Abstract
Preimplantation genetic diagnosis (PGD) is widely applied in reciprocal translocation carriers to increase the chance for a successful live birth. However, reciprocal translocation carrier embryos were seldom discriminated from the normal ones mainly due to the technique restriction. Here we established a clinical applicable approach to identify precise breakpoint of reciprocal translocation and to further distinguish normal embryos in PGD. In the preclinical phase, rearrangement breakpoints and adjacent single nucleotide polymorphisms (SNPs) were characterized by next-generation sequencing following microdissecting junction region (MicroSeq) from 8 reciprocal translocation carriers. Junction-spanning PCR and sequencing further discovered precise breakpoints. The precise breakpoints were identified in 7/8 patients and we revealed that translocations in 6 patients caused 9 gene disruptions. In the clinical phase of embryo analysis, informative SNPs were chosen for linkage analyses combined with PCR analysis of the breakpoints to identify the carrier embryos. From 15 blastocysts diagnosed to be chromosomal balanced, 13 blastocysts were identified to be carriers and 2 to be normal. Late prenatal diagnoses for five carriers and one normal fetus confirmed the carrier diagnosis results. Our results suggest that MicroSeq can accurately evaluate the genetic risk of translocation carriers and carrier screen is possible in later PGD treatment.
Keywords: Reciprocal translocation, Breakpoint, Single nucleotide polymorphism, Preimplantation genetic diagnosis
Highlights
-
•
MicroSeq can rapidly characterize translocation breakpoint sequences and nearby haplotype in translocation carriers.
-
•
Translocation leads to frequent gene disruption in carriers with normal phenotype.
-
•
MicroSeq provides sufficient information for successfully carriers screen in PGD treatment.
Reciprocal translocation carriers with fertility problems could benefit from preimplantation genetic diagnosis (PGD) to select chromosomally-balanced embryos for transfer. Currently, carrier embryos and normal embryos are not routinely discriminated, mainly due to technique restriction. Theoretically, half of these offspring may inherit the infertility phenotype. Here we established a clinically applicable approach using microdissection and next generation sequencing (MicroSeq) to identify precise breakpoints and establish nearby haplotypes of derivative chromosomes. This information was used to successfully distinguish normal embryos from reciprocal translocation carrier embryos in later PGD. Our data also revealed that translocation frequently leads to gene disruption in carriers with normal phenotype.
1. Introduction
Reciprocal translocations are one of the most common abnormalities in chromosomal structure, with an incidence of 1/500 to 1/625 human newborns (Ogilvie and Scriven, 2002). The origin of a reciprocal translocation is associated with inherited or de novo nonhomologous chromosome rearrangements, and exposure to chemicals and radiation (Tucker, 2008). It has been reported that > 6% of reciprocal carriers have a variety of symptoms, such as autism, intellectual disabilities, or congenital abnormalities (Gersen et al., 2013). Most of them resulted from microdeletions, duplications, or gene disruption in carriers (Feenstra et al., 2011, Schneider et al., 2015). In addition, because of quadrivalent formations during meiosis, reciprocal translocation carriers with normal phenotype are likely to produce gametes with unbalanced products, which usually result in recurrent miscarriage and sometimes infertility. In these carriers, the genetic risk of reciprocal translocations should be carefully investigated.
A preimplantation genetic diagnosis (PGD) offers an effective treatment option for reciprocal translocation carriers to minimize the risk and distress of pregnancy loss caused by abnormal chromosomal segregation (Braude et al., 2002). Since the 1990s, fluorescence in situ hybridization (FISH) (Scriven et al., 1998), array-based comparative genomic hybridization (Alfarawati et al., 2011), and next-generation sequencing (NGS) (Tan et al., 2014) have been widely used in PGDs to detect abnormal copy numbers of chromosomal segments. In this scenario, both carrier and normal embryos had the same chance to be selected and result in live births and theoretically half of the offsprings will likely to inherit fertility problems. Furthermore, although reciprocal translocation do not increase the risk of physical or mental disability for most inherited balanced translocations (Gardner et al., 2012), the breakpoint in de novo translocation carriers may interrupt functional genes which might generate harmful effects in later life.
To date, some approaches have been developed to distinguish normal and carrier embryos via PGD. Patient-specific breakpoint-spanning or closely flanking FISH probes could be designed to detect both numerical and structural aberrations in either interphase cells or in polar bodies for PGD (Munne et al., 1998, Weier et al., 1999). Moreover, nuclear transferring of human blastomere into mature bovine or mouse oocytes could help to visualize metaphase chromosomes for full karyotyping in PGD (Verlinsky et al., 2002, Willadsen et al., 1999). However, patient-specific FISH probes design and optimization is time-consuming; and blastomere nuclei conversion needs both sophisticated embryologists for nuclei transfer and special cytogeneticists for single cell metaphase harvest and karyotyping in heterokaryons. More recently, it was reported that SNP array based comprehensive chromosome screening (CCS) was successfully used to distinguish normal from balanced translocation carrier embryos for most carrier couples (Treff et al., 2016). However, parental DNA and at least one unbalanced IVF embryo were necessary for the diagnosis, and the genetic risk of carrier cannot be evaluated since precise breakpoints were not identified. Currently there are no effective methods for all carrier couples to precisely identify breakpoints and/or linkage polymorphism markers for both carrier embryo diagnosis and genetic risk evaluation.
Breakpoint identification has been one of the most interesting fields in cytogenetics for investigating the phenotypic outcomes of reciprocal translocations. Several techniques had been developed to map chromosome breakpoints to the kilobase level (Chen et al., 2008, Gribble et al., 2009, Higgins et al., 2008, Sobreira et al., 2011, Talkowski et al., 2011, Talkowski et al., 2012, Vergult et al., 2014). However, these techniques are time-consuming, expensive, and do not provide information about the breakpoint-linked SNPs for use in later PGDs. Recently, chromosome microdissection followed by NGS has been reported for use in precisely identifying the reciprocal translocation breakpoints at the level of the individual base in a leukemia patient (Jancuskova et al., 2013). Here, we describe the “MicroSeq-PGD” method, which combines previously reported chromosome microdissection technique (Hu et al., 2007) and NGS followed by PGD, to characterize the DNA sequence of the translocation breakpoint and to distinguish between normal and carrier embryos in 8 reciprocal translocation carriers (Fig. 1).
