Summary
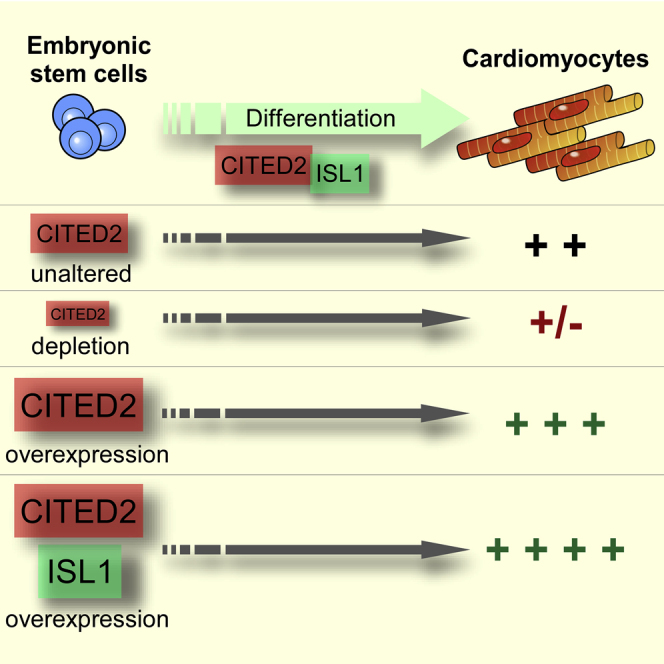
The transcriptional regulator CITED2 is essential for heart development. Here, we investigated the role of CITED2 in the specification of cardiac cell fate from mouse embryonic stem cells (ESC). The overexpression of CITED2 in undifferentiated ESC was sufficient to promote cardiac cell emergence upon differentiation. Conversely, the depletion of Cited2 at the onset of differentiation resulted in a decline of ESC ability to generate cardiac cells. Moreover, loss of Cited2 expression impairs the expression of early mesoderm markers and cardiogenic transcription factors (Isl1, Gata4, Tbx5). The cardiogenic defects in Cited2-depleted cells were rescued by treatment with recombinant CITED2 protein. We showed that Cited2 expression is enriched in cardiac progenitors either derived from ESC or mouse embryonic hearts. Finally, we demonstrated that CITED2 and ISL1 proteins interact physically and cooperate to promote ESC differentiation toward cardiomyocytes. Collectively, our results show that Cited2 plays a pivotal role in cardiac commitment of ESC.
Keywords: transcriptional regulation, cardiac progenitor, Cited2, Isl1, embryonic stem cell, differentiation
Graphical Abstract
Highlights
-
•
Overexpression of CITED2 in ESC promotes cardiogenesis upon differentiation
-
•
Cited2 depletion reduces ESC ability to generate cardiac cells
-
•
Cited2 expression is enriched in cardiac progenitors
-
•
CITED2 and ISL1 cooperate to promote ESC differentiation toward cardiomyocytes
In this article, Bragança and colleagues show that the transcriptional regulator Cited2 is essential for cardiac commitment of mouse embryonic stem cells (ESC), and its depletion in ESC impairs the expression of early mesoderm markers and cardiogenic transcription factors. In addition, they demonstrate that CITED2 and ISL1 proteins interact and cooperate to promote ESC differentiation toward cardiomyocytes.
Introduction
Cardiac morphogenesis results from the specification, differentiation, and migration of spatially and temporally distinct sets of cardiac precursor cells that give rise to the mature cardiac tissue. The first multipotent cardiogenic cells originate from the mesoderm formed at early stages of gastrulation when cells of the epiblast ingress to the primitive streak (Costello et al., 2011, David et al., 2011, Garry and Olson, 2006, Kitajima et al., 2000, Tam et al., 1997). The mesoderm first expresses the markers BRACHYURY and FLK1 and subsequently the cardiogenic marker MESP1 (Bondue et al., 2008, Chan et al., 2013, Ishitobi et al., 2011, Saga et al., 1999). The first (FHF) and second (SHF) heart fields then arise from the cardiogenic mesoderm to ultimately generate the atria, ventricles, and outflow tract of the nascent heart (Cai et al., 2003, Domian et al., 2009, Moretti et al., 2006). Both FHF and SHF progenitors express the transcription factors NKX2.5, TBX5, GATA4, MEF2C, and ISL1, although TBX5 is predominantly present in cells of the FHF, and Isl1 expression is a hallmark of SHF progenitors (Laugwitz et al., 2005, Moretti et al., 2006, Vincent et al., 2010). In mouse, the transcriptional modulator CITED2 is required for normal embryogenesis. Deletion of Cited2 in the epiblast results in embryonic lethality associated with multiple cardiovascular defects (Bamforth et al., 2001, Bamforth et al., 2004, MacDonald et al., 2008, MacDonald et al., 2013, Weninger et al., 2005, Yin et al., 2002). Of important note, however, although Cited2 is expressed in the early mesoderm (Dunwoodie et al., 1998), conditional deletion of Cited2 in BRACHYURY-expressing mesoderm cells or MESP1-expressing cardiogenic mesoderm progenitors did not significantly affect cardiac development (MacDonald et al., 2008). In humans, mutations in the gene encoding CITED2 are associated with congenital heart disease (Chen et al., 2012, Sperling et al., 2005).
The specification and differentiation of cardiac progenitor cells (CPC) and mature cardiovascular cells during the in vitro differentiation of pluripotent stem cells recapitulate the cellular and molecular processes of embryonic development (Blin et al., 2010, Bondue et al., 2008, Bondue et al., 2011, Christoforou et al., 2008, Gai et al., 2009, Kattman et al., 2011, Kouskoff et al., 2005, Laugwitz et al., 2005, Moretti et al., 2006, Van Vliet et al., 2012, Yang et al., 2008). In mouse, an acute Cited2 depletion reduces the self-renewal capacity of most embryonic stem cells (ESC), but a small population of Cited2-null ESC with apparent characteristics of undifferentiated cells adapt to the loss of Cited2 (Kranc et al., 2015, Li et al., 2012). Interestingly, Cited2-null ESC showed an impairment of differentiation, including cardiac commitment (Li et al., 2012). To better understand the role of Cited2 at early stages of mouse ESC differentiation, we here employ Cited2 loss- and gain-of-function approaches to examine the role of Cited2 during cardiac differentiation. Cited2 depletion at the onset of differentiation significantly impairs the expression of Brachyury, Mesp1, Isl1, Gata4, and Tbx5. Conversely, CITED2 overexpression stimulates the expression of these genes in undifferentiated ESC and promotes cardiac lineage commitment and differentiation. We further show that Cited2 expression is highly associated with CPC populations, particularly cardiac progenitors of the SHF. Finally, we show that CITED2 is recruited to the promoter of the Isl1 gene, and provide evidence that the human CITED2 and ISL1 proteins physically interact and synergize to promote cardiogenesis from ESC. Collectively our results show that Cited2 is a key regulator of early cardiac lineage commitment and differentiation of ESC.
