Abstract
Resistance to endocrine therapies remains a significant clinical problem for estrogen receptor-α (ERα)-positive breast cancer. On-target side effects limit therapeutic compliance and use for chemoprevention, highlighting an unmet need for new therapies. Here we present a full-antagonist ligand series lacking the prototypical ligand side chain that has been universally used to engender antagonism of ERα through poorly understood structural mechanisms. A series of crystal structures and phenotypic assays reveal a structure-based design strategy with separate design elements for antagonism and degradation of the receptor and access to a structurally distinct space for further improvements in ligand design. Understanding structural rules that guide ligands to produce diverse ERα-mediated phenotypes has broad implications for the treatment of breast cancer and other estrogen-sensitive aspects of human health including bone homeostasis, energy metabolism, and autoimmunity.
Introduction
Estrogen receptor-α (ERα)-positive breast cancer is initially associated with better prognosis than ERα-negative disease, with improved overall survival at 5 years across age groups1. The majority of breast cancer metastases and fatalities still result from ERα-positive disease since 70–80% of all breast cancer cases are ERα-positive2,3. ERα has been the most successful biomarker in cancer, directing patients to one of several types of endocrine therapy, but de novo and acquired resistance remain significant problems in up to half of patients. Efforts to improve endocrine therapies have been limited by a lack of structural information explaining the agonist activity profiles of selective estrogen receptor modulators (SERMs), distinguishing full antagonists such as fulvestrant from SERMs, or clarifying the contribution of ERα degradation to clinical efficacy of selective estrogen receptor degraders (SERDs) such as GW5638 and new SERDs in clinical trials (NCT02248090, NCT01823835)4,5.
ERα regulates transcription by orchestrating recruitment, dismissal and recycling of coregulator complexes that in turn control the basal transcriptional machinery and catalyze posttranslational modification of histones and other DNA-associated proteins to remodel chromatin and regulate gene expression6–8. Binding of an agonist stabilizes a coactivator-binding surface of the ligand-binding domain (LBD) called activation function-2 (AF2), and also stimulates coactivator recruitment to another site called activation function-1 (AF1) in the AB domains (Supplementary Results, Supplementary Fig. 1a). In the active LBD conformation helix-12 (h12) in the LBD docks across helix-11 and helix-3 (h11 and h3) to form one side of the AF2 surface (see Supplementary Fig. 1b)9,10. Anti-estrogens including SERMs and SERDs have a bulky side chain that protrudes between h3 and h11 to directly relocate h12 (Supplementary Fig. 1c–h), and thereby disrupt the active LBD conformation, block coactivator recruitment, and inhibit expression of mitogenic ERα-target genes9–12. We describe these ER ligands as “direct antagonists” because their side chains directly displace h12 from its agonist-induced position. Minor changes in the orientation and composition of the side chain have been used to fine-tune SERM and SERD activity, including efficacy and on-target side effects13,14.
Here we present the discovery of a series of SERDs lacking a prototypical side chain. These compounds are based on a 7-oxabicyclo[2.2.1]hept-5-ene-sulfonamide (OBHS-N) scaffold, and operate through a structural mechanism that we call “indirect antagonism”. We previously showed that partial agonist activity could be generated by indirectly influencing the docking of h12 against h11, through ligand-induced shifts in h1115–17. We now demonstrate that a similar mechanism can be used to generate a full antagonist profile and induce robust ERα degradation.
Results
OBHS-N ligands are full antagonists
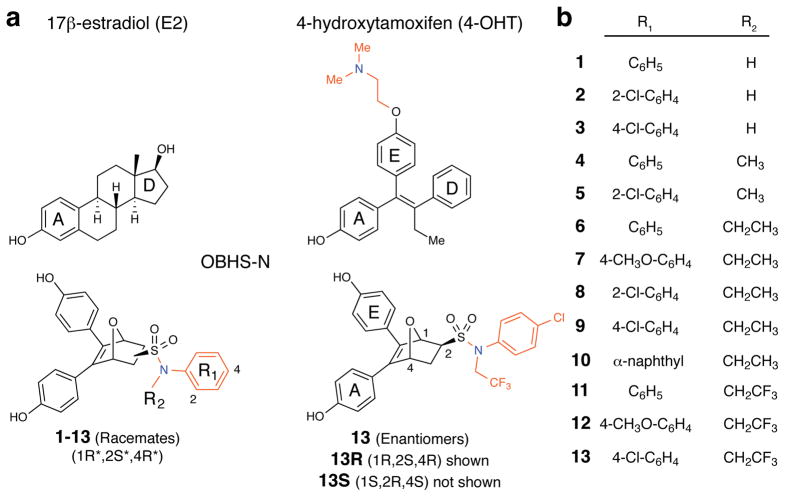
We generated several series of 7-oxabicyclo [2.2.1] hept-5-ene (OBH) compounds containing two phenols, one of which mimics the A-ring of E2 in binding the conserved hydrogen bonding amino acids Glu351 and Arg394, while the other mimics what we call the E-ring in tamoxifen, representing the traditional site for SERM/SERD side chain addition (Fig. 1a). Addition of a phenyl sulfonate led to OBHS compounds that generated a range of partial agonist activities. The parental OBHS compound demonstrated a dissociated phenotype with the strong anti-inflammatory properties of E2, but lacking proliferative or anti-proliferative activity16,17. Here we used a sulfonamide linker to attach the h11-directed phenyl group (R1), which allowed us to add a second functional group (R2) to the nitrogen atom (Fig. 1a), and thereby generate a series of OBHS-N compounds with a variety of h11-directed substitutions (compounds 1–13, Fig. 1b). The Diels-Alder cycloaddition with furan produced exo diastereomers as racemates. We previously published the synthesis, binding affinities and EC50/ IC50 reporter activity for these compounds18, and the dose curves are shown in Supplementary Figure 2.
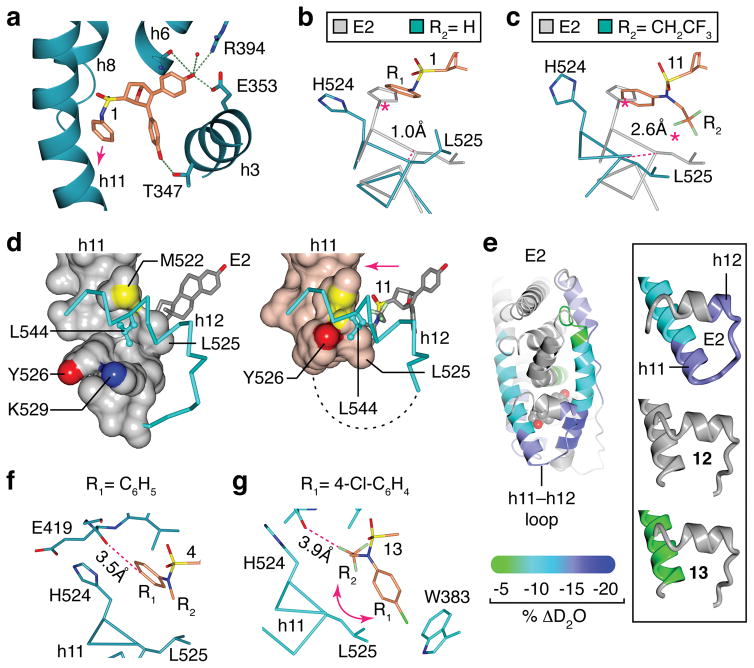
Figure 1. Overview of the ligand-dependent modulation of ERα activity.
