Cells naturally “eat” nanowires, paving way for intracellular sensing devices and photoresponsive therapies.
Keywords: silicon nanowires, dynamics, phagocytosis, active transport, intracellular
Abstract
The ability to seamlessly merge electronic devices with biological systems at the cellular length scale is an exciting prospect for exploring new fundamental cell biology and in designing next-generation therapeutic devices. Semiconductor nanowires are well suited for achieving this goal because of their intrinsic size and wide range of possible configurations. However, current studies have focused primarily on delivering substrate-bound nanowire devices through mechanical abrasion or electroporation, with these bulkier substrates negating many of the inherent benefits of using nanoscale materials. To improve on this, an important next step is learning how to distribute these devices in a drug-like fashion, where cells can naturally uptake and incorporate these electronic components, allowing for truly noninvasive device integration. We show that silicon nanowires (SiNWs) can potentially be used as such a system, demonstrating that label-free SiNWs can be internalized in multiple cell lines (96% uptake rate), undergoing an active “burst-like” transport process. Our results show that, rather than through exogenous manipulation, SiNWs are internalized primarily through an endogenous phagocytosis pathway, allowing cellular integration of these materials. To study this behavior, we have developed a robust set of methodologies for quantitatively examining high–aspect ratio nanowire-cell interactions in a time-dependent manner on both single-cell and ensemble levels. This approach represents one of the first dynamic studies of semiconductor nanowire internalization and offers valuable insight into designing devices for biomolecule delivery, intracellular sensing, and photoresponsive therapies.
INTRODUCTION
Inorganic nanomaterials have emerged as a promising class of materials for interfacing with biological systems (1, 2), with nanoscale devices being used for both fundamental biophysical studies (3–5) and next-generation therapeutics (6, 7). Among these materials, silicon nanowires (SiNWs) are of particular interest because of their excellent electronic properties, distinct one-dimensional (1D) structure, and potential biocompatibility (8). In addition, SiNW synthesis can be controllably tuned to incorporate a diverse set of functionalities, including modulation in structural morphology (9, 10), surface functionalization (11), and composition (12, 13). This allows for the realization of a large library of SiNW-based tools. As a result, SiNWs have been used as a successful nanoscale platform for a variety of applications, including single-cell electrophysiology experiments (14), biomarker detection (15, 16), DNA and drug delivery (17, 18), and protein kinetics studies (19). Despite these successes, there is still much to be understood about the interface between SiNWs and cellular systems, particularly for substrate-independent devices.
To date, SiNW-cell interactions have been primarily studied from the perspective of substrate-bound wires (those wires that are still joined to an underlying bulk material) (3, 20–23) using gravity, electroporation, and/or adhesive forces to access the cell interior (18, 24) or form invaginations (23). Although these configurations allow nanoscale devices to be connected directly to external macroscopic electronics, they lead to bulkier designs, which can be less desirable for certain applications. First, larger devices can exacerbate the mismatch in material properties (such as Young’s modulus, curvature, etc.) between typical inorganic semiconductors and biological samples, leading to poor device integration and increased irritation (25). Second, the incorporation of macroscopic substrates negates many of the benefits of using nanoscale materials in the first place, such as surgery-free device distribution and point-like localized probing. Finally, many of these devices require extensive wiring that extends outside the body, which can be disruptive to existing biological architecture and hinders the use of these devices at the single-cell level. Therefore, a more desirable approach would be to use substrate-independent devices capable of being dispersed in a drug-like manner and of being wholly internalized within a single cell. These materials could act as precursors for future “artificial bionanomachines” (26) or as therapeutic devices and probes that can operate independently at the subcellular level.
Previous work has shown that both label-free (27, 28) and surface-modified (29, 30) SiNWs can be internalized at the single-cell level in a substrate-free manner, making them a promising candidate for fulfilling these criteria. However, little is known about how these devices enter cells, especially in a time-dependent manner. In addition, without the use of labeling reagents to help elucidate the nano-bio interface, studying nanoparticle-cell interactions has been a challenge. However, the use of labeling reagents can also change the device’s surface functionalization, leading to altered surface recognition, making them potentially disruptive to any native nanowire-cell interactions. Yet, this label-free biointerface knowledge is critical in informing future device design and in implementing cell-specific targeting. To expand on this outlook, we demonstrate here that label-free SiNWs can be spontaneously internalized in cellular systems, showing that these materials are primarily endocytosed via a phagocytosis pathway and, once internalized, undergo active intracellular transport, eventually clustering in the perinuclear region. An overview of this can be seen in Fig. 1A, where a cell is depicted as initiating nanowire internalization. To study this process, we have developed a series of label-free tracking protocols for both individual- and ensemble-level dynamics. Finally, we will discuss how using these methods helps fit nanowire internalization into a more familiar biological picture, while raising many exciting opportunities in using nanowires as intracellular nanotherapeutics and diagnostic devices.
Fig. 1. SiNW internalization.
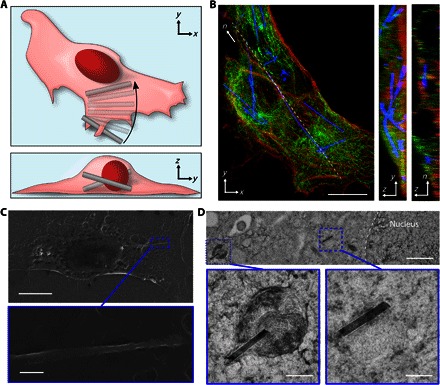
(A) Schematic illustration of SiNW internalization. (B) Confocal fluorescence micrograph of HUVECs (actin, red; tubulin, green) demonstrating SiNW internalization (blue scattering). Maximum projection in the xy plane (left; scale bar, 10 μm), interpolated projection in the yz plane (middle; height, 3.5 μm), and a thin confocal section taken along the dashed line segment n (right; height, 3.5 μm; length, 48.3 μm). (C) SEM micrograph of a HUVEC containing a SiNW [scale bar (top), 10 μm]. The magnified highlighted region indicates that the SiNW is embedded under the cell’s membrane [scale bar (bottom), 300 nm]. (D) TEM micrograph of a HUVEC thin section (~250 nm thick), with higher magnification insets, illustrating the distribution of internalized wires, both in vesicles and in the cytosol [scale bars, 1 μm (top) and 200 nm (insets)].
RESULTS
Serving as the inner lining of blood vessel walls, endothelial cells act as a filtration system between the bloodstream and the rest of the body, helping to regulate the uptake of drugs and clearing apoptotic blood cells and other extracellular materials. To study substrate-free SiNW endocytosis, human umbilical vascular endothelial cells (HUVECs) were selected as a model cell line. Because SiNWs can potentially be distributed in a drug-like fashion, endothelial cells would play a key role in mediating biointegration, with HUVECs, in particular, having been shown to recapitulate many of the features found in native vascular endothelial cells (31).
To show that label-free SiNWs could be internalized by HUVECs, we used both optical and electron microscopy techniques. First, confocal fluorescent microscopy was used to reconstruct 3D volumes containing HUVECs with internalized SiNWs (Fig. 1B). For these experiments, cells were incubated with SiNWs for 24 hours, and the cytoskeleton was labeled using the fluorescent markers tetramethylrhodamine-phalloidin (actin, red) and anti–α-tubulin Alexa Fluor 488 (microtubules, green), with label-free SiNWs visualized using optical scattering (28). The resulting 3D volumes and the line-scan cross section (Fig. 1B, right) were seen to contain SiNWs, demonstrating that SiNWs could be spontaneously internalized by HUVECs.
We next examined these interfaces in greater detail using electron microscopy (EM) techniques. First, HUVECs cultured with SiNWs were chemically fixed, critical point–dried, and then imaged using scanning EM (SEM). The resulting samples were observed to have SiNWs running beneath the cell membrane (Fig. 1C). To show that wires were actually contained within the cell body, cryopreserved thin cell sections were imaged using transmission EM (TEM). To prepare these samples, trypsinized cells were rapidly fixed using high-pressure freezing and processed using freeze substitution techniques (27). The resulting samples were then segmented using an ultramicrotome, yielding thin cell sections (~200 to 300 nm). TEM measurements revealed that internalized SiNWs showed a mixed distribution, with some wires floating free in the cytosol, whereas others were contained in small vesicles (Fig. 1D). This confirmed that SiNWs could be spontaneously internalized by HUVECs without additional surface modification, independent of external mechanical forces or electroporation. In addition, the formation of encapsulation vesicles suggested that the SiNWs entered the cells via an endogenous endocytosis process, rather than through mechanical abrasion (that is, puncturing the cell membrane). It was also observed that multiple SiNWs could be contained within a single larger vesicle, reminiscent of a lysosome (fig. S1), causing SiNWs to become clustered in the perinuclear region. This suggested a dynamic process where SiNWs could be shuttled to a specific cellular regions.