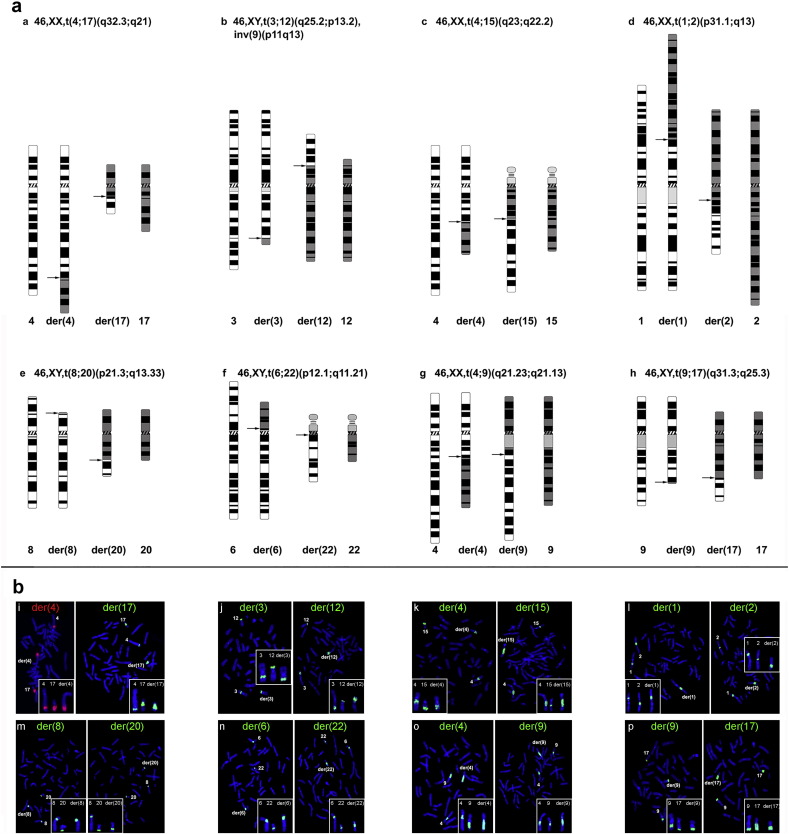
Fig. 1.
Discrimination of normal and carrier embryos via MicroSeq and PGD.
Junction fragments of derivative chromosomes were microdissected and amplified via DOP-PCR. The amplified DNA samples were sequenced using next generation sequencing. Precise breakpoints were determined via long-range PCR. Informative breakpoint-linked SNPs were identified in a bioinformatics analysis and confirmed by sequencing the selected SNPs in the couples. In the normal/carrier embryos diagnosed via PGD-CCS, only the embryos that were identified as positive in the junction-spanning PCR analysis and/or those positive for informative SNPs were predicted to be carrier embryos. The embryos that were negative in the breakpoint and junction-spanning PCR analyses and/or for informative SNPs were predicted to be normal embryos.
2. Materials and Methods
2.1. Study Patients
This clinical diagnostic study was reviewed and approved by the Institutional Review Board (IRB) of the Reproductive and Genetic Hospital of CITIC-Xiangya (LL-SC-SG-2014-013). Metaphase spreads, DNA samples and whole genome amplification products of the patients were used for breakpoint analyses and further PGD. The study was conducted in compliance with the provisions of the Declaration of Helsinki. All patients provided written informed consent before diagnosis. This study included a total of 8 couples at the CITIC-Xiangya Hospital from July 1, 2014 to December 31, 2015. Either member of the couple had reciprocal translocations (excluding Robertsonian translocations, where the breakpoint is adjacent to the centromere). The karyotypes of the couples were determined from G-banded metaphase spreads obtained from peripheral blood using standard techniques.
2.2. Chromosome Microdissection
Chromosome microdissection and PCR amplification of the microdissected DNA were performed as described previously (Hu et al., 2004). Five to eight copies of the region covering the breakpoints were dissected from G-banding metaphase spreads of the patients' peripheral blood samples with glass needles. The dissected DNA fragments were amplified by DOP-PCR with UN1 primer (CCGACTCGAGNNNNNNATGTGG). An initial 6 cycles of PCR (denaturation at 94 °C for 1 min, annealing at 30 °C for 2 min, and extension at 37 °C for 2 min) were performed by adding 0.3 units of T7 DNA polymerase (Sequenase Version 2.0, USB, Cleveland, OH) at each cycle. A conventional PCR using Taq DNA polymerase was then performed for 35 cycles (denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min, and extension at 72 °C for 2 min). Amplified DNA was labeled with spectrum-green or spectrum-red dUTP (Vysis, Downers Grove, IL) via PCR and then hybridized to metaphases from patients to confirm the success of chromosome microdissection.
2.3. Next Generation Sequencing of DNA from Microdissected Chromosomes
Breakpoint mapping was based on parallel sequencing with a paired-end protocol and a bioinformatic analysis using the Integrative Genomics Viewer (Robinson et al., 2011). Briefly, 100 ng of amplified microdissected DNA were fragmented by enzyme digestion and purified to yield fragments of 100–500 bp. P1 adaptor oligonucleotides from Life Tech were ligated on repaired A tailed fragments. About 150–300 bp fragments were separated, purified and enriched by electrophoresis and PCR cycles. Genomic libraries were prepared using the Ion Xpress library kit (Life Technologies Inc., Rockville, MD). Each DNA library was then sequenced on a Life Tech. Ion Proton system with 318 chips as paired-end 200-bp reads. Image analysis and base calling was performed using a Life Tech. 460 Flow system.
Sequence data were cleaned by removing the primer sequences and then were aligned to the reference genome (hg19) using the Integrative Genomics Viewer. Briefly, those sequences that could not be aligned to hg19 or aligned to multiple sites of hg19 were removed. SNPs were compared with dbSNP and 1000 Genomes Project database (http://www.1000genomes.org). The SNPs with a mutation frequency of < 40% were picked as candidate breakpoint-specific SNPs. The cleaned sequence data were then aligned to hg19 with integrative genomics viewer.
2.4. Precise Characterization of Breakpoints
We then synthesized specific primers that were supposed to be near to the breakpoints to amplify the junction fragments with genomic DNA from the translocation carriers. The PCR products were then sequenced to precisely identify the accurate breakpoints by Sanger sequencing.
We also synthesized specific primers to amplify and then sequenced the selected SNPs of the couples. Only the SNPs that were heterozygous in the translocation carriers and homozygous in their normal partner were considered as informative SNPs.