(a) Structures of 17β-estradiol (E2), 4-hydroxytamoxifen (4-OHT), and the OBHS-sulfonamides (OBHS-N). The ring designations A, D and E are given to facilitate comparisons among these structures. In the OBHS-N structures, a helix-11 (h11)-directed aryl group, R1, was attached to the ligand core via a sulfonamide linker, which provided a site to affix a second h11-directed functional group, R2. Locant numbers 1, 2, and 4 for stereochemical designations are on the 7-oxabicyclo[2.2.1]hept-5-ene core (see lower right structure), and locant numbers 2 and 4 on the R1 ring (lower left structure) are the locations of the substituents listed in panel b.
(b) List of OBHS-N analogs (1–13). All 13 analogs have the 1R*,2S*,4R* relative configuration in the 7-oxabicyclo[2.2.1]hept-5-ene core and were prepared and tested as racemates (i.e., a 1:1 mixture of 1R,2S,4R and 1S,2R,4S enantiomers). In addition, both enantiomers of analog 13, designated 13R and 13S, were isolated by chiral HPLC and were also tested individually.
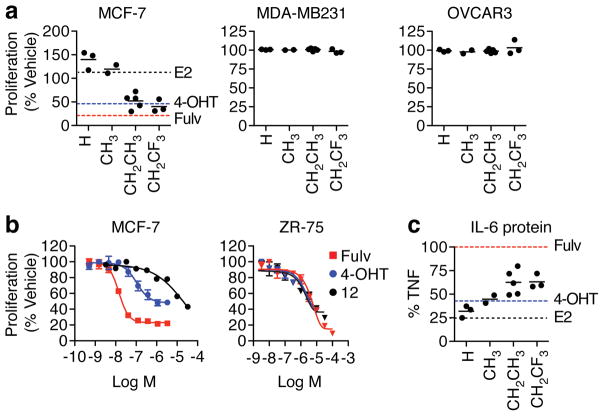
We first evaluated effects of the ligands in cell culture models. The anti-proliferative effect of the compounds was determined by the size of the alkyl R2 group on the sulfonamide linker, with very little effect coming from the type of R1 substitution on the phenyl. Ligands with smaller R2 substituents stimulated proliferation, whereas those with bulkier R2 groups inhibited proliferation of MCF-7 cells as effectively as 4-hydroxytamoxifen (4-OHT) and fulvestrant (Fig. 2a). None of the OBHS-N compounds inhibited proliferation of ER-negative breast cancer (MBA-MB231) cells, or a panel of hormone-independent ovarian cancer cell lines (Fig. 2a, Supplementary Fig. 3a). Dose response curves were measured for 11–13, which were the only OBHS-N analogs that showed both inhibition of proliferation and efficient ERα degradation (shown below). These compounds showed lower potency than 4-OHT or fulvestrant, but similar efficacy in ERα-positive MCF-7 and ZR-75 breast cancer cells (Fig. 2b, Supplementary Fig. 3b). Next, the effects of 12 were examined under non-adherent conditions using a mammosphere formation assay, as non-attachment growth provides a distinct measure of oncogenesis compared to growth on plastic19. Compound 12 inhibited the number of mammospheres formed as effectively as fulvestrant (Supplementary Fig. 3c–d).
Figure 2. OBHS-N analogs exhibit growth-inhibitory and anti-inflammatory effects.
(a) MCF-7, MDA-MB231 and OVCAR3 cells were cultured in steroid free media, treated on days 1 and 4 with 10 μM OBHS-N compounds, and analyzed on day 7. Ligands were grouped by R2 group. Each data point represents the average (mean) number of cells observed in triplicate experiments with a distinct compound, and is shown as a percentage of vehicle-treated cells. Dotted lines represent values for 10 nM estradiol (E2, black), 1 μM 4-OHT (cyan) and 1 μM fulvestrant (Fulv, red).
(b) Dose-dependent suppression of ERα+ breast cancer cell proliferation by tamoxifen, fulvestrant or compound 12. MCF-7 or ZR-75 cells were treated as in panel a, with increasing doses of ligands. Data represents mean ± s.e.m of triplicate values.
(c) Steroid-deprived MCF-7 cells were treated with vehicle, 10 ng/ml TNFα, or TNFα + OBHS-N compounds, E2, 4-OHT or fulvestrant (Fulv) at 10 μM for 24 hr. Media was collected and IL-6 protein levels were measured by AlphaLISA. Each point represents average of n = 3.
Although ERα mediates proliferative signals required for estrogen-dependent breast cancer progression, ERα activity is also associated with a better prognosis, which may relate to inhibition of inflammatory and oncogenic pathways activated by NF-κB. ERα is recruited to cytokine promoters by tethering to NF-κB or AP-1 proteins20–23. We examined inhibition of TNFα-induced secretion of IL-6, a cytokine associated with oncogenesis and worse prognosis in breast cancer. Importantly fulvestrant, which fully antagonizes E2-mediated suppression of IL-623, did not inhibit IL-6 secretion, unlike tamoxifen, which acts as a partial agonist to inhibit IL-6 secretion (Fig. 2c). There was a structure-activity relationship with the OBHS-N compounds where once again, R2 determined the anti-inflammatory response (Fig. 2c). Importantly, the trifluoroethyl OBHS-N compounds profiled as full antagonists with respect to proliferation but show greater suppression of TNFα-induced IL-6 secretion compared to fulvestrant.
Inhibition of the canonical ERα signaling pathway
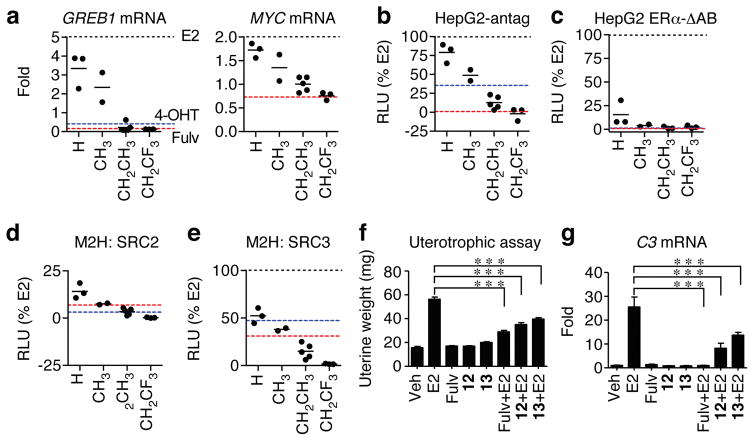
We developed a chemical biology platform equipped with a battery of assays to probe various mechanistic aspects of ER signaling24. Consistent with the observed effects on cell proliferation, the R2 group determined induction of the MYC and GREB1 genes, with the bulkier substitutions engendering inverse agonism comparable to fulvestrant (Fig. 3a). To probe for SERM agonist activity in other cell types, we used well-known model systems where ligands activate a 3xERE-driven luciferase reporter. Full antagonism of ERα was observed with the trifluoroethyl R2 compounds in HepG2 cells (Fig. 3b). Activity of OBHS-N analogs in the proliferation assays and native gene assays closely matched their ERα agonist activities in Ishikawa and HepG2 cells where tamoxifen produces approximately 10% and 40–50% reporter activity respectively, compared to E2 (Supplementary Fig. 3e–f). All OBHS-N analogs behaved as full antagonists or inverse agonists of ERβ 18 (Supplementary Fig. 2). We also repeated the agonist assay in HepG2 cells with the truncated receptor, ERα-ΔAB, which lacks AF1 (Supplementary Fig. 1a). Only the H substituted R2 compounds showed activity in this assay, demonstrating that most of the agonist activity was AF1-dependent (Fig. 3c). Thus, the R2 group of OBHS-N analogs controls AF1-dependent agonist activity, with bulkier substitutions driving full antagonism of ERα-mediated transcription and cell proliferation.
Figure 3. Modulation of ER α signaling by OBHS-N analogs in cells in vitro and in the mouse uterus in vivo.