Although EM studies can provide detailed structural information, they only offer a static view of the internalization process. To further characterize this in a time-dependent fashion, we turned to dynamics studies, examining SiNW endocytosis on both the ensemble and single-cell level, with ensemble dynamics encompassing the large-scale interactions between many cells and many nanowires and single nanowire dynamics encompassing the interactions between individual nanowires and single cells.
Single nanowire dynamics
Individual SiNW internalization dynamics were studied using scatter-enhanced phase-contrast (SEPC) microscopy, which allowed for a clear visualization of both SiNWs and adherent cells (Fig. 2A) (27). For this study, SiNWs were sonicated into growth medium and allowed to settle before they were introduced to HUVECs. Internalization dynamics were then monitored using a custom tracking algorithm, returning the position of each SiNW tip as a function of time (fig. S2). Nanowire dynamics were approximated as being 2D, a reasonable assumption, given the large aspect ratio of the nanowires and the thin volume of the lamellipodium, where transport is initiated. Tracking the nanowire’s tip positions allowed for a precise determination of both the nanowire’s velocity and mode of transport. To correct for stage drift, stationary particles were simultaneously tracked and used to adjust the relative path of the SiNW (fig. S3). Tracking individual SiNWs revealed an active transport process, where nanowires are shuttled to the perinuclear region within ~5 to 30 min of coming into contact with the internalizing cell (see movie S1).
Fig. 2. Active SiNW transport.
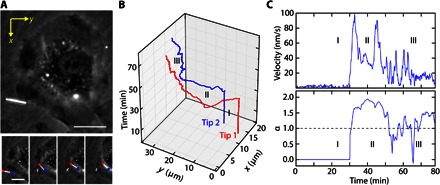
(A) SEPC micrograph of a SiNW before (top) and during (bottom; t = ~7 min per frame) internalization (scale bar, 15 μm), with tips 1 and 2 indicated by red and blue markers, respectively. (B) Path of travel for both tips of the SiNW as a function of time. (C) Instantaneous velocity of the SiNW before (15-frame interval) (I), during (II), and after (III) active transport, with the corresponding rolling MSD diffusivity exponent α, indicating an active transport process. The diffusivity exponent α was obtained over a rolling 30-frame period. All values given are for tip 1 (red).
Briefly summarizing this process, we initially presettled SiNWs onto the underlying substrate, allowing for improved transport quantification by avoiding the large temporal variance associated with the initial SiNW seeding process. Settled nanowires were initially stationary (Fig. 2, B and C, region I), but upon contacting the cell, SiNWs were seen to be “grabbed” (region II), getting shuttled from the lamellipodium to the perinuclear region (Fig. 2, B and C, region II). Here, SiNWs displayed “burst-like” velocities, where the nanowire would be transported in sudden large spurts of speed, punctuated by brief pauses (up to 5 min), during which the SiNWs would display Brownian or restricted diffusion before continuing active transport. Here, SiNW transport begins almost immediately (region II), displaying a relatively high mobility, with a maximum instantaneous velocity of 99.4 nm/s (velocities averaged over a 15-frame interval). In most of the studies, transport trajectories were linear (fig. S4), proceeding along approximately straight tracks; however, in some cases, SiNWs could also undergo rotational rearrangements (fig. S5). After transport, SiNWs would eventually settle around the nucleus (although excluded from the nuclear envelope), displaying only small diffusive movements (Fig. 2B, region III). Similar dynamics were also noted for SiNWs in the absence of protein serum (fig. S6A) and for wires with different configurations (fig. S6B), such as a kinked “L-shaped” wire, with this type of wire having previously been used for biophysical force studies (27). This indicated that a diversity of device configurations are tolerated and that protein opsonization is not critical for nanowire uptake.
To distinguish between different modes of SiNW transport, we used a rolling frame mean-squared displacement (MSD) metric, where the MSD is the average distance that a particle travels as a function of lag time, given by
| (1) |
where Δr, τ, q, and α are the nanowire displacement, lag time, diffusion coefficient, and “diffusivity exponent,” respectively. The diffusivity exponent α can be used as a metric of transport properties, distinguishing between Brownian diffusion (α = 1), restricted diffusion (α < 1), and active transport (α > 1) processes. Values for α were obtained by fitting the Ln-Ln plot of the MSD with a linear regression over a rolling interval, with the slope yielding the relative diffusivity (fig. S7). Using the MSD of the SiNW, different modes of cellular transport can be assessed, providing some insight into the underlying mechanism.
The present case strongly suggests that SiNWs are being treated as cargo by the cell and are being actively shuttled by cellular machinery. First, this is suggested by the relatively high velocities and by the observed active transport (α = 1.94) (Fig. 2C, region II). Although cell motility can also display temporary directional motion (α = 1.6), wire transport coupled to cell motility displayed relatively low velocities (~20 nm/s) (fig. S5B, region II), distinguishing it from motor protein–assisted transport. Second, the burst-like transport displayed here is reminiscent of other motor protein–powered intracellular transport (32). Exploring this behavior in more detail, we used nocodazole as a potent microtubule inhibitor to disrupt retrograde dynein-dependent transport (33). In nocodazole-treated cells, SiNWs displayed considerably lower transport velocities (fig. S8), showing a maximum instantaneous velocity of ~10 nm/s in the present case. In addition, although active transport was observed during the initial internalization step (which is likely an actin-dependent process), no further retrograde transport to the perinuclear region was noted. Instead, the SiNW came to rest ~6.5 μm away from the nuclear envelope after its initial internalization, a sizable distance compared to nontreated SiNWs, which on average ended active transport much closer to the nucleus (0.86 ± 0.63 μm, n = 8; maximum observed distance, ~1.4 μm). This indicates that microtubules and, likely, dynein are critical during SiNW transport. Together with the linear trajectories (fig. S4), retrograde movement (Fig. 2), and previously observed vesicle formation (Fig. 1D), this strongly suggests that cells treat the SiNWs as intracellular cargo. In addition, we noted that, in SiNWs samples that have been co-incubated for longer time scales (~3 days), a “tug-of-war” style of bidirectional motion was also observed (movie S2 and fig. S9). The bidirectional motion indicates a competing retrograde and anterograde transport process and suggests the participation of both kinesin and dynein motor proteins in at least the later stages of the SiNW intracellular interface.
To determine the specific route of endocytosis, we have adopted several strategies. Initially, lysosome tracking was pursued (fig. S10). Because of the relatively large length of SiNWs, they were able to colocalize with multiple lysosomes simultaneously. To measure these dynamics, we first examined the ensemble overlap of SiNWs with lysosomes as a function of incubation time (fig. S10, A and B). Here, we saw that, within 3 hours of co-incubation, ~75% of SiNWs that were overlapping cells also showed colocalization with one or more lysosomes, suggesting a preferential interaction. Next, we examined the dynamics of single wires, observing that individual internalized wires could remain colocalized with individual lysosomes on the hour time scale (fig. S10, C and D), showing highly correlated movement with the lysosome as compared to external particles (fig. S10E). However, because of the relatively large size of the SiNWs, they were often overlapping with multiple lysosomes, making it difficult to distinguish a primary internalization vesicle. In addition, any correlation in movement was difficult to distinguish from the overall cell motility. Therefore, to study the specific route of endocytosis in more detail, we turned to an ensemble model based on the SiNW-cell overlap (fig. S11). Using this method allowed for both a simple-to-implement single–time point measurement to assess cell type–dependent internalization and a dynamic ensemble study based on a 2D random walker model, which lead to identifying a specific mechanism of internalization.
Ensemble nanowire dynamics
We assayed the ensemble SiNW uptake using the rate of SiNW-cell overlap. To achieve this, first, SiNWs were allowed to settle on a substrate before seeding cells. During incubation, cells could then migrate over the surface, picking up SiNWs as they moved. During this process, both dark field (DF) and phase contrast (PC) micrographs were collected to determine the percentage of SiNWs overlapping with cells (Fig. 3). Although not a direct confirmation of internalization, SiNW-cell overlap acts as a reporter of nanowire-cell interactions. In a random noninteracting system, we would expect minimal clustering of the wires and an “overlap reporter” value (β) at unity (β = 1), where β is equal to the percentage of nanowires overlapping with cells at time t (Yt), divided by the cell confluence (Ct), that is, the percentage of area covered by cells, such that β = Yt/Ct. However, in the case of positive SiNW uptake, we would expect to see wires clustered into small regions corresponding to the position of each cell (Fig. 3A), resulting in a larger SiNW-cell overlap reporter value (β > 1). In this regard, the use of overlap values β enables an easy-to-perform optical assay to study a variety of SiNW-cell interactions, including both the cell line and nanowire length dependence on internalization.