2.5. Comprehensive Chromosomal Screening
Preimplantation genetic diagnosis by comprehensive chromosomal screening was performed as previously described (Tan et al., 2014). Briefly, pituitary desensitization was performed using a long luteal Gonadotropin-releasing hormone (GnRH) agonist protocol based on patient situations (Erb and Wakim, 2008). After oocyte retrieval, all eggs were fertilized by intracytoplasmic sperm injection (ICSI). All embryos were cultured in sequential media (G1 and G2, Vitrolife, Goteborg, Sweden) to the blastocyst stage. Approximately 3–8 trophectoderm (TE) cells were aspirated using a biopsy pipette with a 30-μm internal diameter and dissected with a Zilos TK laser (Hamilton Thorne, MA, USA). Biopsied TE cells were then used for whole genome amplification (WGA) via multiple displacement amplification with a REPLI-g Single Cell Kit (Qiagen, Valencia, CA). NGS and comprehensive chromosomal screening was then performed as previously described (Tan et al., 2014).
2.6. Carrier Embryo Diagnosis
The excess WGA products were amplified with junction-spanning-specific primers via PCR, with genomic DNA of translocation carriers serving as the positive control and genomic DNA from healthy donors serving as the negative control. The informative SNPs flanking the breakpoint were amplified via PCR and then sequenced to reduce the risk of recombination. In the normal/carrier embryos diagnosed via PGD-CCS, only the embryos that were identified as positive in the junction-spanning PCR analysis and/or those positive for informative SNPs were predicted to be carrier embryos. The embryos that were negative in the junction-spanning PCR analyses and/or for informative SNPs were predicted to be normal embryos (Fig. 1).
2.7. Blastocyst Vitrification, Warming and Transfer
Blastocysts were vitrified after the biopsy using Kitazato vitrification solution (Kitazato Biopharma Co. Ltd. Shizuoka. Japan) and closed High Security Vitrification straws (Cryo Bio System, France). Each blastocyst was stored in an individual straw. After warming and dilution, blastocysts were cultured in blastocyst medium for 1–2 h. Only chromosomally normal/balanced blastocysts were selected for warming, and the surviving re-expanded blastocysts with high morphological grades were selected for transfer. Luteal support was applied in cryopreserved embryo transfer (CET) cycles. Warmed blastocysts were transferred either 5 days after ovulation during a natural menstrual cycle or 5 days after the initiation of ovulation via progesterone administration. Briefly, 6 mg Estradiol Valerate was started from Day 3 for 10–15 days, then luteal support was applied when a satisfactory endometrial development (thickness ≥ 8 mm) was confirmed with ultrasound. No more than two blastocysts were transferred, and single blastocyst transfer to each patient with well-cryopreserved embryos was recommended.
2.8. Prenatal Diagnosis
Clinical pregnancy was confirmed when an intrauterine gestational sac with a heartbeat was observed via ultrasound examination 30–40 days after embryo transfer. Amniocentesis was performed at 16–18 weeks' gestation age. The amniocentesis fluid samples from fetuses were used for karyotyping to confirm the PGD results.
3. Results
3.1. MicroSeq Analysis
Totally 16 breakpoints for 8 translocation carriers were characterized. Reverse FISH using amplified DNA fragment as probes showed that all microdissected products covered rearrangement breakpoints (Fig. 2). The 16 amplified DNA samples were then sequenced at 1.76–3.59 million reads. We found that 17.3–56.4% of the reads were uniquely aligned to the reference human genome (hg19). The coverage of aligned reads to the dissected regions ranged from 3.6–35.7%. After mapping the same chromosomal sequences from two derivative chromosomes together, the breakpoints of each chromosomal translocation were narrowed down to 24 bp-8.96 kbp (Table 1).
Fig. 2.
Reverse FISH of microdissected DNA.
Reciprocal translocations of the eight patients are illustrated in the upper panel. Reverse FISH using amplified DNA fragment as probes shows that all microdissected products cover rearrangement breakpoints (lower panel).
Table 1.
Summary of breakpoint characterization results of the eight patients
| Patient | Carrier karyotypea | der | chr | No. of mapped sequencing reads | Coverage (%) |
Distance of the nearest NGS mapping reads to breakpoint (bp) | Total_Mutation SNP numbers | Identifying linked to breakpoint informative SNPs number | Breakpoint position from Sanger sequencing | Disrupt gene (break region) | OMIM ID | Gene Ontology |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 46,XX,t(4;17)(q32.3;q21.2) | der(4) | 4 | 50,516 | 3.59 | 2,347 | 58 | 2 | chr4:169,227,027-169,227,031 | DDX60 (Intron 5/37) | 613974 | Nucleic acid binding and hydrolase activity |
