Abstract
Enveloped viruses infect target cells by fusing their membrane with cellular membrane through a process that is mediated by specialized viral glycoproteins. The inefficient and highly asynchronous nature of viral fusion complicates studies of virus entry on a population level. Single virus imaging in living cells has become an important tool for delineating the entry pathways and for mechanistic studies of viral fusion. We have previously demonstrated that incorporation of fluorescent labels into the viral membrane and trapping fluorescent proteins in the virus interior enables the visualization of single virus fusion in living cells. Here, we implement a new approach to non-invasively label the viral membrane glycoproteins through metabolic incorporation of unnatural sugars followed by click-reaction with organic fluorescent dyes. This approach allows for efficient labeling of diverse viral fusion glycoproteins on the surface of HIV pseudoviruses. Incorporation of a content marker into surface-labeled viral particles enables sensitive detection of single virus fusion with live cells.
Keywords: Retrovirus, Vesicular Stomatitis virus, metabolic labeling, unnatural sugar, click chemistry, live cell imaging
1. Introduction
Enveloped viruses infect cells by fusing their envelope membrane with a host cell membrane, a process that culminates in the release of the nucleocapsid into the cytoplasm. Viral envelope glycoprotein-mediated merger of viral and cellular membranes is generally asynchronous and relatively inefficient. As a result of this, most viruses fail to productively enter the cells and are eventually degraded. Single virus imaging has emerged as a powerful tool to delineate the virus entry pathways into host cells in spite of virus heterogeneity and the stochastic nature of viral fusion. A number of virus labeling strategies suitable for imaging single particle entry have been introduced and validated in the recent years (reviewed in (Brandenburg and Zhuang, 2007; Huang and Xie, 2014)). Among these, tagging viral proteins with GFP or other fluorescent proteins and incorporation of lipophilic dyes into the viral membrane have been most widely implemented (see for example (Burdick, Hu, and Pathak, 2013; Ewers et al., 2005; Floyd et al., 2008; Lakadamyali et al., 2003; Lampe et al., 2007; McDonald et al., 2002; Miyauchi et al., 2009)).
In order to identify particles that undergo fusion, which is a prerequisite for the release of the nucleocapsid into the cytosol, viruses must be co-labeled with two distinct viral determinants – one that is released upon fusion (referred to as content marker) and a reference marker that is, at least transiently, retained by a virus or remains at the site of viral fusion (Campbell et al., 2007; Dale et al., 2011; Miyauchi et al., 2009; Padilla-Parra et al., 2013). Retroviruses are ideally suited for introducing a releasable viral content marker into virions, since their Gag polyprotein is cleaved at several locations by the viral protease upon virus maturation (Freed, 2001; Sundquist and Krausslich, 2012). Insertion of a fluorescent protein flanked by a protease cleavage site produces free fluorescent proteins that are released into the cytoplasm upon virus-cell fusion (Dale et al., 2011; Miyauchi et al., 2009; Padilla-Parra et al., 2013). Two strategies have been employed to provide a reference marker for tracking loss of viral content – labeling the retroviral membrane or the viral core (Albanese et al., 2008; Burdick et al., 2013; Campbell et al., 2007; Lampe et al., 2007; McDonald et al., 2002; Melikyan et al., 2005; Miyauchi et al., 2009; Padilla-Parra et al., 2013; Zhou et al., 2012).
Labeling of proteins in living cells is an active area of research. The goal is to achieve non-invasive, site-specific labeling with different photostable fluorescent dyes. The labeling strategies include direct labeling with amine-reactive dyes (for indiscriminate labeling of proteins) and introduction of various tags (biotin, SNAP, Halo, tetracysteine and others) for subsequent site-directed labeling with fluorescent probes (reviewed in (Crivat and Taraska, 2012; Dean and Palmer, 2014)). These approaches have been adapted for labeling of enveloped viruses (Arhel et al., 2006; Joo et al., 2008; Joo et al., 2010; Mengistu et al., 2015; Munro et al., 2014). Successful insertion of fluorescent proteins into viral fusion proteins has also been reported (Lehmann et al., 2005; Nakane, Iwamoto, and Matsuda, 2015). These strategies involve genetic or chemical modifications of proteins that could compromise the function of metastable viral fusion proteins (Munro et al., 2014; Nakane et al., 2015). We therefore sought to label the variable carbohydrate chains of viral glycoproteins – a modification that is well tolerated (Chu, Oum, and Carrico, 2015).
Copper-catalyzed azide-alkyne cycloaddition (CuAAC) (“click chemistry”) has revolutionized the field of bioconjugation (Bertozzi, 2011; Best, 2009). Click chemistry is a highly selective chemical reaction that proceeds in the presence of a wide variety of other functionalities in the complex biological milieu (i.e., is bioorthogonal), and is well suited for the physiological reaction conditions such as aqueous solutions, pH ~7.0 and physiological temperatures. Azide and alkyne moieties are readily introducible into biomolecules due to their small size and biological inertness. CuAAC has been adopted to label living systems (Baskin et al., 2010; Breidenbach et al., 2010; Chang et al., 2009; Prescher and Bertozzi, 2005) with a combination of selective metabolic engineering techniques, which include cellular (Fernandez-Suarez et al., 2007; Luchansky et al., 2003) and viral surfaces (Banerjee et al., 2010; Banerjee et al., 2011a; Banerjee et al., 2011b; Oum and Carrico, 2012). However, the requirement for a cytotoxic cuprous catalyst restricts the utility of CuAAC. Therefore, copper-free “click” reactions, such as the Staudinger ligation of azides with functionalized phosphines, strain-promoted alkyne-azide cycloaddition (SPAAC) and strain-promoted inverse-electron-demand Diels-Alder cycloaddition (SPIEDAC), have been developed (Arumugam et al., 2011; Carpenter, Hausner, and Sutcliffe, 2011; Nikic et al., 2014; Sachin et al., 2012) as alternatives.
Two recent studies (Chu et al., 2015; Zhao et al., 2015) have demonstrated the utility of metabolic incorporation of peracylated azidomannosamine (Ac4ManNAz) into viral glycoproteins. Metabolically labeled glycoproteins on the surface of lentiviral particles were selectively modified using click chemistry in order to retarget pseudoviruses to specific cell types. This approach has also enabled fluorescence labeling of the measles virus glycoproteins (Zhao et al., 2015). Here, we sought to explore the utility of click-labeling azido sugars on diverse viral proteins for single particle imaging in living cells. Ac4ManNAz metabolically incorporated into viral glycoproteins was successfully click-labeled with fluorescent dyes without considerably compromising their function. This labeling strategy combined with incorporation of a viral content marker enables reliable visualization of single viral fusion events.