(a–e) Dotted lines represent values for 10 nM E2 (black), 1 μM 4-OHT (cyan) and 1 μM fulvestrant (Fulv, red).
(a) Steroid-deprived MCF-7 cells were treated with 10 μM OBHS-N analogs for 24 hr. Total mRNA was reverse-transcribed and analyzed by qPCR for GREB1 and MYC expression relative to GAPDH. Values are mean, n = 2.
(b–c) Luciferase assays. HepG2 cells were co-transfected with 3xERE-luc reporter and ERα-WT or ERα-ΔAB expression plasmids were steroid-deprived and treated with 10 μM OBHS-N ligands. For the HepG2 antagonist mode assay, cells were cotreated with 10 nM E2. Values are mean, n = 3.
(d–e) HEK293T cells were co-transfected with a 5xUAS-luc reporter and expression plasmids for full-length ERα-VP16 and Gal4-SRC2/3 and were treated with 10 μM OBHS-N ligands. Values are mean, n = 3.
(f) Uterine weights of ovariectomized mice (n = 4 per group) treated for 4 days with vehicle (Veh), E2 (10 ng), fulvestrant (240 μg), or OBHS-N compounds 12 or 13 alone (240 μg), and in combination with E2. Values are mean ± s.e.m. Statistical significance was determined by ANOVA and Bonferroni’s Multiple Comparison test. *p < 0.05; **p < 0.01;***p < 0.001.
(g) Uteri (n = 4 per group) from animals in f were analyzed for expression the C3 gene. Values are mean ± s.e.m. Statistical significance was determined by ANOVA and Dunnett’s Multiple Comparison test. *p < 0.05; **p < 0.01;***p < 0.001.
To further understand their effects on ERα-mediated transcription, we tested whether OBHS-N analogs modulate events preceding RNA polymerization, such as receptor dimerization, DNA binding and coactivator recruitment. While there was no structure-activity relationship with dimerization or DNA binding (not shown), OBHS-N ligands induced ERα interactions with the coactivators, SRC2 (NCOA2/GRIP1) and SRC3 (NCOA3/AIB1), that were proportional to the size of the R2 group (Fig. 3d–e). OBHS-N ligands with bulkier R2 groups showed less coactivator recruitment than fulvestrant, especially with SRC3 (Fig. 3e). Overall, our panel of biochemical assays indicates that OBHS-N ligands show a graded signaling profile, where antagonism of ERα-mediated coactivator recruitment, transcription and cell proliferation are directly proportional to the size of the R2 group.
To probe for SERM agonist and antagonist activity in vivo, we compared the effects of E2, fulvestrant and OBHS-N compounds 12 and 13 alone and together with E2 on uterine growth in ovariectomized mice. Compounds 12 and 13 were devoid of any agonist activity in the mouse uterus, but each significantly reduced the E2-stimulated uterine growth, as was also observed with fulvestrant (Fig. 3f). Further, compounds 12, 13, and fulvestrant failed to increase the expression of E2-induced genes, complement component C3, lactoferrin, and aquaporin 5, but markedly reduced the E2-stimulated expression levels of these genes (Fig. 3g, Supplementary Fig. 3g). Thus these compounds behave like fulvestrant in both blocking growth of breast cancer cells, and being completely devoid of uterotrophic activity in vivo.
Modulation of ERα degradation
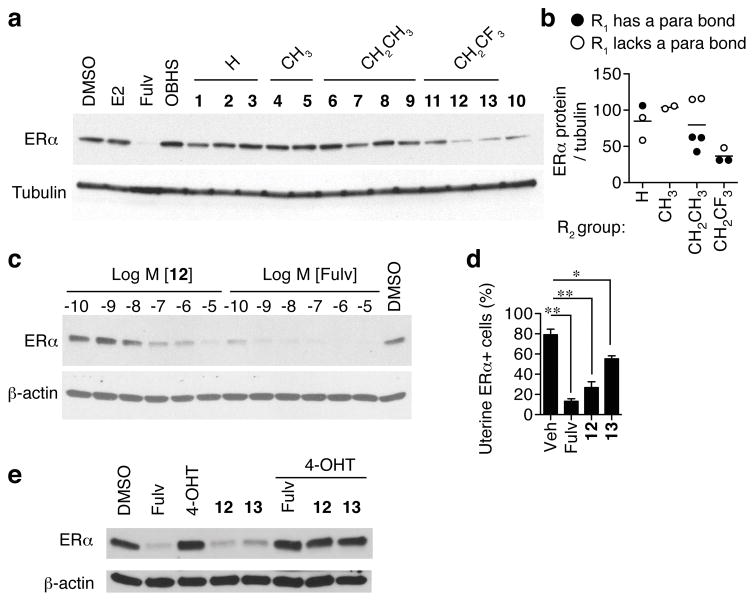
Compounds 10–13 strongly induced receptor degradation, including those with the trifluoroethyl R2 group or the α-napthyl R1 group (Fig. 4a). Compounds with a trifluoroethyl R2 group led to the lowest ERα protein levels, but R1 also had an independent effect. Among OBHS-N ligands with either the ethyl or trifluoroethyl R2 group, compounds containing a para-bond at R1, including 4-chloro-, 4-methoxy- and α-naphthyl- R1 groups, lowered ERα protein levels more than those with an ortho- or meta-substituted aryl group (Fig. 4a–b). Full images of gels are shown in Supplementary Figure 4. A similar efficacy was seen in T47D cells (Supplementary Fig. 5a). ERα degradation was attributable to proteasomal activity, as shown by its blockade with the proteasome inhibitor, MG132 (Supplementary Fig. 5b). Consistent with the affinity of 12 in the 30 nM range18, dose curves in MCF-7 cells showed a requirement for a similar dose of 12 to maximize receptor loss (Fig. 4c). To verify that these compounds cause ERα degradation in vivo, we performed immunohistochemistry on mouse uterine tissue after the uterotrophic assay and counted numbers of ERα+ cells. We found that 12 robustly inhibited ERα expression, as did fulvestrant, while 13 showed a statistically significant but more moderate inhibition (Fig. 4d). Taken together, these results suggest that full antagonism and proteasomal degradation of ERα are partially separable consequences of ligand binding, and are controlled by distinct structural features of ligand-receptor interaction.
Figure 4. Ligand-dependent proteasomal degradation of ERα.
(a) Steroid-deprived MCF-7 cells were treated with 10 μM OBHS-N ligands, OBHS or E2 and 10 nM fulvestrant (Fulv) for 6 hr. Western blot was performed to detect ERα protein levels.
(b) Contributions of the OBHS-N R1 and R2 groups to ERα degradation. Each data point represents the normalized average value for an OBHS-N compound relative to tubulin (n = 2 experiments).
(c) Dose-dependent degradation of ERα protein in MCF-7 cells treated with fulvestrant or OBHS-N compound 12. Cells were treated with indicated doses of compounds for 48 hr and western blot performed as in panel a.
(d) ERα protein expression in the uterus of ovariectomized female mice treated with vehicle, fulvestrant, OBHS-N compounds 12 and 13 as described in Figure 3f. ERα+ cells were quantified by immunohistochemistry (1-way ANOVA, Tukey’s Multiple Comparison Test; *, P < 0.05; **, P < 0.01) (e) Steroid-deprived MCF-7 cells were treated with 100 nM fulvestrant, 10 μM 4-OHT, 10 μM 12, 10 μM 13, 10 μM 4-OHT + 10 nM fulvestrant, 10 μM 4-OHT + 10 μM 12 or 10 μM 4-OHT + 10 μM 13 for 6 hr. Western blot was performed to detect ERα protein levels.