Results
CITED2 Overexpression Promotes ESC Differentiation to Cardiac Lineages
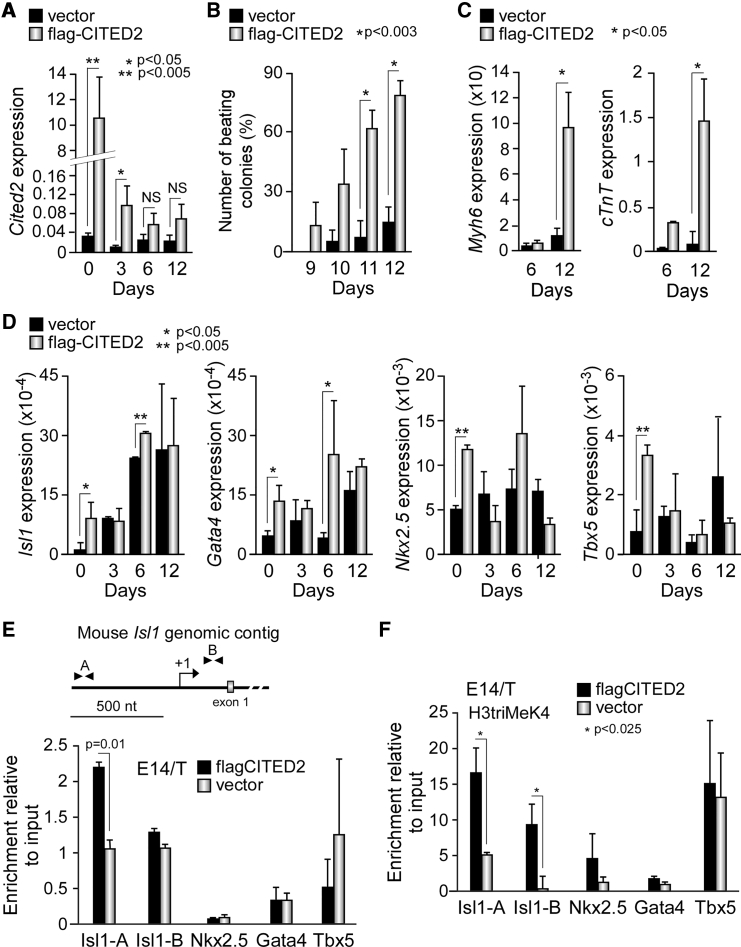
The knockout of Cited2 in mouse ESC has been previously reported to impair cardiomyocyte differentiation (Li et al., 2012). To investigate whether the overexpression of CITED2 promotes ESC differentiation into cardiac lineages, we transfected undifferentiated mouse E14/T ESC with an episomal plasmid expressing a FLAG-tagged human CITED2 (flag-CITED2) or a control vector as previously described (Kranc et al., 2015). Upon differentiation, high levels of flag-CITED2 expression were detected in E14/T ESC (named E14T/flagCITED2 hereafter) at the onset of differentiation (day 0 [D0]) in comparison with the endogenous Cited2 expression in control cells (named E14T/Control hereafter). However, the ectopic expression of flag-CITED2 in cells derived from E14T/flagCITED2 ESC rapidly declined during the first 3 days of differentiation, and returned to control levels by D6 (Figure 1A). An increase in the number of spontaneous contractile clusters (beating foci), marking the occurrence of terminal cardiomyocyte differentiation, was observed in cell cultures derived from E14T/flagCITED2 ESC in comparison with control cells (Figure 1B). Thus, the overexpression of flag-CITED2 significantly promoted cardiomyocyte differentiation, as confirmed by the elevated expression of cardiac structural genes, such as α-cardiac myosin heavy chain (Myh6) and cardiac troponin T (cTnT) transcripts detected in these cells in comparison with control cells (Figure 1C). The vascular endothelial growth factor receptor 2 (VegfR2, a marker of endothelial differentiation) expression was also increased in cells derived from E14T/flagCITED2 ESC, while the expression of the skeletal troponin I (sTnI, a marker of skeletal muscle differentiation) was unaffected by the overexpression of flag-CITED2 (Figure S1A), suggesting that flag-CITED2 overexpression supports ESC specification to cardiovascular cell lineages. To unravel the mechanism by which flag-CITED2 promoted the cardiac differentiation process, we assessed the expression of transcription factors known to play critical roles in the specification of ESC to cardiac lineages, particularly Isl1, Gata4, Nkx2.5, and Tbx5. Surprisingly, the overexpression of flag-CITED2 significantly increased the transcript levels of these factors in undifferentiated ESC (D0), and of Isl1 and Gata4 at D6 of differentiation (Figure 1D). The increase in the expression of Isl1 and Gata4 proteins was also detected in undifferentiated E14/T ESC overexpressing flag-CITED2 (Figure S1B). Since CITED2 is a transcriptional modulator, we hypothesized that flag-CITED2 enhanced Isl1, Gata4, Nkx2.5, and Tbx5 expression by a direct effect on their transcriptional regulatory regions. Therefore, we investigated by chromatin immunoprecipitation (ChIP) assays whether flag-CITED2 was recruited at the promoters of these factors. A significant enrichment of the Isl1 promoter region was detected in E14T/flagCITED2 ESC extracts immunoprecipitated with an anti-flag antibody in comparison with E14T/Control extracts (Figure 1E). No difference of enrichment was observed for the exon 1 of Isl1, or for the Nkx2.5, Gata4, and Tbx5 promoter fragments tested in the same conditions (Figure 1E). Interestingly, flag-CITED2 overexpression also significantly enhanced the occupancy of Isl1 promoter and exon 1 by histones H3 trimethylated on lysine 4 (H3triMeK4), a mark of actively transcribed chromatin (Figure 1F). No enrichment of H3triMeK4 occupancy was detected at the promoters of Nkx2.5, Gata4, and Tbx5 in the same extracts. Although these experiments do not enable us to rule out the presence of flag-CITED2 at other regulatory elements of Nkx2.5, Gata4, and Tbx5 genes, they exposed its presence at the Isl1 promoter. Moreover, the presence of flag-CITED2 at Isl1 regulatory regions was also correlated with an increased recruitment of H3triMeK4 at these regions, suggesting that flag-CITED2 may exert a direct positive effect on the expression of Isl1.
Figure 1.
Ectopic Expression of Human CITED2 Promotes Cardiac Differentiation
(A) Relative expression of flag-Cited2 during differentiation, determined by qPCR in E14/T ESC transfected with a plasmid expressing FLAG-tagged CITED2 (flag-CITED2) or the control empty plasmid (vector). NS, not significant.
(B) Number of colonies with beating foci counted at the indicated days of differentiation in cells treated as described in (A).
(C) Expression levels of Myh6 and cTnT determined by qPCR at D6 and D12 of differentiation in cells treated as described in (A).
(D) Expression of Isl1, Gata4, Nkx2.5, and Tbx5 at D0, D3, D6, and D12 of differentiation in E14/T-derived cell extracts prepared as described in (A).
(E) Top: diagram of the mouse Isl1 genomic contig showing the transcriptional start site (arrow), exon 1 (gray box), and positions of PCR primers (arrow heads) used in ChIP assays. Bottom: enrichment of Isl1 promoter (Isl1-A) and exon 1 (Isl1-B), and Nkx2.5, Gata4, and Tbx5 promoters in extracts of undifferentiated E14/T ESC overexpressing flag-CITED2 or control cells by ChIP assays with anti-FLAG monoclonal antibody.
(F) Enrichment of the genomic regions of Isl1, Nkx2.5, Gata4, and Tbx5 in ChIP assays with extracts and primers described in (E), using an anti-histone H3triMeK4-specific antibody.
Results are presented as the mean ± SEM of three independent biological experiments.