2. Materials and methods
2.1. Reagents and cells
All chemicals were obtained from commercial sources and used without further purification. Bafilomycin A1 and neuraminidase were obtained from Sigma-Aldrich (St Louis, MO). Clickable dyes, Click-iT® Alexa Fluor® 488 DIBO and Click-iT® Alexa Fluor® 647 DIBO alkyne were purchased from Invitrogen (Carlsbad, CA). HEK 293T/17 cells were obtained from ATCC (Manassas, VA). HeLa-derived indicator TZM-bl cells expressing CD4, CXCR4 and CCR5 were obtained from NIH AIDS Reference Reagent Program (donated by Drs. J.C. Kappes and X. Wu (Wei et al., 2002)). TZM-bl/TVA950 cells expressing high levels of the TVA950 receptor for Avian Sarcoma and Leukosis Virus subtype A (ASLV-A) have been described previously (Padilla-Parra et al., 2012a; Padilla-Parra et al., 2012b). CV-1 cells expressing CD4 and CXCR4 were kindly provided by Dr. D. Kabat (Kozak et al., 1997). HEK 293T/17, TZM-bl and CV-1/CD4/CXCR4 were grown in Dulbecco’s Modified Eagle Medium (DMEM, Cellgro, Manassas, VA) supplemented with 10% FBS (HyClone Laboratories, Logan, UT), 100 U/ml penicillin/streptomycin (Gemini Bio-Products, West Sacramento, CA). Growth medium for HEK 293T/17 also contained 0.5 mg/ml G418 sulfate (Cellgro). CV-1/TVA950 cells were grown in DMEM supplemented with 10% Cosmic Calf Serum (HyClone) and 100 U/ml penicillin/streptomycin.
2.2. Pseudovirus production
Fluorescently labeled pseudoviruses bearing HIV-1 HXB2 Env glycoprotein (HXB2pp) were produced as described previously (Demirkhanyan et al., 2012; Desai et al., 2014). Briefly, HEK293T/17 cells were transfected with 1 μg of pR8ΔEnv, 2 μg of HIV-1 Gag-imCherry, 2 μg of YFP-Vpr, 1 μg of pcRev, and 4 μg of pCAGGS-HXB2 (encoding for HXB2 Env) using JetPRIME reagent (Polyplus-transfection SA, NY). Particles pseudotyped with ASLV Env or VSV-G were produced using the above mixture of plasmids in which HXB2 Env vector was replaced with 5 μg of a plasmid expressing either the cytoplasmic tail-deleted ASLV-A Env or 3 μg of PMDG-VSV-G, respectively. At 12 h post-transfection, the transfection medium was removed, and cells were further cultivated in fresh phenol red-free complete growth medium. Virus-containing medium was collected 48 h post-transfection, passed through a 0.45 μm filter, aliquoted and stored at −80 °C. The infectious titer was determined by β-galactosidase or luciferase assays, as described previously (Demirkhanyan et al., 2012; Miyauchi et al., 2009). Briefly, TZM-bl or TZM-bl//TVA950 cells were seeded in 96-well black/clear bottom plates at a density of 1×104 cells/well one day before infection. Cells were spinoculated with freshly diluted viral stocks (1 ng of p24, as measured by ELISA) for 30 min at 3,000 rpm, 4 °C, and cultured in a CO2 incubator for 2 days. The expression levels of luciferase transgene were determined by a top-count luminometer (PerkinElmer TopCount NXT) using the Bright-Glo luciferase assay kit (Promega).
2.3. p24 ELISA assay
First, 96-well plates (Thermo Scientific) were coated with anti-CA 183 (anti-p24) monoclonal antibody diluted l:1000 in PBS (100 μL/well). The plates were incubated at 37 °C overnight, washed with the washing buffer (0.2% Tween-20 in PBS) and blocked with 250 μL PBS/5% FBS. After 1 h at 37 °C, the plates were washed and incubated with viral lysates. The viruses were lysed using 10x lysis buffer (5% Triton-X100 in PBS (pH 7.4) with 0.2% azide) and protease inhibitor cocktail (Complete Mini, Roche) and serial 10-fold dilutions were prepared with sample dilution buffer (10% FBS, 0.5% Triton X-100 in PBS), along with the p24 standards (from Alliance® HIV-I p24 ELISA Kit). One hundred μL of serially diluted samples and standards were transferred to the α-p24 antibody-coated 96-well plates and incubated at 4 °C overnight. The plates were washed and incubated with 100 μL of Human Anti-HIV IgG (NIH AIDS Reagent Program) diluted 1:2000. After 1 h incubation at 37 °C, the plates were washed, and incubated for 1 h at 37 °C with 100 μL of secondary goat anti-human IgG-HRP (Thermo Scientific) diluted 1:5000. After additional washing, the plates were incubated for 30 min at room temperature with 100 μL HRP substrate (TMB Substrate Kit from Thermo Scientific) prepared from a 1:1 mixture of TMB peroxidase substrate and peroxidase solution. The reaction was quenched by H2SO4, and the optical densities at 450 nm were measured by a plate reader (Synergy HT, Bio-TEK).
2.4. Metabolic incorporation of SiaNAz into membrane glycoproteins and copper-free click labeling
SiaNAz (N-azidosialic acid) labeled pseudoviruses were produced by transfection of HEK/293T cells, as described above, except that the culture medium was supplemented with 50 μM of N-azido tetraacetyl mannosamine (Ac4ManNAz, Pierce, product #88904), as described in the text. SiaNAz-labeled pseudoviruses were harvested at 48 h post-transfection and used for click-labeling. Clickable dyes were added to 20 μL of SiaNAz-labeled virus stock to the final concentration of 100 μM, gently mixed and incubated at room temperature for at least 1 h. Excess of unreacted dye and Ac4ManNAz remaining in the medium were removed by passing through a size exclusion column (Centri-Spin 20, Princeton Separations, Adelphia, NJ). The labeled viruses were used immediately or kept in aliquots at −80 °C.
2.5. Single virus imaging and image analysis
Cells grown on glass-bottom Petri dishes (MatTek, MA) were chilled on ice and washed with cold PBS. Predetermined amount of viral suspension (MOI ~0.01) was added to the cells and spinoculated at 4 °C for 20 min, as described previously (Miyauchi et al., 2009). The cells were then washed twice with cold PBS and placed on the stage of a Zeiss LSM 780 confocal microscope. Virus entry was initiated by adding 2.5 ml of pre-warmed imaging buffer and imaged at 37 °C using a C-Apo 40×/1.2NA water-immersion objective. Images were acquired every 7–8 s. The time-lapse images were first independently inspected by two trained individuals to identify particles that lost the content marker, mCherry. Particle trajectories and their mean/total fluorescence intensities were determined using Volocity imaging software (PerkinElmer, MA), as described previously.
