Structured Abstract
To review studies investigating if genetic factors play a role in external apical root resorption (EARR) during orthodontic treatment. Heritability estimation in human sibpairs, comparison of multiple inbred mouse strains, human sib-pair linkage and parents-child trio association studies, and two gene (Il-1b, and P2rx7) knock out mouse models. Heritability for EARR of the maxillary central incisors concurrent with orthodontic treatment is 0.8. DBA/2J, BALB/cJ, and 129P3/J inbred mouse strains are highly susceptible (p < .05) to histological root resorption (RR) associated with orthodontic force (RRAOF), whereas A/J, C57BL/6J and SJL/J mice are resistant. Non-parametric sibling pair linkage analysis identified evidence of linkage (LOD = 2.5; p = 0.02) of EARR with microsatellite D18S64 (tightly linked to TNFRSF11A, also known as RANK). There is significant linkage disequilibrium of IL-1B (p = 0.0003), and OPG (p = 0.003) with EARR. RRAOF increases in Il1b KO (p ≤ 0.013), and increases in P2rx7 KO (p < 0.02) mice compared to wild-type. Genetic factors play a marked role in EARR concurrent with orthodontic force, accounting for one-half to two-thirds of the variation. Two pathways for this may involve: 1) activation control of osteoclasts through the ATP/P2XR7/IL-1B inflammation modulation pathway; and 2) RANK/RANKL/OPG osteoclast activation control. Histological RR occurs and is typically healed. If resorption outpaces healing, then EARR develops. Normal and parafunctional forces, as well as orthodontic forces, may add to or interact with the individual’s susceptibility to pass the threshold of developing EARR.
Keywords: interleukin-1beta, orthodontics, osteoprotegerin, P2XR7, root resorption
Introduction
Root resorption detectable histologically can be a preliminary step toward external apical root resorption (EARR) that is permanent and detectable radiographically. It is believed that when root resorption (RR) exceeds the reparative capacity of cementum, EARR ensues. Exposure of dentin increases the likelihood of osteoclastic attack and EARR, particularly if the tooth is subjected to forces from alternating directions in a parafunctional manner (1).
From 7% to 13% of individuals who have not had orthodontic treatment show some EARR (2, 3), presumably at least in part as a function of occlusal forces. There is an association of EARR in those who have not received orthodontic treatment with missing teeth, increased periodontal probing depths, and reduced crestal bone heights (3). Individuals with bruxism, chronic nail biting, and anterior open bites with concomitant tongue thrust may also show an increased extent of EARR before orthodontic treatment (4). Dental trauma, especially with re-implantation of an avulsed tooth, is also associated with increased EARR (5).
External apical RR is also increased as a pathologic consequence of orthodontic mechanical loading in some patients (6, 7). The amount of orthodontic movement is positively associated with the resulting extent of EARR (8–10). Orthodontic tooth movement, or ‘biomechanics’, has been found to account for approximately one-tenth to one-third of the total variation in EARR (11–13).
Owman-Moll and coworkers showed that individual variation overshadowed the force magnitude and the force type in defining the susceptibility to histological RR associated with orthodontic force (14). Individual variations were considerable regarding both extension and depth of histoloigcal RR within individuals, and these were not correlated to the magnitude of tooth movement achieved (15).
There is considerable individual variation in EARR associated with orthodontic treatment, indicating an individual predisposition and multifactorial etiology (16–21). Heritability estimates have shown approximately half of EARR variation concurrent with orthodontia, and almost two-thirds of maxillary central incisor EARR specifically, can be attributed to genetic variation (21, 22). A retrospective twin study on EARR found evidence for both a genetic and environmental factors influencing EARR (23). In addition, studies in a panel of different inbred mice also supported a genetic component involving multiple genes in histological RR (24, 25).
While there is a relationship between orthodontic force and RR, it is against the backdrop of previously undefined individual susceptibility. Since mechanical forces and other environmental factors do not adequately explain the variation seen among individual expressions of EARR, interest has increased on genetic factors influencing the susceptibility to EARR. The reaction to orthodontic force, including rate of tooth movement, can differ depending on the individual’s genetic background (1, 26).
