Summary
Many solid cancers have an expanded CD44+/hi/CD24−/low cancer stem cell (CSC) population, which are relatively chemoresistant and drive recurrence and metastasis. Achieving a more durable response requires the development of therapies that specifically target CSCs. Recent evidence indicated that inhibiting the SUMO pathway repressed tumor growth and invasiveness, although the mechanism has yet to be clarified. Here, we demonstrate that inhibition of the SUMO pathway repressed MMP14 and CD44 with a concomitant reduction in cell invasiveness and functional loss of CSCs in basal breast cancer. Similar effects were demonstrated with a panel of E1 and E3 SUMO inhibitors. Identical results were obtained in a colorectal cancer cell line and primary colon cancer cells. In both breast and colon cancer, SUMO-unconjugated TFAP2A mediated the effects of SUMO inhibition. These data support the development of SUMO inhibitors as an approach to specifically target the CSC population in breast and colorectal cancer.
Keywords: breast cancer, colon cancer, cancer stem cell, sumoylation, TFAP2A, CD44, MMP14
Highlights
-
•
Sumoylation regulates CD44 and MMP14 expression in basal breast and colon cancer
-
•
SUMO inhibition clears cancer stem cells, repressing invasiveness and tumor growth
-
•
Anacardic acid functions as a SUMO inhibitor to repress cancer stem cells
-
•
TFAP2A mediates anti-tumor effects of SUMO inhibition in breast and colon cancers
Weigel and colleagues provide substantial evidence for developing cancer stem cell-specific therapy based on inhibiting the SUMO pathway. They show that inhibition of sumoylation enzymes by knockdown or small-molecule inhibitors repressed cancer stem cells with loss of CD44 and MMP14, and reduced invasiveness and inhibition of tumor growth. Common SUMO-sensitive mechanisms were dependent upon TFAP2A in breast and colon cancer.
Introduction
Breast cancer subtypes with particular molecular signatures, e.g., HER2+ and basal/triple-negative subtypes, have a worse prognosis with increased rates of recurrence and metastasis, likely due to an expansion of cancer stem cells (CSCs), alternatively referred to as tumor-initiating cells (TICs) (Blick et al., 2010, Park et al., 2010, Ricardo et al., 2011). Breast CSCs are characterized by the markers CD44+/hi/CD24−/low (Al-Hajj et al., 2003, Blick et al., 2010, Ricardo et al., 2011) and by expression of genes that promote epithelial-mesenchymal transition (EMT) (Blick et al., 2010, Mani et al., 2008), which is critical for cancer progression and metastasis (Choi et al., 2013, Sarrio et al., 2008, Sheridan et al., 2006, Thiery, 2002, Tsai and Yang, 2013). Aggressive cancers of other tissues of origin such as thyroid, colorectum, pancreas, and skin also demonstrate expansion of the CD44+/hi CSC population (Dou et al., 2007, Erfani et al., 2016, Jing et al., 2015, Liu and Brown, 2010, Parmiani, 2016). In contrast to the majority of cells in a tumor, CSCs/TICs have the ability to form tumor xenografts (Al-Hajj et al., 2003, Iqbal et al., 2013). Moreover, CSCs are relatively chemoresistant and become enriched after chemotherapy, leading to the theory that CSCs drive cancer recurrence and metastasis (Alamgeer et al., 2014, Iqbal et al., 2013, Lawson et al., 2015, Lee et al., 2011). Improvements in cancer therapy to achieve durable cancer remission or cure will require novel therapies that are cytotoxic to CSCs (Das et al., 2008).
There is growing interest in the role of sumoylation in regulating pathways critical to oncogenesis, cancer growth, and progression (Bettermann et al., 2012). Sumoylation is a process resulting in the reversible binding of a small ubiquitin-like modifier (SUMO) to a lysine residue in the target protein (Geiss-Friedlander and Melchior, 2007). Sumoylation is mediated through a cascade involving an activating enzyme (i.e., SAE1/2), E2-conjugating enzyme (i.e., UBC9), and E3 ligase (i.e., PIAS family) (Bettermann et al., 2012, Hay, 2005). Experimental methods to inhibit the SUMO pathway have relied on elimination of enzymes in the SUMO pathway or use of compounds that inhibit sumoylation enzymes, such as anacardic acid (Fukuda et al., 2009). Sumoylation has profound effects on gene expression, which likely involves post-translational modification of transcription factors by SUMO conjugation (Gill, 2005). EMT, and its converse, mesenchymal-epithelial transition, are regulated by transcription factors, many of whose activity is in turn regulated by SUMO conjugation (Bogachek et al., 2015a). We recently reported that sumoylation of transcription factor activator protein 2α (TFAP2A) in basal breast cancer alters its transcriptional activity and that SUMO-unconjugated TFAP2A acquires activity that results in a profound alteration of the expression profile away from the CSC/EMT phenotype and toward that of the well-differentiated phenotype, clearing of the CD44+/hi/CD24−/low CSC population, and repressing the TIC potential (Bogachek et al., 2014). Treatment of mice with anacardic acid inhibited the outgrowth of basal breast cancer xenografts, demonstrating the proof of principle that small-molecule SUMO inhibitors might form the basis of CSC-specific therapy (Bogachek et al., 2014, Bogachek et al., 2015b). Another recent study reported that knockdown of the SUMO enzyme PIAS1 repressed the TIC breast cancer population through epigenetic chromatin alterations resulting in gene silencing of cyclin D2, estrogen receptor, and WNT5A (Liu et al., 2014). Further studies have reported that knockdown of sumoylation enzymes impaired the outgrowth of colorectal cancer xenografts (He et al., 2015), suggesting the broad application of SUMO inhibitors in cancer therapy.
Several important questions need to be addressed concerning the clinical development of SUMO inhibitors in cancer therapy. First, the role of SUMO inhibitors in repressing the CSC/TIC population needs to be formally demonstrated. Second, the possibility that SUMO inhibitors such as anacardic acid act through off-target effects needs to be eliminated. Third, other carcinoma cell types need to be analyzed to determine whether similar SUMO-sensitive transcriptional mechanisms are operational. In the current study, we sought to address these critical questions by examining mechanisms of CSC maintenance in breast and colorectal cancer models.