3. Results
3.1. Metabolic incorporation of unnatural sugars and click-labeling of viral envelope glycoproteins
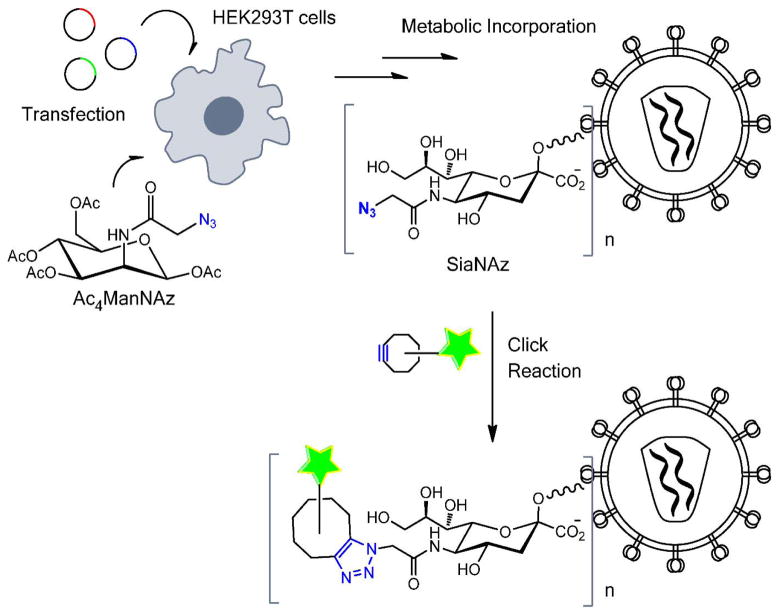
It has been demonstrated that peracylated azidomannosamine (Ac4ManNAz) is metabolized to incorporate into sialoglycoproteins in the form of SiaNAz (Fig. 1) (Chang et al., 2009; Luchansky et al., 2004). Cellular glycoproteins containing SiaNAz can then be labeled by click chemistry. Successful metabolic incorporation of this unnatural sugar into lentiviral particles bearing the Vesicular Stomatitis Virus G glycoprotein (VSV-G) or the Sindbis Virus E1/E2 glycoproteins has been recently reported (Chu et al., 2015). We sought to determine whether the SiaNAz incorporation is tolerated by retroviral fusion proteins. Toward this goal, HIV-based viral particles were pseudotyped with HIV-1 Env (HXB2 strain) and Avian Sarcoma and Leukosis Virus subtype A Env (ASLV Env), as well as with VSV-G, as a control. These pseudoviruses denoted HXB2pp, ASLVpp and VSVpp, respectively, were produced by transfection of 293T cells, as described previously (Desai et al., 2015; Padilla-Parra et al., 2013).
Figure 1.
Illustration of virus glycoprotein labeling by click reaction with a metabolically incorporated unnatural sugar.
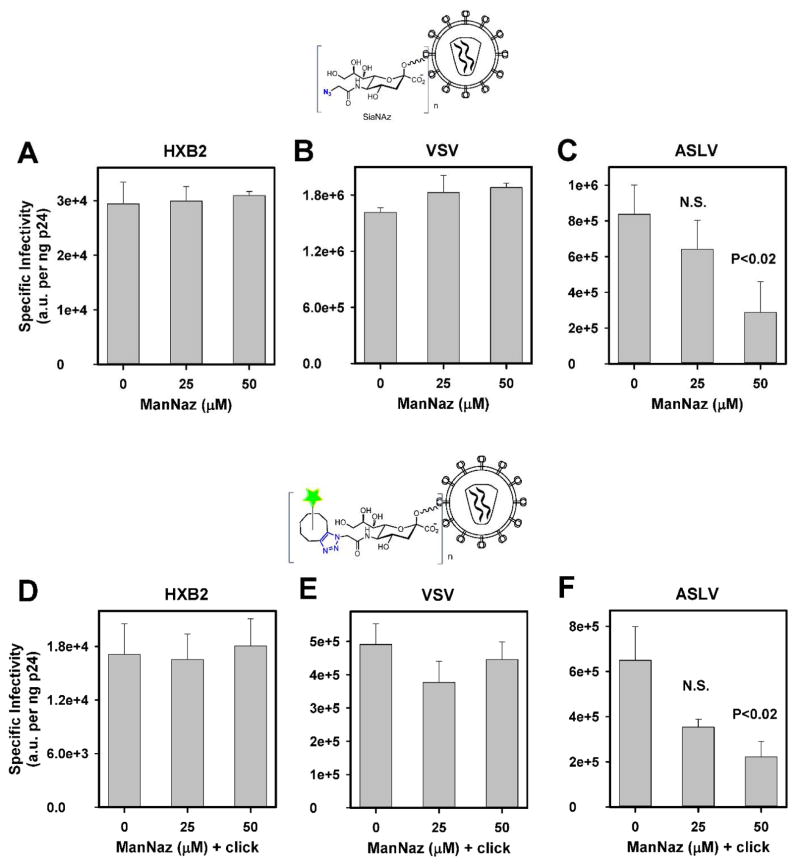
Ac4ManNAz was added at different times post-transfection, and the culture time with this sugar was varied to minimize the effect on the virus infectivity (Suppl. Fig. 1). Viruses were collected, their infectious titers were determined using the indicator TZM-bl cells and normalized to the amount of viral capsid protein (p24), as described in Methods. The highest viral titer was recovered when Ac4ManNAz was added at 12 h post-transfection and maintained for additional 36 h before collecting the virus (Suppl. Fig. 1). We then used this protocol to find the optimal concentration of the unnatural sugar. As shown in Figure 2A–B, production of HXB2pp and VSVpp in the presence of up to 50 μM Ac4ManNAz did not significantly affect the specific infectivity of these pseudoviruses. By contrast, the ASLVpp infectivity was compromised (Fig. 2C, p<0.05). These results thus indicate that incorporation of unnatural sugars does not affect the ability of HIV-1 Env or VSV-G to mediate membrane fusion, but can interfere with the ASLV Env function.
Figure 2. Effects of SiaNAz incorporation and click labeling on pseudovirus infectivity.