Fig. 3. Ensemble SiNW internalization dynamics using SiNW-cell overlap.
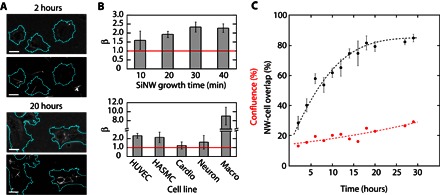
(A) Corresponding PC (top) and DF (bottom) micrographs taken at 2 hours (top set) and 20 hours (bottom set) after HUVEC incubation with SiNWs (scale bars, 25 μm), indicating increased SiNW-cell overlap and clustering in the perinuclear region (artificial cell outline highlighted in teal). (B) Uptake statistics of SiNWs with varying growth lengths (HUVECs) (top) and in multiple cell lines (bottom) after 24 hours, showing that longer wires are more likely to be internalized on the basis of geometric considerations and that cardiomyocytes (Cardio) and DRG neurons (Neuron) do not internalize unmodified SiNWs, whereas J774A.1 macrophages (Macro) show larger uptake rates. Average SiNW length by growth time: 10 min: 9.8 μm; 20 min: 14.5 μm; 30 min: 23.1 μm; 40 min: 31.7 μm. (C) Example ensemble SiNW-cell overlap (black dots) and cell confluence (red dots) trace as a function of time for unmodified SiNWs in HUVECs. Larger-than-confluence overlap indicates SiNW internalization. Expected overlap trend (black line) fit using the 2D random walker model (Dt = 410 μm2/hour, R2 = 0.93). Cell confluence modeled as an exponential fit (red line).
Using this overlap reporter model, we examined the importance of nanowire length on SiNW uptake (Fig. 3B). SiNWs of different lengths were prepared by varying growth time during synthesis, with increased durations resulting in longer wires on average after sonication. For each sample, β was then determined after 24 hours of co-incubation with HUVECs. In all cases, label-free SiNW internalization was observed (β > 1), with longer wires showing a higher rate of overlap with cells (~45% increase in β). Although, at first, this suggests that HUVECs prefer longer SiNWs, this result is consistent with the fact that longer wires are more likely to come into contact with cells. When normalized by the length of the wire, no significant difference was noted between the longest and shortest growth times (Student’s t test, P > 0.9), indicating that SiNW length is not a critical factor in determining which nanowires can be internalized once they are already in contact with a cell. During this time, we also noted that the cells tolerated a range of SiNWs concentrations (fig. S12). Using an MTT colorimetric assay, we examined the effects of SiNW concentration on the cell’s metabolism, with an approximately fourfold increase in concentration, leading to only a mild reduction in the cell’s activity (28 ± 4% reduction after 3 days). Surprisingly, at smaller SiNW concentrations, an increase in the cell’s metabolic activity was observed, as compared to the control samples. Because it has previously been reported that nanowires can temporarily increase reactive oxygen species in cells (34), this metabolic spike is likely the result of some initial oxidative stress; however, the same study showed that, over slightly longer time scales, the nanowires did not elicit any cytotoxic effects, suggesting that a similar behavior may be occuring here. The overlap reporter model also provided insight into cell line–specific internalization.
Along with HUVECs, human aortic smooth muscle cells (HASMCs) were selected as a model muscle system because they can provide responsive contractility and have been previously used for intracellular nanowire force studies (27). In addition, mouse-derived J774A.1 monocyte macrophages were also selected as a model system for professional phagocytes because of their commercial availability, reproducibility as compared to primary macrophages, and facile use. Finally, because both neurons and cardiac cells are of particular interest for bioelectronic applications (35), primary cardiomyocytes and dorsal root ganglia (DRG) neurons from neonatal rat models were selected because of their widely studied properties (36) and common use in bioengineering (37). In the case of both HUVECs and HASMCs, clustering (Fig. 3A) and β values in excess of unity (Fig. 3C) were observed (β = 2.3 ± 0.3 and 2.15 ± 0.6, respectively), suggesting that both cell lines were capable of internalizing label-free SiNWs. J774A.1 cells were also seen to internalize nanowires at high rates (see movie S3), demonstrating clustering and high overlap values after 24 hours (β = 9.0 ± 1.9; Fig. 3C). This approximately fivefold increase in uptake is likely the result of macrophage cells being only partially adherent, allowing them greater cell motility, and is consistent with their role in keeping the bloodstream clear of foreign materials. However, for primary cardiomyocytes and DRG neurons, no clustering was observed and β values did not statistically exceed the baseline (β = 1.3 ± 0.4 and 1.6 ± 0.8, respectively), indicating that neither cell line was capable of internalizing label-free SiNWs (fig. S13). Although the DRG neurons (as determined by β-tubulin III staining) did show slightly elevated β values, this was ascribed primarily to their close association with other nonneuronal cells retained during primary culture, some of which appeared to be able to internalize SiNWs (clustering, β = 3.3 ± 1.5). Collectively, these results suggest heterogeneity in the cell-specific response to SiNWs, namely, that only certain cell types are capable of internalizing label-free wires. This is consistent with the fact that there are many different routes of entry for particles to be internalized (38, 39) but that not all of these endocytosis pathways are expressed across all cell types.
To delineate between different modes of endocytosis, we turned to a dynamic ensemble model, looking at the SiNW-cell overlap as a function of time, a method reminiscent of previous phagokinetics studies (40). To understand the resulting dynamics, we parameterized the system using a 2D random walker model (eq. S1 to S10), which we derived from the equations of Dvoretzky and Erdös (41), with the ensemble percentage of SiNWs that were overlapping cells, Yt, being given by
| (2) |
where t is time, Dt is the effective cell migration constant, Mw is the maximum percentage of SiNWs available for internalization, Acell is the average area covered by a single cell, Ct is the percent cell confluence as a function of time, and B = Mw − C0. This models differs significantly from other drug kinetics studies. In a typical drug delivery model, cells are considered stationary, while drugs are considered mobile, being able to freely diffuse throughout the solution. However, in the present model we consider the settled SiNWs to be stationary, instead assuming the cells are mobile, with cell motility bringing them into contact with new SiNWs as they travel across the culture surface.
This model was observed to be in agreement with experimental values (Fig. 3D), and its use presented several advantages. First, as a relatively constrained system, this model offers distinct physical insights into the ensemble internalization process because all of the terms refer to directly measureable quantities. For instance, the maximum percentage of internalizable wires, Mw, was found to be ~96%, with an average effective cell migration rate of 437 ± 36 μm2/hour (R2 = 0.89, n = 7) as determined for HUVECs using Dt as the single free-fit parameter (linear least-squares regression). Second, this model can be used to create an expected rate of SiNW internalization in the absence of perturbations, potentially providing useful biophysical insights. For example, this model indicates that the presence of initially internalized SiNWs does not significantly affect later cell motility and uptake rates. Because a steady rate of cell migration is assumed, changes in cellular motility are readily observed using this technique. However, HUVECs did not show any significant deviation from the internally predicted uptake rates (fig. S14), suggesting that the presence of initial SiNWs did not greatly affect later internalization events, at least on the single-day time scale. This was corroborated by studying single nanowire dynamics, which showed that secondary internalization events preserved many of the salient features of initial nanowire internalization, such as active transport (maximum α = 1.9, maximum velocity = 87 nm/s) (fig. S15).
In addition, the expected ensemble internalization rate provided by this model can be used to gauge the effect of different treatment regimes on SiNW internalization, using known endocytosis inhibitors to block specific routes of internalization. This strategy allows for a careful examination of the internalization process and can help point toward a specific method of endocytosis (Fig. 4A).
Fig. 4. Mechanistic and morphological studies.