ATP/P2XR7/IL-1B pathway
Clinical association of IL-1B with EARR concurrent with orthodontia
In 2003, Al-Qawasmi et al. identified an IL-1B (+3953/+3954, rs1143634) polymorphism in orthodontically treated individuals as having a role in the genetic influence on EARR. The polymorphism variation was found to account for 15% of the variation in EARR in that sample. Persons in their sample homozygous for the IL-1B allele 1 had a 5.6 fold (95% CI 1.9– 21.2) increased risk of EARR greater than 2 mm as compared with those who are not homozygous for the IL-1 beta allele 1. Data indicate that allele 1 at the IL-1B gene, known to decrease the production of IL-1 cytokine in vivo (27, 28), significantly increases the risk of EARR (29).
Il1b knockout mouse model
A murine model in which RR is induced by orthodontic force was applied to interleukin-1b knockout (Il1b−/−) mice to further investigate the role of interleukin 1-b in RR (30). Thirty-three male mice of the wild-type strain (C57BL/6J,+/+) and the Il1b knockout (B6.129-IL-1BtmChaplin) strain (31) obtained from David Chaplin (University of Alabama at Birmingham, Birmingham, AL, USA) were divided into control or treatment groups. A red Elgiloy® cobalt steel alloy 0.0058 × 0.022 inch open coil spring (Rocky Mountain Orthodontics, Denver, CO, USA) was used to apply the orthodontic force as described previously (24). The number of control (C) or treated (T) mice per group were as follows: wild-type (C = 7, T = 8), and knockout (C = 8, T = 10). Both the control and treated animals were fed a diet of finely milled mouse chow ad libitum to minimize discomfort and appliance distortion in the treated mice. The Wilcoxon rank-sum non-parametric test was used to evaluate differences in the mean root resorption (MRR), root resorption attributable to orthodontic force (RRAOF = force value-baseline value) and tartrate resistant acid phosphatase (TRAP) measures associated with Il1b gene status, sex, or treatment status. Assuming α = 0.05 and using a Bonferroni correction to adjust for four distinct non-parametric tests for each phenotypic hypotheses yielded an adjusted significance level (α*) of 0.0127. Finally, the Spearman correlation coefficient was computed for TRAP and RRAOF to quantify the association between these two variables (α = 0.05).
Nine days into the experiment, the mice in all groups were euthanized. Immediately after that the maxillae were removed, fixed, and demineralized. Paraffin embedded specimens were cut into parasagittal sections of 5 μm thickness. Evaluation of RR on the mesial aspect of the mesial root of the maxillary first molar on eight H&E stained sections was analyzed using light microscopy at X100 magnification as described elsewhere (32). Four additional sections also selected randomly were stained for TRAP and evaluated as before (24).
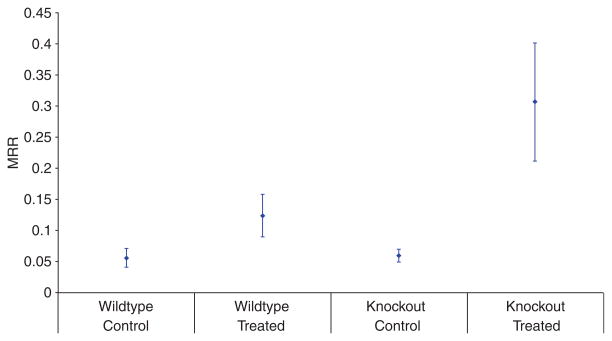
There was no significant difference in MRR (p = 0.64) between the untreated wild-type and untreated knockout mice. There was a significant difference in both the wild-type and knockout animals with treatment compared to their respective controls. There was also a significant difference between the treated wild-type and the treated knockout mice, with the knockout mice having approximately three times the percent MRR of the treated wild-type mice (Fig. 1). Thus, the absence of IL-1β cytokine, a typical mediator of the inflammatory response, increased orthodontically-induced RR. Interestingly, this effect was not mediated by significant changes in the number of TRAP positive cells near the root surface.
Fig. 1.
Root resorption attributed to orthodontic force (RRAOF) for the two groups of treated mice. Each point represents mean RRAOF and the vertical bars represents one standard deviation above and below mean value.
