colonies were much whiter compared to the parental strain before attempting Hsp104-mediated curing, suggesting the colonies have a stronger [PSI+] status and phenotype but the reason for this is unclear. We confirmed that these colonies do not carry extragenic suppressor mutations because treatment with guanadine hydrochloride cured the [PSI+] phenotype as evidence by reversion to a red color (data not shown). Induction of the GAL-Hsp104-DsRed construct for varied time points from 2 h to 72 h also showed consistently less efficient prion curing by the hsp31Δ strain (Fig. 3E). These results demonstrate the presence of Hsp31 is required for optimal prion curing efficiency at varied Hsp104 expression conditions.
Hsp31 Collaborates with Hsp104 to Cure [PSI+] Prion
The decreased efficiency in Hsp104 curing [PSI+] in the absence of Hsp31 lead us to further explore the relationship between these 2 chaperones. Small heat shock proteins Hsp26 and Hsp42 are known to collaborate with Hsp104 in rescuing the polyglutamine toxicity and solubilization of amyloid aggregates.31 We focused on the collaboration of Hsp104 with Hsp31 but it should be noted that a variety of other co-chaperones may also be involved but are beyond the scope of this study. Overall, the results we present herein are indicative of a direct effect of Hsp31 on [PSI+] as outlined in the discussion. We first tested the effect of expressing Hsp31 and Hsp104 together in curing of the [PSI+] prion. Expression of Hsp104 (p426-GPD-Hsp104) in the [PSI+] strain resulted in a curing frequency of 3 % that is lower than that observed in Fig. 3 because the co-existence of 2 different constructs presumably decreases plasmid copy numbers and affects expression levels which we have observed previously when expressing GAL-Sup35-EYFP in combination with other vector plasmids41 and has been noted previously by others.47 Co-expression of Hsp31 and Hsp104 increased the frequency of prion curing to 6 % compared to the respective controls (Fig. 4A). We also tested the collaboration between Hsp104 and Hsp31 in the hsp31Δ strain and detected that co-expression of Hsp104 with Hsp31 was able to cure the [PSI+] prion to a greater extent than individual chaperone expression (Fig. 4B). These results corroborate the inefficient curing in hsp31Δ and establish that Hsp31 is required for optimal Hsp104 activity.
FIGURE 4.

Expression of Hsp31 in combination with Hsp104 increases prion curing. (A) Hsp31 and p426-GPD-Hsp104 were co-transformed in the [PSI+] strain. Vector without inserts served as controls. (B) Quantification of the experiments in panel A. Prion curing was increased from about 2.5% to 6% when Hsp31 was co-expressed with Hsp104 compared to the control strain (*unpaired Student's t-test; p ≤ 0.001, n = 3). (C) [PSI+] hsp31Δ strain harboring plasmids for Hsp104 and Hsp31. (D) Quantification of experiments describe in panel C. The combination of Hsp104 and Hsp31 increased prion curing in the [PSI+] hsp31Δ strain consistent with WT strain in A-B. (** One-way ANOVA; p ≤ 0.001, n = 3). (E) Image of p426-GPD-Hsp42 transformed cells demonstrating curing compared to vector only. (F) Hsp31 and p426-GPD-Hsp42 were co-transformed in the [PSI+] strain and quantified. The combination of Hsp42 and Hsp31 did not increase curing (ns = not significant). Data and images shown are representative of at least 3 independent biological experiments for all panels.
Previously, it was shown that small heat shock protein, Hsp42, collaborates with Hsp26 to prevent [PSI+] prionogenesis by distinct and synergistic mechanisms with Hsp10430 hence we tested the interaction between Hsp42 and Hsp31 on [PSI+] prion curing. Elevated levels of Hsp42 driven by the GPD promoter was sufficient to increase curing of [PSI+] in this strain transformed with a single plasmid (5.4% compared to 1.4%). Introduction of a second vector control plasmid, decreases the curing efficiency of the GPD-HSP42 construct to background levels. We also co-expressed Hsp31 and Hsp42 in the [PSI+] strain and did not detect any increased curing efficiency, in contrast to Hsp31 co-expression with Hsp104 (Fig. 4C). This lack of synergy between these proteins implies that Hsp42 and Hsp31 act at a similar stage of the prion cycle.
Effect of [PSI+] Curing in the hsp31Δ Strain is not Due to Loss of Hsp104 Thermotolerance Function
A possible mechanism for the decrease in [PSI+] prion curing by Hsp104 in the hsp31Δ background is decreased expression or activity of Hsp104. To measure the functional competence of Hsp104 we tested the thermotolerance activity of Hsp104 in exponentially growing cells. We induced Hsp104 expression levels in both strains by incubating the culture at 37°C for 30 minute and then heat shocked at 50°C. The survival of cells after heat shock is dependent on Hsp104 induction, and we observed survival in hsp31Δ was comparable to the isogenic WT [PSI+] strain (Fig. 5A). Moreover, the basal thermotolerance without induction of Hsp104 was not affected by deletion of Hsp31 (Fig. 5A). In addition, we observed a slightly elevated level of Hsp104 in the hsp31Δ strain compared to WT, hence the expression of Hsp104 is not compromised (Fig. 5B). Thus, reduction in [PSI+] prion curing in the hsp31Δ strain is not due to general impairment of Hsp104 activity or expression.
FIGURE 5.