HIV-1 particles pseudotyped with HIV-1 HXB2 Env (A, D), VSV-G (B, E) and ASLV Env (C, F) were produced in 293T cells in the absence or in the presence of varied concentrations of Ac4ManNAz (ManNAz). Virus infectivity was evaluated by a reporter luciferase assay using 1 ng of viral p24, as determined by ELISA, before (A–C) and after (D–F) labeling with indicated concentrations of Alexa488-DIBO, as described in Methods. Data are means and standard deviations from triplicate experiments.
Next, we tested whether click-labeling of SiaNAz-containing viral glycoproteins can interfere with the virus’ ability to infect target cells. Control pseudoviruses and pseudoviruses produced in the presence of varied doses of Ac4ManNAz were labeled with 100 μM of green clickable dye AF488-DIBO (Invitrogen) for 2 h at room temperature followed by removal of excess dye by size-exclusion chromatography. In the absence of click-reactive sugars, this procedure resulted in ~2 fold decrease in specific infectivity of control HXB2pp and VSVpp produced in the absence of Ac4ManNAz, while having a minor effect of the titer of ASLVpp (compare Fig. 2A–C to 2D–F). Thus, mere incubation of viruses with the dye for 2 h in the absence of the click-reaction followed by gel-filtration reduced the viral titer. For viruses containing SiaNAz, the above protocol led to efficient labeling, as assessed by fluorescence microscopy (see below). Importantly, the click reaction itself did not result in additional loss of infectivity of pseudoviruses produced in the presence of varied doses of Ac4ManNAz, as compared to control viruses lacking this sugar (Fig. 2D–F). In other words, under our conditions, cyclo-addition of clickable-dye itself did not compromise the function of diverse viral glycoproteins beyond the effects of metabolic incorporation of Ac4ManNAz or virus handling.
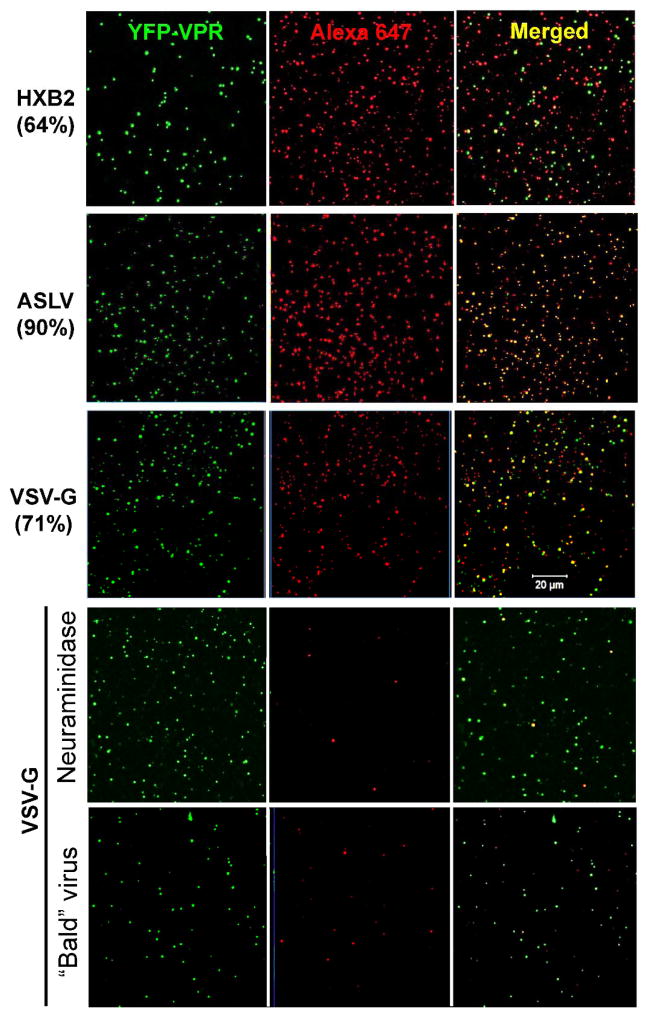
The efficiency of labeling of viral surface glycoproteins using the above optimized protocol was determined by imaging pseudoviruses adhered to poly-lysine coated coverslips. In order to discriminate between labeling of pseudoviruses and cell membrane vesicles that are present in the viral preparations (e.g., (Cantin et al., 2008)), we incorporated the viral core marker, YFP-Vpr, into virions. To avoid spectral overlap between YFP and AF488-DIBO used in Figure 2, these viruses were co-labeled with the far-red AF647-DIBO dye (Fig. 3A). Depending on the virus batch, click-labeling produced detectable red fluorescence in 64–90% of particles containing YFP-Vpr. The labeling efficiency could be further improved by longer incubation times or by increasing the concentration of AF647-DIBO. However, these conditions were avoided, since they adversely affected virus infectivity (data not shown).
Figure 3. Efficiency of pseudovirus labeling with Alexa647-DIBO.
(A) HXB2, VSV-G and ASLV pseudoviruses produced in the presence of 50 μM Ac4ManNAz were labeled with 100 μM Alexa647-DIBO (red), separated from free dye by gel filtration, and adhered to poly-lysine-coated coverslips. Viral cores were labeld with YFP-Vpr (green). The numbers on the left show percent of YFP-Vpr puncta that colocalized with Alexa647-DIBO. (B) Controls for specificity of click-labeling. VSV-G pseudoviruses were co-labeled with YFP-Vpr (green) and Alexa647-DIBO (red) and adhered to a poly-lysine-coated coverslip (top). SiaNAz-labeled virus stock (10 μL) was diluted 1:10 in MES buffer (pH 6.5) containing 20 mU neuraminidase, and the mixture was incubated at room temperature for 1 h prior to adhering to a coverslip (middle). Viruses lacking VSV-G (“bald”) were adhered to a coverslip (bottom). Scale bar 20 μm.
To control for the specificity of click labeling, we produced pseudoviruses lacking viral glycoproteins (“bald” particles) in the presence of 50 μM of Ac4ManNAz and click-labeled those with 100 μM of AF647-DIBO. No significant labeling of these “bald” particles with AF647-DIBO dye was detected (Fig. 3B). Importantly, this result also shows that, in spite of the propensity of HIV-based pseudoviruses to incorporate many cell membrane proteins, the viral glycoproteins (in this case, VGV-G) represent the overwhelming majority of proteins on the surface of particles that incorporate SiaNaz. Therefore, our conditions allow for specific labeling of viral envelope glycoproteins with clickable organic dyes. As an additional control, VSVpp click-labeled with AF647-DIBO were treated with neuraminidase in order to remove sialic acid residues, including the metabolically incorporated SiaNAz, from the surface glycoproteins. This treatment quantitatively removed the SiaNAz from the viral membrane, as evidenced by the lack of effective labeling of these viruses with AF647-DIBO (Fig. 3B). Collectively, these results demonstrate that the cyclo-addition reaction almost exclusively labeled viral surface glycoproteins and that these glycoproteins represent the majority of SiaNAz-containing proteins in the viral membrane.