P2rx7 (P2x7r) knockout mouse model
Viecilli, Katona et al. refined the murine model to bring the force (and the resulting stress) applied to a clinically relevant level, and to evaluate the effect of force and RR using histology, and in three dimensions finite element modeling and μCT imaging, to investigate the effect of orthodontic force in the mouse in which the gene (P2rx7) for purinergic receptor P2X, ligand-gated ion channel, seven has been inactivated (33–35). Similar to the findings in the Il1b knockout mouse, there was no difference at baseline between the wild-type and knockout mice RR, while the application of force resulted in a significant increase in wild-type RR, and in addition a significant (p < 0.02) increase in RR in the knockout mice with force applied over the force applied wild-type mice.
Recently the role of ATP and its cognate receptors, including purinergic receptor P2X, ligand-gated ion channel, seven, in the inflammatory process has been shown to involve the metabolism of apoptic and necrotic tissue (36). Following mechanical trauma damaged cells release ATP that leads to the activation of the receptor on the cell surface of macrophages and some other cell types, which in turn releases interleukin-1 cytokines. The released cytokines affect non-bone marrow derived cells, which, in turn, release chemo-attractants for neutrophils and lymphocytes (37). The neutrophils can act quickly to eliminate apoptotic cells and prevent further necrosis. A failure or decrease in this process results in an attenuated acute inflammatory response that may not resolve, resulting in an overwhelming chronic response with generalized tissue damage (36, 38).
Macrophages from P2rx7 knockout mice are unable to respond to extracellular ATP. P2rx7 knockout mice primed with lipopolysaccharides and challenged with ATP in vivo failed to generate significant levels of interleukin-1 beta (39). The finding that RR increases with force in both Il1b and P2rx7 knockout mice further strengthens the evidence that interleukin-1 beta may in some cases play a role in EARR, and indicates that variation in purinergic receptor P2X, ligand-gated ion channel, 7, as well as other proteins involved in the maturation and release of interleukin-1 beta, could also be a factor. The observation that the RR in both the Il1b and P2rx7 knockout mice is not statistically significant from that seen in their respective wild-type mice before orthodontic force is applied, but does significantly increase with orthodontic force, provides evidence for an interaction between genotype and environmental factors that influence RR.
RANK/RANKL/OPG pathway
Osteoblasts and stromal stem cells express receptor activator of NF-κ B ligand (RANKL), which binds to its receptor activator of nuclear factor-κ B (RANK), on the surface of osteoclasts and their precursors. This regulates the differentiation of precursors into multinucleated osteoclasts and osteoclast activation and survival both normally and in most pathologic conditions associated with increased bone resorption. Osteoprotegerin (OPG, coded for by the TNFRSF11B gene) is secreted by osteoblasts and osteogenic stromal stem cells and protects from excessive bone resorption by binding to RANKL and preventing it from interacting with RANK (40).
Familial expansile osteolysis (FEO, OMIM no. 174810) is a rare, autosomal dominant bone disorder characterized by osteolytic lesions, which develop usually in the long bones during early adulthood, and can also result in spontaneous resorption of teeth and loss of the dentition. FEO is caused by mutations in the TNFRSF11A gene that encodes RANK (41). Based upon this condition and the importance of the RANK/RANKL/OPG pathway in the control of osteoclast activation, TNFRSF11A (RANK) is a candidate gene for EARR.
D18S64, which is tightly linked to TNFRSF11A (RANK)
Non-parametric sibling pair linkage analysis identified evidence of linkage (LOD = 2.5; p = 0.02) of EARR affecting the maxillary central incisor with the microsatellite marker D18S64, which is tightly linked to TNFRSF11A (RANK). This indicates that the TNFRSF11A locus, or another tightly linked gene, is associated with EARR. This was the first genetic data in a clinical setting to suggest that the RANK/RANKL/OPG pathway may be a factor in some patients with EARR (42).
TNFRSF11B (Osteoprotegerin)
Osteoprotegerin knockout mice subjected to orthodontic forces have a significant increase in osteoclasts and lower bone mineral density (43, 44). Increases in OPG production inhibit orthodontic tooth movement in rats with a decrease in osteoclasts (45, 46). In addition, OPG is associated with physiologic and pathologic RR (47, 48). More evidence of the RANK/RANKL/OPG pathway being involved with some cases of EARR comes from a clinical study involving the TNFRSF11B (OPG) gene (49).
