Abstract
Purpose
The majority of prostate cancer mortality can be attributed to metastatic castration-resistant prostate cancer, an advanced stage which remains incurable despite recent advances. The androgen receptor (AR) signaling axis remains active in CRPC. Recent studies suggest that the expression of an AR splice variant, AR-V7, may underlie resistance to abiraterone and enzalutamide. However, controversy exists over the optimal assay. The objective of this study is to develop a fast and sensitive assay for AR splice variants (AR-Vs) in patients.
Materials and Methods
Two approaches were assessed in this study. The first was based on depletion of leukocytes and the second used RNA purified directly from whole blood preserved in PAXgene tubes. Transcript expression was analyzed by quantitative RT-PCR.
Results
Through side-by-side comparison, we concluded that the whole-blood approach was suitable for the detection of AR-Vs. The specificity of the assay was corroborated in a cancer-free cohort. Using the PAXgene assay, samples from a cohort of 46 CRPC patients were analyzed. Overall, AR-V7 and ARv567es were detected in 67.53% and 29.87% of the samples, respectively. Statistical analysis revealed a strong association of AR-V positivity with a history of second-line hormonal therapies.
Conclusions
To our knowledge, this study is the first to demonstrate PAXgene-preserved whole blood can be used to obtain clinically relevant information regarding the expression of two AR-Vs. The data from a CRPC cohort support a role of AR-Vs in resistance to therapies targeting the AR ligand-binding domain.
Keywords: androgen receptor splice variants, castration-resistant prostate cancer, enzalutamide, abiraterone, whole-blood assay
Introduction
Prostate cancer is the second leading cause of cancer mortality in men in the United States. The majority of disease-related deaths can be attributed to metastatic castration-resistant prostate cancer (mCRPC), which is marked by rising serum prostate-specific antigen (PSA) levels approximately 16 months after initial androgen deprivation therapy (ADT). Despite recent advances, mCRPC remains the most critical challenge in the clinical management of prostate cancer.
It is well-accepted that the AR signaling axis plays a critical role in CRPC. Through a number of ligand-dependent and –independent mechanisms, cancer cells adapt to low circulating androgens and maintain activation of AR. Particularly, a number of AR splice variants (AR-Vs) that are devoid of a functional ligand-binding domain (LBD) have been identified1–4. Two major variants, AR-V7 and ARv567es, have been shown to be capable of regulating target gene expression independent of full-length AR (AR-FL)2–5. Recent studies suggest the expression of these AR-Vs underlie resistance to second-line hormonal therapies6,7.
Circulating tumor cells (CTCs) are shed from solid tumors into the circulation. A number of CTC detection technologies have been developed in recent years. To date, the most widely adopted is surface marker-based CTC capturing, which relies on cell surface antigens such as EpCAM, cytokeratin, PSMA, or a combination of these markers8. Alternatively, CTCs can be enriched by depleting hematopoietic cells. For this purpose, CD45, expressed on the surface of all leukocytes and their progenitors9, is commonly used10,11. Yet other studies have demonstrated the validity of using whole-blood-derived RNA from patients for RT-PCR analyses, without CTC selection or enrichment12–14. These studies focused on genes highly expressed in prostate cancer; the results were clinically relevant and highly concordant with CTC enumeration analyses13,14.
In this study, we set out to detect the expression of AR-V7 and ARv567es in the circulation. We evaluated the CD45-based negative selection approach and the whole blood approach. Based on the results, we chose the whole blood approach and analyzed blood samples obtained from a cohort of CRPC patients.
Materials and Methods
RNA extraction from whole blood
From each patient, 5-mL blood was collected into 2 PAXgene Blood RNA Tubes (PreAnalytiX). The tubes were gently inverted and incubated at room temperature for 2–24 h or stored at -20°C before processing. Prior to RNA i solation, frozen samples were brought to room temperature for 2 h and centrifuged at 3,000g for 10 min. RNA isolation was performed using the PAXgene Blood RNA Kit (Qiagen).
Leukocyte depletion
CTC enrichment was performed by using a two-step procedure as described11 with modifications.