3.2. Imaging of click-labeled pseudovirus fusion with target cells
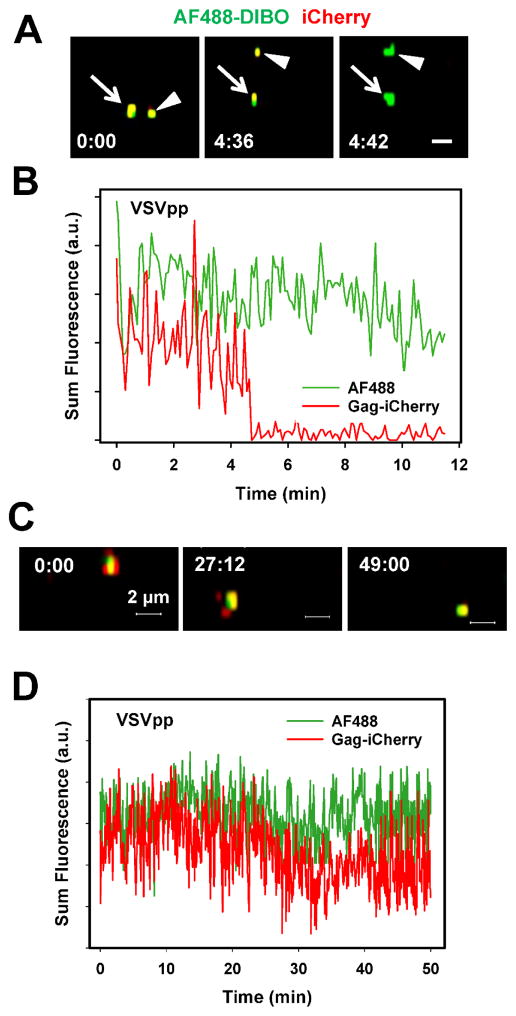
The utility of click-labeling of the viral surface glycoproteins was assessed by live cell imaging of particles co-labeled with AF488-DIBO (green) and a releasable content marker based on the Gag-imCherry (red) construct. Virus-incorporated Gag-imCherry is cleaved by the viral protease producing free mCherry, which is readily released upon viral fusion (Padilla-Parra et al., 2013). Loss of this content marker, but not of the membrane marker AF488-DIBO reports single virus fusion. Since the metabolic incorporation of Ac4ManNAz and click labeling did not considerably compromise VSVpp infectivity and since VSV G mediates much more efficient virus fusion than HXB2 Env (Desai et al., 2015; Padilla-Parra et al., 2013), we used VSV-G pseudoviruses for the initial imaging experiments. A quick loss of mCherry without significant changes in the AF488-DIBO signal (Fig. 4A, B) demonstrates that click-labeled VSVpp was fusion-competent. The AF488-DIBO signal was sufficiently bright and photostable (no significant photobleaching was observed in the course of the experiment, as shown in Fig. 4C, D) to enable reliable tracking of single particles in live cells. Whereas the mCherry signal was lost due to fusion and subsequent diffusion into the cytoplasm, AF488-DIBO retains its punctate appearance due to a limited dilution of this dye upon merger of viral and endosomal membranes.
Figure 4. Fusion of single VSVpp co-labeled with Alexa488-DIBO (green) and Gag-imCherry (red).
(A) Images of two double-labeled VSVpp fusing with endosomes of CV-1-derived cells nearly at the same time, as manifested in change from colocalized yellow color to green color. Scale bar 2 μm. (B) Sum fluorescence intensity profiles for the lower particle in panel A obtained by single particle tracking. (C, D) images and fluorescence profiles for a control double-labeled particle that did not undergo fusion.
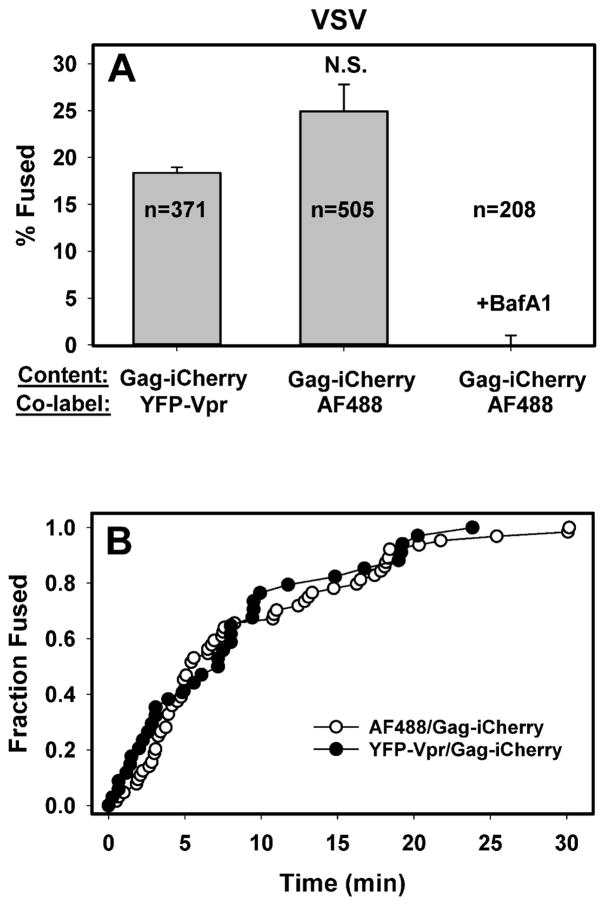
To determine whether click labeling can adversely affect the efficiency or the kinetics of viral fusion, we compared entry/fusion of AF488-DIBO/Gag-imCherry labeled particles to pseudoviruses co-labeled with YFP-Vpr and Gag-imCherry. The latter labeling strategy has been employed by us and others to detect single virus fusion events (Desai et al., 2015; Padilla-Parra et al., 2013). Neither the fraction of double-labeled viruses undergoing fusion (Fig. 5A) nor the kinetics of fusion (Fig. 5B) was diminished by click-labeling. In fact, the extent of fusion was slightly greater than that observed in parallel experiments for YFP-Vpr/Gag-imCherry viruses (Fig. 5A). mCherry release could not be detected in control experiments in the presence of bafilomycin A1 (BafA1, Fig. 5A), which blocks VSV fusion by raising the endosomal pH.