Results
Knockdown of SUMO Pathway Suppressed CD44 and MMP14 and Inhibited Tumorigenesis
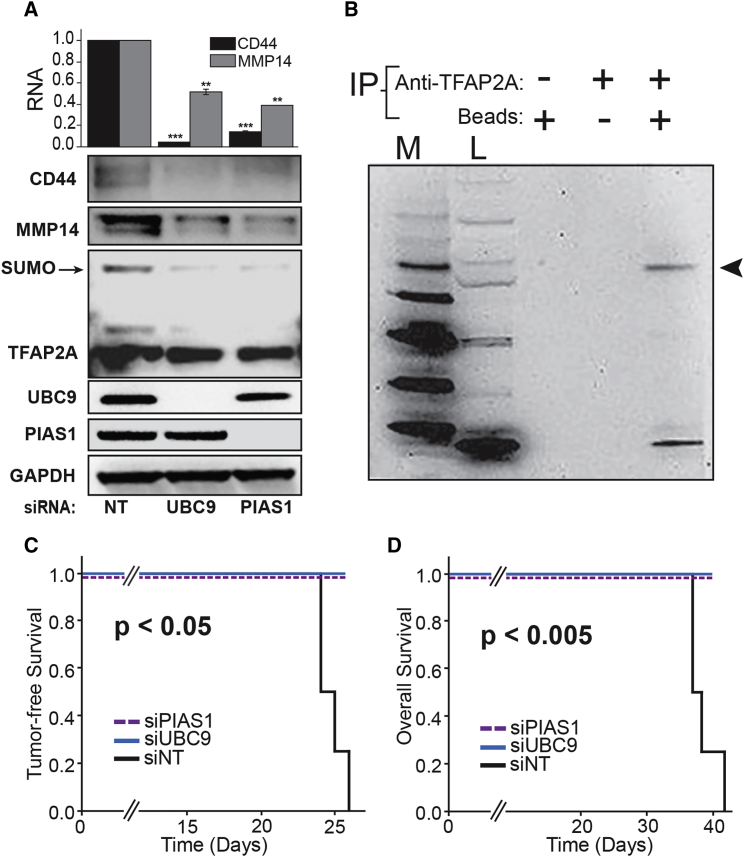
The CSC/EMT phenotype is characterized by the expression of several key drivers of cancer growth, invasion, and metastasis including CD44 and MMP14 (Godar et al., 2008, Hiraga et al., 2013, Yan et al., 2015). Previous studies demonstrated that anacardic acid repressed expression of CD44 and inhibited the outgrowth of basal/triple-negative breast cancer (TNBC) xenografts (Bogachek et al., 2014). We sought to confirm that knockdown of SUMO pathway enzymes replicated the effects of anacardic acid in basal breast cancer. Knockdown of UBC9 and PIAS1 in IOWA-1T basal breast cancer cells repressed expression of CD44 and MMP14 with elimination of SUMO-conjugated TFAP2A (Figure 1A). To clearly demonstrate that the 65-kDa isoform of TFAP2A was SUMO conjugated, we subjected extracts to immunoprecipitation with anti-TFAP2A versus immunoglobulin G (IgG) and resolution by western blot with anti-SUMO-1/2/3 antibody. The identity of the 65-kDa SUMO-conjugated TFAP2A isoform was confirmed (Figure 1B). Since knockdown of the SUMO pathway enzymes repressed the CSC marker CD44, the effect of inhibition of the SUMO pathway on tumorigenesis was examined. Knockdown of SUMO enzymes increased tumor-free survival and overall survival (Figures 1C and 1D), and demonstrated that previously reported effects of anacardic acid on tumorigenesis were likely mediated through SUMO inhibition.
Figure 1.
Inhibition of the SUMO Pathway Repressed CD44, MMP14, and SUMO-Conjugated TFAP2A and Repressed Xenograft Outgrowth
(A) Knockdown (KD) of UBC9 and PIAS1 with siRNA compared with non-targeting (NT) transfection repressed expression of CD44 and MMP14 RNA (top) (∗∗p < 0.01, ∗∗∗p < 0.001) and protein (bottom), and reduced SUMO-conjugated TFAP2A by western blot. Graph of RNA data was from three independent experiments. Relative CD44 protein: NT, 1.0; UBC9 KD, 0.18; PIAS1 KD, 0.28; relative MMP14 protein: NT, 1.0; UBC9 KD, 0.40; PIAS1 KD, 0.26.
(B–D) Immunoprecipitation (IP) (B) with first two lanes showing markers (M) and load (L) and IP of IOWA-1T protein extracts with anti-TFAP2A (+) or IgG control (−) with binding beads (+) or non-binding beads (−) with western blot probed with anti-SUMO-1/2/3 antibodies; arrowhead highlights SUMO-conjugated TFAP2A corresponding to 65 kDa. Free SUMO protein is seen at bottom of lane, likely from degradation of SUMO-conjugated TFAP2A. Knockdown of SUMO enzymes significantly increased tumor-free survival (C) and overall survival (D) of mice with IOWA-1T xenografts (n = 5 animals per experimental group and n = 4 animals per NT group).
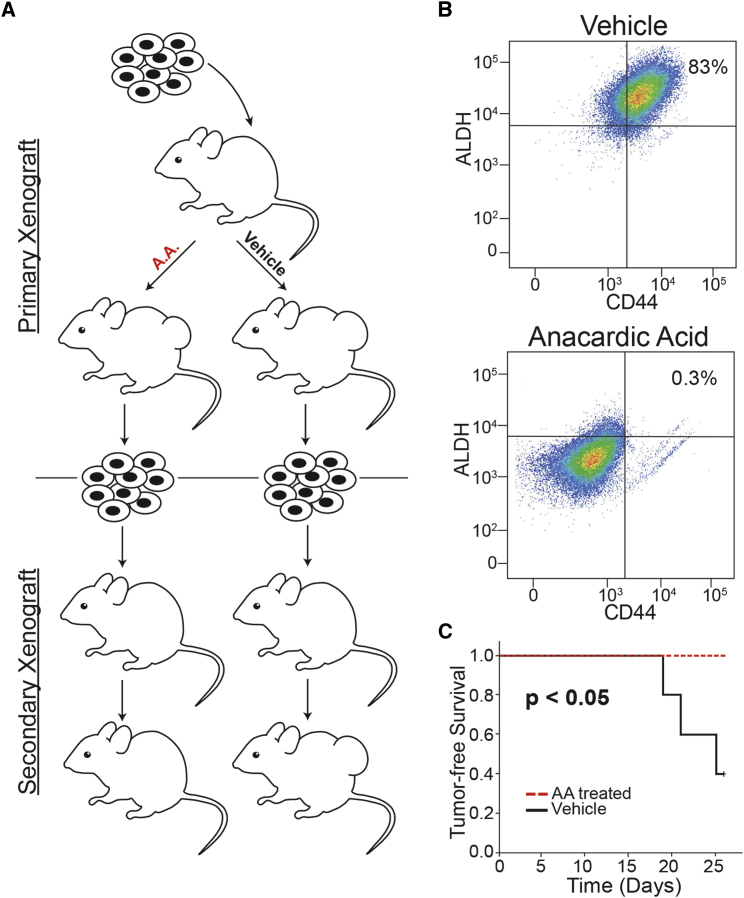
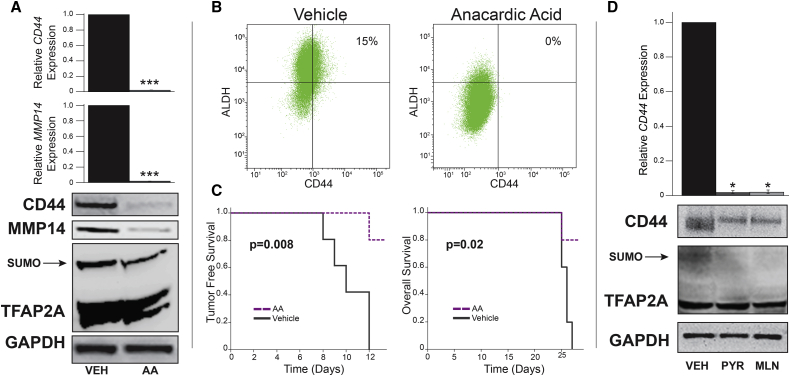
Previous tumorigenesis studies demonstrated a significant effect of SUMO inhibitors to repress the outgrowth of basal breast cancer xenografts (Bogachek et al., 2014, Bogachek et al., 2015b). However, we noticed that after 1 month approximately 40% of the mice treated with anacardic acid developed small xenografts and we hypothesized that this was possibly due to outgrowth of non-CSC. Serial propagation of secondary xenografts has been established as a property of the CSC/TIC population (Patel et al., 2012). Hence, we generated secondary xenografts from tumors that appeared after extended observation in animals treated with anacardic acid (Figure 2A). In addition to CD44, cells staining bright with ALDEFLUOR have been used to characterize breast CSCs (Ricardo et al., 2011). Cell suspensions isolated from xenografts that developed in animals treated with anacardic acid versus vehicle were subjected to fluorescence-activated cell sorting (FACS) analysis with CD44 and ALDEFLUOR, whereby cells isolated from anacardic acid-treated animals demonstrated a near complete loss of the CD44+/hi/ALDH+/hi CSC population compared with tumors arising in vehicle-treated animals (Figure 2B). The CD44+/hi/ALDH+/hi cell phenotype decreased from a baseline of 83% in tumors from vehicle-treated animals to <1% for tumors isolated from animals treated with anacardic acid. Cell suspensions isolated from primary xenografts were re-injected into naive mice. Cells isolated from xenografts that eventually formed in animals gavaged with anacardic acid were not capable of developing secondary xenografts even after extended observation over 6 months, confirming eradication of the CSC/TIC population (Figure 2C).