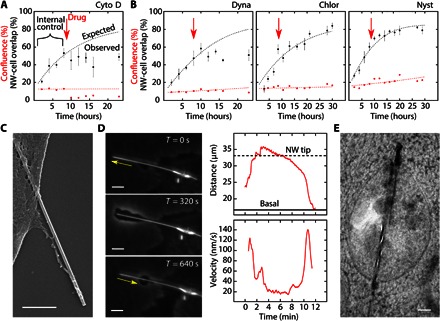
(A) Positive control study of Cyto D (actin inhibitor) showing the SiNW-cell overlap (black dots), cell confluence (red dots), and the expected overlap trend (black line), modeled on the first 8 hours of internalization (internal control) before drug introduction (red arrow). Cell confluence modeled as an exponential fit (red line). (B) Endocytosis inhibitors: dynasore (Dynamin; left), chlorpromazine (Clathrin; middle), and nystatin (Lipid Rafts; right), indicating dynamin’s critical role in SiNW internalization. (C) SEM micrograph showing membrane extension along a SiNW (scale bar, 500 nm). (D) Time-lapse SEPC micrographs of a membrane extending along a SiNW before cellular uptake (left) (scale bar, 5 μm). Distance of the protrusion’s leading edge from the base of the SiNW over time (top right), with the corresponding instantaneous velocities (bottom right). Base membrane and nanowire tip distances given as solid and dotted lines, respectively. Velocities smoothed over an 11-frame interval. (E) TEM micrograph showing a long intercellular SiNW protruding from a vesicle into the cytosol (scale bar, 250 nm).
Mechanistic studies
While using the dynamic SiNW-cell overlap model, an internal control was adopted to help correct for deviations in the rate of nanowire internalization, which was found to be sensitive to the initial seeded cell population (fig. S14A). To establish this internal control, HUVECs were cocultured with SiNWs for a minimum of 7.5 hours before administering endocytosis inhibitors. Then, using Eq. 2, the projected SiNW-cell overlap was determined in the absence of any perturbations. The resulting projection was found to be in agreement with experimentally observed values present in the negative control, where no inhibitors were administered (R2 = 0.91) (fig. S14B), suggesting the validity of this method. To quantitate the significance of a drug’s impact on internalization, a Pearson χ2 test was employed, using the projection from the internal control model as the expected result and the experimentally measured overlaps as the observed trend. In the case of the negative control, a nonsignificant deviation from the projected trend was observed χ2(7, n = 6) = 1.62, P > 0.95 (fig. 14B), with a ΔY29 of only 0.6%, where ΔYt is the difference between the expected and experimentally observed SiNW-cell overlap values after t hours.
As a positive control for drug efficacy, cell migration was halted using the potent actin polymerization inhibitor cytochalasin D (Cyto D), restricting the cells’ motility and their ability to access new SiNWs. This resulted in the abrupt termination of nanowire internalization (ΔY23 = 28.4%), showing significant deviation from the expected SiNW-cell overlap, χ2(4, n = 6) = 35.26, P < 0.01. A similar trend was observed using dynasore (Fig. 4B, left), a cell-permeable dynamin inhibitor, which yields a statistically significant change from the internal control (ΔY23 = 29.2%), χ2(4, n = 12) = 23.5, P < 0.01, indicating that dynamin, a protein that is responsible for regulating membrane curvature and vesicle scission (39), plays a critical role in nanowire internalization. This observation is important because it concretely links SiNW uptake to cell-regulated endocytosis, showing that the nanowires are being actively internalized through a protein-dependent process, rather than passively through mechanical abrasion. To examine this in more detail, both clathrin-dependent and clathrin-independent mechanisms were probed.
Using chlorpromazine as a clathrin blocker (Fig. 4B, middle), no significant change was noted in nanowire internalization (ΔY30 = 6.1%), χ2(6, n = 6) = 1.75, P = 0.94, suggesting a clathrin-independent pathway. This result is surprising, considering that clathrin-coated pits are one of the dominant routes of entry for many spherical inorganic nanoparticles (NPs), including Au NPs (42) and SiO2 NPs (~300 nm) (43). To confirm that clathrin pathways were effectively blocked at the present inhibitor concentrations, fluorescently labeled transferrin was used as a positive control, resulting in observable clathrin inhibition (~40%) (fig. S16). This means that, despite the nanoscale diameter of the SiNWs (20 to 250 nm), cells were able to distinguish the high aspect ratio of the material, suggesting a mechanism of topological sensing where a cell is able to gauge a material’s aspect ratio and thereby delineate its mode of cellular entry. One such pathway that shows shape-sensitive internalization at the nanoscale is caveolae-mediated endocytosis (44). Although the role of caveolae remains controversial (45), we examined this route by administering the drug nystatin (Fig. 4B, right), which disrupts lipid raft and caveolae formation by cholesterol binding (45). However, nystatin showed no significant change in nanowire internalization (ΔY29 = 5.7%), χ2(7, n = 6) = 1.73, P > 0.95.
Another pathway that is particle size–dependent is phagocytosis, because it requires particles to be completely encapsulated before internalization. Reviewing the cell line–dependent internalization, we noted that macrophages, primary phagocytes, were able to readily internalize SiNWs (Fig. 3C). In addition, reagents for blocking phagocytosis, including actin inhibitors such as Cyto D, showed a significant decrease in SiNW uptake (Fig. 4A). Together, this suggested that phagocytosis may play an important role in nanowire endocytosis. However, macrophages can present multiple endocytosis pathways, whereas actin polymerization can restrict cell migration, potentially leading to false positives. Therefore, to study this pathway in more detail, we first examined the cell’s morphology during internalization before using the competitive surface-binding protein annexin V (A5), which can inhibit phagocytosis without restricting cell migration (see table S1 for a summary of endocytosis inhibitor results).
Cell uptake morphology
To determine whether particles were being engulfed during internalization, we examined the morphology of HUVECs using EM and SEPC studies. When examined under SEM, in some cases, cells co-incubated with SiNWs were seen to have membranes extending along the entire length of the wire (Fig. 4C), suggesting initiation of phagocytosis. However, SEM only offers a static view. To supplement this, SEPC was used to observe the real-time dynamics of membrane extension along a single SiNW (Fig. 4D). This was achieved using high–aspect ratio SiNWs (~33 μm in length), with longer wires requiring greater extension distances, thus enabling easier optical characterization. When using these wires, we observed that, starting at the base of the membrane, the cell would extend a protrusion along the entire length of the wire at a maximum rate of ~120 nm/s, eventually reaching past the wire’s tip before pausing briefly and then being retracted back to the basal level at similar speeds (~140 nm/s) (see movie S4 and Fig. 4D). Just after this retraction, the SiNW is then seen to enter the dynamic internalization process, as previously described. This membrane engulfment is phenotypical of phagocytosis, further suggesting this route of internalization.
Surprisingly, during this process, we also observed that, in some cases, the membrane would not initially bring the entire SiNW into the cell but would instead retract as a punctured vesicle before continuing SiNW internalization. This behavior has been noted before in other high–aspect ratio particles, such as carbon nanotubes (46) and silver nanowires (47), and is often referred to as “frustrated phagocytosis.” This partial encapsulation was further confirmed using TEM (Fig. 4E), showing that, for longer SiNWs, a portion of the sample can extend past the internalization vesicle. Because of the nature of thin cell sectioning, only a portion of the SiNW segments was observable under TEM. This means that segments of the SiNWs can extend past the field of view, precluding precise quantification of the percentage of partially or wholly encapsulated SiNWs. The role of frustrated phagocytosis suggests that there is a maximum length at which cells can healthily internalize wires, a process that should be examined in further detail in future studies. Despite this partial encapsulation in some cases, overall, these observations reinforced phagocytosis as a mechanism for SiNW internalization.
A5 as a phagocytosis inhibitor
To confirm phagocytosis as the route of SiNW internalization, A5 was used as a competitive surface-binding inhibitor (48–50), with A5 binding to the surface of the SiNW rather than directly to the cell. Compared to other phagocytosis inhibitors, such as Cyto D (Fig. 4A), that work by polymerizing actin, A5 offered the advantage of not restricting cell migration, which can potentially lead to a false identification using stationary particles. Previously, A5 has been used as a phagocytosis inhibitor by binding to the membranous protein phosphatidylserine, interfering with phagocytosis by screening apoptotic cells (49, 51). A similar approach is pursued here, using the positively charged A5 protein to nonspecifically bind to the surface of the negatively charged SiNW, effectively screening the SiNWs from internalization (Fig. 5A).
Fig. 5. A5 as a phagocytosis inhibitor of SiNWs.