On-target mechanism of action
To show that the OBHS-N compounds are acting through ERα we used competition with control ERα ligands. We first demonstrated that 12 and 13 reversed E2-mediated proliferation (Supplementary Fig. 6a). In both the ERE-luciferase assay and the M2H assay of ERα interaction with SRC3, cells were treated with 1 μM 12 or 13, and inhibition was completely reversed by increasing doses of E2 (Supplementary Fig. 7–8). Similarly, the inverse agonist effect of 12 and 13 on the GREB1 mRNA was fully reversed by increasing doses of E2 (Supplementary Fig. 9a), while the non-ERα target gene, AKT1, was unaffected by ERα ligands including 12 and 13 (Supplementary Fig. 9b). The anti-proliferative effects of 12 and 13 were fully reversed by increasing doses of E2 in the MCF-7 cells (Supplementary Fig. 10). Importantly, the effects on ERα degradation were also reversible. Since E2 also induces degradation, we co-treated MCF-7 cells with 4-OHT, which does not induce degradation and fully reversed the effects of 12 and 13 (Fig. 4e). Thus we did not detect any off-target effects that led to toxicity, and degradation was ERα mediated and through binding to the pocket.
We also recently reported a meta-analysis of 241 compounds from 19 different scaffolds that included the OBHS-N compounds25, and used linear regression to identify which ERα-mediated signal transduction features predicted the proliferative response to the ligands. For some scaffolds, the induction of GREB1 mRNA and recruitment of SRC3 in the mammalian 2-hybrid assay predicted the proliferative response. OBHS-N compounds showed the strongest predictions, and over 90% of their anti-proliferative effect was explained by SRC3 recruitment (r2 = 0.92) and induction of GREB1 (r2 = 0.92)25. Here we found that MYC expression also strongly predicted their proliferative effects on MCF-7 cells (r2 = 0.73, p = 0.0002, F test for non-zero slope)(Supplementary Fig. 6b), supporting an ERα-mediated mechanism of action.
Helix-11 positioning determines ERα antagonism
We obtained crystal structures of the ERα LBD in complex with seven distinct OBHS-N ligands using a surface mutation (Y537S) (Supplementary Table 1, Supplementary Fig. 11a–g) that greatly facilitates crystallization by stabilizing the agonist conformer and allowing soaking of apo crystals16,26–28. This procedure also allows us to crystallize ligands with a range of activity profiles, including partial agonists that induce conformational heterogeneity in h12 and are difficult to crystallize, and to visualize how the range of ligands interact with, and disrupt the active conformation of the receptor23,24,29. The Y537S mutation occurs naturally in hormone-resistant metastatic breast cancer30–33, provide the additional benefit of understanding how ER ligands behave in a clinically relevant context. A caveat of this approach is that the A chains in the dimer display a crystal packing contact on the C-terminus of h11 that limits ligand-induced perturbation of this helix and can alter ligand binding23,24,29. As with our recent meta-analysis34, the following analyses are for the B chains of the structures unless indicated.
In all 7 structures, the B chain bound unambiguously to the (1S,2R,4S) OBHS-N enantiomer (Supplementary Fig. 11), as found in structures bound to the OBHS and S-OBHS series34. Compound 1, where R2 is a hydrogen atom, binds ERα in an orientation with the two phenols mimicking the A-ring and the “E-ring” of tamoxifen to form hydrogen bonds with the receptor (Fig. 5a, Supplementary Fig. 1d). The parental R1 aryl group in 1 bound against h11, strongly clashing with the agonist positioning of His524, and inducing a 1.1 Å shift in the main chain end of h11 (Fig. 5b), altering the packing between h11 and h12 (Supplementary Fig. 12a). With compound 4, the methyl R2 shifted Leu525 even further, inducing a rotation in the h11 C-terminus and further altering the interface between h11 and h12 (Supplementary Fig. 12b).
Figure 5. Structural basis for full antagonism of ERα by OBHS-N analogs.
(a) Detail of the crystal structure of the ERα LBD in complex with 1 is shown as blue ribbons. (PDB 5KCC)
(b–c) Crystal structures of ERα bound to 1 or 11 were superposed with the E2-bound ERα structure. H11 is shown as α-carbon trace. (PDB 3UUD, 5KCC, 5KCW)
(d) Detail of E2- or 11-bound ERα, with h11 drawn as a surface and h12 as blue cylinders. The missing h11–h12 loop in the 11-bound ERα is drawn as dashed line. (PDB 3UUD, 5KCW)
(e) ERα-Y537S LBD was incubated with 3- to 5-fold molar excess of E2, 12 or 13. Exchange of amide hydrogens for deuterium was assessed by HDX mass spectrometry and mapped on the E2- or 13-bound LBD structures. The highlighted regions show ligand-dependent decrease in HDX, which indicates enhanced stability.
(f–g) Crystal structure of the 4-bound ERα LBD shows that the OBHS-N R1 group was accommodated near His524, but para-substituted R1 group of 13 induced a rotation of the amide moiety. This rotated orientation of the amide is further stabilized by an electrostatic interaction between the chlorine atom and Trp383 (PDB 5KCD, 5KD9).
In the compound 11-bound structure, the trifluoroethyl R2 group caused the ligand to bind in a mixture of conformations (Supplementary Fig. 11f), with rotation around the sulfonamide bond, so that the R1 and R2 groups switched positions for a mixture with 57% of the molecules having the R1 group flipped towards h12, as determined by refining the occupancy of both conformers. The positioning of larger R2 groups towards Leu525 clashed even more strongly, inducing a 2.5Å shift in h11, and rendering the C-terminus of h11 completely disordered (Fig. 5c, Supplementary Fig. 12c).
When bound to a full agonist such as E2, h12 Leu544 docks against a hydrophobic groove on h11, formed by Leu525 and the aliphatic regions of Tyr526 and Lys529. In contrast, compound 11 induced a shift in h11 that forced h12 Leu544 out of the hydrophobic groove (Fig. 5d). Thus, our conformation-trapping approach revealed a conformer of h11 that lacks the extensive VDW contacts with h12 required to stabilize the active LBD conformation in solution. Here crystal packing and the Y537S mutation stabilize this active h12 conformer, but in solution, we expect this h12 conformer to be rare, explaining how the ligand-induced positioning of h11 drives full antagonism.
To verify the binding mode of the ligands in solution, we obtained a binding footprint using amide hydrogen/deuterium exchange mass spectroscopy (HDX-MS). This measures perturbations in hydrogen bonding network due to ligand binding by measuring the differences in uptake of solvent deuterium and is reflective of the conformational changes induced by ligand interactions. Using purified ERα-Y537S LBD, E2 induced protection from exchange (decreased D2O uptake) in the h3, h11, and h12 regions (Fig. 5e). Whereas 13 induced moderate protection from exchange in the h11 region while h12 was unperturbed with either 12 or 13. This is consistent with our interpretation of the crystal structures that support lack of stabilization at the end of h11 and the h11–12 loop in the presence of 12 and 13.
Degradation is partially separable from antagonism
We identified a structural feature of the ligand-receptor interaction that drives increased degradation from the para R1 substitutions. With compounds 7, 9 and 13, the para R1 substitutions that contributed to degradation flipped the R2 and aryl group around the sulfonamide linker (Supplementary Fig. 11d–g). This occurred because the para substitution would clash with the backbone carbonyl of Glu419 in the loop between h7 and h8, which prevented the R1 group from binding there (Fig. 5f–g and Supplementary Fig. 12d–e). In the context of an ethyl R2 group, the para-substituted R1 in 7 or 9 induced rotation of the amide moiety, and thereby redirected the R1 group towards Leu525 and helix-12 (Supplementary Fig. 12f–g). Thus rotation of the amide moiety explains why the trifluoroethyl R2 group drives degradation of ERα in the absence of a para-substituted R1 group, and why the para-substituted R1 group further enhances degradation of ERα.