Lysis of red blood cells (RBCs)
Patient blood (10 mL) was collected in sodium citrate tubes (Becton Dickinson) for immediate processing. RBCs were removed by adding a lysis buffer (154 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) at 25 mL buffer/mL blood. After 5 min at room temperature, remaining cells were collected by centrifugation at 300g for 5 min. The pellet was washed twice with the labeling buffer (PBS with 2 mM EDTA, 0.5% bovine serum albumin, Ca2+/Mg2+ free) and resuspended. Cells were counted and the concentration was adjusted to 1 x 108 cells/mL.
Removal of leukocytes
Following RBC lysis, cell suspension was transferred to a round-bottom tube (Becton Dickinson). To every 108 cells, 200 μL of FcR Blocking Reagent (Miltenyi Biotec) and 50 μL of EasySep® CD45 Depletion Cocktail (StemCell) were added and incubated at room temperature for 30 minutes. For immunomagnetic labelling, EasySep® Magnetic Nanoparticles (StemCell) was added at 100 μL/mL and mixed by pipetting. The suspension was incubated at room temperature for 15 minutes and the volume was adjusted to 2.5 mL. The tube was placed into an EasySep® magnet (StemCell) for 10 minutes. Labeled cells (CD45+) were separated by decanting the supernatant (CD45−) into a new tube. RNA was extracted from the CD45+ and CD45− fractions using the RNeasy Mini Kit (Qiagen).
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
RNA samples were quantitated by a Nanodrop 2000 spectrophotometer (Thermo Fisher). Subsequently, 0.5 μg of RNA was reverse-transcribed using SuperScript III (Life Technologies) and random hexamers. The Taqman assay was chosen as the quantitative PCR technology to ensure the specificity of detection. PCR reactions were performed on a CFX96 Real-Time PCR Detection System (Bio-Rad). The primers for AR-FL and AR-V7 are published3 and those for ARv567es are: forward, 5’-CTACTCCGGACCTTACGGGGACATGCG-3’; reverse, 5’-TTGGGCACTTGCACAGAGAT-3’. The probe for all three amplicons was designed in our lab: 5’-AAGAGCCGCTGAAGGGAAACAGAAGTACTTG-3’. Ribosomal protein L30 (RPL30) was used as the housekeeping gene and the assay for RPL30 was from Applied Biosystems. Forty cycles was used as detection limit. The expression levels of the same transcript measured by two approaches were compared by using the 2-ΔCT method15 and adjusted for the difference in starting blood volume.
Subjects
Forty-six CRPC patients were selected and consented via Tulane Cancer Center and/or Urology clinic at Tulane University Hospital. For each patient, 5 mL of blood were collected in PAXgene tubes; when feasible serial samples were collected. A total of 73 blood samples were collected and clinically annotated. A summary of the patient demographics is presented in Table 1. The history of treatment with abiraterone acetate and enzalutamide is summarized in Supplementary Table S1 and the complete treatment history in Supplementary Table S2. Additionally, blood samples from 21 post-radical prostatectomy (post-RP) patients with undetectable PSA (<0.01 ng/mL) were analyzed.
Table 1.
Patient demographics.
| Overall | Caucasians | African Americans | ||
|---|---|---|---|---|
| Patients | n= | 46 | 41 | 5 |
|
| ||||
| Age at diagnosis | 60 (range 43–77) | 61.1 (range 46–77) | 57.8 (range 43–66) | |
|
| ||||
| Total Gleason score | 8 (range 6–9) | 8 (range 6–9) | 7 | |
|
| ||||
| Family history | Yes | 26% (n=12) | 27% (n=11) | 20% (n=1) |
| No | 72% (n=33) | 71% (n=29) | 80% (n=4) | |
| Unknown | 2% (n=1) | 2% (n=1) | ||
|
| ||||
| Median PSA(ng/mL) at sampling | 60.8 (range <0.01–>3000) | 59.7 (range <0.01–3000) | 184.4 (range 48.9–2050) | |
|
| ||||
| Median time from diagnosis to sampling (days) | 1877.5 (range 230–7781) | 1877.5 (range 230–7781) | 2424 (range 691–7080) | |
Statistical analysis
All statistical analyses including descriptive statistics, Student’s T-test and Fischer’s Exact Test were performed with SAS v9.4 (SAS Institute, Cary, NC). All tests are two-tailed with p-values less than the alpha (p≤0.05) were considered statistically significant. Medians and ranges were reported for all continuous variables. Percentages were reported for all categorical variables.