Depletion of Cited2 in ESC Differentiation Affects Cardiac Commitment
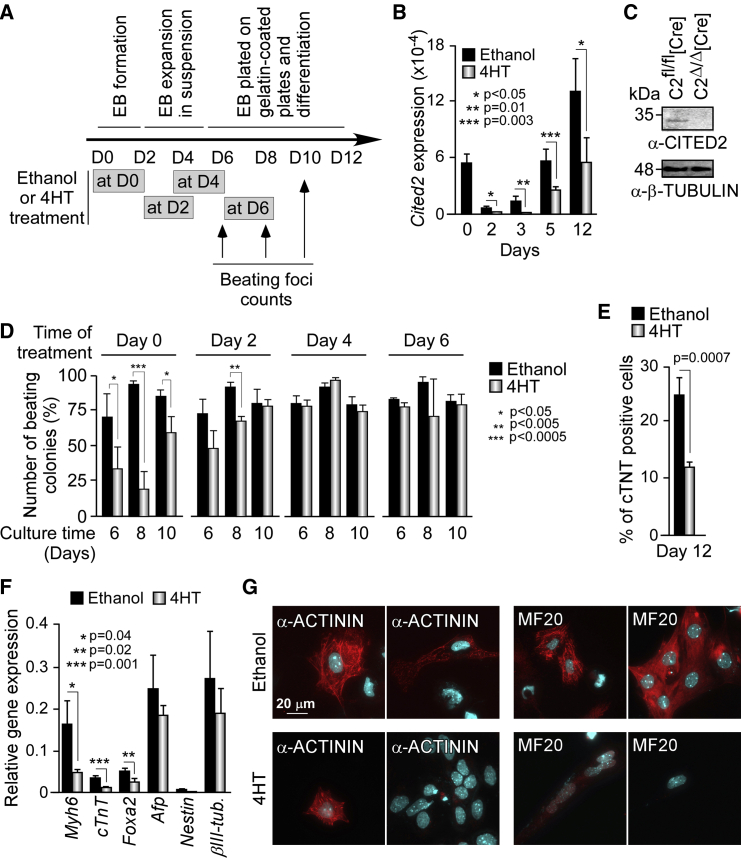
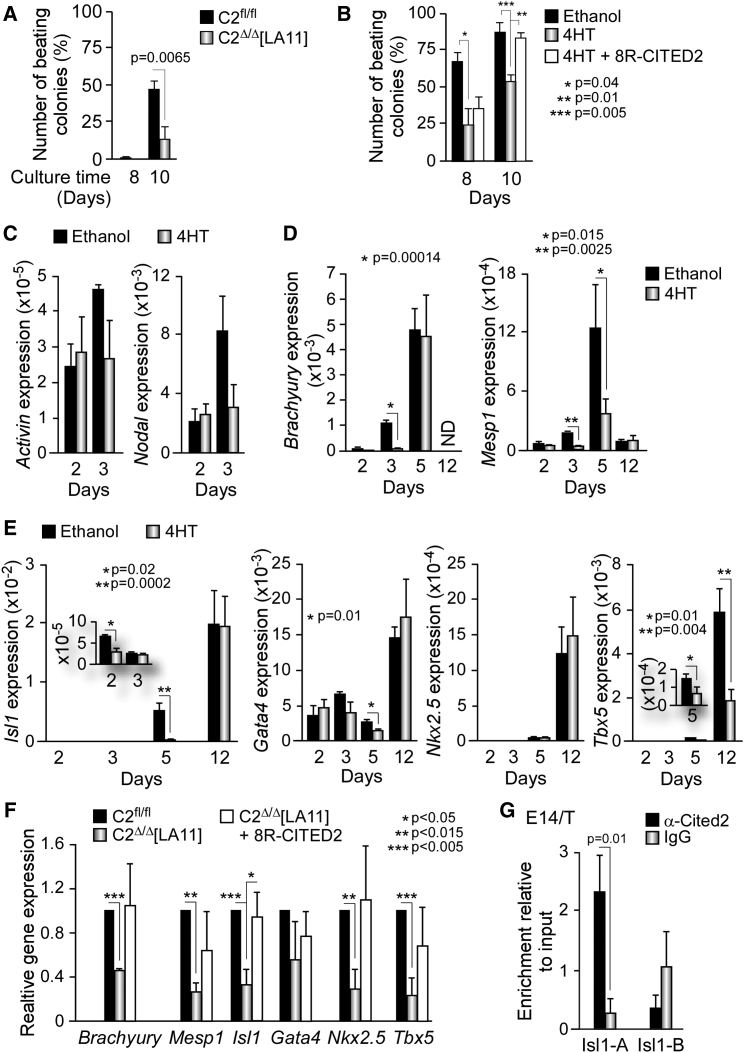
To further investigate the role of Cited2 in cardiac differentiation, we used C2fl/fl[Cre] ESC, which allow the conditional knockout of Cited2 by supplementation of 4-hydroxytamoxifen (4HT) to the culture medium (Kranc et al., 2015). First, we analyzed Cited2 expression kinetics by qPCR during the differentiation of control C2fl/fl[Cre] ESC (Figures 2A and 2B, black bars). The pattern of Cited2 expression was biphasic, with a decrease of the transcript levels from D0 to D2 of differentiation, followed by an elevation from D3 onward (Figure 2B, black bars). A similar expression pattern for Cited2 was also observed in E14T/Control cells (Figure 1A, black bars). Next, we examined Cited2 expression in cells derived from C2fl/fl[Cre] ESC treated with 4HT at D0 of differentiation during 48 hr, and compared it with control cells derived from C2fl/fl[Cre] ESC treated with ethanol used as vehicle (Figures 2A–2C). Cited2 depletion was incomplete during the time course of differentiation since Cited2 transcripts remained detectable at D5 and D12 of differentiation in cells treated with 4HT, although these levels were significantly reduced in comparison with control cells (Figure 2B). Noticeably, the number of beating foci was reduced in cell cultures derived from ESC depleted of Cited2 at D0 compared with control cells (Figure 2D). The decline of Myh6 and cTnT transcripts expression and the reduced number of cells expressing cTNT protein in cultures treated with 4HT supported the requirement of Cited2 expression for ESC differentiation into cardiac lineages (Figures 2E and 2F). To evaluate the expression and organization of sarcomeric proteins in cardiomyocytes, we performed fluorescent immunodetection of the α-ACTININ and MYOSIN HEAVY CHAIN (MF20) proteins in cells at D10 of differentiation derived from C2fl/fl[Cre] ESC treated either with ethanol or 4HT at D0 (Figure 2G). Approximately 25%–30% of the cells derived from control C2fl/fl[Cre] ESC were positively stained for α-ACTININ or MF20 and presented some degree of organization of the sarcomeric apparatus (Figures 2G and S1C). The depletion of Cited2 by treatment of C2fl/fl[Cre] ESC with 4HT resulted in the decline of α-ACTININ and MF20 protein detection, and in the diminution of the number of cells stained for α-ACTININ or MF20 in comparison with the control cells (Figures 2G and S1C). Together, these observations were consistent with the requirement of Cited2 for ESC-derived cardiogenesis. In contrast, Cited2 depletion did not significantly alter the expression of Nestin and βIII-Tubulin (markers of neural cells, ectoderm origin) and the α-fetoprotein (hepatic marker, endoderm origin). Cited2 depletion did result in a reduction of Foxa2 (endoderm and hepatocyte marker) expression at D12 (Figure 2F), consistent with the role played by Cited2 in liver development (Qu et al., 2007). Interestingly, the treatment of C2fl/fl[Cre] ESC with 4HT at D2 of differentiation only mildly affected the emergence of beating clusters, while no alteration was observed after 4HT treatment at D4 or D6 of differentiation in comparison with control cells (Figure 2D). These observations suggest that Cited2 supports ESC specification to cardiomyocyte lineages during the first 2–3 days of differentiation. In agreement with these results, we showed that Cited2-null C2Δ/Δ[LA11] ESC devoid of Cited2 expression (Kranc et al., 2015) have a reduced capacity to generate cardiac contractile clusters when compared to control C2fl/fl ESC (Figure 3A). To confirm that the cardiac differentiation defects observed in Cited2-depleted cells were due to the loss of Cited2 expression, C2fl/fl[Cre] ESC treated with 4HT at D0 were transduced with a human recombinant CITED2 protein (sharing interchangeable functions with the mouse protein in cardiac development [Chen et al., 2012, Kranc et al., 2015]) fused at its N-terminal domain with a stretch of eight arginines (termed 8R-CITED2). Polyarginine peptides are transduction domains that enable proteins to cross the cellular membrane when applied to the culture medium (Lundberg et al., 2003). The purified 8R-CITED2 added into the culture medium crossed the cellular membrane, translocated into the nucleus of Cited2-null ESC, and interfered with the high-affinity binding of overexpressed human CITED2 protein to the CH1 domain of p300 (Freedman et al., 2003), suggesting that 8R-CITED2 is functional (Figure S2). Interestingly, the supplementation of 8R-CITED2 at D2 of differentiation in the culture medium of C2fl/fl[Cre] ESC treated with 4HT at D0 restored the emergence of beating foci to control levels (Figure 3B). These results argue that the cardiogenic defects of Cited2-depleted ESC are caused by the loss of Cited2 expression. Altogether, these observations suggested that Cited2 is important for early events of ESC commitment to cardiac cell lineages.
Figure 2.
Cited2 Is Important for Early Steps of Cardiac Differentiation
(A) Timeline depicting the protocol used for differentiation of mouse ESC from D0 onward. The time of ethanol or 4HT treatment and the days of beating activity assessment are indicated.
(B) Expression of Cited2 determined by qPCR at D0, D2, D3, D5, and D12 of differentiation in cells derived from C2fl/fl[Cre] ESC treated at D0 with 4HT or ethanol for 48 hr.
(C) CITED2 protein levels determined by western blotting in extracts from C2fl/fl[Cre] ESC, 48 hr after incubation with ethanol or 4HT. Loading in each lane was monitored by detection of β-TUBULIN.
(D) Percentage of colonies with beating foci derived from C2fl/fl[Cre] ESC treated with ethanol or 4HT at D0, D2, D4, and D6 of differentiation.
(E) Quantification by flow cytometry of cells expressing cTNT at D12, treated as described in (B). The percentage of cells expressing cTNT over the total number of cells is indicated.
(F) Expression levels determined by qPCR of Myh6, cTnT, Foxa2, α-fetoprotein (Afp), Nestin, and βIII-tubulin (βIII-tub.) at D12 of differentiation in cells treated as described in (B).