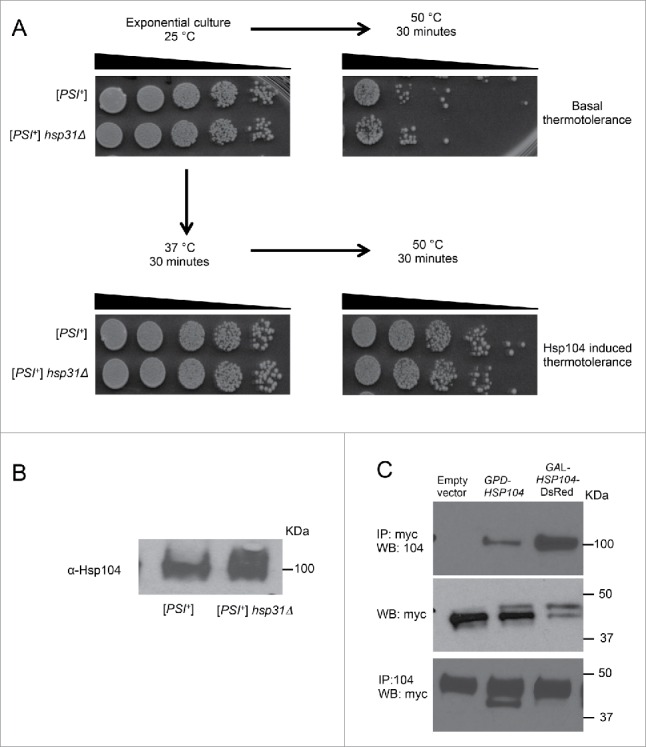
Hsp31 interacts with Hsp104 and deletion of HSP31 does not alter Hsp104's thermotolerance response. (A) HSP31 deletion does not impair Hsp104's function in thermotolerance. Exponentially growing cells of the [PSI+] hsp31Δ and [PSI+] strains were drawn from the culture and decimal serial dilutions were plated onto YPD plates and incubated for 2 d at 30°C in each case. Both strains showed a comparable basal tolerance (top right image) and induced tolerance after pretreatment at 37°C (bottom right image) for 30 min to a 50°C heat shock treatment. Left images are non-treated cultures that serve as control. (B) Endogenous level of Hsp104 was determined in exponentially growing cultures of [PSI+] hsp31Δ and [PSI+] strains in YPD media using Hsp104 specific antibody. (C) Immunoprecipitation of Hsp31 from HSP31-9myc strain with overexpression of Hsp104 either under GPD or GAL promoter, using anti-myc antibody followed by immunoblotting with anti-Hsp104 antibody. Middle panel shows the successful pull down of Hsp31-9myc in all strains using anti-myc antibody. The lower panel Hsp104 was immunoprecipitated using anti-Hsp104 antibody followed by immunoblotting with anti-myc antibody. Equal amount of cells lysates were loaded in each lane.
Hsp104 Physically Interacts with Hsp31
The close collaboration between Hsp104 and Hsp31 prompted us to test the physical association between them. Co-immunoprecipitation followed by western blot analysis in yeast lysates demonstrated that Hsp31 interacts with Hsp104. Immunoprecipitation was performed using HSP31-9myc genomically tagged at the endogenous locus and overexpressing Hsp104 either under the GPD or the GAL promoter. First, Hsp31-9myc was pulled down using anti-myc antibody conjugated to agarose beads from exponentially growing cell lysates. Western blots confirmed the successful pull down of Hsp31-9myc (Fig. 5C; middle panel). The upper panel demonstrates the successful pull-down of Hsp104 in both GPD-HSP104 and GAL-HSP104 expressing lysates but not in the vector control (Fig. 5C). Similar results were obtained with an alternative co-immunoprecipitation approach in which polyclonal anti-Hsp104 antibody and protein G dynabeads were used to pull down Hsp104 followed by western blot analysis with anti-myc antibody to detect Hsp31. This approach confirmed the interactions and also demonstrated that Hsp31 is pulled down with strains having endogenous levels of Hsp104 (Fig. 5C; bottom panel vector control lane). These results demonstrate that Hsp31 physically interacts with Hsp104 using 2 different immunoprecipitation protocols and the Hsp31-Hsp104 interaction is detectable under physiological expression levels.
Hsp31 Together with Hsp104 Antagonizes Prion Dependent Toxicity of Excess Sup35
Overexpression of full length Sup35 or its PrD exhibits toxicity in the [PSI+] strain.48,49 The toxicity of excess Sup35 in [PSI+] or [PSI+] hsp31Δ strain was investigated. Deletion of HSP31 has no effect on Sup35 toxicity (Fig. 6) in contrast to our previous report of increased toxicity when α-Syn is expressed in the hsp31Δ strain background.41 However, elevated levels of Hsp31 expressed from the GPD promoter rescued [PSI+] cells from Sup35 toxicity in both WT and deleted strains. As expected, Hsp104 reduced the toxicity of Sup35 to a greater extent than Hsp31. Strikingly Hsp31, together with Hsp104 strongly reduced the toxicity of Sup35 in the [PSI+] strain (Fig. 6). We also observed the rescue effect of sole expression of Hsp31 or Hsp104 and when expressed together in the [PSI+] hsp31Δ strain. The level of rescue was not as dramatic as in the WT strains suggesting that the expression of endogenous Hsp31 has a role in reducing toxicity in conjunction with heterologous expression of Hsp31 or Hsp104. The role of endogenous Hsp31 is not clear but could be a direct effect of chaperone activity or because autophagy can be impaired in hsp31Δ strains37 which may lead to increased proteotoxicity of Sup35.
FIGURE 6.

Hsp31 and Hsp104 reduce Sup35 prion toxicity. Hsp31, Hsp104 or the indicated combination of both along with GAL-PrD-Sup35-EYFP or full length Sup35 were overexpressed in [PSI+] and [PSI+] hsp31Δ strains. Decimal serial dilutions were plated onto selection plates with 2% glucose that serve as control or 2% galactose to induce the expression. Plates were incubated at 30°C for 3 d before producing the images. Hsp31 or Hsp104 rescued toxicity of GAL-PrD-Sup35-EYFP or full length Sup35 in these strains. Combination of Hsp31 and Hsp104 greatly reduce the toxicity compared to when they are individually expressed.
Hsp31 and Hsp104 Modulate Sup35 Aggregation in [PSI+] Cells
Collaboration of Hsp31 with Hsp104 to reduce Sup35 induced prion toxicity prompted us to investigate whether this activity correlates with protein disaggregation activity of Hsp104. To assess the state of Sup35 in [PSI+] cells harboring Hsp31 and Hsp104 expression plasmids (pAG415-GPD-HSP31-DsRed and p426-GPD-Hsp104 plasmids), we performed sedimentation analysis to determine the ratio of Sup35 in the soluble versus aggregate forms. The individual overexpression of Hsp104 or Hsp31 resulted in a very strong signal in the insoluble pellet fraction. However, the combination of Hsp104 and Hsp31 markedly reduced Sup35 aggregation found in the pellet fraction and increased the amount of Sup35 found in the soluble fraction (Fig. 7A). Soluble Sup35 was very susceptible to proteolysis during processing of the samples as evident by the lower molecular weight species, which is consistent with earlier reports.18,50 These results demonstrate that sole overexpression of Hsp31 does not appear to intervene in the established prion cycle present in a [PSI+] strain but can inhibit aggregate formation and prionogenesis in a [psi-] strain (Fig. 1). However, the results also show that Hsp31 can cooperate with Hsp104 to reduce Sup35 toxicity and simultaneously increase Sup35 solubility.