| 17 | 167,018 | 5.41 | 3,506 | 74 | 0 | |||||||
| der(17) | 4 | 34,792 | 6.35 | 1,514 | 43 | 6 | chr17:40,053,060-4,005,3061 | ACLY (Intron 13/28) | 108728 | Cofactor binding and ATP citrate synthase activity | ||
| 17 | 81,784 | 10.45 | 2,258 | 31 | 4 | |||||||
| 2 | 46,XY,t(3;12)(q25.2;p13.2),inv(9)(p11q13) | der(3) | 3 | 169,432 | 9.89 | 818 | 428 | 3 | chr3:153,121,850-153,121,851 | – | – | – |
| 12 | 175,243 | 19.14 | 614 | 328 | 7 | |||||||
| der(12) | 3 | 349,210 | 12.12 | 1,144 | 130 | 0 | chr12:10,311,870-10,311,877 | OLR1 (Exon 4/4) | 602601 | Carbohydrate binding and low-density lipoprotein receptor activity | ||
| 12 | 206,640 | 20.02 | 1,177 | 100 | 0 | |||||||
| 3 | 46,XX,t(4;15)(q23;q22.2) | der(4) | 4 | 156,036 | 9.66 | 898 | 149 | 2 | chr4:100,891,783-100,891,784 | – | – | – |
| 15 | 215,786 | 9.23 | 206 | 176 | 4 | |||||||
| der(15) | 4 | 440,532 | 9.27 | 561 | 517 | 3 | chr15:60,310,881-60,310,896 | FOXB1 (Intron 1/2) | – | Transcription factor activity, sequence-specific DNA binding and RNA polymerase II transcription factor activity, sequence-specific DNA binding | ||
| 15 | 241,418 | 10.21 | 1,045 | 186 | 5 | |||||||
| 4 | 46,XX,t(1;2)(p31.1;q13) | der(1) | 1 | 735,087 | 23.73 | 223 | 87 | 1 | chr1:70,936,280-70,936,290 | – | – | – |
| 2 | 203,356 | 16.74 | 2,045 | 162 | 3 | |||||||
| der(2) | 1 | 183,725 | 14.14 | 167 | 1,214 | 8 | chr2:112,661,491-112,661,518 | MERTK (Intron 1/18) | 604705 | Transferase activity, transferring phosphorus-containing groups and protein tyrosine kinase activity | ||
| 2 | 158,214 | 18.75 | 1,362 | 358 | 5 | |||||||
| 5 | 46,XY,t(8;20)(p21.3;q13.33) | der(8) | 8 | 215,633 | 30.18 | 250 | 161 | 7 | chr8:22,991,894-22,991,895 | – | – | – |
| 20 | 11,242 | 26.68 | 0 | 8 | 0 | |||||||
| der(20) | 8 | 189,386 | 15.74 | 2,210 | 223 | 4 | chr20:61,754,133-61,754,134 | – | – | – | ||
| 20 | 310,178 | 18.2 | 6,750 | 518 | 7 | |||||||
| 6 | 46,XY,t(6;22)(p12.1;q11.21) | der(6) | 6 | 36,694 | 24.12 | NA | 57 | 0 | – | – | – | – |
| 22 | 89,584 | 10.4 | NA | 50 | 2 | |||||||
| der(22) | 6 | 675,112 | 29.86 | NA | 1,194 | 3 | – | – | – | – | ||
| 22 | 101,853 | 22.44 | NA | 183 | 2 | |||||||
| 7 | 46,XX,t(4;9)(q21.23;q21.13) | der(4) | 4 | 572,409 | 35.73 | 0 | 856 | 9 | chr4:84,877,905-84,877,912 | BC005018 (Intron 7/7) | – | NA |
| 9 | 550,854 | 31.51 | 24 | 1,236 | 8 | |||||||
| der(9) | 4 | 555,651 | 16.84 | 302 | 1,072 | 8 | chr9:75,640,607-75,640,608 | ALDH1A1 (Intron 1/12) | 100640 | Oxidoreductase activity and acyl-CoA dehydrogenase activity | ||
| 9 | 76,442 | 25.86 | 121 | 160 | 4 | |||||||
| 8 | 46,XY,t(9;17)(q31.3;q25.3) | der(9) | 9 | 152,030 | 6.1 | 880 | 102 | 2 | chr9:114,402,958-114,402,959 | DNAJC25 (Intron 1/4) | – | Signal transducer activity |
| 17 | 27,428 | 5.74 | 925 | 15 | 0 | |||||||
| der(17) | 9 | 590,926 | 32.54 | 1,490 | 1,235 | 7 | chr17:76,740,337-76,740,338 | CYTH1 (Intron 1/12) | 182115 | Lipid binding and ARF guanyl-nucleotide exchange factor activity | ||
| 17 | 898,244 | 31.53 | 160 | 1,453 | 8 |
Carrier karyotypes in this table have been revised following the breakpoint mapping information and the original G-band karyotypes are provided in Table S2.
3.2. Breakpoint Characterization
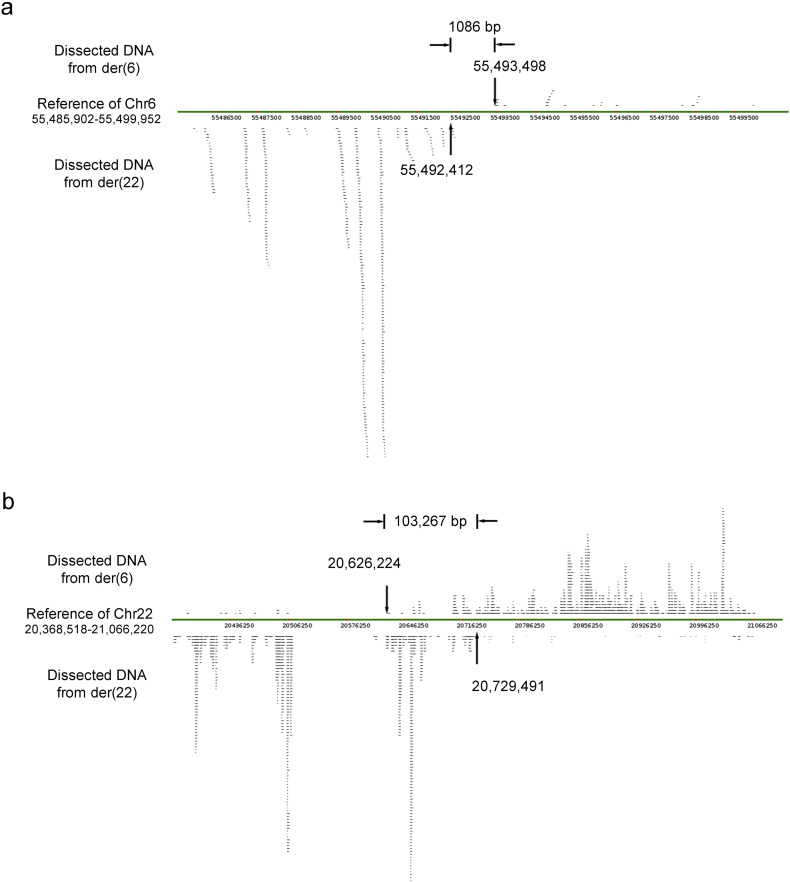
To further characterize the precise sequence of the rearrangement breakpoints, multiple PCR reactions spanning the narrowed-down regions of the derivative chromosomes were performed, and the amplicons were then sequenced to precisely identify the breakpoint sequences at the level of individual bases. We successfully identified 14 breakpoints in 7 patients. Interestingly, translocations in 6/7 patients caused the disruption of 9 genes, including the disease-related genes OLR1 and MERTK (Table 1). Fortunately, further sequencing revealed that no mutation was detected in these 9 genes of their healthy spouses. Most breakpoints were in introns and had microhomologous sequences, deletions or insertions after the rearrangement (Fig. S1). For case 6, with the t(6;22) translocation, the sequence from der(6) and der(22) that mapped to the reference genome of chromosome 6p12.1 had a 1086 bp gap (55,492,412–55,493,498) after alignment. However, the aligned sequence in 22q11.21 was located just inside a complex palindromic region, which resulted in an overlap of 103,267 bp (20,626,224–20,729,491) and a failure of junction-spanning PCR (Fig. S2). Thus, the precise breakpoint in this patient could not be identified.