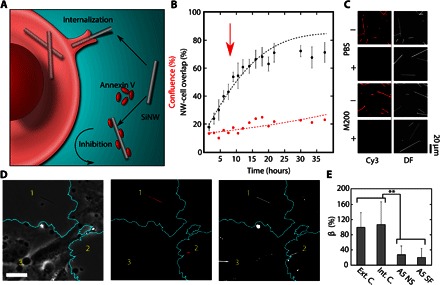
(A) Schematic illustration showing the inhibition mechanism of A5. Unlike inhibitors that target the cell directly, the nonspecific binding of A5 to the negatively charged SiNW surface can screen the SiNW from uptake. (B) A5 inhibitor study, showing a reduced nanowire-cell overlap (black dots) as compared to the expected internalization (black line) (red arrow indicates dosage time). (C) Fluorescent and DF micrographs showing the level of A5-Cy3 absorption onto SiNWs in the presence (+) and absence (−) of serum in PBS and M200 solutions, indicating that the ability of A5 to bind with SiNWs is restricted by the presence of serum proteins. (D) Example micrograph showing the same region in the PC (left), DF (middle), and Cy3 (right, artificial color) channels, indicating that SiNW surfaces modified with Cy3-A5 (1 and 2) are excluded from the cell, whereas the internal controls (3) are endocytosed (artificial cell outline, teal) (scale bar, 30 μm). (E) SiNW-cell overlap in HUVECs after 24 hours of SiNWs with nonspecifically bound A5-Cy3 (A5 NS) and surface-functionalized A5-Cy3 (A5 SF), relative to an external (Ext. C.) and internal (Int. C.) control of unmodified SiNWs (β given as a percentage of external control) (**P < 0.01). DF and Cy3 were background-subtracted using a Gaussian spatial filter uniformly applied to each image.
In this way, A5 was found to be able to inhibit SiNW uptake when introduced during internalization, showing a modest reduction in the percentage of internalized NWs (ΔY38 = −13.4%), χ2(10, n = 6) = 9.38, P = 0.49 (Fig. 5B), reinforcing phagocytosis as the primary route of endocytosis; however, the observed inhibition was rather mild. Increasing the concentration of A5 (from 4 to 16 nM) (fig. S17) led to further suppression in the SiNW-cell overlap (ΔY22 = −17.8%), χ2(5, n = 6) = 8.35, P = 0.14 (see table S1). This confirmed that A5 could inhibit SiNW uptake; however, the limited activity suggested that this method was only partially effective at preventing SiNW uptake. To understand this behavior, we examined how A5 proteins were binding SiNWs in solution.
In the case of apoptotic cells, A5 acts as an inhibitor by ligating exposed phosphatidylserine residues, reducing the rate of phagocytosis by effectively screening the apoptotic cell (52). However, in the case of SiNWs, this binding would be nonspecific, occurring primarily from electrostatic interactions. This means that, while in solution, A5 would be in competition with other positively charged serum proteins to bind the nanowire surface. This competition has been observed before in other systems, with positively charged serum proteins binding nonspecifically in spherical silica particles (53) to form a protein corona. To determine whether these proteins could be interfering with A5 binding, leading to mild inhibition, we examined the SiNW-cell interface.
Protein corona formation was initially studied using optical light scattering. Previously, we have shown that SiNW optical scattering correlates strongly with nanowire diameter and that even small changes on the nanometer scale can result in large optical shifts (28). Using this to monitor material degradation, we also showed that SiNWs exhibited mild decomposition under physiological conditions on the day time scale (27). However, using this technique to examine internalized SiNWs, we actually observed an increase in optical scattering from 25 to 72 hours (~38% increase, P < 0.05), indicating an apparent “increase” in SiNW diameter (fig S18). Although we believe that SiNWs will degrade further over longer time scales, as has been reported in other silicon-based devices (54), this would suggest that, over a shorter time scale, the cells were able to deposit material around the SiNW, corresponding well with the formation of a protein corona. Upon further inspection, this was confirmed using TEM (fig. S18E), with cytosolic wires forming possible “protein sheaths” or elongated protein coronas, with an average protein thickness of 16 ± 10 nm (n = 21) observed for cytosolic wires. These proteins could possibly interfere with A5 binding to the SiNW surface, leading to the observed mild inhibition rates. To determine whether this was the case, we examined the role of serum proteins in the nonspecific binding of A5 to SiNWs.
For the nonspecific A5 binding experiments (Fig. 5C), SiNWs were incubated with cyanine-labeled A5 (A5-Cy3) at 37°C for ~24 hours in the absence and presence of serum proteins (2% fetal bovine serum) in both phosphate-buffered saline (PBS) and M200 medium. In the absence of serum, A5-Cy3 was observed to bind to the SiNW surface in PBS, showing strong fluorescent signal intensity (Fig. 5C), whereas the samples containing serum exhibited only minimal binding, with a relative fluorescent signal intensity of only 0.29 ± 0.03% (background-subtracted using unlabeled SiNWs and normalized by SiNW concentration) (Fig. 5C). A similar trend was observed in M200 medium, with serum-free and serum-rich media showing a relative signal intensity of 87 ± 16% and 0.54 ± 0.04%, respectively, as compared to the PBS-bound A5. These results show that serum is sufficient in restricting the access of A5-Cy3 to the surface of SiNW and supports a competitive model of binding between A5 and other serum proteins. This explains the observed mild inhibition because A5 would have difficulties displacing other proteins from the surface of the SiNWs.
To circumvent this competition and show that surface-bound A5 was enough to disrupt nanowire uptake, we examined SiNWs with either nonspecifically absorbed or surface-functionalized A5-Cy3 (fig. S19 and Fig. 5, D and E). For nonspecifically absorbed samples, SiNWs were first incubated with A5-Cy3 in PBS in the absence of serum for 1 hour at 37°C, whereas for surface-functionalized samples, A5-Cy3 was conjugated to the SiNW surface using an aldehyde-silane modification step (11). In both cases, wires were then sonicated into an M200 solution and allowed to settle before HUVECs were introduced. After 24 hours of co-incubation, samples were then chemically fixed, and the SiNW-cell overlap was monitored optically in the Cy3, PC, and DF channels. After incubation in the serum-containing medium, many of the nonspecifically absorbed SiNWs lost their A5-Cy3 coating; however, those wires that did retain their coating (as determined by fluorescent imaging) showed a significantly reduced rate of overlap with HUVECs (fig. S19), as compared to an external control of unmodified wires (β = ~28% of control; Student’s t test, P < 0.01) (Fig. 5E). This indicated that, when predeposited onto the nanowire surface, A5 was able to effectively disrupt SiNW uptake by screening the nanowires from the internalizing cell.
We further confirmed this by studying the uptake of SiNWs with covalently linked A5-Cy3 (Fig. 5E), which showed a similar reduction in uptake (β = ~20.5% of control; Student’s t test, P < 0.01) (determined using the Cy3 fluorescence overlap of labeled SiNWs with HUVECs). In addition, the use of surface-conjugated A5-Cy3 allowed for an internal control of unmodified wires, which was not available using nonspecifically bound A5, because the electrostatically adsorbed surface coating can be transient and in equilibrium with other unmodified wires. By using specifically modified wires, A5-Cy3 coating could be maintained throughout the entire incubation, allowing for a study of the distribution of labeled versus unlabeled SiNWs within the same solution (Fig. 5D). When compared to an internal control of unmodified SiNWs, A5-Cy3–labeled wires showed highly reduced overlaps (β = ~19.1% of control; Student’s t test, P < 0.01), whereas the internal standards showed no significant difference from external controls (β = 4.1 ± 2.3 and 3.8 ± 1.5, respectively) (Fig. 5E). This offered several valuable insights. First, this indicates that HUVECs are able to discriminate between SiNWs conjugated with A5-Cy3 and those with no surface modification, allowing the cell to selectively determine which nanowires will be internalized. Second, it indicated that the presence of A5-Cy3 SiNWs in solution did not inhibit cell activity in other ways, such as hindering cell motility, because unmodified wires were able to be internalized as normal.
Collectively, this demonstrates that A5 is capable of selectively inhibiting SiNW uptake and, when combined with the high rate of macrophage uptake and cell morphology analysis, indicates that SiNWs are primarily internalized via a phagocytosis mechanism. This mechanism was further confirmed using lovastatin (Lova), a 3-hydroxy-3-methylglutaryl–coenzyme A reductase inhibitor, which has been shown to inhibit phagocytosis uptake by restricting cholesterol production (55). When administered, Lova demonstrated considerable inhibition of SiNW uptake, exhibiting a significant deviation from both an internal [χ2(9, n = 4) = 16.2, P = 0.04, ΔY36 = 22%] and an external control (~52% reduction in additional SiNW uptake after treatment) (fig. S20). This concretely links SiNW uptake with an endogenous endocytosis mechanism, showing that SiNWs are effectively internalized via a phagocytosis pathway.