This study evaluated the association between the single nucleotide polymorphism (SNP) rs2073618 of the TNFRSF11B (OPG) gene and EARR in orthodontically treated patients. This TNFRSF11B SNP is located in the first of five exons, changing the nucleotide (G → C) at position 1181 in the third codon, resulting in the third amino acid changing from lysine (Lys) to asparagine (Asn). This SNP is a factor in bone mineral density and the susceptibility to Paget’s disease of bone (50, 51). Indiana University Institutional Review Board review approved informed consent was obtained from all participants. A total of 135 Caucasian subjects were studied.
Radiographic evaluation of EARR took place by measuring the maxillary central incisors. Pre- and post-treatment occlusal radiographs were scanned and measurements made using Adobe Photoshop CS Version 8.0 (Adobe, Seattle, WA, USA). The rule-of-three formula was used to quantify EARR using the median CEJ (11, 52).
To assess the reproducibility and method error of the radiographic measurements, double measurements were made 2 months apart on 24 randomly selected occlusal radiographs. The error for EARR calculations on the occlusal radiographs was 0.18 mm using the equation Sx = √(ΣD2/2N), where Sx is the error, D is the difference between the double measurement, and N is the number of double measurements (53).
For DNA analysis the inside of the cheek was scraped 10 times with two sterile nylon bristle brushes. The cells obtained underwent DNA isolation with the Puregene method (Gentra Systems, Minneapolis, MN, USA) and were stored at −80°C.
Automated polymerase chain reaction and allelic discrimination using the 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) and Taqman® polymerase probes and primers determined genotypes (G,G; G,C; or C,C).
All analyzes used the greater of the EARR measurements for the two incisors. Stepwise linear regression analysis was employed using treatment time, overjet, overbite, Angle molar classification and sex to identify significant covariates with EARR. A p-value of 0.10 or less was required for retention in the model. Significant covariates were used in all subsequent analyzes. Analyzes were performed categorizing EARR measurements as either affected (EARR ≥ 2 mm) or unaffected (EARR < 2 mm).
The mean age of the patients pre-treatment was 14.6 years (±6.9 SD). The average interval between pretreatment and post-treatment radiographs (treatment time) was 1.6 years (±0.5 SD). Based on Hardy–Weinberg equilibrium the expected and observed counts for OPG genotypes were not significantly different using the chi-squared test (χ21 = 0.043; p = 0.84).
Regression analysis indicated that sex, overjet, overbite, and molar (Angle) classification were poor predictors for EARR. Therefore these covariates were excluded from the linear regression test based on p > 0.10. The length of treatment (p = 0.009) variable was used in subsequent statistical models, and indicated that increases in length of treatment resulted in increased EARR.
Logistic regression of the three genotype groups found the odds ratio was 1.9 with a 95% confidence interval of (1.1, 3.3). That is, for each copy of allele C a person was 1.9 times more likely to be affected. Combining the two least affected genotype groups, the odds ratio was 2.8 with a 95% confidence interval of (1.2, 6.7) indicating a person with a C,C genotype is 2.8 times more likely to be affected than a person with a G,G or G,C genotype. Both the three and two group logistic regression analyzes obtained a statistically significant effect for the OPG genotype (both p = 0.02).
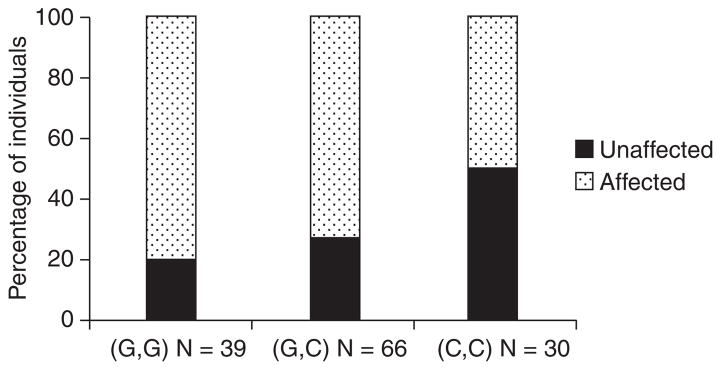
The frequencies for affection status by genotype are in shown in Fig. 2. Using the three group OPG genotypes, a chi-square test yielded (χ21 = 8.5339; p = 0.003); indicating a significant association between affection status and the OPG genotype. Similar results were obtained for the two group genotype test (χ21 = 8.3680; p = 0.004). The data indicates the G1181C OPG polymorphism accounts for approximately 8% of total EARR variation in the sample.