FIGURE 7.
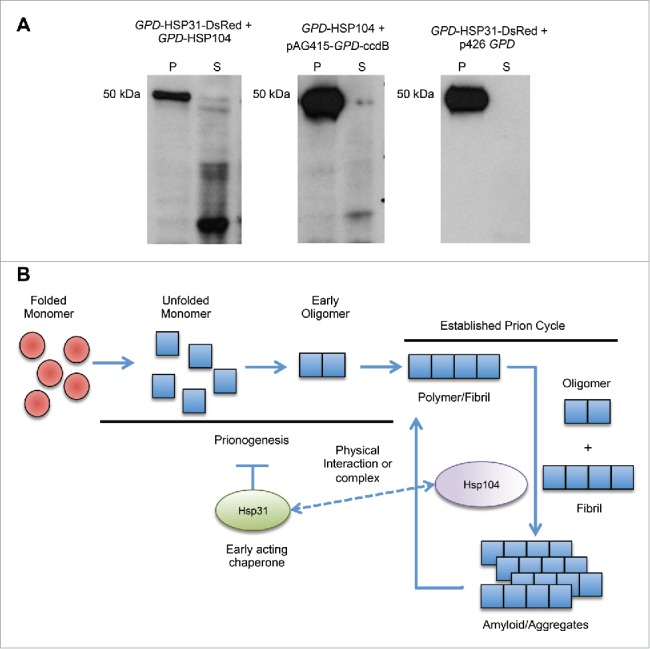
Hsp31 acts early in the prionogenesis process. (A) Hsp31 overexpression together with Hsp104 decreases the aggregation of Sup35 formed by overexpression of GAL-PrD-Sup35-EYFP. Crude lysates of cells expressing Hsp31 (pAG415-GPD-HSP31-DsRed plasmid), Hsp104 (p426-GPD-Hsp104 plasmid) or both together were subjected to sedimentation analysis. Lysates were ultracentrifuged into P (pellet) and S (soluble) fractions and analyzed by immunoblotting using GFP-specific antibody. (B) Model depicting the intervention of Hsp31 during the prionogenesis process but lack of involvement in an established chaperone cycle. Hsp31 and Hsp104 physically interact but it remains to be determined if this interaction is involved in a handoff of substrates.
DISCUSSION
Previous studies have demonstrated Hsp31 is a multitasking protein involved in several cellular pathways ranging from functioning as a glutathione independent methylglyoxalase to stress responder that acts as a molecular chaperone. In this study, we have established the inhibitory role of Hsp31 in Sup35 prion formation and its collaboration with Hsp104 to prevent prion aggregation and toxicity in yeast. In [PSI+] strains, soluble Sup35 protein is depleted into insoluble prion aggregates, hence, become less active resulting in nonsense suppression.49 The stronger [PSI+] prion phenotype is associated with larger amounts of protein aggregates. Overexpression of Sup35 PrD-EYFP in a [psi−] strain efficiently induces de novo [PSI+] prion formation and resulting aggregates, which appear as a peripheral ring associated with the vacuoles. The first step in de novo prion induction is the formation of a single prion seed, also known as a “propagon.”51,52 These seeds sequester the soluble Sup35 and grow at both ends into larger aggregates that appeared as rings or dots under microscopy. Moreover, it has been suggested that not all cells with fluorescent aggregates will transform into [PSI+] prions, rather about 50% of the cells with fluorescent foci will die and some of the aggregate-containing cells may not possess amyloids.53 Overexpression of Hsp31 in a [psi−] strain inhibits Sup35 aggregate formation and this was confirmed by flow cytometry and SDD-AGE. These results validate the previous observation that Hsp31 reduces Sup35 aggregates in the W303 strain.41 In addition, the inhibition of Sup35 aggregation by Hsp31 could result in the inhibition of prion formation, because the [PIN+] element required for prion induction is present in this strain. It should be noted that our experiments did not assess the status of [PIN+] and hence it is possible Hsp31 expression may affect [PIN+] in addition to [PSI+]. In addition, further testing of Hsp31 with weaker [PSI+] variants could result in more robust or sustained curing and warrant further investigation. It should be noted that the 74D-694 strain [PSI+] used in this study had a strong enough phenotype to resist curing by Hsp26 but was moderate at best because the strain color was pink rather than white.
We observed that overexpression of Hsp31 results in a significant reduction in prion induction from Sup35-PrD overexpression. However, reduced prion induction only takes place efficiently when Sup35-PrD was overexpressed for a transient period of time and upon longer expression of Sup35-PrD, Hsp31 was unable to reduce prion formation. Importantly, Hsp31 alone is unable to cure the [PSI+] prion indicating that it has no disaggregase activity. We postulate that the inability of curing but the concomitant ability to prevent the formation of de novo Sup35 SDS-resistant aggregates suggests that Hsp31 acts early in the process of prion oligomerization but once larger oligomers are formed it is not further active in preventing prion propagation (Fig. 7B).
Hsp31 may not participate in modulating an established prion cycle by itself but does appear to have a role in conjunction with Hsp104 because in a strain lacking Hsp31, the level of [PSI+] curing was reduced with Hsp104 overexpression. In addition, we showed Hsp31 overexpression promotes the elimination of [PSI+] by Hsp104. In fact, another small heat shock protein Hsp26 was shown to potentiate protein disaggregation by Hsp104. Interestingly, Hsp26 is only active as a disaggregase when it clusters together with the protein substrate and not after protein aggregation.30 A factor in considering the mechanism of action is that Hsp31 and the human ortholog, DJ-1, have protein deglycase activity.54,55 Prion glycation can occur through the action of glyoxal or methylglyoxal metabolites56 and the animal prion protein, PrPSc, has been reported to be modified at N terminal amino acid residues in clinically infected hamsters.57 Further evidence indicates that glycation can promote the stability of protein aggregates by covalent crosslinking58 suggesting the activity of a deglycase may abrogate prion propogation. Deciphering the contribution of the enzyme activity vs. chaperone function of Hsp31 would delineate the mechanisms of prion modulation by this multi-functional enzyme. We have previously shown that chaperone activity can be independent of the enzyme activity because overexpression of an enzymatically inactive Hsp31 mutant prevents the toxicity of α-synuclein in yeast.41 While there is enough evidence to propose Hsp31 acts early in the process of [PSI+] prion oligomerization process, future investigation of Hsp31 binding to the Sup35 monomer or other early oligomer and the role of deglycation in inhibiting prion induction would assist in elucidating the mechanism of action.