DISCUSSION
Here, we have demonstrated that high–aspect ratio and label-free SiNWs can be internalized through a phagocytosis pathway. To achieve this, we have developed a robust set of methodologies that allow for the dynamic study of nanowire-cell interactions, showing for the first time that nanowires undergo an active transport process. The identification of phagocytosis as the primary mechanism of SiNW internalization is an important step in several ways. First, it demonstrates that nanowire morphology plays an important role in internalization and distinguishes high–aspect ratio nanowires from other nanoparticles, such as nanospheres with similar diameters (43). Second, it helps fit nanowire internalization into a broader, more familiar biological picture. Namely, our results suggest a very specific time-dependent pathway (Fig. 6), where SiNWs are first internalized by a membrane extension engulfing the particles (Fig. 4). From here, the SiNWs undergo active transport to the perinuclear region (Fig. 2), presumably along cytoskeleton filaments, as indicated by the burst-like transport and relatively linear trajectories (fig. S4). This results in the nanowires becoming clustered (Fig. 3A) as they are collected into larger lysosomal compartments (fig. S1). Here, we have carefully examined each of these components, placing SiNW internalization into a larger picture, which may appear familiar from other endocytosis studies (39), but has hitherto been connected to SiNW internalization. In turn, this pathway raises several intriguing possibilities. For instance, it appears that SiNWs undergo an as-yet unstudied release process from the lysosome, resulting in a distribution between the cytosol and vesicles (Fig. 1D). However, at this point, it is not clear whether this release process is the result of mechanical abrasion, because SiNWs appear to be able to puncture vesicle membranes (Fig. 4E), or whether this process is directed by the native cellular architecture. Studying this pathway has important implications in designing and implementing new functional devices in biology and medicine.
Fig. 6. Schematic overview of SiNW internalization.
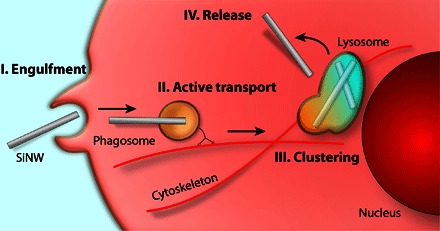
After first coming into contact with the SiNW, the cell membrane extends along the entire length of the SiNW, engulfing the particle. This results in either complete or partial encapsulation of the SiNW into a small vesicle. The SiNW is then transported to the perinuclear region for processing. Eventually, the particle, through a yet unknown process, is able to leave the lysosome and is released to the cytosol.
In addition, more work should be done in examining the exact protein receptors responsible for SiNW uptake. Although we have shown that SiNWs are internalized via a phagocytosis mechanism, within this classification, there are a large number of specific protein receptors, including opsonic, pattern recognition, and apoptotic corpse receptors (56). Although serum-free samples showed that uptake can occur without opsonization, it is possible that multiple receptors could be at play. The inhibition of SiNW uptake by Lova hints that Fc receptors may play an important role because Lova has been linked with a reduction in Fc receptor concentration (55); however, this remains to be clarified. One possible strategy to elucidate this mechanism would be to take a nonphagocytotic cell incapable of internalizing SiNWs, transfect it with the appropriate receptors, and modify it until it was able to uptake the wires. These strategies could be used to provide more cell-specific targeting of devices. Despite the need for further examination, this present study helps pave the way for using SiNWs as a platform for seamlessly integrating electronic devices with biology systems.
As we have previously mentioned, SiNWs are of particular interest for biointegration because of their excellent electronic properties, biocompatibility, and potential to incorporate a diverse set of functionalities, allowing for the realization of a large library of nanowire-based devices, for example, using nanowires as intercellular and intracellular force probes (27). Understanding the internalization pathway of SiNWs offers key insights into the single cell–level interactions and helps inform future device design. For example, one can image designing devices capable of differentiating between different cell types. The fact that neurons and cardiomyocytes rejected SiNWs, whereas other cells were capable of internalizing these materials, implies that SiNW-based therapeutic devices can be designed to specifically target certain cell types over others. In addition, it may be possible to implement a pH- or force-responsive device capable of modulating cellular activity at specific points in the pathway based on vesicle and cytoskeleton interactions. Alternatively, the partial encapsulation, or “frustrated phagocytosis,” of the particles raises the possibility of using the portion of the nanowire that is exposed to the cytosol as a delivery vector for biomolecules, which would traditionally be degraded in the harsh environment of the endosome. Although these are just a few examples, understanding the pharmacokinetics of SiNWs helps pave the way for new and exciting opportunities in seamlessly integrating electronic devices into biological systems. The ability to distribute these devices in a substrate-free manner marks an important milestone in developing active “drug-like” therapeutic and diagnostic techniques for next-generation biomolecule delivery vectors, intracellular electrodes, and photoresponsive therapies.
MATERIALS AND METHODS
Nanowire growth
SiNWs were grown using the vapor-liquid-solid mechanism in a homebuilt chemical vapor deposition system. Silicon growth substrates were first rinsed with acetone and isopropyl alcohol and then dried using N2 gas. Substrates were then oxygen plasma–cleaned (Plasma Etch PS-100LF) for 10 min at 100 W, coated in 1:3 dilute poly-l-lysine for 15 min, and then rinsed with deionized (DI) water. For SiNW growth, Au NPs between 20 and 250 nm (Ted Pella Inc.) were allowed to settle for 15 min and then rinsed once more with DI water before they were dried using N2 gas. SiNW growth conditions were as follows: 480°C, 40 torr, 60 standard cubic centimeters per minute (SCCM) of hydrogen carrier gas, and 2 SCCM of silane. To prepare samples for use with cells, SiNWs were first rinsed with hydrofluoric acid (9.8 %; Sigma-Aldrich) for 10 to 30 s before they were washed in DI water. To sterilize the SiNWs, they were then transferred to a 70% ethanol solution and kept under an ultraviolet lamp for ~30 min. After this, samples were transferred to the appropriate cell medium and sonicated for 7 min to suspend the SiNWs into solution. Typical wafer sizes were 0.5 cm2, with final working medium concentrations of ~5.0 × 106 SiNWs per milliliter. Medium solutions were then transferred to the appropriate container, SiNWs were allowed to settle overnight before the medium was aspirated off, and cell samples were added to the culture.
Cell culture
HUVECs and HASMCs were cultured using the same protocol as previously reported (27). For J774A.1 cells (Sigma-Aldrich), samples were cultured under sterile conditions in Dulbecco’s modified Eagle’s medium with glucose (4.5 g/liter), l-glutamine, and sodium pyruvate (Corning) supplemented with 10% fetal bovine serum and gentamicin solution (0.04 mg/ml) (Sigma-Aldrich). Cell cultures were maintained at 37°C and 5% CO2, with medium exchanged after 24 hours and every 48 hours thereafter. For DRG neuron and cardiomyocyte cultures, see the Supplementary Materials.
Ensemble nanowire studies
The ensemble rate of nanowire internalization was approximated using the SiNW-cell colocalization as a function of time. To achieve this, a series of both PC and DF micrographs were obtained at each time point (fig. S11, A and B, left). Samples were imaged at predetermined locations before viewing to prevent sampling bias. In a typical experiment, eight locations were examined for each time point. These images were then postprocessed using NIH (National Institutes of Health) ImageJ and converted to binary, highlighting the area encompassed by cells and nanowires (fig. S11, A and B, right). For each image, a histogram was obtained, yielding information on cell confluence and nanowire population. Using ImageJ built-in batch processing, a corresponding “AND” image was produced, which contained only those NWs that were overlapping cellular regions. A histogram of this image was then used to determine the relative SiNW-cell overlap. A representative composite overlap image (fig. S11C) shows the population of SiNWs considered outside the cell and those considered overlapping with the cells. For SiNW length studies and alternative cell line studies, SiNWs grown from 100-nm seed Au NPs were used, and overlap reporter values were recorded 24 hours after the cells were seeded. In the length-dependent study, the average length of the wires after sonication, as measured via DF microscopy, was as follows: 10 min: 9.8 μm; 20 min: 14.5 μm; 30 min: 23.1 μm; 40 min: 31.7 μm.
Endocytosis inhibitor studies
For inhibitor studies, SiNWs grown from 100-nm seed Au NPs were sonicated (7 min) into M200 medium with growth supplement (Life Technologies) and then allowed to settle overnight. After settling, HUVECs were introduced at ~15% initial confluence, and drugs were administered after 8 hours of incubation. All drugs were obtained from Sigma-Aldrich and were administered at the following concentrations (dissolved in dimethyl sulfoxide): chlorpromazine, 2.5 μg/ml; nystatin, 50 ng/ml; dynasore, 80 μM; Cyto D, 5 ng/ml; A5, 4 and 16 nM; Lova, 10 μM (NC9907790, Cayman Chemical). Throughout this process, the SiNW-cell overlap was monitored by taking alternating DF and PC micrographs every ~2 hours at random substrate locations (selected before viewing to avoid sampling bias).