Figure 2.
Anacardic Acid Eliminated CSC in IOWA-1T Xenografts
(A) Diagram of experiment examining formation of secondary xenografts. Cells harvested from primary xenografts that formed in animals treated with vehicle versus anacardic acid (A.A.) were re-inoculated into naive animals with no further drug treatment.
(B) FACS analysis with ALDEFLUOR (ALDH) on vertical axis and CD44 (horizontal axis) of cells harvested from primary xenografts from animals treated with vehicle versus anacardic acid; negative controls with no staining control and ALDEFLUOR plus DEAB negative control from vehicle-treated cells is shown in Figure S1.
(C) Tumor-free survival of mice (n = 5 per group) forming secondary xenografts from experiment diagrammed in (A). Tumors from animals treated with anacardic acid (AA) failed to propagate as secondary xenografts; by comparison, tumors from vehicle-treated mice readily formed secondary xenografts.
SUMO Inhibitors Repress CSC Markers, Invasiveness, and Outgrowth of Xenografts
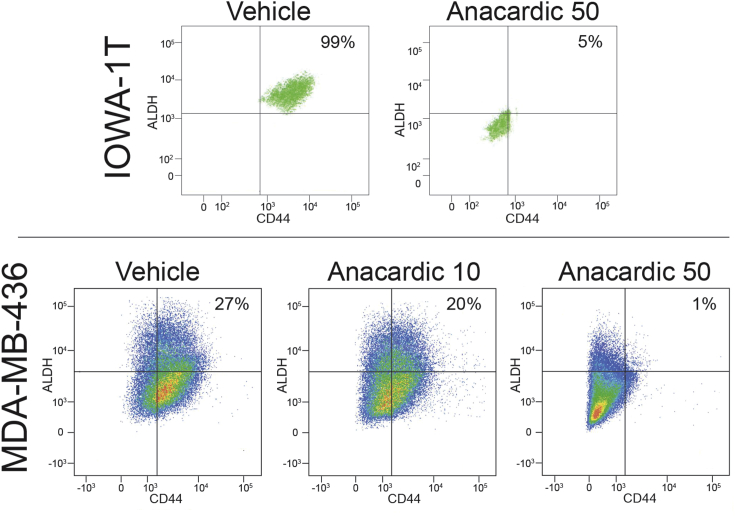
Previous studies in the TNBC cell lines BT-20 and BT-549 demonstrated that SUMO inhibitors effectively eliminated the CD44+/hi/CD24−/low CSC phenotype (Bogachek et al., 2014). Using the CSC markers CD44 and ALDEFLUOR, the effect of anacardic acid was further investigated in TNBC cells. By FACS analysis, anacardic acid significantly reduced the CD44+/hi/ALDH+/hi cell population in IOWA-1T cells from a baseline of 98.7% to 5.5%, which was consistent with a significant reduction of the CSC/TIC population (Figure 3A). Since nearly all of the IOWA-1T cells express CSC phenotypic markers, we tested the MDA-MB-436 cell line, in which approximately 30% of the cells express the CSC phenotype. Anacardic acid treatment resulted in a dose-dependent reduction in the CSC population from a baseline of 27% of the cells expressing CSC markers to 20% at 10 μM anacardic acid and approximately 1% with 50 μM anacardic acid (Figure 3B).
Figure 3.
Anacardic Acid Represses the CSC Phenotype in TNBC
IOWA-1T and MDA-MB-436 TNBC cell lines were treated with anacardic acid and analyzed by FACS analysis for expression of the CD44+/hi/ALDH+/hi CSC phenotype. Staining controls with no antibody and ALDEFLUOR plus DEAB are shown in Figure S2. In IOWA-1T, 50 μM anacardic acid decreased the CD44+/hi/ALDH+/hi CSC population from 98.7% to 5.5%. Similarly, there was a dose-dependent effect of anacardic acid in MDA-MB-436 cells reducing the baseline CD44+/hi/ALDH+/hi CSC population from 27.4% in vehicle-treated cells to 20.3% at 10 μM to 1.15% at 50 μM anacardic acid.
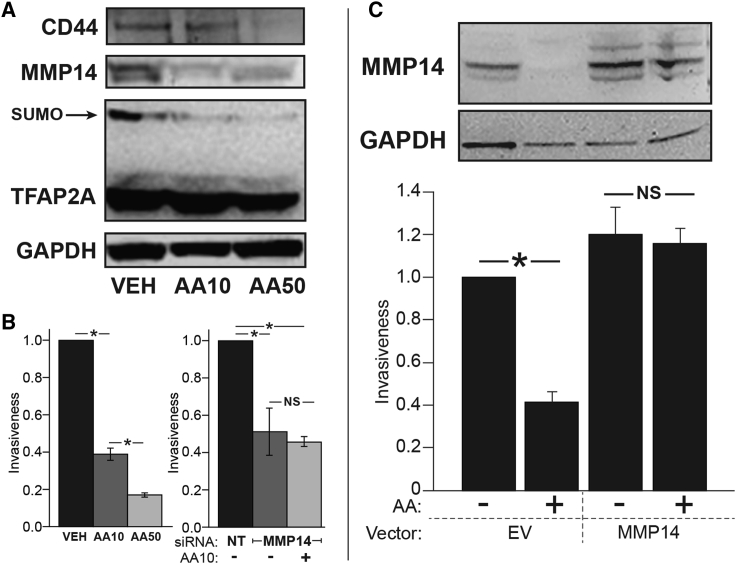
Similarly, anacardic acid repressed CD44 and MMP14 in a dose-dependent fashion with repression of the SUMO-conjugated form of TFAP2A (Figure 4A). Since MMP14 has effects on cancer cell invasion, the effect of anacardic acid on cell invasiveness was assessed. Anacardic acid treatment resulted in a dose-dependent reduction of invasiveness (Figure 4B). To prove that the effect on invasiveness was mediated through repression of MMP14, we manipulated MMP14 expression with anacardic acid treatment. Knockdown of MMP14 similarly reduced cell invasiveness, and knockdown of MMP14 effectively eliminated the ability for anacardic acid to further reduce invasiveness (Figure 4B, right panel). MMP14 was overexpressed by transfection of an expression vector; the expression of MMP14 achieved was approximately twice the baseline expression. MMP14 expression was repressed by anacardic acid in cells transfected with empty vector only. Forced expression of MMP14 slightly increased invasiveness and completely abrogated the effect of anacardic acid. Together, these results indicate that anacardic acid mediated anti-invasion through inhibition of MMP14.
Figure 4.
SUMO Inhibitors Repressed Invasiveness through MMP14
(A) Western blots for CD44, MMP14, and TFAP2A from IOWA-1T cells treated with vehicle (VEH), and anacardic acid at 10 μM (AA10) or 50 μM (AA50).
(B) Dose-dependent change in relative invasiveness of IOWA-1T with drug treatment (left panel): vehicle (VEH), and anacardic acid at 10 μM (AA10) or 50 μM (AA50) (∗p < 0.01); graph of data was from three independent experiments. Right panel: knockdown of MMP14 reduced invasiveness compared with non-targeting (NT) siRNA; knockdown of MMP14 eliminated the effect of anacardic acid at 10 μM (AA10) (∗p < 0.01; NS, not significant); graph of data was from three independent experiments. Error bars represent the SE.