Results
Selection of the whole blood approach for AR-V detection
To identify a protocol for detecting AR-Vs in the blood, we evaluated the whole-blood approach (i.e. the PAXgene approach) and the CTC enrichment approach based on depletion of leukocytes (i.e. the CD45-depletion approach). Ten mCRPC patients who have progressed taxane chemotherapy and have received multiple rounds of abiraterone and/or enzalutamide were identified for this purpose. Blood samples obtained from the same patient were analyzed side-by-side (Supplementary Fig. S1). As shown in Supplementary Table S3, AR-V7 transcripts were detected in 9 out of 10 samples by both methods. However, two samples were found to be positive for ARv567es by the PAXgene approach, but only 1 by the CD45-depletion approach, suggesting that the leukocyte depletion process may lead to a loss of sensitivity. Indeed, the AR-V transcript levels measured by the CD45-depletion approach were consistently lower than those by the PAXgene approach: the estimated decrease for AR-V7 was ~40% (Supplementary Table S4).
The separation of CD45− and CD45+ cells during leukocyte depletion provides an opportunity to investigate the sources of AR transcripts. As shown in Table 2, in the majority of the samples (7 out of 9), the AR-V7 signal was exclusively from the CD45− fraction. In the remaining samples, the levels of AR-V7 measured in the CD45+ fraction were markedly lower than in the CD45− fraction. Similarly, the vast majority of ARv567es transcript was found in the CD45− fraction. In contrast, AR-FL is abundantly expressed in both fractions (Table 2). The presence of AR-FL transcripts in CD45+ cells is consistent with studies documenting AR expression in lymphocytes and macrophages16,17.
Table 2.
Distribution of AR transcripts in the CD45− and CD45+ fractions*.
| ID | AR-V7
|
ARv567es
|
AR-FL
|
|||
|---|---|---|---|---|---|---|
| CD45− | CD45+ | CD45− | CD45+ | CD45− | CD45+ | |
|
|
|
|
|
|
|
|
| 1 | 100.00% | 0.00% | - | - | 46.05% | 53.95% |
| 2 | 100.00% | 0.00% | - | - | 29.21% | 70.79% |
| 3 | 100.00% | 0.00% | - | - | 63.96% | 36.04% |
| 4 | 85.93% | 14.07% | 96.16% | 3.84% | 55.55% | 44.45% |
| 5 | - | - | - | - | 65.27% | 34.73% |
| 6 | 100.00% | 0.00% | - | - | 54.75% | 45.25% |
| 7 | 100.00% | 0.00% | - | - | 20.02% | 79.98% |
| 8 | 91.65% | 8.35% | - | - | 76.71% | 23.29% |
| 9 | 100.00% | 0.00% | - | - | 75.59% | 24.41% |
| 10 | 100.00% | 0.00% | - | - | 31.34% | 68.66% |
Percentage values were calculated from the expression ratio between the two fractions, with a total of 100%.
Collectively, these results suggest that the AR-Vs transcripts in the CD45+ fraction are barely detectable, suggesting that depleting the hematopoietic cells offers little improvement in specificity for the detection of AR-Vs. Furthermore, performing this procedure could lead to a loss of sensitivity.