A5 binding assay
SiNWs were incubated with A5-Cy3 (Sigma-Aldrich, from human placenta) at 37°C in 5% CO2 for ~24 hours in the absence and presence of serum proteins (2% fetal bovine serum) in both PBS and M200 medium. After incubation, samples were rinsed twice in fresh PBS and imaged under the same optical conditions (that is, exposure, light intensity, etc.). Fluorescent signal intensities were quantified using NIH ImageJ. To correct for background scattering, the signal intensity from a control sample containing only SiNWs (that is, no A5-Cy3) was also measured (~9% relative signal intensity). In addition, because the glass substrate could also be stained by the A5 protein, DF images were used as a mask to selectively measure intensities only from the regions containing SiNWs. Reported signal intensities were also normalized by the relative SiNW population. For uptake studies of pretreated wires, a similar process was used, with SiNWs incubated with A5-Cy3 in PBS (37°C in 5% CO2) for 20 to 60 min in the absence of serum. Samples were then rinsed using fresh M200 (with serum), and HUVECs were added and allowed to co-incubate for 24 hours. Samples were then fixed using 4% paraformaldehyde for 20 min and transferred to fresh PBS to avoid autofluorescence before they were imaged under the same conditions.
Supplementary Material
Acknowledgments
We thank P. Bezanilla for use of his facilities during primary tissue culture, B. Hissa for fruitful conversations on endocytosis pathways, K. Koehler for her assistance with confocal microscopy, and J. Jumper for technical advice in modeling 2D random walkers. Funding: This work was supported by the Air Force Office of Scientific Research (AFOSR FA9550-14-1-0175 and FA9550-15-1-0285), the NSF (NSF CAREER, DMR-1254637; NSF MRSEC, DMR 1420709), the Searle Scholars Foundation, and the University of Chicago Start-up Fund. Author contributions: J.F.Z. and B.T. conceived the idea. J.F.Z., R.P., G.M., Y.W., and M.B. carried out the experiments. J.F.Z. performed the data analysis. B.T. supervised the research. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/12/e1601039/DC1
Supplementary Text
table S1. Summary of inhibitor action on SiNW internalization.
fig. S1. Perinuclear SiNW clustering.
fig. S2. SiNW tracking algorithm.
fig. S3. Stage drift controls.
fig. S4. Linear SiNW transport trajectories.
fig. S5. Migration coupled active transport.
fig. S6. Serum-free SiNW internalization.
fig. S7. Example MSD calculation.
fig. S8. Nocodazole-inhibited SiNW transport.
fig. S9. Active tug-of-war SiNW transport.
fig. S10. Lysosome tracking.
fig. S11. SiNW-cell colocalization determination.
fig. S12. Concentration-dependent metabolic activity assay.
fig. S13. Cell line–specific SiNW-cell colocalization.
fig. S14. Ensemble HUVEC control samples.
fig. S15. Multiple internalization events.
fig. S16. Chlorpromazine-positive control.
fig. S17. High-concentration A5 study.
fig. S18. Cytosolic SiNW protein sheath formation.
fig. S19. Prestained A5 SiNWs.
fig. S20. Lova inhibitor study.
movie S1. Single-cell SiNW active transport.
movie S2. Bidirectional active tug-of-war SiNW transport.
movie S3. Macrophage internalization of SiNWs.
movie S4. SiNW membrane engulfment.
REFERENCES AND NOTES
- 1.Silva G. A., Neuroscience nanotechnology: Progress, opportunities and challenges. Nat. Rev. Neurosci. 7, 65–74 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Schroeder A., Heller D. A., Winslow M. M., Dahlman J. E., Pratt G. W., Langer R., Jacks T., Anderson D. G., Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer 12, 39–50 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Hanson L., Zhao W., Lou H.-Y., Lin Z. C., Lee S. W., Chowdary P., Cui Y., Cui B., Vertical nanopillars for in situ probing of nuclear mechanics in adherent cells. Nat. Nanotechnol. 10, 554–562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giraldo J. P., Landry M. P., Faltermeier S. M., McNicholas T. P., Iverson N. M., Boghossian A. A., Reuel N. F., Hilmer A. J., Sen F., Brew J. A., Strano M. S., Plant nanobionics approach to augment photosynthesis and biochemical sensing. Nat. Mater. 13, 400–408 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Yan R., Park J.-H., Choi Y., Heo C.-J., Yang S.-M., Lee L. P., Yang P., Nanowire-based single-cell endoscopy. Nat. Nanotechnol. 7, 191–196 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Chiappini C., De Rosa E., Martinez J. O., Liu X., Steele J., Stevens M. M., Tasciotti E., Biodegradable silicon nanoneedles delivering nucleic acids intracellularly induce localized in vivo neovascularization. Nat. Mater. 14, 532–539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dvir T., Timko B. P., Kohane D. S., Langer R., Nanotechnological strategies for engineering complex tissues. Nat. Nanotechnol. 6, 13–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.L. Marcon, R. Boukherroub, Biocompatibility of semiconducting silicon nanowires, in Semiconducting Silicon Nanowires for Biomedical Applications, J. L. Coffer, Ed. (Elsevier, ed. 1, 2014), pp. 62–85. [Google Scholar]
- 9.Luo Z., Jiang Y., Myers B. D., Isheim D., Wu J., Zimmerman J. F., Wang Z., Li Q., Wang Y., Chen X., Dravid V. P., Seidman D. N., Tian B., Atomic gold–enabled three-dimensional lithography for silicon mesostructures. Science 348, 1451–1455 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Tian B., Xie P., Kempa T. J., Bell D. C., Lieber C. M., Single-crystalline kinked semiconductor nanowire superstructures. Nat. Nanotechnol. 4, 824–829 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patolsky F., Zheng G., Lieber C. M., Fabrication of silicon nanowire devices for ultrasensitive, label-free, real-time detection of biological and chemical species. Nat. Protoc. 1, 1711–1724 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Yang C., Zhong Z., Lieber C. M., Encoding electronic properties by synthesis of axial modulation-doped silicon nanowires. Science 310, 1304–1307 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Tian B., Zheng X., Kempa T. J., Fang Y., Yu N., Yu G., Huang J., Lieber C. M., Coaxial silicon nanowires as solar cells and nanoelectronic power sources. Nature 449, 885–889 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Tian B., Cohen-Karni T., Qing Q., Duan X., Xie P., Lieber C. M., Three-dimensional, flexible nanoscale field-effect transistors as localized bioprobes. Science 329, 830–834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stern E., Vacic A., Rajan N. K., Criscione J. M., Park J., Ilic B. R., Mooney D. J., Reed M. A., Fahmy T. M., Label-free biomarker detection from whole blood. Nat. Nanotechnol. 5, 138–142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie P., Xiong Q., Fang Y., Qing Q., Lieber C. M., Local electrical potential detection of DNA by nanowire–nanopore sensors. Nat. Nanotechnol. 7, 119–125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.VanDersarl J. J., Xu A. M., Melosh N. A., Nanostraws for direct fluidic intracellular access. Nano Lett. 12, 3881–3886 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Shalek A. K., Robinson J. T., Karp E. S., Lee J. S., Ahn D.-R., Yoon M.-H., Sutton A., Jorgolli M., Gertner R. S., Gujral T. S., MacBeath G., Yang E. G., Park H., Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc. Natl. Acad. Sci. U.S.A. 107, 1870–1875 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan X., Li Y., Rajan N. K., Routenberg D. A., Modis Y., Reed M. A., Quantification of the affinities and kinetics of protein interactions using silicon nanowire biosensors. Nat. Nanotechnol. 7, 401–407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim W., Ng J. K., Kunitake M. E., Conklin B. R., Yang P., Interfacing silicon nanowires with mammalian cells. J. Am. Chem. Soc. 129, 7228–7229 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Xu A. M., Aalipour A., Leal-Ortiz S., Mekhdjian A. H., Xie X., Dunn A. R., Garner C. C., Melosh N. A., Quantification of nanowire penetration into living cells. Nat. Commun. 5, 3613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wierzbicki R., Købler C., Jensen M. R. B., Łopacińska J., Schmidt M. S., Skolimowski M., Abeille F., Qvortrup K., Mølhave K., Mapping the complex morphology of cell interactions with nanowire substrates using FIB-SEM. PLOS ONE 8, e53307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson L., Lin Z. C., Xie C., Cui Y., Cui B., Characterization of the cell–nanopillar interface by transmission electron microscopy. Nano Lett. 12, 5815–5820 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Han S.-W., Nakamura C., Kotobuki N., Obataya I., Ohgushi H., Nagamune T., Miyake J., High-efficiency DNA injection into a single human mesenchymal stem cell using a nanoneedle and atomic force microscopy. Nanomedicine 4, 215–225 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Kim D.-H., Song J., Choi W. M., Kim H.-S., Kim R.-H., Liu Z., Huang Y. Y., Hwang K.-C., Zhang Y.-w., Rogers J. A., Materials and noncoplanar mesh designs for integrated circuits with linear elastic responses to extreme mechanical deformations. Proc. Natl. Acad. Sci. U.S.A. 105, 18675–18680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozin G. A., Manners I., Fournier-Bidoz S., Arsenault A., Dream nanomachines. Adv. Mater. 17, 3011–3018 (2005). [Google Scholar]