(C) IOWA-1T cells were transfected with empty vector (EV) or an expression vector for MMP14, treated with anacardic acid (+) or vehicle (−), and assayed for invasiveness. Western blots (top) confirm expression of MMP14 and confirm loss of MMP14 with anacardic acid in cells transfected with EV. Forced expression of MMP14 abrogated the effect of anacardic acid on invasiveness (∗p = 0.0002; NS, not significant); graph of data was from three independent experiments. Error bars represent the SE.
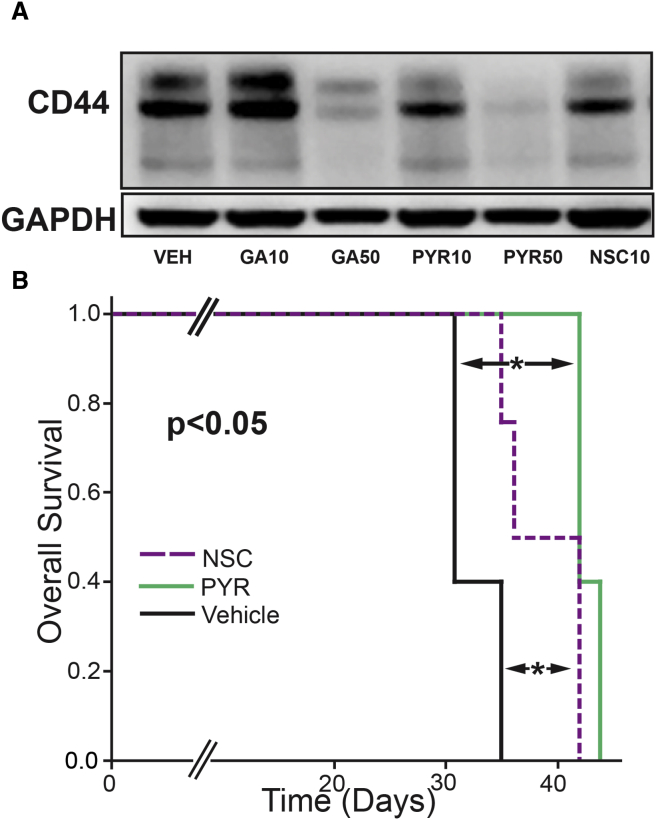
Additional compounds with SUMO inhibitory activity have been described. A number of compounds with activity as E1 and E3 SUMO inhibitors were tested for effects on CD44 expression. As seen in Figure 5A, a number of compounds with SUMO inhibitory activity similarly had the ability to repress expression of CD44 (Figure 5A). NSC and PYR-41 were tested for their ability to repress the outgrowth of xenografts. As seen in Figure 5B, both compounds were able to significantly increase the overall survival of mice inoculated with IOWA-1T xenografts. Although these compounds did not have effects as robust as those of anacardic acid, these data confirm the potential therapeutic effect of targeting several enzymes in the SUMO pathway.
Figure 5.
Panel of SUMO Inhibitors Repressed CD44 and Tumorigenesis
(A) A panel of SUMO inhibitors was tested for the ability to repress CD44: vehicle (VEH); ginkgolic acid at 10 μM (GA10) or 50 μM (GA50); PYR-41 at 10 μM (PYR10) or 50 μM (PYR50); and NSC-207895 at 10 μM (NSC10). Relative CD44 protein: VEH, 1.0; GA10, 1.17; GA50, 0.22; PYR10, 0.77; PYR50, 0.14; NSC10, 0.77.
(B) Median overall survival of mice (n = 5 mice in vehicle and PYR-41; n = 4 mice in NSC group) inoculated with IOWA-1T xenografts were increased to 43 ± 0.5 and 39 ± 2 days with PYR-41 (PYR) and NSC-207895 (NSC), respectively, compared with a vehicle-treated control group 33 ± 1 days (∗p < 0.05).
Effect of SUMO Inhibitors in Colorectal Cancer
Colorectal CSCs are also identified within the CD44+/hi/ALDH+/hi cell population. Anacardic acid efficiently repressed expression of CD44 and MMP14 in HCT116 colon cancer cells, with repression of the SUMO-conjugated TFAP2A isoform and elimination of CD44+/hi/ALDH+/hi cells, reducing the percentage of CD44+/hi/ALDH+/hi cells from 15% to 0% (Figures 6A and 6B). Anacardic acid treatment significantly increased tumor-free survival and overall survival of mice with HCT116 xenografts (Figure 6C). Examining other SUMO inhibitors, PYR-41 and MLN4924 repressed CD44 expression, and similarly repressed SUMO-conjugated TFAP2A (Figure 6D).
Figure 6.
SUMO Inhibitors Repressed CD44 and MMP14 Expression and Inhibited Tumorigenesis in Colon Cancer Cells
(A) RNA (top panel) and western blot (bottom panel) of CD44, MMP14, and TFAP2A in HCT116 cells treated with vehicle (VEH) or 10 μM anacardic acid (AA); graph of RNA data was from three independent experiments. Relative protein for CD44: VEH, 1.0; AA, 0.08. Relative protein for MMP14: VEH, 1.0; AA, 0.13 (∗∗∗p < 0.001).
(B) FACS analysis of CD44 and ALDH in HCT116 cells treated with vehicle or anacardic acid demonstrate elimination of the CD44+/hi/ALDH+/hi CSC population from 15% to 0% with anacardic acid. Negative control staining is shown in Figure S3.
(C) Tumor-free survival and overall survival of mice (n = 5 mice in all groups) inoculated with HCT116 cells pre-treated for 48 hr with anacardic acid (AA) or vehicle.
(D) Expression of CD44 RNA (top) and CD44 and TFAP2A protein (bottom) in HCT116 cells treated in vitro with 10 μM PYR-41 (PYR) or 10 μM MLN-4924 (MLN). Graph of RNA data was from three independent experiments. Relative CD44 protein: VEH, 1.0; PYR, 0.37; MLN, 0.33 (∗p < 0.05).
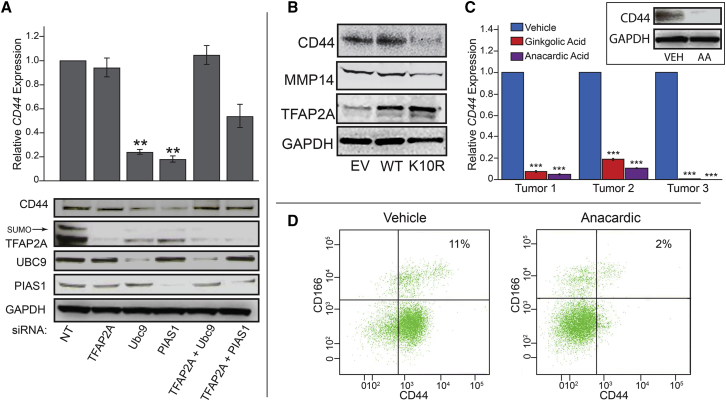
Previous studies in basal breast cancers demonstrated that repression of CD44 by SUMO inhibition was dependent upon TFAP2A (Bogachek et al., 2014). To investigate whether the same mechanism was functional in colon cancer cells, we examined the role of TFAP2A with knockdown of SUMO enzymes. As seen in Figure 7A, knockdown of either UBC9 or PIAS1 repressed CD44 expression in HCT116 cells. Whereas knockdown of TFAP2A alone had no effect on CD44, concurrent knockdown of TFAP2A eliminated the effects of knockdown of the SUMO enzymes on CD44 expression. The site of sumoylation of TFAP2A is at lysine 10 and the TFAP2A mutant K10R is non-sumoylatable (Bogachek et al., 2014, Eloranta and Hurst, 2002). The TFAP2A K10R mutant is transcriptionally functional and demonstrates the ability to regulate patterns of gene expression distinct from wild-type TFAP2A. To further evaluate the role of SUMO-unconjugated TFAP2A, we expressed the SUMO-insensitive K10R mutant of TFAP2A in HCT116 cells and compared it with wild-type TFAP2A. Transfection increased the overall expression of TFAP2A by about a factor of 2 compared with endogenous baseline. As seen in Figure 7B, expression of K10R-TFAP2A repressed expression of CD44 and MMP14, whereas wild-type TFAP2A had no effect compared with transfection of empty vector. These data support the hypothesis that SUMO inhibitors repress MMP14 and CD44 with elimination of the CSC population mediated through the activity of SUMO-unconjugated TFAP2A.