(G) Immunofluorescent detection of α-ACTININ (left panels, red staining) and MYOSIN HEAVY CHAIN MF20 (right panels, red staining) in C2fl/fl[Cre] ESC-derived cells at D10 of differentiation, after treatment for 2 days with ethanol (upper panels) or 4HT (lower panels) at D0. Nuclei were counterstained using DAPI (blue), and cells were examined on at 100× magnification.
Results in (B), (D), (E), and (F) are presented as the mean ± SEM of three independent biological experiments.
Figure 3.
Loss of Cited2 Impairs Expression of Genes Specifying Cardiac Mesoderm
(A) Percentage of colonies with beating foci derived from C2fl/fl and C2Δ/Δ[LA11] ESC at D8 and D10 of differentiation.
(B) Percentage of colonies with beating foci counted at 8 and 10 days after the initiation of differentiation in cell cultures derived from C2fl/fl[Cre] ESC treated with ethanol or 4HT at D0 of differentiation, and with 4HT at D0 of differentiation and supplemented with recombinant 8R-CITED2 protein at D2 of differentiation (4HT + 8R-CITED2).
(C) Expression of Activin A and Nodal determined by qPCR at D2 and D3 of differentiation in cultures derived from C2fl/fl[Cre] ESC treated with 4HT or ethanol at D0 for 48 hr.
(D) Expression of mesoderm markers (Brachyury and Mesp1) at D2, D3, D5, and D12 of differentiation in cells generated from C2fl/fl[Cre] ESC treated as described in (Figure 2B).
(E) Expression of Isl1, Gata4, Nkx2.5, and Tbx5 in cell cultures as described in (D). The inserts for Isl1 and Tbx5 detail the expression of these genes at D5 and D3, respectively.
(F) Relative expression of Brachyury, Mesp1, Isl1, Gata4, Nkx2.5, and Tbx5 determined by qPCR at D5 of differentiation in cultures derived from untreated C2fl/fl and C2Δ/Δ[LA11] ESC, and C2Δ/Δ[LA11] ESC supplemented with the recombinant 8R-CITED2 protein (C2Δ/Δ[LA11] + 8R-CITED2) at D2 of differentiation for 48 hr. Gene expression in C2fl/fl ESC was set to 1.
(G) Enrichment of Isl1 genomic regions in extracts of E14/T ESC-derived cells at D5 by ChIP assays with anti-CITED2 or control (immunoglobulin G) polyclonal antibodies.
Results are presented as the mean ± SEM of three independent biological experiments.
Cited2 Is Necessary for the Expression of Genes Specifying Cardiac Mesoderm and Cardiac Progenitors
During early differentiation, the expression of pluripotency markers Oct4, Sox2, and Nanog was silenced with comparable rates in control and 4HT-treated cells (Figure S1D). Thus, a misregulation of pluripotency factors cannot account for the deficiency in cardiac differentiation of Cited2-depleted ESC. Next, we assessed the expression of genes marking early (cardiac) mesoderm induction such as Activin A, Nodal, Brachyury, and Mesp1 (Figures 3C and 3D). No significant alteration in Nodal and Activin A expression was observed at D2 and D3 in control and D0 Cited2-depleted cell cultures (Figure 3C). In contrast, Brachyury expression was significantly reduced at D3 but restored at D5, and Mesp1 expression was markedly downregulated at D3 and D5 (Figure 3D). The decrease of Brachyury and Mesp1 expression is in agreement with a previous report that had indicated that Cited2 supports mesoderm differentiation (Li et al., 2012) and with our observations suggesting that CITED2 is important during the first 2–3 days of ESC differentiation (Figure 2). In addition, the expression of Isl1, Gata4, and Tbx5 transcripts, as well as the expression of Isl1 and Gata4 proteins, was reduced at D5 of differentiation in Cited2-depleted cells (Figures 3E and S1B). Isl1 expression levels were also lower at D2 while Tbx5 expression remained decreased at D12 (Figure 3E). The expression of Nkx2.5 was unchanged in similar conditions (Figure 3E).
Since the treatment of C2fl/fl[Cre] ESC with 4HT at D0 only achieved a partial knockout of Cited2 (Figure 2B), the remaining Cited2 expression may confound the effects of Cited2 depletion on gene expression. Therefore, to clarify the consequences of Cited2 loss on Brachyury, Mesp1, Isl1, Gata4, Nkx2.5, and Tbx5 expression, we differentiated control C2fl/fl ESC and Cited2-null C2Δ/Δ[LA11] ESC, which are impaired for cardiac differentiation (Figure 3B). In this context Gata4 expression was not altered, but a significant downregulation of Brachyury, Mesp1, Isl1, Nkx2.5, and Tbx5 expression was detected in cultures derived from C2Δ/Δ[LA11] ESC at D5 of differentiation in comparison with control cells (Figure 3F). Interestingly, supplementation of C2Δ/Δ[LA11] ESC-derived cells at D2 of differentiation with 8R-CITED2, stimulated Brachyury, Mesp1, Isl1, Nkx2.5, and Tbx5 expression at D5 of differentiation with Isl1 expression being restored to control levels (Figure 3F). This observation further argues for a mechanistic link between Cited2 and Isl1 expression levels. To determine whether endogenous CITED2 is recruited to the promoter of Isl1 at D5 of differentiation, we performed ChIP using anti-CITED2 and control polyclonal antibodies with cellular extracts prepared from cells derived from E14/T ESC (Figure 3G). Endogenous CITED2 was specifically detected at the promoter region of Isl1 in these cells at D5 of differentiation, suggesting that CITED2 binds to Isl1 promoter in cells derived from ESC. Together, these observations suggested that Cited2 not only supports mesoderm induction but also regulates the expression of key genes during CPC specification including Isl1.
Cited2 Is Expressed in Cardiac Progenitors
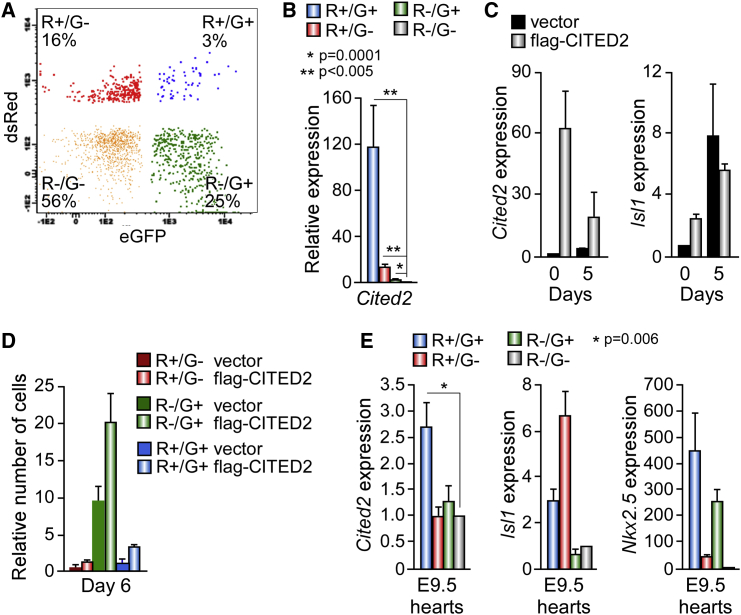
Since Cited2 expression was associated with the expression of CPC markers, such as Isl1, Nkx2.5, Gata4, and Tbx5 (Figures 1D, 3E, and 3F), we sought to determine whether Cited2 was expressed in CPC. For this purpose, we used mouse AD2 ESC harboring the dsRed (R) and eGFP (G) genes under the control of the Mef2c/AHF and Nkx2.5 cardiac-specific enhancers, respectively (Domian et al., 2009). The differentiation of AD2 ESC originates R+/G−, R−/G+, R+/G+, and R−/G− cell populations equivalent to CPC of the pharyngeal mesoderm, FHF CPC, committed ventricular CPC of the SHF, and non-cardiac cells, respectively (Domian et al., 2009). Cited2 expression, assessed by qPCR in AD2-derived cell populations isolated by fluorescence-activated cell sorting at D6 of differentiation, revealed that Cited2 transcripts were enriched in all CPC populations when compared with R−/G− cells, with higher levels of expression observed in R+/G+ cells (Figures 4A and 4B). The expression of Isl1, Nkx2.5, Gata4, and Tbx5 in the distinct CPC populations (Figure S1E) was as previously reported (Domian et al., 2009). To determine whether Cited2 promotes the emergence of CPC, 2 days prior to differentiation we transiently transfected AD2 ESC with either a control vector or the plasmid expressing flag-CITED2 (Figure 4C). The expression of flag-CITED2 stimulated endogenous Isl1 expression at D0, as observed in E14/T cells (Figure 1D), and significantly increased the number of R+/G+ cells derived from AD2 ESC differentiation at D6 (Figure 4D). In addition, the expression profile of Cited2 in cardiac progenitors in vivo was assessed in CPC populations isolated from embryonic day 9.5 (E9.5) hearts of mouse transgenic embryos harboring the dsRed and eGfp genes (Domian et al., 2009). Interestingly, Cited2 expression was enriched in the R+/G+ subpopulation simultaneously expressing Isl1 and Nkx2.5 (Figure 4E). Collectively, these observations indicated that Cited2 is expressed in FHF and SHF CPC derived from ESC.