Sensitivity and specificity of the whole-blood assay
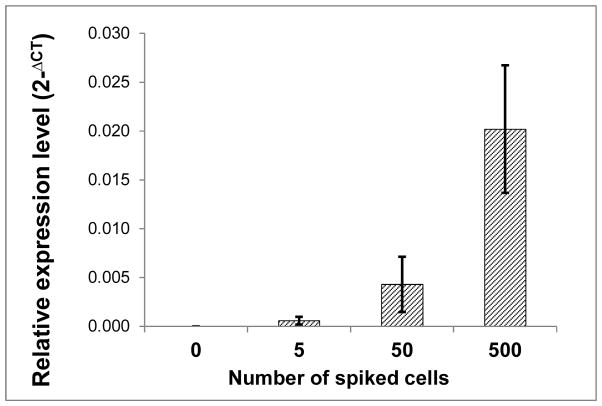
To assess the sensitivity of this assay, 22Rv1 cells (express AR-V7) were spiked into 5 ml blood from a healthy donor. Fig. 1 shows that this assay could detect 5–50 AR-V7+ cells in 5 ml of blood, or 1–10 cells per ml.
Figure 1. Sensitivity of the PAXgene assay for AR-V7 detection.
22RV1 cells were spiked into 5 ml blood from a healthy donor and transferred to PAXgene tubes. RNA extraction and qRT-PCR analysis were performed as described in Materials and Methods.
To evaluate the specificity, we analyzed blood samples from post-RP patients with undetectable PSA. Neither AR-V7 nor ARv567es was detected in this cohort (Table 3). Similarly, neither AR-V was detected in blood from healthy donors (n=5, data not shown). These results suggest that this assay specifically detects AR-Vs expressed in cancer cells.
Table 3.
Detection of AR transcripts in two cohorts.
| Post-RP | mCRPC | P value* | |
|---|---|---|---|
| Age at sampling | 65.05 (range 53–76) | 68.52 (range 47–86) | 0.1559 |
| AR-FL+ | 17/21 (80.95%) | 69/73 (94.52%) | 0.0712 |
| AR-V7+ | 0/21 (0%) | 50/73 (68.49%) | <0.0001 |
| ARv567es + | 0/21 (0%) | 23/73 (31.51%) | 0.0014 |
| AR-V+ | 0/21 (0%) | 53/73 (72.60%) | <0.0001 |
| AR-V7+, ARv567es+ | 0/21 (0%) | 20/73 (27.40%) | 0.0051 |
For dichotomous variables, P-values were calculated by Fisher’s Exact Test. For Age at Sampling, P-value was by the Student’s t-test.
Correlation of AR-Vs expression with prior treatments with abiraterone and enzalutamide
To investigate the clinical relevance of AR-Vs expression in whole blood, we analyzed 73 samples from 46 CRPC patients. In this cohort, 69 (94.52%) were positive for AR-FL, 50 (68.49%) for AR-V7, and 23 (31.51%) for ARv567es (Table 3). More than 70% (53/73) of samples expressed at least one variant, and 27.40% (20/73) expressed both. Notably, the majority of samples expressing ARv567es were also positive for AR-V7 (20/23).
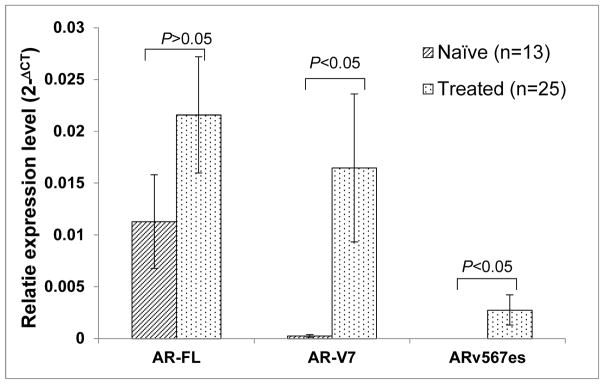
Based on the history of second-line hormonal therapies, viz. abiraterone, ketoconazole, and enzalutamide, the CRPC cohort was categorized into naïve and treated groups. Due to a recent report demonstrating an association of AR-V7 expression with prior treatment with docetaxel18, patients who have received docetaxel or cabazitaxel were excluded. The expression levels of both variants, but not that of AR-FL, were higher in the treated than in the naïve group (Fig. 2). In the treated group, AR-V7 transcripts was expressed in 68% of samples (17/25), as compared to 23.08% (3/13) in the naïve group. Fisher’s Exact Test revealed a strong association of AR-V7 positivity with a history of second-line hormonal therapies (Table 4). Similarly, ARv567es positivity is associated with a history of these therapies (9/25 in treated, 0/13 in naïve). Additionally, patients who have been treated with second-line hormonal therapies are more likely to express one (AR-V+) or both isoforms of AR-Vs (AR-V7+/ARv567es+).