Figure 7.
TFAP2A-Dependent Effect of SUMO Inhibition in Colorectal Cancer Cell Lines and Primary Colorectal Tumors
(A) HCT116 cells were transfected with siRNA to UBC9 or PIAS1 alone or in combination with siRNA to TFAP2A to demonstrate effect on CD44 RNA (∗∗p < 0.01) (top panel) and CD44, TFAP2A, UBC9, and PIAS1 protein (bottom). Graph of RNA data was from three independent experiments. Relative CD44 protein: NT knockdown (KD), 1.0; TFAP2A KD, 0.98; UBC9 KD, 0.48; PIAS1 KD, 0.35; TFAP2A + UBC9 KD, 1.0; TFAP2A + PIAS1 KD, 0.91. Error bars represent the SE.
(B) HCT116 cells were transfected with empty vector (EV), expression vectors for TFAP2A wild-type (WT), or K10R mutant (K10R) and western blot was performed after 96 hr. Relative TFAP2A protein: EV, 1.0; TFAP2A, 1.8; K10R, 2.2. Relative CD44 protein: EV, 1.0; TFAP2A, 1.09; K10R, 0.28. Relative MMP14 protein: EV, 1.0; TFAP2A, 0.98; K10R, 0.49.
(C) CD44 RNA from three tumor isolates treated with ginkgolic or anacardic acid in vitro (∗∗∗p < 0.001). Inset: example of western blot from primary colorectal tumor cells. Relative CD44 protein: vehicle (VEH), 1.0, anacardic acid (AA), 0.02.
(D) FACS analysis of CD44 and CD166/ALCAM expression in primary colon cancer cells treated with anacardic acid or vehicle. Percentage of cells expressing CSC phenotypic markers CD44+/hiCD166+/hi was reduced from 11% to 2% with anacardic acid treatment.
To further the clinical relevance, we treated primary colorectal tumors obtained during surgical resection with SUMO inhibitors in vitro. Treatment with SUMO inhibitors significantly repressed CD44 mRNA and protein expression by western blot (Figure 7C), and suggested similar effects on the CSC population in colorectal cancer. The CSC population was evaluated by FACS analysis using CD44 and the additional colon CSC marker CD166/activated leukocyte cell adhesion molecule (ALCAM) (Sanders and Majumdar, 2011). Anacardic acid reduced the population expressing the CSC phenotypic markers CD44+/hiCD166+/hi from a primary colon cancer isolate from 11% to 2% (Figure 7D). Considering CD44 only, anacardic acid treatment of a primary colon cancer isolate reduced the CD44+/hi population from 80.8% to 8%.
Discussion
The data presented herein substantially expand the potential of developing CSC-targeted therapy based on inhibition of the SUMO pathway. We show that inhibition of sumoylation by knockdown of UBC9 and PIAS1 effectively repressed expression of MMP14 and CD44, reduced invasiveness, and substantially inhibited tumorigenesis in a basal breast cancer model. Similar effects were demonstrated with a variety of small molecules that inhibit different steps in the SUMO pathway. Serial propagation of tumor xenografts as secondary xenografts has been used to identify the CSC/TIC population (Patel et al., 2012), and our finding that small tumors that developed in animals treated with SUMO inhibitor could not be serially transplanted as secondary xenografts is further evidence that SUMO inhibitors functionally eliminated the CSC/TIC population. Interestingly, the data also suggest that SUMO inhibition induces lasting effects on the CSC population that are maintained after stopping the drug. Parallel experiments in a colorectal cancer cell line model and primary colon cancer isolates demonstrated that identical SUMO-sensitive pathways of gene regulation and physiologic response of tumor growth were present and functional. Our findings are in agreement with other studies showing that knockdown of the SUMO pathway enzymes UBC9 and SAE2 in colon cancer cells or PIAS1 in basal breast cancer reduced growth and inhibited tumorigenesis of xenografts (He et al., 2015, Liu et al., 2014).
Our data support a TFAP2A-dependent transcriptional mechanism that is functional in both basal breast and colorectal carcinomas. Previous findings indicated that AP-2 transcription factors regulate the process of EMT and that many transcription factors which induce EMT are regulated by post-translational sumoylation (Bogachek et al., 2015a). In both basal breast cancer (Bogachek et al., 2014) and colorectal cancer (Figure 7), knockdown of TFAP2A abrogated the effects of SUMO inhibition on repression of CD44. Furthermore, the non-sumoylatable K10R TFAP2A mutant was able to repress expression of CD44 and MMP14, whereas overexpression of wild-type TFAP2A failed to repress expression of these genes. The findings lead to the most likely conclusion that SUMO-unconjugated TFAP2A represses CD44 and MMP14 transcription. However, there are other possible mechanisms to account for the findings. First, it should be noted that SUMO inhibition has the potential to reduce the overall expression of TFAP2A (e.g., Figure 7A). Previous studies have shown that knockdown of TFAP2C increases TFAP2A expression, leading to the conclusion that TFAP2C represses TFAP2A expression. Since SUMO-unconjugated TFAP2A acquires transcriptional activity that mimics TFAP2C, it is likely that SUMO-unconjugated TFAP2A autoregulates its own level of expression. This mechanism would account for the reduction of overall TFAP2A expression found with SUMO inhibition. Additionally there may be other transcription factors, some of which may be SUMO sensitive, that are involved in the physiologic findings related to maintenance of the CSC/TIC population. The current findings indicate that additional efforts are needed to elucidate transcriptional mechanisms that maintain the CSC phenotype.
Compelling data exist that CD44 is not merely a marker but that CD44 expression drives the cancer phenotype, inducing a propensity for expanded growth, invasion, and metastasis (Godar et al., 2008, Hiraga et al., 2013). Several approaches have been used to directly target CD44 as a means of inhibiting the CSC population. Anti-CD44 antibodies have been utilized as one means of specifically targeting the CSC population (Arabi et al., 2015, Liu and Jiang, 2006, Molejon et al., 2015). Other approaches to target the CSC population via CD44 have been based on altering transcriptional pathways regulating CD44 expression including Wnt (Yun et al., 2016), FOXP3 (Zhang et al., 2015), SMURF1 (Khammanivong et al., 2014), and Bmi-1 (Yu et al., 2014). Our current findings indicate that SUMO inhibitors can repress expression of CD44 in the CSC population through a TFAP2A-dependent mechanism. In some instances, the effect of repression at the RNA level exceeded the reduction in CD44 protein expression. Although repression of CD44 expression is likely mediated through reduced transcription, activation of cryptic transcriptional initiation and altered RNA splicing may account for differences between the degree of repression noted when comparing RNA and protein reduction. Further work is needed to clarify the mechanism of CD44 repression by TFAP2A, which may repress CD44 transcription directly or could affect CD44 expression through secondary mechanisms.