We eliminated several possible indirect mechanisms for the cooperation of Hsp31 with Hsp104 including the possibility that disruption of Hsp31 might compromise Hsp104 expression and thermotolerance function (Fig. 5). Taking into account that deletion of Hsp31 down regulates the mRNA level of Ssa3, a possible effect of Hsp31 deletion on [PSI+] prion curing by Hsp104 could be due to imbalances between Hsp104 and Hsp70 chaperone as these are required for efficient prion curing. However, this cannot be the case when Hsp31 is co-expressed with Hsp104, as overexpression of Hsp31 does not alter the level of Hsp70 protein.41 An additional mechanism leading to Hsp104-mediated curing is that modulation of Hsp31 expression levels could have affect other co-chaperones, which are known to have a role in prion curing.
We also showed that Hsp31 together with Hsp104 significantly reduced Sup35 prion toxicity. These results indicate that Hsp31 cooperates with Hsp104 to potentiate Sup35 prion disaggregation and thereby prevent toxicity. Intriguingly, previous studies have shown such cooperation between small heat shock proteins and ATP dependent chaperones as Hsp104 and Hsp70.31,38,59 We also observed a significant reduction of Sup35 toxicity by Hsp31 overexpression alone. A number of possible explanations could account for these results such as; Hsp31 might prevent sequestration of soluble Sup35 into already present larger aggregates and therefore inhibit toxicity. In addition, we have previously demonstrated that Hsp31 can reduce oxidative stress in cells41 and this may be the reason for the rescue of toxicity.
Intriguingly, we also found that Hsp31 physically interacts with Hsp104. Hsp104 is known to interact with Ydj1, and co-chaperones of Hsp90; Sti1 and Cpr7.10,26 Interestingly, Sti1 and Cpr7 are not required for Sup35 prion propagation but deletion of either of these reduced the Sup35 prion curing by Hsp104. Hsp31 has been documented to interact with the yeast Hsp90, Hsp82,60 based on affinity-mass spectrometry hence the interaction may involve a bridging chaperone such as Hsp90. However, our data is the first demonstration of Hsp31s interaction with Hsp104 and could mean these chaperones may pass substrates to each other and supports the hypothesis that Hsp31 affects [PSI+] directly. Several studies have shown that Hsp70 interacts with the M-domain of Hsp104.61 In addition, co-chaperones of Hsp90 such as Sti1 and Cpr7 use a TPR motif to interact with an acidic c-terminal tail of Hsp104.62 Hsp31 does not have a TPR motif so further investigation of the possible direct interaction with Hsp104 could uncover a novel motif and mode of interaction. Although the molecular details of the interaction are unknown, the involvement of Hsp31 in the prion modulation process and the apparent close functional cooperation with Hsp104 is an important step in understanding the biological roles of this multi-tasking protein.
MATERIAL AND METHODS
Yeast Strains and Plasmids
[psi−, PIN+] and [PSI+] strains, derivatives of 74D-694 [MATa, his3, leu2, trp1, ura3; suppressible marker ade1-14] were used throughout the study for prion induction, curing and toxicity assays. The W303 HSP31-9myc strain was used for pull down assays. Details of the plasmid, strains and primers used in this study are provided in the tables (Tables 1–3). Deletion of HSP31 in [PSI+] strain was obtained by transforming a PCR product containing the nourseothricin N- acetyl-transferase (NAT) gene flanked by HSP31 homology regions. The primers consisted of 20 nucleotides for amplifying the NAT gene from pFA6a-NATNT2 (Euroscarf), and 50 nucleotides immediately preceding the HSP31 start codon or after the stop codon. The amplified product was integrated into [PSI+] and [psi−, PIN+] strain. Successful integration and deletion was confirmed by diagnostic PCR.
TABLE 1.
List of yeast strains used in the study.
| Strain | Genotype | Source/reference |
|---|---|---|
| [PSI+] 74D-694 | MATa ade 1–14, his3, leu2, trp1, ura3 [PSI+PIN+] | J-C. Rochet |
| W303-1A | MATa can1-100 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 ade2-1 | R. Rothstein |
| [PSI+]-hsp31Δ | W303-1A hsp31Δ::NATMX | This study |
| W303-HSP31-9myc | W303-1A HSP31-9myc::KANMX | This study |
| [psi−] 74D-694 | 74D-694 MATa ade 1–14, his3, leu2, trp1, ura3 [psi−PIN+] | J-C. Rochet |
TABLE 2.
List of plasmids used in the study.
| Plasmids | Type of plasmid | Source/reference |
|---|---|---|
| pAG415GPD-HSP31-dsRed | Yeast, CEN | This study |
| pAG415ccdB-dsRed | Yeast, CEN | Alberti et al.63 |
| pAG424GAL-PrD-Sup35-EYFP | Yeast, 2 µ | Alberti et al.8,63 |
| pLA1-Sup35 | Yeast, CEN | J. Shorter30 |
| p426-GPD | Yeast, 2 µ | J. Shorter30 |
| p426-GPD-Hsp42 | Yeast, 2 µ | J. Shorter30 |
| p426-GPD-Hsp104 | Yeast, 2 µ | J. Shorter30 |
| p2HG-GPD-Hsp104 | Yeast, 2 µ | J-C.Rochet |
| p2HG-GPD | Yeast, 2 µ | J-C.Rochet |
| pAG425-GAL-ccdB-DsRed | Yeast, 2 µ | Addgene |
| pAG425-GAL-Hsp104-DsRed | Yeast, 2 µ | This study |
TABLE 3.