Figure 5. Comparison of the extent and kinetics of fusion of single VSVpp co-labeled with different markers.
(A) The fraction of double-labeled particles that fuse with CV-1-derived cells within 1 h at 37 °C. Pseudoviruses co-labeled with YFP-Vpr/Gag-imCherry or with Alexa488-DIBO/Gag-imCherry were allowed to enter CV-1-derived cells and all fusion events were annotated. Control experiments (rightmost bar) were carried out in the presence of 200 nM Bafilomycin A1. Data are means and standard deviations from 3 independent experiments. (B) Kinetics of fusion of VSVpp co-labeled with either YFP-Vpr/Gag-imCherry or Alexa488-DIBO/Gag-imCherry. The time intervals from shifting to 37 °C to individual fusion (mCherry release) events were determined and plotted as cummulative distributions normalized to the last time point.
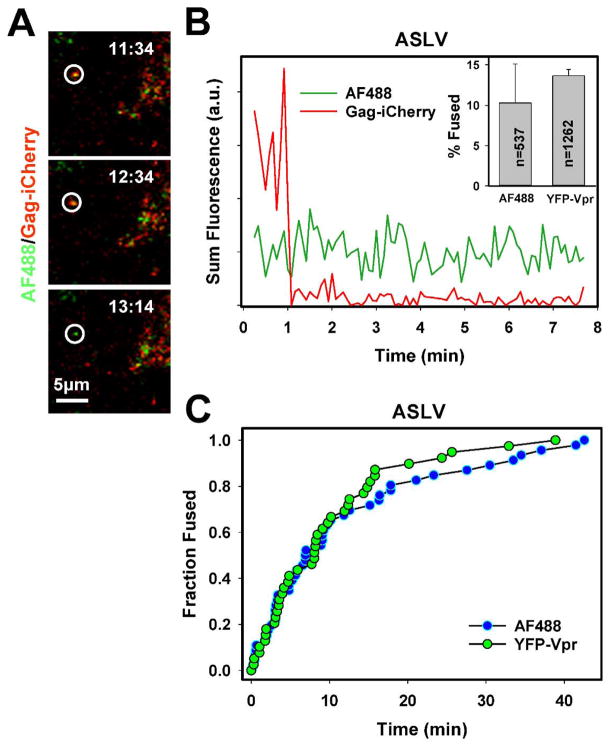
Using the click-labeling of unnatural sugars, we also readily detected single ASLVpp fusion with target cells expressing the TVA950 receptor, which was evident by an abrupt loss of the mCherry signal (Fig. 6A, B and Supplemental Movie 1). The kinetics of single click-labeled ASLVpp fusion was identical to that of pseudoviruses co-labeled with YFP-Vpr and Gag-imCherry (Fig. 6C). In spite of the somewhat reduced infectivity of the click labeled ASLVpp (Fig. 2C), the extent of fusion was comparable to that of YFP-Vpr/Gag-imCherry-labeled viruses (Fig. 6B, inset and (Desai et al., 2015)). The apparent lack of an effect of metabolic labeling on single virus-cell fusion could be due to a large variance between the results of independent imaging experiments, as manifested in the large error bars (Fig. 6B, inset).
Figure 6. Fusion of single ASLVpp co-labeled with Alexa488-DIBO (green) and Gag-imCherry (red).
The image panels (A) and the graph (B) show fusion of single ASLVpp particle with CV-1 cells expressing the TVA950 receptor. Inset to panel B: shows the mean fusion efficiency of pseudoviruses labeled with Alexa488-DIBO/Gag-imCherry and with YFP-Vpr/Gag-imCherry. (C) Kinetics of fusion of single ASLVpp co-labeled with either YFP-Vpr/Gag-imCherry or Alexa488-DIBO/Gag-imCherry. See also Supplemental Movie 1.
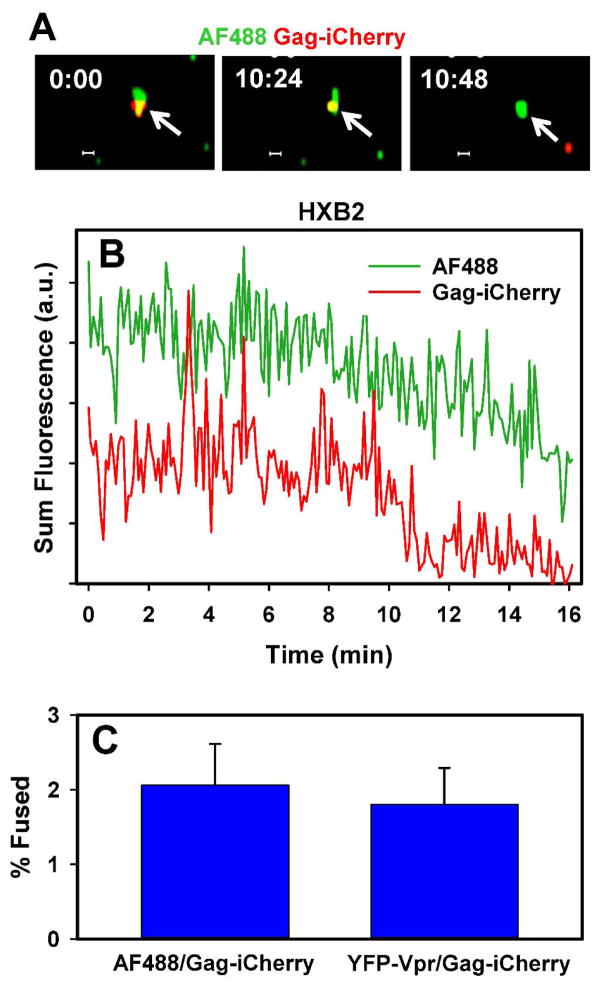
Finally, we examined the ability of click-labeled HXB2pp to fuse with target cells expressing CD4 and CXCR4. In spite of the well-documented low efficiency of HIV-1 Env-mediated fusion (Desai et al., 2015; Miyauchi et al., 2009; Padilla-Parra et al., 2013), single virus imaging revealed that the viral content marker was released from about 2% of AF488-DIBO/Gag-imCherry labeled particles (Fig. 7). This result is in agreement with the previously published data using alternative HXB2pp labeling strategies (Desai et al., 2015; Miyauchi et al., 2009; Padilla-Parra et al., 2013). Thus, click-labeling of unnatural sugars incorporated into the viral surface glycoproteins provides a versatile platform for imaging single virus entry and fusion into target cells.
Figure 7. Analysis of fusion of single HXB2pp co-labeled with Alexa488-DIBO (green) and Gag-imCherry (red).