Fig. S1.
Junction fragment sequences of the seven cases whose breakpoints were identified.
Sequences from two chromosomes are labeled in blue or purple. Arrows show the direction. Deleted sequences are shown in green. Inserted sequences are marked with red and in lower case. Micro-homologies are indicated in bold capital characters. (Chr) chromosome, (der) derivative chromosome.
Fig. S2.
Alignment results of NGS sequences from der(6) and der(22) in case 6.
The sequence from der(6) to der(22) that mapped to the reference genome of chromosome 6 had a 1086 bp gap (55, 492, 412–55, 493, 498) after alignment (a). However, the aligned sequence in 22q11.21 had an overlap of 103.267 kb (20, 626, 224–20, 729, 491) (b).
3.3. Breakpoint Adjacent SNPs Characterization
From the sequence information obtained for the flanking rearrangement breakpoints, we identified 12,564 mutant SNPs in 8 patients, with 8–1453 SNPs per junction fragment (Table 1). We selected 244 SNPs within 10 Mbp from the breakpoint and with a frequency of < 40% in the 1000 Genomes Project database as candidate SNPs for the linkage analysis. The genotypes of selected SNPs in the balanced translocation carriers and their spouses were further analyzed via sequencing. Among them, we found that 124 SNPs were heterogeneous in the translocation carriers and homogenous in their spouses, which means that these were informative SNPs. With these SNPs, we successfully established partial haplotypes near the breakpoints of the derivative chromosomes in all 8 couples, with 2–17 SNPs per derivative chromosome (Table S1).
3.4. Preimplantation Genetic Diagnosis
The 8 couples recruited in this study received 10 PGD treatment cycles. A total of 45 blastocysts were biopsied for WGA. Biopsy TE cells from 44 blastocysts were successfully amplified. After comprehensive chromosome screening via NGS, 26 out of the 44 blastocysts had translocation-related abnormalities, 3 blastocysts showed de novo aneuploidy unrelated to translocation, and the other 15 blastocysts were balanced, indicating that they were either normal or had balanced translocation (Table S2).
We then performed a PCR analysis that spanned the breakpoints, and we amplified the breakpoint-adjacent informative SNPs for sequencing analysis with the rest of the WGA products to distinguish normal blastocysts from blastocysts with balanced translocations. In total, 124 SNPs within 10 Mbp of the rearrangement breakpoints were successfully analyzed. We then performed linkage analysis with the informative SNPs in 44 embryos of 8 patients. The total effective detection frequency is 593. Interestingly, recombinations were only observed at distances > 5 Mbp from the breakpoint, from 0% (0/305) at < 5 Mbp region to 4.45% (13/288) at > 5 Mbp region (Table S3). Thus, informative SNPs within 5 Mbp could be probably used to diagnose carrier embryos. The linkage analysis showed that 13 blastocysts were balanced translocation carriers, and only 2 blastocysts were normal (Table S1, Table S2). A junction-spanning PCR analysis was performed on 12 blastocysts from 7 patients whose rearrangement breakpoints had been successfully identified. Breakpoints were detected in 10 blastocysts, indicating that they were carrier embryos. The other two embryos were negative, indicating normal embryos. The junction-spanning PCR results showed 100% agreement with the previous linkage analysis results.
3.5. Clinical Outcome
Before completing the analysis, 7 patients had already had their frozen embryos transferred. Nine blastocysts were thawed, and 8 blastocysts survived. Seven transfer cycles were performed, and 4 women became pregnant. In one woman, the pregnancy ended at 45 days after the embryo transfer (ET) in her first cycle, but she successfully became pregnant at the second ET. After prenatal diagnosis via amniocentesis, all of the babies were diagnosed as balanced translocation carriers. To date, five healthy babies with balanced translocation have been successfully delivered. These results agree with previous predictions (Table S2). For case 8, the carrier analysis results came out before her frozen embryo was transferred. She had 4 balanced blastocysts: three blastocysts were predicted to be carrier embryos and only one to be normal. After giving informed consent, the patient decided to have the normal blastocyst transferred, and a successful pregnancy was established. A prenatal diagnosis confirmed that the fetus had a normal karyotype (Fig. 3). Furthermore, for all 29 chromosomal unbalanced embryos, existence of derivative chromosomes predicted by junction-spanning PCR and/or breakpoint-adjacent informative SNPs analysis was consistent with that was predicted by PGD-CCS (Table S2). The above results also proved the accuracy of our PGD approach based on MicroSeq technique.
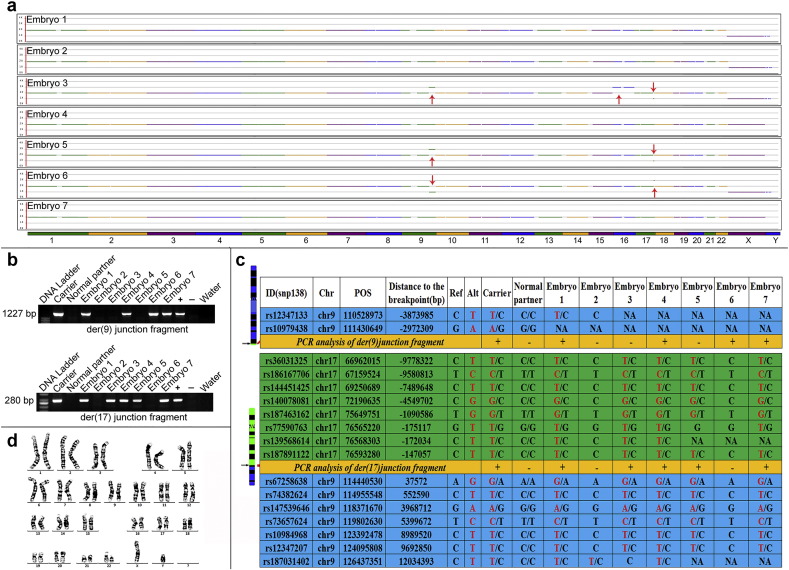
Fig. 3.
PGD and carrier diagnosis of a t(9;17) patient.