List of primers used in the study.
| Gene/description | Forward | Reverse |
|---|---|---|
| hsp31Δ | AAGTACTTCCCACTGGCTAATTACACAGATAAAACTCAAACAAATTTATAATGACATGGAGGCCCAGAATACCC | CTTACATCTATATAGTAGTACAAAGGAAATTCTAATTATCAACCTTTGGCTCACAGTATAGCGACCAGCATTCAC |
| 9myc tagging of HSP31 | TCTGCGCACTCCACTGCCGTAAGATCCATCGACGCTTTAAAAAACCGTACGCTGCAGGTCGAC | TCCTTACATCTATATAGTAGTACAAAGGAAATTCTAATTATCAACCTTTGGCTCAATCGATGAATTCGAGCTCG |
| HSP31 cloning | AAACTCGAGATGGCCCCAAAAAAAGTTTTACTCGC | TTTGCTAGCTCAGTTTTTTAAAGCGTCGATGGATCTTAC |
| HSP31 9myc tag diagnostic | ACAGAGAATTAACGTTACTCATTCC | ATATTTGGATATTGGGGAAACACAT |
| hsp31Δ diagnostic | TTCGTGGTCGTCTCGTACTC | GCAGGGCATGCTCATGTAGA |
Yeast Growth Conditions
We used isogenic [psi−] and [PSI+] derivatives of 74D-694 [MATa, his3, leu2, trp1, ura3; suppressible marker ade1-14. Cells were grown at 30°C on synthetic dextrose medium (SD; 0.7% yeast nitrogen base, 2% glucose) with appropriate amino acid dropout mixture for selection and maintenance of the particular plasmid. Synthetic complete (SC) medium contains 2% raffinose in place of glucose and 2% galactose for induction of genes under the GAL promoter. ¼ YPD solid medium used in the plating assays contains 0.5% yeast extract, 2% peptone, and 2% glucose. Cultures were always maintained in actively growing conditions and OD600 was used to measure the growth rate.
SDD-AGE
[psi−] cells were co-transformed with pAG424-GAL-PrD-Sup35-EYFP and pAG415- GPD-HSP31-DsRed plasmids. Cultures were grown in SD media overnight and induced in SC 2% raffinose + 2% galactose media for 24 h. Prion aggregates were analyzed using SDD-AGE as described previously.41 Briefly, cells were harvested by centrifugation and spheroplasts were generated and lysed in 4 x SDS sample buffer at room temperature for 15 min before loading onto a 1.8 % agarose gel followed by transfer to nitrocellulose membrane. Immunoblots were performed using anti-GFP antibody (Roche; 11814460001); anti-DsRed antibody (Santa Cruz Biotechnology; Sc-33353) and anti-histone H3 antibody (Abcam; ab1791).
Fluorescence Microscopy and Flow-Cytometry
The [psi−] strain was co-transformed with pAG424-GAL-PrD-Sup35-EYFP and pAG415-GPD-HSP31-DsRed. Transformants was selected and re-streaked on SD (-tryptophan -leucine) agar plates. Cells were grown overnight at 30°C in SD (-tryptophan -leucine) medium and PrD-Sup35 expression was induced for 24 h in SC (-tryptophan -leucine) medium with 2% raffinose and 2% galactose. Cells were examined under fluorescence microscopy using a Nikon A1 confocal microscope with a Nikon Plan apochromat 60 X (NA 1.4) oil immersion objective to acquire fluorescence and DIC images and were analyzed using Image J. The identical cultures used in microscopy were also subjected to flow cytometry. After induction cells were collected and washed with PBS, filtered and analyzed for EYFP fluorescence intensity using the Beckman Coulter FC500 flow cytometer with the FL-1 channel. A total of 10,000 events were acquired for each sample and data was analyzed using FlowJo software to calculate median fluorescence intensity.
Sup35 Prion Curing
Curing of [PSI+] was performed by transforming the strain with plasmids; p2HG-GPD-Hsp104, pAG425-GAL-Hsp104, pAG415-GPD-HSP31-DsRed, p426-GPD-Hsp42 and their corresponding vectors. For double transformation both plasmids were co-transformed and selected on double dropout media simultaneously. Transformed cells were grown in the SD medium at 30°C for overnight growth and plated on ¼ YPD plates which were incubated for 3 d at 30°C and shifted for another day at 4°C for colony color development. To score for curing, colonies with red color were counted as [psi−]. Cultures carrying plasmid with the GAL promoter were grown in SC medium before plating on ¼ YPD. Plates were imaged with a scanner and the colonies in the images were analyzed and counted in a blind manner using the colony counting function in Image J. Sectored colonies were counted as cured colonies.
Sup35 Prion Induction
For the induction experiment, at least 3 independent transformants with pAG424-GAL-PrD-Sup35-EYFP and pAG415-GPD-HSP31-DsRed plasmid were grown at 30°C in SD medium overnight, centrifuges and washed 3 times with water, and shifted to SC medium to induce prion formation. Aliquots were withdrawn at 6, 12 and 48 h and diluted to a density of 50,000 cells per 100 μl for plating onto ¼ YPD plate. The plates were then incubated for 3 d at 30°C and another day at 4°C for color development. Percentage of [PSI+] induction was measured as the number of white ([PSI+] colonies) colonies divided by the total number of colonies.
Prion Toxicity Assay
The [PSI+] strain was transformed with pAL1 Sup35 and plasmids expressing Hsp31, Hsp42 and or Hsp104 along with their corresponding vector. After selection on SD plates, cells were grown into 5 ml of SD liquid media with 2 % glucose at 30°C overnight. Cells were harvested and washed 3 times with water and a 5-fold serial dilution was performed with a starting OD600 of 0.8. Diluted samples of 5 μl were spotted onto SD and SC plates with the appropriate dropout selection. Plates were incubated at 30°C for 3 d and imaged with a scanner.
Pull Down Assay
The HSP31-9myc tagged strain was transformed with plasmids pAG425-GAL-Hsp104 or p426-GPD-Hsp104. Cells lysates were prepared in a buffer (50 mM Tris–HCl, 1 mM EDTA, 5 mM DTT, 10% (v/v) glycerol, 0.5 M NaCl, at pH 7.5) with freshly added protease inhibitor cocktail (Roche). Anti-myc antibody-conjugated agarose beads were used to pull down Hsp31 protein by incubating the beads with the lysate at 4°C for 1 h. After the pull down, beads were washed 3 times with PBS-T and bound protein was eluted by boiling the sample in SDS loading buffer before separating the proteins on SDS-PAGE and western blotting using Hsp104 antibody (Abcam; ab69549). Immunoprecipitation was also preformed in reverse by immobilizing Hsp104 antibody on protein G dynabeads (Life Technology) and incubating the total cell lysate with this complex for 1 h. In this case, western blotting was performed using anti-myc antibody (Sigma; M4439).