Figure 4.
CITED2 Is Expressed in Cardiac Progenitors and its Overexpression Promotes Cardiac Progenitor Specification
(A) Representative flow cytometry plots of the R+/G−, R−/G+, R+/G+, and R−/G− cell populations derived from AD2 ESC at D6 of differentiation. Numbers indicate percentage of cells within each gate.
(B) Relative expression of Cited2 determined by qPCR in cell populations derived from AD2 at D7 differentiation. Expression of the indicated genes is reported relative to their expression in non-cardiac cells R−/G− set at 1.
(C) Relative expression of ectopic CITED2 and endogenous Isl1 determined by qPCR at D0 and D5 of differentiation, in AD2 ESC transiently transfected with a plasmid expressing flag-CITED2 or a control vector. flag-Cited2 and Isl1 expressions are relative to their expression in cells transfected with the control vector set at 1.
(D) Quantification, at D6 of differentiation by flow cytometry, of R+/G−, R−/G+, R+/G+, and R−/G− populations derived from AD2 ESC transfected with a plasmid expressing flag-CITED2 or a control vector. Cell numbers are reported relative to background determined at the onset of differentiation, which was set at 1.
(E) Relative expression of Cited2, Isl1, and Nkx2.5 determined by qPCR in embryonic cardiac progenitor populations at E9.5.
Results in (B) and (E) are presented as the mean ± SEM of three independent biological experiments, while results in (C) and (D) are presented as the mean ± SEM of two independent biological experiments each performed in technical triplicates.
CITED2 and ISL1 Proteins Interact and Synergize to Enhance Cardiac Differentiation
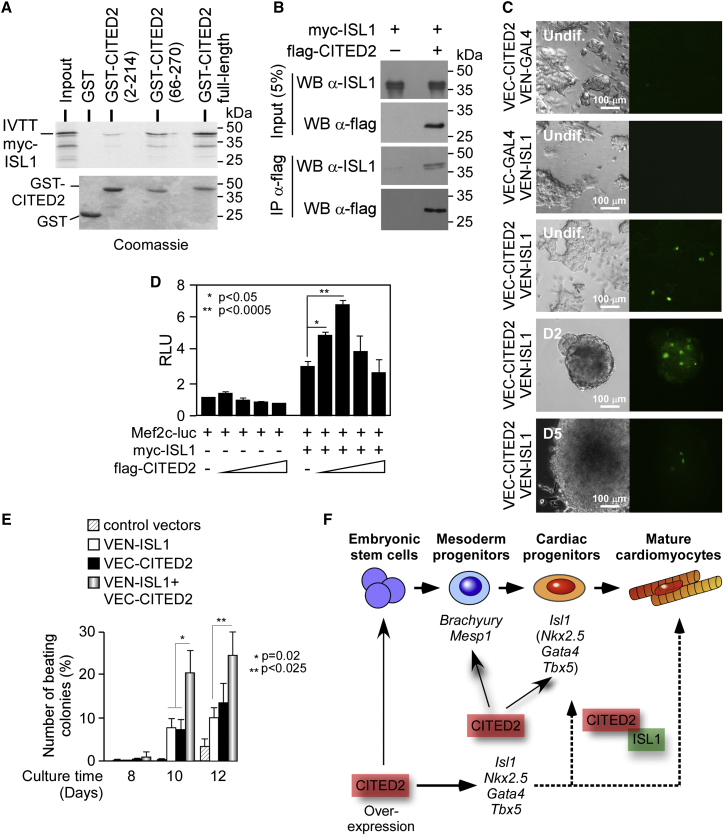
ISL1 is a protein with two LIM domains located at its N-terminal domain, both important for SHF expansion and morphogenetic control of cardiogenesis (Witzel et al., 2012). Since CITED2 binds to the LIM domain of the transcription factor LHX2 (Glenn and Maurer, 1999), we hypothesized that ISL1 and CITED2 proteins might physically interact. We therefore synthesized a 35S-labeled myc-tagged full-length human ISL1 protein (myc-ISL1) by coupled in vitro transcription-translation, and tested for its ability to bind to a glutathione S-transferase (GST)-CITED2 fusion protein (Figure 5A). Myc-ISL1 interacted specifically with GST-CITED2 full-length protein and the C-terminal residues of CITED2 (amino acids 66–270). It also interacted weakly with CITED2 N-terminal residues (amino acids 2–214). To confirm the interaction between CITED2 and ISL1 in cells, we used the anti-FLAG antibody to immunoprecipitate protein extracts from HEK293T cells expressing either myc-ISL1 alone or myc-ISL1 and flag-CITED2. Immunopurified proteins were then analyzed by western blot with anti-ISL1 and anti-FLAG antibodies (Figure 5B). Myc-ISL1 was co-immunoprecipitated with flag-CITED2, implying that ISL1 and CITED2 were in protein complexes in HEK293T cells. We also visualized ISL1-CITED2 interaction in living cells using vectors expressing either ISL1 in fusion with the N-terminal domain of the fluorescent protein VENUS (VEN-ISL1), CITED2 in fusion with the C-terminal domain of VENUS (VEC-CITED2), or control vectors to perform bifluorescence complementation (BiFC) assays (Machado-Oliveira et al., 2015). The co-transfection of VEN-ISL1 and VEC-CITED2 in E14/T cells resulted in a specific detection of fluorescence, confirming that ISL1 and CITED2 associate in these cells (Figures 5C and S3).
Figure 5.
CITED2 Binds to ISL1 and Both Synergize to Enhance Cardiac Differentiation
(A) Binding of 35S-labeled myc-ISL1 protein either to GST alone or GST fused to full-length CITED2 (GST-CITED2, amino acid residues 2–270), the N-terminal fragment GST-CITED2(2–214), or the C-terminal fragment GST-CITED2(66–270). Ten percent of labeled myc-ISL1 used for the binding assay was loaded as the input. Top panel: autoradiogram showing the binding of 35S-labeled ISL1 to GST and GST-CITED2 proteins. Bottom panel: Coomassie blue stain of the gels showing relative amounts of GST and GST-CITED2 proteins.
(B) Whole-cell extracts from HEK293T cells expressing myc-ISL1 alone or in combination flag-CITED2 immunoprecipitated (IP) with an anti-FLAG antibody, and proteins detected by western blot (WB) with anti-ISL1 and anti-FLAG antibodies. Five percent of the input was also loaded.
(C) Interaction between CITED2 and ISL1 visualized by BiFC assays in undifferentiated E14/T ESC. Fluorescence emission (right panels) and morphological aspect of E14/T cells as observed in bright field (left panels) was examined 1 day after transfection and in embryoid bodies at D2 and D5 of differentiation.
(D) Hep3B cells were transiently co-transfected with 40 ng of Mef2c-luc reporter, together with 100 ng of either myc-ISL1 expressing or empty control vector, and increasing amounts of a flag-CITED2-expressing plasmid (0, 50, and 100 ng) combined with the empty plasmid to achieve a total of 400 ng in each condition. The luciferase activity was normalized for the β-GALACTOSIDASE activity conferred by CMV-lacZ (100 ng). Relative luminescence units (RLU) are presented relative to values of the Mef2c-luc transfected with the control vectors set at 1.
(E) Percentage of colonies with beating foci at D8, D10, and D12 in cultures derived from differentiation of E14/T ESC transfected individually or in combination with vectors expressing VEN-ISL1 and VEC-CITED2, or control vectors at D0.