In total, 7 embryos were obtained and analyzed. Based on complete chromosome screening, embryos 1, 2, 4, and 7 were identified as normal/carrier embryos, whereas embryos 3, 5, and 6 were identified as unbalanced embryos (a). Junction-spanning PCR analysis of whole-genome amplification products from embryos showed that embryos 1, 4, and 7 were positive and that embryo 2 was negative (b), indicating that only embryo 2 was a normal embryo, while the other 3 embryos were carriers. SNP analysis confirmed that only embryo 2 was negative for informative SNPs (red) characters in panel (c). The normal blastocyst was transferred after the subject gave informed consent, and a successful pregnancy was established. Prenatal diagnosis confirmed that the fetus had a normal karyotype (d).
4. Discussion
PGD is currently used to exclude chromosomally unbalanced embryos and to ensure a successful live birth for balanced translocation carriers with an unfortunate obstetric history or infertility. Although a variety of techniques have been developed to identify carrier embryos in PGD, the efficiency and consistence performance of these techniques between individuals hampered their clinical applications. In this scenario, embryos with normal karyotypes from translocation carriers cannot be preferentially transferred and one half of the offsprings would theoretically inherit the translocation-bearing chromosomes and may face reproductive difficulties in the future. Moreover, the genetic risk caused by de novo reciprocal translocation was not investigated routinely in phenotypically normal carriers. In the current study, using MicroSeq approach, we were generally able to precisely map rearrangement breakpoints to the level of individual bases. Compare with other available techniques, this approach combines chromosome microdissection and NGS techniques. Precise dissection of chromatin covering breakpoint minimizes the following NGS cost. NGS of amplified dissected DNA can reliably and accurately obtain sequence of junction fragments, which help to precisely identify the breakpoint as well as nearby SNPs. It takes only 5 days for chromosome microdissection, and regular library construction protocol is suitable for the following NGS.
In most cases, we found that multiple genes were disrupted and that some potential pathogenic genes were affected by balanced translocations. Although most interrupted genes in this study are recessive, balanced translocation offsprings may suffer from genetic defects if another allele had pathogenic mutations or deletions. In the 9 disrupted genes from the 8 patients, homozygous loss of function of Acly, DDX60, Fkh5 and Aldh1a1 were associated with lethal in early embryo development (Beigneux et al., 2004),defects in antiviral immune response (Oshiumi et al., 2015), severe growth retardation (Wehr et al., 1997) and sensitivity for retinol toxicity (Molotkov and Duester, 2003) respectively. Moreover, MERTK, a recessive gene that had been associated with retinal dystrophy (McHenry et al., 2004), was disrupted in another patient. Fortunately, in the follow-up study there was no genetic mutation detected in those disrupted genes in the healthy spouse genome. Taking the above observations into consideration, although no discernible increased risk of clinical abnormality was observed in the children with the same balanced translocation karyotype as in their asymptomatic carrier parents (Gardner et al., 2012), our data suggest that potential genetic risks may exist in phenotypically normal de novo translocation carriers. Considering the risks of further reproductive difficulties and the potential genetic risks, it is worthy to characterize and evaluate the breakpoint-affected genes ahead. Nevertheless, the significance of gene interruption should be evaluated and consulted by professional genetic consultants, especially regarding genes of unknown significance and cancer predisposition genes.
In this study, we observed a bias of carrier to non-carrier embryos with a ratio of 13:2. However, limited sample size may account for this bias. Treff and colleagues evaluated 126 balanced embryos in PGD by SNP array based CCS. Among them, 62 (49%) were predicted to be normal embryos and 64 (51%) were predicted as balanced translocation carrier embryos (Treff et al., 2016). Thus, more data and appropriate statistical methods are needed to measure the carrier to non-carrier embryo ratio. Moreover, it is important to inform that a non-carrier embryo is no guarantee in any one PGD cycle, and a carrier embryo could also be transferred if no further genetic risk is detected.
In this study, the number of informative SNPs was highly viable from breakpoint to breakpoint. The amplification efficiency of DOP-PCR may account for this variation. However, there are enough informative SNPs for further PGD for embryos with balanced translocations. Moreover, we only included the SNPs with > 10 × coverage in this study. Since more informative SNPs will minimize the impact of allelic dropout to ensure the PGD accuracy, SNPs with < 10 × coverage could also be tested and included to increase the number of SNPs for further PGD. For Robertsonian translocations, although it is difficult to find exact breakpoints that located in cetromeric region, nearby informative SNPs linkage analysis will help to discriminate normal embryos and embryos with balanced Robertsonian translocations. However, the position of informative SNPs should be carefully investigated to avoid recombination-caused misdiagnosis.
Although the breakpoints in 22q11.21 in one patient were not identified due to interference from complex palindromic sequences, we successfully characterized the SNPs adjacent to the rearrangement breakpoints in all patients. The guidelines of the European Society of Human Reproduction and Embryology suggest that extragenic markers within 1 Mbp of a mutation should be included in PGDs to reduce mistakes caused by recombination, as the probability for recombination within 1 Mbp is low (Harton et al., 2011). However, according to our linkage analysis, data from 44 embryos from reciprocal carriers show that recombination events were not observed within 5 Mbp of the rearrangement breakpoint. It is possible that the increased distance of the homologous chromatids in the center of the quadrivalent reduced the chances of recombination in the flanking sequences of the rearrangement breakpoints. This finding enabled us to obtain more informative SNPs over a relatively wider area for use in PGDs. This also makes a less-detailed MicroSeq sequencing step practical.
In this study, we succeeded in developing a reliable approach for performing linkage and junction-spanning PCR analyses to distinguish normal and carrier embryos in a PGD based on the information provided by a MicroSeq analysis. This approach has been shown to be effective based on the prenatal validation of the initial diagnoses. However, the sensitivity and specificity of this approach should be evaluated in a larger sample size. In addition, this approach may also provide a future means of diagnosing embryos that are carriers of Robertsonian translocations and inversions during PGDs.
The following are the supplementary data related to this article.
Summary of breakpoint ajacent informative SNPs and PGD results of the eight patients and their embryos.
Summary of the PGD results for all embryos of the eight patients.
Summary of breakpoint ajacent informative SNPs linkage analysis in 44 embryos.
Funding Sources
This study was supported by the National Natural Science Foundation of China81372627 (L.H.) and 81222007 (G.L.), National Basic Research Program of China2012CB944901 (G.X.L.) and 2016YFC1000206 (G.L.), Natural Science Foundation of Hunan Province13JJ3038, (L.H.) and the Program for New Century Excellent Talents in University (G.L.). Development of MicroSeq technique was supported by funding of Guangxiu-Gaoxin Life Science Co., Ltd. These funding sources had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the manuscript; and in the decision to submit the paper for publication.