Thermotolerance Assay
WT [PSI+] and [PSI+] hsp31Δ strains were grown in YPD medium starting from OD600 of 0.2 until they reach exponential growth phase after 6 h. Equal number of cells from each strain were incubated at 37°C to induce Hsp104 expression and then heat shocked at 50°C for 20 min. Aliquots were placed on ice before and after heat shock. A portion of cultures were heat shocked at 50°C without Hsp104 induction at 37°C. Samples were collected and 5-fold dilutions were spotted on YPD medium.
Sedimentation Assay
[PSI+] strains harboring Hsp31, Hsp104 and appropriate co-expression vector plasmids were grown for 12 h and centrifuged to collect cells. Cells were washed in water and lysed at 4°C by bead beating in lysis buffer (50 mM Tris HCl pH 7.5, 50 mM NaCl, 2 mM EDTA and 5% Glycerol plus freshly added protease inhibitor cocktail41). Equal volume of cold RIPA buffer (50 mM Tris HCl pH 7.0, 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate and 0.1 % SDS) was added to the lysate and the mixture was vortexed for 10 s. Lysate was centrifuged at 800 rpm for 2 min at 4°C in a Eppendorf microcentrifuge. Lysate supernatant was subjected to ultracentrifugation at 80,000 rpm in a TLA-120.2 rotor for 30 min using an Optima Max-XD Ultracentrifuge (Beckman Coulter). Supernatant was collected and pellet was re-suspended in equal volume of lysis and RIPA buffer. Supernatant and pellet fractions were subjected to SDS-PAGE and immunoblotted using GFP antibody (Roche).
ABBREVIATIONS
- α-Syn
α-Synuclein
- PD
Parkinson disease
- PrD
Prion-forming domain
- HSP
Heat shock protein
- SC
Synthetic complete
- SDD-AGE
Semi-denaturing detergent agarose gel electrophoresis
- SD
Synthetic dextrose
- DsRed
Red fluorescence protein from Discosoma sp
- WT
Wild-type
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors do not have any conflict of interest or financial disclosures.
ACKNOWLEDGMENTS
We thank James Shorter, Martin Duennwald and Jean-Christophe Rochet for providing strains and plasmids.
FUNDING
K.A. was supported by the United States Educational Foundation in Pakistan through a Fulbright fellowship. This publication was made possible, in part, with support from the Indiana Clinical and Translational Sciences Institute funded, in part by grant # UL1TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award and a grant from National Institutes of Health - NIGMS (R01 GM087461). The authors gratefully acknowledge the Purdue University Genomics Core and support from the Purdue University Center for Cancer Research, NIH grant P30 CA023168.
AUTHOR CONTRIBUTIONS
K.A. performed all the experiments and was responsible for data collection. K.A. and T.R.H. conceived the project, designed experiments, interpreted results and wrote the manuscript. C-J.T. was involved in project conception, initiated prion experiments and final writing. T.R.H. supervised the project.
REFERENCES
- [1].Harrison RS, Sharpe PC, Singh Y, Fairlie DP. Amyloid peptides and proteins in review. Rev Physiol Biochem Pharmacol 2007; 159:1-77; PMID:17846922 [DOI] [PubMed] [Google Scholar]
- [2].Wickner RB, Edskes HK, Bateman DA, Kelly AC, Gorkovskiy A, Dayani Y, Zhou A. Amyloid diseases of yeast: prions are proteins acting as genes. Essays Biochem 2014; 56:193-205; PMID:25131596; http://dx.doi.org/ 10.1042/bse0560193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Langkilde AE, Morris KL, Serpell LC, Svergun DI, Vestergaard B. The architecture of amyloid-like peptide fibrils revealed by X-ray scattering, diffraction and electron microscopy. Acta Crystallogr D Biol Crystallogr 2015; 71:882-95; PMID:25849399; http://dx.doi.org/ 10.1107/S1399004715001674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Portillo A, Hashemi M, Zhang Y, Breydo L, Uversky VN, Lyubchenko YL. Role of monomer arrangement in the amyloid self-assembly. Biochim Biophys Acta 2015; 1854:218-28; PMID:25542374; http://dx.doi.org/ 10.1016/j.bbapap.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Manogaran AL, Fajardo VM, Reid RJ, Rothstein R, Liebman SW. Most, but not all, yeast strains in the deletion library contain the [PIN(+)] prion. Yeast 2010; 27:159-66; PMID:20014044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mankar S, Anoop A, Sen S, Maji SK. Nanomaterials: amyloids reflect their brighter side. Nano Rev 2011; 2; PMID:22110868; http://dx.doi.org/ 10.3402/nano.v2i0.6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liebman SW, Chernoff YO. Prions in yeast. Genetics 2012; 191:1041-72; PMID:22879407; http://dx.doi.org/ 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009; 137:146-58; PMID:19345193; http://dx.doi.org/ 10.1016/j.cell.2009.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Helsen CW, Glover JR. A new perspective on Hsp104-mediated propagation and curing of the yeast prion [PSI (+) ]. Prion 2012; 6:234-9; PMID:22561166; http://dx.doi.org/ 10.4161/pri.19913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reidy M, Masison DC. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol Cell Biol 2010; 30:3542-52; PMID:20479121; http://dx.doi.org/ 10.1128/MCB.01292-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bösl B, Grimminger V, Walter S. The molecular chaperone Hsp104–a molecular machine for protein disaggregation. J Struct Biol 2006; 156:139-48; PMID:16563798; http://dx.doi.org/ 10.1016/j.jsb.2006.02.004 [DOI] [PubMed] [Google Scholar]