(F) Model for the role of CITED2 during cardiogenesis of ESC, as supported by the data from our study. CITED2 is required for the normal expression of mesoderm progenitor markers such as Brachyury and Mesp1. In addition, at this stage CITED2 contributes to the expression of Isl1, Nkx2.5, Gata4, and Tbx5, which are CPC markers. The overexpression of CITED2 triggers an increase of Isl1, Nkx2.5, Gata4, and Tbx5 expression in undifferentiated ESC, which may favor cardiac differentiation. Finally, CITED2 and ISL1 proteins physically interact and cooperatively promote cardiac differentiation.
Results in (D) and (E) are presented as the mean ± SEM of three independent biological experiments.
To test the functional significance of CITED2-ISL1 interaction, we co-transfected Hep3B cells with vectors expressing myc-ISL1 and flag-CITED2 together with a reporter construct harboring luciferase under the control of a composite regulatory region containing the Mef2c cardiac-specific enhancer, which is a direct target of Isl1 (Witzel et al., 2012). The transfection of myc-ISL1 vector increased the reporter activity, and this was further enhanced by co-transfection of flag-CITED2 vector (Figure 5D). These observations provide evidence that myc-ISL1 and flag-CITED2 synergize to stimulate the activity of an ISL1-responsive reporter.
To determine whether CITED2 and ISL1 cooperate to promote ESC differentiation toward cardiac cell fate, the number of beating foci originating from undifferentiated E14/T ESC expressing VEC-CITED2 and VEN-ISL1 individually or in combination was evaluated at D8, D10, and D12 of differentiation (Figure 5E). The transfection of VEC-CITED2 or VEN-ISL1 individually led to the emergence of beating foci at similar levels, while the co-transfection of VEC-CITED2 and VEN-ISL1 significantly increased the number of contractile foci at D12. Altogether, these observations provide evidence for a physical and functional interaction between ISL1 and CITED2 that promotes ESC differentiation toward cardiac cell lineages.
Discussion
In this study, we show that Cited2 is required for early cardiac commitment of mouse ESC, in agreement with a previous study (Li et al., 2012). We and others have demonstrated that Cited2 expression promotes mouse ESC self-renewal (Chen et al., 2012, Kranc et al., 2015, Pritsker et al., 2006). Interestingly, we show that the deletion of Cited2 in ESC or depletion of Cited2 at the onset of differentiation impairs cardiac lineage commitment. Remarkably, we determine that endogenous expression of Cited2 transcripts is biphasic during ESC differentiation, starting with a decrease from D0 to D2. This decline of Cited2 expression upon differentiation might be necessary for ESC to switch from a non-permissive to a permissive differentiation state. In a second phase, Cited2 expression increases from D3 of differentiation onward, implying that Cited2 might be required for subsequent differentiation processes. This is corroborated by the rescue of the cardiac differentiation defects in Cited2-depleted ESC with the supplementation of the recombinant CITED2 protein to the cells at D2 of differentiation, a day before the levels of endogenous Cited2 start to increase during differentiation.
The impairment of cardiac differentiation resulting from Cited2 depletion at the onset of ESC differentiation may be due to a consequent downregulation of Brachyury and Mesp1 expression. The decrease of Isl1, Nkx2.5, Gata4, and Tbx5 expression that we observed in Cited2-depleted cells at D5 of differentiation, which corresponds to the time when CPC emerge, might be a consequence of Mesp1 downregulation, since these genes are activated by MESP1 to promote cardiogenesis and CPC specification during cardiac development and ESC differentiation (Bondue et al., 2008, David et al., 2011). On the other hand, we show that Cited2 expression is enriched in CPC, and CITED2 protein is recruited to Isl1 promoter at D5 of differentiation during CPC specification. This suggests that CITED2 may play a role in CPC specification, proliferation, and/or differentiation. In agreement with this hypothesis, flag-CITED2 overexpression promoted both the emergence of CPC and terminal cardiomyocyte differentiation from ESC, and increased Isl1, Nkx2.5, Gata4, and Tbx5 expression in undifferentiated ESC. Fluctuations in the expression of genes encoding transcription factors with capacities to instruct lineage specification have been evidenced in undifferentiated pluripotent ESC and, in accordance with the transcripts expressed, subsets of ESC might be prone to undergo particular differentiation programs (Lanner and Rossant, 2010). The overexpression of CITED2 in undifferentiated ESC might specify and promote CPC and cardiac differentiation by raising the expression of the pro-cardiogenic factors Isl1, Nkx2.5, Gata4, and Tbx5. Interestingly, the transdifferentiation of human dermal fibroblasts into cardiac progenitors by expression of MESP1 and ETS2 has been shown to stimulate the expression of CITED2 (Islas et al., 2012), suggesting that CITED2 may play a role in cardiac progenitor specification and/or functions. Therefore, CITED2 overexpression might be instrumental for specification of CPC and cardiomyocyte differentiation from pluripotent stem cells.
We also provided evidence for a privileged regulatory interaction between Cited2 and Isl1. Indeed, our ChIP assays show that CITED2 is specifically recruited at the Isl1 promoter. Furthermore, supplementation of Cited2-null ESC with CITED2 recombinant protein restores the expression of Isl1 to normal levels at D5 of ESC differentiation. In addition, ESC overexpressing flag-CITED2 are enriched for H3triMeK4 at the Isl1 regulatory elements, which marks transcriptionally active chromatin. The mechanisms by which flag-CITED2 overexpression contribute to the enrichment of H3triMeK4 at the Isl1 loci and increase Gata4 and Nkx2.5 expression in undifferentiated ESC remain to be elucidated. Interestingly, at the protein level we demonstrated that ISL1 and CITED2 interact, delineated the amino acids 66–214 of CITED2 as part of the ISL1 interacting domain, and by transient transfection assays established that CITED2 increased ISL1-mediated Mef2c-enhancer activity and that ISL1 and CITED2 have a synergistic effect on cardiac cells derived from ESC. The exact molecular mechanism by which CITED2 and ISL1 cooperate in cardiac specification remains to be clarified, but it has been recently demonstrated that in mouse embryonic hearts and cardiac progenitors ISL1 interacts with p300 to selectively promote H3 acetylation at the Mef2c promoter (Yu et al., 2013). Since CITED2 interacts strongly with p300, it would be of interest to determine whether CITED2 cooperates with p300 to promote its functional interaction with ISL1.
Altogether, our results indicated that Cited2 contributes to the expression of a subset of pivotal cardiopoietic genes involved in mesoderm and cardiac progenitor specification (Figure 5F). During mouse embryonic development, Cited2 expression was detected in the early mesoderm- and cardiac-derived structures (Dunwoodie et al., 1998), but rather surprisingly a Brachyury/T-Cre or Mesp1-Cre conditional Cited2 knockout only resulted in infrequent and minor heart developmental defects of mouse embryos, while Cited2 knockout in the epiblast consistently caused heart malformations (Bamforth et al., 2001, MacDonald et al., 2008). In the present report, we show that Cited2 depletion at the onset of differentiation causes the most severe impact on cardiac differentiation, and results also in the impairment of Brachyury and Mesp1 expression at D3. Interestingly, the cardiogenic defects due to the loss of Cited2 expression at D0 were reversed by supplementation of the 8R-CITED2 at D2 of differentiation. On the other hand, Cited2 depletion at later time points (D2, D4, and D6) had little or no effect on cardiogenesis. Together, these observations indicate that Cited2 function is important for early commitment of ESC to mesoderm and/or cardiac specification, or at least contribute to the correct expression of Brachyury and Mesp1, or other genes crucial for these processes. Therefore, the lack of a strong phenotype in Brachyury/T-Cre and Mesp1-CreCited2 conditional knockout might be because the depletion of Cited2 in these embryos was triggered after Brachyury and Mesp1 were activated and after the requirement for Cited2. Of particular interest, we show that CITED2 stimulates the expression of Isl1, a marker of SHF cardiac progenitors, and binds to its promoter at the time of CPC specification. We also show that Cited2 expression is enriched in SHF cardiac progenitors derived either from ESC or mouse E9.5 embryonic hearts. Moreover, Cited2-null embryos display a variety of cardiac developmental defects such as ventricular septal defects with overriding aorta or double-outlet right ventricle, outflow tract defects, and transposition of the great arteries, which may result from anomalies of SHF and cardiac neural crest cell progenitors known to express ISL1 (Bamforth et al., 2001, Bruneau, 2013). Therefore, CITED2 and ISL1 may also interact during heart development and play a critical role for the fulfillment of CPC functions in this process. It would be of interest to investigate the contribution of the CITED2-ISL1 interaction in CPC functions both in vitro and in vivo.