Conflict of Interest Statement
None.
Author Contributions
L.H. and G.L. conceived and designed the study; L.H., D.H.C., X.H.W., and W.B.H. performed chromosome microdissection, breakpoint analysis and carrier embryo diagnosis; F.G. and K.L.L. recruited the patients, retrieved oocytes and transferred embryo; Y.Q.T. and P.Y.X. performed comprehensive chromosome screening; C.F.L. performed blastocyst biopsy; T.F. and K.Y. performed next generation sequencing and sequencing data analysis; G.X.L. and G.L. provided overall supervision; and L.H., D.H.C. and G.L. drafted the manuscript, with input from all of the authors.
Acknowledgements
We thank the families for their participation in this research. We also thank all members in PGD team of Reproductive and Genetic Hospital of CITIC–Xiangya and National Engineering and Research Center of Human Stem Cells for their invaluable efforts.
References
- Alfarawati S., Fragouli E., Colls P., Wells D. First births after preimplantation genetic diagnosis of structural chromosome abnormalities using comparative genomic hybridization and microarray analysis. Hum. Reprod. 2011;26:1560–1574. doi: 10.1093/humrep/der068. [DOI] [PubMed] [Google Scholar]
- Beigneux A.P., Kosinski C., Gavino B., Horton J.D., Skarnes W.C., Young S.G. ATP-citrate lyase deficiency in the mouse. J. Biol. Chem. 2004;279:9557–9564. doi: 10.1074/jbc.M310512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braude P., Pickering S., Flinter F., Ogilvie C.M. Preimplantation genetic diagnosis. Nat. Rev. Genet. 2002;3:941–953. doi: 10.1038/nrg953. [DOI] [PubMed] [Google Scholar]
- Chen W., Kalscheuer V., Tzschach A., Menzel C., Ullmann R., Schulz M.H., Erdogan F., Li N., Kijas Z., Arkesteijn G., Pajares I.L., Goetz-Sothmann M., Heinrich U., Rost I., Dufke A., Grasshoff U., Glaeser B., Vingron M., Ropers H.H. Mapping translocation breakpoints by next-generation sequencing. Genome Res. 2008;18:1143–1149. doi: 10.1101/gr.076166.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb T.M., Wakim A.N. GnRH agonist long protocol vs. a single 3-mg gnRH antagonist: a comparison of 2 protocols for pituitary down-regulation in oocyte donor-controlled ovarian hyperstimulation cycles. J. Reprod. Med. 2008;53:331–337. [PubMed] [Google Scholar]
- Feenstra I., Hanemaaijer N., Sikkema-Raddatz B., Yntema H., Dijkhuizen T., Lugtenberg D., Verheij J., Green A., Hordijk R., Reardon W., Vries B., Brunner H., Bongers E., Leeuw N., van Ravenswaaij-Arts C. Balanced into array: genome-wide array analysis in 54 patients with an apparently balanced de novo chromosome rearrangement and a meta-analysis. Eur. J. Hum. Genet. 2011;19:1152–1160. doi: 10.1038/ejhg.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R.J.M., Sutherland G.R., Shaffer L.G., Oxford University Press . Oxford University Press; New York: Oxford: 2012. Chromosome abnormalities and genetic counseling. Oxford monographs on medical genetics 61. (1 online resource (xiv, 634 p.)) [Google Scholar]
- Gersen S.L., Keagle M.B., SpringerLink (Online service) Springer; New York: 2013. The Principles of Clinical Cytogenetics. (Imprint: Springer: 1 online resource (X, 569 p.)) [Google Scholar]
- Gribble S.M., Ng B.L., Prigmore E., Fitzgerald T., Carter N.P. Array painting: a protocol for the rapid analysis of aberrant chromosomes using DNA microarrays. Nat. Protoc. 2009;4:1722–1736. doi: 10.1038/nprot.2009.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harton G.L., De Rycke M., Fiorentino F., Moutou C., SenGupta S., Traeger-Synodinos J., Harper J.C., European Society for Human Reproduction, Embryology (ESHRE) PGD Consortium ESHRE PGD consortium best practice guidelines for amplification-based PGD. Hum. Reprod. 2011;26:33–40. doi: 10.1093/humrep/deq231. [DOI] [PubMed] [Google Scholar]
- Higgins A.W., Alkuraya F.S., Bosco A.F., Brown K.K., Bruns G.A., Donovan D.J., Eisenman R., Fan Y., Farra C.G., Ferguson H.L., Gusella J.F., Harris D.J., Herrick S.R., Kelly C., Kim H.G., Kishikawa S., Korf B.R., Kulkarni S., Lally E., Leach N.T., Lemyre E., Lewis J., Ligon A.H., Lu W., Maas R.L., MacDonald M.E., Moore S.D., Peters R.E., Quade B.J., Quintero-Rivera F., Saadi I., Shen Y., Shendure J., Williamson R.E., Morton C.C. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am. J. Hum. Genet. 2008;82:712–722. doi: 10.1016/j.ajhg.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Sham J.S., Tjia W.M., Tan Y.Q., Lu G.X., Guan X.Y. Generation of a complete set of human telomeric band painting probes by chromosome microdissection. Genomics. 2004;83:298–302. doi: 10.1016/j.ygeno.2003.08.021. [DOI] [PubMed] [Google Scholar]
- Hu L., Sham J.S., Xie D., Wen J.M., Wang W.S., Wang Y., Guan X.Y. Up-regulation of fibroblast growth factor 3 is associated with tumor metastasis and recurrence in human hepatocellular carcinoma. Cancer Lett. 2007;252:36–42. doi: 10.1016/j.canlet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Jancuskova T., Plachy R., Stika J., Zemankova L., Hardekopf D.W., Liehr T., Kosyakova N., Cmejla R., Zejskova L., Kozak T., Zak P., Zavrelova A., Havlikova P., Karas M., Junge A., Ramel C., Pekova S. A method to identify new molecular markers for assessing minimal residual disease in acute leukemia patients. Leuk. Res. 2013;37:1363–1373. doi: 10.1016/j.leukres.2013.06.009. [DOI] [PubMed] [Google Scholar]
- McHenry C.L., Liu Y., Feng W., Nair A.R., Feathers K.L., Ding X., Gal A., Vollrath D., Sieving P.A., Thompson D.A. MERTK arginine-844-cysteine in a patient with severe rod-cone dystrophy: loss of mutant protein function in transfected cells. Invest. Ophthalmol. Vis. Sci. 2004;45:1456–1463. doi: 10.1167/iovs.03-0909. [DOI] [PubMed] [Google Scholar]