Fig. 2.
The percentage of individuals affected (external apical root resorption ≥ 2 mm in at least one maxillary central incisor) by TNFRSF11B Osteoprotegerin (OPG) single nucleotide polymorphism rs2073618 genotype. There is a significant association between affection status and the OPG genotype. (χ21 = 8.5339; p = 0.003).
Conclusions
Genetic factors play a marked role in EARR concurrent with orthodontic force, accounting for one-half to two-thirds of the variation. Two pathways for this may involve: 1) activation control of osteoclasts through the ATP/P2XR7/IL-1B inflammation modulation pathway; and 2) RANK/RANKL/OPG osteoclast activation control pathway. Further research into the association of orthodontic treatment and genetic variation, particularly in the genes that code for proteins involved in the ATP/P2XR7/IL-1B and RANK/RANKL/OPG pathways are likely to further clarify the genetic factors associated with EARR concurrent with orthodontic treatment.
Clinical relevance
External apical root resorption can happen with or without orthodontic treatment. The increased incidence in orthodontic patients can result in the orthodontist being blamed for its occurrence, presumably because of too great a force being placed on the teeth, and or moving the teeth from trabecular bone into more dense cancellous bone. It had been recognized that some patients were more susceptible to EARR than others, and that sometimes this tendency ‘ran in families’. Research in this area indicates that the patient’s genetic makeup has a substantial influence on EARR, indicating that it is a complex trait.
References
- 1.Abass SK, Hartsfield JK., Jr Orthodontics and external apical root resorption. Semin Orthod. 2007;13:246–56. [Google Scholar]
- 2.Rudolph CE. A comparative study in root resorption in permanent teeth. J Am Dent Assoc. 1936;23:822–6. [Google Scholar]
- 3.Harris EF, Robinson QC, Woods MA. An analysis of causes of apical root resorption in patients not treated orthodontically. Quintessence Int. 1993;24:417–28. [PubMed] [Google Scholar]
- 4.Harris EF, Butler ML. Patterns of incisor root resorption before and after orthodontic correction in cases with anterior open bites. Am J Orthod Dentofacial Orthop. 1992;101:112–9. doi: 10.1016/0889-5406(92)70002-R. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson M, Kinirons MJ. Factors affecting the time of onset of resorption in avulsed and replanted incisor teeth in children. Dent Traumatol. 2001;17:205–9. doi: 10.1034/j.1600-9657.2001.170503.x. [DOI] [PubMed] [Google Scholar]
- 6.Brezniak N, Wasserstein A. Root resorption after orthodontic treatment: part 1. literature review. Am J Orthod Dentofacial Orthop. 1993;103:62–6. doi: 10.1016/0889-5406(93)70106-X. [DOI] [PubMed] [Google Scholar]
- 7.Brezniak N, Wasserstein A. Root resorption after orthodontic treatment: part 2. literature review. Am J Orthod Dentofacial Orthop. 1993;103:138–46. doi: 10.1016/S0889-5406(05)81763-9. [DOI] [PubMed] [Google Scholar]
- 8.DeShields RW. A study of root resorption in treated Class II, division I malocclusions. Angle Orthod. 1969;39:231–45. doi: 10.1043/0003-3219(1969)039<0231:ASORRI>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 9.Sharpe W, Reed B, Subtelny JD, Polson A. Orthodontic relapse, apical root resorption, and crestal alveolar bone levels. Am J Orthod Dentofacial Orthop. 1987;91:252–8. doi: 10.1016/0889-5406(87)90455-0. [DOI] [PubMed] [Google Scholar]