Several studies have linked expression of CD44 and MMP14 to the CSC population. Data suggest common mechanisms regulating co-expression of these two genes; blockade of the ERK pathway suppressed expression of CD44 and MMP14 and inhibited invasiveness of cancer cells (Tanimura et al., 2003). A recent study in pancreatic cancer cells has indicated that CD44 regulates MMP14 through Snail (Jiang et al., 2015). Hence, TFAP2A may directly regulate the expression of both CD44 and MMP14, although regulation of MMP14 may alternatively occur through secondary mechanisms. MMP14 is a membrane-associated matrix metalloproteinase that has become an attractive target for cancer therapy due to its role in tumor growth, invasion, and metastasis (Ager et al., 2015, Haage et al., 2014, Nam and Ge, 2015, Ueda et al., 2003). MMP14 induces changes in cell geometry and plays an important role in proteolysis of the extracellular matrix, physiologic processes that are required for normal mammary branching and morphogenesis (Mori et al., 2013), and tumor growth within a three-dimensional matrix (Hotary et al., 2003). Further studies have demonstrated that MMP14 contributes to tumor growth and invasion through a critical role in altering tumor cell shape and establishing a reactive stroma that supports invasion (Vosseler et al., 2009). MMP14 has been shown to facilitate cell invasion and metastasis in a wide variety of cancer types, and its expression is associated with an unfavorable outcome in breast cancer (Jiang et al., 2006), colorectal cancer (Yang et al., 2013), neuroblastoma (Xiang et al., 2015), nasopharyngeal carcinoma (Zhao et al., 2015), small cell lung cancer (Wang et al., 2014), and mammary phyllodes tumors (Kim et al., 2012). The current results provide support for the ability to repress MMP14 expression and affect cell invasion and metastasis through the therapeutic development of SUMO inhibitors.
Conclusions
The findings herein substantially expand the evidence for developing CSC-targeted therapy in basal breast and colorectal cancer by inhibiting the SUMO pathway. Since sumoylation is accomplished through an enzymatic cascade, there is the potential for developing small molecules that inhibit different steps of the SUMO pathway. We have shown that a number of small molecules with known SUMO inhibitory effects can repress MMP14 and CD44, resulting in reduced invasiveness and repression of tumorigenesis. Our data further support a TFAP2A-dependent transcriptional mechanism that is functional in both basal breast cancer and colorectal carcinomas. Further work will be required to determine whether other cancer types respond to SUMO inhibitors and whether these effects are mediated though transcriptional regulation by TFAP2A or other SUMO-sensitive transcriptional mechanisms.
Experimental Procedures
Cell Culture, Transfections, and Drug Treatments
IOWA-1T was derived as described by Bogachek et al., 2015a, Bogachek et al., 2015b and is available through the ATCC. MDA-MB-436 and HCT116 were obtained from the ATCC. Under Institutional Review Board approval, colorectal primary cancer samples were obtained from surgical resection specimens. None of the colon cancer patients received neoadjuvant chemotherapy or radiotherapy prior to surgery. Transfections with small interfering RNAs (siRNAs) and drug treatments were performed as described by Bogachek et al. (2014). Cells were harvested after 72 hr for RNA and 96 hr for protein analysis by western blot and FACS. Cells were plated at 2.5 × 105/10 cm2 and treated with ginkgolic acid (Tocris Bioscience), anacardic acid (Tocris Bioscience), PYR-41 (Selleckchem), NSC-207895 (Selleckchem), or MLN4924 (Selleckchem) at the concentration indicated for 2–4 days and collected for RNA, protein, or FACS analysis. The TFAP2A and K10R mutant expression vectors were constructed and transfected as described previously (Bogachek et al., 2014, McPherson and Weigel, 1999, Schuur et al., 2001).
RNA Analysis
Total RNA was isolated from cell lines using an RNeasy Mini Kit (Qiagen) and cDNA was synthesized from 1 μg of total RNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression was observed with real-time qPCR. TaqMan primers and detection probes for genes were CD44, TFAP2A, UBC9, PIAS1, and MMP14 (Applied Biosystems). Expression values were normalized to the mean of 18S rRNA (Applied Biosystems) as an endogenous control.
Western Blots
Protein was isolated into RIPA buffer (EMD Millipore) supplemented with Halt Protease Inhibitor (Thermo Fisher Scientific). Immunoprecipitations were performed utilizing the Pierce Co-Immunoprecipitation Kit (Thermo Fisher) using anti-TFAP2A antibody (Abcam, ab108311) or rabbit IgG as a control, according to the manufacturer's instructions. Western blots were reacted using the following primary antibodies at their recommended dilutions according to the manufacturer's instructions: CD44 (R&D Systems, BBA10; Abcam, ab51037), GAPDH (Santa Cruz Biotechnology, sc-32233), SUMO-1 (Epitomics, 1563-1), SUMO-2/3 (Abcam, ab109005), MMP14 (Abcam, ab51074), TFAP2A (Abcam, ab108311), UBC9 (Abcam, ab33044), and PIAS1 (Abcam, ab109388). Protein sizes were determined using MagicMark Western Protein Standard (Thermo Fisher).
Flow Cytometry
FACS analysis was performed as previously described (Cyr et al., 2015). For FACS analysis the antibodies CD44 and ALCAM/CD166 (R&D Systems, FAB4948A and FAB6561P) were used. FACS with ALDEFLUOR including diethylaminobenzaldehyde (DEAB) control utilized the ALDEFLUOR Kit (STEMCELL Technologies) according to the manufacturer's instructions. FACS with gating for negative controls is provided in Figures S1–S4.
Invasion Assay
Invasion assay was performed with the Cell Invasion Assay Kit (Chemicon International). A total of 106 cells was loaded into an invasion chamber, and migrated cells were stained and counted in accordance with the manufacturer's instructions. Overexpression of MMP14 was accomplished by transfection of the MMP14 expression vector pCMV6-MMP14 (OriGene). The plasmid was sequenced to confirm the plasmid identity. Negative controls were transfected with empty vector, in parallel.
Xenografts
All animals were cared for in accordance with guidelines established by the University of Iowa Institutional Animal Care and Use Committee. For IOWA-1T xenografts, 2 × 105 cells were flank injected with 10% Matrigel (BD Biosciences). Anacardic acid was administered in 10 mg/kg dosage by gavage daily for 3 weeks. PYR-41 (10 mg/kg) and NSC-207895 (5 mg/kg) were administered by four intraperitoneal injections within 8 days. Single-cell suspensions were prepared from xenografts, and 106 cells were injected into nude mice to form secondary xenografts. For the HCT116 line, 2 × 106 cells were pre-treated with anacardic acid versus vehicle and were flank inoculated as noted above. Tumor-free survival was recorded when tumors were confirmed by two investigators; overall survival was noted when mice died with tumors or when tumors reached 2 cm, at which point mice were euthanized. Statistical comparisons of tumor-free and overall survival were made by log rank (www.r-project.org).
Author Contributions
M.V.B., conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; J.M.P., provision of study material or patients, collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; J.P.d.A., conception and design, provision of study material or patients, collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; A.W.L., data analysis and interpretation, manuscript writing, final approval of manuscript; M.V.K., collection and/or assembly of data, final approval of manuscript; J.R.W., collection and/or assembly of data, final approval of manuscript; V.W.G., administrative support, collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; V.T.W., data analysis and interpretation, final approval of manuscript; R.J.W., conception and design, financial support, administrative support, provision of study materials or patients, data analysis and interpretation, manuscript writing, final approval of manuscript.
Acknowledgments
This work was supported by the NIH grants R01CA183702 (PI: R.J.W.) and T32CA148062 (PI: R.J.W.), and by a generous gift from the Kristen Olewine Milke Breast Cancer Research Fund. J.P.D., A.W.L., and V.T.W. were supported by the NIH grant T32CA148062.
