Abstract
Glioma stem-like cells (GSC) with tumor initiating activity orchestrate the cellular hierarchy in glioblastoma (GBM) and engender therapeutic resistance. Recent work has divided GSC into two subtypes with a mesenchymal (MES) GSC population as the more malignant subtype. In this study, we identify the FOXD1-ALDH1A3 signaling axis as a determinant of the MES GSC phenotype. The transcription factor FOXD1 is expressed predominantly in patient-derived cultures enriched with MES, but not with the proneural (PN) GSC subtype. shRNA-mediated attenuation of FOXD1 in MES GSC ablates their clonogenicity in vitro and in vivo. Mechanistically, FOXD1 regulates the transcriptional activity of ALDH1A3, an established functional marker for MES GSC. Indeed, the functional roles of FOXD1 and ALDH1A3 are likely evolutionally conserved, insofar as RNAi-mediated attenuation of their orthologous genes in Drosophila blocks formation of brain tumors engineered in that species. In clinical specimens of high-grade glioma, the levels of expression of both FOXD1 and ALDH1A3 are inversely correlated with patient prognosis. Lastly, a novel small molecule inhibitor of ALDH we developed, termed GA11, displays potent in vivo efficacy when administered systemically in a murine GSC-derived xenograft model of GBM. Collectively, our findings define a FOXD1-ALDH1A3 pathway in controlling the clonogenic and tumorigenic potential of MES GSC in GBM tumors.
Keywords: glioblastoma, cancer stem cell, stem cell marker, brain cancer, cancer initiating cell, forkhead transcription factors
Introduction
Glioblastoma (GBM) is the most common and fatal primary brain tumor in adults. GBM tumors are resistant to conventional radiation therapy and chemotherapies, making the current available treatments ineffective (1). Intra-tumoral cellular heterogeneity in GBM contributes to tumor aggressiveness and therapy resistance, with glioma stem-like cells (GSCs) at the apex of the hierarchy (1,2). GSCs are poorly differentiated tumor cells with stem cell properties including self-renewal, and are responsible for tumor initiation (3–5). Recently, we and others established patient-derived GSC lines from GBM patients, which contain two distinct and mutually-exclusive GSC subtypes termed Proneural (PN) and Mesenchymal (MES) (4,6). MES GSCs, the more aggressive and radio-resistant subtype, express higher levels of ALDH1A3 than PN GSCs in vitro (6), suggesting that ALDH1A3 is a potential marker of MES GSCs. Therefore, understanding the regulatory pathway(s) controlling ALDH1A3 expression in this cell type would be expected to identify new and relevant therapeutic targets.
The Forkhead family of transcription factors (TFs) regulate a wide variety of cellular functions during development and many are implicated in cancers (7,8). As a member of this family, FOXD1 is preferentially expressed in human embryonic tissues, including kidney and testis, but not in adult tissues (9). Moreover, FOXD1 regulates organogenesis (10–12), especially the commitment to a mesenchymal lineage during organogenesis (13–15). FOXD1’s regulatory role in stemness is further demonstrated by facilitating the reprogramming of mouse embryonic fibroblasts into induced pluripotent stem cells (16). Recent studies have suggested parallels between reprogramming and tumorigenesis, and highlighted the shared transcription factors involved. Similarly, the deregulation of FOXD1 is implicated in the tumorigenesis of cancers of prostate, breast, and clear cell sarcoma of the kidney (17–19). Nonetheless, the physiological roles of FOXD1 in brain cancers and GSCs remain unknown.
In this study, we demonstrate that FOXD1 is critical for the maintenance of MES GSCs, and thus, the tumorigenicity of this subtype of GBM tumors. By exploring our genome-wide expression profiling data, we identified FOXD1 as the most up-regulated Forkhead TF in the patient-derived MES GSC enriched cultures. We found that FOXD1 promotes clonogenicity and tumorigenicity of MES GSCs both in vitro and in vivo by the direct transcriptional regulation of the key molecule ALDH1A3. Subsequently, we verified that the tumorigenicity of FOXD1 is mediated by ALDH1A3. We also proved that FOXD1 and ALDH1A3 are prognostic factors in glioma clinical samples. Finally, we developed novel anti-ALDH small molecule inhibitors and demonstrated effectiveness in vitro and in vivo.
Materials and Methods
Ethics
This study was performed under the supervision of the respective Institutional Animal Care and Use Committees, and the Human Subjects Research protocols were approved by the Institutional Review Board at the University of Alabama at Birmingham (UAB), MD Anderson Cancer Center (MDA) and/or Ohio State University (OSU) as described previously (6,20).
GSC cultures
The glioma (neuro)spheres used in this study were generated at OSU and MDA. Neurosphere cultures from clinical samples were established and characterized as described previously (Supplementary Table S1) (6,20). The detailed source, year of receipt, and culture methods are described in Supplementary information. The unique identities of each glioma neurosphere line were confirmed by short tandem repeat (STR) analysis as described in Supplementary Table S2 (20).
Mouse intracranial xenograft tumor models
The GSC suspension (1×104 cells for MES83 and 2.5×105 cells for MES267) was injected into the brains of nude mice (6-week-old) as previously described (20,21). When neurological symptoms observed, mice were sacrificed and mouse brains were collected for analysis.
Lentivirus transduction
Two independent lentiviral shRNA constructs for knocking down FOXD1 were purchased from Sigma (TRCN0000013970 and TRCN0000230322). For overexpression, cDNAs of FOXD1 (RC220504, Origene), and FOXG1 (RC207964, Origene) were subcloned into a lentiviral vector (Origene, PS100064) according to the manufacturer’s protocol. Lentiviruses were packaged in 293FT cells (from Invitrogen at 2013). The lentivirus transduction was performed as previously described (20).
Neurosphere formation assay
MES83 and MES28 GSCs infected with lentivirus were seeded into 96 well plates at 1, 10, 20, 30, 40, and 50 cells per well. After 7 days for MES83 GSCs and 10 days for MES28 GSCs, the numbers of spheres with diameters greater than 60μm were counted. Data were analyzed as described previously (http://bioinf.wehi.edu.au/software/elda/) (22).
Immunohistochemistry scoring
German immunohistochemical score (GIS) was used to evaluate the expression of FOXD1 and ALDH1A3 (23,24). The detailed procedure is provided in the supplementary information.
Western blot
The detailed procedure is provided in the supplementary information. Original film scans of western blots were shown in Supplementary Fig. S1.
GEO accession numbers
The accession numbers of GEO datasets used in this study are GSE67089, GSE4290, GSE4536, and GSE2223.