- 10.Parker RJ, Harris EF. Directions of orthodontic tooth movements associated with external apical root resorption of the maxillary central incisor. Am J Orthod Dentofacial Orthop. 1998;114:677–83. doi: 10.1016/s0889-5406(98)70200-8. [DOI] [PubMed] [Google Scholar]
- 11.Linge L, Linge BO. Patient characteristics and treatment variables associated with apical root resorption during orthodontic treatment. Am J Orthod Dentofacial Orthop. 1991;99:35–43. doi: 10.1016/S0889-5406(05)81678-6. [DOI] [PubMed] [Google Scholar]
- 12.Baumrind S, Korn EL, Boyd RL. Apical root resorption in orthodontically treated adults. Am J Orthod Dentofacial Orthop. 1996;110:311–20. doi: 10.1016/s0889-5406(96)80016-3. [DOI] [PubMed] [Google Scholar]
- 13.Horiuchi A, Hotokezaka H, Kobayashi K. Correlation between cortical plate proximity and apical root resorption. Am J Orthod Dentofacial Orthop. 1998;114:311–8. doi: 10.1016/s0889-5406(98)70214-8. [DOI] [PubMed] [Google Scholar]
- 14.Owman-Moll P, Kurol J, Lundgren D. Continuous versus interrupted continuous orthodontic force related to early tooth movement and root resorption. Angle Orthod. 1995;65:395–401. doi: 10.1043/0003-3219(1995)065<0395:CVICOF>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 15.Kurol J, Owman-Moll P, Lundgren D. Time-related root resorption after application of a controlled continuous orthodontic force. Am J Orthod Dentofacial Orthop. 1996;110:303–10. doi: 10.1016/s0889-5406(96)80015-1. [DOI] [PubMed] [Google Scholar]
- 16.Massler M, Malone AJ. Root resorption in human permanent teeth: a Roentgenographic study. Am J Orthod. 1954;40:619–33. [Google Scholar]
- 17.Massler M, Perreault JG. Root resorption in the permament teeth of young adults. J Dent Child. 1954;21:158–64. [Google Scholar]
- 18.Reitan K. Some factors determining the evaluation of forces in orthodontics. Am J Orthod. 1957;43:32–45. [Google Scholar]
- 19.Newman WG. Possible etiologic factors in external root resorption. Am J Orthod. 1975;67:522–39. doi: 10.1016/0002-9416(75)90298-5. [DOI] [PubMed] [Google Scholar]
- 20.Sameshima GT, Sinclair PM. Predicting and preventing root resorption: part I. diagnostic factors. Am J Orthod Dentofacial Orthop. 2001;119:505–10. doi: 10.1067/mod.2001.113409. [DOI] [PubMed] [Google Scholar]
- 21.Harris EF, Kineret SE, Tolley EA. A heritable component for external apical root resorption in patients treated orthodontically. Am J Orthod Dentofacial Orthop. 1997;111:301–9. doi: 10.1016/s0889-5406(97)70189-6. [DOI] [PubMed] [Google Scholar]
- 22.Hartsfield JK, Everett ET, Al-Qawasmi RA. Genetic factors in external apical root resorption and orthodontic treatment. Crit Rev Oral Biol Med. 2004;15:115–22. doi: 10.1177/154411130401500205. [DOI] [PubMed] [Google Scholar]
- 23.Ngan DC, Kharbanda OP, Byloff FK, Darendeliler MA. The genetic contribution to orthodontic root resorption: a retrospective twin study. Aust Orthod J. 2004;20:1–9. [PubMed] [Google Scholar]
- 24.Al-Qawasmi RA, Hartsfield JK, Jr, Everett ET, Weaver MR, Foroud TM, Faust DM, et al. Root resorption associated with orthodontic force in inbred mice: genetic contributions. Eur J Orthod. 2006;28:13–9. doi: 10.1093/ejo/cji090. [DOI] [PubMed] [Google Scholar]
- 25.Abass S, Hartsfield JK, Jr, Al-Qawasmi R, Everett ET, Foroud T, Roberts WE. Inheritance of susceptibility to root resorption associated with orthodontic force in mice. Am J Orthod Dentofacial Orthop. 2008;134:742–50. doi: 10.1016/j.ajodo.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 26.Iwasaki LR, Crouch LD, Nickel JC. Genetic factors and tooth movement. Semin Orthod. 2008;14:135–45. [Google Scholar]