The dramatic shifts in h11 were prevented in the A chains of the structures due to a 3.7 Å contact between the backbone of h11 with the backbone of h1 in the symmetry-related molecule (Supplementary Fig. 13a–b). This artificial restraint of h11 in the A chain prevented accommodation of the para-substituted R groups of compounds 7 and 9 near Leu525 as seen in the B chain (Supplementary Fig. 11d–e, Supplementary Fig. 13c). Instead, the (1R,2S,4R) enantiomer of compounds 7 and 9 bound the A chain with R groups positioned further away from h11 Leu525 than the (1S,2R,4S) enantiomers in the B chain. This also induced a shift in the ligand core that prevented H-bonding with Thr347 (Supplementary Fig. 13d), suggesting a lower receptor-binding affinity for the (1R,2S,4R) enantiomers. The (1S,2R,4S) enantiomer of compound 13 bound both the A and B chains, but the R groups were disordered in the A chain (Supplementary Fig. 13e), as seen with the (1R,2S,4R) enantiomer of 7 (Supplementary Fig. 11d). Importantly, His524 of the A chain rotated back into the pocket, while Phe425 rotated away from the ligand, suggesting that the R groups were oriented towards h7 and h8 (Supplementary Fig. 13e), as we previously visualized with a 17α-substituted E2 ligand27.
We were able to separate the enantiomers of 13 into two well-resolved peaks (designated 13R for the 1R,2S,4R, and 13S for the 1S,2R,4S enantiomers, Fig. 1) through HPLC (see online methods). The two compounds had a ~20-fold difference in relative binding affinity towards ERα (with relative binding affinities (RBAs) being 0.961 and 0.054, with respect to E2, with an RBA of 100). The difference between the enantiomers was more profound in reporter assays where the higher affinity compound (2nd peak, presumed to be 13S) displayed an IC50 of 30 nM, while the lower affinity compound (1st peak, presumed to be 13R) only inhibited at 10 μM (Supplementary Fig. 13f–g). Surprisingly, the lower affinity compound partially activated ERα in the agonist mode assay (Supplementary Fig. 13g), which was not apparent with the racemate (Fig. 3b) in which the potent full antagonist activity of the higher affinity enantiomer dominates the weak partial agonism of the low affinity enantiomer.
Antagonism of constitutively active ERα-Y537S
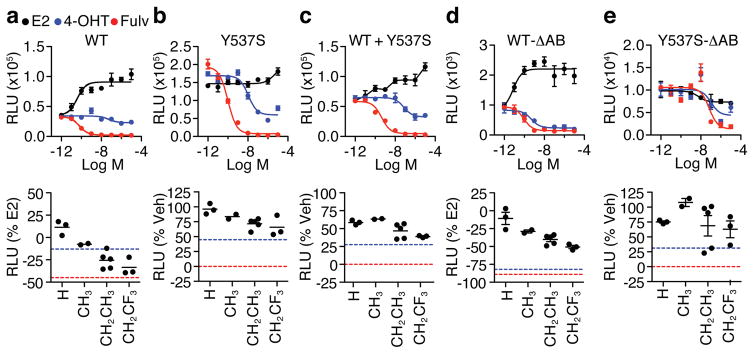
Sequencing of tumors from metastatic hormone-resistant patient samples revealed mutations in the ESR1 gene, with Y537S being the most predominant30–33. We used 3xERE-driven luciferase reporter assays to compare ligand activities mediated by ERα-WT, ERα-Y537S and a mixture of both receptors (ERα-WT+Y537S), which mimics the clinically relevant setting of mixed alleles. The compounds showed very similar profiles in single-dose screen in HeLa cells (Fig. 6a) and dose-response screen in HEK293T cells (Supplementary Fig. 14a) transfected with the wild type receptor, which were also very similar to results with the other cell types described above. Fulvestrant reduced Y537S and WT+Y537S transcriptional activities to almost background levels, while tamoxifen had a partial inhibitory effect. The OBHS-N compounds also displayed partial inhibitory effects that were once again driven by bulkier substitutions at R2 (Fig. 6b–c and Supplementary Fig. 14b–c). Furthermore, the compounds were inactive or inverse agonists on the truncated wild type receptor, ERα-ΔAB, but showed considerable activity through ERα-Y537S-ΔAB (Fig. 6d–e and Supplementary Fig. 14d–e). This indicates that in solution, the ERα-Y537S mutation stabilizes the active LBD conformation a substantial fraction of the time when bound to OBHS-N analogs or tamoxifen, while the much bulkier side chain of fulvestrant is able to dislocate h12 from the active position, even in the presence of the mutation. Examination of the structure of 13-bound ERα-Y537S demonstrates that the E-ring phenol and the chlorobenzyl R1 ring make close contacts with L536 in the h11–h12 loop and L540 in h12 (Supplementary Fig. 15). This suggests that further extension of these moieties will be required to block the active LBD conformer in the presence of the ERα-Y537S mutation.
Figure 6. Antagonism of ERα-Y537S.
(a–e) HeLa cells were co-transfected with the 3xERE-luc reporter and the indicated ERα expression plasmids. The next day, cells were treated for 24 hr with increasing doses of E2, 4-OHT or fulvestrant (Fulv), top panels, or 10 μM OBHS-N compounds, bottom panels. Dose curve data is shown as mean ± s.e.m. Each single-dose data represents the mean (n = 3). Also see Supplementary Figure 14 for compound dose curves in 293T cells.
Discussion
Antagonism of ERα for treatment of breast cancer is complicated by the tissue- and pathway-selective signaling activities of the ligands, and by multiple mechanisms for the development of resistance35,36. Our findings demonstrate that the current SERM/SERD approach9–11 is not the only structural mechanism to antagonize the receptor16–18. We identified a structure-based design strategy where the degree of h11 dislocation is controlled in a graded fashion largely by the R2 substituents of the sulfonamides to determine the level of inhibition of the classical ERα signaling pathway, including recruitment of SRC2 and SRC3, inhibition of GREB1 and MYC expression, and ERα-mediated antiproliferative effects. Importantly, full antagonism was also reflected in the uterotrophic assay, where compounds 12 and 13 did not induce proliferation or uterine expression of canonical ERα target genes but efficiently antagonized E2, showing that they are biologically active in vivo. While the most efficacious compounds were comparable to fulvestrant in most assays, several showed improved profiles with respect to low ERα–SRC3 interaction, and inhibition of IL-6 secretion. Most notably, these levels of activity were achieved through ligand-LBD interactions that are distinct from those of SERMs and fulvestrant-like SERDs.
For the most efficacious compound, 13, we separated the enantiomers and showed that one of them (presumed to be the 1S,2R,4S enantiomer 13S) accounts for its full antagonist activity, as the other (presumed to be the 1R,2S,4R enantiomer 13R) had much higher IC50 value and elicits a partial agonist activity that was not apparent in the racemate. This is due to the positioning of the R groups further away from h11, which also lowers its binding affinity towards ERα by shifting the core and preventing H-bonding with h3 Thr347. For pre-clinical development, separation of enantiomers will be necessary as the enantiomers may have differences in 1) on-target effects, 2) receptor-binding affinity, and 3) in vivo off-target effects, any of which could be different from those of the corresponding racemic mixtures.
Deletion analysis identifies which domains are responsible for signaling. Deletion of ERα domains in mice revealed, for example, that AF1 is necessary for E2-induced uterine proliferation but not vascular protection37,38. Ligand-dependent control of AF1 is crucial for antagonism of the wild type receptor. We demonstrate that indirect antagonism via h11 dislocation is sufficient to inhibit AF1 and generate a full antagonist/SERD profile. Our data suggests that the agonist conformer is partially populated by the ERα-Y537S mutation in the presence of either the sulfonamides or tamoxifen, as shown by the enhanced activity compared to fulvestrant in the ERα-Y537S-ΔAB assay where activity is driven by AF2. This supports the idea that antagonism of the mutant involves two structurally distinct steps i.e. displacement of the active h12 conformer and control of AF1 activity. Tamoxifen did not fully antagonize suggesting that the side chain can be accommodated in the mutant active conformer.