The image panels (A) and the graph (B) show fusion (mCherry release) of single HXB2pp particle with CV-1 cells expressing CD4 and CXCR4. (C) The extent of fusion (mean and standard deviation from 3 independent experiments).
4. Discussion
We have demonstrated that click-labeling of sugar moieties of viral glycoproteins is an efficient and generalizable method for labeling the viral membrane without considerably compromising the virus’ ability to productively infect target cells. Importantly, this approach is compatible with single virus imaging and should also be compatible with super-resolution imaging of single virions by STORM (stochastic optical reconstruction microscopy). Compared to other strategies, such as viral lipid labeling or labeling of surface proteins with amine-reactive dyes, metabolic incorporation of sugars and click reaction are less invasive and yield robust labeling of nearly all viral particles. Importantly, click-labeling proceeded efficiently in serum-containing growth medium without the need to concentrate or purify the virus. Under our conditions, amine-labeling and lipid-dye labeling reduced specific infectivity and produced overwhelming background signals in live cells, thus precluding single particle tracking (data not shown).
Surprisingly, metabolic incorporation of SiaNAz diminished infectivity of ASLV Env, but not HIV-1 Env or VSV-G. Such differential sensitivity of viral glycoproteins could be due to differences in glycosylation sites and/or folding pathways. Further optimization of the ASLV Env labeling protocol, including lowering the concentration of Ac4ManNAz and/or the labeling time, should help to minimize the adverse effect on this protein’s function. Future experiments will reveal whether the above labeling strategy provides a sufficiently stable reference marker for post-fusion endosomes which would allow identification and tracking of the released viral cores in the cytoplasm based on the spatial separation of a membrane and core markers (Padilla-Parra et al., 2012a).
Supplementary Material
Highlights.
Unnatural sugar, Ac4ManNAz, can be efficiently incorporated into viral glycoproteins without significantly compromising their functional activity
Copper-free click labeling of viral glycoproteins containing unnatural sugars with organic fluorophores does not affect their ability to mediate membrane fusion
Click labeling of viral surface glycoproteins combined with incorporation of a genetically engineered fluorescent protein, which provides a releasable viral content marker, enables the visualization of single virus entry and fusion in living cells
Acknowledgments
The authors wish to thank the NIH AIDS Reagent Program for TZM-bl cells (donated by Drs. J.C. Kappes and X. Wu). This work was supported by the NIH R01 grant AI053668 and Pittsburgh Center for HIV Protein Interactions (P50GM082251) Collaboration Development Fund to G.B.M.
Abbreviations
- Ac4ManNAz
peracylated azidomannosamine
- ASLV
Avian Sarcoma and Leukosis Virus
- Env
viral envelope glycoprotein
- SiaNaz
N-azidosialic acid
- VSV
Vesicular Stomatitis Virus
- YFP
yellow fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albanese A, Arosio D, Terreni M, Cereseto A. HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PloS one. 2008;3:e2413. doi: 10.1371/journal.pone.0002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhel N, Genovesio A, Kim KA, Miko S, Perret E, Olivo-Marin JC, Shorte S, Charneau P. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Methods. 2006;3:817–24. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- Arumugam S, Chin J, Schirrmacher R, Popik VV, Kostikov AP. [18F]Azadibenzocyclooctyne ([18F]ADIBO): A biocompatible radioactive labeling synthon for peptides using catalyst free [3+2] cycloaddition. Bioorganic & Medicinal Chemistry Letters. 2011;21:6987–6991. doi: 10.1016/j.bmcl.2011.09.126. [DOI] [PubMed] [Google Scholar]
- Banerjee PS, Ostapchuk P, Hearing P, Carrico I. Chemoselective Attachment of Small Molecule Effector Functionality to Human Adenoviruses Facilitates Gene Delivery to Cancer Cells. J Am Chem Soc. 2010;132:13615–13617. doi: 10.1021/ja104547x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PS, Ostapchuk P, Hearing P, Carrico IS. Unnatural Amino Acid Incorporation onto Adenoviral (Ad) Coat Proteins Facilitates Chemoselective Modification and Retargeting of Ad Type 5 Vectors. J Virol. 2011a;85:7546–7554. doi: 10.1128/JVI.00118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PS, Zuniga ES, Ojima I, Carrico IS. Targeted and armed oncolytic adenovirus via chemoselective modification. Bioorganic & medicinal chemistry letters. 2011b;21:4985–4988. doi: 10.1016/j.bmcl.2011.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR. Visualizing enveloping layer glycans during zebrafish early embryogenesis. Proceedings of the National Academy of Sciences. 2010;107:10360–10365. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertozzi CR. A decade of bioorthogonal chemistry. Acc Chem Res. 2011;44:651–3. doi: 10.1021/ar200193f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–84. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- Brandenburg B, Zhuang X. Virus trafficking - learning from single-virus tracking. Nature reviews Microbiology. 2007;5:197–208. doi: 10.1038/nrmicro1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenbach MA, Gallagher JE, King DS, Smart BP, Wu P, Bertozzi CR. Targeted metabolic labeling of yeast N-glycans with unnatural sugars. Proceedings of the National Academy of Sciences. 2010;107:3988–93. doi: 10.1073/pnas.0911247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick RC, Hu WS, Pathak VK. Nuclear import of APOBEC3F-labeled HIV-1 preintegration complexes. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4780–9. doi: 10.1073/pnas.1315996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Perez O, Melar M, Hope TJ. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 2007;360:286–93. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin R, Diou J, Belanger D, Tremblay AM, Gilbert C. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J Immunol Methods. 2008;338:21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Carpenter RD, Hausner SH, Sutcliffe JL. Copper-free click for PET: Rapid 1, 3-dipolar cycloadditions with a fluorine-18 cyclooctyne. ACS medicinal chemistry letters. 2011;2:885–889. doi: 10.1021/ml200187j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Metabolic Labeling of Sialic Acids in Living Animals with Alkynyl Sugars. Angewandte Chemie International Edition. 2009;48:4030–4033. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Oum YH, Carrico IS. Surface modification via strain-promoted click reaction facilitates targeted lentiviral transduction. Virology. 2015;487:95–103. doi: 10.1016/j.virol.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Crivat G, Taraska JW. Imaging proteins inside cells with fluorescent tags. Trends Biotechnol. 2012;30:8–16. doi: 10.1016/j.tibtech.