Drosophila stock
The following transgenic Drosophila flies were used: repoGAL4 UAS-GFP/TM3, Sb, UAS-ptenRNAi, UAS-RasV12/Cyo, UAS-aldhRNAi/Cyo, ptcGAL4 UAS-GFP/Cyo, UAS-fd59ARNAi, and UAS-fd59A. The Drosophilas for all experiments were incubated at 25°C.
Immunofluorescence, imaging, and quantification in Drosophila samples
Immunofluorescence was performed as described (25) and the images were captured using the Olympus Fluoview 1000 confocal microscope.
Statistical analysis
Data are presented as mean ± SD. The number of replicates for each experiment is stated in the figure legend. Statistical differences between and among groups were determined by two tailed t-test and one-way analysis of variance (ANOVA) followed by Dunnett’s post-test, respectively. The statistical significance of Kaplan–Meier survival plot was determined by log-rank analysis. Statistical analysis was performed by Microsoft Excel 2013 and Graphpad Prism 6.0, unless mentioned otherwise in the figure legend. P<0.05 was considered as statistically significant.
Additional details about the materials and methods are available in the supplementary information.
Results
FOXD1 and FOXG1 exhibit inverse expression pattern in GSCs
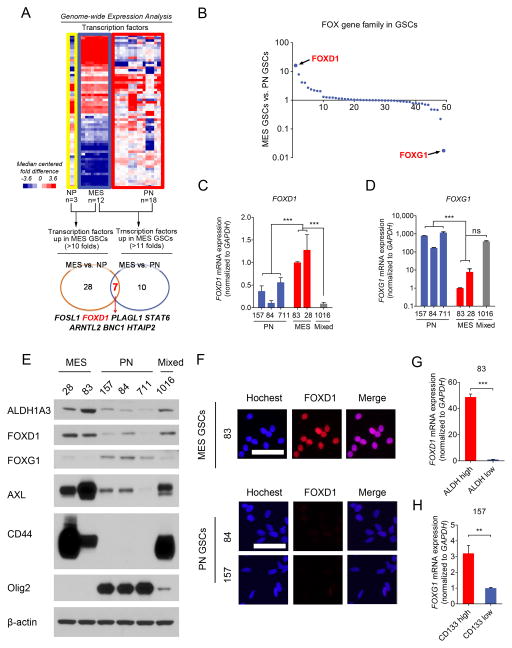
To identify the TFs critical for the GSC phenotype, we explored our dataset (GSE67089 (6)) with 30 patient-derived glioma sphere cultures and 3 human fetal brain-derived sphere cultures (normal spheres) for the expression levels of all TFs (1,988 genes (26)). Twenty eight TFs were found up-regulated in MES glioma spheres by more than 10 fold compared to normal neural progenitor cells. Ten TFs were at higher levels in MES glioma spheres by more than 11 fold than the levels of these TFs in PN glioma spheres. Among them, seven TFs were overlapped, including FOSL1, FOXD1, PLAGL1, STAT6, ARNTL2, BNC1, and HTAIP2 (Fig. 1A).
Figure 1. FOXD1 is a key MES GSC transcription factor.
A. Genome-wide transcriptome microarray analysis (GSE67089) shows that FOXD1 is one of seven upregulated transcription factors in MES gliomaspheres when compared to Neural Progenitors (NPs) (>10 fold) and PN glioma spheres (>11 fold).
B. mRNA expression levels of the Forkhead TF family members in the GSE67089 dataset reveal that FOXD1 and FOXG1 are the highest expressed genes in MES and PN glioma spheres, respectively.
C, D. RT-qPCR analyses of FOXD1 (C) (MES vs. PN, P<0.001, n=3; MES vs. mixed, P<0.001, n=3, one way ANOVA) and FOXG1 (D) (MES vs. PN, P=0.0025, n=3; MES vs. mixed, P=0.2537, n=3, one way ANOVA) mRNA in the indicated glioma spheres.
E. Western blot analyses of ALDH1A3, FOXD1, FOXG1, CD44, AXL and Olig2 expression in the indicated glioma spheres. β-actin serves as a loading control.
F. Immunocytochemistry analyses of FOXD1 in MES83, PN84, and PN157 glioma spheres. Hoechst is used for nuclear staining. Bar, 50μm.
G. RT-qPCR analyses of FOXD1 mRNA in ALDHhigh and ALDHlow cells derived from MES83 glioma spheres. (P<0.001, n=3, t-test)
H. RT-qPCR analyses of FOXG1 mRNA in CD133high and CD133low cells derived from PN157 glioma spheres. (P=0.0017, n=3, t-test)
Forkhead TFs are involved in deciding cell fates during development and tumorigenesis (7,16). Our previous study demonstrated the requirement of one Forkhead TF, FOXM1, for the survival and proliferation of oncogenic but not normal neural stem cells (27–29). Therefore, we focused on the Forkhead TFs. FOXD1 was the most up-regulated MES-associated gene in the Forkhead family, whereas FOXG1 was the most up-regulated gene in PN glioma spheres (Fig. 1B and Supplementary Fig. S2A, B). This was validated by RT-qPCR (Fig. 1C and D) and western blot (Fig. 1E) with six patient-derived GSC-enriched cultures. In western blot assays, ALDH1A3, CD44, and AXL were included as MES markers while OLIG2 as a PN marker. Similarly, immunocytochemistry exhibited that glioma sphere line MES83 has a higher expression of nuclear FOXD1 than PN84 and PN157 (Fig. 1F). We then investigated whether the stem cell fraction in MES and PN glioma spheres express FOXD1 and FOXG1, respectively. Our previous studies identified CD133 as a PN GSC marker while ALDH1A3 as MES GSC marker (4,6,27). As expected, FOXD1 was almost exclusively expressed in the ALDHhigh subfraction of MES83 cells, but not in ALDHlow subpopulations (Fig. 1G and Supplementary Fig. S2C). In contrast, the CD133high subpopulation had significantly higher expression of FOXG1 than CD133low subpopulations in the PN157 glioma spheres (Fig. 1H and Supplementary Fig. S2D). To extend this observation, we performed bioinformatics analysis using another clinical data (IVY GBM Atlas Project database) (http://glioblastoma.alleninstitute.org/). GBM is known to display intra-tumoral cellular heterogeneity. We investigated RNAseq data that was collected from different regions of GBM tumor tissues that exhibit distinct cellular subtypes including PN and MES tumor cells. This analysis showed an inverse pattern of the expression levels of FOXG1 and FOXD1 in different regions (Supplementary Fig. S2E and F). In contrast, there were no significant differences in either FOXD1 or FOXG1 among the GBM subtypes in the TCGA dataset (Supplementary Fig. S2G). The TCGA dataset measures the average expression levels of genes. This maybe the possible reason why the expression level of either FOXG1 or FOXD1 failed to show a significant difference among the four subtypes of GBM. Collectively, these data suggest that FOXD1 and FOXG1 exhibit inverse expression profiles in vitro: FOXD1 is elevated in MES, while FOXG1 is elevated in PN GSCs.