- [12].Masison DC, Reidy M. Yeast prions are useful for studying protein chaperones and protein quality control. Prion 2015; 9:174-83; PMID:26110609; http://dx.doi.org/ 10.1080/19336896.2015.1027856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sweeny EA, Shorter J. Mechanistic and structural insights into the prion-disaggregase activity of Hsp104. J Mol Biol 2015; 428:1870-85; PMID:26608812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Newnam GP, Birchmore JL, Chernoff YO. Destabilization and recovery of a yeast prion after mild heat shock. J Mol Biol 2011; 408:432-48; PMID:21392508; http://dx.doi.org/ 10.1016/j.jmb.2011.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995; 268:880-4; PMID:7754373; http://dx.doi.org/ 10.1126/science.7754373 [DOI] [PubMed] [Google Scholar]
- [16].Mokry DZ, Abrahão J, Ramos CH. Disaggregases, molecular chaperones that resolubilize protein aggregates. An Acad Bras Cienc 2015; 87:1273-92; PMID:26312418; http://dx.doi.org/ 10.1590/0001-3765201520140671 [DOI] [PubMed] [Google Scholar]
- [17].Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol 2001; 21:4656-69; PMID:11416143; http://dx.doi.org/ 10.1128/MCB.21.14.4656-4669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 1996; 15:3127-34; PMID:8670813 [PMC free article] [PubMed] [Google Scholar]
- [19].Park YN, Zhao X, Yim YI, Todor H, Ellerbrock R, Reidy M, Eisenberg E, Masison DC, Greene LE. Hsp104 overexpression cures Saccharomyces cerevisiae [PSI+] by causing dissolution of the prion seeds. Eukaryot Cell 2014; 13:635-47; PMID:24632242; http://dx.doi.org/ 10.1128/EC.00300-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Park YN, Morales D, Rubinson EH, Masison D, Eisenberg E, Greene LE. Differences in the curing of [PSI+] prion by various methods of Hsp104 inactivation. PLoS One 2012; 7:e37692; PMID:22719845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Reidy M, Masison DC. Modulation and elimination of yeast prions by protein chaperones and co-chaperones. Prion 2011; 5:245-9; PMID:22052352; http://dx.doi.org/ 10.4161/pri.17749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mogk A, Kummer E, Bukau B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front Mol Biosci 2015; 2:22; PMID:26042222; http://dx.doi.org/ 10.3389/fmolb.2015.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Reidy M, Sharma R, Shastry S, Roberts BL, Albino-Flores I, Wickner S, Masison DC. Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS Genet 2014; 10:e1004720; PMID:25329162; http://dx.doi.org/ 10.1371/journal.pgen.1004720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Romanova NV, Chernoff YO. Hsp104 and prion propagation. Protein Pept Lett 2009; 16:598-605; PMID:19519517; http://dx.doi.org/ 10.2174/092986609788490078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sporn ZA, Hines JK. Hsp40 function in yeast prion propagation: Amyloid diversity necessitates chaperone functional complexity. Prion 2015; 9:80-9; PMID:25738774; http://dx.doi.org/ 10.1080/19336896.2015.1020268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J 2008; 27:2712-24; PMID:18833196; http://dx.doi.org/ 10.1038/emboj.2008.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Allen KD, Wegrzyn RD, Chernova TA, Müller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 2005; 169:1227-42; PMID:15545639; http://dx.doi.org/ 10.1534/genetics.104.037168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Masison DC, Kirkland PA, Sharma D. Influence of Hsp70s and their regulators on yeast prion propagation. Prion 2009; 3:65-73; PMID:19556854; http://dx.doi.org/ 10.4161/pri.3.2.9134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kirkland PA, Reidy M, Masison DC. Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 2011; 188:565-77; PMID:21555396; http://dx.doi.org/ 10.1534/genetics.111.129460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol 2012; 10:e1001346; PMID:22723742; http://dx.doi.org/ 10.1371/journal.pbio.1001346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem 2005; 280:23869-75; PMID:15845535; http://dx.doi.org/ 10.1074/jbc.M502854200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J 2004; 23:638-49; PMID:14749732; http://dx.doi.org/ 10.1038/sj.emboj.7600080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol 2011; 195:617-29; PMID:22065637; http://dx.doi.org/ 10.1083/jcb.201106037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Haslbeck M, Walke S, Stromer T, Ehrnsperger M, White HE, Chen S, Saibil HR, Buchner J. Hsp26: a temperature-regulated chaperone. EMBO J 1999; 18:6744-51; PMID:10581247; http://dx.doi.org/ 10.1093/emboj/18.23.6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Amm I, Norell D, Wolf DH. Absence of the yeast Hsp31 chaperones of the DJ-1 superfamily perturbs cytoplasmic protein quality control in late growth phase. PLoS One 2015; 10:e0140363; PMID:26466368; http://dx.doi.org/ 10.1371/journal.pone.0140363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sajjad MU, Green EW, Miller-Fleming L, Hands S, Herrera F, Campesan S, Khoshnan A, Outeiro TF, Giorgini F, Wyttenbach A. DJ-1 modulates aggregation and pathogenesis in models of Huntington's disease. Hum Mol Genet 2014; 23:755-66; PMID:24070869; http://dx.doi.org/ 10.1093/hmg/ddt466 [DOI] [PubMed] [Google Scholar]
- [37].Miller-Fleming L, Antas P, Pais TF, Smalley JL, Giorgini F, Outeiro TF. Yeast DJ-1 superfamily members are required for diauxic-shift reprogramming and cell survival in stationary phase. Proc Natl Acad Sci U S A 2014; 111:7012-7; PMID:24706893; http://dx.doi.org/ 10.1073/pnas.1319221111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem 2005; 280:23861-8; PMID:15843375; http://dx.doi.org/ 10.1074/jbc.M502697200 [DOI] [PubMed] [Google Scholar]
- [39].Devine MJ, Plun-Favreau H, Wood NW. Parkinson's disease and cancer: two wars, one front. Nature reviews Cancer 2011; 11:812-23; PMID:22020207; http://dx.doi.org/ 10.1038/nrc3150 [DOI] [PubMed] [Google Scholar]
- [40].Cao J, Lou S, Ying M, Yang B. DJ-1 as a human oncogene and potential therapeutic target. Biochemical pharmacology 2015; 93:241-50; PMID:25498803; http://dx.doi.org/ 10.1016/j.bcp.2014.11.012 [DOI] [PubMed] [Google Scholar]