- 27.Pociot F, Molvig J, Wogensen L, Worsaae H, Nerup J. A TaqI polymorphism in the human interleukin-1 beta (IL-1 beta) gene correlates with IL-1 beta secretion in vitro. Eur J Clin Invest. 1992;22:396–402. doi: 10.1111/j.1365-2362.1992.tb01480.x. [DOI] [PubMed] [Google Scholar]
- 28.di Giovine FS, Cork MJ, Crane A, Mee JB, Duff GW. Novel genetic association of an IL-1B gene variation a+3953 with IL-1B protein production and psoriasis [abstract] Cytokine. 1995;7:606. [Google Scholar]
- 29.Al-Qawasmi RA, Hartsfield JK, Jr, Everett ET, Flury L, Liu L, Foroud TM, et al. Genetic predisposition to external apical root resorption. Am J Orthod Dentofacial Orthop. 2003;123:242–52. doi: 10.1067/mod.2003.42. [DOI] [PubMed] [Google Scholar]
- 30.Al-Qawasmi RA, Hartsfield JK, Hartsfield JK, Jr, Everett ET, Weaver MR, Foroud TM, et al. Root resorption associated with orthodontic force in IL-1beta knockout mouse. J Musculoskelet Neuronal Interact. 2004;4:383–5. [PubMed] [Google Scholar]
- 31.Shornick LP, De Togni P, Mariathasan S, Goellner J, Strauss-Schoenberger J, Karr RW, et al. Mice deficient in IL-1beta manifest impaired contact hypersensitivity to trinitrochlorobenzone. J Exp Med. 1996;183:1427–36. doi: 10.1084/jem.183.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu LH, Lee K, Imoto S, Kyomen S, Tanne K. Histological and histochemical quantification of root resorption incident to the application of intrusive force to rat molars. Eur J Orthod. 1999;21:57–63. doi: 10.1093/ejo/21.1.57. [DOI] [PubMed] [Google Scholar]
- 33.Viecilli RF, Katona TR, Chen J, Hartsfield JK, Jr, Roberts WE., Jr Orthodontic mechanotransduction and the role of the P2X7 receptor. Am J Orthod Dentofacial Orthop. 2009 doi: 10.1016/j.ajodo.2008.10.018. in press. [DOI] [PubMed] [Google Scholar]
- 34.Viecilli RF, Katona TR, Chen J, Hartsfield JK, Jr, Roberts WE. Three-dimensional mechanical environment of orthodontic tooth movement and root resorption. Am J Orthod Dentofacial Orthop. 2008;133:791, e11–26. doi: 10.1016/j.ajodo.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 35.Viecilli RF, Katona TR, Chen J, Hartsfield JK, Jr, Roberts WE. Comparison of dentoalveolar morphology in WT and P2X7R KO mice for the development of biomechanical orthodontic models. Anat Rec. 2009;292:292–8. doi: 10.1002/ar.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lister MF, Sharkey J, Sawatzky DA, Hodgkiss JP, Davidson DJ, Rossi AG, et al. The role of the purinergic P2X7 receptor in inflammation. J Inflamm (Lond) 2007;4:5–18. doi: 10.1186/1476-9255-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Brosnan CF. Regulation of immune response by P2X7 receptor. Crit Rev Immunol. 2006;26:499–513. doi: 10.1615/critrevimmunol.v26.i6.30. [DOI] [PubMed] [Google Scholar]
- 38.Labasi JM, Petrushova N, Donovan C, McCurdy S, Lira P, Payette MM, et al. Absence of the P2X7 receptor alters leukocyte function and attenuates an inflammatory response. J Immunol. 2002;168:6436–45. doi: 10.4049/jimmunol.168.12.6436. [DOI] [PubMed] [Google Scholar]
- 39.Solle M, Labasi J, Perregaux DG, Stam E, Petrushova N, Koller BH, et al. Altered cytokine production in mice lacking P2X(7) receptors. J Biol Chem. 2001;276:125–32. doi: 10.1074/jbc.M006781200. [DOI] [PubMed] [Google Scholar]
- 40.Boyce BF, Xing L. The RANKL/RANK/OPG pathway. Curr Osteoporos Rep. 2007;5:98–104. doi: 10.1007/s11914-007-0024-y. [DOI] [PubMed] [Google Scholar]