FOXD1 expression is associated with poorer prognosis in glioma patients
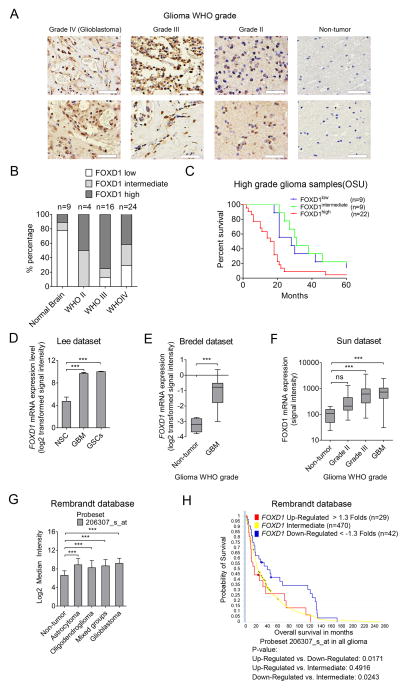
We then examined whether FOXD1 expression is associated with the histopathological grade of glioma and patient prognosis. With immunohistochemistry, 44 glioma tumor tissues with varying grades and nine adjacent normal brain tissues were analyzed. Intense immunostaining, indicating FOXD1 expression, was observed in the nuclei of high-grade glioma tissues (Grade II: 2 of 4; Grade III: 12 of 16; Grade IV: 10 of 24) but rarely in normal brain tissues (1 of 9) (Fig. 2A and B). The survival periods of patients with high levels of FOXD1 were significantly shorter than those with FOXD1 intermediate expression (Fig. 2C, P=0.0039). Inversely, patients with low FOXD1 expression exhibited better prognosis than those with high expression (Fig. 2C, P=0.0324). Consistently, GBM-derived spheres expressed higher FOXD1 than normal brain-derived spheres (GSE4536, Fig. 2D (30)). Similar results were obtained from two other datasets (GSE2223, Fig. 2E (31); and GSE4290, Fig. 2F (32)), and from the Rembrandt database (Fig. 2G). The elevated expression of FOXD1 was also associated with poorer survival in Rembrandt database (Fig. 2H). Altogether, these data suggest that the expression of FOXD1 is elevated in high-grade glioma and is a clinically-relevant target in GBM.
Figure 2. FOXD1 expression is clinically relevant in high-grade gliomas.
A, B. Representative immunohistochemical images (A) and analyses (B) of FOXD1 in WHO grade IV (Glioblastoma), grade III glioma, grade II glioma, and non-tumor brain samples. Bar, 50 μm. (Non-tumor n=9, grade II n=4, grade III n=16, grade IV n=24).
C. Kaplan-Meier analyses evaluating the correlation between FOXD1 protein expression and survival of 40 high grade glioma patients (FOXD1 high vs. low, P=0.0324; FOXD1 high vs. intermediate, P=0.0039; FOXD1 intermediate vs. low, P=0.2896, log rank test).
D–F. Analyses of the indicated GEO datasets show a higher expression of FOXD1 in glioma than in Neural Stem Cell (NSC) samples and non-tumor tissues. D. Lee dataset (GSE4536; NSCs n=3, GBM n=22, GSCs n=20; P<0.001, one way ANOVA, probe set 206307_s_at). E. Bredel dataset (GSE2223; Non-tumor n=4, GBM n=29; P<0.0001, t test, probe set 1876). F. Sun dataset (GSE4290, Non-tumor n=23, grade II n=45, grade III n=31, GBM n=81; P< 0.0001, one-way ANOVA, probe set 206307_s_at).
G. Analysis of the Rembrandt data shows a higher FOXD1 mRNA expression in astrocytoma (n=148), oligodendroglioma (n=67), mixed groups (n=11), and in GBM samples (n=228) than non-tumor samples (n=28) (probe set 206307_s_at).
H. Analysis of the Rembrandt database indicates the inverse correlation between FOXD1 mRNA expression and post-surgical survival of glioma patients (P=0.0171, FOXD1 Up-Regulated > 1.3-Fold, n=29 vs. FOXD1 Down-Regulated < −1.3-Fold, n=42; P=0.0243, Down-Regulated < −1.3-Fold, n=42 vs. Intermediate n=470, probe set: 206307_s_at).
FOXD1 is required for the clonogenicity of MES GBM in vitro and in vivo
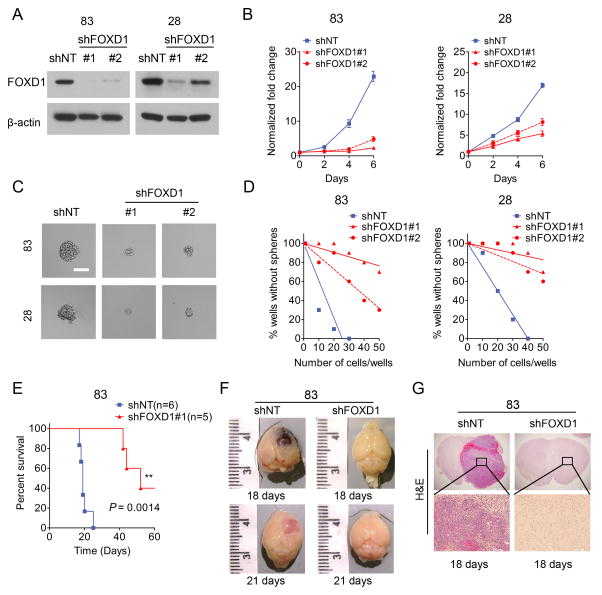
To investigate the physiological role of FOXD1 in MES GSCs, we knocked down the expression of FOXD1 using two different lentiviral shRNA vectors in MES glioma sphere lines (MES83 and MES28) (4,6); a non-targeting shRNA (shNT) was used as a negative control. Western blotting showed that both shRNA clones were capable of knocking down FOXD1, with clone #1 having higher efficiency (Fig. 3A). As a result, MES glioma spheres with FOXD1 knocked-down (KD) grew significantly slower (33) than control cells (Fig. 3B). Moreover, the sphere forming capacity of these cells was dramatically decreased (Fig. 3C and D). Of note, the extent to which cell growth and clonogenicity were inhibited was more prominent with shFOXD1#1 than shFOXD1#2, which was consistent with their effects on FOXD1 KD. Our previous study demonstrated that MES, not PN GSCs depended more on glycolysis (6). As expected, lactic acid levels as a measure of glycolysis were largely reduced in MES83 cells transduced with shFOXD1#1, indicating that FOXD1 may be involved in the regulation of glycolysis of MES GSCs (Supplementary Fig. S3). To determine whether FOXD1 is essential for tumorigenicity in MES glioma spheres in vivo, we used orthotopic xenografts into mouse brains. Injection of MES83 spheres into the striatum of immunocompromised mouse brains resulted in lethal tumors within 30 days (6,34). Although the cells transduced with the lentiviral shNT construct did not show altered tumorigenesis or subsequent mouse survival, xenografting of the FOXD1 silenced MES glioma spheres diminished the proportion of tumor formation (3 of 5 mice) and prolonged survival periods (Fig. 3E). Histologically, the tumors in the control mice were highly vascularized GBM-like brain tumors with central necrosis, and developed by day 18 after transplantation (Fig. 3F and G).