- Molotkov A., Duester G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J. Biol. Chem. 2003;278:36085–36090. doi: 10.1074/jbc.M303709200. [DOI] [PubMed] [Google Scholar]
- Munne S., Morrison L., Fung J., Marquez C., Weier U., Bahce M., Sable D., Grundfeld L., Schoolcraft B., Scott R., Cohen J. Spontaneous abortions are reduced after preconception diagnosis of translocations. J. Assist. Reprod. Genet. 1998;15:290–296. doi: 10.1023/A:1022544511198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie C.M., Scriven P.N. Meiotic outcomes in reciprocal translocation carriers ascertained in 3-day human embryos. Eur. J. Hum. Genet. 2002;10:801–806. doi: 10.1038/sj.ejhg.5200895. [DOI] [PubMed] [Google Scholar]
- Oshiumi H., Miyashita M., Okamoto M., Morioka Y., Okabe M., Matsumoto M., Seya T. DDX60 is involved in RIG-I-dependent and independent antiviral responses, and its function is attenuated by virus-induced EGFR activation. Cell Rep. 2015;11:1193–1207. doi: 10.1016/j.celrep.2015.04.047. [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Puechberty J., Ng B.L., Coubes C., Gatinois V., Tournaire M., Girard M., Dumont B., Bouret P., Magnetto J., Baghdadli A., Pellestor F., Genevieve D. Identification of disrupted AUTS2 and EPHA6 genes by array painting in a patient carrying a de novo balanced translocation t(3;7) with intellectual disability and neurodevelopment disorder. Am. J. Med. Genet. A. 2015;167:3031–3037. doi: 10.1002/ajmg.a.37350. [DOI] [PubMed] [Google Scholar]
- Scriven P.N., Handyside A.H., Ogilvie C.M. Chromosome translocations: segregation modes and strategies for preimplantation genetic diagnosis. Prenat. Diagn. 1998;18:1437–1449. [PubMed] [Google Scholar]
- Sobreira N.L.M., Gnanakkan V., Walsh M., Marosy B., Wohler E., Thomas G., Hoover-Fong J.E., Hamosh A., Wheelan S.J., Valle D. Characterization of complex chromosomal rearrangements by targeted capture and next-generation sequencing. Genome Res. 2011;21:1720–1727. doi: 10.1101/gr.122986.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski M.E., Ernst C., Heilbut A., Chiang C., Hanscom C., Lindgren A., Kirby A., Liu S., Muddukrishna B., Ohsumi T.K., Shen Y., Borowsky M., Daly M.J., Morton C.C., Gusella J.F. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am. J. Hum. Genet. 2011;88:469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski M.E., Ordulu Z., Pillalamarri V., Benson C.B., Blumenthal I., Connolly S., Hanscom C., Hussain N., Pereira S., Picker J., Rosenfeld J.A., Shaffer L.G., Wilkins-Haug L.E., Gusella J.F., Morton C.C. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N. Engl. J. Med. 2012;367:2226–2232. doi: 10.1056/NEJMoa1208594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Yin X., Zhang S., Jiang H., Tan K., Li J., Xiong B., Gong F., Zhang C., Pan X., Chen F., Chen S., Gong C., Lu C., Luo K., Gu Y., Zhang X., Wang W., Xu X., Vajta G., Bolund L., Yang H., Lu G., Du Y., Lin G. Clinical outcome of preimplantation genetic diagnosis and screening using next generation sequencing. Gigascience. 2014;3:30. doi: 10.1186/2047-217X-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treff N.R., Thompson K., Rafizadeh M., Chow M., Morrison L., Tao X., Garnsey H., Reda C.V., Metzgar T.L., Neal S., Jalas C., Scott R.T., Jr., Forman E.J. SNP array-based analyses of unbalanced embryos as a reference to distinguish between balanced translocation carrier and normal blastocysts. J. Assist. Reprod. Genet. 2016 doi: 10.1007/s10815-016-0734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker J.D. Low-dose ionizing radiation and chromosome translocations: a review of the major considerations for human biological dosimetry. Mutat. Res. 2008;659:211–220. doi: 10.1016/j.mrrev.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Vergult S., Van Binsbergen E., Sante T., Nowak S., Vanakker O., Claes K., Poppe B., Van der Aa N., van Roosmalen M.J., Duran K., Tavakoli-Yaraki M., Swinkels M., van den Boogaard M.J., van Haelst M., Roelens F., Speleman F., Cuppen E., Mortier G., Kloosterman W.P., Menten B. Mate pair sequencing for the detection of chromosomal aberrations in patients with intellectual disability and congenital malformations. Eur. J. Hum. Genet. 2014;22:652–659. doi: 10.1038/ejhg.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinsky Y., Cieslak J., Evsikov S., Galat V., Kuliev A. Nuclear transfer for full karyotyping and preimplantation diagnosis for translocations. Reprod. BioMed. Online. 2002;5:300–305. doi: 10.1016/s1472-6483(10)61836-6. [DOI] [PubMed] [Google Scholar]
- Wehr R., Mansouri A., de Maeyer T., Gruss P. Fkh5-deficient mice show dysgenesis in the caudal midbrain and hypothalamic mammillary body. Development. 1997;124:4447–4456. doi: 10.1242/dev.124.22.4447. [DOI] [PubMed] [Google Scholar]
- Weier H.U., Munne S., Fung J. Patient-specific probes for preimplantation genetic diagnosis of structural and numerical aberrations in interphase cells. J. Assist. Reprod. Genet. 1999;16:182–191. doi: 10.1023/A:1020360706317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willadsen S., Levron J., Munne S., Schimmel T., Marquez C., Scott R., Cohen J. Rapid visualization of metaphase chromosomes in single human blastomeres after fusion with in-vitro matured bovine eggs. Hum. Reprod. 1999;14:470–475. doi: 10.1093/humrep/14.2.470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of breakpoint ajacent informative SNPs and PGD results of the eight patients and their embryos.
Summary of the PGD results for all embryos of the eight patients.
Summary of breakpoint ajacent informative SNPs linkage analysis in 44 embryos.