Published: December 1, 2016
Footnotes
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.11.001.
Supplemental Information
References
- Ager E.I., Kozin S.V., Kirkpatrick N.D., Seano G., Kodack D.P., Askoxylakis V., Huang Y., Goel S., Snuderl M., Muzikansky A. Blockade of MMP14 activity in murine breast carcinomas: implications for macrophages, vessels, and radiotherapy. J. Natl. Cancer Inst. 2015;107:djv017. doi: 10.1093/jnci/djv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamgeer M., Ganju V., Kumar B., Fox J., Hart S., White M., Harris M., Stuckey J., Prodanovic Z., Schneider-Kolsky M.E. Changes in ALDEHYDE DEHYDROGENASE-1 expression during neoadjuvant chemotherapy predict outcome in locally advanced breast cancer. Breast Cancer Res. 2014;16:R44. doi: 10.1186/bcr3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi L., Badiee A., Mosaffa F., Jaafari M.R. Targeting CD44 expressing cancer cells with anti-CD44 monoclonal antibody improves cellular uptake and antitumor efficacy of liposomal doxorubicin. J. Control Release. 2015;220:275–286. doi: 10.1016/j.jconrel.2015.10.044. [DOI] [PubMed] [Google Scholar]
- Bettermann K., Benesch M., Weis S., Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett. 2012;316:113–125. doi: 10.1016/j.canlet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- Blick T., Hugo H., Widodo E., Waltham M., Pinto C., Mani S.A., Weinberg R.A., Neve R.M., Lenburg M.E., Thompson E.W. Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. J. Mammary Gland Biol. Neoplasia. 2010;15:235–252. doi: 10.1007/s10911-010-9175-z. [DOI] [PubMed] [Google Scholar]
- Bogachek M.V., Chen Y., Kulak M.V., Woodfield G.W., Cyr A.R., Park J.M., Spanheimer P.M., Li Y., Li T., Weigel R.J. Sumoylation pathway is required to maintain the basal breast cancer subtype. Cancer Cell. 2014;25:748–761. doi: 10.1016/j.ccr.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogachek M.V., De Andrade J.P., Weigel R.J. Regulation of epithelial-mesenchymal transition through SUMOylation of transcription factors. Cancer Res. 2015;75:11–15. doi: 10.1158/0008-5472.CAN-14-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogachek M.V., Park J.M., De Andrade J.P., Kulak M.V., White J.R., Wu T., Spanheimer P.M., Bair T.B., Olivier A.K., Weigel R.J. A novel animal model for locally advanced breast cancer. Ann. Surg. Oncol. 2015;22:866–873. doi: 10.1245/s10434-014-4174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Lee H.J., Jang M.H., Gwak J.M., Lee K.S., Kim E.J., Kim H.J., Lee H.E., Park S.Y. Epithelial-mesenchymal transition increases during the progression of in situ to invasive basal-like breast cancer. Hum. Pathol. 2013;44:2581–2589. doi: 10.1016/j.humpath.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Cyr A.R., Kulak M.V., Park J.M., Bogachek M.V., Spanheimer P.M., Woodfield G.W., White-Baer L.S., O'Malley Y.Q., Sugg S.L., Olivier A.K. TFAP2C governs the luminal epithelial phenotype in mammary development and carcinogenesis. Oncogene. 2015;34:436–444. doi: 10.1038/onc.2013.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Srikanth M., Kessler J.A. Cancer stem cells and glioma. Nat. Clin. Pract. Neurol. 2008;4:427–435. doi: 10.1038/ncpneuro0862. [DOI] [PubMed] [Google Scholar]
- Dou J., Pan M., Wen P., Li Y., Tang Q., Chu L., Zhao F., Jiang C., Hu W., Hu K. Isolation and identification of cancer stem-like cells from murine melanoma cell lines. Cell Mol. Immunol. 2007;4:467–472. [PubMed] [Google Scholar]
- Eloranta J.J., Hurst H.C. Transcription factor AP-2 interacts with the SUMO-conjugating enzyme UBC9 and is sumoylated in vivo. J. Biol. Chem. 2002;277:30798–30804. doi: 10.1074/jbc.M202780200. [DOI] [PubMed] [Google Scholar]
- Erfani E., Roudi R., Rakhshan A., Sabet M.N., Shariftabrizi A., Madjd Z. Comparative expression analysis of putative cancer stem cell markers CD44 and ALDH1A1 in various skin cancer subtypes. Int. J. Biol. Markers. 2016;31:e53–e61. doi: 10.5301/jbm.5000165. [DOI] [PubMed] [Google Scholar]
- Fukuda I., Ito A., Hirai G., Nishimura S., Kawasaki H., Saitoh H., Kimura K., Sodeoka M., Yoshida M. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem. Biol. 2009;16:133–140. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Godar S., Ince T.A., Bell G.W., Feldser D., Donaher J.L., Bergh J., Liu A., Miu K., Watnick R.S., Reinhardt F. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haage A., Nam D.H., Ge X., Schneider I.C. Matrix metalloproteinase-14 is a mechanically regulated activator of secreted MMPs and invasion. Biochem. Biophys. Res. Commun. 2014;450:213–218. doi: 10.1016/j.bbrc.2014.05.086. [DOI] [PubMed] [Google Scholar]
- Hay R.T. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- He X., Riceberg J., Pulukuri S.M., Grossman S., Shinde V., Shah P., Brownell J.E., Dick L., Newcomb J., Bence N. Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PLoS One. 2015;10:e0123882. doi: 10.1371/journal.pone.0123882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga T., Ito S., Nakamura H. Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Res. 2013;73:4112–4122. doi: 10.1158/0008-5472.CAN-12-3801. [DOI] [PubMed] [Google Scholar]
- Hotary K.B., Allen E.D., Brooks P.C., Datta N.S., Long M.W., Weiss S.J. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- Iqbal J., Chong P.Y., Tan P.H. Breast cancer stem cells: an update. J. Clin. Pathol. 2013;66:485–490. doi: 10.1136/jclinpath-2012-201304. [DOI] [PubMed] [Google Scholar]
- Jiang W.G., Davies G., Martin T.A., Parr C., Watkins G., Mason M.D., Mansel R.E. Expression of membrane type-1 matrix metalloproteinase, MT1-MMP in human breast cancer and its impact on invasiveness of breast cancer cells. Int. J. Mol. Med. 2006;17:583–590. [PubMed] [Google Scholar]
- Jiang W., Zhang Y., Kane K.T., Collins M.A., Simeone D.M., di Magliano M.P., Nguyen K.T. CD44 regulates pancreatic cancer invasion through MT1-MMP. Mol. Cancer Res. 2015;13:9–15. doi: 10.1158/1541-7786.MCR-14-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing F., Kim H.J., Kim C.H., Kim Y.J., Lee J.H., Kim H.R. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int. J. Oncol. 2015;46:1582–1588. doi: 10.3892/ijo.2015.2844. [DOI] [PubMed] [Google Scholar]
- Khammanivong A., Gopalakrishnan R., Dickerson E.B. SMURF1 silencing diminishes a CD44-high cancer stem cell-like population in head and neck squamous cell carcinoma. Mol. Cancer. 2014;13:260. doi: 10.1186/1476-4598-13-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.E., Kim J.H., Lee K.H., Choi Y.D., Lee J.S., Lee J.H., Nam J.H., Choi C., Park M.H., Yoon J.H. Stromal matrix metalloproteinase-14 expression correlates with the grade and biological behavior of mammary phyllodes tumors. Appl. Immunohistochem. Mol. Morphol. 2012;20:298–303. doi: 10.1097/PAI.0b013e318235a132. [DOI] [PubMed] [Google Scholar]