Figure 3. FOXD1 regulates MES GSC growth both in vitro and in vivo.
A. Western blot analyses of MES83 and MES28 glioma spheres transduced with shRNA targeting FOXD1 (shFOXD1#1 or shFOXD1#2) or a non-targeting control (shNT).
B. In vitro growth assay shows that shRNAs targeting FOXD1 (shFOXD1#1 and shFOXD1#2) inhibit cell proliferation of MES83 and MES28 glioma spheres (P<0.0001, n=6, one-way ANOVA).
C. Representative images of MES83 and MES28 glioma spheres transduced with shRNA targeting FOXD1. shNT serves as a control. Bar, 60μm.
D. In vitro clonogenicity assays (limiting dilution neurosphere formation assays) indicate that FOXD1 shRNA decreases clonogenicity of MES83 and MES28 cells (MES83 P<0.001, and MES28 P<0.001, ELDA analyses).
E. Kaplan-Meier analysis of nude mice harboring intracranial tumors derived from MES83 GSCs transduced with shNT (n=6) or shFOXD1#1 (n=5). (P=0.0014, with log-rank test)
F, G. Representative images of brains (F) and H&E stained brain sections (G) of mice after intracranial transplantation of MES83 glioma spheres transduced with shNT or shFOXD1#1. Bar, 1 mm (G, upper panel) and 100 μm (G, lower panel).
FOXD1 regulates ALDH1A3 transcription in MES glioma spheres
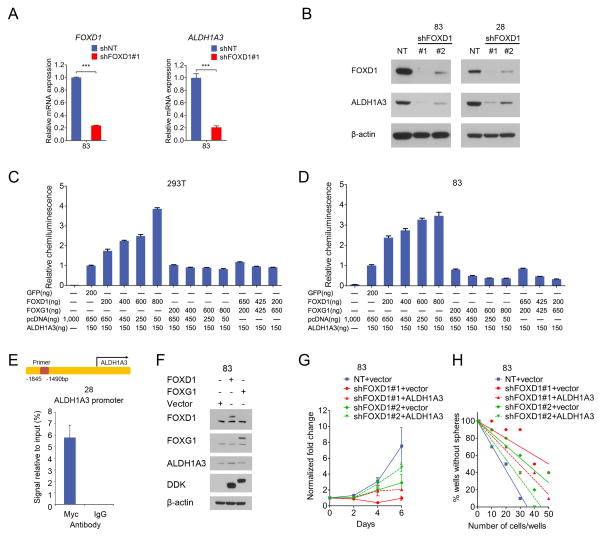
Because FOXD1 is almost exclusively expressed in ALDHhigh MES GBM cells, not in ALDHlow cells (Fig 1G), we examined whether FOXD1 transcriptionally regulates the ALDH1A3 gene, thereby orchestrating the stem cell properties in MES GSCs. Indeed, FOXD1 silencing in the MES83 glioma spheres significantly decreased the ALDH1A3 mRNA and protein levels (Fig. 4A and B). Since there is a common Forkhead TF binding motif in the promoter region of the ALDH1A3 gene (Supplementary Fig. S4A), we hypothesized that FOXD1 activated ALDH1A3 transcription. Thus, we performed a luciferase assay to measure ALDH1A3 promoter activity upon exogenous expression of FOXD1 in 293T cells and MES83 spheres (Fig. 4C and D). As expected, ectopically expressed FOXD1 increased ALDH1A3 reporter activity. Since FOXD1 and FOXG1 exhibit mutually-exclusive expression patterns (Fig. 1B–D), we assumed that FOXG1 may counteract FOXD1 activity in MES GSCs. Intriguingly, co-expression of FOXG1 and FOXD1 in MES83 spheres counter-acted ALDH1A3 reporter activities driven by FOXD1 alone (Fig. 4C and D). Further, the transcriptional regulation of ALDH1A3 by FOXD1 was confirmed by chromatin immunoprecipitation PCR, which indicated that FOXD1 directly binds to the promoter region of the ALDH1A3 gene in MES28 glioma spheres (Fig. 4E). In addition, the overexpression of FOXD1 in MES83 increased the expression of ALDH1A3 (Fig. 4F). To investigate the physiological role of the FOXD1-ALDH1A3 axis in MES GSCs, we evaluated whether ALDH1A3 overexpression rescues the phenotypes of MES glioma spheres induced by FOXD1 KD. The reduced in vitro cell growth and neurosphere formation by FOXD1 KD were partially, yet not completely, restored by the overexpression of ALDH1A3, but not by the control vector (Fig. 4G, H and Supplementary Fig. S4B). On the other hand, FOXD1 overexpression alone did not induce any noticeable changes in the PN spheres, at least in the expressions of representative markers (AXL, CD44, and Olig2) (Supplementary Fig. S4C). We also analyzed dataset (GSE67089) in our previous publication (6) for the expression levels of FOXD1 with all the members of ALDH family. We consistently found that FOXD1 is co-expressed with ALDH1A3, but not other ALDH members, in MES but not in PN subtype spheres (Supplementary Fig. S4D). Collectively, these data suggest that FOXD1 is required, but not sufficient, for the establishment of the MES phenotype in GSCs.
Figure 4. ALDH1A3 is a functional MES GSC marker and is transcriptionally regulated by FOXD1.
A. RT-qPCR analyses of ALDH1A3 and FOXD1 mRNA in MES83 glioma spheres transduced with shFOXD1#1 or shNT (P<0.001, n=3, with t-test).
B. Western blot analyses of ALDH1A3 in MES83 and MES28 glioma spheres transduced with shFOXD1#1, shFOXD1#2 or shNT. β-actin serves as a loading control.