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale BM, McNerney GP, Thompson DL, Hubner W, de Los Reyes K, Chuang FY, Huser T, Chen BK. Cell-to-cell transfer of HIV-1 via virological synapses leads to endosomal virion maturation that activates viral membrane fusion. Cell host & microbe. 2011;10:551–62. doi: 10.1016/j.chom.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean KM, Palmer AE. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat Chem Biol. 2014;10:512–23. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirkhanyan LH, Marin M, Padilla-Parra S, Zhan C, Miyauchi K, Jean-Baptiste M, Novitskiy G, Lu W, Melikyan GB. Multifaceted mechanisms of HIV-1 entry inhibition by human alpha-defensin. The Journal of biological chemistry. 2012;287:28821–28838. doi: 10.1074/jbc.M112.375949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014;10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TM, Marin M, Sood C, Shi J, Nawaz F, Aiken C, Melikyan GB. Fluorescent protein-tagged Vpr dissociates from HIV-1 core after viral fusion and rapidly enters the cell nucleus. Retrovirology. 2015;12:88. doi: 10.1186/s12977-015-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers H, Smith AE, Sbalzarini IF, Lilie H, Koumoutsakos P, Helenius A. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15110–5. doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat Biotech. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd DL, Ragains JR, Skehel JJ, Harrison SC, van Oijen AM. Single-particle kinetics of influenza virus membrane fusion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15382–7. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO. HIV-1 replication. Somat Cell Mol Genet. 2001;26:13–33. doi: 10.1023/a:1021070512287. [DOI] [PubMed] [Google Scholar]
- Huang LL, Xie HY. Progress on the labeling and single-particle tracking technologies of viruses. The Analyst. 2014;139:3336–46. doi: 10.1039/c4an00038b. [DOI] [PubMed] [Google Scholar]
- Joo KI, Lei Y, Lee CL, Lo J, Xie J, Hamm-Alvarez SF, Wang P. Site-specific labeling of enveloped viruses with quantum dots for single virus tracking. ACS nano. 2008;2:1553–62. doi: 10.1021/nn8002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo KI, Tai A, Lee CL, Wong C, Wang P. Imaging multiple intermediates of single-virus membrane fusion mediated by distinct fusion proteins. Microsc Res Tech. 2010;73:886–900. doi: 10.1002/jemt.20841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak SL, Platt EJ, Madani N, Ferro FE, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–82. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9280–5. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe M, Briggs JA, Endress T, Glass B, Riegelsberger S, Krausslich HG, Lamb DC, Brauchle C, Muller B. Double-labelled HIV-1 particles for study of virus-cell interaction. Virology. 2007;360:92–104. doi: 10.1016/j.virol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. The Journal of cell biology. 2005;170:317–25. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchansky SJ, Argade S, Hayes BK, Bertozzi CR. Metabolic Functionalization of Recombinant Glycoproteins†. Biochemistry. 2004;43:12358–12366. doi: 10.1021/bi049274f. [DOI] [PubMed] [Google Scholar]
- Luchansky SJ, Hang HC, Saxon E, Grunwell JR, Yu C, Dube DH, Bertozzi CR. Constructing Azide-Labeled Cell Surfaces Using Polysaccharide Biosynthetic Pathways. In: Yuan CL, Reiko TL, editors. Methods Enzymol. Academic Press; 2003. pp. 249–272. [DOI] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. The Journal of cell biology. 2002;159:441–52. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan GB, Barnard RJ, Abrahamyan LG, Mothes W, Young JA. Imaging individual retroviral fusion events: from hemifusion to pore formation and growth. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8728–33. doi: 10.1073/pnas.0501864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengistu M, Ray K, Lewis GK, DeVico AL. Antigenic properties of the human immunodeficiency virus envelope glycoprotein gp120 on virions bound to target cells. PLoS Pathog. 2015;11:e1004772. doi: 10.1371/journal.ppat.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–44. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, 3rd, Kwong PD, Blanchard SC, Mothes W. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–63. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane S, Iwamoto A, Matsuda Z. The V4 and V5 Variable Loops of HIV-1 Envelope Glycoprotein Are Tolerant to Insertion of Green Fluorescent Protein and Are Useful Targets for Labeling. The Journal of biological chemistry. 2015;290:15279–91. doi: 10.1074/jbc.M114.628610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikic I, Plass T, Schraidt O, Szymanski J, Briggs JA, Schultz C, Lemke EA. Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew Chem Int Ed Engl. 2014;53:2245–9. doi: 10.1002/anie.201309847. [DOI] [PubMed] [Google Scholar]
- Oum YH, Carrico IS. Altering Adenoviral Tropism via Click Modification with ErbB Specific Ligands. Bioconj Chem. 2012;23:1370–1376. doi: 10.1021/bc200477z. [DOI] [PubMed] [Google Scholar]
- Padilla-Parra S, Marin M, Gahlaut N, Suter R, Kondo N, Melikyan GB. Fusion of Mature HIV-1 Particles Leads to Complete Release of a Gag-GFP-Based Content Marker and Raises the Intraviral pH. PloS one. 2013;8:e71002. doi: 10.1371/journal.pone.0071002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Parra S, Marin M, Kondo N, Melikyan GB. Synchronized Retrovirus Fusion in Cells Expressing Alternative Receptor Isoforms Releases the Viral Core into Distinct Sub-cellular Compartments. PLoS Pathog. 2012a;8:e1002694. doi: 10.1371/journal.ppat.1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Parra S, Matos PM, Kondo N, Marin M, Santos NC, Melikyan GB. Quantitative imaging of endosome acidification and single retrovirus fusion with distinct pools of early endosomes. Proceedings of the National Academy of Sciences of the United States of America. 2012b;109:17627–32. doi: 10.1073/pnas.1211714109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- Sachin K, Jadhav VH, Kim E-M, Kim HL, Lee SB, Jeong H-J, Lim ST, Sohn M-H, Kim DW. F-18 Labeling Protocol of Peptides Based on Chemically Orthogonal Strain-Promoted Cycloaddition under Physiologically Friendly Reaction Conditions. Bioconj Chem. 2012;23:1680–1686. doi: 10.1021/bc3002425. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Krausslich HG. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrobial agents and chemotherapy. 2002;46:1896–905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Cai L, Adogla EA, Guan H, Lin Y, Wang Q. Labeling of Enveloped Virus via Metabolic Incorporation of Azido Sugars. Bioconjug Chem. 2015;26:1868–72. doi: 10.1021/acs.bioconjchem.5b00310. [DOI] [PubMed] [Google Scholar]
- Zhou P, Zheng Z, Lu W, Zhang F, Zhang Z, Pang D, Hu B, He Z, Wang H. Multicolor labeling of living-virus particles in live cells. Angew Chem Int Ed Engl. 2012;51:670–4. doi: 10.1002/anie.201105701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.