- 41.Hughes AE, Ralston SH, Marken J, Bell C, MacPherson H, Wallace RG, et al. Mutations in TNFRSF11A, affecting the signal peptide of RANK, cause familial expansile osteolysis. Nat Genet. 2000;24:45–8. doi: 10.1038/71667. [DOI] [PubMed] [Google Scholar]
- 42.Al-Qawasmi RA, Hartsfield JK, Jr, Everett ET, Flury L, Liu L, Foroud TM, et al. Genetic predisposition to external apical root resorption in orthodontic patients: linkage of chromosome-18 marker. J Dent Res. 2003;82:356–60. doi: 10.1177/154405910308200506. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki T. Differentiation and functions of osteoclasts and odontoclasts in mineralized tissue resorption. Microsc Res Tech. 2003;61:483–95. doi: 10.1002/jemt.10370. [DOI] [PubMed] [Google Scholar]
- 44.Oshiro T, Shiotani A, Shibasaki Y, Sasaki T. Osteoclast induction in periodontal tissue during experimental movement of incisors in osteoprotegerin-deficient mice. Anat Rec. 2002;266:218–25. doi: 10.1002/ar.10061. [DOI] [PubMed] [Google Scholar]
- 45.Kanzaki H, Chiba M, Takahashi I, Haruyama N, Nishimura M, Mitani H. Local OPG gene transfer to periodontal tissue inhibits orthodontic tooth movement. J Dent Res. 2004;83:920–5. doi: 10.1177/154405910408301206. [DOI] [PubMed] [Google Scholar]
- 46.Dunn MD, Park CH, Kostenuik PJ, Kapila S, Giannobile WV. Local delivery of osteoprotegerin inhibits mechanically mediated bone modeling in orthodontic tooth movement. Bone. 2007;41:446–55. doi: 10.1016/j.bone.2007.04.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Low E, Zoellner H, Kharbanda OP, Darendeliler MA. Expression of mRNA for osteoprotegerin and receptor activator of nuclear factor kappa beta ligand (RANKL) during root resorption induced by the application of heavy orthodontic forces on rat molars. Am J Orthod Dentofacial Orthop. 2005;128:497–503. doi: 10.1016/j.ajodo.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 48.Fukushima H, Kajiya H, Takada K, Okamoto F, Okabe K. Expression and role of RANKL in periodontal ligament cells during physiological root-resorption in human deciduous teeth. Eur J Oral Sci. 2003;111:346–52. doi: 10.1034/j.1600-0722.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- 49.Shank SB, Shank K, Caudill R, Foroud T, Wetherill L, Weaver M, et al. Evaluation of SNPs in orthodontic patients with root resorption. J Dent Res. 2007;86(Spec Iss A):abstract #1922. URL http://iadr.confex.com/iadr/2007orleans/techprogram/abstract_92981.htm. [Google Scholar]
- 50.Daroszewska A, Hocking LJ, McGuigan FE, Langdahl B, Stone MD, Cundy T, et al. Susceptibility to Paget’s disease of bone is influenced by a common polymorphic variant of osteoprotegerin. J Bone Miner Res. 2004;19:1506–11. doi: 10.1359/JBMR.040602. [DOI] [PubMed] [Google Scholar]
- 51.Zhao HY, Liu JM, Ning G, Zhao YJ, Zhang LZ, Sun LH, et al. The influence of Lys3Asn polymorphism in the osteoprotegerin gene on bone mineral density in Chinese postmenopausal women. Osteoporos Int. 2005;16:1519–24. doi: 10.1007/s00198-005-1865-9. [DOI] [PubMed] [Google Scholar]
- 52.Brezniak N, Goren S, Zoizner R, Dinbar A, Arad A, Wasserstein A, et al. A comparison of three methods to accurately measure root length. Angle Orthod. 2004;74:786–91. doi: 10.1043/0003-3219(2004)074<0786:ACOTMT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 53.Dahlberg G. Statistical Methods for Medical and Biological Students. London: George Allen & Unwin Ltd; 1940. [Google Scholar]