C, D. Luciferase assays with 293T cells (C) and MES83 glioma spheres (D) co-transfected with an ALDH1A3 promoter reporter plasmid together with overexpression vectors for the indicated genes (n=3).
E. ChIP-qPCR assay using Myc antibody or control IgG in MES28 glioma spheres transfected with a FOXD1 (Myc-DDK-tagged) plasmid shows the binding of FOXD1 on the ALDH1A3 promoter (P<0.001, n=3, t-test).
F. Western blot analyses of the indicated proteins in MES83 glioma spheres transduced with the overexpression plasmids encoding FOXD1, FOXG1, or empty vector.
G, H. ALDH1A3 overexpression partially restores the in vitro proliferation (G, P<0.001, one way ANOVA) and neurosphere formation capacities (H, P<0.001, ELDA analysis) which are inhibited by shFOXD1#1 or #2 in MES83 glioma spheres.
FOXD1 and ALDH1A3 are evolutionarily conserved genes that contribute to glial neoplasms
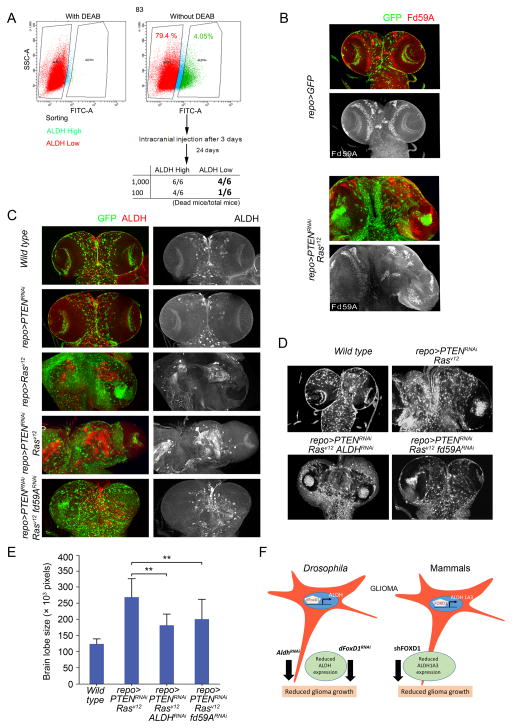
Previously, we reported that ALDHhigh MES glioma sphere cells have higher clonogenic potential in vitro than ALDHlow cells in MES GBM (6). To compare the in vivo tumorigenic abilities of ALDHhigh and ALDHlow cells in MES glioma spheres, we injected ALDHhigh or ALDHlow MES83 cells into mouse brains at two different cell numbers (100 and 1,000 cells). When 100 ALDHlow or ALDHhigh cells were injected into mouse brains, only one of six mice injected with ALDHlow cells died by 24th day after transplantation. In contrast, four of six mice in the ALDHhigh group died due to tumor burden. Similar results were observed when 1,000 tumor cells were injected (Fig. 5A), indicating that the tumor initiating cells reside in the minor subset of MES GBM cells with elevated ALDH1 activity (approximately 7% of the MES83 cells).
Figure 5. RNA interference-mediated silencing of FD59A and ALDH attenuates growth of Drosophila glial neoplasia.
A. Upper panel, ALDEFLUOR assay in MES83 glioma spheres with or without ALDH1 inhibitor DEAB. Lower panel, frequencies of tumor formation of ALDH1high and ALDH1low cell populations of MES83 glioma spheres in mice.
B. Expression of Fd59A (Drosophila ortholog of FOXD1) (red, grey) in the Drosophila CNS derived from larvae of repoGAL4 UASGFP and repoGAL4 UASGFP UASPTENRNAi UASRasv12. Glial cells are marked by GFP (green).
C. Expression levels of ALDH (red, grey) in the larval CNS of repoGAL4 UASGFP (Wild type), repoGAL4 UASGFP UASPTENRNAi, repoGAL4 UASGFP UASRasv12, repoGAL4 UASGFP UASPTENRNAi UASRasv12 and repoGAL4 UASGFP UASPTENRNAi UASRasv12 fd59ARNAi.
D. The effects of ALDHRNAi and fd59ARNAi on the growth of glial neoplasms of repoGAL4 UASPTENRNAi UASRasv12 larvae. Both RepoGAL4 UASGFP (Wild-type) and repoGAL4 UASPTENRNAi UASRasv12 samples are included for comparison.
E. The quantification of brain tumor volume (brain lobe size in pixels) from the indicated larvae (repoGAL4 UASPTENRNAi UASRasv12 ALDHRNAi vs. repoGAL4 UASPTENRNAi UASRasv12, P=0.008, n=3, one way ANOVA; repoGAL4 UASPTENRNAi UASRasv12 fd59ARNAi vs. repoGAL4 UASPTENRNAi UASRasv12, P=0.007, n=3, one way ANOVA).
F. The schematic diagram depicts that ALDH and fd59A (dFOXD1) are evolutionarily conserved genes contributing tumorigenesis of glial neoplasms.
Next, we examined the functional role of FOXD1 and ALDH1A3 in Drosophila. In flies, the orthologs of FOXD1and ALDH1A3 are fd59A (35) and DmALDH (36), respectively. fd59A is highly expressed in the Drosophila larval (embryonic) Central Nervous System (CNS) (35). We used recently established Drosophila glioma model involving co-activation of oncogenic Ras and PI3K pathways by the GAL4 UAS system (repoGAL4 UASPTENRNAi UASRasV12), which causes the formation of invasive glial neoplasms that mimic human gliomas (37). In the larval glial neoplasms derived from the repoGAL4 UASPTENRNAi; UASRasV12 larvae, Fd59A and ALDH levels were substantially upregulated compared to the normal CNS (repoGAL4 UASGFP) (Fig. 5B and C). To investigate if the formation of the glial neoplasms in Drosophila requires ALDH, we transduced UASALDHRNAi into Drosophila glioma tumors. Elimination of ALDH resulted in reduction in glioma growth by 1.7-fold (Fig. 5D and E, P=0.008). This anti-tumor effect of ALDH silencing was also observed in the mosaic models of Drosophila eye cancer (Supplementary Fig. S5). Next, we investigated the effects of downregulation of Fd59A in these cancers. As shown in Fig. 5D and E, downregulation of Fd59A resulted in a reduction in glioma growth (P=0.007) with a concomitant downregulation of ALDH levels (Fig. 5C). Taken together, these data suggest that FOXD1 and ALDH1A3 are evolutionarily conserved genes that contribute to CNS tumorigenesis (Fig. 5F).