It is not clear whether ERα degradation contributes to efficacy of full antagonists. E2 also leads to efficient degradation of ERα, which is required for activation and proliferative signals39. Antagonist-induced loss of ERα occurs through different mechanisms and involves ERα translocation to an insoluble fraction of the nuclear matrix followed by proteasomal degradation40,41, or by binding outside the pocket to induce the unfolded protein response42. We showed that degradation with OBHS-N compounds is mediated by the proteasome and blocked by 4-OHT. It is also noteworthy that degradation is tissue-selective, as shown by the discovery of SERM/SERD hybrids such as GW563843 that degrade ERα in breast cancer but are bone sparing. In a survey of FDA-approved ERα antagonists, degradation was found to correlate with inhibition of MCF-7 breast cancer viability4. However, overexpression of ERα can overwhelm the degradation machinery, but in this context fulvestrant still fully antagonizes proliferation and reporter activity44,45.
While different SERDs may use different structural mechanisms to induce degradation46, our data further suggests that some of these mechanisms are separable from antagonism, at least within the sulfonamide series. This structurally distinct mechanism, through indirect antagonism rather than involvement of the prototypical SERM/SERD side chain, provides new opportunities for producing biologically distinct molecules for breast cancer prevention and treatment.
ONLINE METHODS
Chemical Synthesis
The synthesis of compounds 1–13 has been described previously 18.
Enantiomer separation
Enantiomers of analog 13, designated 13R and 13S, were separated by a semi-preparative Regis (S,S) WHELK-O1 25cm × 10.0mm column at a temperature of 20°C, eluted with 70:30 hexane: IPA at a flow rate of 1 ml/min. The HPLC was monitored using a Waters 2487 Dual wavelength detector. After separation on the semipreparative column, purity was tested on an analytical (R,R) WHELK-O2 column using the same solvents. All solvents were HPLC grade. HPLC peaks are shown in Supplementary Figure 16.
Cell culture
HepG2, MCF-7, MDA-MB231 and ZR-75 cells were maintained at 37 °C and 5% CO2 in media consisting of Dulbecco’s minimum essential media (DMEM) (Cellgro by Mediatech, Inc., Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Hyclone by Thermo Scientific, South Logan, UT), and 1% penicillin/streptomycin/neomycin antibiotic mixture (PSN), 1% Glutamax (Gibco by Invitrogen) and 1% non essential amino acids (NEA) (Cellgro). Steroid deprivation was achieved by culturing cells for 24–48 hours in media composed of phenol red-free DMEM supplemented with 10% charcoal/dextran-stripped FBS, 1% Glutamax, 1%NEA and 1% PSN. Cell lines were obtained from ATCC and tested monthly for mycoplasma.
Luciferase assay
Luciferase assay to determine ER activity were performed as previously described24. To this end, HepG2 cells were transfected with a 3xERE-driven luciferase reporter and ERα constructs using FugeneHD reagent (Roche Applied Sciences, Indianapolis IN). The cells were then transferred to 384-well plates in steroid free media and stimulated with ER ligands 24 hours later. Luciferase activity as measured after 18–24 hours using Britelite™ reagent (PerkinElmer Inc., Waltham, MA).
Mammalian 2-hybrid assay
Human embryonic kidney 293T cells were transfected with an upstream activating sequence (UAS)-driven luciferase reporter, a wild type human ERα construct fused to the activation domain of the herpesvirus protein VP16, and a bait consisting of wild type SRC2 or SRC3 fused to the DNA-binding domain of GAL4. Luciferase activity was measured after 24 hours of stimulation with various ER ligands.
IL-6 assay
MCF-7 cells were steroid deprived and stimulated with 10 ng/ml TNFα alone or in combination with the ER ligands overnight in 384-well format. Media conditioned by these cells was analyzed using the AlphaLISA human IL-6 kit (PerkinElmer), according to manufacturer’s directions.
Cell proliferation assay
MCF-7 cells were grown on 384-well plates in steroid-free media at a density of 1000 cells/well and stimulated with ER ligands. MDA-MB231, and ZR-75 cells were growing on 384-well plates in phenol red-free normal growth media at a density of 1000 cells/well and stimulated with ER ligands. After 7 days, cell proliferation was measured using CellTiter-Glo Luminescent Cell Viability Assay kit (Promega, Madison, WI) according to manufacturer’s instructions. Data was normalized using a standard curve for respective cell lines.
Mammosphere formation assay
Breast cancer cells were seeded at single cell density in low attachment plates in media described by Dontu et al.47. Media was supplemented with 1% methyl cellulose to prevent cellular aggregation 48. After 7 days, the number of mammospheres ≥ 75μm in diameter was determined.
Mouse uterotrophic and gene expression assays
All animals were maintained in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals, and all procedures were approved by the University of Illinois Institutional Animal Care and Use Committee. Female C57BL/6 mice (Harlan /NCI Laboratories, Frederick, MD) were ovariectomized at 8 weeks of age, and after 18 days, mice (4 per group) were injected sc with different ligands in ethanol-corn oil mix at a ratio of 1:9 for four days. At 24 hours after the last injection, mice were euthanized, and the uterus was removed and the wet weight measured. Uterine RNA was also obtained and used for determination of the expression of E2-regulated genes by qPCR, as done previously49.
Western blot
Cells were grown in 6 well plates in steroid free media and treated with compounds for either 6 or 48 hr. Then the cells were washed once with PBS and lysed in ice-cold RIPA buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% NP40, 0.5% Sodium deoxycholate, 1 mM EDTA and 0.1% SDS). Equal amounts of protein were loaded on Any kD™ Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-rad, Hercules, CA) and transferred onto PVDF membranes (Thermo Scientific, Rockford, IL). Membranes were blocked with 5% nonfat dry milk in PBS-T, then probed with ERα-H222 (1:1000 dilution; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and anti-β-actin or tubulin (1:10,000 dilution; Sigma-Aldrich Corp). After washing in TBS-T, membranes were incubated with their appropriate HRP conjugated secondary antibodies (Santa Cruz Biotechnology) and developed using an ECL detection system (GE Healthcare Bio-Sciences, Pittsburg, PA)
Quantitative RT-PCR
MCF-7 cells were plated in 384-well plates in 25 μl in steroid free medium at a density of 2,500 cells/well. Three days later, cells were stimulated with compounds for 1 h (c-MYC) and 24 h GREB1. Total RNA was extracted with RNAgem Tissue Plus RNA extraction kit (Zygem). Cells were washed once with PBS and lysed with 7.5 μl RNAgem extraction reagent at 75 °C for 5 min. Genomic DNA was removed with DNASEI treatment for 5 min at 37 °C and stopped by incubating at 75 °C. Five microliters of this mixture were used for cDNA synthesis in a total volume of 10 μl using a High-Capacity cDNA Reverse Transcription Kit (Life Technologies).
The cDNA was diluted three times, and 4 μl of diluted cDNA was used in a qPCR reaction using TaqMan Gene Expression Master Mix (Life Technologies) in a total reaction volume of 10 μl. The qPCR reaction was carried out in a duplex format using predesigned real-time qPCR assays (Life Technologies) with GAPDH primers with a VIC probe as an endogenous control and target gene (c-MYC and GREB1) primers with FAM probe. The reactions were run on an ABI 7900 HT real-time PCR system, and the fold change relative to vehicle-treated samples was calculated using the ΔΔCt method.