- Lawson D.A., Bhakta N.R., Kessenbrock K., Prummel K.D., Yu Y., Takai K., Zhou A., Eyob H., Balakrishnan S., Wang C.Y. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.E., Kim J.H., Kim Y.J., Choi S.Y., Kim S.W., Kang E., Chung I.Y., Kim I.A., Kim E.J., Choi Y. An increase in cancer stem cell population after primary systemic therapy is a poor prognostic factor in breast cancer. Br. J. Cancer. 2011;104:1730–1738. doi: 10.1038/bjc.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jiang G. CD44 and hematologic malignancies. Cell Mol. Immunol. 2006;3:359–365. [PubMed] [Google Scholar]
- Liu J., Brown R.E. Immunohistochemical detection of epithelial mesenchymal transition associated with stemness phenotype in anaplastic thyroid carcinoma. Int. J. Clin. Exp. Pathol. 2010;3:755–762. [PMC free article] [PubMed] [Google Scholar]
- Liu B., Tahk S., Yee K.M., Yang R., Yang Y., Mackie R., Hsu C., Chernishof V., O'Brien N., Jin Y. PIAS1 regulates breast tumorigenesis through selective epigenetic gene silencing. PLoS One. 2014;9:e89464. doi: 10.1371/journal.pone.0089464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson L.A., Weigel R.J. AP2alpha and AP2gamma: a comparison of binding site specificity and transactivation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 1999;27:4040–4049. doi: 10.1093/nar/27.20.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molejon M.I., Tellechea J.I., Moutardier V., Gasmi M., Ouaissi M., Turrini O., Delpero J.R., Dusetti N., Iovanna J. Targeting CD44 as a novel therapeutic approach for treating pancreatic cancer recurrence. Oncoscience. 2015;2:572–575. doi: 10.18632/oncoscience.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H., Lo A.T., Inman J.L., Alcaraz J., Ghajar C.M., Mott J.D., Nelson C.M., Chen C.S., Zhang H., Bascom J.L. Transmembrane/cytoplasmic, rather than catalytic, domains of Mmp14 signal to MAPK activation and mammary branching morphogenesis via binding to integrin beta1. Development. 2013;140:343–352. doi: 10.1242/dev.084236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam D.H., Ge X. Direct production of functional matrix metalloproteinase-14 without refolding or activation and its application for in vitro inhibition assays. Biotechnol. Bioeng. 2015;113:717–723. doi: 10.1002/bit.25840. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Lee H.E., Li H., Shipitsin M., Gelman R., Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin. Cancer Res. 2010;16:876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmiani G. Melanoma cancer stem cells: markers and functions. Cancers (Basel) 2016;8 doi: 10.3390/cancers8030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G.K., Yee C.L., Terunuma A., Telford W.G., Voong N., Yuspa S.H., Vogel J.C. Identification and characterization of tumor-initiating cells in human primary cutaneous squamous cell carcinoma. J. Invest. Dermatol. 2012;132:401–409. doi: 10.1038/jid.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo S., Vieira A.F., Gerhard R., Leitao D., Pinto R., Cameselle-Teijeiro J.F., Milanezi F., Schmitt F., Paredes J. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011;64:937–946. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- Sanders M.A., Majumdar A.P. Colon cancer stem cells: implications in carcinogenesis. Front Biosci. (Landmark Ed.) 2011;16:1651–1662. doi: 10.2741/3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrio D., Rodriguez-Pinilla S.M., Hardisson D., Cano A., Moreno-Bueno G., Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- Schuur E.R., McPherson L.A., Yang G.P., Weigel R.J. Genomic structure of the promoters of the human estrogen receptor-alpha gene demonstrate changes in chromatin structure induced by ap2gamma. J. Biol. Chem. 2001;276:15519–15526. doi: 10.1074/jbc.M009001200. [DOI] [PubMed] [Google Scholar]
- Sheridan C., Kishimoto H., Fuchs R.K., Mehrotra S., Bhat-Nakshatri P., Turner C.H., Goulet R., Jr., Badve S., Nakshatri H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura S., Asato K., Fujishiro S.H., Kohno M. Specific blockade of the ERK pathway inhibits the invasiveness of tumor cells: down-regulation of matrix metalloproteinase-3/-9/-14 and CD44. Biochem. Biophys. Res. Commun. 2003;304:801–806. doi: 10.1016/s0006-291x(03)00670-3. [DOI] [PubMed] [Google Scholar]
- Thiery J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Tsai J.H., Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J., Kajita M., Suenaga N., Fujii K., Seiki M. Sequence-specific silencing of MT1-MMP expression suppresses tumor cell migration and invasion: importance of MT1-MMP as a therapeutic target for invasive tumors. Oncogene. 2003;22:8716–8722. doi: 10.1038/sj.onc.1206962. [DOI] [PubMed] [Google Scholar]
- Vosseler S., Lederle W., Airola K., Obermueller E., Fusenig N.E., Mueller M.M. Distinct progression-associated expression of tumor and stromal MMPs in HaCaT skin SCCs correlates with onset of invasion. Int. J. Cancer. 2009;125:2296–2306. doi: 10.1002/ijc.24589. [DOI] [PubMed] [Google Scholar]
- Wang Y.Z., Wu K.P., Wu A.B., Yang Z.C., Li J.M., Mo Y.L., Xu M., Wu B., Yang Z.X. MMP-14 overexpression correlates with poor prognosis in non-small cell lung cancer. Tumour Biol. 2014;35:9815–9821. doi: 10.1007/s13277-014-2237-x. [DOI] [PubMed] [Google Scholar]
- Xiang X., Zhao X., Qu H., Li D., Yang D., Pu J., Mei H., Zhao J., Huang K., Zheng L. Hepatocyte nuclear factor 4 alpha promotes the invasion, metastasis and angiogenesis of neuroblastoma cells via targeting matrix metalloproteinase 14. Cancer Lett. 2015;359:187–197. doi: 10.1016/j.canlet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Yan T., Lin Z., Jiang J., Lu S., Chen M., Que H., He X., Que G., Mao J., Xiao J. MMP14 regulates cell migration and invasion through epithelial-mesenchymal transition in nasopharyngeal carcinoma. Am. J. Transl. Res. 2015;7:950–958. [PMC free article] [PubMed] [Google Scholar]
- Yang B., Gao J., Rao Z., Shen Q. Clinicopathological and prognostic significance of alpha5beta1-integrin and MMP-14 expressions in colorectal cancer. Neoplasma. 2013;60:254–261. doi: 10.4149/neo_2013_034. [DOI] [PubMed] [Google Scholar]
- Yu J., Lu Y., Cui D., Li E., Zhu Y., Zhao Y., Zhao F., Xia S. miR-200b suppresses cell proliferation, migration and enhances chemosensitivity in prostate cancer by regulating Bmi-1. Oncol. Rep. 2014;31:910–918. doi: 10.3892/or.2013.2897. [DOI] [PubMed] [Google Scholar]
- Yun E.J., Zhou J., Lin C.J., Hernandez E., Fazli L., Gleave M., Hsieh J.T. Targeting cancer stem cells in castration-resistant prostate cancer. Clin. Cancer Res. 2016;22:670–679. doi: 10.1158/1078-0432.CCR-15-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Xu Y., Hao Q., Wang S., Li H., Li J., Gao Y., Li M., Li W., Xue X. FOXP3 suppresses breast cancer metastasis through downregulation of CD44. Int. J. Cancer. 2015;137:1279–1290. doi: 10.1002/ijc.29482. [DOI] [PubMed] [Google Scholar]
- Zhao J., Kong Z., Xu F., Shen W. A role of MMP-14 in the regulation of invasiveness of nasopharyngeal carcinoma. Tumour Biol. 2015;36:8609–8615. doi: 10.1007/s13277-015-3558-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.