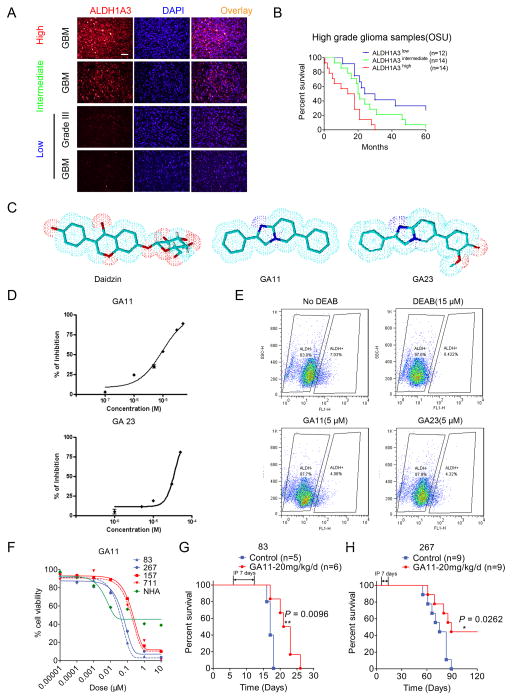
ALDH1A3 expression indicates poor prognosis of post-surgical glioma patients
To interrogate whether ALDH1A3 is a clinically relevant biomarker, we analyzed 40 high-grade glioma patient samples (Fig. 6A). The log rank test showed that the glioma patients with higher ALDH1A3 protein expression has a significantly shorter post-surgical survival periods compared to either the intermediate or low ALDH1A3 protein expression groups (Fig. 6B, P=0.0285, ALDH1A3high vs. ALDH1A3intermediate; and P=0.0016, ALDH1A3high vs. ALDH1A3low). Additionally, the data from Rembrandt database demonstrates a similar pattern (Supplementary Fig. S6). These data indicate the clinical significance of ALDH1A3.
Figure 6. The novel ALDH small molecule inhibitor GA11 attenuates MES GSC growth both in vitro and in vivo.
A. Representative immunofluroescence images of ALDH1A3 in 40 high grade glioma samples. DAPI is used for nuclear labeling. Bar, 50 μm.
B. Kaplan-Meier analysis of ALDH1A3 expression indicates the negative correlation between ALDH1A3 protein expression and survival in high grade glioma patients. (ALDH1A3 high vs. ALDH1A3 intermediate, P=0.0285, ALDH1A3 high vs. ALDH1A3 low, P=0.0016, and ALDH1A3 intermediate vs. ALDH1A3 low, P=0.1769, with log rank test).
C. The comparison of the essential core structure of the naturally occurring ALDH inhibitor, daidzin, and the structures of synthesized novel imidazo [1,2-a] pyrimidine ALDH inhibitors, GA11 and GA23.
D. Log-dose response analysis of GA11 (upper panel) and GA23 (lower panel) in yeast.
E. Flow cytometry analyses using ALDEFLUOR indicate that both GA11 and GA23 (5μM, 30 min) inhibit ALDH activity in MES83 glioma spheres.
F. Log-dose response analyses of the effects of GA11 on the viabilities of MES83, MES267, PN157, PN711, glioma spheres and NHA cells.
G, H. Treatment with GA11 (intraperitoneal injection, 20mg/kg for 7 days from day seven) prolongs survival periods of mice bearing MES83-derived intracranial tumors (G) (P=0.0096, with log rank test) and those of mice with MES267-derived intracranial tumors (H) (P=0.0262, with log rank test).
The novel ALDH1 inhibitor GA11 has anti-GBM effects in vitro and in vivo
Based on the inhibitory effects of FOXD1 silencing on the MES GSC-derived mouse brain tumors and of ALDH1A3 silencing on the Drosophila brain cancer model, we sought to design novel, clinically efficacious small molecule inhibitors selectively targeting the ALDH1 activity for GBM therapies. The natural product daidzin and its congeners have been reported to inhibit ALDH1 (38). To design clinically applicable analogs with better pharmacokinetic properties (39), we identified the imidazo [1,2-a] pyrimidine heterocyclic core as the essential scaffold. We added two planar, aromatic and lipophilic areas to this scaffold in positions two and six of the nucleus, thereby generating the novel small molecules GA11 and GA23 (Fig. 6C and Supplementary Fig. S7A). The structural assessment confirmed that the replacement of glucopyranose portion of daidzin with an aromatic ring reduces both hydrogen bond forming potential and rotatable bonds, resulting in smaller molecules with a lower polar surface area and limited flexibility. This modification retained the core structure of daidzin essential for inhibiting ALDH1 activity, while establishing a drug-like profile, i.e. a more favorable prediction of the blood-brain barrier (BBB) penetration. Indeed, an in silico evaluation confirmed that the physical-chemical properties of these novel compounds were fully consistent with those required for BBB penetration (Supplementary Table S3) (40). In addition, the molecular weights of GA11 and GA23 (close to 310, the mean value of marketed CNS drugs) are significantly lower than that of daidzin. Moreover, the H-bonding potential of these novel compounds is commensurate to that of successful CNS drug candidates. Indeed, the sum of heteroatoms capable of hydrogen bonding, like nitrogen (N) and oxygen (O), is less than five, conferring on the molecules a high probability of entering the CNS. Additionally, the rotatable bond count is less than eight, and the limited flexibility of the molecules should promote passive permeation through BBB. Lowering both molecular weight and H-bonding potential have a significant effect on the polar surface area (PSA) of the compounds, which is a key factor in determining the extent of BBB penetration. Unlike daidzin, the novel compounds have PSA values lower than the stringent cut-off 60–70 Å, which characterizes commercial CNS drugs (40).
To validate the inhibitory effects, we first determined the enzymatic activity of yeast ALDH incubated with GA11 and GA23 in an in vitro enzymatic assay (30% of the ALDH sequences in yeast and eukaryotes are conserved (41)). As expected, both GA11 and GA23 inhibited the ALDH enzymatic activity with IC50 values in the micro molar range (Fig. 6D). GA11 was more potent. Further, both GA11 and GA23 inhibited human ALDH1 demonstrated by a marked reduction of ALDHhigh cellular populations in MES83 glioma spheres treated with GA11 and GA23 (Fig. 6E). As a result, both GA11 and GA23 inhibited the growth of MES glioma spheres. In contrast, PN glioma spheres were relatively more resistant to these two compounds (Fig. 6F and Supplementary Fig. S7B–D). More importantly, systemic treatment of MES83 and MES267-based mouse brain tumors with GA11 significantly attenuated tumor growth, thereby extending the survival of tumor bearing mice compared to the vehicle treated counterparts (Fig. 6G, H and Supplementary Fig. S7E).