Experimental Procedures
All mice were cared for within the Animal Care facilities of the Massachusetts General Hospital under the supervision of an active and functioning Subcommittee on Research Animal Care (SRAC), which serves as the Institutional Animal Care and Use Committee (IACUC) as required by the Public Health Service (PHS) Policy on Humane Welfare Regulations.
Embryonic Stem Cells, Culture Conditions, and Isolation of Cardiac Progenitor Populations
Apple D2 (AD2), C2fl/fl, C2Δ/Δ[LA11], C2fl/fl[Cre], and E14/T mouse ESC lines were described previously, and were cultured on gelatin-coated plates in undifferentiating medium supplemented with LIF (Chambers et al., 2003, Domian et al., 2009, Kranc et al., 2015). All ESC lines were differentiated using the hanging-drop method in medium containing 20% fetal bovine serum without LIF supplementation (differentiation medium).
Flow Cytometry Analysis
For detection of cardiac troponin I type 3 (cTNT), cells were fixed with 0.5% paraformaldehyde for 20 min at room temperature, blocked, and permeabilized with PBS containing 0.5% BSA and 0.1% saponin for 5 min at 4°C, washed with blocking solution, and incubated for 1 hr at 4°C with a monoclonal anti-mouse cTNT antibody (NB110-2546, Novus Biologicals) at a 1:800 dilution followed by 1 hr of incubation at 4°C with a secondary goat anti-mouse immunoglobulin G conjugated with Alexa 488 (A21202, Life Technologies) used at 1:2,000 dilution. Flow cytometry analyses were performed on a FACSCalibur (BD Biosciences) operating at 488 nm excitation with standard emission filters. Baseline of noise fluorescence was established with cells incubated only with the secondary antibody.
Real-Time qPCR
Total RNA isolation, cDNA synthesis, and qPCR assays were carried out as previously described (Kranc et al., 2015) with the primers listed in Table S1. The primer set designated Cited2 detects both mouse endogenous Cited2 and human exogenous flag-CITED2. For the selection of reliable reference genes, the expression of three reference genes, hprt, Gapdh, and 18S was tested in samples prepared from C2fl/fl[Cre] and E14/T ESC at different time points of the differentiation (Figure S3E). Gapdh and 18S showed a very consistent expression across the differentiation process of both cell types. We opted for the normalization of gene expression levels to Gapdh, as previously performed (Kranc et al., 2015).
Immunochemistry
Immunocytochemistry was performed with C2fl/fl[Cre] ESC treated with ethanol or 4HT at D0 and differentiated for 10 days. Western blotting assays were performed using 20 μg of whole-cell lysates prepared from the indicated mouse ESC as previously described (Kranc et al., 2015).
Chromatin Immunoprecipitation Assays
ChIP experiments and enrichment of target genomic elements by qPCR were performed as previously described (Kranc et al., 2015) using the primers listed in Table S1.
Production and Transduction of the Recombinant 8R-CITED2 Protein
Full-length human CITED2 cDNA and an oligonucleotide encoding eight arginines (8R) were cloned into the pGEX6P1 vector (GE Healthcare Life Sciences) to express a chimeric protein consisting of the GST in fusion with the 8R domain and CITED2 (termed GST-8R-CITED2, Figure S2). Newly constructed plasmids were validated by sequencing and the expression of fusion proteins tested by western blot (Figure S2). Details of plasmid construction and protein purification are available upon request.
Protein Interactions and Plasmids
For in vitro binding assays, GST-CITED2 fusion proteins and in vitro translated myc-ISL1 were prepared as previously described (Machado-Oliveira et al., 2015). Myc-ISL1 expression plasmid was constructed in pcDNA3 (Invitrogen) with an amino-terminal myc epitope tag. Plasmids expressing flag-CITED2 and pPyCAGIP were previously described (Chen et al., 2012). For BiFC assays, plasmids expressing VEC-CITED2, VEN-p300CH1, VEC-GAL4, VEN-GAL4, and the VEN vector were described elsewhere (Machado-Oliveira et al., 2015). The plasmid VEN-ISL1 expressing VEN (amino acid residues 1–155 of the VENUS fluorescent protein) in fusion with ISL1 was obtained by subcloning ISL1 cDNA fragment of the Myc-ISL1 expression plasmid into the VEN vector in frame with the VEN domain. All newly constructed plasmids were validated by sequencing and the expression of fusion proteins tested by western blot (Figure S3). Details of plasmid construction are available upon request. For co-immunoprecipitation assays, ∼0.4 mg of whole-cell extracts from HEK293T cells transfected with flag-CITED2 expression vector alone or together with myc-ISL1 vector were used.
Reporter Assays
Hep3B cells, a human hepatocellular carcinoma cell line with low levels of endogenous CITED2, were plated in 24-well plates at 2.5 × 104 cells per well and transfected the following day using Lipofectamine 2000 (Invitrogen) with Mef2c-luc reporter (Witzel et al., 2012) and expression vectors. CMV-lacZ plasmid was co-transfected in all experiments, and both LUCIFERASE and β-GALACTOSIDASE activities measured as previously described (Kranc et al., 2015).
Statistical Analysis
Statistical significance was determined by two-tailed Student's t tests assuming unequal variance. p Values of <0.05 were considered statistically significant.
Author Contributions
I.P.-L., conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; A.C.M., D.V.O., J.M.A.S., R.N., E.G., A.M., A.M.v.D.V., G.M.O., collection and/or assembly of data; G.F., conception and design, provision of equipment and reagents; I.B., conception and design, collection and/or assembly of data, financial support; J.B., conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. A.C.M., D.V.O., and J.M.A.S. contributed equally.
Acknowledgments
This work is supported by national Portuguese funding through FCT, Fundação para a Ciência e a Tecnologia, project UID/BIM/04773/2013 CBMR, projects PEst-OE/EQB/LA0023/2013 and PTDC/SAU-ENB/111702/2009 to J.B., U01HL100408, NIH/NHLBI to I.D., and SFRH/BPD/74807/2010 to G.M.O. I.P.L. thanks the Biomedical Sciences PhD program and FCT for funding (SFRH/BD/62054/2009). J.M.A.S. is a PhD student of the ProRegeM - PhD Program in Mechanisms of Disease and Regenerative Medicine of the University of Algarve and New University of Lisbon financed by the FCT. We thank Shoumo Bhattacharya (University of Oxford) for CITED2 expressing vectors, Austin Smith (University of Cambridge) for E14/T cells and pPyCAGIP vector, Gergana Dobreva (Max Planck Institute for Heart and Lung Research) for Mef2c-luc reporter, Claudia Florindo, head of the Microscopy Facility (University of Algarve), and José A. Belo (New University of Lisbon) for reagents.
Published: November 3, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.10.002.