X-ray crystallography
The ERα Y537S LBD was produced in E. coli, crystallized, and soaked in various ligands as previously described16,27. Note that IPTG induction at 25 ºC for 6 hr produced protein that was readily crystallizable, while overnight induction at 18C produced a very small amount of truncated protein that interfered with crystallization. Data was collected at the Stanford Synchrotron Radiation Lightsource (Beamline 11-1; Temp. = 100 K; wavelength = 1.0 Å), and scaled using HKL3000 software50. Structures were solved via molecular replacement using the PHENIX software suite51. The protein components of the genistein-bound ER structure (PDB ID: 2QA8)16 were used as the starting model. The models were subject to extensive combinatorial refinement as previously described52 rebuilding using COOT53. Graphical images were made with CCP4MG54. Data collection and refinement statistics are shown in supplementary table 1. Additional refinement statistics and Molprobity scores55,56 are presented below.
| 1 (5KCC) | 4 (5KCD) | 5 (5KCE) | 7 (5KCF) | 9 (5KCT) | 10 (5KCU) | 11 (5KCW) | 13 (5KD9) | |
|---|---|---|---|---|---|---|---|---|
| Clashscore, all atoms | 2.7 | 3.9 | 3.8 | 4.2 | 4.6 | 4.8 | 4.2 | 4.1 |
| Ramachandran outliers (%) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Ramachandran favored (%) | 97.4 | 99.2 | 98.3 | 98.5 | 99.2 | 98.9 | 98.9 | 98.3 |
| Side chain outliers (%) | 0.3 | 0.7 | 0.5 | 0.0 | 0.2 | 0.0 | 0.0 | 0.5 |
| Molprobity score | 1.16 | 1.17 | 1.17 | 1.20 | 1.23 | 1.25 | 1.21 | 1.20 |
Hydrogen/Deuterium exchange mass spectrometry (HDX-MS)
Solution-phase amide HDX was carried out on our automated system as described previously 57. Briefly, for the on-exchange reaction, 5 μl of 10 μM apo-ERα or the ligand bound ERα was diluted to 25 μl with D2O-containing HDX buffer and incubated at 4 °C for 10, 30, 60, 900 or 3,600 sec. Following on-exchange, the reaction was quenched with 25 μl 1% (v/v) TFA in 5 M urea and 50 mM TCEP. Samples were passed over an immobilized pepsin column (prepared in house) at 50 μl min−1 (0.1% v/v TFA, 15 °C 58. Peptic peptides were first trapped onto a C8 trap cartridge for desalting (Hypersil Gold, Thermo Fisher) and then eluted with a linear gradient [4% (w/v) CH3CN to 40% (w/v) CH3CN, 0.3% (w/v) formic acid over 5 min, at 4 °C] from a 1 mm × 50 mm C18 HPLC column (Hypersil Gold, Thermo Fisher) and subjected to electrospray ionization directly coupled to a high resolution Orbitrap mass spectrometer (Q Exactive, Thermo Fisher). MS/MS peptide identification was carried out using Mascot (Matrix Science) and manually verified to filter the false positives (using a decoy database) and to include only the highest scoring peptides for each charge state. For on-exchange experiments, the percent deuterium uptake for each peptide at various time points was calculated using HDX workbench software59. To measure the difference in exchange rates between the Apo and ligand bound Y537S receptor, we calculated the average percentage deuterium uptake for the apo ERα following 10, 30, 60, 900 and 3600 sec of on-exchange. From this value, we subtracted the average percent deuterium uptake measured for ligand bound ERα. The percent difference is color coded according to the heat map legend indicated in the figure. Each HDX experiment is carried out in triplicates.
Statistics
The uterotrophic weight data was analyzed with ANOVA and Bonferonni’s Multiple Comparison test. For uterotrophic gene expression statistical significance was determined by ANOVA and Dunnett’s Multiple Comparison test. Significance for linear regression was determined with an F test for non-zero slope.
PDB
Structures were deposited in the Protein Data Bank with the following codes: 5KCC, 5KCD, 5KDE, 5KCF, 5KCT, 5KCU, 5KCW, 5KD9.
Supplementary Material
Acknowledgments
Research support from the National Institutes of Health (PHS 5R37DK015556 to J.A.K.; 5R33CA132022, 5R01DK077085 to K.W.N.; 1U01GM102148 to K.W.N and P.R.G., and 5R01CA130932 to J.F.), The Breast Cancer Research Foundation (to B.S.K.), BallenIsles Men’s Golf Association (to J.C.N.), Frenchman’s Creek Women for Cancer Research (to S.S.), Susan G. Komen for the Cure® (PDF12229484 to IK); and the National Natural Science Foundation of China (81172935, 81373255, 81573279), Hubei Province’s Outstanding Medical Academic Leader Program (to H.-B.Z.)
Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
Competing financial interest statement
The authors declare no competing financial interests.
Accession codes
Structures were deposited in the Protein Data Bank with the following codes: 5KCC, 5KCD, 5KDE, 5KCF, 5KCT, 5KCU, 5KCW, 5KD9.
Author contributions
S.S. and J.C.N. contributed equally to this work. S.S. and J.C.N. designed and performed (experiments, and wrote the manuscript; N.E.B., V.C., and J.N. performed in vitro experiments; P.R.G., S.N., V.D. and D.G. designed, performed and interpreted HDX based experiments; J.F. and I.K. contributed mammosphere assays; B.S.K., N.B. and Y.Z. designed and contributed experiments including in vivo experiments in mice; H.B.Z., J.A.K., and J.M. designed, synthesized and performed chemical analysis of compounds; B.S.K. and J.A.K. contributed in writing and revising of the manuscript; K.W.N. designed experiments and wrote the manuscript.