Figure 2. Elevated expression of AR-Vs in mCRPC patients that have been treated with second-line hormonal therapies.
The naïve group are defined as patients who have never received treatments with abiraterone acetate, ketoconazole, or enzalutamide, and the treated group are patients who have received these treatments and progressed. Statistics analysis was performed using the Student’s t-test (2-tail).
Table 4.
AR-V positivity correlates with prior history of second-line hormonal treatments.
| Naïve | Treated | P-value by Fisher’s Exact Test | |
|---|---|---|---|
| AR-FL+ | 12/13 | 24/25 | 1.0000 |
| AR-V7+ | 3/13 | 17/25 | 0.0156 |
| ARv567es + | 0/13 | 9/25 | 0.0159 |
| AR-V+ | 3/13 | 18/25 | 0.0062 |
| AR-V7+/ARv567es+ | 0/13 | 8/25 | 0.0335 |
Discussion
As noted by many, development of predictive biomarkers are critical to optimal clinical decision-making. The study by Antonarakis et al7 clearly represents a step forward in this direction. Based on this ground-breaking study, the AR-V7 transcript in CTCs predicts resistance to both abiraterone and enzalutamide. However, a potential shortcoming of this assay is the dependence on the selection of EpCAM+ or Her2+ CTCs19, which likely represent only a fraction of the entire CTC population. In addition, the use of an epithelial marker (EpCAM) may exclude the population that have undergone epithelial-to-mesenchymal transition (EMT). This is particularly relevant since studies have shown AR-V7 and ARv567es promote EMT20–23 in prostate cancer cells, raising the concern that AR-V-expressing CTCs may not be captured efficiently by epithelial markers. Furthermore, it is clear from studies of CTCs using immuno-detection methodologies that not all patients have detectable CTCs24, thus limiting the number of patients that can have these AR-V7 assays performed.
To avoid the problems associated with CTC positive selection, we tested a whole-blood-based approach and a negative selection approach by depleting lymphocytes. A comparison of these approaches led us to conclude that the whole blood approach performed at least as well as the CD45-depletion approach, with regard to the sensitivity of AR-V detection (Supplementary Table S3). The expression of AR-Vs was found predominantly in the CD45- population (Table 2), suggesting that depleting CD45+ cells is unnecessary. Furthermore, the variants were detected at lower levels by the CTC-enrichment protocol (Supplementary Table S4), possibly due to the lack of RNA preservation during the enrichment process. In a few cases, we did detect variants in the CD45+ fraction (Table 2). This was most likely a result of cross contamination, rather than expression of the variants by CD45+ cells per se. This is supported by data from the post-RP cohort that neither AR-V was detectable in whole-blood-derived RNA (Table 3). Cross contamination could be caused by non-specific binding of the CD45 antibodies, aberrant expression of CD45 by cancer cells25,26, or the formation of lymphocytes/cancer cells microemboli27. Regardless, this observation suggests the CTCs could be lost during negative selection.
Recently, Steinestel and colleagues reported using AR-V7 and AR mutations in CTCs to guide therapy-switch in patients18. The estimated overall benefit of the molecularly-informed treatment decision was 27% over the uninformed18. This study, along with that of Antonarakis7, highlights the importance of incorporating AR-V profiling into individualized treatment decision-making. Despite that our assay is not CDC-based, the findings are in line with these CDC-based studies, supporting a role of AR-V7 expression in resistance to second-line hormonal therapies.