Supplemental Information
References
- Bamforth S.D., Bragança J., Eloranta J.J., Murdoch J.N., Marques F.I.R., Kranc K.R., Farza H., Henderson D.J., Hurst H.C., Bhattacharya S. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat. Genet. 2001;29:469–474. doi: 10.1038/ng768. [DOI] [PubMed] [Google Scholar]
- Bamforth S.D., Bragança J., Farthing C.R., Schneider J.E., Broadbent C., Michell A.C., Clarke K., Neubauer S., Norris D., Brown N.A. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat. Genet. 2004;36:1189–1196. doi: 10.1038/ng1446. [DOI] [PubMed] [Google Scholar]
- Blin G., Nury D., Stefanovic S., Neri T., Guillevic O., Brinon B., Bellamy V., Rücker-Martin C., Barbry P., Bel A. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J. Clin. Invest. 2010;120:1125–1139. doi: 10.1172/JCI40120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A., Lapouge G., Paulissen C., Semeraro C., Lacovino M., Kyba M., Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Bondue A., Tannler S., Chiapparo G., Chabab S., Ramialison M., Paulissen C., Beck B., Harvey R., Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J. Cell Biol. 2011;192:751–765. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau B.G. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb. Perspect. Biol. 2013;5:a008292. doi: 10.1101/cshperspect.a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C.-L., Liang X., Shi Y., Chu P.-H., Pfaff S.L., Chen J., Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chan S.S.-K., Shi X., Toyama A., Arpke R.W., Dandapat A., Iacovino M., Kang J., Le G., Hagen H.R., Garry D.J. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. 2013;12:587–601. doi: 10.1016/j.stem.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-m., Bentham J., Cosgrove C., Braganca J., Cuenda A., Bamforth S.D., Schneider J.E., Watkins H., Keavney B., Davies B. Functional significance of SRJ domain mutations in CITED2. PLoS One. 2012;7:e46256. doi: 10.1371/journal.pone.0046256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforou N., Miller R.A., Hill C.M., Jie C.C., McCallion A.S., Gearhart J.D. Mouse ES cell–derived cardiac precursor cells are multipotent and facilitate identification of novel cardiac genes. J. Clin. Invest. 2008;118:894–903. doi: 10.1172/JCI33942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I., Pimeisl I.-M., Drager S., Bikoff E.K., Robertson E.J., Arnold S.J. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 2011;13:1084–1091. doi: 10.1038/ncb2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R., Jarsch V.B., Schwarz F., Nathan P., Gegg M., Lickert H., Franz W.-M. Induction of MesP1 by Brachyury(T) generates the common multipotent cardiovascular stem cell. Cardiovasc. Res. 2011;92:115–122. doi: 10.1093/cvr/cvr158. [DOI] [PubMed] [Google Scholar]
- Domian I.J., Chiravuri M., van der Meer P., Feinberg A.W., Shi X., Shao Y., Wu S.M., Parker K.K., Chien K.R. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326:426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwoodie S.L., Rodriguez T.A., Beddington R.S.P. Msg1 and Mrg1, founding members of a gene family, show distinct patterns of gene expression during mouse embryogenesis. Mech. Dev. 1998;72:27–40. doi: 10.1016/s0925-4773(98)00011-2. [DOI] [PubMed] [Google Scholar]
- Freedman S.J., Sun Z.-Y.J., Kung A.L., France D.S., Wagner G., Eck M.J. Structural basis for negative regulation of hypoxia-inducible factor-1a by CITED2. Nat. Struct. Mol. Biol. 2003;10:504–512. doi: 10.1038/nsb936. [DOI] [PubMed] [Google Scholar]
- Gai H., Leung E., Costantino P., Aguila J., Nguyen D., Fink L., Ward D., Ma Y. Generation and characterization of functional cardiomyocytes using induced pluripotent stem cells derived from human fibroblasts. Cell Biol. Int. 2009;33:1184–1193. doi: 10.1016/j.cellbi.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Garry D.J., Olson E.N. A common progenitor at the heart of development. Cell. 2006;127:1101–1104. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- Glenn D.J., Maurer R.A. MRG1 binds to the LIM domain of Lhx2 and may function as a coactivator to stimulate glycoprotein hormone alpha -subunit gene expression. J. Biol. Chem. 1999;274:36159–36167. doi: 10.1074/jbc.274.51.36159. [DOI] [PubMed] [Google Scholar]
- Ishitobi H., Wakamatsu A., Liu F., Azami T., Hamada M., Matsumoto K., Kataoka H., Kobayashi M., Choi K., Nishikawa S.-i. Molecular basis for Flk1 expression in hemato-cardiovascular progenitors in the mouse. Development. 2011;138:5357–5368. doi: 10.1242/dev.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas J.F., Liu Y., Weng K.-C., Robertson M.J., Zhang S., Prejusa A., Harger J., Tikhomirova D., Chopra M., Iyer D. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc. Natl. Acad. Sci. USA. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kitajima S., Takagi A., Inoue T., Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- Kouskoff V., Lacaud G., Schwantz S., Fehling H.J., Keller G. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc. Natl. Acad. Sci. USA. 2005;102:13170–13175. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranc K.R., Oliveira D.V., Armesilla-Diaz A., Pacheco-Leyva I., Matias A.C., Escapa A.L., Subramani C., Wheadon H., Trindade M., Nichols J. Acute loss of Cited2 impairs Nanog expression and decreases self-renewal of mouse embryonic stem cells. Stem Cells. 2015;33:699–712. doi: 10.1002/stem.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanner F., Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- Laugwitz K.-L., Moretti A., Lam J., Gruber P., Chen Y., Woodard S., Lin L.-Z., Cai C.-L., Lu M.M., Reth M. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Ramirez-Bergeron D.L., Dunwoodie S.L., Yang Y.-C. Cited2 controls pluripotency and cardiomyocyte differentiation of murine embryonic stem cells through Oct4. J. Biol. Chem. 2012;287:29088–29100. doi: 10.1074/jbc.M112.378034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg M., Wikstrom S., Johansson M. Cell surface adherence and endocytosis of protein transduction domains. Mol. Ther. 2003;8:143–150. doi: 10.1016/s1525-0016(03)00135-7. [DOI] [PubMed] [Google Scholar]
- MacDonald S.T., Bamforth S.D., Chen C.-M., Farthing C.R., Franklyn A., Broadbent C., Schneider J.E., Saga Y., Lewandoski M., Bhattacharya S. Epiblastic Cited2 deficiency results in cardiac phenotypic heterogeneity and provides a mechanism for haploinsufficiency. Cardiovasc. Res. 2008;79:448–457. doi: 10.1093/cvr/cvn101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald S.T., Bamforth S.D., Bragança J., Chen C.-M., Broadbent C., Schneider J.E., Schwartz R.J., Bhattacharya S. A cell-autonomous role of Cited2 in controlling myocardial and coronary vascular development. Eur. Heart J. 2013;34:2557–2567. doi: 10.1093/eurheartj/ehs056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Oliveira G., Guerreiro E., Matias A.C., Facucho-Oliveira J., Pacheco-Leyva I., Bragança J. FBXL5 modulates HIF-1α transcriptional activity by degradation of CITED2. Arch. Biochem. Biophys. 2015;576:61–72. doi: 10.1016/j.abb.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Moretti A., Caron L., Nakano A., Lam J.T., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martin-Puig S. Multipotent embryonic Isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Pritsker M., Ford N.R., Jenq H.T., Lemischka I.R. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2006;103:6946–6951. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Lam E., Doughman Y.Q., Chen Y., Chou Y.T., Lam M., Turakhia M., Dunwoodie S.L., Watanabe M., Xu B. Cited2, a coactivator of HNF4alpha, is essential for liver development. EMBO J. 2007;26:4445–4456. doi: 10.1038/sj.emboj.7601883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y., Miyagawa-Tomita S., Takagi A., Kitajima S., Miyazaki J.i., Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Sperling S., Grimm C.H., Dunkel I., Mebus S., Sperling H.-P., Ebner A., Galli R., Lehrach H., Fusch C., Berger F. Identification and functional analysis of CITED2 mutations in patients with congenital heart defects. Hum. Mutat. 2005;26:575–582. doi: 10.1002/humu.20262. [DOI] [PubMed] [Google Scholar]
- Tam P.P., Parameswaran M., Kinder S.J., Weinberger R.P. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: the role of ingression and tissue movement during gastrulation. Development. 1997;124:1631–1642. doi: 10.1242/dev.124.9.1631. [DOI] [PubMed] [Google Scholar]
- Van Vliet P., Wu S.M., Zaffran S., Pucéat M. Early cardiac development: a view from stem cells to embryos. Cardiovasc. Res. 2012;96:352–362. doi: 10.1093/cvr/cvs270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S.D., Buckingham M.E. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr. Top Dev. Biol. 2010;90:1–41. doi: 10.1016/S0070-2153(10)90001-X. [DOI] [PubMed] [Google Scholar]
- Weninger W.J., Floro K.L., Bennett M.B., Withington S.L., Preis J.I., Barbera J.P., Mohun T.J., Dunwoodie S.L. Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development. 2005;132:1337–1348. doi: 10.1242/dev.01696. [DOI] [PubMed] [Google Scholar]
- Witzel H.R., Jungblut B., Choe C.P., Crump J.G., Braun T., Dobreva G. The LIM protein ajuba restricts the second heart field progenitor pool by regulating Isl1 activity. Dev. Cell. 2012;23:58–70. doi: 10.1016/j.devcel.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M., Henckaerts E., Bonham K., Abbott G.W., Linden R.M. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yin Z., Haynie J., Yang X., Han B., Kiatchoosakun S., Restivo J., Yuan S., Prabhakar N.R., Herrup K., Conlon R.A. The essential role of Cited2, a negative regulator for HIF-1α, in heart development and neurulation. Proc. Natl. Acad. Sci. USA. 2002;99:10488–10493. doi: 10.1073/pnas.162371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Kong J., Pan B., Sun H., Lv T., Zhu J., Huang G., Tian J. Islet-1 may function as an assistant factor for histone acetylation and regulation of cardiac development-related transcription factor Mef2c expression. PLoS One. 2013;8:e77690. doi: 10.1371/journal.pone.0077690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.