References
- 1.Yu KD, Wu J, Shen ZZ, Shao ZM. Hazard of breast cancer-specific mortality among women with estrogen receptor-positive breast cancer after five years from diagnosis: implication for extended endocrine therapy. J Clin Endocrinol Metab. 2012;97:E2201–9. doi: 10.1210/jc.2012-2423. [DOI] [PubMed] [Google Scholar]
- 2.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9:R6. doi: 10.1186/bcr1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennecke H, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–7. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 4.Lai A, et al. Identification of GDC-0810 (ARN-810), an Orally Bioavailable Selective Estrogen Receptor Degrader (SERD) that Demonstrates Robust Activity in Tamoxifen-Resistant Breast Cancer Xenografts. J Med Chem. 2015;58:4888–904. doi: 10.1021/acs.jmedchem.5b00054. [DOI] [PubMed] [Google Scholar]
- 5.Wardell SE, Nelson ER, Chao CA, Alley HM, McDonnell DP. Evaluation of the pharmacological activities of RAD1901, a selective estrogen receptor degrader. Endocr Relat Cancer. 2015 doi: 10.1530/ERC-15-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–44. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 7.Metivier R, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 8.Cicatiello L, et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol Cell Biol. 2004;24:7260–74. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzozowski AM, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–8. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 10.Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–37. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 11.Pike AC, et al. Structural insights into the mode of action of a pure antiestrogen. Structure. 2001;9:145–53. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 12.Wu YL, et al. Structural basis for an unexpected mode of SERM-mediated ER antagonism. Mol Cell. 2005;18:413–24. doi: 10.1016/j.molcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Willson TM, et al. Dissection of the molecular mechanism of action of GW5638, a novel estrogen receptor ligand, provides insights into the role of estrogen receptor in bone. Endocrinology. 1997;138:3901–11. doi: 10.1210/endo.138.9.5358. [DOI] [PubMed] [Google Scholar]
- 14.Laxmi YR, et al. Anti-breast cancer potential of SS1020, a novel antiestrogen lacking estrogenic and genotoxic actions. Int J Cancer. 2010;127:1718–26. doi: 10.1002/ijc.25167. [DOI] [PubMed] [Google Scholar]
- 15.Shiau AK, et al. Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism. Nat Struct Biol. 2002;9:359–64. doi: 10.1038/nsb787. [DOI] [PubMed] [Google Scholar]
- 16.Nettles KW, et al. NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Nat Chem Biol. 2008;4:241–7. doi: 10.1038/nchembio.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Y, et al. Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands. ChemMedChem. 2012;7:1094–100. doi: 10.1002/cmdc.201200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu M, et al. Bicyclic core estrogens as full antagonists: synthesis, biological evaluation and structure-activity relationships of estrogen receptor ligands based on bridged oxabicyclic core arylsulfonamides. Org Biomol Chem. 2012;10:8692–700. doi: 10.1039/c2ob26531a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kastrati I, Canestrari E, Frasor J. PHLDA1 expression is controlled by an estrogen receptor-NFkappaB-miR-181 regulatory loop and is essential for formation of ER+ mammospheres. Oncogene. 2015;34:2309–16. doi: 10.1038/onc.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cvoro A, et al. Distinct roles of unliganded and liganded estrogen receptors in transcriptional repression. Mol Cell. 2006;21:555–64. doi: 10.1016/j.molcel.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Nettles KW, et al. CBP Is a dosage-dependent regulator of nuclear factor-kappaB suppression by the estrogen receptor. Mol Endocrinol. 2008;22:263–72. doi: 10.1210/me.2007-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinenov Y, Gupte R, Rogatsky I. Nuclear receptors in inflammation control: repression by GR and beyond. Mol Cell Endocrinol. 2013;380:55–64. doi: 10.1016/j.mce.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nwachukwu JC, et al. Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network. Elife. 2014;3:e02057. doi: 10.7554/eLife.02057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan S, et al. Ligand-binding dynamics rewire cellular signaling via estrogen receptor-alpha. Nat Chem Biol. 2013;9:326–32. doi: 10.1038/nchembio.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nwachukwu JC, et al. Predictive features of ligand-specific signaling through the estrogen receptor. Mol Syst Biol. 2016;12:864. doi: 10.15252/msb.20156701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson KE, Choi I, Gee A, Katzenellenbogen BS, Katzenellenbogen JA. Altered ligand binding properties and enhanced stability of a constitutively active estrogen receptor: evidence that an open pocket conformation is required for ligand interaction. Biochemistry. 1997;36:14897–905. doi: 10.1021/bi971746l. [DOI] [PubMed] [Google Scholar]
- 27.Nettles KW, et al. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 2007;8:563–8. doi: 10.1038/sj.embor.7400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delfosse V, et al. Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes. Proc Natl Acad Sci U S A. 2012;109:14930–5. doi: 10.1073/pnas.1203574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruning JB, et al. Coupling of receptor conformation and ligand orientation determine graded activity. Nat Chem Biol. 2010;6:837–43. doi: 10.1038/nchembio.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeselsohn R, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20:1757–67. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–30. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson DR, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toy W, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–45. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nwachukwu JC, et al. Predictive features of ligand-specific signaling through the estrogen receptor. Mol Syst Biol. 2016;12:864. doi: 10.15252/msb.20156701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patani N, Martin LA. Understanding response and resistance to oestrogen deprivation in ER-positive breast cancer. Mol Cell Endocrinol. 2014;382:683–94. doi: 10.1016/j.mce.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 36.Kuske B, et al. Endocrine therapy resistance can be associated with high estrogen receptor alpha (ERalpha) expression and reduced ERalpha phosphorylation in breast cancer models. Endocr Relat Cancer. 2006;13:1121–33. doi: 10.1677/erc.1.01257. [DOI] [PubMed] [Google Scholar]
- 37.Billon-Gales A, et al. The transactivating function 1 of estrogen receptor alpha is dispensable for the vasculoprotective actions of 17beta-estradiol. Proc Natl Acad Sci U S A. 2009;106:2053–8. doi: 10.1073/pnas.0808742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abot A, et al. The AF-1 activation function of estrogen receptor alpha is necessary and sufficient for uterine epithelial cell proliferation in vivo. Endocrinology. 2013;154:2222–33. doi: 10.1210/en.2012-2059. [DOI] [PubMed] [Google Scholar]
- 39.Bhatt S, Xiao Z, Meng Z, Katzenellenbogen BS. Phosphorylation by p38 mitogen-activated protein kinase promotes estrogen receptor alpha turnover and functional activity via the SCF(Skp2) proteasomal complex. Mol Cell Biol. 2012;32:1928–43. doi: 10.1128/MCB.06561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan M, Bigsby RM, Nephew KP. The NEDD8 pathway is required for proteasome-mediated degradation of human estrogen receptor (ER)-alpha and essential for the antiproliferative activity of ICI 182,780 in ERalpha-positive breast cancer cells. Mol Endocrinol. 2003;17:356–65. doi: 10.1210/me.2002-0323. [DOI] [PubMed] [Google Scholar]
- 41.Lupien M, et al. Raloxifene and ICI182,780 increase estrogen receptor-alpha association with a nuclear compartment via overlapping sets of hydrophobic amino acids in activation function 2 helix 12. Mol Endocrinol. 2007;21:797–816. doi: 10.1210/me.2006-0074. [DOI] [PubMed] [Google Scholar]
- 42.Andruska ND, et al. Estrogen receptor alpha inhibitor activates the unfolded protein response, blocks protein synthesis, and induces tumor regression. Proc Natl Acad Sci U S A. 2015;112:4737–42. doi: 10.1073/pnas.1403685112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connor CE, et al. Circumventing tamoxifen resistance in breast cancers using antiestrogens that induce unique conformational changes in the estrogen receptor. Cancer Res. 2001;61:2917–22. [PubMed] [Google Scholar]
- 44.Wardell SE, Marks JR, McDonnell DP. The turnover of estrogen receptor alpha by the selective estrogen receptor degrader (SERD) fulvestrant is a saturable process that is not required for antagonist efficacy. Biochem Pharmacol. 2011;82:122–30. doi: 10.1016/j.bcp.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wardell SE, Nelson ER, Chao CA, McDonnell DP. Bazedoxifene exhibits antiestrogenic activity in animal models of tamoxifen-resistant breast cancer: implications for treatment of advanced disease. Clin Cancer Res. 2013;19:2420–31. doi: 10.1158/1078-0432.CCR-12-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wittmann BM, Sherk A, McDonnell DP. Definition of functionally important mechanistic differences among selective estrogen receptor down-regulators. Cancer Res. 2007;67:9549–60. doi: 10.1158/0008-5472.CAN-07-1590. [DOI] [PubMed] [Google Scholar]
- 47.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–59. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frasor J, et al. Response-specific and ligand dose-dependent modulation of estrogen receptor (ER) alpha activity by ERbeta in the uterus. Endocrinology. 2003;144:3159–66. doi: 10.1210/en.2002-0143. [DOI] [PubMed] [Google Scholar]
- 50.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nwachukwu JC, et al. Improved crystallographic structures using extensive combinatorial refinement. Structure. 2013;21:1923–30. doi: 10.1016/j.str.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Debreczeni JE, Emsley P. Handling ligands with Coot. Acta Crystallogr D Biol Crystallogr. 2012;68:425–30. doi: 10.1107/S0907444912000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–94. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chalmers MJ, et al. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–14. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 58.Busby SA, Chalmers MJ, Griffin PR. Improving digestion efficiency under H/D exchange conditions with activated pepsinogen coupled columns. Int J Mass Spectrometry. 2007;259:130–139. [Google Scholar]
- 59.Pascal BD, et al. HDX workbench: software for the analysis of H/D exchange MS data. J Am Soc Mass Spectrom. 2012;23:1512–21. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.