- [41].Tsai CJ, Aslam K, Drendel HM, Asiago JM, Goode KM, Paul LN, Rochet JC, Hazbun TR. Hsp31 Is a Stress Response Chaperone That Intervenes in the Protein Misfolding Process. J Biol Chem 2015; 290:24816-34; PMID:26306045; http://dx.doi.org/ 10.1074/jbc.M115.678367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zondler L, Miller-Fleming L, Repici M, Gonçalves S, Tenreiro S, Rosado-Ramos R, Betzer C, Straatman KR, Jensen PH, Giorgini F, et al.. DJ-1 interactions with α-synuclein attenuate aggregation and cellular toxicity in models of Parkinson's disease. Cell Death Dis 2014; 5:e1350; PMID:25058424; http://dx.doi.org/ 10.1038/cddis.2014.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997; 147:507-19; PMID:9335589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP, Belenkiy SM. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol 2000; 35:865-76; PMID:10692163; http://dx.doi.org/ 10.1046/j.1365-2958.2000.01761.x [DOI] [PubMed] [Google Scholar]
- [45].Aslam K, Hazbun TR. Hsp31, a member of the DJ-1 superfamily, is a multitasking stress responder with chaperone activity. Prion 2016; 10:103-11; PMID:27097320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wickner RB, Bezsonov E, Bateman DA. Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc Natl Acad Sci U S A 2014; 111:E2711-20; PMID:24938787; http://dx.doi.org/ 10.1073/pnas.1409582111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Couthouis J, Rebora K, Immel F, Berthelot K, Castroviejo M, Cullin C. Screening for toxic amyloid in yeast exemplifies the role of alternative pathway responsible for cytotoxicity. PLoS One 2009; 4:e4539; PMID:19262694; http://dx.doi.org/ 10.1371/journal.pone.0004539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Vishveshwara N, Bradley ME, Liebman SW. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol Microbiol 2009; 73:1101-14; PMID:19682262; http://dx.doi.org/ 10.1111/j.1365-2958.2009.06836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pezza JA, Villali J, Sindi SS, Serio TR. Amyloid-associated activity contributes to the severity and toxicity of a prion phenotype. Nat Commun 2014; 5:4384; PMID:25023996; http://dx.doi.org/ 10.1038/ncomms5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kochneva-Pervukhova NV, Alexandrov AI, Ter-Avanesyan MD. Amyloid-mediated sequestration of essential proteins contributes to mutant huntingtin toxicity in yeast. PLoS One 2012; 7:e29832; PMID:22253794; http://dx.doi.org/ 10.1371/journal.pone.0029832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Byrne LJ, Cole DJ, Cox BS, Ridout MS, Morgan BJ, Tuite MF. The number and transmission of [PSI] prion seeds (Propagons) in the yeast Saccharomyces cerevisiae. PLoS One 2009; 4:e4670; PMID:19262693; http://dx.doi.org/ 10.1371/journal.pone.0004670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics 2003; 165:23-33; PMID:14504215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Arslan F, Hong JY, Kanneganti V, Park SK, Liebman SW. Heterologous aggregates promote de novo prion appearance via more than one mechanism. PLoS Genet 2015; 11:e1004814; PMID:25568955; http://dx.doi.org/ 10.1371/journal.pgen.1004814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Richarme G, Mihoub M, Dairou J, Bui LC, Leger T, Lamouri A. Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J Biol Chem 2015; 290:1885-97; PMID:25416785; http://dx.doi.org/ 10.1074/jbc.M114.597815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mihoub M, Abdallah J, Gontero B, Dairou J, Richarme G. The DJ-1 superfamily member Hsp31 repairs proteins from glycation by methylglyoxal and glyoxal. Biochem Biophys Res Commun 2015; 463:1305-10; PMID:26102038; http://dx.doi.org/ 10.1016/j.bbrc.2015.06.111 [DOI] [PubMed] [Google Scholar]
- [56].Panza G, Dumpitak C, Birkmann E. Influence of the maillard reaction to prion protein aggregation. Rejuvenation Res 2010; 13:220-3; PMID:20370497; http://dx.doi.org/ 10.1089/rej.2009.0954 [DOI] [PubMed] [Google Scholar]
- [57].Choi YG, Kim JI, Jeon YC, Park SJ, Choi EK, Rubenstein R, Kascsak RJ, Carp RI, Kim YS. Nonenzymatic glycation at the N terminus of pathogenic prion protein in transmissible spongiform encephalopathies. J Biol Chem 2004; 279:30402-9; PMID:15084583; http://dx.doi.org/ 10.1074/jbc.M400854200 [DOI] [PubMed] [Google Scholar]
- [58].Vicente Miranda H, Outeiro TF. The sour side of neurodegenerative disorders: the effects of protein glycation. J Pathol 2010; 221:13-25; PMID:20186922; http://dx.doi.org/ 10.1002/path.2682 [DOI] [PubMed] [Google Scholar]
- [59].Walter GM, Smith MC, Wisén S, Basrur V, Elenitoba-Johnson KS, Duennwald ML, Kumar A, Gestwicki JE. Ordered assembly of heat shock proteins, Hsp26, Hsp70, Hsp90, and Hsp104, on expanded polyglutamine fragments revealed by chemical probes. J Biol Chem 2011; 286:40486-93; PMID:21969373; http://dx.doi.org/ 10.1074/jbc.M111.284448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al.. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 2005; 120:715-27; PMID:15766533; http://dx.doi.org/ 10.1016/j.cell.2004.12.024 [DOI] [PubMed] [Google Scholar]
- [61].Doyle SM, Genest O, Wickner S. Protein rescue from aggregates by powerful molecular chaperone machines. Nat Rev Mol Cell Biol 2013; 14:617-29; PMID:24061228; http://dx.doi.org/ 10.1038/nrm3660 [DOI] [PubMed] [Google Scholar]
- [62].Abbas-Terki T, Donze O, Briand PA, Picard D. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol Cell Biol 2001; 21:7569-75; PMID:11604493; http://dx.doi.org/ 10.1128/MCB.21.22.7569-7575.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Alberti S, Gitler AD, Lindquist S. A suite of Gateway® cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 2007; 24:913-9; PMID:17583893; http://dx.doi.org/ 10.1002/yea.1502 [DOI] [PMC free article] [PubMed] [Google Scholar]