- 27.Zimmerman J. F., Murray G. F., Wang Y., Jumper J. M., Austin J. R., Tian B., Free-standing kinked silicon nanowires for probing inter- and intracellular force dynamics. Nano Lett. 15, 5492–5498 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Zimmerman J. F., Murray G. F., Tian B., Optical determination of silicon nanowire diameters for intracellular applications. J. Phys. Chem. C 119, 29105–29115 (2015). [Google Scholar]
- 29.Zhang W., Tong L., Yang C., Cellular binding and internalization of functionalized silicon nanowires. Nano Lett. 12, 1002–1006 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Lee J.-H., Zhang A., You S. S., Lieber C. M., Spontaneous internalization of cell penetrating peptide-modified nanowires into primary neurons. Nano Lett. 16, 1509–1513 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Park H.-J., Zhang Y., Georgescu S. P., Johnson K. L., Kong D., Galper J. B., Human umbilical vein endothelial cells and human dermal microvascular endothelial cells offer new insights into the relationship between lipid metabolism and angiogenesis. Stem Cell Rev. 2, 93–102 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Wang B., Kuo J., Granick S., Bursts of active transport in living cells. Phys. Rev. Lett. 111, 208102 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Lakadamyali M., Rust M. J., Babcock H. P., Zhuang X., Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 100, 9280–9285 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cacchioli A., Ravanetti F., Alinovi R., Pinelli S., Rossi F., Negri M., Bedogni E., Campanini M., Galetti M., Goldoni M., Lagonegro P., Alfieri R., Bigi F., Salviati G., Cytocompatibility and cellular internalization mechanisms of SiC/SiO2 nanowires. Nano Lett. 14, 4368–4375 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Spira M. E., Hai A., Multi-electrode array technologies for neuroscience and cardiology. Nat. Nanotechnol. 8, 83–94 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Lee S., Leach M. K., Redmond S. A., Chong S. Y. C., Mellon S. H., Tuck S. J., Feng Z.-Q., Corey J. M., Chan J. R., A culture system to study oligodendrocyte myelination processes using engineered nanofibers. Nat. Methods 9, 917–922 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capulli A. K., MacQueen L. A., Sheehy S. P., Parker K. K., Fibrous scaffolds for building hearts and heart parts. Adv. Drug Deliv. Rev. 96, 83–102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conner S. D., Schmid S. L., Regulated portals of entry into the cell. Nature 422, 37–44 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Doherty G. J., McMahon H. T., Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Albrecht-Buehler G., The phagokinetic tracks of 3T3 cells. Cell 11, 395–404 (1977). [DOI] [PubMed] [Google Scholar]
- 41.A. Dvoretzky, P. Erdös, Some problems on random walk in space, in Proceedings of the Second Berkeley Symposium on Mathematical Statistics and Probability (Univ. California Press, 1951), pp. 353–367. [Google Scholar]
- 42.Chithrani B. D., Chan W. C. W., Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 7, 1542–1550 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Blechinger J., Bauer A. T., Torrano A. A., Gorzelanny C., Bräuchle C., Schneider S. W., Uptake kinetics and nanotoxicity of silica nanoparticles are cell type dependent. Small 9, 3970–3980 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Agarwal R., Singh V., Jurney P., Shi L., Sreenivasan S. V., Roy K., Mammalian cells preferentially internalize hydrogel nanodiscs over nanorods and use shape-specific uptake mechanisms. Proc. Natl. Acad. Sci. U.S.A. 110, 17247–17252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Roy C., Wrana J. L., Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 6, 112–126 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Brown D. M., Kinloch I. A., Bangert U., Windle A. H., Walter D. M., Walker G. S., Scotchford C. A., Donaldson K., Stone V., An in vitro study of the potential of carbon nanotubes and nanofibres to induce inflammatory mediators and frustrated phagocytosis. Carbon 45, 1743–1756 (2007). [Google Scholar]
- 47.Schinwald A., Donaldson K., Use of back-scatter electron signals to visualise cell/nanowires interactions in vitro and in vivo; frustrated phagocytosis of long fibres in macrophages and compartmentalisation in mesothelial cells in vivo. Part. Fibre Toxicol. 9, 34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan X., Krahling S., Smith D., Williamson P., Schlegel R. A., Macrophage surface expression of annexins I and II in the phagocytosis of apoptotic lymphocytes. Mol. Biol. Cell 15, 2863–2872 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie R., Gao C., Li W., Zhu J., Novakovic V., Wang J., Ma R., Zhou J., Gilbert G. E., Shi J., Phagocytosis by macrophages and endothelial cells inhibits procoagulant and fibrinolytic activity of acute promyelocytic leukemia cells. Blood 119, 2325–2334 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Callahan M. K., Halleck M. S., Krahling S., Henderson A. J., Williamson P., Schlegel R. A., Phosphatidylserine expression and phagocytosis of apoptotic thymocytes during differentiation of monocytic cells. J. Leukoc. Biol. 74, 846–856 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Callahan M. K., Williamson P., Schlegel R. A., Surface expression of phosphatidylserine on macrophages is required for phagocytosis of apoptotic thymocytes. Cell Death Differ. 7, 645–653 (2000). [DOI] [PubMed] [Google Scholar]
- 52.van Engeland M., Nieland L. J., Ramaekers F. C., Schutte B., Reutelingsperger C. P. M., Annexin V-affinity assay: A review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry 31, 1–9 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Tenzer S., Docter D., Kuharev J., Musyanovych A., Fetz V., Hecht R., Schlenk F., Fischer D., Kiouptsi K., Reinhardt C., Landfester K., Schild H., Maskos M., Knauer S. K., Stauber R. H., Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 8, 772–781 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Hwang S.-W., Tao H., Kim D.-H., Cheng H., Song J.-K., Rill E., Brenckle M. A., Panilaitis B., Won S. M., Kim Y.-S., Song Y. M., Yu K. J., Ameen A., Li R., Su Y., Yang M., Kaplan D. L., Zakin M. R., Slepian M. J., Huang Y., Omenetto F. G., Rogers J. A., A physically transient form of silicon electronics. Science 337, 1640–1644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loike J. D., Shabtai D. Y., Neuhut R., Malitzky S., Lu E., Husemann J., Goldberg I. J., Silverstein S. C., Statin inhibition of Fc receptor–mediated phagocytosis by macrophages is modulated by cell activation and cholesterol. Arterioscler. Thromb. Vasc. Biol. 24, 2051–2056 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Flannagan R. S., Jaumouillé V., Grinstein S., The cell biology of phagocytosis. Annu. Rev. Pathol. 7, 61–98 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/12/e1601039/DC1
Supplementary Text
table S1. Summary of inhibitor action on SiNW internalization.
fig. S1. Perinuclear SiNW clustering.
fig. S2. SiNW tracking algorithm.
fig. S3. Stage drift controls.
fig. S4. Linear SiNW transport trajectories.
fig. S5. Migration coupled active transport.
fig. S6. Serum-free SiNW internalization.
fig. S7. Example MSD calculation.
fig. S8. Nocodazole-inhibited SiNW transport.
fig. S9. Active tug-of-war SiNW transport.
fig. S10. Lysosome tracking.
fig. S11. SiNW-cell colocalization determination.
fig. S12. Concentration-dependent metabolic activity assay.
fig. S13. Cell line–specific SiNW-cell colocalization.
fig. S14. Ensemble HUVEC control samples.
fig. S15. Multiple internalization events.
fig. S16. Chlorpromazine-positive control.
fig. S17. High-concentration A5 study.
fig. S18. Cytosolic SiNW protein sheath formation.
fig. S19. Prestained A5 SiNWs.
fig. S20. Lova inhibitor study.
movie S1. Single-cell SiNW active transport.
movie S2. Bidirectional active tug-of-war SiNW transport.
movie S3. Macrophage internalization of SiNWs.
movie S4. SiNW membrane engulfment.