The findings presented herein have several limitations. First, this is a cross-sectional study, rather than a prospective study. A prospective study with longitudinal evaluation of patients is needed to establish expression of AR-Vs in whole blood as a biomarker of treatment responsiveness. Second, the cells expressing the AR transcripts are not assessable by this assay. As a result, questions such as whether AR-FL and AR-Vs are co-expressed in the same cells could not be properly addressed. These questions are clinically significant as AR-Vs have been shown to heterodimerize with AR-FL28 and facilitate its nuclear translocation29. Third, there is no tissue confirmation using direct biopsies of tumors.
Despite the shortcomings, the study presented herein is novel. To the best of our knowledge, this is the first report of AR-V7 detection using a whole-blood assay, which also detected ARv567es in a significant proportion of patients. Eliminating the CTC selection process reduces the hands-on time and improves the sensitivity of the assay, allowing more patients to benefit from such assays. Indeed, the percentage of AR-V7+ patients in this study was higher than both CTC-based assays (68.49% here vs 50%/49%)7,18. The small volume of blood needed and the stability of RNA in PAXgene tubes make it practical to incorporate this assay into routine patient monitoring. In view of the potential benefits, it is necessary to further evaluate and refine this assay by a prospective study with an adequate cohort.
Supplementary Material
Table S1. Abiraterone and enzalutamide treatment summaries in the CRPC cohort (n=46).
Table S2. Treatment history at the time of sample collection CRPC patients (including serial samples).
Table S3. Ct values for AR transcripts detected by two approaches.
Table S4. Relative expression of AR-V transcripts by CD45-depletion approach as compared to the PAXgene approach.
Supplementary Figure S1. Diagram of the experimental design to compare the PAXgene approach with the CD45-depleteion approach.
Acknowledgments
Grant support: ACS RSG-07-218-01-TBE, DOD W81XWH-12-1-0275 and W81XWH-14-1-0480, Louisiana BoR LEQSF(2012-15)-RD-A-25, NIH/NIGMS 5P20GM103518-10, Oliver Sartor Prostate Cancer Research Fund.
Glossary
- AR-FL
full-length androgen receptor
- CRPC
castration-resistant prostate cancer
- CTC
circulating tumor cells
- mCRPC
metastatic castration-resistant prostate cancer
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dehm SM, Schmidt LJ, Heemers HV, et al. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Z, Yang X, Sun F, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun S, Sprenger CC, Vessella RL, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu R, Lu C, Mostaghel EA, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Chan SC, Brand LJ, et al. Androgen Receptor Splice Variants Mediate Enzalutamide Resistance in Castration-Resistant Prostate Cancer Cell Lines. Cancer Res. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonarakis ES, Lu C, Wang H, et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harouaka R, Kang Z, Zheng S-Y, et al. Circulating tumor cells: Advances in isolation and analysis, and challenges for clinical applications. Pharmacol Ther. 2014;141:209–221. doi: 10.1016/j.pharmthera.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas ML. The Leukocyte Common Antigen Family. Annu Rev Immunol. 1989;7:339–369. doi: 10.1146/annurev.iy.07.040189.002011. [DOI] [PubMed] [Google Scholar]
- 10.Lara O, Tong X, Zborowski M, et al. Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Exp Hematol. 2004;32:891–904. doi: 10.1016/j.exphem.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Lang JC, Balasubramanian P, et al. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol Bioeng. 2009;102:521–534. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gandhok NK, Looney S, Koochekpour S, et al. Relationships between reverse transcriptase-polymerase chain reaction for prostate specific antigen, survival, and various prognostic laboratory factors in patients with hormone refractory prostate cancer. Urol Oncol Semin Orig Investig. 2005;23:163–167. doi: 10.1016/j.urolonc.