Discussion
GBMs display intra-tumoral cellular heterogeneity; within the same tumor different cell populations respond differently to therapies (3,42,43). GSCs are at the apex of the cellular hierarchy (3,42). Thus, understanding how GSCs are maintained may improve the efficacies of current therapeutic strategies. Recent evidence suggests that GSCs are subclassified into two subtypes with MES GSCs being more therapy-resistant (4,6). Therefore, identification of MES GSC regulatory molecules has the potential to lead to novel and efficacious GBM therapeutics. Our study (Fig. 5A) is the first demonstrating that ALDH1high GBM cells are more tumorigenic in vivo than the ALDH1low counterparts, indicating that ALDH1 is a specific marker for MES GSCs. This is further supported by analyses of clinical glioma samples (Fig. 6B). To develop chemotherapeutics targeting MES GSC, we synthesized a novel class of imidazo [1,2-a] pyridine derivatives called GA11 and GA23 as ALDH1 inhibitors. These compounds were designed based on the conserved structural traits of the known natural occurring inhibitors including daidzin. In addition to fulfilling pharmacophore needs, these novel compounds possess a good hydrophilic-lipophilic balance, which allows for better penetration of the BBB. In principle, both these features make GA11, in particular, an attractive drug candidate to target GSCs in GBM tumors. This structural prediction was validated in a cell-free kinase assay, which proved the ability of GA11 to inhibit the target enzyme ALDH in yeast (Fig. 6D) and further, an in vitro ALDEFLUOR assay showed the substantial decline of human ALDH1 activity in GA11-treated MES glioma spheres (Fig. 6E). Consistently, GA11 inhibited in vitro glioma sphere proliferation and in vivo xenograft growth in mouse brains (Fig 6F and G, H, respectively). Further pre-clinical evaluation of GA11 and its analogs is currently underway for clinical development of anti-ALDH1 therapeutics for GBM.
Another novel set of findings in this study is that ALDH1A3 is transcriptionally regulated by FOXD1 (Fig. 4E). The ALDH1A3 signaling in cancers is likely evolutionally conserved based on the fact that ALDH1A3 silencing in vivo displayed substantial suppression of the genetically-engineered Drosophila brain cancers (Fig. 5D, E). Interestingly, this FOXD1-mediated ALDH1A3 transcriptional activation was counter-acted by another Forkhead family member, FOXG1 in the MES GSC-enriched cultures (Fig. 4C, D). This data is supported by the presence of a Forkhead TF consensus sequence in the ALDH1A3 promoter region. These data suggest that MES GSCs hijack the molecular mechanism for normal development (e.g. mouse retina) to promote their tumor growth (44). It is not clear, however, whether these two Forkhead TFs compete for the DNA binding in the ALDH1A3 promoter or if FOXG1 indirectly influences FOXD1 signaling in MES GSCs. Additionally, the restoration of ALDH1A3 by exogenous expression did not fully rescue the defects in MES glioma sphere growth caused by FOXD1 silencing (Fig. 4G), suggesting that additional undetermined oncogenic mechanisms are likely involved in MES GBM. Further studies are needed to fully clarify the mechanisms by which FOXD1 maintains the MES GSCs phenotype and thus, GBM tumorigenesis and therapy-resistance.
In conclusion, this study describes the upregulation of ALDH1A3 and FOXD1 in clinical glioma samples and establishes their functional roles in the maintenance of MES GSCs, and therefore MES GBM tumorigenesis. Furthermore, this study provided the first evidence supporting the fact that elevated FOXD1 expression is a negative prognostic factor in glioma, and establishes a role for FOXD1 directly regulating ALDH1A3 transcription in MES GSCs. Taken together, the FOXD1-ALDH1A3 axis is critical for tumor initiation in MES GSCs, therefore providing possible new molecular targets for the treatment of GBM and other ALDH1-activated cancers.
Supplementary Material
Acknowledgments
Financial support
This study is supported by P01CA163205, R01NS083767, R01NS087913, and R01CA183991 (I. Nakano). P. Cheng was supported by The First Hospital of China Medical University. I. Waghmare was supported in part by the Graduate Program at the University of Dayton. M. Kango-Singh was supported in part by Start-up funds from the University of Dayton.
We thank all the members in the Nakano laboratory for constructive discussion for this study. We also thank Amber K. O’Connor for English language editing for this study.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed by the authors.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. The New England journal of medicine. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Venere M, Fine HA, Dirks PB, Rich JN. Cancer stem cells in gliomas: identifying and understanding the apex cell in cancer’s hierarchy. Glia. 2011;59:1148–54. doi: 10.1002/glia.21185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer cell. 2013;24:331–46. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waghmare I, Roebke A, Minata M, Kango-Singh M, Nakano I. Intercellular cooperation and competition in brain cancers: lessons from Drosophila and human studies. Stem cells translational medicine. 2014;3:1262–8. doi: 10.5966/sctm.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110:8644–9. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nature reviews Genetics. 2009;10:233–40. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakano I. Transcription factors as master regulator for cancer stemness: remove milk from fox? Expert review of anticancer therapy. 2014;14:873–5. doi: 10.1586/14737140.2014.940324. [DOI] [PubMed] [Google Scholar]
- 9.Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. The EMBO journal. 1994;13:5002–12. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatini V, Huh SO, Herzlinger D, Soares VC, Lai E. Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of Winged Helix transcription factor BF-2. Genes & development. 1996;10:1467–78. doi: 10.1101/gad.10.12.1467. [DOI] [PubMed] [Google Scholar]
- 11.Thackray VG. Fox tales: regulation of gonadotropin gene expression by forkhead transcription factors. Molecular and cellular endocrinology. 2014;385:62–70. doi: 10.1016/j.mce.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carreres MI, Escalante A, Murillo B, Chauvin G, Gaspar P, Vegar C, et al. Transcription factor Foxd1 is required for the specification of the temporal retina in mammals. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:5673–81. doi: 10.1523/JNEUROSCI.0394-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fetting JL, Guay JA, Karolak MJ, Iozzo RV, Adams DC, Maridas DE, et al. FOXD1 promotes nephron progenitor differentiation by repressing decorin in the embryonic kidney. Development. 2014;141:17–27. doi: 10.1242/dev.089078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung C, Linn G, Chow YH, Kobayashi A, Mittelsteadt K, Altemeier WA, et al. Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. American journal of respiratory and critical care medicine. 2013;188:820–30. doi: 10.1164/rccm.201212-2297OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gumbel JH, Patterson EM, Owusu SA, Kabat BE, Jung DO, Simmons J, et al. The forkhead transcription factor, Foxd1, is necessary for pituitary luteinizing hormone expression in mice. PloS one. 2012;7:e52156. doi: 10.1371/journal.pone.0052156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koga M, Matsuda M, Kawamura T, Sogo T, Shigeno A, Nishida E, et al. Foxd1 is a mediator and indicator of the cell reprogramming process. Nature communications. 2014;5:3197. doi: 10.1038/ncomms4197. [DOI] [PubMed] [Google Scholar]