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Helo P, Cronin AM, Danila DC, et al. Circulating Prostate Tumor Cells Detected by Reverse Transcription-PCR in Men with Localized or Castration-Refractory Prostate Cancer: Concordance with CellSearch Assay and Association with Bone Metastases and with Survival. Clin Chem. 2009;55:765–773. doi: 10.1373/clinchem.2008.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danila DC, Anand A, Schultz N, et al. Analytic and Clinical Validation of a Prostate Cancer–Enhanced Messenger RNA Detection Assay in Whole Blood as a Prognostic Biomarker for Survival. Eur Urol. 2014;65:1191–1197. doi: 10.1016/j.eururo.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Z, Shackleton CHL, Pahwa S, et al. Prominent sex steroid metabolism in human lymphocytes. Mol Cell Endocrinol. 1998;138:61–69. doi: 10.1016/s0303-7207(98)00052-5. [DOI] [PubMed] [Google Scholar]
- 17.Smithson G, Couse JF, Lubahn DB, et al. The Role of Estrogen Receptors and Androgen Receptors in Sex Steroid Regulation of B Lymphopoiesis. J Immunol. 1998;161:27–34. [PubMed] [Google Scholar]
- 18.Steinestel J, Luedeke M, Arndt A, et al. Detecting predictive androgen receptor modifications in circulating prostate cancer cells. Oncotarget. 2015:5. doi: 10.18632/oncotarget.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todenhöfer T, Hennenlotter J, Feyerabend S, et al. Preliminary experience on the use of the Adnatest® system for detection of circulating tumor cells in prostate cancer patients. Anticancer Res. 2012;32:3507–3513. [PubMed] [Google Scholar]
- 20.Liu G, Sprenger C, Sun S, et al. AR Variant ARv567es Induces Carcinogenesis in a Novel Transgenic Mouse Model of Prostate Cancer. Neoplasia N Y N. 2013;15:1009–1017. doi: 10.1593/neo.13784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cottard F, Asmane I, Erdmann E, et al. Constitutively Active Androgen Receptor Variants Upregulate Expression of Mesenchymal Markers in Prostate Cancer Cells. PLoS ONE. 2013;8:e63466. doi: 10.1371/journal.pone.0063466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun F, Chen H, Li W, et al. Androgen Receptor Splice Variant AR3 Promotes Prostate Cancer via Modulating Expression of Autocrine/Paracrine Factors. J Biol Chem. 2014;289:1529–1539. doi: 10.1074/jbc.M113.492140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong D, Sethi S, Li Y, et al. Androgen receptor splice variants contribute to prostate cancer aggressiveness through induction of EMT and expression of stem cell marker genes. The Prostate. 2015;75:161–174. doi: 10.1002/pros.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galletti G, Portella L, Tagawa ST, et al. Circulating Tumor Cells in Prostate Cancer Diagnosis and Monitoring: An Appraisal of Clinical Potential. Mol Diagn Ther. 2014;18:389–402. doi: 10.1007/s40291-014-0101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngo N, Patel K, Isaacson PG, et al. Leucocyte common antigen (CD45) and CD5 positivity in an “undifferentiated” carcinoma: a potential diagnostic pitfall. J Clin Pathol. 2007;60:936–938. doi: 10.1136/jcp.2006.044750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danbara M, Yoshida M, Kanoh Y, et al. Flow cytometric detection of small cell lung cancer cells with aberrant CD45 expression in micrometastatic bone marrow. Jpn J Clin Oncol. 2009;39:771–775. doi: 10.1093/jjco/hyp088. [DOI] [PubMed] [Google Scholar]
- 27.Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells—biology and biomarkers. Nat Rev Clin Oncol. 2014;11:129–144. doi: 10.1038/nrclinonc.2013.253. [DOI] [PubMed] [Google Scholar]
- 28.Xu D, Zhan Y, Qi Y, et al. Androgen Receptor Splice Variants Dimerize to Transactivate Target Genes. Cancer Res. 2015;75:3663–3671. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao B, Qi Y, Zhang G, et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646–1656. doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Abiraterone and enzalutamide treatment summaries in the CRPC cohort (n=46).
Table S2. Treatment history at the time of sample collection CRPC patients (including serial samples).
Table S3. Ct values for AR transcripts detected by two approaches.
Table S4. Relative expression of AR-V transcripts by CD45-depletion approach as compared to the PAXgene approach.
Supplementary Figure S1. Diagram of the experimental design to compare the PAXgene approach with the CD45-depleteion approach.