- 17.Karlsson J, Holmquist Mengelbier L, Ciornei CD, Naranjo A, O’Sullivan MJ, Gisselsson D. Clear cell sarcoma of the kidney demonstrates an embryonic signature indicative of a primitive nephrogenic origin. Genes, chromosomes & cancer. 2014;53:381–91. doi: 10.1002/gcc.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Heul-Nieuwenhuijsen L, Dits NF, Jenster G. Gene expression of forkhead transcription factors in the normal and diseased human prostate. BJU international. 2009;103:1574–80. doi: 10.1111/j.1464-410X.2009.08351.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhao YF, Zhao JY, Yue H, Hu KS, Shen H, Guo ZG, et al. FOXD1 promotes breast cancer proliferation and chemotherapeutic drug resistance by targeting p27. Biochemical and biophysical research communications. 2015;456:232–7. doi: 10.1016/j.bbrc.2014.11.064. [DOI] [PubMed] [Google Scholar]
- 20.Kim SH, Ezhilarasan R, Phillips E, Gallego-Perez D, Sparks A, Taylor D, et al. Serine/Threonine Kinase MLK4 Determines Mesenchymal Identity in Glioma Stem Cells in an NF-kappaB-dependent Manner. Cancer cell. 2016;29:201–13. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng P, Phillips E, Kim SH, Taylor D, Hielscher T, Puccio L, et al. Kinome-wide shRNA screen identifies the receptor tyrosine kinase AXL as a key regulator for mesenchymal glioblastoma stem-like cells. Stem cell reports. 2015;4:899–913. doi: 10.1016/j.stemcr.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Remmele W, Schicketanz KH. Immunohistochemical determination of estrogen and progesterone receptor content in human breast cancer. Computer-assisted image analysis (QIC score) vs. subjective grading (IRS) Pathology, research and practice. 1993;189:862–6. doi: 10.1016/S0344-0338(11)81095-2. [DOI] [PubMed] [Google Scholar]
- 24.Hutterer M, Knyazev P, Abate A, Reschke M, Maier H, Stefanova N, et al. Axl and growth arrest-specific gene 6 are frequently overexpressed in human gliomas and predict poor prognosis in patients with glioblastoma multiforme. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:130–8. doi: 10.1158/1078-0432.CCR-07-0862. [DOI] [PubMed] [Google Scholar]
- 25.Verghese S, Bedi S, Kango-Singh M. Hippo signalling controls Dronc activity to regulate organ size in Drosophila. Cell Death Differ. 2012;19:1664–76. doi: 10.1038/cdd.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–52. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano I, Joshi K, Visnyei K, Hu B, Watanabe M, Lam D, et al. Siomycin A targets brain tumor stem cells partially through a MELK-mediated pathway. Neuro-oncology. 2011;13:622–34. doi: 10.1093/neuonc/nor023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi K, Banasavadi-Siddegowda Y, Mo X, Kim SH, Mao P, Kig C, et al. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem cells. 2013;31:1051–63. doi: 10.1002/stem.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minata M, Gu C, Joshi K, Nakano-Okuno M, Hong C, Nguyen CH, et al. Multi-kinase inhibitor C1 triggers mitotic catastrophe of glioma stem cells mainly through MELK kinase inhibition. PloS one. 2014;9:e92546. doi: 10.1371/journal.pone.0092546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 31.Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer research. 2005;65:8679–89. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 32.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Nakano I, Kornblum HI. Methods for analysis of brain tumor stem cell and neural stem cell self-renewal. Methods Mol Biol. 2009;568:37–56. doi: 10.1007/978-1-59745-280-9_4. [DOI] [PubMed] [Google Scholar]
- 34.Cheng P, Phillips E, Kim SH, Taylor D, Hielscher T, Puccio L, et al. Kinome-wide shRNA Screen Identifies the Receptor Tyrosine Kinase AXL as a Key Regulator for Mesenchymal Glioblastoma Stem-like Cells. Stem cell reports. 2015;4:899–913. doi: 10.1016/j.stemcr.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HH, Frasch M. Survey of forkhead domain encoding genes in the Drosophila genome: Classification and embryonic expression patterns. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;229:357–66. doi: 10.1002/dvdy.10443. [DOI] [PubMed] [Google Scholar]
- 36.Chakravorty S, Wajda MP, Vigoreaux JO. Courtship song analysis of Drosophila muscle mutants. Methods. 2012;56:87–94. doi: 10.1016/j.ymeth.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Read RD, Cavenee WK, Furnari FB, Thomas JB. A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS genetics. 2009;5:e1000374. doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao GY, Li DJ, Keung WM. Synthesis of potential antidipsotropic isoflavones: inhibitors of the mitochondrial monoamine oxidase-aldehyde dehydrogenase pathway. Journal of medicinal chemistry. 2001;44:3320–8. doi: 10.1021/jm0101390. [DOI] [PubMed] [Google Scholar]
- 39.Del Turco S, Sartini S, Sentieri C, Saponaro C, Navarra T, Dario B, et al. A novel 2,3-diphenyl-4H-pyrido[1,2-a]pyrimidin-4-one derivative inhibits endothelial cell dysfunction and smooth muscle cell proliferation/activation. European journal of medicinal chemistry. 2014;72:102–9. doi: 10.1016/j.ejmech.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 40.Pajouhesh H, Lenz GR. Medicinal chemical properties of successful central nervous system drugs. NeuroRx : the journal of the American Society for Experimental NeuroTherapeutics. 2005;2:541–53. doi: 10.1602/neurorx.2.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saigal D, Cunningham SJ, Farres J, Weiner H. Molecular cloning of the mitochondrial aldehyde dehydrogenase gene of Saccharomyces cerevisiae by genetic complementation. Journal of bacteriology. 1991;173:3199–208. doi: 10.1128/jb.173.10.3199-3208.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan Q, Li Q, Liu S, Ning N, Zhang X, Xu Y, et al. Concise Review: Targeting Cancer Stem Cells Using Immunologic Approaches. Stem cells. 2015;33:2085–92. doi: 10.1002/stem.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes & development. 2015;29:1203–17. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian NM, Pratt T, Price DJ. Foxg1 regulates retinal axon pathfinding by repressing an ipsilateral program in nasal retina and by causing optic chiasm cells to exert a net axonal growth-promoting activity. Development. 2008;135:4081–9. doi: 10.1242/dev.023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.